Abstract
Advances in the sequencing and the analysis of the genomes of both modern and ancient peoples have facilitated a number of breakthroughs in our understanding of human evolutionary history. These include the discovery of interbreeding between anatomically modern humans and extinct hominins; the development of an increasingly detailed description of the complex dispersal of modern humans out of Africa and their population expansion worldwide; and the characterization of many of the genetic adaptions of humans to local environmental conditions. Our interpretation of the evolutionary history and adaptation of humans is being transformed by analyses of these new genomic data.
Archaeological and palaeontological data have shaped our understanding of the events that led to the emergence and spread of anatomically modern humans. However, in general, these data can not be used to determine the genetic relationships between different people or groups of people. Although archaeological research has been highly successful in elucidating the spread of cultures, its ability to determine whether the spread of a culture occurred by the movement of people or ideas is often limited. The inclusion of analyses of genomic data from modern or ancient people facilitates the direct determination of the genealogical relationships between humans as well as the elucidation of migration routes, diversification events and genetic admixture among various groups.
In the 1980s, the advent of genomic data from modern humans enabled theories of the origins of humans to be tested directly. Studies of mitochondrial DNA (mtDNA) and other simple markers led to a number of important evolutionary insights. Most importantly, the first analysis of global human mtDNA sequence variation1 led to acceptance of the out-of-Africa model, a hypothesis proposing that modern humans originated in Africa, from where the population expanded outwards. An alternative hypothesis, known as the multiregional model, which suggests that anatomically modern humans evolved simultaneously in multiple locations, facilitated by gene flow that results from ongoing migration between locations, was widely rejected.
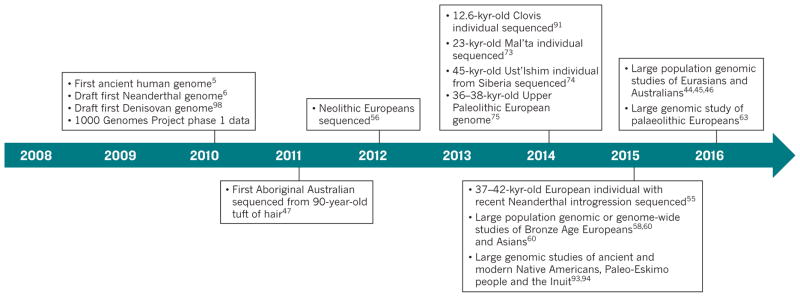
Phylogenetic trees of human mtDNA sequences have a root in Africa that is consistent with the out-of-Africa model. However, mtDNA reflects only female inheritance and, because it does not recombine, mtDNA has the information content of only a single genetic marker. There is a considerable chance that a phylogenetic tree inferred from only one marker is not representative of the overall genomic pattern and history of human evolution2. To rigorously test complex models of human history, analysis of the nuclear genome is also needed. Consequently, a number of questions were left unanswered by the analysis of markers from mtDNA (and the Y chromosome) performed in the 1980s, 1990s and 2000s. For example, it was unclear if gene flow had occurred between anatomically modern humans and other hominins2. The origins of Native Americans were also debated heavily3 and the relative importance of the movement of people and ideas in cultural transitions, such as the emergence and spread of agriculture4, remained unclear for Europe and other regions of the world. In the past ten years, considerable advances in DNA sequencing and methods for the enrichment and extraction of ancient DNA enabled studies to address many of these outstanding questions about human history and evolution (Fig. 1). Such technological advances have allowed researchers to sequence genomes from the remains of ancient humans5 and other hominins6 that died thousands of years ago. The addition of both temporal and geographic aspects to genome sequencing, by including samples from a wide range of historical times and locations, has provided fresh insights into human evolutionary history.
Figure 1. Timeline of important milestones in human evolutionary genomics.
A large number of studies have contributed important insights into human history using genomic data; those of particular influence in terms of the data or data analyses that they present are shown.
In this Review, we provide an overview of the most important insights into human evolutionary history that have been facilitated by obtaining and sequencing many human genomes. In some cases, the analysis of new genomic data has helped to establish further evidence for mainstream theories that were previously supported by palaeontological and archaeological evidence. In other cases, such analysis has led to the discovery of entirely new insights that could not have been predicted on the basis of existing data.
Origins in Africa
The earliest evidence for anatomically modern humans comes from fossils located in Ethiopia that can be dated to about 150,000–190,000 years (150–190 kyr) ago7,8. Beyond Africa, fossil evidence of anatomically modern humans has been reported as early as about 100 kyr ago in the Middle East9 and about 80 kyr ago in southern China10. However, other hominins, such as Neanderthals, which disappeared from the fossil record about 40 kyr ago11 (Fig. 2), have been found throughout Eurasia as far back as 400 kyr.
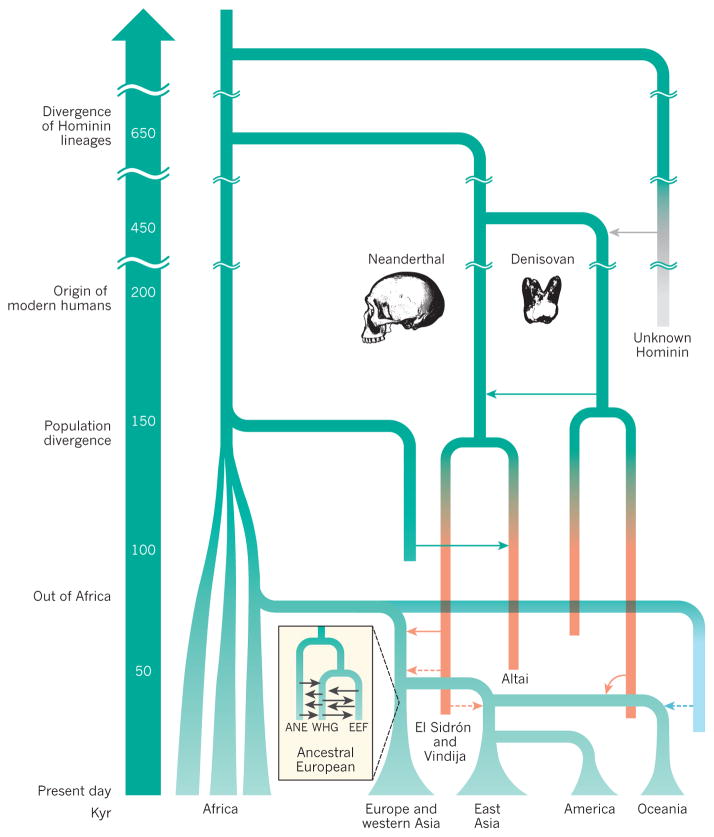
Figure 2. Simplified model of human evolutionary history.
Relationships between contemporary populations and the approximate times at which they diverged are shown. These include important well established (solid lines) and tentative (dashed lines) admixture events between groups of modern humans and between modern and archaic humans. The model also shows the potential small proportion of ancestry in Oceanic populations that is derived from an early out-of-Africa migration (turquoise). Studies of ancient DNA can provide high-resolution insights into the history of populations and have revealed that present-day Europeans comprise admixture between three ancestral groups57 (inset). ANE, ancient north Eurasian; EEF, early European farmer; WHG, west European hunter-gatherer.
Consistent with the evidence that the root of the human mtDNA phylogenetic tree is in Africa, initial studies of genomic diversity indicated that Africans have the highest levels of diversity among any living population12 as well as extensive population substructure; a study of genome-wide microsatellite DNA variation in more than 3,000 Africans identified 14 ancestral population clusters that correlate broadly with geography, culture and language13. Genome-wide single nucleotide polymorphism (SNP) genotyping studies largely supported these observations13–18. The findings of these and other studies indicate that African populations have maintained a large and subdivided structure throughout their evolutionary history16,19 and that the deepest splits between human populations lie in sub-Saharan Africa18,20. There is also evidence of both ancient and modern migration events across sub-Saharan Africa, as well as extensive admixture in the region13. The migration event that most shaped the genomic landscape of Africa was the movement of Bantu-language-speaking populations from their homeland in the highlands of Nigeria and Cameroon into much of sub-Saharan Africa in the past 4 kyr (Figs 2 and 3) and their subsequent admixture with and possible replacement of indigenous hunter-gatherer populations13. Other important migration events include the migration of pastoralist populations, which comprise farmers raising livestock, from their southern Sudanese homeland to eastern and central Africa about 7 kyr ago, and the migration of agropastoralists (who engage in both raising livestock and growing crops) from Ethiopia to Kenya and Tanzania about 5 kyr ago13.
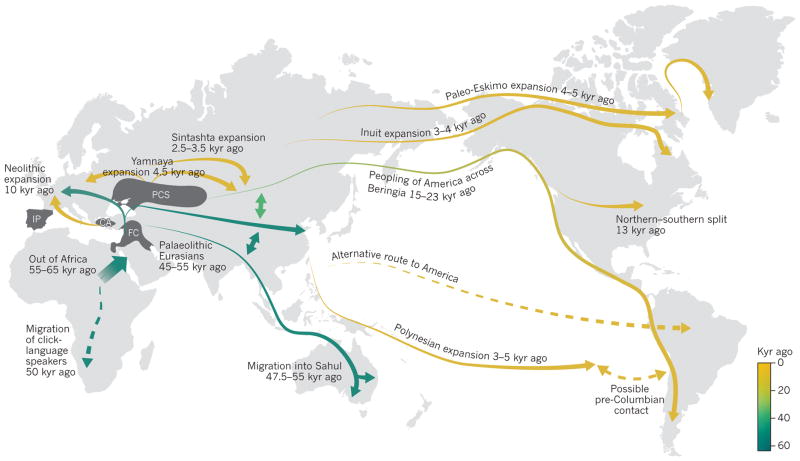
Figure 3. Major human migrations across the world inferred through analyses of genomic data.
Some migration routes remain under debate. For example, there is still some uncertainty regarding the migration routes used to populate the Americas. Genomic data are limited in their resolution to determine paths of migration because further population movements, subsequent to the initial migrations, may obscure the geographic patterns that can be discerned from the genomic data. Proposed routes of migration that remain controversial are indicated by dashed lines. CA, Central Anatolia; FC, Fertile Crescent; IP, Iberian Peninsula; PCS, Pontic–Caspian steppe.
Analysis of whole-genome sequencing and SNP array data found that the genetic lineages of click-language-speaking San populations of southern Africa capture the deepest split between populations of humans, with their divergence estimated to have occurred around 160–110 kyr ago15,18,20–22 (Fig. 2). However, genetic markers with uniparental inheritance and linguistic studies suggest that click-language-speaking hunter-gatherer populations may originally have been more widespread and were replaced in areas other than southern Africa or, alternatively, that they may have originated in eastern Africa and then migrated to southern Africa in the past 50 kyr13,23. Indeed, other hunter-gatherer populations that speak languages that use clicks, including the Hadza people and the Sandawe people, currently reside in Tanzania in eastern Africa, although they display limited genomic affinity with the San people of southern African15,18.
The exact origin of anatomically modern humans in Africa remains unknown, mainly because of the scarcity of fossil and archaeological data in the tropical regions of the continent. However, a multiregional origin of modern humans in Africa24, in which modern features evolved in a fragmented manner in several areas connected by gene flow, is still possible, especially given the opportunity for migration and admixture across the continent. Indeed, there is evidence for the admixture of anatomically modern humans with archaic populations in Africa25–27. The characterization of genomes from individuals who lived in Africa more than 10 kyr ago is challenging because the environment of the samples from which DNA is extracted, including the local climatic conditions, is not favourable to the preservation of genetic material. However, the statistical analysis of whole-genome sequencing data from geographically diverse hunter-gatherer populations provides evidence of archaic human lineages that have undergone introgression (the exchange of genetic material through interbreeding) and that diverged from modern human lineages as long ago as 1.2 Myr or 1.3 Myr25,27,28 or as recently as 35 kyr ago26. The degree of archaic admixture in Africa therefore remains controversial, and many ongoing efforts are aiming to resolve this question.
Out of Africa and the meeting with Neanderthals
The dispersal of anatomically modern humans out of Africa (Figs 2 and 3), a notable event in human evolutionary history, left a strong signature on the genetic variation of all non-African populations, including lower levels of diversity29 and higher levels of linkage disequilibrium30. However, the number, the geographic origin and migratory routes and the timing of major dispersals remains elusive. For instance, there is evidence to support the origins of modern humans in eastern, central and southern Africa18,31–33, single and multiple dispersals out of Africa7,34–36, a north or south dispersal route37,38 and estimates for the timing of dispersals occurring about 50 kyr–100 kyr ago20,39–43. Three studies44–46 that leverage fresh, high-quality whole-genome sequencing data from geographically diverse individuals from more than 270 locations worldwide help to resolve some of these questions. They also point to the occurrence of a single out-of-Africa dispersal in which all contemporary non-African peoples branched off from the same ancestral population that left Africa, possibly with minor genetic contributions from an earlier modern human migration wave into Oceania46. Furthermore, on leaving Africa, modern humans may have immediately separated into two waves of dispersal44. As proposed in refs 36 and 47, one wave led ultimately to the founding of Australasia and New Guinea and the other contributed to the ancestry of present-day mainland Eurasians. However, the exact routes of migration in the early diversification of people outside Africa remain a topic of research and controversy.
It is now clear that the ancestors of all contemporary non-African people encountered, and admixed with, Neanderthals48. All non-African individuals studied so far contain around 2% Neanderthal ancestry49–51, suggesting that admixture mostly occurred shortly after the dispersal of anatomically modern humans from Africa, which is consistent with a single–dispersal-based out-of-Africa model (Figs 2 and 3). On the basis of patterns of linkage disequilibrium, the date of hybridization has been estimated to be approximately 50–65 kyr ago52, and knowledge of the timing of admixture with Neanderthals helps to bound estimates of when the ultimately successful out-of-Africa dispersal occurred.
Estimates of the proportion of Neanderthal ancestry that persists in the genomes of modern humans51,53 points towards a more complex history of interaction between Neanderthals and modern humans. Specifically, East Asian people have about 20% more Neanderthal sequences compared to European people, which may reflect the effects of natural selection, the occurrence of further admixture events in the ancestors of present-day East Asians after the population split from Europeans50,53,54 (Fig. 2) or the dilution of Neanderthal ancestry in Europeans owing to admixture with populations that did not contain considerable levels of Neanderthal sequences. Strikingly, SNP genotyping in an early modern human from Romania who lived about 40 kyr ago provided further evidence that introgression occurred at several times and locations in Eurasia55, although the individual did not contribute detectable ancestry to present-day populations. Recent studies50 have suggested a more complex admixture history than previously thought, and we caution that our understanding of admixture models is fluid at present and that further demographic models are also compatible with the observed.
The peopling of Europe
European populations are likely to be composed of three or more genetic components, some of which entered Europe at different times56–61 (Figs 2 and 3). The first anatomically modern humans lived in Europe as early as 43 kyr ago11,62. These early Paleolithic Europeans have probably made little genetic contribution to the European people of today61 as there is evidence of turnover in the genetic composition of Europeans before the Last Glacial Maximum (LGM), possibly in relation to climate oscillations63 — although the exact contributions from early Europeans is still under debate61.
Around 11 kyr ago, after the LGM had passed, a new way of life based on animal husbandry, agriculture and sedentarism — and known as a Neolithic lifestyle — started to emerge in several subregions of the Fertile Crescent64 (Fig. 3). Analyses of ancient DNA showed that this population of farmers expanded from Central Anatolia into Europe; however, other regions of the Fertile Crescent contributed only limited genetic material to the early European farmers61. They reached the Iberian Peninsula (Fig. 3) roughly 7 kyr ago and arrived in Britain and Scandinavia about 6 kyr ago61. Genomic data from the remains of Neolithic humans have shown that this process was driven by the mass migration of groups of farmers61 and the assimilation of local hunter-gatherers65, demonstrating that the Neolithic way of life spread across Europe through the migration of people rather than solely as an idea or a culture. The Neolithic lifestyle helped to increase the size of populations, as seen in the estimates of effective population sizes that were generated from genomic data66, although archaeological data suggests that the health of the individuals who lived as farmers was sometimes poor as there were ample signs of malnutrition and caries66,67.
Another wave of migration into Europe, which introduced the third European genetic component, occurred during the late Neolithic period and the early Bronze Age. Herders from the Pontic–Caspian steppe who belonged to the Yamnaya culture were involved in a migration to central Europe about 4.5 kyr ago58,60. The herders themselves were descendants of various hunter-gatherer groups from (modern) Russia58 and the Caucasus68. This migration was probably linked to conquests and technological innovations such as horseback riding and may have spread Indo-European languages to Europe58,60, although some linguistics researchers suggest that these languages were already spoken by Neolithic farmers69. Clearly, the late Neolithic period and the Bronze Age were dynamic times that led to the spread of the genetic material of the steppe herders across western and northern Europe58,60.
The three main genetic components of modern-day European populations reflect the contributions of hunter-gatherers to the recolonization of Europe after the LGM, the migration of Neolithic farmers from Anatolia to Europe and the late-Neolithic period and Bronze Age migration to Europe from the east. These components can explain much of the genetic diversity found in present-day Europe65. For example, the Neolithic genetic component seems to be most dominant in southern European populations such as the Sardinian people56,57,66. Genetic variation among modern-day Europeans is strongly correlated with geography70,71 and shows a gradient of decreasing diversity with increasingly northern latitudes72. Although the main components of genetic diversity were introduced into Europe in separate waves of migration, subsequent processes of gene flow that were limited by geography have shaped the present genetic landscape. Culture and lifestyle were therefore more important determinants of genomic differentiation and similarity in many periods during Prehistoric Europe than geography61.
The peopling of Asia and Oceania
Most evidence indicates that Asia was colonized through at least two early waves of migration. One wave included the ancestors of Australians and the Papuan people and the other included other ancestors of East Asians, with admixing between the two47 (Figs 2 and 3), although other evidence suggests that there was only one dispersal event45. However, the details of how Asia was first colonized remain largely unknown. Two early modern human genomes from Asia have been sequenced. The first genome came from an individual of the Mal’ta-Buret’ culture of southern central Siberia who lived about 24 kyr ago73 and shows a strong genetic affiliation to both western Eurasians and Native Americans but a weaker affiliation to East Asians and Siberians, implying that there was a very different geographic distribution of genetic signatures during the Upper Palaeolithic period in comparison to the present day. The second genome, which came from an individual who lived in the Ust’-Ishim region of western Siberia74 about 45 kyr ago, shows almost equal genetic affinity with western Eurasians, East Asians74 and Aboriginal Australians when differences in Denisovan admixture are accounted for44. Together with evidence from the 36–38-kyr-old genome of the Kostenki 14 individual from European Russia75, showing a close affinity to contemporary western Eurasians but not East Asians, this points to the occurrence of a divergence between East Asians and western Eurasians around 36–45 kyr ago. A study that included low-coverage sequencing of the genomes of 101 ancient humans from across Bronze Age Eurasia60 showed that two later population expansions into central Asia from Europe and western Asia resulted in mixing with and locally replacing the Mal’ta-like hunter-gatherers. The first event was an expansion of Yamnaya herders into Asia about 5 kyr ago, which occurred at the same time as the Yamnaya expansion into Europe. Subsequently, between 2.5 and 3.5 kyr ago, Yamnaya people in central Asia (forming the Afanasievo culture) were locally replaced by individuals from the Sintashta culture60, who moved in from the Urals and Europe and admixed with East Asians.
Archaeological evidence shows that humans were present in Oceania around 47.5–55 kyr ago76,77. Morphological variation in ancient human skeletons from Australia has been used to propose that there were at least two independent migrations to the ancient continent of Sahul, which is comprised of modern Australia, New Guinea and Tasmania78. Similar claims have been put forward on the basis of linguistic data and lithic technology or the introduction of domesticated species such as the dingo36. However, the only extensive population genomic study so far on Aboriginal Australians and Papuans44 finds evidence for only a single founding event in Sahul, which was followed by a divergence of the Papuan and Aboriginal Australian ancestral population and further genetic diversification in the Aboriginal Australian population that could have coincided with environmental changes such as desertification. Aboriginal Australians therefore seem to have been living in a high level of isolation until only relatively recent times.
A study79 of genome-wide SNP data from modern people in Oceania confirmed archaeological predictions that Polynesians, who are distributed across a triangle of islands in the South Pacific that is bounded by Rapa Nui (also known as Easter Island) to the east, represent an expansion into Oceania of individuals with mixed Melanesian and East Asian ancestry. The Melanesian ancestry was added to the original East Asian ancestry after the initial Polynesian expansion had begun80. Whether Polynesians reached the Americas and admixed with Native Americans during their eastward expansion that ended about 1 kyr ago remains controversial. A genetic study of ancient chicken remains from South America supports this scenario81 but has also been questioned82. Genome sequencing of the remains of humans from Brazil that date to around 1650, and therefore pre-date the recorded trade of Polynesian slaves to South America83, shows that the individuals are closely related to contemporary Polynesians. These data potentially provide further support for early contact between Polynesians and Native Americans but they could also be the result of the European-mediated transportation of people. More convincing are the results of a genome-wide study of the modern-day inhabitants of Easter Island84, which provided statistical support for Native American admixture that can be dated to 1280–1495, several hundred years before Europeans reached the islands in 1722. However, only evidence of Polynesian and Native American admixture in human remains that pre-date colonization in the Americas would settle the debate.
The peopling of the Americas
The oldest most widely accepted evidence of humans in the Americas dates to about 15–14 kyr ago85, and widespread settlement of the Americas appeared with the emergence of the Clovis complex (around 12.6–13 kyr ago), which is the earliest well-characterized archaeological assemblage in the Americas. However, until around 13 kyr ago, much of North America was covered by a large ice sheet, which would have made it difficult for people to move from Beringia (now northeastern Siberia and northwestern North America) to the southern parts of the Americas. After the ice melted, a roughly 1,500 km interior ice-free corridor formed3. Metagenomic analyses of lake cores from Canada86 have estimated that this corridor first became biologically viable around 12.6 kyr ago, which makes it an unlikely early route for the southward migration of pre-Clovis and Clovis groups of people, although a study of bison is in disagreement87. How and when the earliest ancestors of Americans crossed the Pleistocene ice sheets into southern North America is unknown, as is whether movement of the pre-Clovis and Clovis groups represents the same migration. However, a movement towards the south along the west coast of North America that occurred more than 14 kyr ago, and that was possibly followed by southerly or northerly back-migrations through the interior, seems to be the most plausible scenario (Fig. 3).
On the basis of cranial morphology and lithic analysis, it has been proposed that early Americans were not direct ancestors of contemporary Native Americans, but instead were related to Australo-Melanesians, Polynesians, the Ainu people of Japan or Europeans who were later replaced or assimilated by ancestors of Native Americans from Siberia88–90. However, several genomic studies have largely rejected these models. In 2014, the oldest and the only Clovis-associated human genome from the Americas (found in Montana, United States), which belonged to an individual who lived about 12.6 kyr ago, was published91. Analyses suggested that the Clovis population from which the genome came was directly ancestral to many contemporary Native Americans. Similarly, analysis of the genome sequence of the roughly 9.5-kyr-old Kennewick Man skeleton found in the state of Washington in the United States92, which was thought to be closely related to the Ainu and Polynesians on the basis of cranial morphology, determined that he was most closely related to contemporary Native Americans. Moreover, populations that were considered to be relicts of an early migration into the Americas and closely related to Australo-Melanesians have been shown to be genetically related to contemporary Native Americans93,94.
Estimates of the time of divergence between Siberians and Native Americans, based on whole-genome sequences, point to the formation of the Native American gene pool as early as around 23 kyr ago93, which lends further support to the early entrance of ancestors of Native Americans into the Americas. When the accepted dates for the earliest archaeological sites in the Americas are taken into consideration, ancestors of Native Americans could have remained in isolation until around 8 kyr ago in Siberia or Beringia, following the split from their Siberian ancestors, before moving eastwards into the Americas. Although modern Siberians are the closest relatives of Native Americans outside of the Americas, genome sequencing of a 24-kyr-old Mal’ta skeleton73 suggests that Native Americans are derived from a mixture of populations that are related to the Mal’ta lineage as well as one or more unknown East-Asian lineages. Because the Clovis-associated genome and contemporary Native Americans contain similar amounts of the Mal’ta genetic signature (14–38%), the admixture event happened more than 12.6 kyr ago. However, whether it took place inside or outside the Americas remains unclear.
In Native Americans, genomic data have been used to locate a basal division that can be dated to about 14–13 kyr ago73,91. The southern branch includes groups of Amerindian-language-speaking people and the northern branch includes groups of Athabascan-language-speaking people as well as other groups that speak languages such as Cree or Algonquin. Divergence estimates based on analyses of whole-genome sequencing data suggest that both groups diversified from Siberians concurrently, implying that there was only one founding event for both Amerindian and Athabascan populations that was followed by subsequent gene flow from Asia93. Whether the divergence between the two Native American branches took place in Siberia or the north or south of the American ice sheets is still under debate, and the analysis of further ancient genomes will be needed to resolve this. Similarly, it remains undetermined whether the discovery of the Australo-Melanesian signature in some contemporary Brazilian Native Americans (Fig. 3) can be attributed to gene flow at a later time93 or an unknown early founding population94. So far, no studies of the genomes of ancient humans from the Americas have shown this genetic signature.
The Inuit of the American Arctic have been shown to originate from a migration separate to that of other Native Americans95,96. However, it has long been discussed whether the first people to inhabit the Arctic, the now extinct Paleo-Eskimo culture, which appeared about 5 kyr ago in the Americas, represent the ancestors of the present-day Inuit or an independent founder population from Siberia96 (Fig. 3). Sequencing of DNA from a 4-kyr-old tuft of hair from Greenland5 showed that the population the individual belonged to had migrated from Siberia to the North American Arctic independently of the Native American and Inuit migrations97. The group then survived in the Arctic for about 4 kyr by reinventing their subsistence strategies and technology but were eventually replaced by the Inuit around 700 yr ago.
The meeting with Denisovans
As well as the Neanderthals, at least one other type of archaic human — the enigmatic Denisovans — lived in Eurasia when the first modern humans started to appear on the continent. Little is known about the morphology and distribution of Denisovans, who are known only from the genome sequences of a finger bone and three teeth that were found in the Denisova Cave in Siberia98–100. They are most closely related to Neanderthals, with a genetic differentiation that is similar to the deepest splits between modern humans48 but an estimated time of divergence that possibly dates back 200–400 kyr48. Denisovans have many peculiarities; for instance, they may carry genetic material (obtained through admixture) from individuals related to earlier types of humans (Fig. 2), possibly Homo erectus48. Arguably, Denisovans can be considered to be the eastern or southern end of a spectrum of archaic humans that lived in Eurasia (and possibly beyond), with Neanderthals representing the western end.
Similar to Neanderthals, Denisovans interbred with anatomically modern humans. About 3–6% of the genome of some groups of people, including Melanesians in Oceania, can be traced to a Denisovan-like ancestor48,98 (Fig. 2). Continental southeast Asians carry genetic material on the order of 0.1–0.3% that can be traced to Denisovans48,101. The genomes of both Neanderthals and Denisovans have been subjected to genetic selection on introgression. Most selection in humans seems to be directed against the introgressed DNA, because there is a paucity of introgressed DNA near functional regions of the genome51. Furthermore, large genomic regions that are depleted of both Neanderthal and Denisovan sequences have been identified102, which is consistent with the rapid purging of deleterious sequences. However, some of the introgressed DNA might have helped humans to adapt to the local environment, such as the adaptation to high altitudes in Tibetan people103.
Studies of the first reliable genome data from archaic humans proposed two punctuated and very specific events for admixture between anatomically modern humans and archaic humans6,98. Since then, we have learned that such admixture is much more common, with multiple events taking place between various groups of modern and archaic humans48,50,101,103 in both directions104 (Fig. 2). At present, it is unclear whether the Denisovan introgression into Melanesians and Australians occurred in Australasia or Asia, as the ancestors of modern Australasians migrated across the continent. If it had occurred in Asia, present-day Asians would be mostly descended from other groups that arrived during subsequent waves of migration. Similarly, it is unknown whether the Denisovan admixture in East Asians is a result of the same admixture event or events that affected Australasians.
Human adaptation to new environments
The analysis of genetic data can inform us about not only the evolutionary history of humans, but also how natural selection has affected our species. As humans spread, first within Africa and subsequently to the rest of the world, they encountered new environmental conditions that induced selective regimes, including extreme cold in much of the Americas and Eurasia during the last ice age, altered exposure to sunlight and pathogens not previously encountered. Cultural innovations such as improved methods of hunting and fishing and the development of plant and animal domestication also induced new environmental conditions, including changes in diet. Genome-sequencing data sets have provided an opportunity to systematically scan the human genome for regions that have been targeted by selection. By taking advantage of the extensive resources that are now available for the analysis of human genomes, it has been possible to determine the specific functions of individual genetic variants that were targeted by selection, thereby providing a link between the evidence of selection that was discovered through the analysis of DNA sequences and the role of these sequences in adaptation to local environments. Furthermore, studies of ancient DNA now enable the direct observation of changes in allele frequency through time105,106.
Adaptation to the local environment
One of the most obvious changes in the environment that humans encountered as they migrated out of Africa was a reduction in exposure to sunlight at higher latitudes. Populations that live near the equator have dark skin to protect against skin damage and the photolysis of folate by ultraviolet radiation107. However, ultraviolet radiation also has an important role in catalysing the production of vitamin D, which is essential for skeletal development and health. In populations that live at higher latitudes, and therefore receive lower doses of ultraviolet radiation, having lighter skin is thought to be an advantage that enables more efficient vitamin D production. Selection that favours lighter skin pigmentation in higher latitudes has affected several genes, including MC1R, SLC24A5 and SLC45A2 (also known as MATP)108,109.
Another example of the adaptation of humans to local environments is found in populations that live in hypoxic (low oxygen) environments at high altitude in regions such as Tibet. A study by Beall110 showed that Tibetans who have adapted to life at high altitudes exhibit modified regulation of red-blood-cell production in response to hypoxia. Genomic studies have suggested that this adaptation is driven by changes in allele frequency in two genes in the hypoxia response pathway: EPAS1 and EGLN1 (refs 111–114).
Changes in diet, particularly those that are associated with the emergence of new hunting technologies or agricultural practices, have also had a considerable impact on the human genome. The best known example is selection for lactase persistence (the avoidance of lactose intolerance), which affects regulation of the gene LCT among dairy farming populations in Europe115 and Africa116,117. Similarly, genes in the FADS family, the products of which catalyse the synthesis of poly-unsaturated fatty acids, seem to have been under selection during several transitions towards or away from a vegetarian diet in humans118–121.
However, the most important driver of local adaptation is probably the local pathogenic environment122; examples of this include the genes that encode the major histocompatibility complex123, which presents foreign peptides for recognition by immune cells, and the β-globin locus and the gene G6PD in Africans, in which specific mutations provide protection against malaria for heterozygous carriers124,125. (A more comprehensive review of adaptation to local environments can be found in ref. 126).
Lessons from the genomic analysis of natural selection
Several lessons can be learned from genome-wide studies of selection in humans. First, although most of the initial scans for selection proposed that selection acts immediately on new mutations, it is becoming clear that in many cases, selection acts on standing variation — that is, alleles that were present for some time before they became favoured127. Furthermore, this variation has in many cases been introgressed by interbreeding with other hominins128,129. There is a growing list of genetic variants that were identified as introgressed from Neanderthals and Denisovans that have been favoured in anatomically modern humans by natural selection129. For example, the adaptive EPAS1 haplotype in Tibetans seems to be introgressed from Denisovans, and the selection on the major histocompatibility complex genes and the gene MC1R is probably facilitated by introgression from Neanderthals130,131. As humans migrated out of Africa and encountered new environments, introgression with other hominins that had already adapted to these environments seems to have been an important factor in facilitating rapid acceleration. This might be particularly true for genes related to immunity and defence against infection, as anatomically modern humans probably encountered pathogenic agents that could jump from other hominins, and to which humans did not yet have immunity.
Second, much of the selection that has affected the human genome has been in response to changes in the environment that were induced by people. These include changes in diet that were driven by cultural innovations and an increase in the pathogen load of the population owing to changes in social structure and the emergence of cities132. As we modify our environment, the resulting changes in conditions induce new selective pressures. Biological evolution and cultural evolution are therefore intimately linked.
Third, a close relationship often exists between genetic variants that have been under selection and those that have a strong influence on human health. Studies of human evolution are therefore of increasing relevance for medical genetics. For example, variants found to be selected for by adaptation to high altitudes provide a model for studying hypertension133. Similarly, variants shown to be under selection in relation to dietary adaptation, such as those in the gene family FADS, may facilitate the development of genomics-informed personalized diets.
Fourth, the genes that show the greatest difference in allele frequency between continental groups (indigenous Africans, Europeans, Americans and Australians) are enriched for associations with visible traits such as skin, hair and eye pigmentation103. An interesting consequence is that the geographic groups are more different from each other in terms of pigmentation than they are, on average, at the level of the genome. Humans from various parts of the world are therefore more genetically similar than might be predicted on the basis of observed hair colour, skin colour or other visible traits.
Last, genetic variants with large effects, such as those that influence eye colour, hair colour or lactase persistence, are unusual. Instead, such traits seem to be highly complex and may be influenced by many loci across the genome. Unlike selection at individual sites, polygenic adaptation — the result of selection acting on complex traits — can occur rapidly, even in a few generations, but the footprints of selection at individual loci are extremely weak and cannot be detected by standard methods134. The best characterized example of polygenic adaptation in humans is selection for an adaptive increase in the height of northern Europeans135, a smaller increase in the height of Europeans compared to non-Europeans136 and a decrease in the height in Sardinians137. Selection for height may have had small effects on loci across most of the human genome138. Polygenic adaptation may also have influenced a variety of other morphological traits, including increases in size of the head and body of infants in northern Europe138. It is probable that a considerable component of selection in humans is polygenic and is yet to be discovered by studies that scan for genomic regions that are under selection.
Challenges for human evolutionary genomics
Evolutionary and demographic inference based on genomic data from humans has often been the subject of considerable debate. Human evolution is a natural experiment that has been repeated only once, and the inference of past demography is therefore a historical science, with inherent limitations. Furthermore, the ancestral geographic locations of humans cannot be deduced directly from the genomes of modern humans; instead, they are typically inferred indirectly from the locations in which samples are found. Genetic analyses can inform knowledge of the ancestral relationships between individuals, but ancestral DNA is not inscribed with geographic locations. Also, the dating of divergence or admixture events is only as good as the clock by which dating is measured. Rates of mutation across the human genome and between individuals may be variable, and there has been considerably controversy regarding the mutations rates that should be used in demographic studies. Estimates of these rates vary depending on the methodology that is used for estimation, although a consensus rate of 0.5 × 109 base pairs per year has emerged in the past few years43. It is possible that this estimate will be revised further, which would affect the dating of events inferred by genomic data in previous studies. Despite these limitations, advances in the analysis of both modern and ancient human genomes have changed many aspects of our understanding of human evolution.
The mounting evidence for interbreeding between Neanderthals, Denisovans and anatomically modern humans has put a focus on the role of introgression in human evolution. Such studies have found that anatomically modern humans did not evolve with complete independence from other hominins outside Africa, as proposed by the strictest versions of the out-of-Africa model that were often subscribed to after the publication of mtDNA evidence almost 30 years ago1. Levels of interbreeding suggest that the true model of human evolution — sometimes referred to as the leaky replacement model — lies between the multi-regional model and the out-of-Africa model139. In fact, the levels of Neanderthal admixture in modern humans are probably strong underestimates of the amount of admixture that was present at the time of introgression55,140.
There is considerable evidence that substantial amounts of Neanderthal DNA was purged by selection, through genetic incompatibilities51 or simply because the high level of inbreeding in Neanderthals caused the Neanderthal DNA to be enriched with deleterious mutations. A linear fit to the change in the proportion of Neanderthal admixture suggested that the original proportion, at the time at which admixture took place, was probably about 5%63. However, selection does not decrease the proportion of admixture linearly through time, and a more detailed mathematical analysis showed that the proportion of Neanderthal DNA in humans might originally have been as high as 10%140. As the effective population size of Neanderthals may have been around 10% of the human population of the time48, the observed proportion of introgression (when selection is taken into account) suggests that humans and Neanderthals could have mixed in proportion with their effective population sizes140. In other words, Neanderthals may not have become extinct because they lacked suitable ecological adaptations or through competition or warfare with humans. Instead, they may simply have been absorbed into the human species. Testing absorption models more directly, for example by sequencing older human remains from Eurasia, may finally resolve the much-debated disappearance of Neanderthals. Similar questions remain for Denisovans. For example, what is the contribution of purifying selection (negative selection against deleterious mutations) to present-day levels and patterns of Denisovan-like ancestry in Australasians? What was the geographic distribution of Denisovans? And where did the introgression between humans and Denisovans occur? The identification and genomic analysis of further Denisovan samples may help to answer these questions.
Future studies of archaic hominin admixture are likely to reveal further twists in the story of human evolution. Perhaps of greatest interest is genomic data from under-sampled regions of the world, which may help to refine evolutionary theories, including the question of whether there are further, as-yet uncharacterized, lineages of archaic humans. Indeed, preliminary data from contemporary African populations suggest that gene flow occurred with at least one other archaic hominin lineage25–27.
For over a century, researchers have discussed whether the biological and cultural variation that exists between groups of modern humans evolved as a result of the long-term isolation of populations, with innovation occurring through the spread of ideas141, or whether genetic variation and cultural changes were driven by human migration and the mixing of populations142. From the analyses of genomic data obtained so far, it is clear that the migration and admixture of populations has played a much larger part in generating genetic and cultural patterns of diversity than was previously thought56–58,60,66, although some groups have remained in isolation for a long period of time44,97. The genomic studies also suggest that the geographic distribution of genetic signatures in many modern populations were established relatively late in human history60. Yet for many regions of the world, especially Asia and Africa, our understanding of the historical events that led to the current distribution of genetic and cultural variation is still very fragmented.
The study of ancient human genomes has progressed rapidly, and in the past few years we have moved from studies of single genomes to population genomic studies that include hundreds of ancient individuals. Although this has provided a wealth of insights into human evolutionary genomics, this field is still in its infancy. Continued efforts to sequence and analyse the genomes of both modern and ancient humans, with a focus on under-sampled areas of the world, will help us to form a more complete picture of the events that have shaped the cultural and genetic variation of contemporary humans.
Acknowledgments
This work was supported in part by: US National Institutes of Health (NIH) grant R01GM110068 (J.M.A.); the European Research Council, the Knut and Alice Wallenberg Foundation and the Swedish Research Council (M.J.); the Danish National Research Foundation and the Lundbeck Foundation and KU2016 initiative (E.W.); and NIH grants R01GM116044 (R.N.), 1R01DK104339-01 and 1R01GM113657-01 (S.T.). We also thank S. Moon, S. Tucci, A. Sapfo-Malaspinas, M. Raghavan and M. W. Pedersen for discussions or for help in preparing the figures.
Footnotes
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this paper at go.nature.com/2hzojzv.
References
- 1.Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC. African populations and the evolution of human mitochondrial DNA. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 2.Nordborg M. On the probability of Neanderthal ancestry. Am J Hum Genet. 1998;63:1237–1240. doi: 10.1086/302052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meltzer DJ. First Peoples in a New World. University of California Press; 2009. [Google Scholar]
- 4.Ammerman AJ, Biagi P. The Widening Harvest. Archaeological Institute of America; 2003. [Google Scholar]
- 5.Rasmussen M, et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–762. doi: 10.1038/nature08835. This paper presents the first whole-genome sequence of an ancient human and demonstrates that there is a lack of genetic continuity between Arctic populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. This paper reports the first whole-genome sequence of a Neanderthal and supplies the first convincing evidence for introgression between humans and Neanderthals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White TD, et al. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423:742–747. doi: 10.1038/nature01669. [DOI] [PubMed] [Google Scholar]
- 8.McDougall I, Brown FH, Fleagle JG. Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature. 2005;433:733–736. doi: 10.1038/nature03258. [DOI] [PubMed] [Google Scholar]
- 9.Grün R, et al. U-series and ESR analyses of bones and teeth relating to the human burials from Skhul. J Hum Evol. 2005;49:316–334. doi: 10.1016/j.jhevol.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Liu W, et al. The earliest unequivocally modern humans in southern China. Nature. 2015;526:696–699. doi: 10.1038/nature15696. [DOI] [PubMed] [Google Scholar]
- 11.Higham T, et al. The timing and spatiotemporal patterning of Neanderthal disappearance. Nature. 2014;512:306–309. doi: 10.1038/nature13621. [DOI] [PubMed] [Google Scholar]
- 12.Rosenberg NA, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. This study demonstrates that the human population structure worldwide is correlated with geography and human history. [DOI] [PubMed] [Google Scholar]
- 13.Tishkoff SA, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. This article is a comprehensive investigation of genetic variation in Africa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Busby GB, et al. Admixture into and within sub-Saharan Africa. eLife. 2016;5:e15266. doi: 10.7554/eLife.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickrell JK, et al. The genetic prehistory of southern Africa. Nature Commun. 2012;3:1143. doi: 10.1038/ncomms2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurdasani D, et al. The African Genome Variation Project shapes medical genetics in Africa. Nature. 2015;517:327–332. doi: 10.1038/nature13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bryc K, et al. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci USA. 2010;107:786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlebusch CM, et al. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science. 2012;338:374–379. doi: 10.1126/science.1227721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beltrame MH, Rubel MA, Tishkoff SA. Inferences of African evolutionary history from genomic data. Curr Opin Genet Dev. 2016;41:159–166. doi: 10.1016/j.gde.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gronau I, Hubisz MJ, Gulko B, Danko CG, Siepel A. Bayesian inference of ancient human demography from individual genome sequences. Nature Genet. 2011;43:1031–1034. doi: 10.1038/ng.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlebusch CM, Soodyall H. Extensive population structure in San, Khoe, and mixed ancestry populations from southern Africa revealed by 44 short 5-SNP haplotypes. Hum Biol. 2012;84:695–724. doi: 10.3378/027.084.0603. [DOI] [PubMed] [Google Scholar]
- 22.Veeramah KR, et al. An early divergence of KhoeSan ancestors from those of other modern humans is supported by an ABC-based analysis of autosomal resequencing data. Mol Biol Evol. 2012;29:617–630. doi: 10.1093/molbev/msr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tishkoff SA, et al. History of click-speaking populations of Africa inferred from mtDNA and Y chromosome genetic variation. Mol Biol Evol. 2007;24:2180–2195. doi: 10.1093/molbev/msm155. [DOI] [PubMed] [Google Scholar]
- 24.Stringer C. Human evolution: out of Ethiopia. Nature. 2003;423:692–695. doi: 10.1038/423692a. [DOI] [PubMed] [Google Scholar]
- 25.Hammer MF, Woerner AE, Mendez FL, Watkins JC, Wall JD. Genetic evidence for archaic admixture in Africa. Proc Natl Acad Sci USA. 2011;108:15123–15128. doi: 10.1073/pnas.1109300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh P, et al. Model-based analyses of whole-genome data reveal a complex evolutionary history involving archaic introgression in Central African Pygmies. Genome Res. 2016;26:291–300. doi: 10.1101/gr.196634.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lachance J, et al. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell. 2012;150:457–469. doi: 10.1016/j.cell.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh P, et al. Whole-genome sequence analyses of Western Central African Pygmy hunter-gatherers reveal a complex demographic history and identify candidate genes under positive natural selection. Genome Res. 2016;26:279–290. doi: 10.1101/gr.192971.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramachandran S, et al. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jakobsson M, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 31.Clark JD, et al. Stratigraphic, chronological and behavioural contexts of Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423:747–752. doi: 10.1038/nature01670. [DOI] [PubMed] [Google Scholar]
- 32.Pickrell JK, et al. Ancient west Eurasian ancestry in southern and eastern Africa. Proc Natl Acad Sci USA. 2014;111:2632–2637. doi: 10.1073/pnas.1313787111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henn BM, et al. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci USA. 2011;108:5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lahr MM, Foley RA. Towards a theory of modern human origins: geography, demography, and diversity in recent human evolution. Am J Phys Anthropol. 1998;27(suppl):137–176. doi: 10.1002/(sici)1096-8644(1998)107:27+<137::aid-ajpa6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 35.Groucutt HS, et al. Rethinking the dispersal of Homo sapiens out of Africa. Evol Anthropol. 2015;24:149–164. doi: 10.1002/evan.21455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pugach I, Delfin F, Gunnarsdottir E, Kayser M, Stoneking M. Genome-wide data substantiate Holocene gene flow from India to Australia. Proc Natl Acad Sci USA. 2013;110:1803–1808. doi: 10.1073/pnas.1211927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed FA, Tishkoff SA. African human diversity, origins and migrations. Curr Opin Genet Dev. 2006;16:597–605. doi: 10.1016/j.gde.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 38.Quintana-Murci L, et al. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nature Genet. 1999;23:437–441. doi: 10.1038/70550. [DOI] [PubMed] [Google Scholar]
- 39.Gravel S, et al. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci USA. 2011;108:11983–11988. doi: 10.1073/pnas.1019276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris K, Nielsen R. Inferring demographic history from a spectrum of shared haplotype lengths. PLoS Genet. 2013;9:e1003521. doi: 10.1371/journal.pgen.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schiffels S, Durbin R. Inferring human population size and separation history from multiple genome sequences. Nature Genet. 2014;46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scally A, Durbin R. Revising the human mutation rate: implications for understanding human evolution. Nature Rev Genet. 2012;13:745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 44.Malaspinas AS, et al. A genomic history of Aboriginal Australia. Nature. 2016;538:207–214. doi: 10.1038/nature18299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mallick S, et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538:201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pagani L, et al. Genomic analyses inform on migration events during the peopling of Eurasia. Nature. 2016;538:238–242. doi: 10.1038/nature19792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rasmussen M, et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prüfer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wall JD, et al. Higher levels of neanderthal ancestry in East Asians than in Europeans. Genetics. 2013;194:199–209. doi: 10.1534/genetics.112.148213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vernot B, Akey JM. Complex history of admixture between modern humans and Neandertals. Am J Hum Genet. 2015;96:448–453. doi: 10.1016/j.ajhg.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sankararaman S, et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sankararaman S, Patterson N, Li H, Pääbo S, Reich D. The date of interbreeding between Neandertals and modern humans. PLoS Genet. 2012;8:e1002947. doi: 10.1371/journal.pgen.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vernot B, Akey JM. Resurrecting surviving Neandertal lineages from modern human genomes. Science. 2014;343:1017–1021. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 54.Kim BY, Lohmueller KE. Selection and reduced population size cannot explain higher amounts of Neandertal ancestry in East Asian than in European human populations. Am J Hum Genet. 2015;96:454–461. doi: 10.1016/j.ajhg.2014.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu Q, et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature. 2015;524:216–219. doi: 10.1038/nature14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skoglund P, et al. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science. 2012;336:466–469. doi: 10.1126/science.1216304. The first multi-individual study of ancient genomes; it supports the idea that migration drove the Neolithic transition in Europe. [DOI] [PubMed] [Google Scholar]
- 57.Lazaridis I, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haak W, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. This study uses SNP data from 69 Europeans who lived 3–8 kyr ago to point to the origin of Indo-European languages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gamba C, et al. Genome flux and stasis in a five millennium transect of European prehistory. Nature Commun. 2014;5:5257. doi: 10.1038/ncomms6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Allentoft ME, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. This study uses the sequencing of 101 ancient genomes to identify population movements in the Bronze Age. [DOI] [PubMed] [Google Scholar]
- 61.Günther T, Jakobsson M. Genes mirror migrations and cultures in prehistoric Europe — a population genomic perspective. Curr Opin Genet Dev. 2016;41:115–123. doi: 10.1016/j.gde.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Benazzi S, et al. Early dispersal of modern humans in Europe and implications for Neanderthal behaviour. Nature. 2011;479:525–528. doi: 10.1038/nature10617. [DOI] [PubMed] [Google Scholar]
- 63.Fu Q, et al. The genetic history of Ice Age Europe. Nature. 2016;534:200–205. doi: 10.1038/nature17993. By sequencing the genomes of 51 Eurasian people who lived 7–45 kyr ago, this study finds turnover in the population composition of Europeans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Asouti E, Fuller DQ. A contextual approach to the emergence of agriculture in Southwest Asia. Curr Anthropol. 2013;54:299–345. [Google Scholar]
- 65.Günther T, et al. Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc Natl Acad Sci USA. 2015;112:11917–11922. doi: 10.1073/pnas.1509851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skoglund P, et al. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science. 2014;344:747–750. doi: 10.1126/science.1253448. [DOI] [PubMed] [Google Scholar]
- 67.Cohen MN, Armelagos GJ, editors. Paleopathology at the Origins of Agriculture. Academic; 1984. [Google Scholar]
- 68.Jones ER, et al. Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nature Commun. 2015;6:8912. doi: 10.1038/ncomms9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bouckaert R, et al. Mapping the origins and expansion of the Indo-European language family. Science. 2012;337:957–960. doi: 10.1126/science.1219669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Novembre J, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. This study identifies fine-scale population structure in Europe that is highly correlated with geography. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lao O, et al. Correlation between genetic and geographic structure in Europe. Curr Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 72.Auton A, et al. Global distribution of genomic diversity underscores rich complex history of continental human populations. Genome Res. 2009;19:795–803. doi: 10.1101/gr.088898.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Raghavan M, et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 2014;505:87–91. doi: 10.1038/nature12736. This study sequences the genome of a 24-kyr-old Eurasian individual and shows that modern Native Americans are the descendants of a population admixed between Western Eurasians (represented by the sequenced sample) and an ancestral East Asian population. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fu Q, et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514:445–449. doi: 10.1038/nature13810. This study sequences DNA from a 45 kyr old Siberian person who was more closely related to modern Asians than to modern Europeans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seguin-Orlando A, et al. Genomic structure in Europeans dating back at least 36,200 years. Science. 2014;346:1113–1118. doi: 10.1126/science.aaa0114. [DOI] [PubMed] [Google Scholar]
- 76.Clarkson C, et al. The archaeology, chronology and stratigraphy of Madjedbebe (Malakunanja II): a site in northern Australia with early occupation. J Hum Evol. 2015;83:46–64. doi: 10.1016/j.jhevol.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 77.O’Connell JF, Allen J. The process, biotic impact, and global implications of the human colonization of Sahul about 47,000 years ago. J Archaeol Sci. 2015;56:73–84. [Google Scholar]
- 78.Thorne AG, Wolpoff MH. Regional continuity in Australasian Pleistocene hominid evolution. Am J Phys Anthropol. 1981;55:337–349. doi: 10.1002/ajpa.1330550308. [DOI] [PubMed] [Google Scholar]
- 79.Wollstein A, et al. Demographic history of Oceania inferred from genome-wide data. Curr Biol. 2010;20:1983–1992. doi: 10.1016/j.cub.2010.10.040. [DOI] [PubMed] [Google Scholar]
- 80.Skoglund P, et al. Genomic insights into the peopling of the Southwest Pacific. Nature. 2016;538:510–513. doi: 10.1038/nature19844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Storey AA, et al. Radiocarbon and DNA evidence for a pre-Columbian introduction of Polynesian chickens to Chile. Proc Natl Acad Sci USA. 2007;104:10335–10339. doi: 10.1073/pnas.0703993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gongora J, et al. Indo-European and Asian origins for Chilean and Pacific chickens revealed by mtDNA. Proc Natl Acad Sci USA. 2008;105:10308–10313. doi: 10.1073/pnas.0801991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Malaspinas AS, et al. Two ancient human genomes reveal Polynesian ancestry among the indigenous Botocudos of Brazil. Curr Biol. 2014;24:R1035–R1037. doi: 10.1016/j.cub.2014.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moreno-Mayar JV, et al. Genome-wide ancestry patterns in Rapanui suggest pre-European admixture with Native Americans. Curr Biol. 2014;24:2518–2525. doi: 10.1016/j.cub.2014.09.057. [DOI] [PubMed] [Google Scholar]
- 85.Jenkins DL, et al. Clovis age Western Stemmed projectile points and human coprolites at the Paisley Caves. Science. 2012;337:223–228. doi: 10.1126/science.1218443. [DOI] [PubMed] [Google Scholar]
- 86.Pedersen MW, et al. Postglacial viability and colonization in North America’s ice-free corridor. Nature. 2016;537:45–49. doi: 10.1038/nature19085. [DOI] [PubMed] [Google Scholar]
- 87.Heintzman PD, et al. Bison phylogeography constrains dispersal and viability of the Ice Free Corridor in western Canada. Proc Natl Acad Sci USA. 2016;113:8057–8063. doi: 10.1073/pnas.1601077113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.González-José R, et al. Craniometric evidence for Palaeoamerican survival in Baja California. Nature. 2003;425:62–65. doi: 10.1038/nature01816. [DOI] [PubMed] [Google Scholar]
- 89.Stanford DJ, Bradley BA. Across Atlantic Ice. University of California Press; 2012. [Google Scholar]
- 90.Owsley DW, Jantz RL, editors. Kennewick Man. Texas A&M University Press; 2014. [Google Scholar]
- 91.Rasmussen M, et al. The genome of a Late Pleistocene human from a Clovis burial site in western Montana. Nature. 2014;506:225–229. doi: 10.1038/nature13025. This study sequences a 12.6 kyr old individual from the Clovis culture, finding that people from this culture are ancestors of modern Native Americans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rasmussen M, et al. The ancestry and affiliations of Kennewick Man. Nature. 2015;523:455–458. doi: 10.1038/nature14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Raghavan M, et al. Genomic evidence for the Pleistocene and recent population history of Native Americans. Science. 2015;349:aab3884. doi: 10.1126/science.aab3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Skoglund P, et al. Genetic evidence for two founding populations of the Americas. Nature. 2015;525:104–108. doi: 10.1038/nature14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reich D, et al. Reconstructing Native American population history. Nature. 2012;488:370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gilbert MTP, et al. Paleo-Eskimo mtDNA genome reveals matrilineal discontinuity in Greenland. Science. 2008;320:1787–1789. doi: 10.1126/science.1159750. [DOI] [PubMed] [Google Scholar]
- 97.Raghavan M, et al. The genetic prehistory of the New World Arctic. Science. 2014;345:1255832. doi: 10.1126/science.1255832. [DOI] [PubMed] [Google Scholar]
- 98.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. This study reports the draft Denisovan genome sequence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sawyer S, et al. Nuclear and mitochondrial DNA sequences from two Denisovan individuals. Proc Natl Acad Sci USA. 2015;112:15696–15700. doi: 10.1073/pnas.1519905112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stringer CB, Barnes I. Deciphering the Denisovans. Proc Natl Acad Sci USA. 2015;112:15542–15543. doi: 10.1073/pnas.1522477112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Skoglund P, Jakobsson M. Archaic human ancestry in East Asia. Proc Natl Acad Sci USA. 2011;108:18301–18306. doi: 10.1073/pnas.1108181108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vernot B, et al. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science. 2016;352:235–239. doi: 10.1126/science.aad9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huerta-Sánchez E, et al. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 2014;512:194–197. doi: 10.1038/nature13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kuhlwilm M, et al. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature. 2016;530:429–433. doi: 10.1038/nature16544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wilde S, et al. Direct evidence for positive selection of skin, hair, and eye pigmentation in Europeans during the last 5,000 y. Proc Natl Acad Sci USA. 2014;111:4832–4837. doi: 10.1073/pnas.1316513111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mathieson I, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jablonski NG, Chaplin G. Human skin pigmentation as an adaptation to UV radiation. Proc Natl Acad Sci USA. 2010;107:8962–8968. doi: 10.1073/pnas.0914628107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lamason RL, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 109.Norton HL, et al. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol Biol Evol. 2007;24:710–722. doi: 10.1093/molbev/msl203. [DOI] [PubMed] [Google Scholar]
- 110.Beall CM. Andean, Tibetan, and Ethiopian patterns of adaptation to high-altitude hypoxia. Integr Comp Biol. 2006;46:18–24. doi: 10.1093/icb/icj004. [DOI] [PubMed] [Google Scholar]
- 111.Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Beall CM, et al. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci USA. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Simonson TS, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- 114.Bigham A, et al. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6:e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bersaglieri T, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tishkoff SA, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nature Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ranciaro A, et al. Genetic origins of lactase persistence and the spread of pastoralism in Africa. Am J Hum Genet. 2014;94:496–510. doi: 10.1016/j.ajhg.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mathias RA, et al. Adaptive evolution of the FADS gene cluster within Africa. PLoS ONE. 2012;7:e44926. doi: 10.1371/journal.pone.0044926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ameur A, et al. Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am J Hum Genet. 2012;90:809–820. doi: 10.1016/j.ajhg.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fumagalli M, et al. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science. 2015;349:1343–1347. doi: 10.1126/science.aab2319. [DOI] [PubMed] [Google Scholar]
- 121.Kothapalli KSD, et al. Positive selection on a regulatory insertion–deletion polymorphism in FADS2 influences apparent endogenous synthesis of arachidonic acid. Mol Biol Evol. 2016;33:1726–1739. doi: 10.1093/molbev/msw049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fumagalli M, et al. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 2011;7:e1002355. doi: 10.1371/journal.pgen.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Traherne JA. Human MHC architecture and evolution: implications for disease association studies. Int J Immunogenet. 2008;35:179–192. doi: 10.1111/j.1744-313X.2008.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Piel FB, et al. Global distribution of the sickle cell gene and geographical confirmation of the malaria hypothesis. Nature Commun. 2010;1:104. doi: 10.1038/ncomms1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fan S, Hansen MEB, Lo Y, Tishkoff SA. Going global by adapting local: a review of recent human adaptation. Science. 2016;354:54–59. doi: 10.1126/science.aaf5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hamblin MT, Di Rienzo A. Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am J Hum Genet. 2000;66:1669–1679. doi: 10.1086/302879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Simonti CN, et al. The phenotypic legacy of admixture between modern humans and Neandertals. Science. 2016;351:737–741. doi: 10.1126/science.aad2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Racimo F, Sankararaman S, Nielsen R, Huerta-Sanchez E. Evidence for archaic adaptive introgression in humans. Nature Rev Genet. 2015;16:359–371. doi: 10.1038/nrg3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Abi-Rached L, et al. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 2011;334:89–94. doi: 10.1126/science.1209202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ding Q, et al. Neanderthal origin of the haplotypes carrying the functional variant Val92Met in the MC1R in modern humans. Mol Biol Evol. 2014;31:1994–2003. doi: 10.1093/molbev/msu180. [DOI] [PubMed] [Google Scholar]
- 132.Karlsson EK, Kwiatkowski DP, Sabeti PC. Natural selection and infectious disease in human populations. Nature Rev Genet. 2014;15:379–393. doi: 10.1038/nrg3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bigham AW, Lee FS. Human high-altitude adaptation: forward genetics meets the HIF pathway. Genes Dev. 2014;28:2189–2204. doi: 10.1101/gad.250167.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 2010;20:R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Turchin MC, et al. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nature Genet. 2012;44:1015–1019. doi: 10.1038/ng.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Berg JJ, Coop G. A population genetic signal of polygenic adaptation. PLoS Genet. 2014;10:e1004412. doi: 10.1371/journal.pgen.1004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zoledziewska M, et al. Height-reducing variants and selection for short stature in Sardinia. Nature Genet. 2015;47:1352–1356. doi: 10.1038/ng.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Field Y, et al. Detection of human adaptation during the past 2,000 years. Science. 2016;354:760–764. doi: 10.1126/science.aag0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gibbons A. A new view of the birth of Homo sapiens. Science. 2011;331:392–394. doi: 10.1126/science.331.6016.392. [DOI] [PubMed] [Google Scholar]
- 140.Harris K, Nielsen R. The genetic cost of Neanderthal introgression. Genetics. 2016;203:881–891. doi: 10.1534/genetics.116.186890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Renfrew C. Before Civilization: Radiocarbon Revolution and Prehistoric Europe. Pimlico; 1973. [Google Scholar]
- 142.Childe VG. The Dawn of European Civilization. Kegan Paul; 1925. [Google Scholar]