Abstract
The human chemokine interleukin-8 (IL-8; CXCL8) is a key mediator of innate immune and inflammatory responses. This small, soluble protein triggers a host of biological effects upon binding and activating CXCR1, a G protein-coupled receptor, located in the cell membrane of neutrophils. Here, we describe 1H-detected magic angle spinning solid-state NMR studies of monomeric IL-8 (1-66) bound to full-length and truncated constructs of CXCR1 in phospholipid bilayers under physiological conditions. Cross-polarization experiments demonstrate that most backbone amide sites of IL-8 (1-66) are immobilized and that their chemical shifts are perturbed upon binding to CXCR1, demonstrating that the dynamics and environments of chemokine residues are affected by interactions with the chemokine receptor. Comparisons of spectra of IL-8 (1-66) bound to full-length CXCR1 (1-350) and to N-terminal truncated construct NT-CXCR1 (39-350) identify specific chemokine residues involved in interactions with binding sites associated with N-terminal residues (binding site-I) and extracellular loop and helical residues (binding site-II) of the receptor. Intermolecular paramagnetic relaxation enhancement broadening of IL-8 (1-66) signals results from interactions of the chemokine with CXCR1 (1-350) containing Mn2+ chelated to an unnatural amino acid assists in the characterization of the receptor-bound form of the chemokine.
Introduction
More than 800 G protein-coupled receptors (GPCRs) are encoded in the human genome. These membrane proteins have seven transmembrane helices and each binds to one or multiple extracellular molecular signals, typically small molecules, but in some cases proteins. Their activation triggers many physiological processes through interactions with intracellular proteins. The structures, dynamics, and functions of GPCRs are of keen interest to both physiology and medicine, especially because they are receptors for a large fraction of the drugs in current use and provide fertile ground for the targeted development of new drugs (1). Although there has been significant progress in recent years in the characterization of static structures of crystalline GPCRs at low temperatures (2, 3, 4), relatively little is known about their binding to, and activation by, naturally occurring agonists, especially proteins, under physiological conditions where both agonist and receptor dynamics are likely to play important roles. Here we examine the interactions between a chemokine agonist and its receptor in the near-native environment of hydrated phospholipid bilayers by NMR spectroscopy.
About 50 chemokines and 20 chemokine receptors constitute an important part of the innate immune system (5). Not only do these proteins play key roles in the defense against microbial infections, but they are also associated with responses to inflammatory diseases such as arthritis, asthma, and cancer (6). The chemokine interleukin-8 (IL-8; CXCL8) binds to and activates the GPCR CXCR1 in the cell membranes of neutrophils, which then interact with G-proteins, arrestins, and other intracellular proteins to initiate biochemical responses of the cell. IL-8 and CXCR1 serve as a prototypical chemokine system because they are the first of their types to be discovered and are among the best characterized (7).
IL-8 is a small soluble protein with 72 residues. Its structure, determined by solution NMR spectroscopy (8) and x-ray crystallography (9), is characterized by having a flexible N-terminal segment containing a conserved Glu-Leu-Arg (ELR) motif just before the characteristic CXC arrangement of cysteine residues (X represents another type of amino acid) that participate in the two disulfide bonds that stabilize the tertiary fold of the three β-strands and single α-helical segment. Most CXC chemokines have between 70 and 130 residues with substantial variation of sequences. However, they share the characteristic three-dimensional chemokine fold first described for IL-8, which suggests that there may be commonality in how they interact with their receptors.
At the high concentrations used in NMR spectroscopy and x-ray crystallography, most chemokines form noncovalent dimers or larger oligomers. IL-8 has been shown to exist and function both as a monomer and dimer (10, 11, 12). The monomer binds to the N-terminal domain of CXCR1 with higher affinity than the dimer (13, 14, 15), and is believed to be the biologically active form of the protein. Modification, mutation, or truncation of residues at the dimer interface stabilize the monomeric form of IL-8 (12, 16). The structure of monomeric IL-8 (1-66), obtained by truncation of the last six C-terminal residues, using solution NMR is shown in Fig. 1 A (17). Overall, the structure of monomeric IL-8 is very similar to that of each subunit in the wild-type IL-8 (1-72) dimer (18).
Figure 1.
Structures of the chemokine IL-8 and its receptor CXCR1. (A) Structure of the monomeric chemokine IL-8 (1-66) in aqueous solution (PDB: 5WDZ). (B) Structure of the chemokine receptor CXCR1 (1-350) in phospholipid bilayers (PDB: 2LNL). Extracellular N-terminal and intracellular C-terminal residues of CXCR1 involved in ligand and G-protein interactions, respectively, are not included in the Figure because they are mobile on the timescales of the NMR experiments used to determine the structure of the receptor. To see this figure in color, go online.
CXCR1 and CXCR2 were the first chemokine receptors to be identified and cloned (19), and were originally referred to as IL-8RA (20) and IL-8RB (21), respectively. They share ∼80% sequence identity. Most of the differences are in the N- and C-terminals, which bind chemokines and G-proteins, respectively. Both receptors have high affinity for IL-8, with a low nanomolar dissociation constant (22, 23). Finding that two similar but distinct receptors bind the same chemokine, and that CXCR1 almost exclusively binds to IL-8 whereas CXCR2 also binds to several other chemokines, provided an early indication of the complexity of the chemokine defense system. At present, the structures of six chemokine receptors have been reported: the crystal structures of CXCR4 (24, 25), CCR2 (26), CCR5 (27), CCR9 (28), and viral US28 (29), and our NMR structure of CXCR1 in lipid bilayers (23), which is shown in Fig. 1 B. Although none of the structures represent complexes of a receptor with one of its native chemokine agonists, the structures of receptors bound to viral chemokines provide information that is complementary to the many mutagenesis and binding studies of their interactions. There have been many solution NMR and other biophysical studies of IL-8 interacting with polypeptides whose sequences correspond to N-terminal residues of CXCR1 (13, 14, 15, 16, 30, 31, 32, 33). The current model for chemokine-receptor interactions involves two binding sites on the receptor. Residues in both the N-terminal domain (binding site-I) and in one or more extracellular loops (binding site-II) of CXCR1 have been shown to interact with IL-8 (15, 33, 34, 35, 36).
Solid-state NMR spectroscopy enables studies of the structure and dynamics of membrane proteins in near-native phospholipid bilayer environments (37, 38, 39, 40). There are several examples where solid-state NMR methods have been used to characterize the structures of ligands bound to GPCRs (41, 42, 43, 44) as well as GPCRs themselves. Most prior studies of membrane proteins or their ligands have involved the direct detection of signals from labeled 13C or 15N sites; this continues to be a fruitful approach and we applied it in our earlier studies of CXCR1 (23, 33, 45). However, dramatic improvements in sensitivity have been obtained by the implementation of 1H-detected magic angle spinning (MAS) solid-state NMR methods (46, 47), especially when fast MAS is combined with perdeuteration of the samples (46, 48, 49, 50, 51). This approach is being applied to an increasing number of membrane proteins (52, 53, 54, 55).
Here, we describe the effects on IL-8 when it binds to CXCR1 in phospholipid bilayers using 1H-detected MAS solid-state NMR. We prepared uniformly 2H and 15N labeled monomeric IL-8 (1-66) (17), and examined its interactions with unlabeled full-length CXCR1 (1-350), which contains both binding site-I and binding site-II, and an N-terminal truncated construct of CXCR1, NT-CXCR1 (39-350), which lacks binding site-I associated with the N-terminal region but retains binding site-II associated with the extracellular loops. Changes observed in spectra of IL-8 (1-66) upon binding CXCR1 (1-350) and NT-CXCR1 (39-350) identify chemokine residues involved in the interactions with CXCR1. In addition, we incorporated the metal-chelating unnatural amino acid 2-amino-3-(8-hydroxyquinolin-3-yl)propanoic acid dihydrochloride (HQA) into full-length CXCR1, expanding upon earlier work (56), to observe intermolecular paramagnetic relaxation enhancement (PRE) between IL-8 (1-66) and CXCR1 (1-350). This provides additional information about the structures and spatial arrangement of the complexed proteins.
Materials and Methods
Incorporation of HQA
The unnatural amino acid HQA was purchased from BOC Sciences (http://www.bocsci.com). The replacement of tryptophan by HQA at position 10 in the sequence of full-length CXCR1 (1-350) was performed as previously described for a different construct of CXCR1 (56). The complete replacement of tryptophan by HQA was confirmed by NMR and fluorescence spectroscopies. Purification and refolding procedures of HQA-containing CXCR1 (W10HQA CXCR1) were identical to those applied to the wild-type protein (23).
Sample preparation
Monomeric IL-8 (1-66) containing residues of 1-66 of the wild-type human protein was expressed and purified using the protocol described previously (57). Uniformly 2H- and 15N-labeled samples were obtained by growing bacteria in BioExpress cell growth media (U-2H, 98%; U-15N, 98%) and deuterium oxide (2H, 99.9%) (Cambridge Isotope Laboratories, Tewksbury, MA; http://www.isotope.com). High deuteration levels (>90%) were verified by comparing the 1H solution NMR spectrum of perdeuterated IL-8 (1-66) to that of an unlabeled sample. The proteoliposome samples of 1,2-dimyristoyl-sn-glycero-3-phosphocholine lipid bilayers containing full-length CXCR1 (1-350) or N-terminal truncated construct NT-CXCR1 (39-350) were prepared as described previously (23, 58). The lipid/protein molar ratio was ∼300:1.
NMR experiments
The solution NMR experiments were performed at 40°C on a Bruker 600 MHz spectrometer equipped with 5 mm triple-resonance cryoprobe with z-axis gradient. The concentration of monomeric IL-8 (1-66) was 50 μM in 20 mM HEPES buffer (pH 7.3). One-dimensional 15N-edited 1H NMR spectra of uniformly 15N-labeled monomeric IL-8 (1-66) were obtained by signal-averaging 128 transients in the absence and presence of CXCR1-containing proteoliposomes at a molar ratio of 1:1.
The samples for solid-state NMR experiments contained 30–50 μg of uniformly 2H- and 15N-labeled IL-8 (1-66) in ∼3 μL of 20 mM HEPES buffer in 100% 1H2O. Spectra were obtained in the absence and presence of various unlabeled CXCR1 constructs in liposomes at a receptor to IL-8 (1-66) molar ratio of 1:1. Samples were prepared by adding an appropriate amount of isotopically labeled IL-8 (1-66) in solution to proteoliposomes containing unlabeled CXCR1 constructs in 5 mL of 20 mM HEPES buffer (pH 7.3). The mixtures were incubated for 30 min at room temperature before ultracentrifugation for 20–40 h at 645,000 × g at 15°C. Approximately 3 μL of fully hydrated proteoliposomes containing IL-8 (1-66) bound to the receptor was transferred into a 1.3 mm MAS rotor. For the PRE experiments, a 200-fold molar excess of MnCl2 was added to the proteoliposome samples before ultracentrifugation.
Solid-state NMR experiments were performed at 900 MHz (21.2 Tesla) on a Bruker Avance III HD spectrometer equipped with a triple-resonance 1.3 mm MAS probe. The sample-spinning rate was controlled to 60 kHz (±2 Hz). The probe temperature was maintained at 8°C using nitrogen cooling gas at −30°C and a flow rate of 1500 L/h; the actual sample temperature, as monitored by the resonance frequency of the water signal, was estimated to be 30°C due to frictional heating.
Two-dimensional 1H-detected 1H-15N chemical shift correlation spectra were acquired using either a 1H-15N heteronuclear single quantum correlation (HSQC) pulse sequence via double insensitive nuclei enhanced by polarization transfers (INEPT) (59) with presaturation for water signal suppression, or via double cross-polarization (CP) transfers (46, 56) with multiple intense solvent suppression intended for sensitive spectroscopic investigation of protonated proteins instantly (47). 15N globally optimized alternating phase rectangular pulse (60) decoupling with irradiation of 22.6 kHz was applied during 1H signal acquisition. In these pulse sequences, the hard π/2 pulses had nutation frequencies of 100 and 55.6 kHz for 1H and 15N, respectively. The delay τ was set to 1/4 J (2.5 ms) for INEPT transfers. The contact time was 1 ms for the first CP transfer from 1H to 15N, and 0.4 ms for the second CP transfer from 15N to 1H. Two-dimensional INEPT correlation spectra were acquired using 64 complex time-domain points with a total acquisition time of 10 ms in the indirect 15N chemical shift dimension, and 2048 complex time-domain points with a total acquisition time of 56.8 ms in the directly detected 1H chemical shift dimension. Two-dimensional CP correlation spectra were acquired using 64 complex time-domain points with a total acquisition time of 10 ms in the indirect 15N chemical shift dimension, and 1024 complex time-domain points with a total acquisition time of 11.3 ms in the directly detected 1H chemical shift dimension. The relaxation delay for all experiments was 1.2 s. The NMR data were processed using TopSpin 3.5 (http://www.bruker.com) and the structures were visualized using PyMol (http://www.pymol.org).
Results
Immobilization of IL-8 bound to CXCR1 in phospholipid bilayers
Although IL-8 exists as a homodimer in solution (8), monomeric IL-8 has been shown to bind to CXCR1 with similar or higher affinity than the dimer (13, 14). Truncation of the last six C-terminal residues results in a stable monomeric form of IL-8 (1-66) (34). Studies of IL-8 (1-66) avoid potential complications due to dimer-to-monomer interconversion of either the free or bound forms of IL-8.
Binding of IL-8 (1-66) to full-length CXCR1 (1-350) is tight enough that IL-8 (1-66) can be found in the insoluble proteoliposome pellet along with CXCR1 (1-350) after ultracentrifugation (Fig. 2 A, lane 3). Similar results were obtained for IL-8 (1-66) binding to NT-CXCR1 (39-350), where the residues responsible for the receptor’s N-terminal binding site are missing (data not shown). However, no IL-8 (1-66) was detected in the pellet obtained by ultracentrifugation of a sample of the protein suspended in the presence of empty liposomes. This indicates that IL-8 does not interact with or partition to phospholipids. Purified uniformly 15N-labeled IL-8 (1-66) (Fig. 2 A, lane 2) in aqueous (1H2O) buffer yields a well-resolved 15N-edited 1H solution NMR spectrum of the amide region (Fig. 2 B), which is similar to that obtained from the wild-type dimeric IL-8 (1-72) (17, 33). The binding of IL-8 (1-66) to CXCR1 (1-350) in phospholipid bilayers at a molar ratio of 1:1 (Fig. 2 A, lane 3) results in the broadening of all IL-8 (1-66) 1H amide signals beyond detection by solution NMR (Fig. 2 C). This demonstrates that the global reorientation of IL-8 (1-66) is highly restricted by binding to CXCR1 embedded in phospholipid bilayers. Similar results were observed for the interaction of wild-type dimeric IL-8 (1-72) with the receptor under the same sample and spectroscopic conditions (data not shown). Notably, signals from those residues of wild-type dimeric IL-8 (1-72) not directly involved in the interaction with CXCR1 (1-350) could be observed in solution NMR spectra when the protein complexes are in rapidly reorienting isotropic bicelles, but not when they are in slowly and anisotropically reorienting lipid environments, such as magnetically aligned bicelles or proteoliposomes (33). The slow global reorientation of IL-8 complexed with CXCR1 in phospholipid bilayers severely compromises the application of solution NMR methods to these samples. However, it is possible to use solid-state NMR methods to investigate IL-8 bound to CXCR1 in phospholipid bilayers. Fig. 3 compares two-dimensional 1H-15N chemical shift correlation spectra of IL-8 (1-66) obtained under various experimental conditions. The use of uniformly 2H- and 15N-labeled IL-8 (1-66), where the perdeuteration facilitates the suppression of 1H-1H dipolar couplings, is essential to obtain these high-resolution MAS spectra.
Figure 2.
(A) SDS-PAGE analysis of IL-8 (1-66), CXCR1 (1-350), and their 1:1 complex. Lane M: Protein molecular weight marker. Lane 1: CXCR1 (1-350) in phospholipid bilayers. Lane 2: IL-8 (1-66) in aqueous buffer. Lane 3: pellet and Lane 4: supernatant, of a 1:1 molar ratio mixture of IL-8 (1-66) and CXCR1 (1-350) in phospholipid bilayers after 20 h of ultracentrifugation at 645,000 × g. Lane 3 demonstrates that IL-8 (1-66) and CXCR1 (1-350) are in the resulting pellet and that neither protein is in the supernatant, indicating that they form a complex in the bilayers. (B and C) One-dimensional 15N-edited 1H solution NMR spectra of uniformly 15N-labeled IL-8 (1-66) obtained at 600 MHz and 40°C. (B) IL-8 (1-66) alone in aqueous solution. (C) 1:1 IL-8 (1-66):CXCR1 (1-350) complex in phospholipid bilayers. The 1H solution NMR signals of IL-8 (1-66) are broadened beyond detection by the slow reorientation of the chemokine bound to CXCR1 in phospholipid bilayers. To see this figure in color, go online.
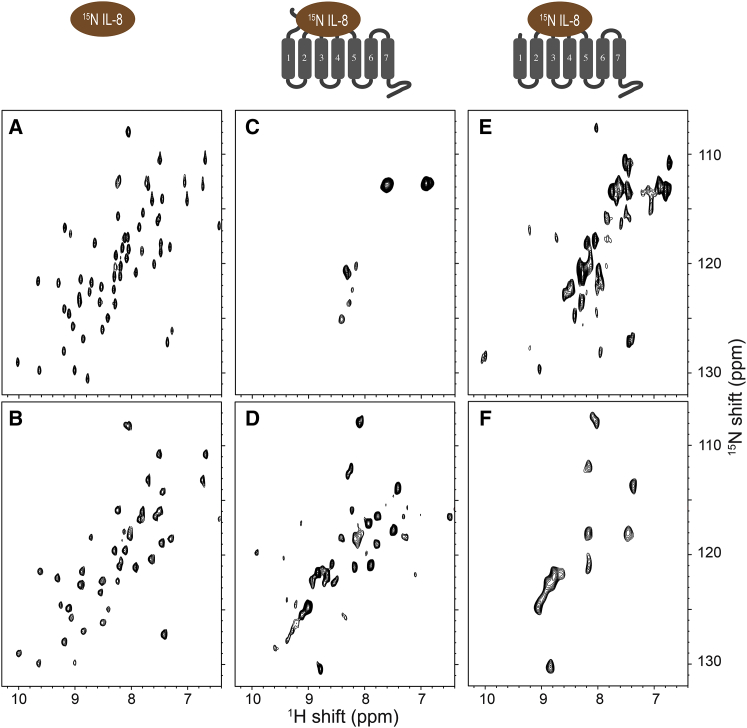
Figure 3.
Two-dimensional 1H-detected 1H-15N correlation NMR spectra of uniformly [2H, 15N-HN] labeled IL-8 (1-66). The cartoons at the top represent the samples: IL-8 (1-66) alone (left), the IL-8 (1-66):CXCR1(1-350) complex (center), and the IL-8 (1-66):NT-CXCR1(39-350) complex (right). (A) HSQC solution NMR spectrum of IL-8 (1-66) alone in aqueous solution obtained at 600 MHz and 40°C. (B–F) 1H-detected 60 kHz MAS solid-state NMR spectra of IL-8 (1-66) in the absence (B) and presence (C–F) of CXCR1 constructs in phospholipid bilayers obtained at 900 MHz and 30°C. (B) IL-8 (1-66) alone in aqueous solution. (C and D) 1:1 IL-8 (1-66):CXCR1 (1-350) complex in phospholipid bilayers. (E and F) 1:1 IL-8 (1-66):NT-CXCR1 (39-350) in phospholipid bilayers. (A–C and E) were obtained with INEPT and (D and F) with CP magnetization transfers, enabling the separate observation of signals from mobile (A–C and E) and immobile (D and F) sites. To see this figure in color, go online.
Fast-exchanging amide hydrogens of IL-8
A two-dimensional 1H-15N HSQC NMR spectrum of uniformly 15N labeled IL-8 (1-66) in aqueous solution is shown with black contours in Figs. 3 A and 4 A. This spectrum was obtained with the sample in a standard 5 mm OD solution NMR sample tube that was not spun. All the amide resonances have been assigned to specific residues, with the exception of the mobile C-terminal residue, 66, whose amide hydrogen exchanges rapidly with water at pH 7.3 (17). A spectrum of the same solution sample obtained in a 1.3 mm rotor spinning at 60 kHz using the same pulse sequence, with minor modifications of the τ delay for INEPT transfers and the use of continuous wave irradiation (presaturation) for water signal suppression, is shown with black contours in Fig. 3 B and with red contours in Fig. 4 A. The observed line widths of amide 1H resonances of IL-8 (1-66) in aqueous solution obtained by conventional solution NMR and obtained by 60 kHz MAS solid-state NMR are ∼30 and 50 Hz, respectively.
Figure 4.
(A) Comparison of one-dimensional 15N-edited 1H NMR and two-dimensional 1H-detected 1H-15N correlation NMR spectra of uniformly 15N-labeled IL-8 (1-66) in aqueous solution obtained by solution NMR (black contours) and solid-state 60 kHz MAS NMR (red contours), respectively. The amide resonances absent in the solid-state MAS NMR spectrum due to fast exchange with water are marked. (B) The fast-exchanging sites in the N-terminal and three loop regions of IL-8 (1-66) are colored in green in the protein structure. To see this figure in color, go online.
The solution NMR and solid-state NMR spectra in Fig. 4 are very similar with extensive overlap of chemical shifts. Importantly, no signals could be observed using CP transfer, indicating that IL-8 (1-66) is not sedimented but remains soluble undergoing isotropic reorientation in solution at 60 kHz MAS, which produces >106 g centrifugal force toward the rotor wall. Presaturation for water suppression in the solid-state MAS spectrum results in missing or very weak intensity signals from solvent-accessible residues (1–5, 11–15, 18, 20, 33, 35, 37, 44, 45, and 57) compared to the solution NMR spectrum obtained with the WATERGATE pulse sequence (61) for water suppression. This result was verified by amide hydrogen-deuterium exchange measurements on IL-8 (1-66) in solution (17).
The residues with fast-exchanging amide hydrogens are represented in green in the IL-8 (1-66) structure (Fig. 4 B); they are mainly located in the N-terminal and three loop regions (N-loop, 30s loop, and 40s loop) This result is in agreement with the previously measured amide nitrogen relaxation rates of a monomeric IL-8 mutant in aqueous solution, in which a high degree of mobility was observed for the unstructured N-terminal region, and reduced mobility for several residues in the loop regions (residues 11, 12, 33, 35, 45, and 57) (30).
IL-8 interaction with CXCR1 in phospholipid bilayers
Comparison of MAS solid-state NMR spectra obtained with CP and INEPT magnetization transfers enables the qualitative characterization of residue-specific local dynamics of IL-8 (1-66) bound to CXCR1 in phospholipid bilayers (Figs. 3, 5, and 7). Signals from immobilized residues are observed in the spectra obtained with CP transfers whereas signals from mobile residues are observed in the spectra obtained with INEPT transfers. 1H-detected 1H-15N correlation solid-state MAS NMR spectra of 2H and 15N labeled IL-8 (1-66) bound to unlabeled full-length CXCR1 (1-350) show that signals from most of the IL-8 (1-66) amide backbone sites could be observed in the CP spectrum (Fig. 3 D (black contours), and Fig. 5 A (red contours). In contrast, only five backbone and a pair of side-chain signals were observed in the INEPT spectrum (Fig. 3 C (black contours), and Fig. 5 B (green contours). Motionally averaged signals of IL-8 (1-66) bound to CXCR1 in phospholipid bilayers were observed in the MAS solid-state NMR spectrum (Figs. 3 C and 5 B), but not in the solution NMR spectrum (Fig. 2 C) because the severe broadening from the dipole-dipole interactions is suppressed by the combination of fast MAS and perdeuteration. Several residues, including those from the C-terminal helical region, which are not observed in either CP or INEPT solid-state NMR experiments, are marked in gray in Fig. 5 C. The C-terminal helical region of IL-8 (1-66) does not show evidence of interaction with the receptor. It is known to be responsible for association with the sulfate groups of glycosaminoglycans (62), and local motions might occur on an intermediate timescale, such that neither CP nor INEPT transfers yield signals (63).
Figure 5.
(A and B) 1H-detected 1H-15N correlation solid-state 60 kHz MAS NMR spectra of [2H, 15N-HN] labeled IL-8 (1-66) bound to unlabeled CXCR1 (1-350) in phospholipid bilayers obtained with (A) CP (red contours) and (B) INEPT (green contours) magnetization transfers. The solid-state MAS NMR spectra are overlaid on the solution NMR HSQC spectrum (Figs. 3A and 4A) of free IL-8 (1-66) (gray contours). Assigned signals are marked. (C) Structure of IL-8 (1-66) with the dynamics of residues in IL-8 (1-66) bound to CXCR1 (1-350) designated by colors. Red represents immobile sites (red contours, A) and green represents mobile sites (green contours, B). Signals from residues in the gray regions are either unassigned or undetected in the solid-state NMR spectra. To see this figure in color, go online.
Figure 7.
(A and B) 1H-detected 1H-15N correlation solid-state 60 kHz MAS NMR spectra of [2H, 15N-HN] labeled IL-8 (1-66) bound to unlabeled NT-CXCR1 (39-350) in phospholipid bilayers obtained with (A) CP (red contours) and (B) INEPT (green contours) magnetization transfers. The solid-state MAS NMR spectra are overlaid on the solution NMR HSQC spectrum (Figs. 3A and 4A) of free IL-8 (1-66) (gray contours). Assigned signals are marked. (C) Structure of IL-8 (1-66) with the dynamics of residues in IL-8 (1-66) bound to NT-CXCR1 (39-350) designated by colors. Red represents immobile sites (red signals, A) and green represents mobile sites (green signals, B). Signals from residues in the gray regions are either unassigned or undetected in the solid-state NMR spectra. To see this figure in color, go online.
The mobile and fast-exchanging IL-8 (1-66) sites in the loops (green sites in Fig. 4 B) were immobilized along with the three β-strands upon interaction with CXCR1 (red sites in Fig. 5 C). Several fast-exchanging residues (12, 18, 20, 44, 45) in the N-loop and the 40s loop of IL-8 (1-66) contribute to its primary binding site (13, 14, 15, 16, 30, 31, 32, 33, 64). Significant chemical shift perturbations of resonances of IL-8 (1-66) upon interaction with CXCR1 (1-350) indicate the locations of the IL-8 (1-66) residues involved in binding. Chemical shift perturbations of five well-resolved resonances of IL-8 (1-66), Gln8, and Lys20 in the downfield (11–12 ppm) region and Phe17, Cys34, and Val58 in the upfield (5.5–6.5 ppm) region are compared in Fig. 6. Phe17 and Val58 resonances were significantly perturbed by both CXCR1 (1-350) in phospholipid bilayers and the soluble peptide ND-CXCR1 (1-38) in aqueous buffer, whereas Cys34 was not affected. This result is consistent with solution NMR data on interactions of both monomers and dimers of IL-8 with various N-terminal CXCR1 peptides containing only binding site-I of the receptor (15). The Gln8 resonance was slightly perturbed by both receptor constructs, but the Lys20 resonance was only perturbed by ND-CXCR1 (1-38) and not by CXCR1 (1-350), suggesting that the IL-8 (1-66) interaction with CXCR1 (1-350) is different from that with ND-CXCR1 (1-38).
Figure 6.
Downfield (A) and upfield (B) regions of 1H-15N correlation spectra of IL-8 (1-66). Signals in black contours are from a solution NMR HSQC spectrum of free IL-8 (1-66); blue contours are from a solution NMR HSQC spectrum of IL-8 (1-66) bound to ND-CXCR1 (1-38) (17); and red contours are from a solid-state MAS NMR spectrum of IL-8 (1-66) bound to CXCR1 (1-350). Assigned residues are marked. To see this figure in color, go online.
IL-8 interaction with NT-CXCR1 in phospholipid bilayers
IL-8 (1-66) binds to NT-CXCR1 (39-350), which lacks the N-terminal 38 residues of the receptor that contribute to binding site-I, but retains binding site-II associated with extracellular loops of the receptor. Significant differences were observed between the 1H-15N correlation spectra of IL-8 (1-66) bound to NT-CXCR1 (39-350) (Figs. 3, E and F and 7, A and B) and to full-length CXCR1 (1-350) (Figs. 3, C and D and 5, A and B). From the IL-8 (1-66) and NT-CXCR1 (39-350) complex, ∼11 backbone amide signals can be observed in the CP spectrum (Fig. 3 F, black contours, and Fig. 7 A, red contours) compared with ∼18 signals in the INEPT spectrum (Fig. 3 E, black contours, and Fig. 7 B, green contours). Out of the 60 expected backbone amide signals, 31 are not detected in either the CP or the INEPT spectra. The corresponding residues are shown in gray on the structure in Fig. 7 C. The observed CP signals are located primarily in the 30s loop, 40s loop, and β3 strand (Fig. 7 C, red). The chemical shift perturbations of IL-8 (1-66) resonances by interactions with NT-CXCR1 (39-350) in the CP spectrum are generally smaller than those observed for interactions with full-length CXCR1 (1-350), possibly indicating differences in the nature of the interactions with the extracellular loops of the receptor (65, 66).
Long-range distance restraints from intermolecular PRE of HQA-incorporated CXCR1
The genetic incorporation of the metal-chelating unnatural amino acid HQA (67) into membrane proteins enables the measurement of intra- and intermolecular distance restraints by enabling PRE NMR experiments (56). The signal intensities in the one-dimensional MAS solid-state 15N-edited 1H NMR spectrum of IL-8 (1-66) bound to W10HQA CXCR1 (1-350) (Fig. 8 A) were reduced by the broadening effects of the bound paramagnetic Mn2+ ion on nearby 1H nuclei (Fig. 8 B). Control experiments show that the presence of free manganese ions in solution has negligible effect on the spectrum of IL-8 (1-66) bound to wild-type CXCR1 (1-350) (Fig. 8, C and D). Most likely this is because the CP transferred 1H signals of IL-8 (1-66), which result from the immobilization of its residues due to interactions with CXCR1 (1-350), are shielded from the free manganese ions.
Figure 8.
One-dimensional 15N-edited 1H-detected 60 kHz MAS solid-state NMR spectra of [2H, 15N-HN] labeled IL-8 (1-66) bound to unlabeled CXCR1 obtained with CP magnetization transfer. The cartoons at the top represent the samples: 1:1 IL-8 (1-66): W10HQA CXCR1 (1-350) complex (left), 1:1 IL-8 (1–66):CXCR1 (1-350) complex (center), and IL-8 (1-66):NT-CXCR1 (39–350) complex (right). The star indicates the location of the metal-chelating unnatural amino acid HQA at residue 10 in the N-terminal domain of CXCR1. (A and B) 1:1 IL-8 (1-66): W10HQA CXCR1 (1-350) complex. (C and D) 1:1 IL-8 (1-66):CXCR1 (1-350) complex. (E) 1:1 IL-8 (1-66):NT-CXCR1 (39-350) complex. (A), (C), and (E) were obtained in the absence of MnCl2. (B) and (D) were obtained in the presence of MnCl2. To see this figure in color, go online.
The spectrum of labeled IL-8 (1-66) bound to unlabeled NT-CXCR1(39-350) (Fig. 8 E), in which the observed signals are primarily from the immobilized IL-8 (1-66) residues interacting with the extracellular loops of the receptor, is similar to that of IL-8 (1-66) bound to W10HQA CXCR1 in the presence of manganese ions (Fig. 8 B). This suggests that the intermolecular PREs resulting from the Mn2+ bound to residue 10 of CXCR1 selectively broaden the IL-8 (1-66) signals from residues near binding site-I of CXCR1.
Discussion
Residues in the N-terminal domain (binding site-I) and in extracellular loops (binding site-II) of CXCR1 interact with IL-8 (13, 14, 16, 30, 31, 32, 33, 35, 66, 68, 69). Specific residues in the N-terminal domain of CXCR1 contribute to both the selectivity and the affinity of the receptor for IL-8. However, much less is known about how IL-8 interacts with the extracellular loops of CXCR1, or how the binding of IL-8 activates the receptor and triggers its biological functions. 1H-detected fast MAS solid-state NMR experiments on isotopically labeled IL-8 (1-66) bound to unlabeled constructs of CXCR1 provide new information about the nature of the interactions of IL-8 with CXCR1 in phospholipid bilayers.
IL-8 binds to synthesized and expressed polypeptides with sequences corresponding to the N-terminal region of CXCR1 that contains binding site-I. The KD values for IL-8 interacting with only binding site-I are in the submicromolar to submillimolar range (15), which is significantly weaker than for binding to full-length CXCR1, which includes both binding site-I and binding site-II (KD = 1–5 nM) (45). In particular, the binding affinity of IL-8 (1-66) for 1TM-CXCR1 (1-72), which includes N-terminal residues (binding site-I) and the first transmembrane helix of CXCR1, is 12.5 μM in nanodiscs, which is similar to that for the soluble N-terminal domain alone [ND-CXCR1 (1-38)] in aqueous buffer (17). Notably, not only IL-8 (1-66) bound to full-length CXCR1 (1-350), which includes binding site-I and binding site-II, but also IL-8 (1-66) bound to the shorter constructs 1TM-CXCR1 (1-72), which includes only binding site-I, and NT-CXCR1 (39-350), which includes only binding site-II, are copelleted by ultracentrifugation. Thus, ligand-receptor complexes in phospholipid bilayers with as low as micromolar binding affinities are copelleted by ultracentrifugation, demonstrating that the complexes are stable and well-suited for NMR experiments. All of the results are consistent with “slow exchange” of the ligand (IL-8) and the receptor (CXCR1) on the relevant NMR timescales.
GPCRs undergo conformational changes as a result of ligand binding and their dynamics are hypothesized to play an important role in the transmission of signals across membranes (70). The N- and C-terminal residues of the apo-receptor display evidence of mobility in several different sample preparations and NMR experiments. However, it has also been established that they play roles in the interactions with the extracellular ligand IL-8 and intracellular G-proteins, respectively (23, 58), and these residues become ordered upon interaction with the ligands.
The data summarized in Fig. 9 provide an opportunity to focus on the changes in the structure and dynamics of the chemokine IL-8 that result from interactions with its receptor CXCR1. The data in Fig. 9, rows C–F, are derived from the spectra in Fig. 3; they show that IL-8 (1-66) undergoes significant changes in dynamics upon interaction with full-length CXCR1 (1-350) and the shorter N-terminal truncated construct NT-CXCR1 (39-350). Previously, the dynamics of monomeric and dimeric IL-8 have been described in the absence and presence of N-terminal CXCR1 peptides by solution NMR (30); the N-terminus of IL-8, including the conserved ELR motif, is disordered and dynamic, whereas the three loops connecting three β−strands and one C-terminal α-helix are well-structured, even though their amide hydrogens undergo facile exchange with water (the data in row B of Fig. 9 are derived from Fig. 4). The IL-8 residues that directly interact with various N-terminal CXCR1 constructs, including ND-CXCR1 (1-38) and 1TM-CXCR1 (1-72), are well defined (15, 30). The residues in the N-loop and 40s loop of IL-8 contribute to its major binding sites and display some evidence of local motions (14, 15, 31, 33). In combination with the solid-state NMR data obtained on IL-8 (1-66) bound to CXCR1 (1-350) and NT-CXCR1 (39-350) in phospholipid bilayers, this suggests that the dynamics of free and bound IL-8 may play important roles in receptor binding selectivity as well as activation.
Figure 9.
Summary of IL-8 (1-66) interactions with CXCR1 (1-350), NT-CXCR1 (39-350), and ND-CXCR1 (1-38). Row A: Nonexchanging amide sites in D2O (17). Row B: Fast-exchanging amide sites in H2O from Fig. 4. (Rows C and D: Immobile and mobile sites by interaction with CXCR1 (1-350), respectively, from Fig. 5. Rows E and F: Immobile and mobile sites by interaction with NT-CXCR1 (39-350), respectively, from Fig. 7. Row G: Most perturbed sites of chemical shifts by interaction with ND-CXCR1 (1-38) (17). To see this figure in color, go online.
Significant changes in IL-8 (1-66) dynamics occur upon binding to wild-type CXCR1 (1-350). A majority of the residues in IL-8 (1-66), including those in the flexible loop regions, are immobilized upon interaction with CXCR1 (1-350). In addition, residue-specific chemical shift perturbations are observed (Fig. 5 A). Although many of the resonance assignments of bound IL-8 (1-66) could be obtained by direct comparison to the spectra of unbound IL-8 (1-66), several signals were significantly perturbed by the interaction and independent assignment experiments on the bound form of IL-8 (1-66) will be needed to map out all of the site-specific chemical shift perturbations. This should be feasible using recently developed 1H-detected fast MAS triple resonance solid-state NMR experiments (52).
The solid-state NMR INEPT spectrum in Fig. 3 C demonstrates that five backbone amide sites are mobile in IL-8 (1-66) bound to CXCR1 (1-350); the signals from these sites are superimposable on those from the ELR residues of IL-8 (1-66) free in solution as shown in Fig. 5 B. The signals of Glu4 and Leu5 are absent in the solution NMR INEPT spectrum of unbound IL-8 (1-66) due to their rapid exchange with water (Fig. 4 A). As a result, their presence in the solid-state NMR INEPT spectrum of bound IL-8 (1-66) demonstrates that these residues are protected from solvent exchange by the interactions of IL-8 with CXCR1. The N-terminal ELR motif of IL-8 has been proposed to interact with the extracellular regions of CXCR1 and trigger receptor activation (36). However, no chemical shift perturbations are observed, and the mobile ELR signals in bound IL-8 (1-66) suggest that the ELR motif does not interact directly with extracellular or transmembrane helical regions of CXCR1, which is consistent with our previous data on the interactions between wild-type IL-8 dimer and full-length CXCR1 in oriented lipid bilayers (33).
Since the N-terminal truncated form of the receptor is missing binding site-I, NMR spectra of the complex of IL-8 (1-66) and NT-CXCR1 (39-350) are informative about the interactions of IL-8 with binding site-II. The relatively small number of IL-8 (1-66) residues that are immobilized by binding are located mainly within the 30s and 40s loops of IL-8 (1-66). They are likely to be involved in direct interactions with residues in extracellular loops of CXCR1. However, most IL-8 residues undergo sufficient motional averaging to yield signals from INEPT experiments in the absence of binding site-I (Fig. 7). Thus, IL-8 interacts with both binding site-I and binding site-II of CXCR1. Other efforts to characterize the two-site model of IL-8 binding to CXCR1 have revealed that the N-loop of IL-8 is the major determinant for CXCR1 activation (71). In contrast, the N-terminus of IL-8 (ELR and CXC) is essential for CXCR2 activation (72), which suggests that this approach has the potential to address the selectivity of chemokines for different receptors.
1H-detected fast MAS solid-state NMR enabled the characterization of IL-8 (1-66)-CXCR1 complexes in phospholipid bilayers, providing insight into the molecular events associated with the first step of the CXCR1-mediated signaling cascade. These studies also served to demonstrate that site-specific incorporation of metal-chelating unnatural amino acid HQA into CXCR1 in combination with high-resolution 1H-detected fast MAS solid-state NMR provide site-specific PRE-derived intermolecular distance restraints in ligand-receptor complex. Thus, solid-state NMR can provide measurements of both intra- and intermolecular distances along with the identification of residues directly involved in binding to specific sites on both the ligand and receptor. Demonstrated here on ligand binding to a GPCR, the approach has the potential to be applicable to a broad range of protein complexes in membranes and other biological supramolecular assemblies.
Author Contributions
S.J.O. and S.H.P. designed the research. S.H.P., S.B., J.R., and A.A.D.A. performed the experiments. S.H.P., S.B., J.R., A.A.D.A., and S.J.O. analyzed the data. and S.J.O. and S.H.P. wrote the manuscript.
Acknowledgments
We thank Dr. Chin Wu for assistance with the NMR instrumentation.
This research was supported by grants P41EB002031, RO1GM066978, and R35GM122501 from the National Institutes of Health, and utilized the Biomedical Technology Resource Center for NMR Molecular Imaging of Proteins at the University of California, San Diego.
Editor: Charles Deber.
References
- 1.Jacobson K.A. New paradigms in GPCR drug discovery. Biochem. Pharmacol. 2015;98:541–555. doi: 10.1016/j.bcp.2015.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katritch V., Cherezov V., Stevens R.C. Structure-function of the G protein-coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang Y.L., Khoshouei M., Sexton P.M. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature. 2017;546:118–123. doi: 10.1038/nature22327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y., Sun B., Skiniotis G. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature. 2017;546:248–253. doi: 10.1038/nature22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexander S.P., Davenport A.P., Davies J.A., CGTP Collaborators The concise guide to PHARMACOLOGY 2015/16: G protein-coupled receptors. Br. J. Pharmacol. 2015;172:5744–5869. doi: 10.1111/bph.13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bendall L. Chemokines and their receptors in disease. Histol. Histopathol. 2005;20:907–926. doi: 10.14670/HH-20.907. [DOI] [PubMed] [Google Scholar]
- 7.Baggiolini M. CXCL8 - The First Chemokine. Front. Immunol. 2015;6:285. doi: 10.3389/fimmu.2015.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clore G.M., Appella E., Gronenborn A.M. Three-dimensional structure of interleukin 8 in solution. Biochemistry. 1990;29:1689–1696. doi: 10.1021/bi00459a004. [DOI] [PubMed] [Google Scholar]
- 9.Baldwin E.T., Weber I.T., Gronenborn A.M. Crystal structure of interleukin 8: symbiosis of NMR and crystallography. Proc. Natl. Acad. Sci. USA. 1991;88:502–506. doi: 10.1073/pnas.88.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burrows S.D., Doyle M.L., Porter D. Determination of the monomer-dimer equilibrium of interleukin-8 reveals it is a monomer at physiological concentrations. Biochemistry. 1994;33:12741–12745. doi: 10.1021/bi00209a002. [DOI] [PubMed] [Google Scholar]
- 11.Das S.T., Rajagopalan L., Rajarathnam K. Monomeric and dimeric CXCL8 are both essential for in vivo neutrophil recruitment. PLoS One. 2010;5:e11754. doi: 10.1371/journal.pone.0011754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowman H.B., Fairbrother W.J., Hébert C.A. Monomeric variants of IL-8: effects of side chain substitutions and solution conditions upon dimer formation. Protein Sci. 1997;6:598–608. doi: 10.1002/pro.5560060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernando H., Chin C., Rajarathnam K. Dimer dissociation is essential for interleukin-8 (IL-8) binding to CXCR1 receptor. J. Biol. Chem. 2004;279:36175–36178. doi: 10.1074/jbc.C400283200. [DOI] [PubMed] [Google Scholar]
- 14.Ravindran A., Joseph P.R., Rajarathnam K. Structural basis for differential binding of the interleukin-8 monomer and dimer to the CXCR1 N-domain: role of coupled interactions and dynamics. Biochemistry. 2009;48:8795–8805. doi: 10.1021/bi901194p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph P.R., Rajarathnam K. Solution NMR characterization of WT CXCL8 monomer and dimer binding to CXCR1 N-terminal domain. Protein Sci. 2015;24:81–92. doi: 10.1002/pro.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernando H., Nagle G.T., Rajarathnam K. Thermodynamic characterization of interleukin-8 monomer binding to CXCR1 receptor N-terminal domain. FEBS J. 2007;274:241–251. doi: 10.1111/j.1742-4658.2006.05579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berkamp S., Park S.H., Opella S.J. Structure of monomeric Interleukin-8 and its interactions with the N-terminal Binding Site-I of CXCR1 by solution NMR spectroscopy. J. Biomol NMR. 2017 doi: 10.1007/s10858-017-0128-3. Published online November 15, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajarathnam K., Clark-Lewis I., Sykes B.D. 1H NMR solution structure of an active monomeric interleukin-8. Biochemistry. 1995;34:12983–12990. doi: 10.1021/bi00040a008. [DOI] [PubMed] [Google Scholar]
- 19.Rollins B.J. Where the confusion began: cloning the first chemokine receptors. J. Immunol. 2009;183:2893–2894. doi: 10.4049/jimmunol.0990065. [DOI] [PubMed] [Google Scholar]
- 20.Holmes W.E., Lee J., Wood W.I. Structure and functional expression of a human interleukin-8 receptor. Science. 1991;253:1278–1280. doi: 10.1126/science.1840701. [DOI] [PubMed] [Google Scholar]
- 21.Murphy P.M., Tiffany H.L. Cloning of complementary DNA encoding a functional human interleukin-8 receptor. Science. 1991;253:1280–1283. doi: 10.1126/science.1891716. [DOI] [PubMed] [Google Scholar]
- 22.Nasser M.W., Raghuwanshi S.K., Richardson R.M. Differential activation and regulation of CXCR1 and CXCR2 by CXCL8 monomer and dimer. J. Immunol. 2009;183:3425–3432. doi: 10.4049/jimmunol.0900305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park S.H., Das B.B., Opella S.J. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 2012;491:779–783. doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin L., Kufareva I., Handel T.M. Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science. 2015;347:1117–1122. doi: 10.1126/science.1261064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu B., Chien E.Y., Stevens R.C. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Y., Qin L., Handel T.M. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature. 2016;540:458–461. doi: 10.1038/nature20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan Q., Zhu Y., Wu B. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science. 2013;341:1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oswald C., Rappas M., Marshall F.H. Intracellular allosteric antagonism of the CCR9 receptor. Nature. 2016;540:462–465. doi: 10.1038/nature20606. [DOI] [PubMed] [Google Scholar]
- 29.Burg J.S., Ingram J.R., Garcia K.C. Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science. 2015;347:1113–1117. doi: 10.1126/science.aaa5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kendrick A.A., Holliday M.J., Eisenmesser E.Z. The dynamics of interleukin-8 and its interaction with human CXC receptor I peptide. Protein Sci. 2014;23:464–480. doi: 10.1002/pro.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skelton N.J., Quan C., Lowman H. Structure of a CXC chemokine-receptor fragment in complex with interleukin-8. Structure. 1999;7:157–168. doi: 10.1016/S0969-2126(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 32.Clubb R.T., Omichinski J.G., Gronenborn A.M. Mapping the binding surface of interleukin-8 complexed with an N-terminal fragment of the type 1 human interleukin-8 receptor. FEBS Lett. 1994;338:93–97. doi: 10.1016/0014-5793(94)80123-1. [DOI] [PubMed] [Google Scholar]
- 33.Park S.H., Casagrande F., Opella S.J. Interactions of interleukin-8 with the human chemokine receptor CXCR1 in phospholipid bilayers by NMR spectroscopy. J. Mol. Biol. 2011;414:194–203. doi: 10.1016/j.jmb.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark-Lewis I., Schumacher C., Moser B. Structure-activity relationships of interleukin-8 determined using chemically synthesized analogs. Critical role of NH2-terminal residues and evidence for uncoupling of neutrophil chemotaxis, exocytosis, and receptor binding activities. J. Biol. Chem. 1991;266:23128–23134. [PubMed] [Google Scholar]
- 35.Leong S.R., Kabakoff R.C., Hébert C.A. Complete mutagenesis of the extracellular domain of interleukin-8 (IL-8) type A receptor identifies charged residues mediating IL-8 binding and signal transduction. J. Biol. Chem. 1994;269:19343–19348. [PubMed] [Google Scholar]
- 36.Rajagopalan L., Rajarathnam K. Structural basis of chemokine receptor function--a model for binding affinity and ligand selectivity. Biosci. Rep. 2006;26:325–339. doi: 10.1007/s10540-006-9025-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown L.S., Ladizhansky V. Membrane proteins in their native habitat as seen by solid-state NMR spectroscopy. Protein Sci. 2015;24:1333–1346. doi: 10.1002/pro.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das B.B., Park S.H., Opella S.J. Membrane protein structure from rotational diffusion. Biochim. Biophys. Acta. 2015;1848(1 Pt B):229–245. doi: 10.1016/j.bbamem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen S.K., Bertelsen K., Vosegaard T. Solid-state NMR methods for oriented membrane proteins. Prog. Nucl. Magn. Reson. Spectrosc. 2015;88-89:48–85. doi: 10.1016/j.pnmrs.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Opella S.J. Solid-state NMR and membrane proteins. J. Magn. Reson. 2015;253:129–137. doi: 10.1016/j.jmr.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez J.J., Shukla A.K., Glaubitz C. The structure of the neuropeptide bradykinin bound to the human G-protein coupled receptor bradykinin B2 as determined by solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2008;47:1668–1671. doi: 10.1002/anie.200704282. [DOI] [PubMed] [Google Scholar]
- 42.Luca S., White J.F., Baldus M. The conformation of neurotensin bound to its G protein-coupled receptor. Proc. Natl. Acad. Sci. USA. 2003;100:10706–10711. doi: 10.1073/pnas.1834523100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verhoeven M.A., Creemers A.F., de Groot H.J. Ultra-high-field MAS NMR assay of a multispin labeled ligand bound to its G-protein receptor target in the natural membrane environment: electronic structure of the retinylidene chromophore in rhodopsin. Biochemistry. 2001;40:3282–3288. doi: 10.1021/bi0023798. [DOI] [PubMed] [Google Scholar]
- 44.Whittaker C.A., Patching S.G., Middleton D.A. Ligand orientation in a membrane-embedded receptor site revealed by solid-state NMR with paramagnetic relaxation enhancement. Org. Biomol. Chem. 2015;13:2664–2668. doi: 10.1039/c4ob02427c. [DOI] [PubMed] [Google Scholar]
- 45.Park S.H., Prytulla S., Opella S.J. High-resolution NMR spectroscopy of a GPCR in aligned bicelles. J. Am. Chem. Soc. 2006;128:7402–7403. doi: 10.1021/ja0606632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marchetti A., Jehle S., Pintacuda G. Backbone assignment of fully protonated solid proteins by 1H detection and ultrafast magic-angle-spinning NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2012;51:10756–10759. doi: 10.1002/anie.201203124. [DOI] [PubMed] [Google Scholar]
- 47.Zhou D.H., Shah G., Rienstra C.M. Proton-detected solid-state NMR spectroscopy of fully protonated proteins at 40 kHz magic-angle spinning. J. Am. Chem. Soc. 2007;129:11791–11801. doi: 10.1021/ja073462m. [DOI] [PubMed] [Google Scholar]
- 48.Andreas L.B., Le Marchand T., Pintacuda G. High-resolution proton-detected NMR of proteins at very fast MAS. J. Magn. Reson. 2015;253:36–49. doi: 10.1016/j.jmr.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Nieuwkoop A.J., Franks W.T., Oschkinat H. Sensitivity and resolution of proton detected spectra of a deuterated protein at 40 and 60 kHz magic-angle-spinning. J. Biomol. NMR. 2015;61:161–171. doi: 10.1007/s10858-015-9904-0. [DOI] [PubMed] [Google Scholar]
- 50.Park S.H., Yang C., Mueller L.J. Resolution and measurement of heteronuclear dipolar couplings of a noncrystalline protein immobilized in a biological supramolecular assembly by proton-detected MAS solid-state NMR spectroscopy. J. Magn. Reson. 2013;237:164–168. doi: 10.1016/j.jmr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reif B. Ultra-high resolution in MAS solid-state NMR of perdeuterated proteins: implications for structure and dynamics. J. Magn. Reson. 2012;216:1–12. doi: 10.1016/j.jmr.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 52.Barbet-Massin E., Pell A.J., Pintacuda G. Rapid proton-detected NMR assignment for proteins with fast magic angle spinning. J. Am. Chem. Soc. 2014;136:12489–12497. doi: 10.1021/ja507382j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dannatt H.R., Taylor G.F., Watts A. 13C- and 1H-detection under fast MAS for the study of poorly available proteins: application to sub-milligram quantities of a 7 trans-membrane protein. J. Biomol. NMR. 2015;62:17–23. doi: 10.1007/s10858-015-9911-1. [DOI] [PubMed] [Google Scholar]
- 54.Mance D., Sinnige T., Weingarth M. An efficient labelling approach to harness backbone and side-chain protons in (1) H-detected solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2015;54:15799–15803. doi: 10.1002/anie.201509170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward M.E., Ritz E., Ladizhansky V. Proton detection for signal enhancement in solid-state NMR experiments on mobile species in membrane proteins. J. Biomol. NMR. 2015;63:375–388. doi: 10.1007/s10858-015-9997-5. [DOI] [PubMed] [Google Scholar]
- 56.Park S.H., Wang V.S., Opella S.J. Paramagnetic relaxation enhancement of membrane proteins by incorporation of the metal-chelating unnatural amino acid 2-amino-3-(8-hydroxyquinolin-3-yl)propanoic acid (HQA) J. Biomol. NMR. 2015;61:185–196. doi: 10.1007/s10858-014-9884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rajagopalan L., Rajarathnam K. Ligand selectivity and affinity of chemokine receptor CXCR1. Role of N-terminal domain. J. Biol. Chem. 2004;279:30000–30008. doi: 10.1074/jbc.M313883200. [DOI] [PubMed] [Google Scholar]
- 58.Park S.H., Casagrande F., Opella S.J. Local and global dynamics of the G protein-coupled receptor CXCR1. Biochemistry. 2011;50:2371–2380. doi: 10.1021/bi101568j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morris G.A., Freeman R. Enhancement of nuclear magnetic resonance signals by polarization transfer. J. Am. Chem. Soc. 1979;101:760–762. [Google Scholar]
- 60.Shaka A.J., Barker P.B., Freeman R. Computer-optimized decoupling scheme for wideband applications and low-level operation. J. Magn. Reson. (1969). 1985;64:547–552. [Google Scholar]
- 61.Piotto M., Saudek V., Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 62.Kuschert G.S., Hoogewerf A.J., Sanderson P.N. Identification of a glycosaminoglycan binding surface on human interleukin-8. Biochemistry. 1998;37:11193–11201. doi: 10.1021/bi972867o. [DOI] [PubMed] [Google Scholar]
- 63.Nowacka A., Bongartz N.A., Topgaard D. Signal intensities in 1H-13C CP and INEPT MAS NMR of liquid crystals. J. Magn. Reson. 2013;230:165–175. doi: 10.1016/j.jmr.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 64.Williams G., Borkakoti N., Price L. Mutagenesis studies of interleukin-8. Identification of a second epitope involved in receptor binding. J. Biol. Chem. 1996;271:9579–9586. doi: 10.1074/jbc.271.16.9579. [DOI] [PubMed] [Google Scholar]
- 65.Wu L., Ruffing N., Qin S. Discrete steps in binding and signaling of interleukin-8 with its receptor. J. Biol. Chem. 1996;271:31202–31209. doi: 10.1074/jbc.271.49.31202. [DOI] [PubMed] [Google Scholar]
- 66.Barter E.F., Stone M.J. Synergistic interactions between chemokine receptor elements in recognition of interleukin-8 by soluble receptor mimics. Biochemistry. 2012;51:1322–1331. doi: 10.1021/bi201615y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee H.S., Spraggon G., Wang F. Genetic incorporation of a metal-ion chelating amino acid into proteins as a biophysical probe. J. Am. Chem. Soc. 2009;131:2481–2483. doi: 10.1021/ja808340b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hébert C.A., Chuntharapai A., Horuk R. Partial functional mapping of the human interleukin-8 type A receptor. Identification of a major ligand binding domain. J. Biol. Chem. 1993;268:18549–18553. [PubMed] [Google Scholar]
- 69.Suetomi K., Rojo D., Navarro J. Identification of a signal transduction switch in the chemokine receptor CXCR1. J. Biol. Chem. 2002;277:31563–31566. doi: 10.1074/jbc.M204713200. [DOI] [PubMed] [Google Scholar]
- 70.Latorraca N.R., Venkatakrishnan A.J., Dror R.O. GPCR dynamics: structures in motion. Chem. Rev. 2017;117:139–155. doi: 10.1021/acs.chemrev.6b00177. [DOI] [PubMed] [Google Scholar]
- 71.Prado G.N., Suetomi K., Navarro J. Chemokine signaling specificity: essential role for the N-terminal domain of chemokine receptors. Biochemistry. 2007;46:8961–8968. doi: 10.1021/bi7004043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sarmiento J., Shumate C., Navarro J. Diverging mechanisms of activation of chemokine receptors revealed by novel chemokine agonists. PLoS One. 2011;6:e27967. doi: 10.1371/journal.pone.0027967. [DOI] [PMC free article] [PubMed] [Google Scholar]