ABSTRACT
Feruloyl esterases (FAEs) are key enzymes required for the production of ferulic acid from agricultural biomass. Previously, we identified and characterized R18, an FAE from Streptomyces cinnamoneus NBRC 12852, which showed no sequence similarity to the known FAEs. To determine the region involved in its catalytic activity, we constructed chimeric enzymes using R18 and its homolog (TH2-18) from S. cinnamoneus strain TH-2. Although R18 and TH2-18 showed 74% identity in their primary sequences, the recombinant proteins of these two FAEs (recombinant R18 [rR18] and rTH2-18) showed very different specific activities toward ethyl ferulate. By comparing the catalytic activities of the chimeras, a domain comprised of residues 140 to 154 was found to be crucial for the catalytic activity of R18. Furthermore, we analyzed the crystal structure of rR18 at a resolution of 1.5 Å to elucidate the relationship between its activity and its structure. rR18 possessed a typical catalytic triad, consisting of Ser-191, Asp-214, and His-268, which was characteristic of the serine esterase family. By structural analysis, the above-described domain was found to be present in a loop-like structure (the R18 loop), which possessed a disulfide bond conserved in the genus Streptomyces. Moreover, compared to rTH2-18 of its parental strain, the TH2-18 mutant, in which Pro and Gly residues were inserted into the domain responsible for forming the R18 loop, showed markedly high kcat values using artificial substrates. We also showed that the FAE activity of TH2-18 toward corn bran, a natural substrate, was improved by the insertion of the Gly and Pro residues.
IMPORTANCE Streptomyces species are widely distributed bacteria that are predominantly present in soil and function as decomposers in natural environments. They produce various enzymes, such as carbohydrate hydrolases, esterases, and peptidases, which decompose agricultural biomass. In this study, based on the genetic information on two Streptomyces cinnamoneus strains, we identified novel feruloyl esterases (FAEs) capable of producing ferulic acid from biomass. These two FAEs shared high similarity in their amino acid sequences but did not resemblance any known FAEs. By comparing chimeric proteins and performing crystal structure analysis, we confirmed that a flexible loop was important for the catalytic activity of Streptomyces FAEs. Furthermore, we determined that the catalytic activity of one FAE was improved drastically by inserting only 2 amino acids into its loop-forming domain. Thus, differences in the amino acid sequence of the loop resulted in different catalytic activities. In conclusion, our findings provide a foundation for the development of novel enzymes for industrial use.
KEYWORDS: Streptomyces, biomass, ferulic acid, feruloyl esterase
INTRODUCTION
Ferulic acid (FA) has antioxidant and anti-inflammatory properties (1) and moderates oxidative stress and inflammation in Alzheimer's disease (2) and allergy (3). One of the most important applications of feruloyl esterase (FAE) is in catalyzing the release of FA, which is used as an additive in functional foods in the food industry, from plant cell walls. FAE is also useful in bioenergy production. Its ability to degrade plant cell walls is important and useful for the production of sugars, which are subsequently converted to biofuels. The ability of FAEs to degrade plant materials has been extensively studied. In cell walls, FA dimers enable cross-linking between hemicellulose molecules (4), leading to the low digestibility of polysaccharides for bioenergy production. Transgenic plants expressing an FAE isolated from Aspergillus niger showed decreased levels of cinnamic acid esters in hemicellulose (5, 6). Furthermore, the cell walls of transgenic wheat coexpressing FAE and xylanase were shown to release a large amount of sugars such as xylose and arabinose (6).
FAEs are present in Aspergillus spp. (7–10). These FAEs are classified into four subgroups, namely, classes A, B, C, and D, according to their amino acid sequences and substrate specificities (10). We previously identified two new esterases (R18 and R43) from a Streptomyces esterase library. These esterases showed FAE activity using FA and other cinnamic acid esters as the substrates (11). Both Streptomyces FAEs released FA and diferulic acid from biomass, especially defatted rice bran, in combination with Streptomyces xylanase and arabinofuranosidase (12). Streptomyces FAEs are categorized as class D FAEs because they release diferulic acid; however, their primary sequences showed no homology with those of other Aspergillus FAEs.
We aimed to understand the mechanism by which Streptomyces FAEs recognize hydroxycinnamic acid esters. Recently, we obtained a draft genome sequence of Streptomyces cinnamoneus TH-2. From the genome sequencing data, we identified an R18 homolog (TH2-18). We expressed in Escherichia coli the genes that encode the FAEs R18 and TH2-18. Subsequently, we compared the optimum pHs, optimum temperatures, and thermal stabilities of these enzymes. Furthermore, we investigated their substrate specificities using ethyl ferulate, methyl ferulate, and methyl esters of other hydroxycinnamic acids as the substrates. To identify the amino acid residue(s) responsible for the catalytic activity of R18, we constructed R18 and TH2-18 chimeras by repeat-length-independent broad-spectrum (RIBS) in vivo DNA shuffling (13). Next, we investigated the kinetic properties of these chimeric FAEs. We also analyzed the crystal structure of R18 to determine the relationship between its activity and its structure.
RESULTS
Expression in E. coli BL21(DE3) of the genes that encode R18 and TH2-18.
We identified a homolog of the gene that encodes R18, namely, the gene that encodes TH2-18, from the genome sequencing data for S. cinnamoneus TH-2. TH2-18 is comprised of 381 amino acids. The signal sequence located in the N terminus of R18 was determined in a previous study (11), and that of TH2-18 was determined by using the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/). It was predicted that 36 amino acids located in the N terminus formed the signal sequence of TH2-18. R18 and TH2-18 shared 74% amino acid sequence similarity (Fig. 1). Next, R18 and TH2-18 were cloned into the pET28a vector and expressed in E. coli BL21(DE3) as recombinant proteins. The signal sequences of R18 and TH2-18 were deleted for the expression of recombinant proteins (recombinant R18 [rR18] and rTH2-18) in E. coli. Purified rTH2-18 was determined to have a molecular mass of 38 kDa, which was similar to that of rR18 (Fig. 2A). The FAE activity of rTH2-18 using 4 mM ethyl ferulate as the substrate was markedly lower than that of rR18 (rR18, 22.1 mU/mg; rTH2-18, 1.45 mU/mg).
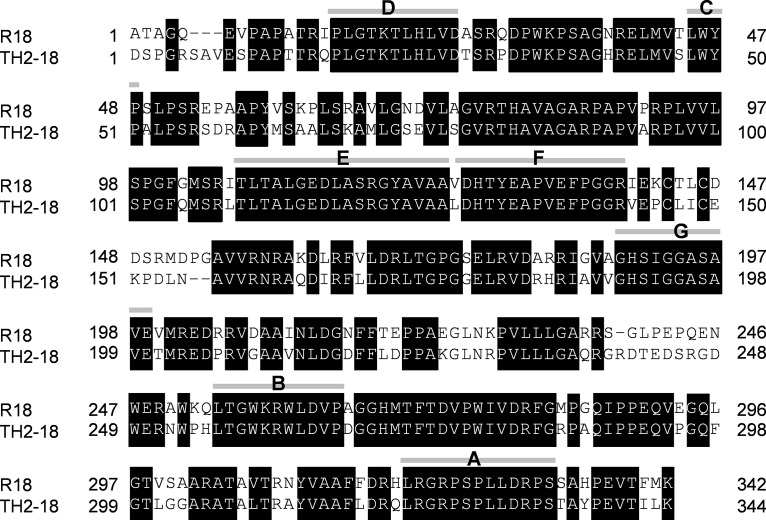
FIG 1.
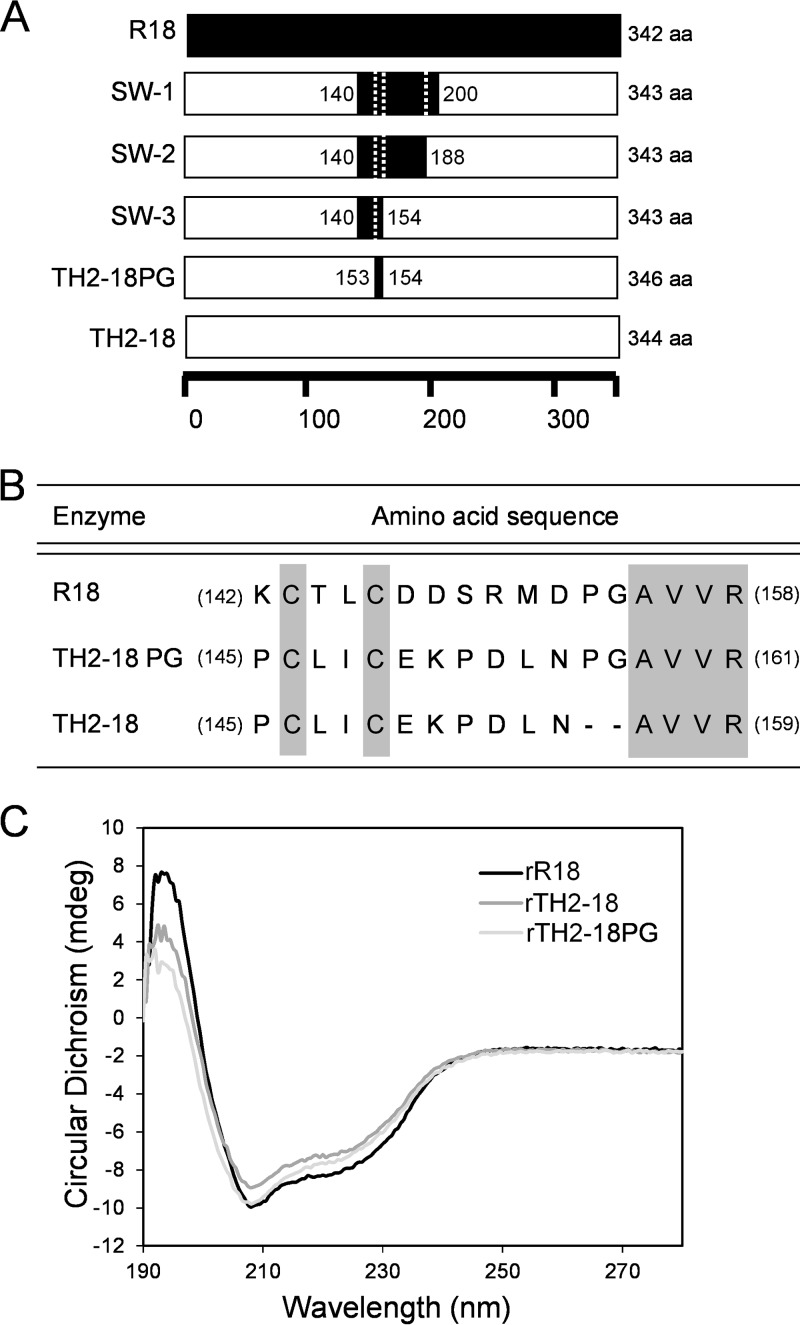
Amino acid sequence alignment of R18 and TH2-18. Shown are primary structures of R18 and TH2-18 without their signal peptides. Boxes and gray bars represent identical amino acids and the areas of recombination between R18 and TH2-18 in chimeric gene construction, respectively. The names of chimeras are indicated in capital letters.
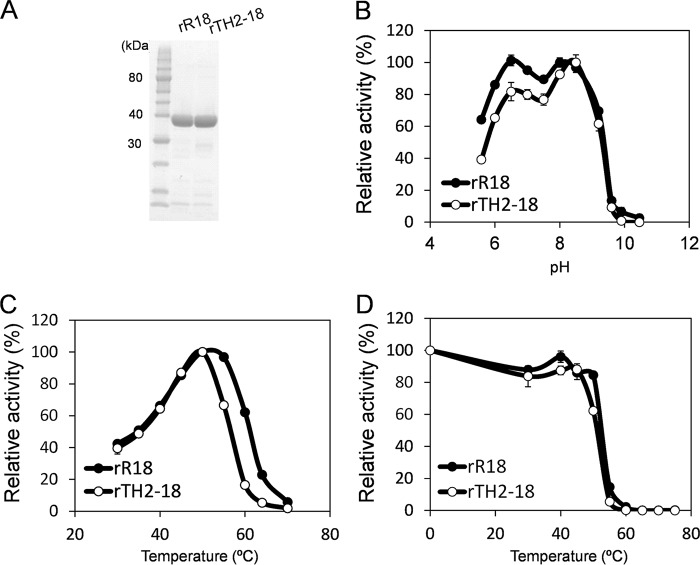
FIG 2.
Characterization of FAE activities of recombinant R18 and TH2-18. (A) SDS-PAGE analysis of rR18 and rTH2-18. Lane 1, protein standard; lane 2, rR18; lane 3, rTH2-18. Proteins (10 μg) were loaded into the respective lanes. (B to D) Effects of temperature (B), pH (C), and thermostability (D) on FAE activities of rR18 and rTH2-18. Values represent the averages of data from three independent experiments. Error bars represent standard deviations.
Characterization of rR18 and rTH2-18.
We investigated the FAE activities of rR18 and rTH2-18 at different pHs and temperatures. The FAE activity of rR18 was measured at pH 5.5 to 10.5. The optimum pHs were observed to be 6.5 and 8 (Fig. 2B). rR18 activity was also measured at 30°C to 60°C. The optimum temperature was observed to be 45°C (Fig. 2C). The FAE activity of rTH2-18 was measured at pH 5.5 to 10.5, and pH 8.5 was found to be the optimum pH for this enzyme (Fig. 2B). rTH2-18 activity was also measured at 30°C to 60°C, and the optimal temperature was observed to be 50°C (Fig. 2C). Both R18 and TH2-18 were completely inactivated by heating at 60°C for 30 min (Fig. 2D). The Km of rTH2-18 was 9.66 mM, which was 2.6 times higher than that of rR18 (Table 1). Additionally, the kcat of rTH2-18 was 0.27 min−1, which was one-sixth of that of rR18 (Table 1).
TABLE 1.
Kinetic parameters of the wild-type and chimeric Streptomyces FAEsa
| Enzyme | Substrate | Mean Km (mM) ± SD | Mean kcat (min−1) ± SD | Relative kcat | kcat/Km (min−1·mM−1) |
|---|---|---|---|---|---|
| rR18 | EF | 3.72 ± 0.6 | 1.70 ± 0.12 | 1 | 0.46 |
| MF | 5.59 ± 0.9 | 2.53 ± 0.18 | 1.488 | 0.457 | |
| MS | 2.56 ± 0.41 | 0.10 ± 0.02 | 0.059 | 0.037 | |
| rR18S191A | EF | ND | ND | ND | ND |
| rR18D214A | EF | ND | ND | ND | ND |
| rR18H268A | EF | ND | ND | ND | ND |
| rR18ΔPG | EF | ND | ND | ND | ND |
| rTH2-18 | EF | 9.66 ± 1.12 | 0.27 ± 0.02 | 0.159 | 0.028 |
| MF | 4.44 ± 0.53 | 0.16 ± 0.01 | 0.094 | 0.037 | |
| MS | 1.63 ± 0.14 | 0.05 | 0.029 | 0.036 | |
| rTH2-18PG | EF | 2.77 ± 0.14 | 0.87 ± 0.09 | 0.512 | 0.317 |
| MF | 4.13 ± 0.46 | 4.38 ± 0.45 | 2.576 | 1.06 | |
| MS | 2.77 ± 0.14 | 4.39 ± 0.44 | 2.582 | 2.56 | |
| Chimera A | EF | 3.8 ± 0.59 | 1.21 ± 0.14 | 0.71 | 0.319 |
| Chimera F | EF | 4.14 ± 0.81 | 0.54 ± 0.07 | 0.318 | 0.131 |
| Chimera G | EF | ND | ND | ND | ND |
| SW-1 | EF | 3.52 ± 0.31 | 0.35 ± 0.01 | 0.206 | 0.099 |
| SW-2 | EF | 3.66 ± 0.23 | 0.49 ± 0.02 | 0.288 | 0.134 |
| SW-3 | EF | 2.81 ± 0.14 | 0.87 ± 0.03 | 0.512 | 0.31 |
EF, ethyl ferulate; MF, methyl ferulate; MS, methyl sinapinate; ND, not determined.
Substrate specificities of rR18 and rTH2-18.
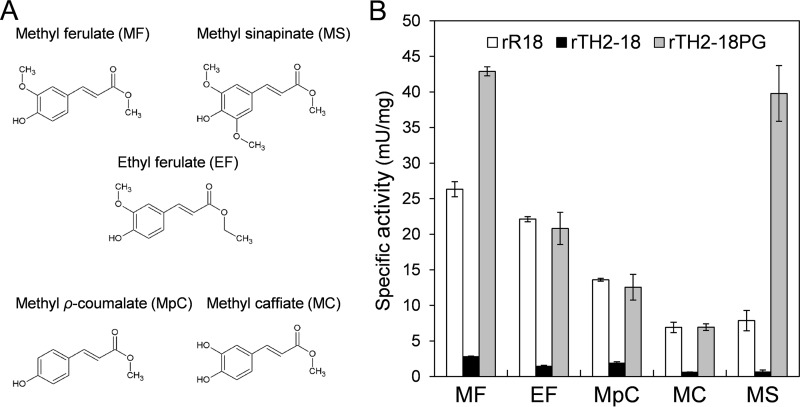
To compare the substrate specificities of rR18 and rTH2-18, ethyl ferulate, methyl ferulate, methyl p-coumarate, methyl caffeate, and methyl sinapinate were used as the substrates (Fig. 3A). Among the five hydroxycinnamic acid esters used as the substrates, rR18 showed the highest activity with methyl ferulate (26.3 mU/mg) (Fig. 3B), which was in agreement with previously reported values (14). Similarly, rTH2-18 showed the highest activity (2.79 mU/mg) (Fig. 3B) with methyl ferulate as the substrate. rTH2-18 showed the second highest activity with methyl p-coumarate as the substrate (1.87 mU/mg) (Fig. 3B). Thus, rTH2-18 showed ∼10-fold-lower FAE activity than did rR18.
FIG 3.
Structures of substrates and substrate specificities of rR18, rTH2-18, and rTH2-18PG. (A) Structures of substrates used in this study. (B) Substrate specificities of rR18, rTH2-18, and rTH2-18PG. Substrates were used at a concentration of 4 mM for the assay. Values represent the averages of data from three independent experiments. Error bars represent standard deviations.
FAE activities of chimeric FAEs.
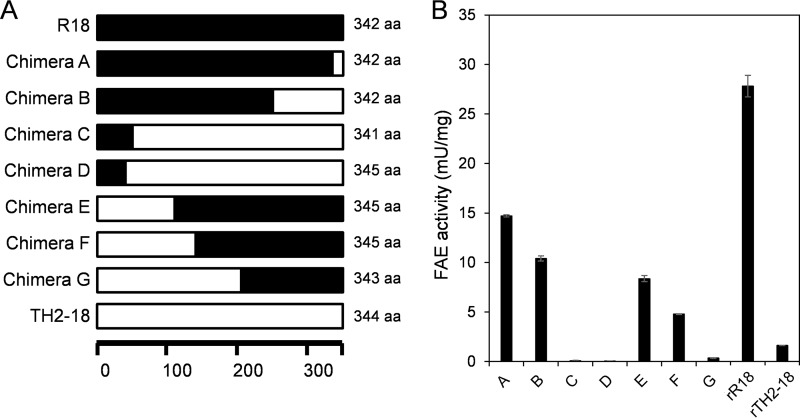
We analyzed the chimeric FAEs constructed by using R18 and TH2-18 to identify the amino acid residue(s) responsible for the catalytic activity of Streptomyces FAEs. For the construction of chimeric FAE genes, we performed the RIBS in vivo DNA shuffling method (13). We obtained four chimeric genes (chimeras A, B, C, and D) whose N-terminal sequences were obtained from the gene that encodes R18 and three chimeric genes (chimeras E, F, and G) whose N-terminal sequences were obtained from the gene that encodes TH2-18 (Fig. 1 and 4A). FAE activity was confirmed by using recombinant chimeric proteins, which were expressed in E. coli BL21(DE3) and purified by using Talon resin (TaKaRa Bio, Kusatsu, Japan). Among the seven chimeric proteins, the FAE activities of chimeras A, B, E, and F were intermediate between those observed for rR18 and those observed for rTH2-18 (Fig. 4B). The FAE activities of chimeras C, D, and G were markedly lower than that of rTH2-18 (Fig. 4B). Upon comparison of the FAE activities between these two groups, it was evident that the chimeric proteins that had the TH2-18 fragment between positions F and G showed lower FAE activities (Fig. 1 and 4B). Notably, the sequences from R18 and TH2-18 were considerably different. The kinetic parameters of chimeras A, F, and G were investigated. The Km values of chimeras A and F were 0.71 and 0.318 times higher than that of rR18, respectively (Table 1). The kcat/Km of chimera G could not be calculated (Table 1). On the basis of these results, we focused on the difference between the activities of chimeras F and G.
FIG 4.
Construction and FAE activities of rR18 and rTH2-18 chimeras. (A) Schematic illustration of the primary structures of rR18, rTH2-18, and the eight chimeras used in this study. The regions marked in black and white represent the sequences of R18 and TH2-18, respectively. aa, amino acids. (B) FAE activities in chimeric FAE lysates. Ethyl ferulate (4 mM) was used as the substrate. Values represent the averages of data from three independent experiments. Error bars represent standard deviations.
FAE activities of the TH2-18 sandwich chimeras.
To identify the amino acid residue(s) important for FAE activity, we constructed chimeric genes using the chimera F gene sequence and the 609-bp sequence from C-terminal region of the gene that encodes TH2-18. We constructed two new chimeric genes, encoding SW-1 and SW-2, which had sequences derived from residues between Ile-140 and Val-200 in R18 and between residues Ile-140 and Ala-188 in TH2-18, respectively (Fig. 5A). Using ethyl ferulate as the substrate, SW-1 and SW-2 showed Km values similar to that of rR18 (Table 1). Furthermore, we constructed another TH2-18 chimeric gene, encoding SW-3, which had the sequence derived from between residues Ile-140 and Gly-154 of R18 (Fig. 5A). The Km of SW-3 was similar to that of rR18. However, it showed higher FAE activity than that of rTH2-18 (Km = 2.81 ± 0.14; kcat = 0.87 ± 0.03) (Table 1).
FIG 5.
Construction of the sandwich-type chimeras and in vitro mutagenesis of rTH2-18. (A) Schematic illustration of the primary structures of four sandwich-type chimeras. The regions marked in black and white represent the sequences of R18 and TH2-18, respectively. (B) Amino acid sequences of chimeric positions in R18, TH2-18, and TH2-18PG. (C) CD spectra of rR18, rTH2-18, and rTH2-18PG.
Characterization of rTH2-18PG.
The R18 protein sequence from residues Ile-140 to Gly-154 consisted of 15 amino acids, including 2 Cys residues (Cys-143 and Cys-146). The corresponding region of TH2-18 consisted of 13 amino acids, including 2 Cys residues (Cys-146 and Cys-149) (Fig. 5B). Thus, we analyzed the role of the amino acid sequence between residues Asp-147 and Pro-154 in regulating FAE activity. We constructed rTH2-18PG, a mutant in which Pro and Gly residues were inserted between residues Asn-152 and Ala-153 in TH2-18, to understand the effect of these two amino acid residues on FAE activity (Fig. 5B). Before evaluating the activity of rTH2-18PG, we performed circular dichroism (CD) spectroscopy and observed that the CD spectra of rR18, rTH2-18, and rTH2-18PG showed similar patterns (Fig. 5C). rTH2-18PG showed high FAE activity (20.8 mU/mg) using ethyl ferulate (4 mM) as the substrate (Fig. 3B). Notably, its FAE activity was similar to that of rR18 (22.1 mU/mg). In addition, methyl ferulate, methyl p-coumarate, methyl caffeate, and methyl sinapinate were also used as the substrates. rTH2-18PG showed the highest activity (42.9 mU/mg) in the presence of methyl ferulate (4 mM) as the substrate (Fig. 3B). Interestingly, its activity was 1.6- and 14-fold higher than those of rR18 and rTH2-18, respectively. rTH2-18PG showed the second highest FAE activity (39.8 mU/mg) in the presence of methyl sinapinate (4 mM) as the substrate (Fig. 3B). Its activity was 5- and 63.7-fold higher than those of rR18 and rTH2-18, respectively. The kinetic parameters of rTH2-18PG were investigated (Table 1). Km values of rTH2-18PG using methyl ferulate and methyl sinapinate as the substrates were 4.13 and 2.77 mM, respectively, whereas their kcat values were 4.38 and 4.39 min−1, respectively (Table 1). In particular, the kcat values of rTH2-18PG using these substrates were approximately 27.4- and 87.8-fold higher than that of rTH2-18. These results suggested that the insertion of Pro and Gly affected the kcat values, but not the Km values, when methyl ferulate and methyl sinapinate were used as the substrates. In contrast, rR18ΔPG, a mutant in which the Pro and Gly residues in the rR18 loop were removed, lost its FAE activity.
FA production from biomass using TH2-18 and TH2-18PG.
Since large amounts of enzymes were required for hydrolyzing the natural substrates, we produced recombinant proteins in a hyperexpression system using Streptomyces lividans and pTONA5a (14) (Fig. 6A). The recombinant enzymes expressed in Streptomyces, namely, srR18, srTH2-18, and srTH2-18PG, showed specific activities of 16 ± 0.23, 1.74 ± 0.04, and 16.1 ± 0.27 mU/mg, respectively, using ethyl ferulate as the substrate. srR18, srTH2-18, and srTH2-18PG could release FA from corn bran, a natural substrate (Fig. 6B). The amounts of FA released did not differ between srR18 and srTH2-18; however, the amount released by srTH2-18PG was 1.6 times higher than the amounts released by srR18 and srTH2-18. These results indicated that the insertion of Pro and Gly residues enhanced FAE activity not only toward the artificial substrates but also toward the natural substrate.
FIG 6.
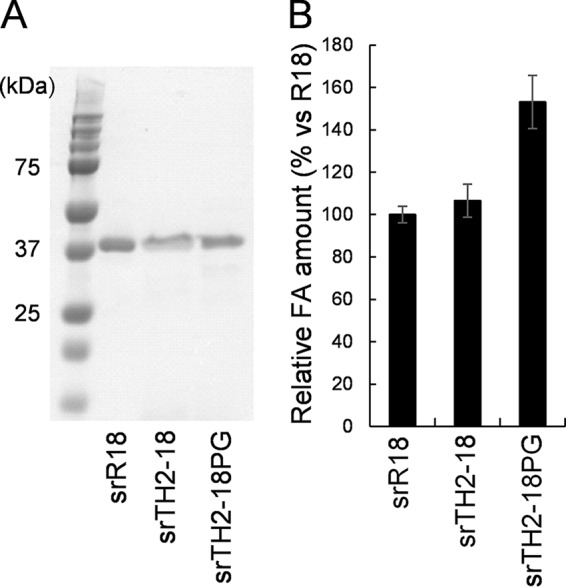
Expression and FA production (using corn bran) of Streptomyces recombinant FAEs. (A) SDS-PAGE analysis of srR18, srTH2-18, and srTH2-18PG (lane 1, protein standard; lane 2, srR18; lane 3, srTH2-18; lane 4, srTH2-18PG). Proteins (10 μg) were loaded into each lane. (B) Production of FA by srR18, srTH2-18, and srTH2-18PG using corn bran as the substrate. Values represent the averages of data from three independent experiments. Error bars represent standard deviations.
Crystal structure of rR18.
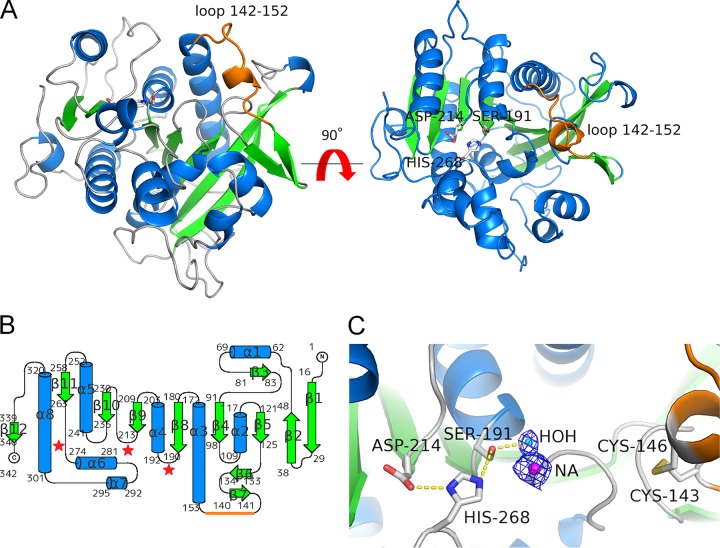
Crystal structures have been reported for the FAEs from A. niger and Aspergillus oryzae (15–17). rR18 did not show any sequence similarities to the known FAEs. Therefore, we determined the crystal structures of ligand-free rR18 and ethyl ferulate-cocrystallized rR18 at resolutions of 1.5 and 2.3 Å, respectively. Ethyl ferulate was not modeled in the structure because of its poor electron density. In the ligand-free and ethyl ferulate-cocrystallized structures, no marked structural changes were observed (root mean square deviation of 0.28 Å for 339 Ca atoms). Therefore, we described the crystal structure of ligand-free rR18 only. A structural overview of rR18 is provided in Fig. 7A.
FIG 7.
Crystal structure of rR18. (A) Structure of apo-rR18. Shown are ribbon representations with α-helices, β-strands, and coils marked in cyan, green, and gray, respectively. The catalytic triad residues (Ser-191, Aps-214, and His-268) are represented as sticks, and the loop (residues 142 to 152) is represented as an orange line. (B) Topology diagram of rR18 showing secondary structure elements (α, helices; β, strands). (C) Closeup view of the rR18 active site. The catalytic triad residues and disulfide bridge between Cys-143 and Cys-146 are shown in a stick representation, with the oxygen atom in red, the nitrogen atom in blue, sulfur atoms in yellow, and carbon atoms in white. A sodium ion in the catalytic site of rR18 is represented as NA.
The protein scaffold of R18 was an α/β hydrolase fold, which is typically observed in other GXSXG lipases and serine proteases. It consisted of one major 9-stranded mixed β-sheet, three minor β-sheet arrangements, and eight helices. A topological diagram of rR18 is shown in Fig. 6B. The structure of rR18 was submitted to the DALI website to obtain structural homologs (18). Human plasma platelet activation factor (PAF) (PDB accession no. 3D5E) was the top hit, with a Z-score of 34.3, a Cα root mean square deviation of 2.8 Å, and an identity of 22%, in a subset of 307 amino acids that were aligned (19).
The active site of rR18 contained the catalytic triad of Ser-191, Asp-214, and His-268. Ser-191 was located at the N terminus of the α-helix (α4) and on the conserved GXSXG motif that was also found in other serine proteases and lipases. The other two catalytic triad residues were appropriately positioned to activate nucleophilic residue Ser-191 for catalysis. Asp-214 was located on the C-terminal end of a β-sheet (β9), and His-268 was located at the N terminus of an α-helix (α6) (Fig. 7C). Point mutations involving the catalytic triad resulted in rR18 with an S-to-A change at position 191 (rR18S191A), rR18D214A, and rR18H268A, which did not show any FAE activity.
Based on structural analysis, the domain from residues Lys-142 to Asp-152 formed a loop structure (the R18 loop) that resulted from disulfide bridge formation between Cys-143 and Cys-146 (Fig. 7).
DISCUSSION
In this study, we revealed that the R18 loop plays an important role in mediating FAE activity by comparing R18, TH2-18, and seven chimeric enzymes between R18 and TH2-18 and analyzing crystal structures. The chimeras (SW-1, SW-2, and SW-3), which possessed the R18 loop and a part of the α3 helix (residues 153 to 177), showed higher enzymatic activities than that of wild-type TH2-18 (Table 1). The loop structure was important for the activity of some enzymes. For example, a residue present in the loop contributed toward maintaining the topological specificity of Streptomyces omiyaensis trypsin (20). A loop mutation in lactonase from Geobacillus kaustophilus increased the specific activity of this enzyme toward organophosphate pesticides (21). A loop in mannanases from an Actinomyces species helped in recognizing galactosyl substrates, such as linear or branched mannans (22). Our kinetic analysis also indicated the importance of the loop structure in mediating Streptomyces FAE activity.
In addition, we analyzed the crystal structure of rR18 and compared it with the crystal structures of homologous proteins that have been reported previously, including FAE-A from A. niger in complex with ethyl ferulate (PDB accession no. 1UWC), ligand-free FAE-B from A. oryzae (PDB accession no. 3WMT), and the feruloyl esterase domain of xylanase Z (FAE-XynZ) from Clostridium thermocellum in complex with FA (PDB accession no. 1JT2) (15, 17, 23) (Fig. 8). The structures of rR18 and FAE-XynZ were superimposable in the region containing main β-sheets and the positions of two catalytic triad residues, Cys and His, but not for the position of Asp. Interestingly, the position of Asp-214 was identical to that found in human PAF in plasma. An analogous stereochemistry of a human pancreatic lipase was studied, which showed a similar “migration” of the catalytic Asp residue occurring due to an apparently unrelated evolutionary event (24). The crystal structure of a complex between FAE-XynZ and FA has been reported (23) (Fig. 8B). Superimposition of the main β-sheets present in both rR18 and FAE-XynZ enabled the direct extrapolation of the position of the FA molecule docked into the active site of rR18 (Fig. 8A). A careful inspection of this putative complex revealed the residues that might be involved in substrate interactions. A minor rotation of the aromatic ring in FA is required to prevent close contact between ferulic 3-OCH3 and Pro-153, although the ring could be buried in a hydrophobic environment with Phe-101, Met-151, Pro-153, Ile-192, and Phe-218. The crystal structure of rR18 showed a rather wide hydrophobic pocket formed by the α5 helix (residues 241 to 252) that seemed suitable for accommodating the diferulic acid moiety in the catalytic cavity. In agreement with this, our previous work demonstrated diferulic acid production from defatted rice bran using R18 (11).
FIG 8.
Comparative view of the active sites of homologous proteins in complex with ferulic acid. (A) The product (ferulic acid) binding model of rR18 was obtained by structural superposition with the feruloyl esterase domain of xylanase Z (FAE-XynZ) from Clostridium thermocellum in complex with ferulic acid (PDB accession no. 1JT2). Residues in the neighborhood of the ligand are shown as sticks. The loop (residues 142 to 152) is indicated in orange. (B) FAE-XynZ from C. thermocellum (PDB accession no. 1JT2). (C) FAE from Aspergillus niger (PDB accession no. 1UWC).
The disulfide bridge between Cys-143 and Cys-146 generated an extremely tight turn (from Thr-144 to Asp-147). It maintained the R18 loop (between the β7 strand and the α7 helix) close to the active site, forming the hydrophobic pocket (Fig. 5). When rR18 was treated with 5 mM dithiothreitol for 15 min, the residual FAE activity was only 20% of that of nontreated rR18 (data not shown). The CXXC motif in R18 and TH2-18 was conserved in other R18 homologs (see Fig. S1 in the supplemental material). The disulfide bridge in the CSXDXHC motif was reported to directly affect the catalytic triad in A. oryzae FAE-B (AoFaeB) and other identified serine proteases (17). Compared to the CSXDXHC motif in AoFaeB, the CXXC motif in the R18 loop was distant from the catalytic triad in Streptomyces FAEs, thereby suggesting that the CXXC motif mediated the interaction between FAEs and an aromatic ring of their substrates.
The structures of rTH2-18, rTH2-18PG, and R18ΔPG were modeled by using the Spanner server (http://sysimm.ifrec.osaka-u.ac.jp/spanner/), with the structure of rR18 as a template. The predicted structures were superimposed on the structure of rR18. The loops of rR18 and predicted loop structures were compared (Fig. S2). The disulfide bridge present between two Cys residues was conserved in the R18 and TH2-18 loops; however, the C terminus of the bridge was not conserved in length or amino acid sequence. The R18 and predicted TH2-18PG loops were located closer to the active site than the TH2-18 (Fig. S2A and S2B) and R18ΔPG (Fig. S2C) loops in the respective molecules. Notably, the R18 loop was located above the position of the TH2-18 loop. These results indicated that rR18 had a slightly deeper and tighter hydrophobic pocket than TH2-18 due to the presence of the R18 loop. We suggest that in Streptomyces FAEs, the R18 loop is suitable for accommodating substrates such as methyl ferulate and ethyl ferulate.
Surprisingly, TH2-18 and R18 released FA from corn bran at similar rates (Fig. 6B); however, using artificial substrates, the activity of rTH2-18 was considerably lower than that of rR18. This disagreement remained unanswered. In the future, in order to explain this contradiction, we will attempt to understand the crystal structure of TH2-18 with its substrate.
In conclusion, we found a loop structure spanning residues Lys-142 to Asp-152 in R18, a Streptomyces FAE, which possessed a disulfide bridge between Cys-143 and Cys-146. We demonstrated that the loop plays an important role in mediating the catalytic activity of this enzyme. In the future, we will investigate the degradation of plant biomass by mutant Streptomyces FAEs.
MATERIALS AND METHODS
Bacterial strains and plasmids.
S. cinnamoneus strain TH-2 (formerly S. septatus TH-2) (25) was the source of the gene that encodes TH2-18. E. coli MK1019 (lacI3 ΔlacZ lacY+ ΔfliC ssb-3 rpsL+) (13) was used as the host for the construction of chimeric FAEs. E. coli BL21(DE3) cells were used for the expression of recombinant proteins. S. lividans strain 1326, harboring a pTONA5a derivative, was used for expression.
Plasmid pCR-Blunt II-TOPO (Kmr; Thermo Fisher Scientific, Carlsbad, CA) was used for cloning of the Streptomyces FAE genes. Plasmid pET28a carrying an N-terminal His tag (Kmr; Merck Japan) was used for expression in E. coli. Plasmid pTONA5a (Kmr and thiostrepton resistant) was used as a shuttle vector between E. coli and S. lividans, as reported previously by Hatanaka et al. (14), and used for expression in S. lividans 1326 cells.
Cloning and expression of Streptomyces FAE genes in E. coli BL21(DE3) and S. lividans.
The S. cinnamoneus gene that encodes TH2-18 (GenBank accession no. LC145308) was identified by performing a BLAST search of the NCBI nucleotide database using the R18 sequence as the query (GenBank accession no. AB921569). The gene was amplified by using GXL polymerase (TaKaRa Bio, Kusatsu, Japan). The amplified DNA sequences of the genes that encode R18 and TH2-18 were cloned by using the Zero Blunt TOPO PCR cloning kit (Thermo Fisher Scientific, Carlsbad, CA). Next, the identity of the cloned sequences in the recombinant vectors was confirmed. The recombinant TOPO vector was subjected to restriction digestion to release the FAE gene fragments. These fragments were then ligated into the pET-28a vector. FAEs were expressed in E. coli BL21(DE3). For expression, the culture was incubated at 30°C with shaking at 150 rpm for 24 h. The expressed recombinant FAEs (rR18 and rTH2-18) were purified by using Talon resin (TaKaRa Bio).
Protein expression in S. lividans strain 1326 was performed by transforming the Streptomyces protoplast with the pTONA5a vector containing the gene of interest (14). The transformants inoculated in PG medium (2% glucose, 0.8% potassium phosphate, 0.05% magnesium chloride, 0.5% yeast extract, and 0.5% polypeptone) were incubated at 30°C with shaking at 150 rpm for 5 days. Extracellular recombinant FAEs were precipitated by adding 60% ammonium sulfate to the mixture and centrifuging the mixture at 30,000 × g for 15 min. The pellets were freeze-dried and used for further analysis.
Enzyme assays.
For FAE activity assays, ethyl ferulate was used as the substrate. The reaction mixture consisted of 5 μl of 1 mg/ml enzyme, 4 mM ethyl ferulate, and 50 mM Tris maleate buffer in a total reaction volume of 50 μl. The reaction mixtures for rR18 and rTH2-18 were each incubated for 60 min at 50°C. The same reaction conditions were used for assaying the activities of chimeric and mutant FAEs. One unit of enzyme activity was defined as the amount of enzyme that released 1 μmol of FA per min. Enzyme activities using other hydroxycinnamate esters, such as methyl ferulate, methyl caffeate, methyl p-coumarate, and methyl sinapinate, as the substrates were also analyzed. All assays were performed under the same reaction conditions as the ones described above. The phenolic compounds released during the reaction were measured by using high-performance liquid chromatography (HPLC) (14). Components of the reaction mixture were separated by HPLC using a Symmetry C18 column (3.5 mm by 2.1650 mm; Waters, Milford, MA, USA) maintained at 40°C. Separation was performed within 5 min, using a linear gradient of 0.1% formic acid in water containing acetonitrile (10 to 60%), at a flow rate of 0.3 ml/min. The separated compounds, namely, FA, caffeic acid, p-coumaric acid, and sinapic acid, were detected at 322 nm.
Construction of chimeric and mutant genes using R18 and TH2-18 gene sequences.
RIBS in vivo DNA shuffling was performed as described previously (13, 20). The genes encoding R18 and TH2-18 were inserted into a pET28a vector in tandem, and a cassette containing the gentamicin resistance (Gmr) gene and E. coli rpsL+ was inserted between the genes encoding R18 and TH2-18. rpsL+ encodes the ribosomal protein S12, which is the target for streptomycin. This vector construct was transformed into E. coli MK1019 cells, and transformed colonies were selected on the basis of resistance to kanamycin and gentamicin. The colonies were cultured overnight in Luria-Bertani (LB) medium containing 50 μg/ml kanamycin. Subsequently, the cultures were spread onto LB agar plates containing 50 μg/ml each of kanamycin and streptomycin. The kanamycin- and streptomycin-resistant colonies were screened for their sensitivity to gentamicin, which was due to a deletion of the cassette and the formation of chimeric genes. Plasmid DNAs harboring chimeric FAEs were isolated from the cultures of gentamicin-sensitive colonies. The sandwich-type chimeric gene constructs were prepared by using an In-Fusion HD cloning kit in accordance with the manufacturer's instructions (TaKaRa Bio). Site-directed mutagenesis was performed as described previously (22). The gene that encodes TH2-18PG was amplified with the following primers: 5′-TGAACCCCGGGGCCGTCGTGCGGAACCGG-3′ and 5′-ACGGCCCCGGGGTTCAGGTCCGGCTTCTC-3′ (the inserted codons for Pro and Gly are underlined). The genes that encode the R18 mutants R18S191A, R18D214A, R18H268A, and R18ΔPG were also constructed by site-directed mutagenesis using the following primers: 5′-CGGCGGATCGGCGTCGCCGGGCACGCCATCGGC −3′ and 5′-CTCCACCGCGCTCGCGCCGCCGATGGCGTGCCC-3′ for the gene that encodes R18S191A, 5′-TCAACCTCGCCGGCAACTTCTTCACCGA-3′ and 5′-AAGTTGCCGGCGAGGTTGATCGCGGCGTC-3′ for the gene that encodes R18D214A, 5′-CCGGCGGCGCCATGACGTTCACCGACGTCC3′ and 5′-AACGTCATGGCGCCGCCGGCGGGCACGTCC-3′ (the changed codons for Ala are underlined) for the gene that encodes R18H268A, and 5′-GATGGATGCCGTCGTGCGCAACCGG and 5′-ACGACGGCATCCATCCGGCTGTCGTC-3′ for the gene that encodes R18ΔPG.
Circular dichroism spectroscopy.
Secondary structures of proteins were analyzed by CD spectroscopy (Chirascan-plus; Applied Photophysics, Leatherhead, UK). The proteins were dissolved in 100 mM Tris-HCl (pH 8) to obtain a final concentration of 1 mg/ml. Spectra were acquired at 20°C by using a cuvette (path length, 2 mm).
rR18 crystallization and data collection.
Ligand-free rR18 was crystallized by the sitting-drop vapor diffusion method at 19.85°C. Briefly, 0.5 μl of a protein solution (31.4 mg/ml, dissolved in 0.1 M Tris-HCl [pH 8]) was mixed with 0.5 μl of a precipitation solution containing 0.2 M sodium acetate trihydrate, 0.1 M sodium cacodylate trihydrate, and 15% (wt/vol) polyethylene glycol 8000. A drop was equilibrated against 100 μl of the precipitation solution. Crystals grew after 1 to 2 weeks. The crystals were cryoprotected by briefly soaking them in a solution of mother liquor supplemented with 20% (vol/vol) glycerol. The ethyl ferulate-cocrystallized crystals were obtained by mixing 0.5 μl of the same protein solution with 5 mM ethyl ferulate and 0.5 μl of precipitation solution (containing 0.17 M ammonium sulfate, 0.085 M sodium cacodylate trihydrate [pH 6.5], 25.5% [wt/vol] polyethylene glycol 8000, and 15% [vol/vol] glycerol). The drop was equilibrated against 100 μl of the precipitation solution. The crystals were flash-frozen directly under a nitrogen stream.
A native data set was collected at beamline 44XU of SPring-8 (Hyogo, Japan), using a ligand-free rR18 crystal. A single-wavelength anomalous diffraction (SAD) data set was collected at beamline 1A (BL-1A) at the Photon Factory Synchrotron facility (Tsukuba, Japan) with an ethyl ferulate-cocrystallized crystal. The obtained diffraction data were indexed and processed by using XDS software (26).
Crystal structure solution.
Sulfur SAD phasing was performed by using the 2.3-Å-resolution data set of ethyl ferulate-cocrystallized rR18 to determine the phase of the structure. Eight sulfur sites (including one disulfide bond) in the crystallographic asymmetric unit were identified and used for generating the initial phases by using the software programs SHELX C/D/E (27) and HKL2MAP (28). The medium-resolution initial phase did not provide sufficient information for the initial model. The initial phase was then refined in the nonisomorphous native data set using data at a resolution of 1.5 Å with SHELX E. Within 20 cycles, the program traced and automatically built the main chains in 80% of the model. Refinement was performed by using REFMAC5 software (29, 30). A summary of data collection, phasing, and refinement of the crystal structures is shown in Table 2.
TABLE 2.
Crystallographic data and refinement statisticsb
| Parameter | Value for structure |
|
|---|---|---|
| Native (ligand free) | Cocrystallized with ethyl ferulate | |
| Crystal | ||
| Space group | P212121 | P212121 |
| Unit cell dimensions a, b, c (Å) | 52.9, 70.9, 87.7 | 53.3, 71.3, 87.7 |
| Beamline | SPring-8 beamline 44XU | Photon Factory BL-1A |
| Data collection | ||
| Wavelength (Å) | 0.9 | 2.7 |
| Resolution (Å) | 50–1.5 (1.54–1.50) | 55.3–2.40 (2.47–2.4) |
| Completeness (%) | 99.9 (99.8) | 93.9 (87.9) |
| Redundancy | 3.8 | 30.6 |
| I/σI | 14.7 (2.3) | 42.9 (10.6) |
| Rmerge | 5.3 (60.4) | 7.2 (19.1) |
| Phasing statistics | ||
| No. of sulfur sites | 8 | |
| Figure of merita | 0.367 (0.692) | |
| Refinement statistics | ||
| Rwork/Rfree | 17.9/20 | 18.1/22 |
| RMSD bond lengths (Å) | 0.006 | 0.008 |
| RMSD bond angles | 1.183 | 1.355 |
Figure of merit after SAD phasing at 3.0 Å (after phase extension to 1.50 Å).
Values in parentheses are for the highest-resolution shells. RMSD, root mean square deviation.
Statistical analysis.
The significance of the differences observed in mean amounts of FA produced and FAE activity between enzymes or treatments was assessed by Student's t test. Differences were considered to be statistically significant at a P value of <0.05.
Accession number(s).
The crystal structures of ligand-free rR18 and ethyl ferulate-cocrystallized rR18 have been deposited in the PDB under accession no. 5YAL and 5YAE, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank the beamline staff of beamline 44XU at SPring-8 and BL-1A at the Photon Factory for their support during data collection.
Research conducted at the Photon Factory was supported by the Platform for Drug Discovery, Informatics, and Structural Life Science project. This work was supported by the Tojuro Iijima Foundation for Food Science and Technology.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02300-17.
REFERENCES
- 1.Mathew S, Abraham TE. 2004. Ferulic acid: an antioxidant found naturally in plant cell walls and feruloyl esterases involved in its release and their applications. Crit Rev Biotechnol 24:59–83. doi: 10.1080/07388550490491467. [DOI] [PubMed] [Google Scholar]
- 2.Yan JJ, Cho JY, Kim HS, Kim KL, Jung JS, Huh SO, Suh HW, Kim YH, Song DK. 2001. Protection against beta-amyloid peptide toxicity in vivo with long-term administration of ferulic acid. Br J Pharmacol 133:89–96. doi: 10.1038/sj.bjp.0704047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sin Singer Brugiolo A, Carvalho Gouveia AC, de Souza Alves CC, de Castro E Silva FM, Esteves de Oliveira É, Ferreira AP. 2017. Ferulic acid supresses [sic] Th2 immune response and prevents remodeling in ovalbumin-induced pulmonary allergy associated with inhibition of epithelial-derived cytokines. Pulm Pharmacol Ther 45:202–209. doi: 10.1016/j.pupt.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Buanafina MMDO. 2009. Feruloylation in grasses: current and future perspectives. Mol Plant 2:861–872. doi: 10.1093/mp/ssp067. [DOI] [PubMed] [Google Scholar]
- 5.Buanafina MM, Langdon T, Hauck B, Dalton S, Morris P. 2008. Expression of a fungal ferulic acid esterase increases cell wall digestibility of tall fescue (Festuca arundinacea). Plant Biotechnol J 6:264–280. doi: 10.1111/j.1467-7652.2007.00317.x. [DOI] [PubMed] [Google Scholar]
- 6.Harholt J, Bach IC, Lind-Bouquin S, Nunan KJ, Madrid SM, Brinch-Pedersen H, Holm PB, Scheller HV. 2010. Generation of transgenic wheat (Triticum aestivum L.) accumulating heterologous endo-xylanase or ferulic acid esterase in the endosperm. Plant Biotechnol J 8:351–362. doi: 10.1111/j.1467-7652.2009.00490.x. [DOI] [PubMed] [Google Scholar]
- 7.Faulds CB, Williamson G. 1995. Release of ferulic acid from wheat bran by a ferulic acid esterase (FAE-III) from Aspergillus niger. Appl Microbiol Biotechnol 43:1082–1087. doi: 10.1007/BF00166929. [DOI] [PubMed] [Google Scholar]
- 8.Koseki T, Takahashi K, Fushinobu S, Iefuji H, Iwano K, Hashizume K, Matsuzawa H. 2005. Mutational analysis of a feruloyl esterase from Aspergillus awamori involved in substrate discrimination and pH dependence. Biochim Biophys Acta 1722:200–208. doi: 10.1016/j.bbagen.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 9.Koseki T, Furuse S, Iwano K, Matsuzawa H. 1998. Purification and characterization of a feruloylesterase from Aspergillus awamori. Biosci Biotechnol Biochem 62:2032–2034. doi: 10.1271/bbb.62.2032. [DOI] [PubMed] [Google Scholar]
- 10.Koseki T, Fushinobu S, Ardiansyah, Shirakawa H, Komai M. 2009. Occurrence, properties, and applications of feruloyl esterases. Appl Microbiol Biotechnol 84:803–810. doi: 10.1007/s00253-009-2148-8. [DOI] [PubMed] [Google Scholar]
- 11.Uraji M, Arima J, Inoue Y, Harazono K, Hatanaka T. 2014. Application of two newly identified and characterized feruloyl esterases from Streptomyces sp. in the enzymatic production of ferulic acid from agricultural biomass. PLoS One 9:e104584. doi: 10.1371/journal.pone.0104584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uraji M, Kimura M, Inoue Y, Kawakami K, Kumagai Y, Harazono K, Hatanaka T. 2013. Enzymatic production of ferulic acid from defatted rice bran by using a combination of bacterial enzymes. Appl Biochem Biotechnol 171:1085–1093. doi: 10.1007/s12010-013-0190-6. [DOI] [PubMed] [Google Scholar]
- 13.Mori K, Mukaihara T, Uesugi Y, Iwabuchi M, Hatanaka T. 2005. Repeat-length-independent broad-spectrum shuffling, a novel method of generating a random chimera library in vivo. Appl Environ Microbiol 71:754–760. doi: 10.1128/AEM.71.2.754-760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatanaka T, Onaka H, Arima J, Uraji M, Uesugi Y, Usuki H, Nishimoto Y, Iwabuchi M. 2008. pTONA5: a hyperexpression vector in Streptomycetes. Protein Expr Purif 62:244–248. doi: 10.1016/j.pep.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 15.McAuley KE, Svendsen A, Patkar SA, Wilson KS. 2004. Structure of a feruloyl esterase from Aspergillus niger. Acta Crystallogr D Biol Crystallogr 60:878–887. doi: 10.1107/S0907444904004937. [DOI] [PubMed] [Google Scholar]
- 16.Hermoso JA, Sanz-Aparicio J, Molina R, Juge N, González R, Faulds CB. 2004. The crystal structure of feruloyl esterase A from Aspergillus niger suggests evolutive functional convergence in feruloyl esterase family. J Mol Biol 338:495–506. doi: 10.1016/j.jmb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki K, Hori A, Kawamoto K, Thangudu RR, Ishida T, Igarashi K, Samejima M, Yamada C, Arakawa T, Wakagi T, Koseki T, Fushinobu S. 2014. Crystal structure of a feruloyl esterase belonging to the tannase family: a disulfide bond near a catalytic triad. Proteins 82:2857–2867. doi: 10.1002/prot.24649. [DOI] [PubMed] [Google Scholar]
- 18.Holm L, Rosenström P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38:545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samanta U, Bahnson BJ. 2008. Crystal structure of human plasma platelet-activating factor acetylhydrolase: structural implication to lipoprotein binding and catalysis. J Biol Chem 283:31617–31624. doi: 10.1074/jbc.M804750200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uesugi Y, Usuki H, Iwabuchi M, Hatanaka T. 2009. The role of Tyr71 in Streptomyces trypsin on the recognition mechanism of structural protein substrates. FEBS J 276:5634–5646. doi: 10.1111/j.1742-4658.2009.07256.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, An J, Yang GY, Bai A, Zheng B, Lou Z, Wu G, Ye W, Chen HF, Feng Y, Manco G. 2015. Active site loop conformation regulates promiscuous activity in a lactonase from Geobacillus kaustophilus HTA426. PLoS One 10:e0115130. doi: 10.1371/journal.pone.0115130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumagai Y, Yamashita K, Tagami T, Uraji M, Wan K, Okuyama M, Yao M, Kimura A, Hatanaka T. 2015. The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition. FEBS J 282:4001–4014. doi: 10.1111/febs.13401. [DOI] [PubMed] [Google Scholar]
- 23.Schubot FD, Kataeva IA, Blum DL, Shah AK, Ljungdahl LG, Rose JP, Wang BC. 2001. Structural basis for the substrate specificity of the feruloyl esterase domain of the cellulosomal xylanase Z from Clostridium thermocellum. Biochemistry 40:12524–12532. doi: 10.1021/bi011391c. [DOI] [PubMed] [Google Scholar]
- 24.Schrag JD, Winkler FK, Cygler M. 1992. Pancreatic lipases: evolutionary intermediates in a positional change of catalytic carboxylates? J Biol Chem 267:4300–4303. [PubMed] [Google Scholar]
- 25.Hatanaka T, Negishi T, Kubota-Akizawa M, Hagishita T. 2002. Purification, characterization, cloning and sequencing of phospholipase D from Streptomyces septatus TH-2. Enzyme Microb Technol 31:233–241. doi: 10.1016/S0141-0229(02)00121-7. [DOI] [Google Scholar]
- 26.Kabsch W. 2010. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr 66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheldrick GM. 2008. A short history of SHELX. Acta Crystallogr A 64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 28.Pape T, Schneider TR. 2004. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J Appl Crystallogr 37:843–844. doi: 10.1107/S0021889804018047. [DOI] [Google Scholar]
- 29.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pannu NS, Murshudov GN, Dodson EJ, Read RJ. 1998. Incorporation of prior phase information strengthens maximum-likelihood structure refinement. Acta Crystallogr D Biol Crystallogr 54:1285–1294. doi: 10.1107/S0907444998004119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.