Abstract
We investigated CD25 expression in older (≥60 years) patients with new acute myelogenous leukemia treated with decitabine and plerixafor. Patients resistant to therapy or survival ≤1 year had significantly higher percentages of CD25pos myeloid blasts in baseline bone marrow. CD25pos patients had an increased odds of resistance compared to CD25neg patients (p=.015). In univariate analysis, we found CD25pos patients had inferior survival compared to CD25neg (p=.002). In patients with intermediate risk cytogenetics, CD25pos status stratified patients associating with inferior survival (p=.002). In multivariable analysis, CD25 and TP53 mutations trended towards predicting remission to therapy but were not predictive of survival. Only remission status, ASXL1 and TET2 mutations were found to independently predict overall survival (OS). We conclude CD25 expression identifies patients at risk for resistance to hypomethylating chemotherapy but does not independently predict OS in an older AML population treated with decitabine and plerixafor
Keywords: CD25, AML, decitabine, prognostication, drug resistance
Introduction
Acute myelogenous leukemia (AML) is a molecularly heterogeneous disease associated with poor outcome in older patients. There is evidence that leukemic stem cell (LSC) interactions with the microenvironment promote chemo resistance, enabling LSC populations to survive and cause disease relapse after induction therapy [1,2]. For this reason, identification of predictive biomarkers in the LSC and myeloid blast population are an important priority. Interleukin-2 receptor α (IL2Ra or CD25) expression has been demonstrated on leukemic blasts and is a marker for chemotherapy-resistant LSCs [3]. Several reports have shown that AML blasts expressing CD25 predict poor survival and early treatment failure [4–8]. Expression of CD25 on CD34+ blasts has also been investigated in patients with myelodysplastic syndromes (MDS) after treatment with the hypomethylating agent azacitidine, demonstrating inferior survival compared to CD25 negative counterparts [9]. In addition, CD25 expression on AML blasts can stratify patients within intermediate risk cytogenetic categories and is associated with LSC gene expression signatures [5]. In the reported studies including AML patients, most patients were ≤60 years and treated with conventional chemotherapeutic agents.
Older patients with AML frequently have comorbidities making them unfit for standard induction chemotherapy which contributes to their inferior outcomes compared to younger counterparts [10]. Hypomethylating agents, such as decitabine are approved for the treatment of MDS and have shown efficacy in treating older patients with AML [11–13]. While hypomethylating induction has become a standard of care for elderly patients deemed unfit for standard induction chemotherapy, its effectiveness in older patients who are CD25pos and the prognostic impact of the marker itself, are not well established in these older patients. Therefore, we retrospectively analyzed baseline CD25 expression in newly diagnosed AML patients ≥60 years, enrolled in a phase I/II clinical trial (NCT01352650) combining the CXCR4 antagonist plerixafor with decitabine, and correlated expression levels with clinical outcomes.
Methods
Patient selection
This trial was registered in www.clinicaltrials.gov as NCT01352650 and was approved by the Institutional Review Board of Weill Cornell Medical College. The study was performed in accordance with the Declaration of Helsinki and all subjects provided written informed consent. The study population enrolled patients ≥60 years old with newly diagnosed, pathologically confirmed AML, as defined by World Health Organization criteria [14]. Two subjects <60 years old who were deemed ineligible for intensive induction therapy were granted exceptions to enroll in the protocol. Patients with acute promyelocytic leukemia or favorable risk cytogenetics according to the European Leukemia Net (ELN) criteria were excluded from participation [15]. For purposes of this study, all risk classifications were performed using ELN criteria. Patients were treated with decitabine alone and in combination with plerixafor during alternating cycles. Plerixafor was provided by Genzyme, Inc., which was later acquired by Sanofi Oncology (Cambridge, MA).
Treatment schedule
Subjects were treated with up to four induction cycles of decitabine, with or without plerixafor. Decitabine was administered as an intravenous infusion of 20 mg/m2 over one hour on days 1–10. Plerixafor was administered at three dose level cohorts. Cohorts 1, 2, and 3 received 320, 540, and 810 µg/kg of plerixafor intravenously on days 1–5, respectively. Cohorts were further subdivided into group A or B: group A subjects received plerixafor during even-numbered cycles and group B received plerixafor during odd-numbered cycles. Patients with a clinical response went on to receive monthly maintenance cycles consisting of five days of decitabine with plerixafor during alternate maintenance cycles.
Patient samples
Diagnostic bone marrow (BM) samples from 69 de novo AML patients, ≥60 years of age or younger patients deemed unfit for intensive induction therapy were collected and stored as specified in the clinical trial protocol. Day 0 (D0) BM aspirate samples were retrospectively assessed for baseline CD25 expression on leukemic blasts. Morphology and cytogenetics were evaluated at Weill Cornell Medical Center as part of routine clinical care.
Response assessment
Responses were determined using the International Working Group criteria [16]. For this study, patients were considered to have responsive disease if they achieved a complete remission (CR) or CR with incomplete peripheral count recovery (CRi) as defined by working group criteria.
CD25 expression analysis
CD25 expression was evaluated on leukemic cells using flow cytometry. Briefly, BM samples were thawed and stained for surface antibodies. Cells were stained with the following antibody fluorochrome conjugates: CD45-allophycocyanin-Hilite®. 7-BD (APC-H7) (clone 2D1; BD), CD34-phycoerythrin with cyanin-5 (PECy5) (clone 581; BD), CD133-allophycocyanin (APC) (clone 293C3; MACS Miltenyi Biotec, Bergisch Gladbach, Germany), CD33-fluorescein isothiocyanate (FITC) (clone P67.6; BD), CD25-peridinin-chlorophyll proteins cyanin-5.5 (PerCP-Cy5.5) (M-A251; BD Pharmingen, San Jose, CA), HLA-DR-V500 (clone G46-6; BD Horizon, San Jose, CA), and CD117-PECy7 (clone 104D2, Biolegend, San Diego, CA) for 15 minutes. Cells were washed twice in cold phosphate buffered saline (PBS) and suspended in 0.5% FBS in PBS. Cells were analyzed on a BD-LSRII flow cytometer and instruments calibrated with BD Cytometer Setup and Tracking beads prior to sample run and data collection. Flow cytometry data were analyzed using FlowJo® software (FlowJo, LLC, Ashland, OR). Populations were initially gated on CD45dim/SSClo characteristics. Subsequently, CD25 expression was evaluated on CD34+ cells within that blast population. Patients were considered CD25 positive (CD25pos) when ≥10% of gated CD34pos cells were also CD25pos, otherwise they were classified as CD25 negative (CD25neg).
Mutational profiling
Enrolled patients underwent mutational profiling for FLT3-ITD, using clinically available targeted next generation sequencing (NGS), performed by the University of Rochester Medical Center (URMC, Rochester, NY). A research-only NGS analysis was also performed using a 29-gene myeloid malignancy targeted amplicon enrichment panel (RainDance Technologies, Billerica, MA) inclusive of ASXL1, BCOR, BRAF, CBL, DNMT3A, ETV6, EZH2, FLT3-TKD, GATA1, GATA2, IDH1, IDH2, JAK2, KIT, KRAS, MPL, NPM1, NRAS, PHF6, PTPN11, RUNX1, SF1, SF3B1, SRSF2, TET2, TP53, U2AF1, WT1, and ZRSR2. Amplicons were sequenced using an Illumina MiSeq (v3 chemistry) with 251-bp paired end reads. Quality-and adapter-trimmed reads passing stringent quality control were aligned to human reference genome (GRCh37) plus decoy sequences (hs37d5) using the BWA-MEM algorithm [17]. A median average coverage depth of 2755 × (1549×–4148×) was achieved across all samples with a median of >98% (95.2%–99.0%) of targeted bases achieving >1000× depth. Single nucleotide variants (SNVs) and small insertion-deletions (indels) were identified using the VarDict algorithm [18] excluding both primer and low complexity regions. Single nucleotide polymorphisms (SNPs) with no known clinical impact of global minor allele frequency (MAF)>0.25% (dbSNP132) were filtered from subsequent analysis. Variant annotation was performed using SnpEff 4.1 [19].
Statistical analysis
Descriptive statistics (mean, standard deviation, median, interquartile range, frequency, and percent) for patient characteristics and clinical variables were calculated for the entire cohort as well as by CD25 positive/negative group status. The two-sample t-test/Wilcoxon’s rank-sum test or the chi-square/Fisher’s exact tests were used to compare the CD25 groups on continuous or discrete variables, respectively. Odds ratios (OR) were calculated to examine the association between CD25 group status and (1) death within 1 year and (2) resistance to therapy. Kaplan–Meier’s survival analysis and the log-rank test were used to estimate and compare the median overall survival (OS) time and associated 95% confidence intervals between the CD25 groups. Further, Kaplan–Meier’s analysis was performed to estimate OS in the CD25 groups stratified cytogenetic risk category (intermediate versus unfavorable).
Multivariable logistic regression using stepwise selection was used to determine a subset of variables independently associated with remission. All variables of clinical interest (i.e. CD25 status [binary, >10% of AML blasts versus ≤10%], LDH, cytogenetic risk [intermediate versus unfavorable], remission status and 29 binary mutational profiles) were used as candidate variables (entry criteria=p≤.20, stay criteria=p≤.10). Adjusted ORs and associated 95% confidence intervals were calculated for the final multivariable logistic regression model.
Cox’s proportional hazards regression with stepwise variable selection (both forward and backward) was used to specify a subset of variables that independently predicted OS (entry and stay criteria as above). Adjusted hazards ratios (HR) and associated 95% confidence intervals were calculated for the final multivariable survival model. The variables in the final model were assessed for collinearity and statistical interaction. The proportional hazards assumption was tested by visual inspection of the Kaplan–Meier plots. All p values are two-sided with statistical significance evaluated at the .05 alpha level.
All statistical analyses were performed with SAS Version 9.4 (SAS Institute, Cary, NC). Graphs were made using Prism software (GraphPad Software, San Diego, CA).
Results
Patient characteristics
Fifty-eight of the 69 enrolled patients in the phase I/II clinical trial had an available D0 BM aspirate and were included in the analysis. Of those 58 patients, 20 (34.5%) were found to be CD25pos based on flow cytometry results performed on BM aspirates. CD25pos patients tended to be older, more commonly male, presented with a higher peripheral white blood cell count, and have a higher BM blast percentage, though none of these parameters were found to be significant in univariate modeling. Both baseline lactate dehydrogenase levels and intermediate risk cytogenetics were noted to be significantly different between CD25pos patients compared to CD25neg patients. Results of baseline patient characteristics are summarized in Table 1.
Table 1.
Baseline patient characteristics.
| Baseline characteristics | All patients | CD25+ | CD25− | p value |
|---|---|---|---|---|
| N (%) | 58 (100) | 20 (34.5) | 38 (65.5) | |
| Age | 73 | 75 | 71 | p=.065 |
| Mean range | (56–87) | (62–87) | (56–82) | |
| Sex, n (%) | ||||
| Male | 33 (57) | 15 (75%) | 18 (47.3) | p=.054 |
| Female | 25 (43) | 5 (25%) | 20 (52.7) | |
| WBC (×1000/µL) | 5.9 | 19 | 5 | p=.088 |
| Median (IQR) range | (0.6–122.0) | (0.8–122) | (1.1–42) | |
| LDH (IU/L) | 284 | 360.5 | 270 | p=.040 |
| Median (IQR) range | (79–1901) | (136–1384) | (79–1901) | |
| Bone marrow blast percentage – mean (SD) | 50.0 (26.5) | 54.2 (31.6) | 47.7 (23.6) | p=.386 |
| Range | (4–74) | (4–95) | (6–85) | |
| Cytogenetics, n (%) | ||||
| Favorable | 4 (7) | 1(5) | 3(8) | |
| Intermediate | 33 (57%) | 15 (75) | 18 (47) | p=.047 |
| Unfavorable | 21 (36.2) | 4 (20) | 17 (44) | |
| Transplanta status, n (%) | ||||
| Yes | 17 (29.3) | 5 (25) | 12 (31.6) | p=.764 |
| No | 41 (70.7) | 15(75) | 26 (68.4) |
All transplants were allogeneic.
n: number; WBC: white blood cell count; IQR: interquartile range; SD: standard deviation.
Mutational analysis
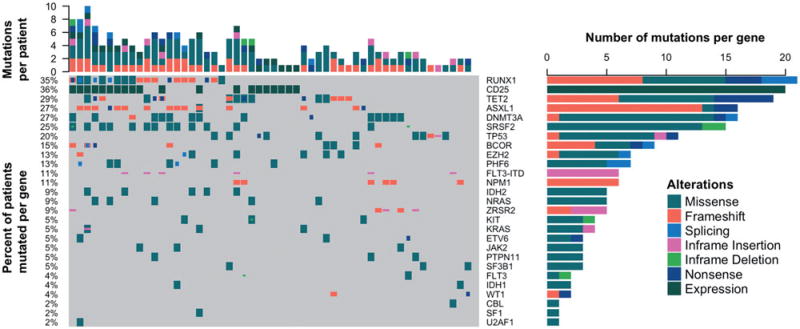
Of the 58 evaluated patients, 55 (95%) patients were evaluated at diagnosis with targeted amplicon research only NGS. Additionally, 49 patients (84%) underwent NGS mutational profiling using commercially available targeted gene sequencing of FLT-3. The heat map with combined results of the different methods and mutational prevalence of recurrently mutated genes are outlined in Figure 1. We found no significant associations of recurrent genetic mutations based on CD25 status, but did note an association with RUNX1 mutations and CD25pos status that trended towards significance (p=.06). Notably, we found no associations with NPM1, DMNT3A, or FLT3-ITD alterations, contrary to previous reports demonstrating associations with three genes and CD25pos status [5].
Figure 1.
CD25 expression status and mutational profile of 55 sequenced patients. Patients underwent next generation sequencing using a targeted exome panel of 30 genes recurrently mutated in AML. Mutations were called if the variant allele frequency was 4%. Each column represents a single patient's profile while the top axis depicts the total number of mutations per patient. Gene names are represented on the right axis and the percent of patients in the cohort with that mutation depicted on the left axis. The specific type of genetic alteration is noted in the figure legend.
CD25 expression, TP53 mutations and response to therapy
Of the 58 patients with baseline BM specimens, 55 patients were evaluable for response. Twenty seven patients (49%) achieved a response defined as CR or CRi with the majority, 23 patients (40%) achieving a CR. Only four (23.5%) of CD25pos patients achieved a remission. As a univariate risk factor for response, CD25 expression was associated with a 79% reduction in achieving a remission (OR 0.21, 95% CI 0.06–0.73, p=.015). In our multivariable analysis taking into account baseline characteristics, cytogenetic risk and mutational landscape, CD25 expression narrowly missed independent significance predicting remission with a 69% reduction in probability of achieving a CRi or better with an adjusted OR 0.27 (95% CI 0.07–1.03, p=.055). In the same multivariable analysis, TP53 mutations had a protective effect with an adjusted OR of 4.25, which also narrowly missed independent significance predicting remission to induction therapy (95% CI 0.94–19.12, p=.06). The association between decitabine sensitivity and TP53 mutational status has been observed previously and our results are consistent with those previous results [20]. Lastly, we found that CD25 status had no effect on mobilization with plerixafor, 38% of CD25pos mobilized compared to 44% of CD25neg patients, p=.72.
CD25 expression, 1 year survival and disease resistance
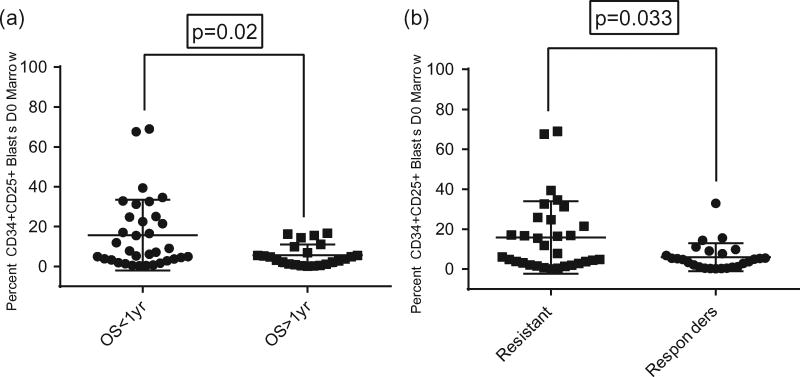
We next explored baseline percentages of CD25pos marrow blasts and found that patients who lived less than 1 year had significantly higher percentages of CD25pos CD34pos blasts in baseline BM samples (median percentage of 7.5% versus 4.2%, respectively, p=.02) (Figure 2(a)). We also found a significant difference in the median baseline percentage of CD25pos CD34pos blasts in patients who were resistant to induction versus those with responsive disease, with higher percentages of CD25pos cells in resistant patients (median 7.8% versus 4.7% respectively, p=.03) (Figure 2(b)).
Figure 2.
(a) Percentage of CD25pos blasts in D0 bone marrow samples is higher in patients with survival less than one year. (b) Percentage of CD25pos blasts in D0 bone marrow samples is higher in patients with resistance to decitabine and plerixafor induction.
We also evaluated the odds of death within one year of diagnosis and odds of resistant disease to decitabine and plerixafor based on CD25 expression status. Patients defined as CD25pos had an OR of death within one year three times higher than CD25neg patients, though the association only trended towards significance (OR 3.0, 95% CI 0.91–9.91, p=.072). In addition, we found CD25pos patients had nearly five times the odds of resistance to decitabine therapy compared to their CD25neg counterparts (OR 4.94, 95% CI 1.36–18.21, p=.015).
Overall survival analysis
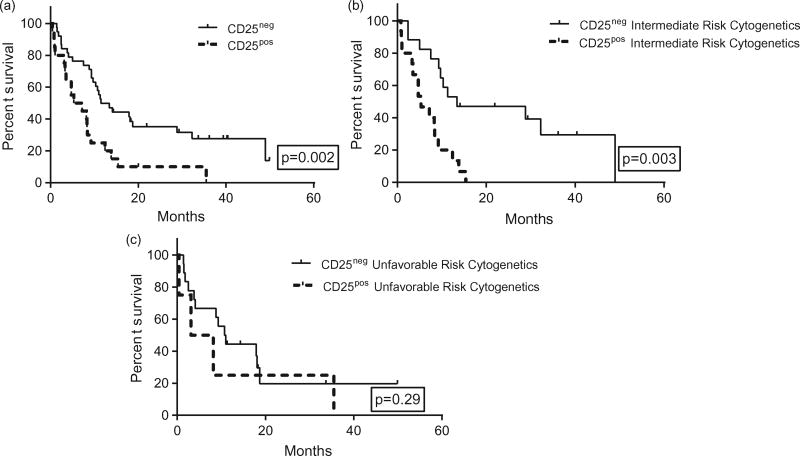
Comparing CD25pos to CD25neg patients, we found a statistically significant difference in OS, with inferior outcomes seen in CD25pos patients. The median OS for CD25pos patients was 6.3 months compared to 11.5 months for CD25neg patients (p=.002) (Figure 3(a)). We observed that CD25 expression can stratify patients with intermediate risk cytogenetics, with CD25pos patients having a median OS of 5.3 months versus 13.4 months for CD25neg patients (p=.003) (Figure 3(b)). In patients with unfavorable cytogenetics, we found no differences in OS based on CD25 expression status, though CD25pos patients with unfavorable cytogenetics lived nearly half as long as CD25neg counterparts 5.7 months versus 10.9 months, respectively (p=.29) (Figure 3(c)).
Figure 3.
(a) Increased CD25 expression associates with inferior overall survival. (b) Increased CD25 expression risk stratifies patients with intermediate cytogenetics but not unfavorable cytogenetics. (c) Increased CD25 expression risk stratifies patients with intermediate cytogenetics but not unfavorable cytogenetics.
In multivariable regression using a stepwise selection analysis, controlling for baseline characteristics, cytogenetics, mutational landscape, and remission, we found that mutations in ASXL1 and TET2 independently predicted poorer OS. Alternatively, achieving a remission was independently associated with improved OS (Table 2). CD25 expression while in a univariate model was associated with worse OS (HR 2.02, 95% CI 1.06–3.82, p=.03), it did not prove to be an independent predictor of OS in multivariable analysis (Table 2).
Table 2.
Univariate and multivariate analyses for overall survival.
| Hazard ratio | 95% CI | p value | ||
|---|---|---|---|---|
| Variable (univariate) | ||||
| CD25 (pos versus neg) | 2.02 | 1.06 | 3.82 | .03 |
| Variable (multivariable) | ||||
| Remission (CR/CRi versus none) | 0.32 | 0.16 | 0.64 | .001 |
| ASXL1 (mutated versus wild type) | 2.47 | 1.09 | 5.64 | .030 |
| TET2 (mutated versus wild type) | 2.30 | 1.14 | 4.63 | .020 |
Pos: positive; neg: negative; CR: complete remission; Cri: complete remission with incomplete recovery.
Discussion
The majority of reports demonstrating poor outcome in AML patients with increased CD25 expression, have generally been from cohorts of younger patients treated with conventional cytotoxic chemotherapeutic regimens. There is very little data associating CD25 expression status and outcome in older populations treated with less intensive hypomethylating agents.
In this report, we demonstrate that CD25 expression correlates with chemotherapy resistance in older patients treated with decitabine, combined with the CXCR4 antagonist plerixafor, on a phase I/II single institution clinical trial. We report that patients with high levels of CD25pos CD34pos AML blasts (>10%) at baseline, represent over one third (34%) of the patient cohort. Similar to previous studies, we found CD25 positivity was associated with intermediate risk cytogenetics [4–6]. We noted an increased odds of resistance to hypomethylating induction therapy for CD25pos patients, while patients with documented resistance to therapy or survival less than 1 year had significantly higher percentages of CD25pos CD34pos blasts in their BM at diagnosis. Patients who were found to be CD25pos had inferior survival compared to CD25neg patients in univariate analysis and CD25 status could identify patients with intermediate risk cytogenetics to be in a higher risk category. Nevertheless, in multivariable modeling, CD25 status did not independently predict inferior OS and only showed a trend towards predicting remission to hypomethylating based induction.
It is known that the interleukin-2 (IL-2) alpha is a key regulator of lymphocyte proliferation, specifically regulatory T cells, and is critical for the maintenance of immune tolerance [21,22]. The IL-2 receptor, which is a combination receptor incorporating CD25 as one subunit, is responsible for intracellular signaling upon binding IL-2 and signals through STAT dependent mechanisms [23]. While the function of IL-2 and CD25 signaling is normal on lymphocytes, it is aberrantly regulated on myeloid leukemia cells [24].
While the association of increased CD25 expression with poor outcome has been documented, the mechanisms have remained elusive. In chronic myelogenous leukemia (CML), it is has been shown that CD25 expression identifies a subset of CML LSCs, which are stimulated by and associate with IL-2pos cells in the BM microenvironment. This suggests niche’s ability to nurture LSCs through a functional signaling receptor [25]. In AML, CD25 is also expressed in LSCs, though its function is likely to be different than in CML as studies have demonstrated that CD25pos AML lacks the additional IL2 receptor components CD122 and CD132 in addition to lacking an IL2 signaling signature [5].
CD25 transcription is regulated under the control of STAT transcription factors, specifically STAT5 [26]. Recently, it has been shown that PIM kinase inhibitors (PIMKi) can target CD25 positive AML cells through suppression of STAT5 and MYC [27] and suggests CD25 expression on AML cells may represent a surrogate marker for subsets of leukemia cells with underlying STAT5 activation [27]. In another recent report, a small number (n=12) of primary AML samples, with high to intermediate CD25 expression, had poor survival. Approximately 42–50% had constitutive activation of STAT5 or accompanying survival pathways such as pAKT or pERK [28]. While experiments with AML cell lines have shown that intrinsic resistance to PIMKis is common, reexamination of the activity of these agents in CD25 positive AML may be warranted.
In addition to potential constitutive activation of STAT pathways, CD25 expression has been associated with the presence of FLT-3ITD and co-expression of the two markers predicts worse survival than mutated FLT3 expression alone [5,6,29]. In our study, we did not demonstrate a significant association with FLT3 or any other underlying recurrent molecular defect. It is important to note there were several patients whose FLT3 mutational status was unknown. Our population was older with a median age of 73, significantly older than those in the studies previously reported. Mutations in FLT3 are less common in the elderly and may explain why we did not see this frequently observed association [30].
Previous reports demonstrated that CD25 is an independent prognostic indicator in patients with intermediate risk cytogenetics, but that CD25 expression failed to demonstrate significance when restricted to patients >60 years or older [7]. In a more recent report, restricted to older AML patients >60 years of age, CD25 expression was found to independently predict inferior survival in those patients who were treated with less intensive cytotoxic regimens combining low dose cytarabine and aclarubicin [31]. Our study of elderly patients treated with decitabine, shows that CD25 expression correlates with poor survival and outcome on a univariate analysis but does not demonstrate independent significance in multivariable analysis for OS. This discrepancy between the two studies may be related to different induction agents, or small numbers in our cohort compared to the most recent report with nearly double the number of patients.
In conclusion, we demonstrate that CD25 expression associates with poor outcome in a cohort of 58 newly diagnosed older or unfit AML patients, treated with decitabine and plerixafor on a phase I/II clinical trial. We determined patients with AML and >10% CD25pos CD34pos blasts in the BM at diagnosis define a subpopulation of patients with poor OS in univariate analysis and increased odds of resistance to hypomethylating therapy. Overall, these results are consistent with those previously reported and highlight the need to further understand the role of CD25 expression in AML blasts and study patients in a prospective manner with targeted therapies aimed at disrupting the underlying activated pathways.
Acknowledgments
Funding
PC and GA were supported by the following grant: Clinical and Translational Science Center (UL1-TR000457-06). DCH is supported by the Leukemia and Lymphoma Society (LLS 6453-13). MLG and GJR are supported by the following grants (LLS 6453-12, LLS 6427-13; R01 CA102031), MLG is also supported by funds from the Irma T Hirschl Trust.
Footnotes
Potential conflict of interest: Disclosure forms provided by the authors are available with the full text of this article online at https://doi.org/10.1080/10428194.2017.1352089.
There are no other relevant disclosures to declare by any of the authors.
References
- 1.Tabe Y, Konopleva M. Role of microenvironment in resistance to therapy in AML. Curr Hematol Malig Rep. 2015;10:96–103. doi: 10.1007/s11899-015-0253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felipe Rico J, Hassane DC, Guzman ML. Acute myelogenous leukemia stem cells: from bench to bedside. Cancer Lett. 2013;338:4–9. doi: 10.1016/j.canlet.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saito Y, Kitamura H, Hijikata A, et al. Identification of therapeutic targets for quiescent, chemotherapyresistant human leukemia stem cells. Sci Transl Med. 2010;2:17ra9. doi: 10.1126/scitranslmed.3000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terwijn M, Feller N, van Rhenen A, et al. Interleukin-2 receptor alpha-chain (CD25) expression on leukaemic blasts is predictive for outcome and level of residual disease in AML. Eur J Cancer. 2009;45:1692–1699. doi: 10.1016/j.ejca.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 5.Gonen M, Sun Z, Figueroa ME, et al. CD25 expression status improves prognostic risk classification in AML independent of established biomarkers: ECOG phase 3 trial, E1900. Blood. 2012;120:2297–2306. doi: 10.1182/blood-2012-02-414425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerny J, Yu H, Ramanathan M, et al. Expression of CD25 independently predicts early treatment failure of acute myeloid leukaemia (AML) Br J Haematol. 2013;160:262–266. doi: 10.1111/bjh.12109. [DOI] [PubMed] [Google Scholar]
- 7.Nakase K, Kita K, Kyo T, et al. Prognostic relevance of cytokine receptor expression in acute myeloid leukemia: interleukin-2 receptor alpha-chain (CD25) expression predicts a poor prognosis. PLoS ONE. 2015;10:e0128998. doi: 10.1371/journal.pone.0128998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikegawa S, Doki N, Kurosawa S, et al. CD25 expression on residual leukemic blasts at the time of allogeneic hematopoietic stem cell transplant predicts relapse in patients with acute myeloid leukemia without complete remission. Leuk Lymph. 2016;57:1375–1381. doi: 10.3109/10428194.2015.1099644. [DOI] [PubMed] [Google Scholar]
- 9.Miltiades P, Lamprianidou E, Vassilakopoulos TP, et al. Expression of CD25 antigen on CD34+ cells is an independent predictor of outcome in late-stage MDS patients treated with azacitidine. Blood Cancer J. 2014;4:e187. doi: 10.1038/bcj.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tawfik B, Pardee TS, Isom S, et al. Comorbidity, age, and mortality among adults treated intensively for acute myeloid leukemia (AML) J Geriatr Oncol. 2016;7:24–31. doi: 10.1016/j.jgo.2015.10.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kantarjian HM, Thomas XG, Dmoszynska A, et al. Multicenter, randomized, open-label, phase III trial of decitabine versus patient choice, with physician advice, of either supportive care or low-dose cytarabine for the treatment of older patients with newly diagnosed acute myeloid leukemia. JCO. 2012;30:2670–2677. doi: 10.1200/JCO.2011.38.9429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie EK, Feldman EJ, Christos PJ, et al. Decitabine in patients with newly diagnosed and relapsed acute myeloid leukemia. Leuk Lymph. 2013;54:2003–2007. doi: 10.3109/10428194.2012.762093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blum W, Garzon R, Klisovic RB, et al. Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proc Natl Acad Sci USA. 2010;107:7473–7478. doi: 10.1073/pnas.1002650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swerdlow SH, Cancer IAfRo, Organization WH. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon: International Agency for Research on Cancer; 2008. [Google Scholar]
- 15.Dohner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 16.Cheson BD, Bennett JM, Kopecky KJ, et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. JCO. 2003;21:4642–4649. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 17.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 2013:3. [Google Scholar]
- 18.Lai Z, Markovets A, Ahdesmaki M, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44:e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Baets G, Van Durme J, Reumers J, et al. SNPeffect 4.0: on-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res. 2012;40:D935–D939. doi: 10.1093/nar/gkr996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welch JS, Petti AA, Miller CA, et al. TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N Engl J Med. 2016;375:2023–2036. doi: 10.1056/NEJMoa1605949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zorn E, Nelson EA, Mohseni M, et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood. 2006;108:1571–1579. doi: 10.1182/blood-2006-02-004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malek TR, Bayer AL. Tolerance, not immunity, crucially depends on IL-2. Nat Rev Immunol. 2004;4:665–674. doi: 10.1038/nri1435. [DOI] [PubMed] [Google Scholar]
- 23.Frank DA, Robertson MJ, Bonni A, et al. Interleukin 2 signaling involves the phosphorylation of Stat proteins. Proc Natl Acad Sci USA. 1995;92:7779–7783. doi: 10.1073/pnas.92.17.7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padros MR, Salamone MC, Zunszain PA, et al. Differential expression of CD25 and HC2 antigens on subtypes of acute myeloid leukemias. Eur J Haematol. 1989;42:436–440. doi: 10.1111/j.1600-0609.1989.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi CI, Takubo K, Kobayashi H, et al. The IL-2/CD25 axis maintains distinct subsets of chronic myeloid leukemia-initiating cells. Blood. 2014;123:2540–2549. doi: 10.1182/blood-2013-07-517847. [DOI] [PubMed] [Google Scholar]
- 26.Lecine P, Algarte M, Rameil P, et al. Elf-1 and Stat5 bind to a critical element in a new enhancer of the human interleukin-2 receptor alpha gene. Mol Cell Biol. 1996;16:6829–6840. doi: 10.1128/mcb.16.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Z, Wang A, Zhang W, et al. PIM inhibitors target CD25-positive AML cells through concomitant suppression of STAT5 activation and degradation of MYC oncogene. Blood. 2014;124:1777–1789. doi: 10.1182/blood-2014-01-551234. [DOI] [PubMed] [Google Scholar]
- 28.Garg S, Shanmukhaiah C, Marathe S, et al. Differential antigen expression and aberrant signaling via PI3/AKT, MAP/ERK, JAK/STAT, and Wnt/beta catenin pathways in Lin−/CD38−/CD34+ cells in acute myeloid leukemia. Eur J Haematol. 2015;96:309–317. doi: 10.1111/ejh.12592. [DOI] [PubMed] [Google Scholar]
- 29.Angelini DF, Ottone T, Guerrera G, et al. A leukemiaassociated CD34/CD123/CD25/CD99+ immunophenotype identifies FLT3-mutated clones in acute myeloid leukemia. Clin Cancer Res. 2015;21:3977–3985. doi: 10.1158/1078-0432.CCR-14-3186. [DOI] [PubMed] [Google Scholar]
- 30.Schneider F, Hoster E, Schneider S, et al. Age-dependent frequencies of NPM1 mutations and FLT3-ITD in patients with normal karyotype AML (NK-AML) Ann Hematol. 2012;91:9–18. doi: 10.1007/s00277-011-1280-6. [DOI] [PubMed] [Google Scholar]
- 31.Fujiwara SI, Muroi K, Yamamoto C, et al. CD25 as an adverse prognostic factor in elderly patients with acute myeloid leukemia. Hematology. 2017;22:347–353. doi: 10.1080/10245332.2016.1276240. [DOI] [PubMed] [Google Scholar]