Abstract
Adduction of an electrophile to privileged sensor proteins and the resulting phenotypically dominant responses are increasingly appreciated as being essential for metazoan health. Functional similarities between the biological electrophiles and electrophilic pharmacophores commonly found in covalent drugs further fortify the translational relevance of these small-molecule signals. Genetically encodable or small-molecule-based fluorescent reporters and redox proteomics have revolutionized the observation and profiling of cellular redox states and electrophile–sensor proteins, respectively. However, precision mapping between specific redox-modified targets and specific responses has only recently begun to be addressed, and systems tractable to both genetic manipulation and on-target redox signaling in vivo remain largely limited. Here we engineer transgenic Caenorhabditis elegans expressing functional HaloTagged fusion proteins and use this system to develop a generalizable light-controlled approach to tagging a prototypical electrophile–sensor protein with native electrophiles in vivo. The method circumvents issues associated with low uptake/distribution and toxicity/promiscuity. Given the validated success of C. elegans in aging studies, this optimized platform offers a new lens with which to scrutinize how on-target electrophile signaling influences redox-dependent life span regulation.
The purpose of this communication is to delineate a protocol for ultimately studying the consequences of on-target electrophile signaling in Caenorhabditis elegans. We describe several C. elegans lines amenable to this procedure and show that single-protein targeting with diffusible reactive signaling electrophiles can be achieved in live worms with no observable negative effects. We have shown similar results in cultured cells1−5 and fish.6 These results in C. elegans, one of the most powerful and genetically tractable systems for the study of stress and aging,7,8 represent a key step in developing model systems for interrogating simultaneously the genetics and biochemistry of electrophile signaling.
Electrophilic pharmaceuticals9,10 as well as endogenous electrophilic signals11,12 are key factors in disease treatment and signaling regulation/fitness promotion, respectively. The burgeoning interest in both facets of electrophiles has established reactive electrophilic species (RES), particularly sp2 carbon-based electrophiles that are commonly prevalent in modern covalent drugs and native electrophilic signals,10 as important areas of study. With a growing appreciation of being able to identify ligandable cysteines,13 the emergence of successful covalent drugs,10,14 and the role of precision endogenous RES signaling,15 the most recent years have witnessed no letup in the push to identify new sensors/signaling pathways.
Numerous powerful technologies for interrogating targets of electrophilic drugs/biomolecules are now available.16 Unfortunately, many of these protocols are restricted to cultured cells and/or offer limited spatiotemporal resolution. Many also require extensive downstream processing. Few methods can easily report on phenotypic outputs directly attributable to one modification. Most methods rely upon bolus dosing of RES to a specific specimen, with little regard for metabolic processing, tissue/organelle segregation, or off-target effects. These issues present significant impediments to progress in the field.
Functional impacts of canonical enzyme-mediated post-translational modifications (PTMs), such as phosphorylation and ubiquitination, have benefited from the combination of small-molecule-based tools17,18 and classical genetic/biochemical approaches such as targeted knockdown, mutagenesis, and phenotypic screening.19,20 Indeed, the synergy between biochemistry/chemical biology and genetics/cell biology is arguably magnified in redox signaling.15,16,21,22 This is because many RES are toxic at elevated concentrations and promiscuous, engage with specific proteins without mediator enzymes, and can be converted to numerous products.11 Thus, genetic approaches alone cannot address precision mapping of nonenzymatic RES signal input on a single target to specific signaling output.15 Indeed, balancing the electrophile dose, time, target, and locale to mirror physiologic signaling in vivo has proven to be difficult.
We recently developed a chemistry-driven tool, called T-REX (targetable reactive electrophiles and oxidants), to begin to rigorously investigate electrophilic signaling (Figure S1).1−6 We achieved target-specific RES labeling by tethering a photocaged RES to the commercial-protein-tag HaloTag fused to a protein of interest (POI). Light exposure released unfettered RES on demand,23 giving first refusal to the Halo-fused POI. Thus, if the POI is a sensor protein, RES labeling occurs: if the POI is not a sensor, this minute amount of RES (stoichiometric to Halo-POI) that is unleashed is metabolized or averaged over the whole proteome. This method constitutes a stringent assay for RES sensing, and because little RES is released, it does not perturb the cell significantly. Built-in controls account for off-target effects and/or artifactual modifications/responses.4,6 We recently adapted this platform to transient expression in larval zebrafish,6 a popular vertebrate research model, and uncovered conserved signaling nodes that interchange RES-based non-enzyme-mediated PTM and conventional enzyme-regulated PTMs at a single-POI-precision scale.10,15 This model provided new insights into redox signaling performed by specific Akt-kinase isoforms in developing fish but is currently incompatible with adult fish, restricting studies on aging. It would thus be adventitious if T-REX were shown to work in a model with a shorter life span that retains transparency throughout its life. C. elegans is one system that fits these requirements.
Establishing T-REX in C. elegans requires that three criteria be met: (1) transgenic lines expressing Halo fused to a specific RES-sensitive POI, (2) Halo must be active in live worms and be able to be labeled by the photoactivatable precursors, and (3) delivery of RES to the POI must be possible in live worms.
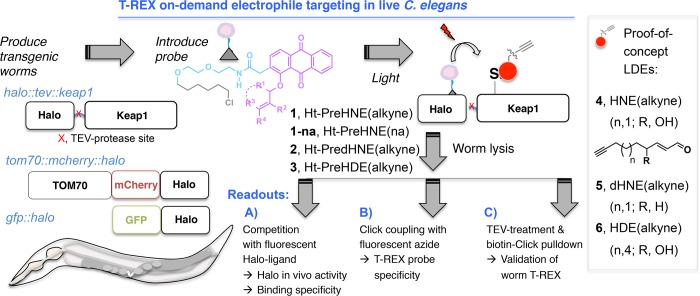
Using standard protocols, we generated several transgenic animals [pHSP16::tom70::mcherry::halo; pHSP16::halo::tev::keap1, where “tev” is a recognition site for the tobacco etch virus (TEV) protease; and pHSP16::gfp::halo] that upregulate Halo protein upon heat shock (HS) (Figure 1). We used the dominant genetic marker pmec7::mrfp that gives constitutive expression in six touch-receptor neurons to mark transgenic worms.24 The HS promoter (HSP16p)25 was chosen because it is inducible and gives a high level of expression of many transgenes. Importantly, the HS promoter can give a high level of expression without the need for codon optimization or C. elegans introns. We chose human Keap1 protein (keap1 hereafter) because it is an established RES sensor26 and validated to be kinetically privileged to react with various RES signals delivered under T-REX-driven electrophile-limited conditions,1−5 and there are few bona fide RES sensors in C. elegans to serve as a robust proof of concept. Expression was validated by both Western blot (Figure 2A and Figure S2) and, in the case of tom70::mcherry::halo, fluorescence (Figure 2B). Halo blot showed that the level of expression of tom70::mcherry::halo was approximately 5-fold higher than that of halo::tev::keap1 (Figure S2B).
Figure 1.
C. elegans T-REX platform. Indicated transgenic Halo worms following transgene induction were treated with bioinert photocaged precursors (1–3) to specific bioactive lipid-derived electrophiles (LDEs) (HNE 4, dHNE 5, and HDE 6).3 At a user-defined time, photouncaging liberates the respective alkyne-functionalized LDEs (4–6) and proximity enhancement21 enables labeling of a quintessential electrophile–sensor protein Keap1 (see also Figure S1A). Halo functionality, probe specificity, and LDE differential modification efficiency on target are assessed by three independent readouts.
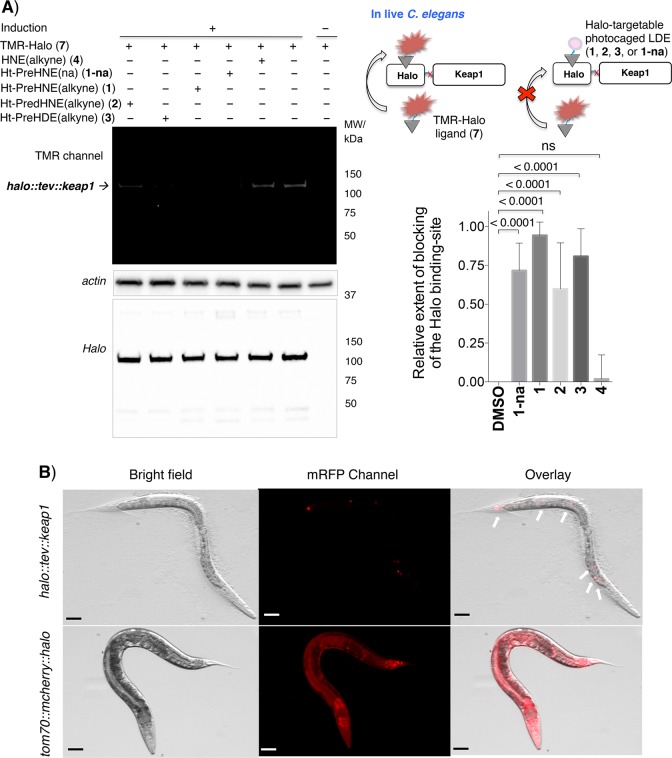
Figure 2.
(A) Photocaged precursors selectively bind to functional HaloTag expressed in live worms. The schematic at the top right shows the blocking experiment (readout A, Figure 1). See the Supporting Information for the detailed procedure. The left panel shows representative data analyzed by in-gel fluorescence (top) and Western blot (bottom) using anti-actin and anti-Halo antibodies, respectively, that confirm protein loading and inducible halo::tev::keap1 expression (halo::tev::keap1 full construct molecular weight of ∼105 kDa). See also Figure S2 for additional replicates. In the bottom right panel, the ratios of the fluorescence signal to the anti-Halo Western blot signal were normalized to dimethyl sulfoxide within each independent gel before averaging across multiple replicates. Errors designate the standard deviation (n = 8 independent biological replicates). (B) Fluorescence images of the heat shock-induced live worms further confirm transgene expression. Fluorescence of tom70-MLS-localized mcherry::halo is visible throughout the worm (bottom row). While halo::tev::keap1 does not itself feature a fluorescent marker, it is co-expressed with a constitutive dominant marker (mec7p::mrfp), which displays fluorescence localized to touch-receptor neurons (top row, white arrows). Scale bars are 50 μm.
Next, we evaluated if the Halo protein could selectively bind our photocaged probes in vivo (readout A, Figure 1). We have reported a fleet of probes that can deliver various endogenous bioactive lipid-derived electrophiles (LDEs) to Keap1 upon light exposure in mammalian cells.3,4 As proofs of principle in extending the platform to C. elegans, we chose precursors to 4-hydroxynonenal (HNE) 4, nonenal (or dehydroxy-hydroxynonenal) (dHNE) 5, and 4-hydroxydodecenal (HDE) 6 (Ht-PreHNE 1, Ht-PredHNE 2, and Ht-PreHDE 3, respectively) (Figure 1 and Figure S1A), all of which give similar levels of HaloTag conjugation in mammalian cells, and similar levels of Keap1-targeted LDEylation following light exposure.3 These photocaged precursors were produced via Williamson ether synthesis, using anthraquinone and a corresponding Grignard-generated allylic bromide.3 Using an optimized protocol, worms were treated with 30 μM probe for 6 h in liquid medium with OP50 bacteria (food) and then washed three times. After this point, worms were lysed by being frozen–thawed/vortexed with beads. Halo protein activity in lysates was assayed by treatment of normalized lysates with TMR-Halo 7 (or FAM-Halo 8), ligands that react irreversibly with Halo (Figure S1B, Figure 2A, and Figure S2). FAM-Halo was used to quantify labeling of tom70::mcherry::halo due to bleed-through of residual mCherry interfering with TMR in the gel (Figure S2B). The reaction of Halo with the TMR or FAM-Halo ligand gives a 1:1 complex that can be resolved via sodium dodecyl sulfate–polyacrylamide gel electrophoresis, giving a fluorescent band that migrates like the unlabeled Halo fusion protein.
Bolus HNE 4 (30 μM, 1 h) treatment did not affect Halo activity in vivo (Figure 2A and Figure S2C). However, pretreatment with Ht-PreHNE/Ht-PredHNE/Ht-PreHDE reduced the intensity of the TMR (or FAM) signal relative to those of untreated controls (Figure 2A and Figure S2). These ligands all contained an alkyne appendage, to facilitate direct detection by Click reaction,27 although conjugation of halo::tev::keap1 by Ht-PreHNE (with no alkyne functionalization) (1-na) was similar regardless of the presence of alkyne functionalization (Figure 2A and Figure S2C). Surprisingly, there were subtle differences between Halo conjugation of these probes, but the occupancy was >50% for each (Figure 2A, inset). Worms were not negatively affected by this protocol. Using Click chemistry, we validated this result of Halo occupancy by photocaged probes in vivo (readout B, Figure 1). Using a protocol similar to that described above, lysates were exposed to Click conditions with FAM-azide (Figure S3). A similar extent of Halo conjugation was observed.
Our final aim in this communication was to evaluate whether T-REX-targeted delivery of LDE to Keap1 could occur upon light exposure in live C. elegans. We first evaluated the in vivo photouncaging efficiency of the Ht-PreHNE covalently bound to Halo by measuring the time-dependent loss of the alkyne label from gfp::halo (Figure 1) preconjugated to Ht-PreHNE 1, enabled by Click coupling in lysates after the exposure of live worms to light (Figure S4). The half-life of HNE release in vivo under these conditions was on par with that previously assessed in cultured cells (t1/2 < 1–2 min).3
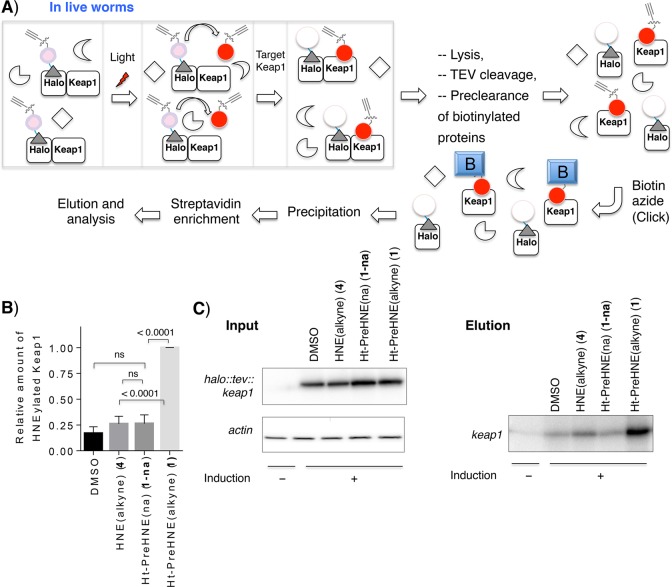
Because the level of expression of the halo:: tev::keap1 transgene was relatively low, we opted to determine the targeted HNEylation efficiency to Keap1 using biotin/streptavidin enrichment (readout C, Figure 1) that gives a signal:noise ratio that is higher than that of the Click assay with azido dyes in worm lysates. In this biotin pull-down protocol, C. elegans expressing halo::tev::keap1 are treated with the desired photocaged LDE precursor for 6 h, and after washout, the LDE is delivered to Keap1 by photouncaging. C. elegans are then lysed, and halo::tev::keap1 is then separated into the two domains (halo and keap1 using TEV protease); lysates are precleared with streptavidin agarose to remove biotinylated proteins, and biotin is conjugated to alkynated proteins using biotin-azide Click coupling. After precipitation, and resolubilization, biotinylated proteins are enriched using streptavidin agarose beads. Using this assay (Figure 3 and Figure S5), keap1 was significantly enriched relative to controls in which (i) Ht-PreHNE (with no alkyne functionalization) (1-na) was used in place of alkyne-functionalized Ht-PreHNE 1 under otherwise identical conditions and (ii) no compound treatment was performed.
Figure 3.
T-REX concept of proximity-targeted on-demand release of HNE in situ under electrophile-limited conditions that selectively HNEylate Keap1 protein expressed in live C. elegans. (A) Schematic of optimized biotin-Click pull-down conditions compatible with the C. elegans lysate (readout C, Figure 1). “B” indicates biotin. See the methods in the Supporting Information for details. (B) The inset shows quantitation of the extent of HNEylated Keap1 under the indicated conditions [n ≥ 5 independent biological replicates (Figure S5); errors indicate the standard error of the mean]. Bulk HNE exposure and T-REX give different extents of HNEylation on Keap1, suggesting that uptake/metabolism is a more significant variable in living model organisms than in living cells where HNEylation efficiencies are largely found to be comparable between the two conditions.3 (C) Representative data set for input and elution samples. Actin serves as a loading control. Induction designates heat shock for transgene expression. Also see Figure S5.
We were also keen to evaluate how our protocol compared to bolus dosing. We thus evaluated the amount of keap1 pulled down by Ht-PreHNE 1 and HNE 4 treatment. Strikingly, treatment with HNE 4 (30 μM), even over 1 h, did not HNEylate keap1 as significantly as that achieved under T-REX involving an only 5 min incubation post light-driven HNE release within the proximity of halo::tev::keap1 in vivo [Figure 3 and Figure S2A (inset)]. These data demonstrate that T-REX likely bypasses absorption, distribution, and metabolism effects of reactive electrophiles and directs the delivery of the HNE to a specific protein, demonstrating that potential LDE sensors could be identified using this method.
In conclusion, we have established a protocol that will enable us to ultimately interrogate on-target electrophile signaling in a key model organism in which in vivo chemical biology is under-represented. Our protocol is robust: photocaged LDE probes (1–3) give high-occupancy conjugation of the Halo protein functionally expressed in vivo, and targeted HNEylation can be accomplished at a user-defined time. The protocol can also be extended to POIs that lack accessible antibodies, by epitope tagging, for instance. On the basis of our recent work in cultured cells1−5 and zebrafish,6 we can use this procedure to either validate sensors in vivo or measure the ramifications of downstream signaling at a specific protein, specifically with an eye on interrogating the precision redox events of conserved importance in metazoan life span.
Acknowledgments
Professor Jun (Kelly) Liu, Mr. Steve Sammons, and the Liu laboratory (Cornell University) are acknowledged for the worm genetic technique transfer, early guidance, and the use of the microinjection facility, and Sanjna L. Surya and Alexandra Van Hall-Beauvais (Aye lab) are for acknowledged TEV-protease preparation.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.7b00642.
Methods and additional observations and data (PDF)
Author Contributions
Y.A. designed and oversaw the project. M.J.C.L. generated transgenic worm strains. M.J.C.L., D.A.U., S.C., and J.A.H. maintained worm cultures. M.J.C.L., D.A.U., and S.C. performed all experiments except time-dependent photoliberation studies, which were performed by J.A.H. H.-Y.L., Y.Z., and Y.W. performed chemical syntheses. Y.A. wrote the paper with assistance from M.J.C.L., S.C., and D.A.U. M.J.C.L., D.A.U., and S.C. are co-lead authors. H.-Y.L. and Y.Z. contributed equally to this work.
Research instrumentation, supplies, and personnel are partly or fully supported by a National Science Foundation (NSF) CAREER grant (CHE-1351400), a Sloan Fellowship (FG-2016-6379), and a Beckman Young Investigator award, an ONR Young Investigator award (N00014-17-1-2529), a National Institutes of Health (NIH) New Innovator award (1DP2GM114850) (to Y.A.); a Cornell University Graduate School Fellowship and a NSF Graduate Research Fellowship (DGE-1650441) (to D.A.U.); an American Chemical Society Division of Organic Chemistry Summer Undergraduate Research Fellowship and Barry Goldwater Scholarship (to S.C.); a NIH CBI Fellowship (to J.A.H.) (National Institute of General Medical Sciences Grant T32-GM008500; Principal Investigator, H. Lin); NSF MRI (CHE-1531632; Principal Investigator, Y.A.) for Cornell University core-facility NMR instrumentation; and NIH Grant 1S10RR025502 (Principal Investigator, R. M. Williams) for the Cornell Imaging Center.
The C. elegans strains generated in this work are pending deposition in the CGC as of the time of publication.
The authors declare no competing financial interest.
Supplementary Material
References
- Fang X.; Fu Y.; Long M. J.; Haegele J. A.; Ge E. J.; Parvez S.; Aye Y. (2013) J. Am. Chem. Soc. 135, 14496–14499. 10.1021/ja405400k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez S.; Fu Y.; Li J.; Long M. J.; Lin H. Y.; Lee D. K.; Hu G. S.; Aye Y. (2015) J. Am. Chem. Soc. 137, 10–13. 10.1021/ja5084249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. Y.; Haegele J. A.; Disare M. T.; Lin Q.; Aye Y. (2015) J. Am. Chem. Soc. 137, 6232–6244. 10.1021/ja5132648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez S.; Long M. J.; Lin H. Y.; Zhao Y.; Haegele J. A.; Pham V. N.; Lee D. K.; Aye Y. (2016) Nat. Protoc. 11, 2328–2356. 10.1038/nprot.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M. J. C.; Lin H.-Y.; Parvez S.; Zhao Y.; Poganik J. R.; Huang P.; Aye Y. (2017) Cell Chem. Biol. 24, 944–957.e7. 10.1016/j.chembiol.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M. J.; Parvez S.; Zhao Y.; Surya S. S.; Wang Y.; Zhang S.; Aye Y. (2017) Nat. Chem. Biol. 13, 333–338. 10.1038/nchembio.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoshechkin I.; Sternberg P. W. (2007) Nat. Rev. Genet. 8, 518–532. 10.1038/nrg2105. [DOI] [PubMed] [Google Scholar]
- Volovik Y.; Marques F. C.; Cohen E. (2014) Methods 68, 458–464. 10.1016/j.ymeth.2014.04.014. [DOI] [PubMed] [Google Scholar]
- Prosperini L.; Pontecorvo S. (2016) Ther. Clin. Risk Manage. 12, 339–350. 10.2147/TCRM.S85099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M. J. C.; Aye Y. (2017) Cell Chem. Biol. 24, 787. 10.1016/j.chembiol.2017.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer F. J.; Cipollina C.; Freeman B. A. (2011) Chem. Rev. 111, 5997–6021. 10.1021/cr200131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs A. T.; Marnett L. J. (2010) Acc. Chem. Res. 43, 673–683. 10.1021/ar900286y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backus K. M.; Correia B. E.; Lum K. M.; Forli S.; Horning B. D.; González-Páez G. E.; Chatterjee S.; Lanning B. R.; Teijaro J. R.; Olson A. J.; Wolan D. W.; Cravatt B. F. (2016) Nature 534, 570–574. 10.1038/nature18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewett M. M.; Xie J.; Zaro B. W.; Backus K. M.; Altman A.; Teijaro J. R.; Cravatt B. F. (2016) Sci. Signaling 9, rs10. 10.1126/scisignal.aaf7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M. J.; Aye Y. (2016) Chem. Res. Toxicol. 29, 1575–1582. 10.1021/acs.chemrestox.6b00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M. J.; Poganik J. R.; Ghosh S.; Aye Y. (2017) ACS Chem. Biol. 12, 586–600. 10.1021/acschembio.6b01148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A. C.; Crews C. M. (2017) Nat. Rev. Drug Discovery 16, 101–114. 10.1038/nrd.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrant M. K.; Cole P. A. (2009) Annu. Rev. Biochem. 78, 797–825. 10.1146/annurev.biochem.78.070907.103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A.; Buzko O.; Heyeck-Dumas S.; Jung I.; Kraybill B.; Liu Y.; Shah K.; Ulrich S.; Witucki L.; Yang F.; Zhang C.; Shokat K. M. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 577–606. 10.1146/annurev.biophys.29.1.577. [DOI] [PubMed] [Google Scholar]
- Aghajan M.; Jonai N.; Flick K.; Fu F.; Luo M.; Cai X.; Ouni I.; Pierce N.; Tang X.; Lomenick B.; Damoiseaux R.; Hao R.; Del Moral P. M.; Verma R.; Li Y.; Li C.; Houk K. N.; Jung M. E.; Zheng N.; Huang L.; Deshaies R. J.; Kaiser P.; Huang J. (2010) Nat. Biotechnol. 28, 738–742. 10.1038/nbt.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson B. C.; Chang C. J. (2011) Nat. Chem. Biol. 7, 504–511. 10.1038/nchembio.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer T. F.; Garcia F. J.; Onak C. S.; Carroll K. S.; Chang C. J. (2015) Annu. Rev. Biochem. 84, 765–790. 10.1146/annurev-biochem-060614-034018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M. J.; Poganik J. R.; Aye Y. (2016) J. Am. Chem. Soc. 138, 3610–3622. 10.1021/jacs.5b12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M. (1994) Curr. Biol. 4, 443. 10.1016/S0960-9822(00)00098-1. [DOI] [Google Scholar]
- Stringham E. G.; Dixon D. K.; Jones D.; Candido E. P. (1992) Mol. Biol. Cell 3, 221–233. 10.1091/mbc.3.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. D.; Dinkova-Kostova A. T. (2014) Trends Biochem. Sci. 39, 199–218. 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Kolb H. C.; Finn M. G.; Sharpless K. B. (2001) Angew. Chem., Int. Ed. 40, 2004–2021. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.