Abstract
Transcriptome-wide screens of peripheral blood during the onset and development of posttraumatic stress disorder (PTSD) indicate widespread immune dysregulation. However, little is known as to whether biological sex and the type of traumatic event influence shared or distinct biological pathways in PTSD. We performed a combined analysis of five independent PTSD blood transcriptome studies covering seven types of trauma in 229 PTSD and 311 comparison individuals to synthesize the extant data. Analyses by trauma type revealed a clear pattern of PTSD gene expression signatures distinguishing interpersonal (IP)-related traumas from combat-related traumas. Co-expression network analyses integrated all data and identified distinct gene expression perturbations across sex and modes of trauma in PTSD, including one wound-healing module downregulated in men exposed to combat traumas, one IL-12-mediated signaling module upregulated in men exposed to IP-related traumas, and two modules associated with lipid metabolism and mitogen-activated protein kinase activity upregulated in women exposed to IP-related traumas. Remarkably, a high degree of sharing of transcriptional dysregulation across sex and modes of trauma in PTSD was also observed converging on common signaling cascades, including cytokine, innate immune, and type I interferon pathways. Collectively, these findings provide a broad view of immune dysregulation in PTSD and demonstrate inflammatory pathways of molecular convergence and specificity, which may inform mechanisms and diagnostic biomarkers for the disorder.
Introduction
Posttraumatic stress disorder (PTSD) is a debilitating disorder that develops after exposure to a traumatic event and increases vulnerability to adverse health outcomes. The estimated lifetime prevalence of PTSD is ~5–6% in men and ~10–12% in women (Kessler, 1995), and even higher among recent war-veterans with estimates as high as ~20% (Ramchand et al, 2010). Although extensive work has identified putative risk factors that are associated with PTSD (DiGanji et al, 2013), the identification of discrete diagnostic biomarkers for the disorder remains elusive. Heterogeneity in susceptibility to PTSD suggests that the response of an individual to trauma may depend on biological sex as well as the type of adverse event (eg, early life adversity, violence, assault, accidents, combat). These factors, in turn, may determine downstream consequences, such as perturbation of biological pathways, making it unlikely that a valid, singular biomarker will be specific to all PTSD cases.
Research into the mechanisms underlying the onset and development of PTSD converge on hypothalamic–pituitary–adrenal (HPA) axis and immune system functioning (Daskalakis et al, 2016; Cohen et al, 2016). As such, several studies have examined pro-inflammatory cytokines and glucocorticoid activity in peripheral blood mononuclear cells or lymphocytes in PTSD cases to build more effective models for identifying molecular factors underlying PTSD. These studies were reviewed by Passos et al (2015), who summarized that increases in C-reactive protein (CRP), interleukin 6 (IL-6), tumor necrosis factor alpha (TNF-α), interleukin 1 beta (IL-1β), and interferon gamma (IFN-γ) all underlie the onset and emergence of PTSD symptoms. However, despite the evident effects of biological sex on incidence rates of PTSD, very few of the reviewed studies examined sex differences in psychobiological or inflammatory responses to trauma. Moreover, the majority of these reports centered their analysis around pre-determined targets, limiting the ability to identify novel genes and molecular pathways relevant to the pathophysiology of PTSD.
Transcriptome-wide screens of peripheral immune cells from individuals with PTSD have extended findings from candidate gene studies through systems-wide exploration of immune system dysregulation in response to PTSD. Segman et al (2005) first reported on transcriptomic differences in peripheral blood from trauma survivors with PTSD on the day of emergency room (ER) visit and 4 months later, implicating dysregulation of transcriptional enhancers and immune activating genes. We later described blood-based transcriptomic signatures implicating sex differences in cytokine pathways activated in PTSD from a population of individuals exposed to various traumatic backgrounds (Neylan et al, 2011). In a separate and non-overlapping study, we also identified transcriptomic differences in central nervous system development and immune tolerance induction pathways in PTSD cases, both with and without a history of childhood maltreatment, implicating trauma-related dimorphism (Mehta et al, 2013). Parallel lines of research have identified candidate prognostic and diagnostic blood-based gene expression classifiers in war-veterans with PTSD (Glatt et al, 2015; Tylee et al, 2015), that were largely associated with dysregulated innate immune function both prior to and following PTSD development (Breen et al, 2015). Collectively, while these data-driven approaches have formed the foundation of ongoing work to build PTSD biomarkers, they have yet to be widely replicated. Inconsistencies may be attributable to various clinical factors (eg, comorbidity, medication) and technical factors (eg, various technologies and statistical methods used to evaluate data). More importantly, these studies are often severely underpowered and model gene expression changes in the context of an explicitly defined biological sex or trauma type. As such, a critical remaining question is how biological pathways in peripheral blood overlap across sex and modes of trauma in PTSD, and how this information may inform the search for more verifiable diagnostic biomarkers for the disorder.
The primary goal of the current investigation was to synthesize the existing data from transcriptome-wide gene expression studies in PTSD and to clarify their relevance to PTSD pathophysiology, while explicitly modeling sex- and trauma-related differences. To do so, we performed a mega-analysis of five independent transcriptome-wide peripheral blood studies covering seven types of trauma in 229 PTSD and 311 comparison individuals. To address our goals, a standardized multistep analytic approach was used that we have reviewed in the context of blood-based biomarker discovery in neuropsychiatric disorders (Breen et al, 2016), and that we have also applied to other transcriptome-wide mega-analyses (Hess et al, 2016; Tylee et al, 2016). To this end, our analyses specifically sought to: (1) determine the relatedness of PTSD gene expression signatures across different types of trauma; (2) identify candidate genes, pathways and co-regulatory networks in PTSD and determine if such alterations are distinct between different biological sex and trauma types; and (3) construct diagnostic blood-based gene expression classifiers to differentiate PTSD cases from trauma-exposed control individuals and clarify the potential clinical utility of peripheral blood gene expression.
Materials and methods
Literature Search and Study Criteria
To systematically identify relevant studies for our combined mega-analysis, we performed a literature search (SCOPUS) and microarray database searches (NCBI GEO and EMBL-EBI ArrayExpress) for transcriptome-wide studies of whole blood- or leukocyte-based gene expression in PTSD. Studies were included if they met the following criteria: (i) cross-sectional post-trauma studies published between 2005 and 2015; (ii) contained individuals meeting structured diagnostic criteria for PTSD (eg, DSM, PCL); (iii) also contained a trauma-exposed healthy control group. Studies were excluded on the bases of: (i) using qualitative real-time PCR or immunoassays as a means to investigate a targeted panel of candidate genes; (ii) investigating mechanisms in lymphoblastoid cell lines, skin fibroblast cultures, serum, and plasma; and (iii) secondary data integration analyses and review papers were also excluded. Five studies met these criteria for which raw gene expression data and clinical covariates (age, sex, ethnicity, and trauma type) were available, and one additional study from which data were unavailable (Sarapas et al, 2011). Using these five studies, seven trauma-specific case–control bio-sets were curated by parsing individuals provided by Neylan et al, 2011 into two separate sub-groups (ie, combat- and assault-related traumas) and Mehta et al, 2013 into two separate sub-groups (ie, childhood- and IP-related traumas). All data were obtained from either the corresponding authors of the original studies or from publicly available data repositories. For diagnostic criteria, we relied on those used at each study site, some of which were based on clinician assessments and others based on standardized screening tool. There was no additional filtering of subjects based on medical comorbidities beyond what was described on the original studies.
Gene Expression Data Processing and Quality Control
All statistical analyses were conducted in the statistical package R. Data from each study were processed, normalized, and quality treated independently (see Supplementary Figure 1 for workflow). Briefly, when multiple microarray probes mapped to the same HGNC symbol, the probe with the highest average expression across all samples was used for further analysis. Normalized data were inspected for outlying samples using unsupervised hierarchical clustering of samples and principal component analysis to identify potential outliers outside two SDs from these grand averages. Combat batch correction (Leek et al, 2015) was applied to remove systematic sources of variability other than case/control status, such as technical, clinical, or demographic factors both within each study (as necessary), and then across all studies using common gene symbols, forming the bases for subsequent mega-analytic case–control groups. The frequencies of circulating immune cells were estimated for each individual in each study using Cibersort cell-type de-convolution (https://cibersort.stanford.edu/) (Newman et al, 2015). See Supplementary File for full details on these data processing steps.
Differential Gene Expression Analyses
Trauma-specific analyses
Differential gene expression (DGE) was performed independently for each of the seven case–control groups using the limma package (Ritchie et al, 2015) to detect relationships between diagnostic status and gene expression levels. The covariates age and sex were included in all models to adjust for their potential confounding influence on gene expression between main group effects. To determine the relatedness of DGE signatures across the seven trauma-specific groups, each gene list was converted into a matrix of binary gene presence/absence calls with respect to each group and a Jaccard coefficient was applied to create a gene-based phylogeny, as previously described (Diaz-Beltran et al, 2016).
Mega-analytic analyses
To increase statistical power and test for biological sex- and trauma-specific gene signatures, individual samples from different experiments were combined based on (i) biological sex and (ii) type of trauma to form three large mega-analytic case–control groups. First, common gene symbols across all available samples were identified (ngenes=4062). Then, Combat batch correction (Leek et al, 2015) was applied to remove systematic sources of variability other than case/control status, such as technical factors (eg, difference technologies, duration of time from PTSD onset to blood acquisition), clinical factors (eg, comorbidities), or demographic factors (eg, ethnicity) across all individual studies. Finally, DGE analysis was performed for each mega-analytic case–control group controlling for effects of age, and unless otherwise specified, the significance threshold was a nominal P-value <0.05. This nominally significant P-value was used to yield a reasonable number of genes to include within functional annotation and gene network analyses.
Weighted Gene Co-Expression Network Analysis
Weighted gene co-expression network analysis (WGCNA) (Langfelder and Horvath, 2008) was used to build signed co-expression networks using a total of 4062 genes found in common across all experiments (see Supplementary Information for details). Two broad analyses were performed. First, a series of module preservation analyses sought to determine whether PTSD development influences the underlying gene co-regulatory patterns, as being preserved or disrupted, compared to controls, and vice versa. For these analyses, module preservation was assessed using a permutation-based preservation statistic, Zsummary, implemented within WGCNA with 500 random permutations of the data (Langfelder et al, 2011). Zsummary takes into account the overlap in module membership as well as the density and connectivity patterns of genes within modules. A Zsummary score <2 indicates no evidence of preservation, 2<Zsummary<10 implies weak preservation and Zsummary >10 suggests strong preservation. Second, to increase confidence and power to detect biologically meaningful modules, a consensus network was built to use all available samples. Once modules were identified from the consensus network, modules were assessed for significant associations to PTSD diagnostic status, sex, and mode of trauma. Singular value decomposition of each module’s expression matrix was performed and the resulting module eigengene (ME), equivalent to the first principal component, was used to represent the overall expression profiles for each module. Differential analyses of MEs was performed using Bayes ANOVA (Kayala and Baldi, 2012) (parameters: conf=12, bayes=1, winSize=5), comparing between diagnostic status, sex, and mode of trauma, correcting P-values for multiple comparisons with post hoc Tukey tests.
Functional Annotation and Protein Interaction Networks
The ToppFunn module of ToppGene Suite software (Chen et al, 2015) was used to assess enrichment of gene ontology (GO) terms using a one-tailed hyper geometric distribution with family-wise false discovery rate (FDR) at 5%. GO semantic similarity analysis was used to assess shared/unique gene content among GO terms using the GoSemSim semantic similarity R package (Yu et al, 2015), and default semantic contribution factors (‘is_a’ relationship: 0.8 and ‘part_of’ relationship: 0.5). Second, gene modules were tested for overrepresentation of PTSD genome-wide association study (GWAS) signatures obtained from the DisGenNet database (Pinero et al, 2015), retrieved using the disease-term query ‘PTSD’. Third, DGE signatures were used to build direct protein–protein interaction (PPI) networks, which can reveal key genes/transcription factors mediating the regulation of multiple target genes. PPIs were obtained from the STRING database (Franceschini et al, 2012) with a signature query of DGE lists from the mega-analytic case–control comparisons. We used a combined STRING score of >0.4 (ie, medium-to-high confidence interactions). For visualization, the STRING network was imported into CytoScape (Shannon et al, 2003).
Construction of PTSD Blood-Based Diagnostic Classifiers
BRB-Array Tools supervised classification methods (Simon et al, 2007) were used to construct gene expression classifiers. Three models were specified to distinguish PTSD cases from controls relative to: (1) men exposed to combat trauma, (2) men exposed to IP traumas, and (3) women exposed to IP traumas. Each model consisted of three steps. First, to ensure a fair comparison, all genes in the training data with P<0.05 were subjected to classifier construction, respective for each mega-analytic case–control group. This heuristic rule of thumb approach was used to cast a wide net to catch all potentially informative genes, while false-positives would be pared off by subsequent optimization and cross-validation steps. Second, classifiers composed of different numbers of genes were constructed by recursive feature elimination (RFE). RFE provided feature selection, model fitting, and performance evaluation via identifying the optimal number of features with maximum predictive accuracy. Third, the ability for RFE to predict group outcome was assessed by support vector machines (SVM) and compared to four different multivariate classification methods (ie, diagonal linear discriminant analysis (DLDA), nearest centroid (NC), first-nearest neighbors (1NN), three-nearest neighbors (3NN)). For each of the three models, classification accuracies are reported for both the training data (70% of data) and the completely withheld test data (30% of data) as area under the receiver operating curve (AUC).
Statistical Power and Sample Size Computation
We estimated the expected discovery rate (EDR), a multi-test equivalent to power, and sample size at a fixed number of biological replicates (n) and type I error rate (α) using the PowerAtlas software (Page et al, 2006). This sample size calculation method is based on studies of the distribution of P-values from DGE analyses from microarray studies controlling for EDR. For evaluating which n is best suited for a future study, we set the average probability of detecting an effect (EDR) to be >0.8 and α=0.05.
Code and Data Availability
Computational code and quality controlled gene expression data are available upon request to the corresponding author and can also be directly downloaded at https://github.com/BreenMS/PTSD-blood-transcriptome-mega-analysis.
Results
Literature Search and Data Curation
A total of five cross-sectional PTSD studies met our criteria (Patients and Methods) for which raw gene expression data and clinical covariates were available (Table 1). From these five studies, seven trauma-specific case–control groups were derived, including three groups exposed to combat traumas, one group exposed to assault traumas, one group exposed to childhood-related traumas, one group exposed to ER accident-related traumas, and one group exposed to ‘other’ IP-related traumas, which could not be explicitly defined. These seven trauma-specific groups were later combined to form three large mega-analytic case–control groups, aimed at explicitly modeling for sex- and trauma-related differences (Table 1, bottom), including: (i) men exposed to combat traumas (nPTSD=85, nControl=84, kgenes=10 112); (ii) men exposed to IP traumas (nPTSD=45, nControl=67, kgenes=4378); and (iii) women exposed to IP traumas (nPTSD=99, nControl =160, kgenes=4378).
Table 1. Blood-Based Transcriptome-Wide Studies of Posttraumatic Stress Disorder Included in the Mega-Analysis.
| Study | Data type | PTSD (n) | Controls (n) | % Female | Age (years) | Sample type | Predominant ancestry, % | Genes analyzed | Additional sample informationa |
|---|---|---|---|---|---|---|---|---|---|
| Breen et al, 2015 GSE64814 | Poly-A enriched, 50 bp paired-end sequencing on Illumina Hi-Seq 2000 | 46 combat | 46 combat | 0% | 23.3±3.22 | Isolated leukocytes | European, 55.5% | 13 944 | Marine Resilience Study (MRS) sample of US. Marines who served a 7-month combat deployment. Blood was drawn 3 months post-deployment for each participant. PTSD symptoms were assessed at this time using CAPS and diagnosis was determined using DSM-IV criteria for partial or full PTSD |
| Tylee et al, 2015 GSE63878 | Affymetrix Hu-Gene 1.0 ST Array | 24 combat | 24 combat | 0% | 22.2±3.1 | Isolated leukocytes | European, 72.0% | 22 772 | MRS sample of US. Marines who served 7–9-month combat deployment. Blood was drawn 3 months post-deployment for each participant. PTSD symptoms were assessed and diagnostic status was determined using the CAPS |
| Mehta et al, 2013 | Illumina HT-12 version 3.0 | 54 childhood 67 other IP | 38 childhood 166 other IP | 73.6% | 42.0±12.6 | Whole blood | African American, 88.9% | 9193 | General medical community-based sample (Atlanta, Georgia) selected for traumatic exposure. Blood samples were obtained years after trauma exposure. PTSD symptoms were assessed using the PSS and diagnosis was determined by applying DSM-IV criteria to PSS items; control subjects were negative for current or lifetime PTSD |
| Neylan et al, 2011 | CodeLink Human Whole Genome BioArrays | 15 combat 14 assault | 14 combat 15 assault | 26.9% | 30.0±6.0 | Isolated CD14+ monocytes | European, 56.7% | 17 988 | Recruited through Veterans Affairs Medical Center PTSD Outpatient Program (San Francisco, California) and through community fliers. Included both assault and combat-related traumas. Blood samples were obtained years after trauma exposure. Symptoms were assessed and diagnostic status was determined using the CAPS |
| Segman et al, 2005 | Affymetrix Human Genome-U95A | 9 ER-trauma | 8 ER-trauma | 37.5% | 31.1±11.4 | Isolated mononuclear cells | Jewish ancestry, 100% | 9668 | Acute trauma exposure in emergency room setting. Samples collected at 4 months after trauma onset. PTSD cases met DSM-IV criteria for PTSD 4 months post trauma |
| Total: 5 studies | 229 | 311 | |||||||
| Combat trauma, men | Mega-analytical group 1 | 85 | 84 | 0% | 24.4±4.7 | Combined | European, 63.6% | 10 112 | |
| IP trauma, men | Mega-analytical group 2 | 45 | 67 | 0% | 41.1±12.8 | Combined | African American 69.7% | 4378 | |
| IP trauma, women | Mega-analytical group 3 | 99 | 160 | 100% | 39.5±12.3 | Combined | African American 76.1% | 4378 |
Abbreviations: CAPS, Clinician administered PTSD Scale; CD14+, cluster of differentiation 14-positive; DSM-IV, Diagnostic and Statistical Manual of Mental Disorders-IV; IP, interpersonal trauma.
We attempted to include available information pertaining to each sample, with particular emphasis on the type of trauma exposure, the time point of biological sample acquisition, and the diagnostic criteria used for each study. All details were obtained from the referenced articles.
Between-Trauma Comparisons
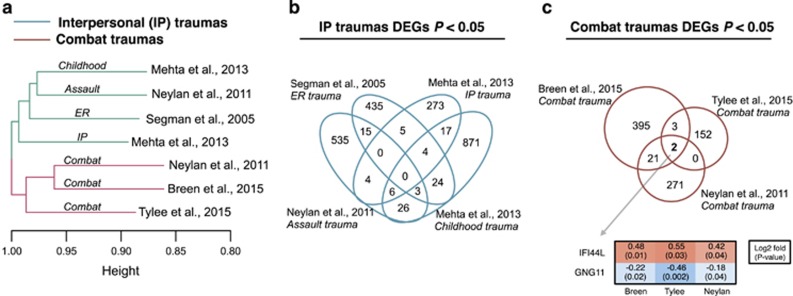
Following standardized data pre-processing procedures (see Patients and Methods and Supplementary File), the proportions of circulating immune cells were estimated for all individuals since complete cell counts with leukocyte differentials were not available. Comparative analyses of the estimated immune cell-type proportions showed no significant differences between PTSD cases and controls in any of the seven trauma-specific case–control groups (Supplementary Table 1), suggesting that cell-type frequencies would not confound downstream analyses. Subsequently, to determine the overall relatedness of the trauma-specific groups, seven lists of covariate adjusted DGE signatures were generated, then converted into a binary matrix of gene presence/absence calls. Distance-based clustering with pair-wise similarity was measured via Jaccard coefficient. A high degree of DGE similarity formed two distinct branches that clustered the five IP trauma groups how from the three combat trauma groups (Figure 1a). Pair-wise overlaps of the seven DGE lists were used to further quantify this result (Figure 1b and c) and identified a number of significant overlaps between: childhood and assault traumas (∩=35, P=0.05); childhood and ER-related traumas (∩=31, P=0.03); assault and ER-related traumas (∩=18, P=0.04); ER-related traumas and IP-related traumas (∩=9, P=0.03); and combat traumas between Breen et al, and Neylan et al, (∩=23, P=6.9e−9) (Supplementary Table 2). No genes were consistently differentially expressed across all five IP trauma groups (Figure 1b), although two genes showed consistent but weak effects across all three combat trauma groups (Figure 1c); interferon-induced protein 44 like (IFI44L) was overexpressed, while G protein subunit gamma 11 (GNG11) was underexpressed in PTSD cases relative to controls. Notably, no gene in any of the above comparisons survived FDR P<0.05.
Figure 1.
Concordance of differential gene expression (DGE) analyses across seven trauma-specific case–control groups. (a) Jaccard clustering of PTSD DGE signatures from the seven trauma-specific case–control groups. Overlap of PTSD DGE signatures found in common across (b) interpersonal (IP) traumas and (c) combat traumas. For combat-related traumas, interferon-induced Protein 44 like (IFI44L) was consistently overexpressed and G protein subunit gamma 11 (GNG11) was consistently underexpressed in PTSD cases relative to control individuals. All analyses were adjusted for age and sex. Supplementary Table 2 contains full lists of overlapping gene symbols.
Mega-Analytic Comparisons
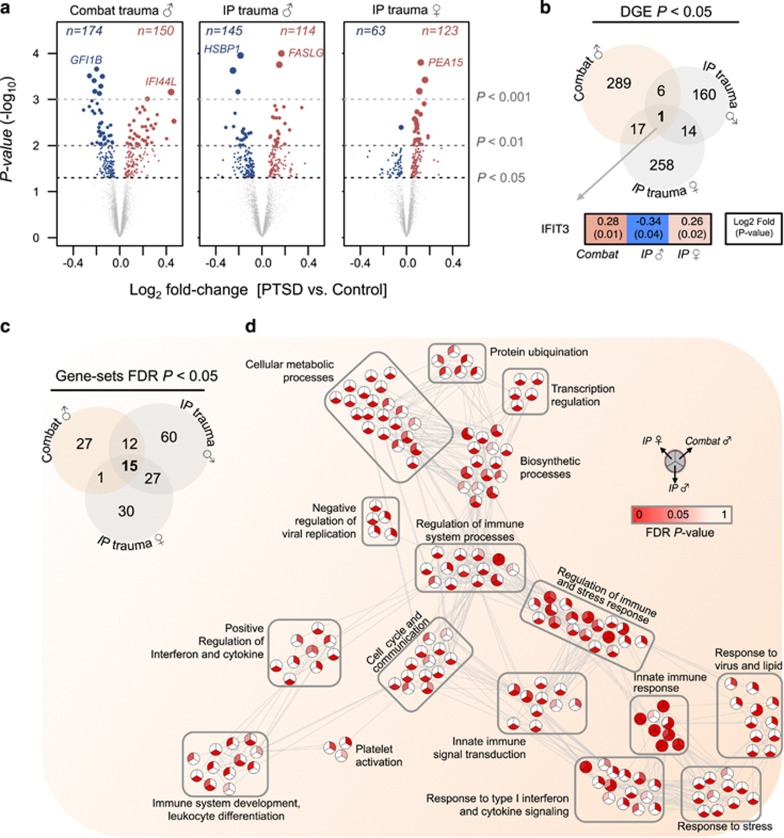
To increase statistical power, these seven trauma-specific case–control groups were combined to form three large mega-analytic case–control groups designed to explicitly model for differences in sex and modes of trauma (ie, combat and IP traumas) and DGE lists were generated for each comparison (Supplementary Table 3). Comparatively equal numbers of over- and underexpressed genes were observed in men exposed to combat traumas (nup=150, ndown=174) and men exposed to IP traumas (nup=114, ndown=145), while women exposed to IP traumas displayed significantly more overexpressed genes than underexpressed genes (nup=123, ndown=63, P=1.2e−05; two-tailed proportions test) (Figure 2a). DGE indicated small, but significant, gene overlaps between men exposed to combat traumas and women exposed to IP traumas (∩=18, P=1.1e−08), men exposed to combat traumas and men exposed to IP traumas (∩=7, P=0.04), and between men and women exposed to IP traumas (∩=15, P=2.3e−08) (Figure 2b). Notably, one gene, interferon-induced protein with tetratricopeptide repeats 3 (IFIT3), was significantly differentially expressed in all comparisons, albeit with different directions of change. Overall, DGE signatures were associated with PTSD diagnosis and not with any other factors, including age, ancestry, study site, or estimated cell-type frequencies (Supplementary Figure 2).
Figure 2.
Differential gene expression (DGE) and gene ontology (GO) analyses for the three mega-analytic case–control comparisons. (a) Volcano plots compare the extent of log2 fold-change and −log10 P-value significance for DGE signatures in all three comparisons. The top over and under expressed genes are labeled for each comparison. Overlap of (b) significant DGE signatures and (c) enriched GO gene sets for all comparisons are displayed. (d) Relatedness of all significantly dysregulated gene sets by semantic similarity. Nodes represent GO terms and edges represent semantic similarity >0.5 (high degree of gene overlap). Nodes are split into thirds and shaded by FDR P-value significance for each mega-analytic group.
Though few genes were differentially expressed in all comparisons, functional annotation of these DGE signatures indicated a high degree of overlap of commonly perturbed biological processes between men exposed to combat- and IP-related traumas (∩=27, P=2.1e−14), men exposed to combat traumas and women exposed to IP traumas (∩=16, P=3.3e−12), and between men and women exposed to IP-related traumas (∩=42, P=1.3e−137) (Figure 2c). In addition to several common biological processes, numerous unique gene sets were also identified for each comparison (Supplementary Table 4) suggesting that differences in sex and trauma types may impact distinct biological processes. However, in exploring the semantic similarity between these distinct gene sets, we identified a series of relevant, biologically meaningful interactions, positioning each distinct biological process as a component of a broad ‘stress response system’ (Figure 2d). To support this observation, we tested whether candidate genes that are dysregulated together indeed interact with each other at the protein level. A significant overrepresentation of direct protein interactions was identified for each DGE list, and a union of all three networks was constructed (Supplementary Figure 3). Notably, IFIT3 demonstrated protein-level interactions with partners across all three networks, among other interferon proteins such as IFI44L, IRF7, IFI44, IFI35, IFIT4, and IRF4. The network generated from men exposed to combat traumas featured several genes with a high degree of connectivity involved in type I interferon signaling and antiviral responses, including DDX58, IFIH1, IFIT1/2, MX1, RSAD2, STAT1, and members of the OAS gene family. Comparably, genes related to men with a history of IP trauma formed a relatively unique network with the most highly connected genes included EZR, H2AFZ, IMPDH2, JUND, STAT5B, and SYK. The network generated for women with a history of IP trauma, consisted of ABL1, ATM, TNF, and UBB, which demonstrated the highest degree of connectivity.
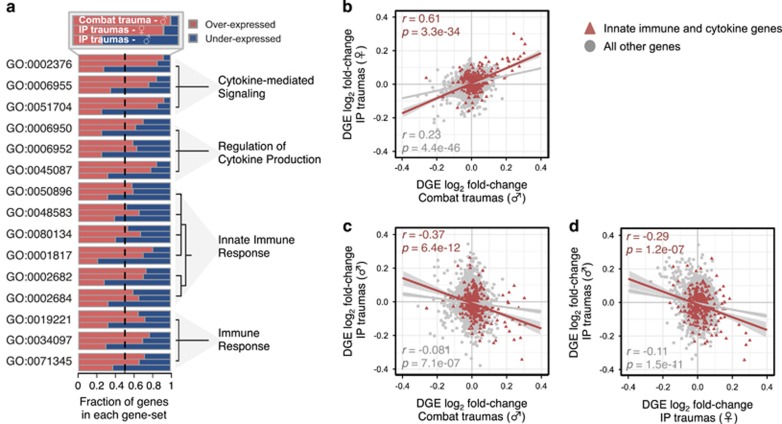
Markedly, of the 15 biological processes found at the intersection of all comparisons (Figure 2c), all terms were strongly enriched for innate immune responses, cytokine signaling, and cytokine production (Figure 3a). Surprisingly, the genes within each of these gene sets were predominantly overexpressed among men exposed to combat traumas and women exposed to IP traumas, but were underexpressed in men exposed to IP traumas. To further quantify this observation, the concordance of transcriptome-wide DGE patterns was calculated among the three mega-analytic case–control comparisons first constraining to all genes specific to innate immune or cytokine signaling and then using all remaining genes (Figure 3b–d). Positive associations in changes of innate immune and cytokine genes were observed between men exposed to combat traumas and women exposed to IP traumas (r=0.61, P=3.3e−34), and negative associations were observed between men exposed to combat and IP traumas (r=−0.37, P=6.4e−12), and between men and women exposed to IP traumas (r=−0.29, P=1.2e−12). Next, we sought to determine whether these biological processes were specific to PTSD or found in common with other neuropsychiatric disorders including major depression, schizophrenia, bipolar disorder, and autism spectrum disorder, by implementing series of cross-disorder overlap comparisons, at both the individual gene and GO level (Supplementary Figure 4; Supplementary Table 4). Indeed, the majority of innate immune and cytokine signatures were more strongly related to a universal diagnosis of PTSD across differences in biological sex and modes of traumas, rather than in these other disorders.
Figure 3.
Concordance of transcriptome-wide innate immune and cytokine signatures. (a) Semantic similarity for all 15 common GO term pairs (left) clustered by hierarchical clustering method with the fraction of over- and underexpressed genes for each GO term (middle) as well as common ancestors (right). Log2 fold changes (PTSD vs controls) were used in a series of pair-wise correlations among the three mega-analytic case–control groups and focused on innate immune and cytokine genes relative to all other genes. (b) A positive association between PTSD-affected men exposed to combat traumas and PTSD-affected women exposed to IP traumas. Negative associations between PTSD-affected men exposed to IP traumas and (c) PTSD-affected men exposed to combat traumas and (d) PTSD-affected women exposed to IP traumas. Pearson correlation coefficients were applied.
Stratified Gene Co-Expression Module Preservation Analyses
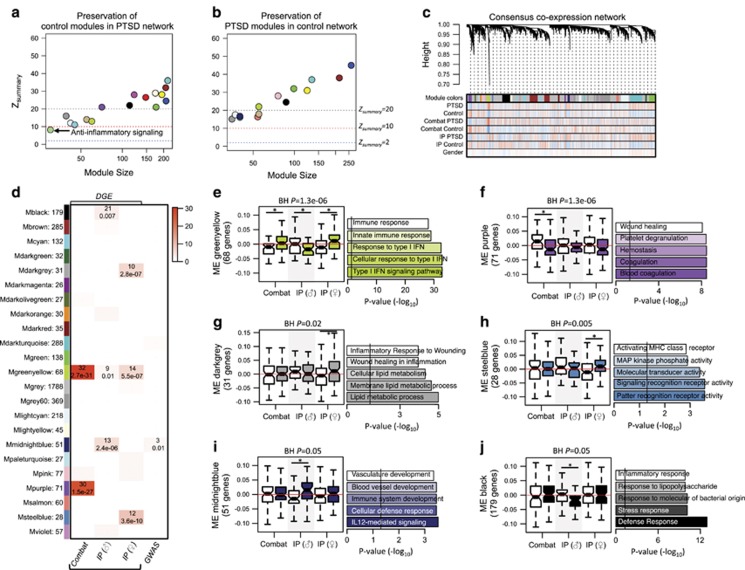
WGCNA was used to assess the extent of module preservation by integrating all PTSD cases compared to all control individuals using a permutation-based preservation statistic (Zsummary, Patients and Methods). This analysis identifies large differences in gene co-regulatory patterns as being disrupted in PTSD cases relative to controls, or vice versa (Figure 4a and b). In control individuals, 16 modules were identified and one module implicated in anti-inflammatory signaling was weakly preserved (Zsummary=8.6) in PTSD cases, including genes IL10, TNFSF14, and LILRB1. In the reverse approach, 14 modules were identified across PTSD cases and all were highly preserved within control individuals, indicating that major changes in the underlying gene–gene connectivity may not be a basis for the pathology of PTSD. A separate series of between trauma type comparisons were also performed (see Supplementary File and Supplementary Figure 5 for details).
Figure 4.
Consensus weighted gene co-expression network analysis (WGCNA). (a) Module preservation identified one module in control individuals with weak preservation in PTSD cases. (b) All PTSD modules were preserved in control samples. (c) Hierarchical clustering tree (dendrogram) of the consensus network and all samples comprising 4062 genes. Each line represents a gene (leaf) and each low-hanging cluster represents a group of co-expressed genes with similar network connections (branch) on the tree. The first band underneath the tree indicates the 23 detected modules and subsequent bands indicate gene-trait correlation when red indicates a strong relationship and blue indicates a strong negative relationship. (d) Gene modules were enriched for DGE signatures and PTSD GWAS signatures curated from DisGenNet database (x-axis). The number of genes within each colored gene module are depicted (y-axis). The top number in each cell indicates the number of genes overlapping and the bottom number indicates the P-value significance of overlap using the Fisher’s exact test. (e–j) A Bayes ANOVA was used on ME values to test for significance between case–control status across different biological sex and trauma types and * indicates BH P<0.05, implying significant PTSD differences. For each module, the top five most significant biological processes and/or pathways are reported.
Consensus Gene Co-Expression Network Analyses
Subsequently, we used the higher confidence and completeness of a consensus network by combining all individuals across the three mega-analytic case–control groups (Figure 4c). This analysis identified 23 co-expression modules, which were tested for enrichment of DGE signatures and PTSD-related GWAS signals (Figure 4d). Module eigenvalues (MEs) were then subjected to a Bayes ANOVA testing to compare the extent of module expression differences between diagnostic status, sex, and type of trauma (Figure 4e–j). A greenyellow module (68 genes) implicated in type I interferon-mediated signaling cascades and enriched with differentially expressed genes from all three mega-analytic comparisons was significantly overexpressed in PTSD-affected men exposed to combat traumas and PTSD-affected women exposed to IP traumas, but was underexpressed in PTSD-affected men exposed to IP traumas. A purple module (71 genes) implicated in blood coagulation and wound healing was underexpressed in PTSD-affected men exposed to combat traumas. A darkgrey module (31 genes) enriched with inflammatory response to wounding and cellular lipid membrane metabolic process, and a steelblue module (28 genes) implicated in intracellular pattern recognition receptor signaling and mitogen-activated protein kinase (MAPK) phosphate activity were both overexpressed among PTSD-affected women exposed to IP traumas. A midnight blue module (51 genes) enriched for IL-12-mediated signaling and vascular development was overexpressed among PTSD-affected men exposed to IP traumas. This module also contained three potential PTSD risk-related genes (FASLG, IFN-γ, RORA) previously identified through GWAS, reflecting greater than chance overlap (P=0.012). A black module (179 genes) implicated in immune response and response to bacterial lipopolysaccharide was underexpressed among PTSD-affected men exposed to IP traumas.
PTSD Blood-Based Diagnostic Classifier Evaluation
Supervised class prediction methods were used to directly assess the putative clinical utility of blood-based gene expression measurements for objective PTSD diagnosis, as well as to identify any discriminant gene(s) that may have been overlooked in our previous analyses. Three models were specified to distinguish PTSD cases from control individuals for each mega-analytic group. Classification accuracies were reported on the training data (70% of data) as well as independently withheld test data (30% of data), and SVM learning consistently outperformed the other methods (Supplementary Figure 6). First, when distinguishing between PTSD cases and control individuals exposed to combat traumas, classification accuracies reached 81% in the training data and 60% in the withheld test data when the expression of 40 genes was used with SVM classification. Second, when separating PTSD-affected men from controls individuals exposed to IP traumas, classification accuracy reached 77% in the training data and 65% on the withheld test data when the expression of 60 genes was used with SVM. Third, when separating PTSD-affected women from controls individuals exposed to IP traumas, classification accuracy reached 67% in the training data and 58% on the withheld test data when the expression of 25 genes as used with SVM; two common genes were selected by both IP-related trauma models across men and women (GNB5 and DGCR14). Further details regarding these analyses are provided in Patients and Methods and Supplementary Table 6.
Statistical Power and Sample Size Estimation
To inform the design of future cross-sectional studies in PTSD, we estimated the expected discovery rate (EDR; a multi-test equivalent to power) and sample size for each mega-analytic case–control group using lists of P-values derived from DGE analyses. Effect sizes estimated from these data were assumed to be fixed with a nominal error rate α=0.05 and several different sample sizes (n) were evaluated. To determine how many case and control samples need to be included in a future study, we set the threshold of inclusion to EDR>0.8. Overall, the sample size needed to reach a power of 0.8 for men exposed to IP trauma was n of 700, for women exposed to IP trauma was n of 7000, while men exposed to combat trauma was n of 10 000 (Supplementary Table 7).
Discussion
To our knowledge, this is the largest transcriptome-wide analysis of PTSD conducted to date. Our combined mega-analysis covered 229 PTSD cases and 311 comparison individuals, enabling us to increase statistical power and to explicitly test whether differences in sex and trauma type play a role in perturbing common or distinct molecular pathways in PTSD. A battery of statistical tests were applied and we report several findings. First, re-analyses of seven trauma-specific case–control groups revealed a high degree of relatedness among IP-related traumas separate from combat-related traumas. Second, once individual samples were combined to form three mega-analytic case–control groups, we observed unique PTSD DGE signatures in men exposed to combat traumas, men exposed to IP traumas and women exposed to IP traumas, which all converged on shared biological processes of innate immune, cytokine, and type I interferon signaling. Third, stratified network analysis identified low module preservation between control individuals exposed to different traumas, but high module preservation between PTSD cases exposed to the same types of trauma, suggesting that the underlying molecular response to different trauma types may be more homogenous in PTSD cases. Fourth, one anti-inflammatory module within control individuals was weakly preserved across all PTSD cases, indicating the disruption/absence of anti-inflammatory gene co-regulation within PTSD cases. Fifth, upon integrating all data to construct one consensus network, numerous sex and trauma-specific modules were identified, and one module implicated in innate immunity and type I interferon signaling was significantly associated to PTSD in all three mega-analytic groups. Sixth, supervised multivariate classification methods constructed diagnostic PTSD classifiers for each mega-analytic group with moderate-to-low classification accuracies on withheld test data. Taken together, our analyses indicate that while small-to-moderate effect sizes are the standard for cross-sectional post-trauma studies of PTSD, our findings consistently converge on similar downstream inflammatory pathways irrespective of sex and the type of traumatic event.
A novel finding was the small, but significant, between-trauma overlap of DGE signatures indicating the existence of a trauma-specific and across-trauma convergent gene regulation and signaling (Figure 1). Indeed, we previously tested the hypothesis that differences in trauma may impact the stress response in PTSD and we identified distinct gene expression signatures between PTSD cases with and without a history of childhood maltreatment that also converged on similar cellular processes (Mehta et al, 2013). Here, we extend upon these results and position them among a broad background of individuals exposed to a range of different traumatic events. Notably, not one of the seven trauma-specific case–control comparisons resulted in a gene passing FDR P-value <0.05, indicating the underpowered nature of the observed data. Thus, to increase statistical power, this initial result provided enough empirical support to combine the data to form three mega-analytic case–control groups, enabling us to explicitly model for differences in sex and trauma.
Our central finding was the identification of largely unique DGE perturbations specific to each mega-analytic case–control group, which converged on common biological processes of innate immune, cytokine, and type I interferon signaling cascades (Figure 2). Our consensus network approach validated and refined this result through identification of a discrete co-regulated gene module (68 genes) implicated in innate immune and type I interferon signaling that displayed divergent expression patterns across sex and traumas in PTSD (Figure 4e–j). This result was previously reported in PTSD cases exposed to combat trauma (Breen et al, 2015), and is now replicated across a larger sample. With respect to potential mechanisms, some of the earliest observed effects of inflammatory cytokines in PTSD underline their impact on the HPA axis, via negative feedback regulation (Michopoulos et al, 2015). Enhanced negative feedback regulation of the HPA axis function is a hallmark of PTSD and is reflected by increased responsiveness to glucocorticoids as manifested by decreased cortisol concentrations following dexamethasone administration (Michopoulos et al, 2015; Yehuda et al, 2002). Inflammatory cytokines have also been shown to access the brain and interact with virtually every pathophysiological domain relevant to PTSD, including neurotransmitter metabolism, neuroendocrine function, and neural plasticity (Felger and Lotrich, 2013). In doing so, peripheral cytokine signals activate relevant brain cell types that serve to amplify central inflammatory responses and conserved behavioral responses. Notably, we also found evidence for biological processes significantly overrepresented in only one of the three mega-analytic groups, suggesting that sex and type of trauma may influence different molecular pathways in PTSD (Figure 2c). However, in determining the relatedness between each of these distinct biological responses, we found that all dysregulated biological processes interacted and collectively formed a biologically meaningful ‘stress response system’ (Figure 2d). Indeed, a dysfunctional HPA axis–immune interface has been previously associated with similar immune and metabolic disturbances, including cell cycle, altered cytokine balance, blood coagulation, and lipid and metabolic processes (Silverman et al, 2013; Silverman and Sternberg, 2012; Elenkov et al, 2000). Under this standard, each dysregulated biological process represents one piece of a larger stress response system, irrespective of sex and trauma, that ultimately converges on shared inflammatory pathways.
Some of the observed PTSD-related effects involved lower expression levels for cytokine-related genes, specifically in men exposed to IP trauma compared to women, and men exposed to combat trauma (Figure 3). Differences in inflammatory cytokine levels in trauma survivors with PTSD have previously been reported (Gill et al, 2009). Discrepancies may potentially reflect different degrees of HPA axis response suppression to glucocorticoid activation (Freidenberg et al, 2010), or alternatively, differences associated with the duration since the traumatic experience as well as other confounding factors. One study examining the levels of inflammatory markers within a refugee population with PTSD postulated that differences could partly be explained by a variable environmental component associated with less antigen exposure (Söndergaard et al, 2004). We also acknowledge that gene expression perturbations may be fundamentally different in controls with prolonged exposure to conflict zones compared to controls exposed to IP traumas, which may be influencing these results. To address this issue, a series of pair-wise co-expression preservation analyses were performed using matched controls exposed to different types of trauma (Supplementary Figure 5). These analyses indicated that gene co-expression modules involved in processes other than inflammatory signatures differed on the basis of exposure to different traumatic events.
Distinct gene expression perturbations were also identified, including (i) one wound-healing module downregulated in men with a history of combat trauma, (ii) two modules implicated in lipid processes and MAPK activity upregulated in women exposed to IP-related trauma, and (iii) one IL-12-mediated signaling module upregulated in men exposed to IP-related trauma. Regarding to the first distinction (i), research highlights the role of platelets in innate and adaptive immune responses and suggest that platelet activation and reactivity is dysregulated by mental stress. A stress response involving blood platelets has also been shown to be a critical biomarker of hemostatic, thrombotic, and inflammatory perturbations (Pacák and Palkovits, 2001). Notably, this result is also a re-affirmation of our previous finding indicating decreased wound healing and blood coagulation in war-veterans with PTSD (Breen et al, 2015), now replicated in a larger cohort of samples. Regarding the second distinction (ii), the MAPK pathway functions as a mediator of cellular stress, including inflammation, by also modulating the levels of glucocorticoid receptor phosphorylation, ultimately leading to differences in cellular transcriptional activity (Galliher-Beckley et al, 2011; Reul, 2014). Since signaling cascades, such as the MAPK, couple to numerous receptors for stress-related neurotransmitters and neuropeptides (Whitaker et al, 2014), future work should determine more precisely which neurotransmitter signaling systems are driving the observed traumatic stress- and sex-induced changes. Regarding the final distinction (iii), the IL-12 signaling pathway initiates innate and adaptive immune responses in part by promoting NK cell toxicity as well as the differentiation of naive CD4+ T cells into T helper 1 cells, and induces the production of IFN-γ, which is also a member of this gene module. Here, upregulation of IL-12 signaling in men exposed to IP trauma indicates immune priming that may promote inflammation. With respect to sex dimorphism, increased percentage of IL-12 producing monocytes and lymphocytes in response to physiological concentrations of testosterone has been reported in men compared to women (Posma et al, 2014). Taken together, these distinct biological perturbations all align with a common pro-inflammatory pathology across sex and modes of trauma in PTSD. However, a great deal of research is needed to further delineate the precise mechanisms, as well as cause and effect relationships, underlying these inflammatory signatures and sex disparities.
Our study also has several limitations. First, sample size and power estimates indicated that our enhanced sample size is still underpowered and a substantially larger number of biological replicates are needed (Supplementary Table 7). These estimates are echoed by our inability to identify multiple test corrected DGE signatures and to construct accurate diagnostic blood-based PTSD classifiers, all of which reported low-to-moderate classification accuracies on withheld test data. Second, it is likely that our results may be influenced by clinical heterogeneity (eg, medical comorbidity, medication) among PTSD cases, potentially contributing to the diminished power. While our effort to carefully measure the contribution of potential confounding factors on our gene expression results demonstrated that DGE signatures were associated with PTSD diagnosis and not with any other factors (eg, age, ancestry, study site, cell-type frequencies), other unmeasured factors may also influence the results. Additionally, to fully understand the contribution of ancestry to gene expression variation, future studies may consider integrating principal components of ancestry from paired GWAS data if available. Third, we were unable to distinguish biological sex differences for combat-related traumas due to a large ascertainment bias of men with a history of combat exposure. Fourth, though our estimates of cell-type fractions implied no differences between case–control groups, we are unable to determine gene expression changes specific to any particular cell type. Finally, as the combined data are cross-sectional with considerable variation in the amount of time from PTSD onset to blood sample acquisition, we are unable to determine whether expression differences represent expression signature of past or current PTSD, or it is a marker of pre-trauma vulnerability to PTSD development. Similarly, some traumas are defined by event (eg, combat, assault) but others are defined by time (eg, childhood) or place (eg, ER). Likewise, both childhood and ER traumas might be due to similar trauma types, including assaults, which are unknown in the current study and may effect the interpretation of our results.
In sum, these data provide evidence for shared inflammatory profiles in peripheral blood gene expression across sex and modes of trauma in PTSD as evident by transcriptional dysregulation and co-expression on processes of innate immune, cytokine, and type I interferon signaling. Moreover, the fact that several unique biological processes were also affected across sex and trauma types that ultimately formed components of a broader stress response system, underscore a shared underlying molecular pathology. While existing animal and cellular work support the sex-dependent effects specific to MAPK and IL-12 signaling modules, further research is needed to delineate cause and effect relationships. Collectively, these findings may have implications for identifying objective diagnostic biomarkers, disease mechanisms, and therapeutic interventions in immune disturbances for PTSD.
Funding and disclosure
The Marine Resiliency Study (MRS) was supported by VA Health Service Research and Development project no. SDR 09-0128, the Marine Corps, and the Navy Bureau of Medicine and Surgery (DGB) and MRS-II (DGB, VBR) and its Demonstration Project (CMN) by the Naval Medical Research Center's Advanced Medical Development program (Naval Medical Logistics Command Contract #N62645-11-C-4037). MRS-II acknowledges support from the administrative core, A Patel, A De La Rosa, members of the MRS-II Team, and the Veterans Medical Research Foundation (VMRF). JDB is co-inventor of two patents for genes associated with PTSD (no. 9243293 and 20120039812). The authors declare no conflict of interest.
Acknowledgments
We gratefully acknowledge the individual volunteers and their families, as well as the Marine and Navy volunteers for their military service and for their participation in these studies. Additionally, we thank Segman and colleagues (2005), as well as others, who have placed their data in the public domain. We thank the Marine and Navy volunteers for their military service and for their participation in this study. We also thank Dr Anna Tocheva for further critical reading/assessment of our manuscript.
Author contributions
DST and SJG assembled all the data from independent sources. MSB designed the research question, analyzed the data, and wrote the manuscript with DST. All remaining authors contributed to the generation of the original data and provided critical reading/assessment of the current manuscript.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Breen MS, Maihofer AX, Glatt SJ, Tylee DS, Chandler SD, Tsuang MT et al (2015). Gene networks specific for innate immunity define post-traumatic stress disorder. Mol Psychiatry 20: 1538–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen MS, Stein DS, Baldwin D (2016). Systematic review of blood transcriptome profiling in neuropsychiatric disorders: guidelines for biomarker discovery. Hum Psychopharmacol 31: 373–381. [DOI] [PubMed] [Google Scholar]
- Chen J, Bardes E, Aronow B, Jegga A (2015). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37: W305–W311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Janicki-Deverts D, Doyle WJ, Miller GE, Frank E, Rabin BS et al (2016). Chronic stress, glucocorticoid receptor resistance, inflammation, and disease risk. Proc Natl Acad Sci USA 109: 5995–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalakis N, Cohen H, Nievergelt CM, Baker DG, Buxbaum JD, Russo SJ et al (2016). New translational perspectives for blood-based biomarkers of PTSD: from glucocorticoid to immune mediators of stress susceptibility. Exp Neurol 284: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Beltran L, Esteban F, Wall D (2016). A common molecular signature in ASD gene expression: following Root 66 to autism. Transl Psychiatry 6: e705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGangi J, Gomez D, Mendoza L, Jason LA, Keys CB, Koenen KC (2013). Pretrauma risk factors for posttraumatic stress disorder: a systematic review of the literature. Clin Psychol Rev 33: 728–744. [DOI] [PubMed] [Google Scholar]
- Elenkov I, Wilder R, Chrousos G, Vizi E (2000). The sympathetic nerve—an integrative interface between two supersystems: the brain and the immune system. Pharmacol Rev 52: 595–638. [PubMed] [Google Scholar]
- Felger J, Lotrich F (2013). Inflammatory cytokines in depression: neurobiological mechanisms and therapeutic implications. Neuroscience 246: 199–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A et al (2012). STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41: D808–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freidenberg B, Gusmano R, Hickling EJ, Blanchard EB, Bremner JD, Frye C (2010). Women with PTSD have lower basal salivary cortisol levels later in the day than do men with PTSD: a preliminary study. Physiol Behav 99: 234–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliher-Beckley A, Williams J, Cidlowski J (2011). Ligand-independent phosphorylation of the glucocorticoid receptor integrates cellular stress pathways with nuclear receptor signaling. Mol Cell Biol 31: 4663–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill J, Saligan L, Woods S, Page G (2009). PTSD is associated with an excess of inflammatory immune activities. Perspect Psychiatr Care 45: 262–277. [DOI] [PubMed] [Google Scholar]
- Glatt SJ, Tylee DS, Chandler SD, PAzol J, Nievergelt CM, Woelk CH et al (2015). Blood-based gene-expression predictors of PTSD risk and resilience among deployed marines: a pilot study. Am J Med Genet B Neuropsychiatr Genet 162: 313–326. [DOI] [PubMed] [Google Scholar]
- Hess J, Tylee DS, Barve R, de Jong S, Ophoff RA, Kumarasinghe N et al (2016). Transcriptome-wide mega-analyses reveal joint dysregulation of immunologic genes and transcription regulators in brain and blood in schizophrenia. Schizophr Res 176: 114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayala M, Baldi P (2012). Cyber-T web server: differential analysis of high-throughput data. Nucleic Acids Res 40: W553–W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler R (1995). Posttraumatic stress disorder in the national comorbidity survey. Arch Gen Psychiatry 52: 1048. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Luo R, Oldham M, Horvath S (2011). Is my network module preserved and reproducible? PLoS Comput Biol 7: e1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J, Johnson W, Parker H, Jaffe A, Storey J (2015). The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28: 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, Pace TW et al (2013). Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc Natl Acad Sci USA 110: 8302–8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michopoulos V, Norrholm S, Jovanovic T (2015). Diagnostic biomarkers for posttraumatic stress disorder: promising horizons from translational neuroscience research. Biol Psychiatry 78: 344–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A, Long Liu A, Green MR, Gentles AJ, Feng W, Xu Y et al (2015). Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12: 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neylan T, Sun B, Rempel H, Ross J, Lenoci M, O’Donovan A et al (2011). Suppressed monocyte gene expression profile in men versus women with PTSD. Brain Behav Immun 25: 524–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacák K, Palkovits M (2001). Stressor specificity of central neuroendocrine responses: implications for stress-related disorders. Endocr Rev 22: 502–548. [DOI] [PubMed] [Google Scholar]
- Page GP, Edwards JW, Gadbury GL, Yelisetti P, Wang J, Trivedi P et al (2006). The PowerAtlas: a power and sample size atlas for microarray experimental design and research. BMC Bioinformatics 7: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos IC, Vasconcelos-Moreno MP, Costa LG, Kunz M, Brietzke E, Quevedo J et al (2015). Inflammatory markers in post-traumatic stress disorder: a systematic review, meta-analysis, and meta-regression. Lancet Psychiatry 2: 1002–1012. [DOI] [PubMed] [Google Scholar]
- Pinero J, Queralt-Rosinach N, Bravo A, Deu-Pons J, Bauer-Mehren A, Baron M et al (2015). DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database 2015: bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posma E, Moes H, Heineman M, Faas M (2014). The effect of testosterone on cytokine production in the specific and non-specific immune response. Am J Reprod Immunol 52: 237–243. [DOI] [PubMed] [Google Scholar]
- Ramchand R, Schell TL, Karney BR, Osilla KC, Bruns RM, Caldarone LB (2010). Disparate prevalence estimates of PTSD among service members who served in Iraq and Afghanistan: possible explanations. J Trauma Stress 23: 59–68. [DOI] [PubMed] [Google Scholar]
- Reul J (2014). Making memories of stressful events: a journey along epigenetic, gene transcription, and signaling pathways. Front Psychiatry 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W et al (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarapas C, Cai G, Bierer LM, Golier JA, Galea S, Ising M et al (2011). Genetic markers for PTSD risk and resilience among survivors of the World Trade Center attacks. Dis Markers 30: 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segman R, Shefi N, Goltser-Dubner T, Friedman N, Kaminski N, Shalev AY (2005). Peripheral blood mononuclear cell gene expression profiles identify emergent post-traumatic stress disorder among trauma survivors. Mol Psychiatry 10: 425–425. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M, Pearce B, Biron C, Miller A (2013). Immune modulation of the hypothalamic-pituitary-adrenal (HPA) axis during viral infection. Viral Immunol 18: 41–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M, Sternberg E (2012). Glucocorticoid regulation of inflammation and its functional correlates: from HPA axis to glucocorticoid receptor dysfunction. Ann N Y Acad Sci 1261: 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y (2007). Analysis of gene expression data using BRB-Array tools. Cancer Inform 2: 11–17. [PMC free article] [PubMed] [Google Scholar]
- Söndergaard H, Hansson L, Theorell T (2004). The inflammatory markers C-reactive protein and serum amyloid A in refugees with and without posttraumatic stress disorder. Clin Chim Acta 342: 93–98. [DOI] [PubMed] [Google Scholar]
- Tylee DS, Chandler SD, Nievergelt CM, Liu X, Pazol J, Woelk CH et al (2015). Blood-based gene-expression biomarkers of post-traumatic stress disorder among deployed marines: a pilot study. Psychoneuroendocrinology 51: 472–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tylee DS, Jess JL, Quinn TP, Barve R, Huang H, Zhang-James Y et al (2016). Blood transcriptomic comparison of individuals with and without autism spectrum disorder: a combined-samples mega-analysis. Am J Med Genet B Neuropsychiatr Genet 9999: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker A, Gilpin N, Edwards S (2014). Animal models of post-traumatic stress disorder and recent neurobiological insights. Behav Pharmacol 25: 398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Halligan S, Grossman R, Golier J, Wong C (2002). The cortisol and glucocorticoid receptor response to low dose dexamethasone administration in aging combat veterans and holocaust survivors with and without posttraumatic stress disorder. Biol Psychiatry 52: 393–403. [DOI] [PubMed] [Google Scholar]
- Yu G, Li F, Qin Y, Bo X, Wu Y, Wang S (2015). GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26: 976–978. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.