Abstract
Insulin resistance is a key mediator of obesity-related cardiometabolic disease, yet the mechanisms underlying this link remain obscure. Using an integrative genomic approach, we identify 53 genomic regions associated with insulin resistance phenotypes (higher fasting insulin adjusted for BMI, lower HDL cholesterol and higher triglycerides) and provide evidence that their link with higher cardiometabolic risk is underpinned by an association with lower adipose mass in peripheral compartments. Using these 53 loci, we show a polygenic contribution to familial partial lipodystrophy-type 1, a severe form of insulin resistance, and highlight shared molecular mechanisms between common/mild and rare/severe insulin resistance. Population-level genetic analyses combined with experiments in cellular models implicate CCDC92, DNAH10 and L3MBTL3 as previously unrecognised molecules influencing adipocyte differentiation. Our findings support the notion that limited storage capacity of peripheral adipose tissue is an important aetiological component in insulin-resistant cardiometabolic disease and highlight genes and mechanisms underpinning this link.
Introduction
Insulin resistance, usually defined as an impaired ability of insulin to maintain normal glucose metabolism and initially manifested by higher levels of circulating insulin, is positively associated with adiposity and is a key mediator of the link between obesity and its adverse impact on metabolic and cardiovascular disease.1–8 Given the current global epidemic of metabolic disease, there is an urgent need for improved understanding of the mechanisms that link over-nutrition to insulin resistance in the general population.7–10
Among individuals stratified on the basis of overall adiposity, there is considerable variation in the extent of adverse metabolic sequelae,11 demonstrating the importance of other factors in the aetiology of insulin resistance and its complications. Indeed, while insulin resistance often coexists with obesity, severe forms of insulin resistance develop without obesity or in association with generalized or regional lack of adipose tissue, i.e. lipodystrophy.12 In lipodystrophies,13 it has been proposed that the impaired capacity of peripheral adipose tissue to expand under the challenge of a positive energy balance leads to lipid accumulation at ectopic sites (e.g. liver, skeletal muscle, pancreas) and eventually to overt diabetes.12,14 The notion of “adipose overflow” or “limited adipose tissue expandability”15–19 is supported (i) by the metabolic disturbances seen in rare, monogenic lipodystrophies and their dramatic amelioration in response to dietary calorie restriction20 or leptin replacement,21,22 and (ii) by a series of elegant rodent studies including those in which adipose tissue capacity was expanded by fat transplantation in lipodystrophic mice23 or by over-expressing adiponectin,24 or where partially lipodystrophic mice were energetically challenged by rendering them leptin deficient.25,26 However, the relevance of this model to the general population remains uncertain.
Some initial human genetic insights into more prevalent forms of insulin resistance are available. Genome-wide studies of gold-standard measures of insulin resistance have been limited by sample size,27 but multiple genomic loci have been associated with fasting insulin levels, a widely-measured marker of insulin resistance.28,29 A subset of these loci were associated with higher triglycerides and lower high-density lipoprotein (HDL) cholesterol,28 which are hallmarks of insulin resistance.13,30 These loci were later validated for their association with insulin resistance,31 suggesting that the combined association with this triad of phenotypes could help identify specific genetic determinants of insulin resistance.
Given the availability of large-scale genome-wide association data on lipid traits and fasting insulin,28,29,32 we undertook an integrative genomic approach to characterise genetic and molecular mechanisms underpinning insulin resistance at a given level of adiposity and its role in cardiometabolic disease in the general population.
Results
Associations with insulin resistance phenotypes at 53 independent genomic regions
We combined genome-wide association results for fasting insulin adjusted for body mass index,22,23 HDL-cholesterol and triglyceride levels32 from up to 188,577 individuals to identify loci associated with a phenotypic pattern indicative of insulin resistance (Online Methods, Supplementary Figures 1-2 and Supplementary Table 1). After aligning the association results of ~2.4 million single nucleotide polymorphisms (SNPs) to the insulin-raising allele, 630 SNPs from 53 1Mb-genomic regions were associated with higher fasting insulin, higher triglycerides and lower HDL-cholesterol (p<0.005 for each phenotype, expected probability of association under null hypothesis p=3.1 x 10-08; Online Methods, Supplementary Figures 3-4). These 53 genomic regions included 10 loci previously implicated in insulin resistance,31 and an additional 43 loci (Supplementary Table 2). A subset of 25 of the 53 loci had previously been associated with HDL cholesterol or triglyceride levels at genome-wide significance,32 while 28 had not (Supplementary Table 2). We first investigated the associations of these loci in a completely independent sample of 6,101 individuals and found that genetic risk scores comprising the 53 lead SNPs were strongly associated with higher fasting insulin, higher triglycerides and lower HDL-cholesterol (Supplementary Figure 5). We next asked whether these variants were associated with “gold-standard” measures of insulin sensitivity. Having a greater number of risk-alleles from the 53-SNP, 43-SNP or 28-SNP (excluding loci previously implicated in insulin resistance and lipid traits, respectively) genetic scores was strongly associated with (a) lower insulin sensitivity measured by euglycemic clamp or insulin suppression test in 2,764 individuals27 (p-value for 53-SNP genetic score = 4.3 x 10-06; Table 1) and (b) lower insulin sensitivity index in 4,769 individuals with a frequently-sampled oral glucose tolerance test33 (p-value for 53-SNP genetic score = 7.3 x 10-10; Table 1).
Table 1. Association with “gold-standard” insulin resistance measures, type 2 diabetes and coronary heart disease of genetic scores comprising lead polymorphisms at identified loci.
Results are displayed for genetic scores comprising either (a) the lead SNP at each of the 53 associated loci or (b) the lead SNP at each of the 43 additional loci identified in this study after removing 10 previously implicated in insulin resistance or (c) the lead SNP at each of the 28 loci not previously associated with levels of HDL cholesterol or triglycerides.
| Exposure | Outcome | Sample size, N or N cases and N controls | Beta or odds ratio | SE or 95% CI | p-value |
|---|---|---|---|---|---|
| Association with “gold-standard” measures of insulin sensitivity | |||||
| 53-SNP score | Insulin sensitivitya | 2,764 | -0.09 | 0.019 | 4.3 x 10-06 |
| 43-SNP score | -0.08 | 0.022 | 4.6 x 10-04 | ||
| 28-SNP score | -0.09 | 0.022 | 2.6 x 10-05 | ||
| 53-SNP score | Insulin sensitivity indexb | 4,769 | -0.10 | 0.016 | 7.3 x 10-10 |
| 43-SNP score | -0.08 | 0.018 | 4.6 x 10-05 | ||
| 28-SNP score | -0.09 | 0.027 | 0.0010 | ||
| Association with disease endpoints | |||||
| 53-SNP score | Type 2 diabetes | 45,836 cases and 230,358 controls | 1.12 | 1.11, 1.14 | 9.2 x 10-61 |
| 43-SNP score | 1.09 | 1.08, 1.11 | 7.6 x 10-29 | ||
| 28-SNP score | 1.11 | 1.09, 1.13 | 1.9 x 10-25 | ||
| 53-SNP score | Coronary heart disease | 63,746 cases and 130,681 controls | 1.05 | 1.04, 1.06 | 1.8 x 10-13 |
| 43-SNP score | 1.04 | 1.03, 1.06 | 5.7 x 10-08 | ||
| 28-SNP score | 1.04 | 1.02, 1.06 | 1.2 x 10-05 | ||
Abbreviations: N, number of participants; SE, standard error; CI, confidence interval; SNP, single nucleotide polymorphism. Beta coefficients are in standardised units per standard deviation of the 53-SNP genetic score (i.e. 4.5 alleles); odds ratios are per standard deviation of the 53-SNP genetic score (i.e. 4.5 alleles). The association with insulin sensitivity is from 2,764 participants of the GENESIS consortium27 and the association with the insulin sensitivity index is from 4,769 participants of the MAGIC consortium who underwent a frequently sampled oral glucose tolerance test (OGTT)33; the association with type 2 diabetes is from the InterAct, DIAGRAM and UK Biobank studies; the association with coronary heart disease is from the CARDIoGRAM and the C4D consortia.
In MAGIC, insulin sensitivity index (ISI) = 10,000/√ (fasting plasma glucose (mg/dl)×fasting insulin×mean glucose during OGTT (mg/dl)×mean insulin during OGTT).33
In GENESIS, insulin sensitivity was measured by clamp or insulin suppression test using study-specific parameters (e.g. glucose disposal or M-value) which were then standardised before meta-analysis.27
Genetic predisposition to insulin resistance via the 53 loci confers higher risk of cardiometabolic disease but lower levels of peripheral adiposity
We next investigated associations of the 53 genomic regions with a range of continuous metabolic traits and disease outcomes. In 45,836 cases and 230,358 controls, the 53-SNP genetic score was associated with a higher risk of type 2 diabetes (odds ratio [OR] per standard deviation [SD] of the genetic score [i.e. ~4.5 alleles], 1.12; 95% confidence interval [CI], 1.11-1.14; p=9.2 x 10-61; Table 1). In studies with available individual-level data, we saw no difference in associations across sex or body mass index (BMI) strata (Supplementary Table 3). Genetically-predicted insulin resistance was also associated with a higher risk of coronary heart disease (Table 1). The associations with type 2 diabetes (OR, 1.10; p=9.0 x 10-32) and coronary heart disease (OR, 1.04; p=9.7 x 10-07) remained after removing 13 loci that were previously shown to be associated with either of the two diseases at genome-wide significance.34,35 Association estimates were also consistent after removing the 25 loci previously associated with HDL cholesterol or triglyceride levels at genome-wide significance (Table 1). We also observed an association with coronary heart disease in 5,369 cases of coronary heart disease and 106,969 controls from the UK Biobank study not previously included in genome-wide discovery analyses of insulin or lipid traits (OR, 1.09; p=5.3 x 10-09). Individually, 30 of the 53 lead SNPs were associated with higher type 2 diabetes risk (p<0.05; Supplementary Table 4), including a novel association at genome-wide significance for rs718314 near ITPR2 (OR per allele, 1.06; p=6.8 x 10-09). We found an enrichment of loci associated with higher risk of both type 2 diabetes and coronary heart disease, including those encompassing the proximal insulin signalling INSR, IRS1 and PIK3R1 genes (11/53 loci associated with both diseases at p<0.05; two-tailed binomial probability of observing this proportion of loci by chance p=8.5 x 10-22; Supplementary Table 5).
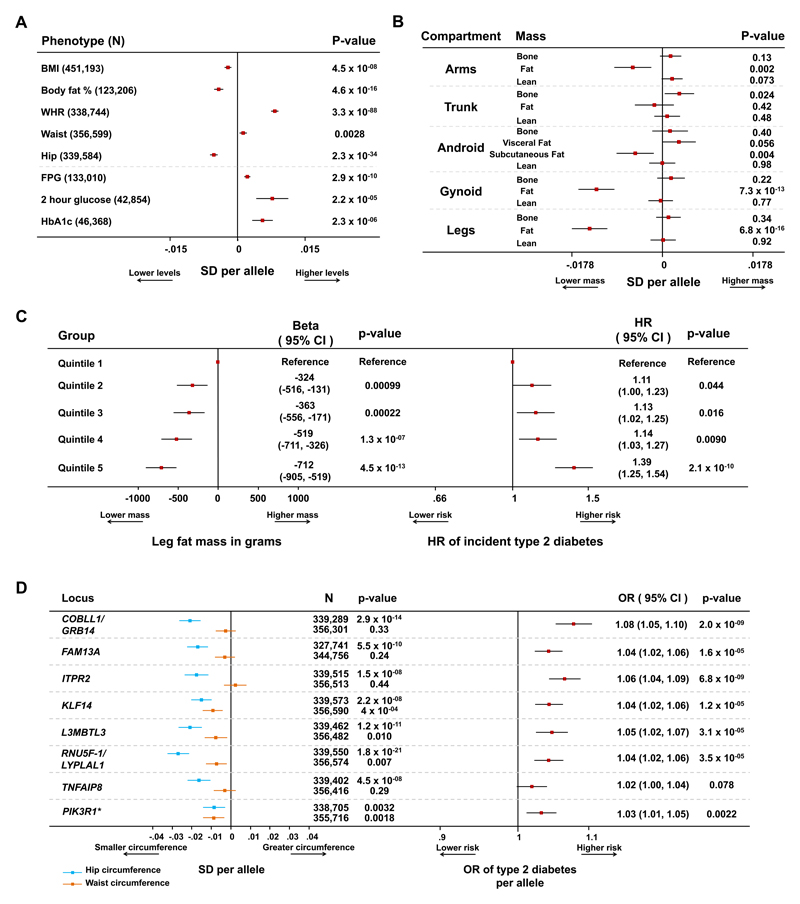
While insulin resistance is often considered secondary to higher adiposity, at the 53 loci we observed associations with lower body fat percentage, BMI and hip circumference (Figure 1A, Supplementary Figures 6 and 7). The larger magnitude of association with measures of body fat rather than with glycaemic phenotypes is consistent with a primary effect of many variants on adipose tissue mass (Figure 1A, Supplementary Figures 6 and 7).
Figure 1. Combined associations with detailed anthropometry and metabolic disease risk at the 53 genomic loci.
Panel A: association of the 53-SNP genetic score with anthropometric and glycaemic traits in meta-analyses of genetic association studies. Body mass index (BMI), waist-to-hip ratio (WHR), waist and hip circumference data are from the GIANT consortium and the UK Biobank study. Body fat percentage data are from the UK Biobank, EPIC-Norfolk and Fenland studies. Fasting plasma glucose (FPG), 2 hour glucose and HbA1c data are from the MAGIC consortium. Squares with error bars represent the per-allele beta coefficients in standard deviation units and their 95% confidence intervals. Panel B: association of genetic scores with compartmental body masses. Data are from 12,848 participants of the Fenland and EPIC-Norfolk studies who underwent a DEXA scan. Squares with error bars represent the per-allele beta coefficients in standard deviation units and their 95% confidence intervals. Panel C: association with lower levels of leg fat mass and higher hazard of incident type 2 diabetes by quintiles of the 53-SNP genetic risk scores. Associations are reported for individuals in the exposed category compared with the bottom quintile (reference category). Associations with leg fat mass are from 9,747 participants of the Fenland study and are reported on the left. Associations with incident type 2 diabetes are from 7,420 incident cases and 9,267 controls of the InterAct study and are reported on the right. Squares represent the beta coefficients in grams of leg fat mass (left plot) or the hazard ratio (HR) for incident type 2 diabetes (right plot) in each category compared with the lowest quintile. Error bars represent the 95% confidence intervals of these estimates. Panel D: associations of individual lead SNPs at eight loci with waist, hip circumference (left) and type 2 diabetes (right). Loci were selected on the basis of their genome-wide significant association with hip circumference or body fat percentage (i.e. PIK3R1). Waist and hip circumference analyses are from a meta-analysis of the GIANT and UK Biobank studies. Type 2 diabetes analyses are from a meta-analysis of the DIAGRAM, InterAct and UK Biobank studies. Squares with error bars represent the per-allele beta coefficients in standard deviation units of waist and hip circumference (left plot) or the per-allele odds ratio (OR) of type 2 diabetes (right plot). Error bars represent the 95% confidence intervals of these estimates. *Detailed associations at the PIK3R1 locus, which was primarily associated with lower body fat percentage, are presented in Supplementary Figure 9.
By follow-up studies using dual-energy X-ray absorptiometry (DEXA) measures in 12,848 individuals, we found that the most marked association of the genetic score was with lower levels of gynoid and leg fat mass (Figure 1B). Individuals in the highest quintile of the 53-SNP genetic score had an average of 712 grams less leg fat mass compared to individuals in the bottom quintile (Figure 1C), which accounted for the majority of the overall body fat association (Supplementary Figure 8). The association with lower levels of leg fat was accompanied by a higher hazard of incident type 2 diabetes (Figure 1C). In 9,150 participants from the EPIC-Norfolk cohort who gained weight during a median follow-up of 3.7 years, carrying a greater number of the 53 risk alleles was inversely associated with change in hip circumference, adjusted for the amount of weight gained (i.e. individuals carrying more alleles were less likely to deposit extra mass in their gluteal region for a given increase in body mass; β in cm of hip circumference per SD of genetic score, -0.07; p=0.027; Supplementary Note). Overall, these association analyses suggest that individuals genetically predisposed to insulin resistance via the 53 loci have a relative inability to expand their peripheral fat compartment when challenged by a positive energy balance and that this incapacity results in higher cardiometabolic disease risk. We also found that the 53-SNP genetic score was associated with higher levels of alanine aminotransferase and gamma-glutamyl transferase (Supplementary Table 6), which suggests that the failure to store lipid in gluteofemoral adipose tissue may be accompanied by hepatic lipid deposition.
The 53-SNP genetic score was associated with greater waist circumference (Figure 1A), but not with trunk adipose tissue (Figure 1B), indicating that the association with body fat distribution and cardiometabolic disease was largely driven by the association with lower levels of peripheral adipose tissue (Figure 1 and Supplementary Figures 6-8). Among the 53 lead SNPs, 17 were within 500kb of a waist-to-hip ratio (WHR) associated SNP36 (Supplementary Figure 6). Consistent with DEXA analyses, the associations with WHR at this subset of overlapping loci were largely driven by an association with lower hip circumference, rather than higher waist circumference (Supplementary Figure 6 and Supplementary Table 4).
Our large-scale meta-analyses allowed the investigation of individual-SNP associations with both adiposity and metabolic risk. At eight of the 53 loci, the lead SNP was associated with lower total body fat percentage or hip circumference at genome-wide significance (p<5 x 10-08), including a novel association of the insulin-raising G allele of rs4976033 near PIK3R1 with lower body fat percentage (p=3.0 x 10-09; see Figure 1D and Supplementary Figure 9). Seven of the eight adiposity-lowering alleles at these loci were associated with a higher risk of type 2 diabetes (p<0.05; Figure 1D).
Role of common variants in the genetic basis of a severe form of lipodystrophy
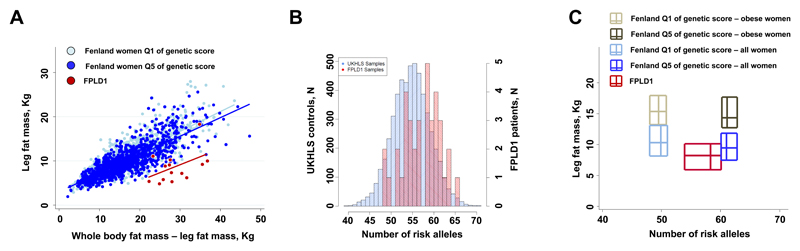
Given the strong association with insulin resistance but lower levels of peripheral adiposity, we hypothesized that the polygenic predisposition to insulin resistance imparted by the 53 loci might contribute to the pathogenesis of familial partial lipodystrophy-type 1 (FPLD1). When compared to women from the population-based Fenland study, women diagnosed with FPLD1 displayed markedly lower levels of leg fat mass for a given fat mass in the rest of the body (Figure 2A). Whilst the name of the condition implies Mendelian inheritance, using exome sequencing in 9 FPLD1 cases we did not identify likely candidate causal genes (Online Methods and Supplementary Table 7). When compared with 5,296 unrelated women from the UKHLS study in a case-control analysis, 37 patients with FPLD1 had a higher burden of the 53 risk alleles (OR per SD of genetic score in logistic regression analyses adjusted for age and the first 10 genetic principal components, 1.70; 95% CI, 1.21-2.39; p=0.0021, Figure 2B; ppermutation=0.0020, see Online Methods). Also, the phenotypes observed in FPLD1 patients in comparison to obese women from the Fenland study mirrored the association pattern of the 53-SNP genetic score (Supplementary Table 8). FPLD1 women had a more severe leg fat phenotype compared to that expected from the relationship between the 53 SNP score and leg fat mass in the Fenland study (Figure 2C), suggesting that additional genetic and environmental factors contribute to determining this extreme phenotype.
Figure 2. Associations at the 53 genomic loci with familial partial lipodystrophy type 1 (FPLD1).
Panel A: distribution of leg fat mass as a function of the fat mass of the rest of the body (from DEXA) in women of the Fenland study at the extreme quintiles (Q) of the 53-SNP genetic score and in 14 FPLD1 subjects. Q1 represents a low genetic burden, whereas Q5 a high genetic burden. Lines of fit are plotted for each group. Panel B: histograms of the distribution of risk alleles in the FPLD1 subjects and in control women from the UKHLS study. Panel C: bi-dimensional box plots of the distribution of leg fat mass as a function of the distribution of the number of risk alleles in women of the Fenland study at the extreme quintiles (Q) of the 53-SNP genetic score and in FPLD1 subjects. Q1 represents a low genetic burden, whereas Q5 a high genetic burden. Each rectangle represents a group of individuals. For each dimension, the two sides of the rectangle represent the interquartile range and the central line the median. Data for obese women from Fenland were plotted to show the relationship between genetic risk and levels of leg fat in a group of women with a similar body mass index to that of FPLD1 patients.
Prioritisation of putative effector genes, cell-types and tissues
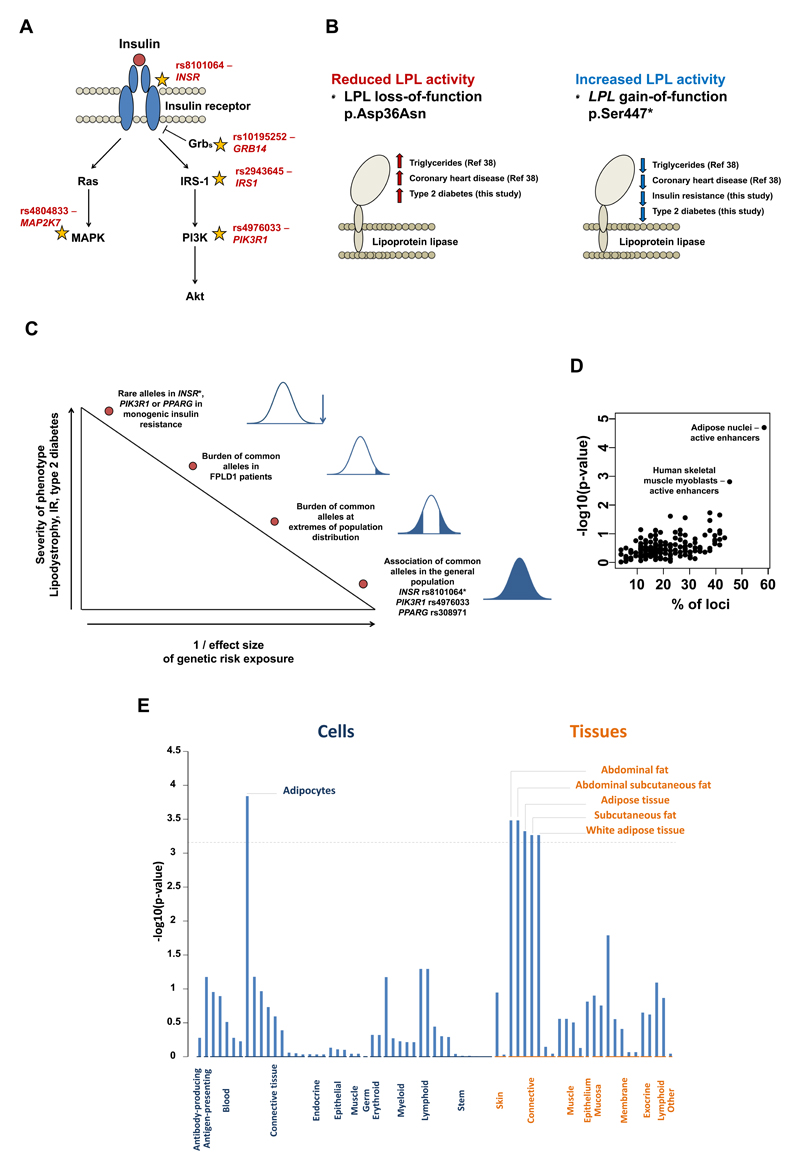
We prioritised putative effector genes at the 53 loci by integrating data about physical proximity, expression quantitative trait locus (eQTL) mapping, functional annotation and previous knowledge about genes causing monogenic forms of insulin resistance (summarised in Supplementary Table 2; see also Supplementary Tables 9-11 for details). Putative effector genes included five with well-established roles in proximal insulin signalling (Supplementary Table 2 and Figure 3A). Other candidates included LPL, encoding the key lipolysis regulator lipoprotein lipase (Supplementary Table 2 and Figure 3B). The insulin-lowering, minor allele of rs1011685 (near LPL) is on the same haplotype (r2=1) as a gain-of-function,37 protein-truncating allele in LPL (p.Ser447*; rs328; minor-allele frequency [MAF], 9.9%). The p.Ser447* gain-of-function variant was recently reported to be associated with lower risk of coronary heart disease,38 while an independent loss-of-function missense variant of LPL39 (p.Asp36Asn; rs1801177; MAF, 1.9%) was associated with higher risk.38 Here, we found that the p.Ser447* gain-of-function variant is associated with greater insulin sensitivity, lower fasting glucose, lower levels of liver markers and protection from type 2 diabetes (OR per allele, 0.93; p=1.6 x 10-05; Figure 3B and Supplementary Figure 10). Conversely, the p.Asp36Asn loss-of-function variant in LPL is associated with a higher risk of type 2 diabetes (OR per allele, 1.11; p=0.0086; Figure 3B and Supplementary Figure 10). Thus, recent findings of an allelic series of LPL variants implicating lipoprotein lipase as a putative therapeutic target in heart disease38 are now complemented by a directionally consistent observation for type 2 diabetes, compatible with a role for impaired lipoprotein lipase-mediated lipolysis in insulin resistance and type 2 diabetes.
Figure 3. Putative effector genes, tissues and cell types.
Panel A: schematic representation of some established components of the insulin signalling pathway with stars reporting the location in the pathway of putative effector genes, with their respective lead single nucleotide polymorphism listed. Panel B: associations of gain- and loss-of-function genetic variants in the LPL gene with type 2 diabetes. The reference number in parenthesis refers to the study reporting the association with triglycerides and coronary heart disease (see reference number 38 of this manuscript).38 Panel C: summary of evidence about links between genetic variants, lipodystrophy, insulin resistance, and type 2 diabetes at different levels of the population phenotypic distribution. *Rare syndromes caused by autosomal dominant INSR mutations are not usually associated with lipodystrophy and the INSR rs8101064 polymorphism is not associated with body fat percentage. Panel D: overlap of the 53 loci (lead SNPs plus proxy variants in r2>0.8) with chromatin state annotations from the NIH Roadmap. Panel E: DEPICT’s annotation of cell types and tissues on the basis of expression patterns in 37,427 human microarray samples. The y-axis represents the –log10(p-value) for enrichment of signal in a cell or tissue type attributed by DEPICT. The horizontal broken line represents the multiple-test corrected threshold of statistical significance (Bonferroni p=0.00072).
Among the 53 loci, three contained genes at which rare mutations have been previously implicated in severe monogenic forms of insulin resistance (i.e. PPARG, PIK3R1, INSR; Figure 3C), which is more than what expected by chance given the prevalence of monogenic insulin resistance genes in the genome (observed percentage 0.54% [3 out of a total of 553 genes in the 53 loci], expected percentage 0.064%, two-tailed binomial p=0.0056). The PIK3R1 gene encodes regulatory subunits of a critical kinase involved in proximal insulin signalling and rare, loss-of-function mutations in this gene are associated with SHORT syndrome, a dysmorphic condition characterised by short stature, partial lipodystrophy and insulin resistance.40–43 To date, such mutations have been identified in few families worldwide and data from Exome Aggregation Consortium show this gene to have decreased tolerance of missense variation (Z=2.42) and to be particularly intolerant of loss-of-function mutations (pLI=1; Exome Aggregation Consortium, Cambridge, MA, URL: http://exac.broadinstitute.org accessed 22nd March 2016). Here, we provide evidence that a common variant near PIK3R1, which accounts for almost half of all alleles in the general population (MAF=49%), is associated with subtle effects on insulin resistance, lower body fat percentage, higher risk of type 2 diabetes and coronary heart disease (Supplementary Figure 9). This association pattern partially overlaps with that reported for PIK3R1 mutations and SHORT syndrome (Supplementary Table 12). We also found that the common rs8101064 allele T in INSR, encoding the insulin receptor, was associated with insulin resistance and higher risk of type 2 diabetes (OR per allele, 1.08; p=0.020), but not with body fat percentage (p=0.16), consistent with the fact that patients with heterozygous loss-of-function mutations in the INSR are frequently insulin resistant but are not commonly lipodystrophic.13
We assessed the overlap of lead SNPs and their proxies (r2>0.8) with functional regulatory annotations across 98 cell types from the NIH Epigenome Roadmap (Online Methods) and, consistent with phenotypic associations, identified substantial overlap with adipose tissue active enhancer elements (31 of 53 loci overlapped adipose tissue active enhancer elements; observed percentage=58.4% of loci, expected=30.1%, binomial p=2.1 x 10-05; Online Methods and Figure 3D). Furthermore, combined pathway analyses with integration of large-scale gene expression data44 implicated adipocytes as likely effector cell-type underlying observed associations (Figure 3E). In subcutaneous adipocyte eQTL data from 1,064 individuals of the EUROBATS and GTex projects (Online Methods and Supplementary Table 10), we observed evidence of eQTL associations with nearby genes (p<10-06) at 21 loci including 14 with supportive evidence of co-localisation of lead phenotypic and eQTL associations (Online Methods and Supplementary Table 10).
Experimental validation of putative effector genes in cellular adipogenesis models
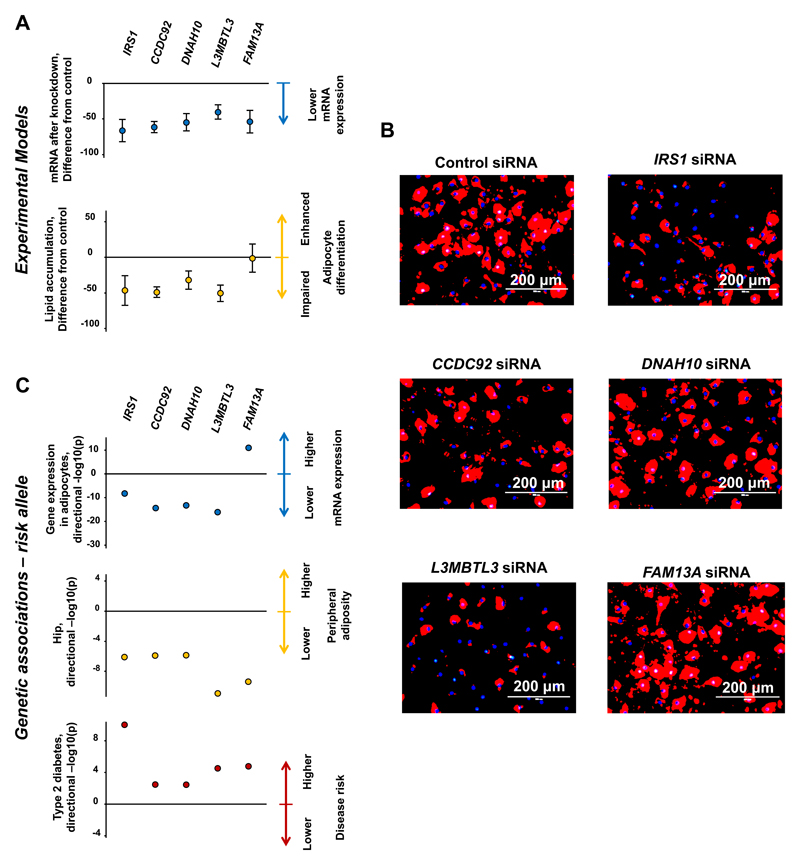
We sought to experimentally validate the role of five putative effector genes (IRS1, CCDC92, DNAH10, L3MBTL3 and FAM13A) across four loci which showed associations with (a) expression of nearby transcripts in subcutaneous adipocytes, (b) lower peripheral adiposity and (c) higher metabolic disease risk (Supplementary Note, Supplementary Figure 11, Supplementary Tables 10 and 13). We used short interfering RNA (siRNA) to reduce mRNA levels for these five genes in OP9-K cells, an adipocyte model which shows rapid differentiation in response to adipogenic stimuli.45 Knockdown of the IRS1, CCDC92, DNAH10 and L3MBTL3 genes significantly reduced both mRNA of the target gene (Figure 4A top graph) and lipid accumulation (Figure 4A bottom graph and Figure 4B). These results were directionally consistent with the association of the insulin-raising alleles at these loci with lower expression of these genes in subcutaneous adipocytes and with lower levels of peripheral fat (Figure 4C). Knockdown of FAM13A reduced mRNA (Figure 4A top graph), but did not significantly affect pre-adipocyte lipid accumulation (Figure 4A bottom graph and Figure 4B). In contrast to the other four genes, the risk allele at FAM13A was associated with higher mRNA expression of this gene in subcutaneous adipocytes (Figure 4C).
Figure 4. Experimental knockdown of putative effector genes in cellular adipogenesis models and comparisons with phenotypic associations.
Panel A: results of experimental knockdown in OP9-K cells. Full circles represent the difference of the means from knockout experiments of a given gene compared with control experiments (n=4-7). Error bars represent the 95% confidence intervals of the difference of the means. Top graph: effect on mRNA levels of knockdown experiments of target genes using short interfering RNA (siRNA) in OP9-K cells. The two-tailed t-test p-values for differences in means were: IRS1, p=4.6 x 10-06; CCDC92, p=2.7 x 10-09, DNAH10, p=2.4 x 10-06; L3MBTL3, p=4.6 x 10-06; FAM13A, p=2.4 x 10-05. Bottom graph: effect on lipid accumulation in siRNA knockdown experiments. The two-tailed t-test p-values for differences in means were: IRS1, p=0.0047; CCDC92, p=1.2 x 10-05, DNAH10, p=0.0027; L3MBTL3, p=0.00013; FAM13A, p=0.92. Panel B: illustrative images showing florescence microscopy from lipid accumulation experiments. Red indicates adipored staining of neutral lipid, blue is hoechst staining of nuclei. Panel C: Association of the risk (insulin-raising) allele of the lead single nucleotide polymorphism in or near each of the putative effector genes with (a) the expression of the corresponding gene in subcutaneous adipocytes in the EUROBATS project (top graph in the panel); (b) hip circumference in a meta-analysis of GIANT and UK Biobank (mid graph); and (c) type 2 diabetes in a meta-analysis of InterAct, DIAGRAM and UK Biobank (bottom graph). Full circles represent the –log10(p-value) for the association of the insulin-raising allele multiplied by the direction of the beta coefficient (i.e. a “directional” –log10(p)). For graphic display purposes, the –log10(p-value) for the association with type 2 diabetes of the rs2943645-T allele near IRS1 is represented as 10 instead of 16.9.
Discussion
Our data implicate the impaired capacity to adequately expand the peripheral adipose tissue compartments in human insulin resistance and related disease at the population level. These results substantially augment existing evidence31,36,46–49 by clarifying the extent and relevance of adipose tissue dysfunction to cardiometabolic disease and by providing novel mechanistic insights into its underpinning biology.
Our results are consistent with the existence of dozens of genomic regions at which common genetic variation affects cardiometabolic disease risk via subtle “lipodystrophy-like” mechanisms. While these common genetic mechanisms have individually small effects, their cumulative effect is large and relevant to a large fraction of the population. For instance, we observed ~700 grams difference in leg fat mass between the top and bottom 20% of the population distribution of risk alleles. We also show a polygenic contribution to an extreme phenotype, referred to as FPLD1 or Köbberling-type lipodystrophy, illustrating the contribution of common alleles to severe forms of insulin resistance. At given loci (e.g. PIK3R1), we found that genetic variants at the two extremes of the allele frequency spectrum result in corresponding consequences at the extremes of the phenotypic severity spectrum. These findings strongly concur with the notion that molecular and pathophysiologic mechanisms first described in severe forms of lipodystrophic insulin resistance are relevant to the general population. While a centripetal distribution of body fat is a well-recognised risk factor for metabolic and cardiovascular disease,50–54 there is considerable confusion about the underlying mechanisms and relative importance of lower peripheral fat versus higher central adiposity. Whilst supportive of a role for central fat accumulation, different lines of evidence from this study suggest a role for impaired peripheral fat deposition in insulin-resistant cardiometabolic disease in the general population. These include strong associations with gluteo-femoral adiposity, overlap with regulatory regions in adipose tissue and with genomic loci known to cause lipodystrophies, as well as functional characterisation in adipocytes. These findings provide evidence from large-scale human genetics studies which add to a body of research about the links between subcutaneous and lower-body adipocyte phenotypes and a favourable metabolic profile.19,55–57
By combining population-scale association studies with eQTL data from adipose tissue and experimental evidence from murine cellular models, we provide evidence that specific risk loci influence adipose gene expression resulting in impaired adipogenesis, reduced peripheral fat depots and ultimately increased cardiometabolic disease risk. For the L3MBTL3, DNAH10 or CCDC92 genes, evidence presented in this study provides the first link with impaired adipocyte differentiation capacity. L3MBTL3 recognises methylated lysine residues on histone tails58 and previous genome-wide anthropometric studies have implicated this locus in adult height and length at birth.59–61 At the chromosome 12q24 locus, our analyses are consistent with the implication of both CCDC92 and DNAH10 in impaired adipogenesis. CCDC92 is a coiled coil domain protein which interacts with proteins in the centriole/ciliary interface.62 DNAH10 encodes one of the heavy chains of the dynein arms of the motile cilia and it is, therefore, surprising that its product appears to have cell autonomous effects on adipocyte biology. However, an essential splice site mutation in DNAH10 has previously been reported to co-segregate with abnormal circulating HDL-cholesterol levels in a family,63 while the locus containing both CCDC92 and DNAH10 has been associated with circulating levels of large HDL particles,64 further supporting an unexpected role for these proteins in metabolism.
Our results have preventive and therapeutic implications for cardiovascular and metabolic disease. First, they suggest that attempts to develop pharmacological agents acting on the molecular mechanisms of obesity are likely to reduce cardiometabolic risk if they reduce calorie intake (e.g. by acting on appetite) or reduce ectopic fat deposition in tissues such as the liver, muscle and pancreas, but not if they impair adipogenesis or peripheral fat deposition. Agents that promote adipocyte differentiation and increase peripheral adipose mass via action on the peroxisome proliferator-activated receptor gamma have powerful antidiabetic actions,65,66 and in some cases have a beneficial effect on cardiovascular outcomes,67,68 although some of these agents have been reported to have an adverse cardiovascular safety profile.69 An early but vital challenge in the translation of genetic findings towards therapeutic insight is the ability to identify likely effector transcripts underlying genetic associations. In the current study, we identify putative effectors of genetic associations and the tissues in which they operate. We also demonstrate that these genetic variants often affect the risk of type 2 diabetes and coronary disease in a consistent direction, which suggests that targeting these pathways may satisfy current regulatory requirements that anti-diabetic agents should not be associated with unacceptable cardiovascular risk.70 This is particularly true of findings from both gain- and loss-of-function variants in the LPL gene and risk of type 2 diabetes, which are directionally consistent with those previously reported for the same mutations and risk of heart disease.38 Notably, the directional concordance for risk of heart disease and type 2 diabetes is in contrast to genetic evidence for other lipid-lowering agents (e.g. cholesterol-lowering variants near the molecular target of statins).71 In the context of a growing body of evidence linking lipolysis and heart disease risk,38,72–75 these data suggest that enhancing lipoprotein lipase activity may also become a viable preventive or therapeutic strategy in type 2 diabetes.
In interpreting the results of this study, it is important to note that combining multiple genetic-association analyses to gain insights about a latent unmeasured phenotype (i.e. insulin resistance) is not immediately comparable with a univariate genome-wide association study of a trait (e.g. fasting insulin). However, we validated these genetic variants as being strongly associated with “gold-standard” measures of insulin sensitivity, with multiple insulin-resistance related diseases, including a severe form of insulin-resistant partial lipodystrophy, and showed overlap with monogenic insulin resistance genes. Thus, approaches to leverage additional sources of evidence to prioritise genomic variation (such as multiple phenotypes or putative functional class76) represent a powerful use of extant genetic association results to advance understanding of biology previously intractable to conventional strategies.
Our results were based on genome-wide analyses of fasting insulin adjusted for BMI,28,29 and we did not identify loci with a primary effect on higher adiposity and secondary association with insulin resistance (e.g. FTO). Our approach was more likely to identify loci influencing insulin resistance at a given level of adiposity. Prompted by the strong association pattern of the genetic scores and variants, we focused on evaluation of mechanisms linking lower levels of peripheral adiposity with insulin resistance. The importance of adipose function as a prominent driver of common insulin resistance is highlighted by the observation that half of all variants associated with fasting insulin at genome-wide significance (without adjustment for BMI) in a previous study28 were included in our genetic score. However, our results do not preclude the presence nor diminish the importance of other mechanisms underlying insulin resistance.5,77 Indeed, the associations we observe of the genetic score with central fat, visceral fat and liver enzymes would be further strengthened after adjustment for overall adiposity. While we conclude that our findings implicate a primary effect on impaired adipose function and a secondary effect on insulin resistance, we cannot entirely exclude the possibility of the reverse, nor that there are pleiotropic contributions to the associations.
Conclusions
Collectively, our findings support the notion that limited capacity of peripheral adipose tissue to store surplus energy is implicated in human insulin resistance and related cardiometabolic disease in the general population. Furthermore, we highlight putative effector genes, tissues and mechanisms underpinning this link.
Online Methods
Study design
We integrated the results of genome-wide analyses on insulin and lipid phenotypes with the aim of identifying genetic variants associated with an insulin resistance phenotypic pattern (Supplementary Figures 1-4). We then investigated the mechanistic links of genetic variation at 53 identified genomic regions with cardiometabolic diseases by integrating analyses of: (a) cardiometabolic traits and outcomes from up to 451,193 individuals; (b) detailed continuous metabolic traits from 12,848 deeply-phenotyped individuals; (c) genetic and clinical features from 37 individuals diagnosed with familial partial lipodystrophy type 1; (d) gene expression from over 100 separate eQTL datasets and (e) siRNA mediated knockdown of putative effector genes in experimental adipogenesis models (Supplementary Table 1 and Supplementary Figures 1-2).
Participating studies
Lists of phenotypes, participating studies and sample sizes for each analysis are in Supplementary Table 1 and Supplementary Figures 1-2. Details about participants and cohorts with individual-level genotype data are in Supplementary Table 14. Ethical approvals were obtained at each study site and informed consent was obtained from all participants.
The Fenland study is a population-based cohort study of 12,435 participants without diabetes born between 1950 and 1975. Participants were recruited from general practice surgeries in Cambridge, Ely and Wisbech (United Kingdom) and underwent detailed metabolic phenotyping and genome-wide genotyping.
EPIC-Norfolk is a prospective cohort study of 25,639 individuals aged between 40 and 79 and living in the Norfolk county in the United Kingdom78 at recruitment. EPIC-Norfolk is a constituent cohort of the European Prospective Investigation of Cancer (EPIC).79 A total of 3,101 participants with available dual energy X-ray absorptiometry (DEXA) were included in analyses of detailed anthropometry, while 9,150 participants were included in analyses of change in hip or waist circumference in individuals who gained weight during follow-up.
EPIC-InterAct is a case-cohort study nested within the EPIC study, a cohort study of 519,978 European participants.80 During an average of 8 years of follow-up, 12,403 individuals who were free of diabetes at baseline were identified incident type 2 diabetes cases.80 InterAct has also defined a randomly-selected subcohort of 16,154 individuals free of diabetes at baseline.80 Data on 15,357 individuals with available genotyping and not overlapping with DIAGRAM34 were included.
UK Biobank is a population-based cohort study of ~500,000 people aged between 40-69 years who were recruited in 2006-2010 from several centres across the United Kingdom.81 Associations with prevalent type 2 diabetes were estimated in 111,016 individuals (4,586 cases and 106,430 controls) of the initial UK Biobank dataset. We also used the UK Biobank data for anthropometry analyses and for a sensitivity analysis of prevalent coronary heart disease (i.e. self-reported myocardial infarction or angina) in 5,369 cases and 106,969 controls.
The United Kingdom Household Longitudinal Study (UKHLS; also known as Understanding Society) is a longitudinal panel survey of 40,000 households representative of the population of the United Kingdom. Participants were surveyed annually since 2009 and contributed information relating to their socioeconomic circumstances, attitudes, and behaviours via a computer assisted interview. For a subset of individuals who took part in a nurse health assessment, blood samples were taken and genomic DNA analysed.
In addition to individual-level genotyping data, we used genome-wide meta-analyses results on a variety of cardiometabolic traits and disease endpoints (Supplementary Table 1 and Supplementary Figures 1-2).
Detailed anthropometric analyses
In the Fenland and EPIC-Norfolk studies, body composition was determined by dual-energy X-ray absorptiometry (DEXA) using a Lunar Prodigy advanced fan beam scanner (GE Healthcare, Bedford, UK) using the encore software version 14.10.022 (GE Healthcare, Bedford UK). Participants were scanned by trained operators using standard imaging and positioning protocols. The coefficient of variation for scanning precision, calculated from 30 consecutive scans, was 2% for total fat mass. The enCORE software was used to demarcate regional boundaries. All the images were manually processed by one trained researcher, who corrected demarcations according to a standardized procedure. Boundaries of body regions are described in details in the Supplementary Note. In the UK Biobank study, body fat percentage was estimated by bio-impedance using the Tanita BC418MA body composition analyser.
Association of genetic variants with insulin resistance phenotypes
A dyslipidaemic pattern with higher triglyceride levels and lower HDL cholesterol is considered characteristic of the clinical presentation of insulin resistance 13 and has been used to specifically identify individuals with insulin resistance.30 In a previous large-scale genome-wide discovery of genetic determinants of fasting insulin levels, among 19 fasting insulin-associated loci we identified ten that were strongly associated with higher triglycerides and lower HDL cholesterol.28 This pattern of association was used to refine loci which were then validated as being associated with “gold-standard” measures of insulin resistance.31 Loci associated with insulinaemia but not with lipid traits included TCF7L2,28 which is primarily implicated in insulin secretion, rather than resistance.82 These findings suggested that the combined association with this triad of phenotypes could help identify specific genetic determinants of insulin resistance.
With this background, we systematically triangulated the results of the association of ~2.4M single nucleotide polymorphisms (SNPs) with fasting insulin adjusted for body mass index (FIadjBMI; from up to 108,557 participants of the MAGIC consortium),28,29 HDL-cholesterol, and triglycerides (from up to 188,577 participants of the Global Lipids Genetics Consortium)32 using publicly available genome-wide results (Supplementary Figures 1-4). For FIadjBMI analyses, we used metabochip association results28 when available. We aligned alleles across the three phenotypes such that the effect allele was the insulin-raising allele. We took forward for further analysis all SNPs associated with higher FIadjBMI, higher triglycerides and lower HDL cholesterol at p<0.005 for each of the three traits. The prior probability for association of a given SNP with the three traits and in the pre-specified directional concordance under the null hypothesis corresponds to 0.005*0.0025*0.0025 = 3.1 x 10-08. In each 1 Mb locus, we retained the lead SNP for association with fasting insulin for further analysis (Supplementary Figure 4).
Fasting insulin analyses adjusted for BMI were chosen because we were interested in identifying genetic determinants of insulin resistance for a given level of adiposity. It has been proposed that adjusting genetic association analyses for covariates such as BMI might result in a bias known as “collider bias”.83,84 Therefore, we assessed the association of the 53 lead SNPs for a bias in the identification of variants primarily associated with BMI and artificially associated with fasting insulin, but found no evidence on such bias (Supplementary Figure 12).
Statistical methods
We studied the association of individual SNPs and of genetic scores with continuous metabolic traits and endpoints. Where individual level genotype data was available, the associations of genetic variants or scores (i.e. the sum of effect alleles) with outcomes were estimated using multivariable linear, logistic or Cox regression models. For result-level association data, we used the inverse-variance weighted method developed by Burgess et al., assigning a weight of 1 to each SNP, to approximate the association of an unweighted genetic risk score.85 Statistical analyses were conducted using STATA v14.1 (StataCorp, College Station, Texas 77845 USA), R v3.2.2 (The R Foundation for Statistical Computing), and METAL.29 All p-values presented in the study are two-tailed p-values.
Analyses in a severe form of partial lipodystrophy and insulin resistance
We studied the clinical and genetic characteristics of 37 women with familial partial lipodystrophy type 1 (FPLD1; also called Köbberling-type familial partial lipodystrophy).
All cases were referred to the insulin resistance/lipodystrophy specialist centre led by Drs Semple, Savage and O’Rahilly in Cambridge. FPLD1 is currently a clinical diagnosis used to describe predominantly women with selective paucity of limb adipose tissue, central obesity, severe insulin resistance, and a higher risk of type 2 diabetes.86,87 As some women with lipodystrophy due to loss-of-function mutations in PPARG manifest a similar phenotype (i.e. FPLD3), mutations in this gene were excluded in all FPLD1 cases included in this study. The biochemical and anthropometric phenotype of the 37 FPLD1 patients was compared with that of female participants of the Fenland study. In these analyses we compared the phenotypes of the 37 FPLD1 patients with those of all Fenland study women (Figure 2) and with those of obese Fenland study women (Figure 2 and Supplementary Table 8), who have a similar BMI to that of FPLD1 women.
To understand the genetic basis of FPLD1, we carried out exome sequencing analyses in 18 individuals from 9 pedigrees with FPLD1 without identifying any clear candidate mutations or genes. Sequencing, variant calling and annotation were performed as described previously (Family 2;88 Families 1 and 3–9 as part of the UK10K project).89 Calls were annotated with 1000 Genomes allele frequencies and the NCBI dbSNP database build 132 (ftp://ftp.ncbi.nih.gov/snp/organisms/human_9606/). Variants were defined as potentially functional if they were non-synonymous, resulted in loss or gain of a stop codon or a frameshift, or occurred within essential splice sites. Those unlikely to have a functional impact were removed, as were all variants found with a MAF >1% in individuals of European descent from the 1000 Genomes Phase 1v3 (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20110521/) or the NHLBI ESP Exomes (URL: http://evs.gs.washington.edu/EVS/; January 2012). Further filtering was then implemented to retain only variants in genes seen in multiple patients.
We compared the burden of the 53-SNP genetic score in the 37 FPLD1 patients with that of 5,296 unrelated control women from UKHLS. Genome-wide genotyping of UKHLS women was performed using the Illumina Infinium HumanCoreExome-12 v1.0 BeadChip. Genotype calling was performed using the Illumina GenCall software. Genome-wide genotyping of FPLD1 patients was performed using the Illumina® Infinium CoreExome-24v1.0 chip. Prior to imputation, the following quality control criteria were applied for exclusion of SNPs in PLINK90 (version 1.07): (1) minor allele frequency <0.01; (2) Hardy-Weinberg equilibrium p<1 x 10-06; (3) call rate <99%; (4) differential missingness between cases and controls p<1 x 10-06; (5) SNPs showing differential genotyping between the CoreExome-24v1.0 and CoreExome-12v1.0 chips. Samples were excluded prior to imputation in PLINK based on the following criteria: (1) call rate <95%; (2) autosomal heterozygosity >3 standard deviations from the mean; (3) pairwise identity by descent was calculated and one individual was removed for every pair of individuals with a pi-hat >0.05, with preference given to retaining female samples; (4) identity assessed by the concordance between the genome-wide and Fluidigm® genotypes at 24 sites (excluding individuals with concordance <90%); (5) ethnic outliers based on a principal components analysis. Imputation was performed using the 1000 Genomes Phase 3 reference panel using SHAPEIT291 (version 2.r644) and IMPUTE292 (version 2.3.1). FPLD1 cases and UKHLS controls were imputed together. In an analysis of the genetic principal components derived from genome-wide genotyping (defined based on the combined sample), the 37 FPLD1 patients clustered with UKHLS control women (Supplementary Figure 13). Association analyses were performed in R by logistic regression adjusting for age and the first 10 genetic principal components. We also derived a permutation-based null comparator by performing 100,000 permutations of randomly selecting 53 SNPs (>1Mb apart) from genome-wide analyses of FPLD1 status adjusted for age and principal components and performing summary statistic Mendelian randomisation.85 Of 100,000 iterations, 201 had a p-value less than our observed association (ppermutation= 2.01 x 10-03).
Prioritisation of putative effector genes
We sought to determine the putative effector genes at the 53 loci. We combined information on (a) physical proximity, (b) eQTL data from over 100 repositories, (c) functional and regulatory annotations and (d) previous knowledge about causal genes for monogenic forms of insulin resistance.
For physical proximity analysis, we reviewed genes within a 1 Mb-window of each lead SNP and generated regional association plots using LocusZoom.93 For eQTL analysis, we analysed both publicly available and unpublished datasets (see below). For functional annotation, we used the gene and the tissue/cell type prioritisation functions of the integrative software DEPICT,44 in order to gain insights about putative effector genes, tissues and cell types. We also looked for nonsynonymous variants in linkage disequilibrium with the lead SNP (r2>0.8) in European ancestry populations using Haploreg v3.94
We assessed the overlap of identified loci with chromatin state definitions of active enhancers and active promoters for 98 cell types from the NIH Epigenome Roadmap project, including a total of 196 genomic annotations. For each of the 53 lead SNPs, we identified the set of proxy variants in high linkage disequilibrium (CEU r2>0.8) using SNAP95 and defined a ‘locus’ as a lead SNP plus its proxies. We then calculated the number of loci where at least one variant at the locus overlapped a given annotation. We determined enrichment in the overlap at the 53 loci compared to an expected distribution built from randomly selected matched loci. First, we identified all variants with genome-wide significant association (p<5 x 10-08) to any trait in Europeans from the GWAS catalog.96 We then pruned this set of variants (using a CEU r2 threshold of 0.1), resulting in a list of independent trait-associated variants. For each of these variants, we constructed a background ‘locus’ as the set of proxy variants in high linkage disequilibrium (CEU r2<0.8). For each of the 53 loci, we then selected a locus from the background set matched on total number of proxy variants, total genomic distance covered, and distance of the midpoint to the closest gene transcription start site. We then re-calculated the overlap of each annotation in the set of matched background loci. We obtained the expected overlap for each annotation by averaging over 1 million permuted background locus sets. We then tested for enrichment of each annotation with a binomial test using the observed number of overlapping loci, total number of loci and expected percentage of overlapping loci.
We compiled a list of 13 genes (PPARG, INSR, PIK3R1, TBC1D4, LMNA, PLIN1, AKT2, CIDEC, AGPAT2, BSCL2, CAV1, PTRF, PCYT1A) known to cause monogenic forms of insulin resistance from the literature13 and used that to look for overlap with genes in identified regions. Two experts in the clinical care of patients with monogenic insulin resistance (Drs Semple and Savage) reviewed the curated list.
Analysis of eQTLs in multiple tissues
Using a curated collection of over 100 separate eQTL datasets, we queried whether the 53 lead SNPs or their proxies (r2>0.8) were associated with transcript expression in a wide range of tissues. Proxy SNPs in linkage disequilibrium in European ancestry populations were identified using SNAP.95 For this study, we considered all associations below a p-value cut-off of 1 x 10-06. A general overview of a subgroup of >50 eQTL datasets has been published,97 with specific citations for the >100 datasets included in the current query provided in the Supplementary Note.
Specific analysis of eQTLs in subcutaneous adipose tissue
We analysed in depth the association of lead SNPs with gene expression in subcutaneous adipose tissue using two subcutaneous adipose tissue cis-eQTL datasets. The first dataset was generated by the EUROBATS consortium and consists of samples from well-phenotyped healthy female twins (n=766) with eQTLs derived as described previously.98 We also used the subcutaneous adipose tissue data (n=298) generated by the GTEx consortium (version 6),99 which were obtained from www.gtexportal.org on 20/11/15. GTEx results were limited to GENCODE “protein_coding” and “lincRNA” biotype transcripts, and only variants with a minor allele frequency >0.01 were used. Linkage disequilibrium statistics between the index SNP (lead SNP for fasting insulin at the locus) and the most significant expression SNP for the gene were calculated in PLINK 1.9 using 1000 Genomes phase 1 version 3 European ancestry samples.90 We also assessed the regulatory trait concordance (RTC) value for SNPs associated with gene expression in adipose tissue, in order to assess the likelihood of co-localisation of signal with the lead eQTL signal at that region.100 In brief, if the index variant and the eQTL do tag the same causal variant, it is expected that removing the genetic effect of the index variant will have a significant consequence on the eQTL association. To this end, the RTC method assesses the likelihood of a shared functional effect between a GWAS variant and an eQTL by quantifying the change in the statistical significance of the eQTL after correcting for the genetic effect of the index variant and comparing its correction impact to that of all other SNPs in the interval. We considered an RTC of ≥ 0.8 or high linkage disequilibrium between the lead eQTL SNP and trait-associated SNP (r2>0.8) to be supporting evidence of co-localisation.
Functional studies in mouse OP9-K cells
We sought to experimentally validate candidate causal genes at loci associated with lower levels of peripheral adiposity, higher risk of type 2 diabetes and with gene expression in subcutaneous adipose tissue (Supplementary Note and Supplementary Figure 11). We studied the effects of gene knockdown using siRNA on adipogenic differentiation in murine OP9-K cell lines. The mouse OP9-K cell line used in this study is a model suitable for mid-throughput screening of genes influencing adipogenesis. OP9-K cells are clonal cells derived from mouse stromal OP9 cells obtained from the bone marrow, that accumulate large lipid-droplets after 72 hours of adipogenic stimulation.45 OP9-K cells were grown and differentiated using oleic acid-containing differentiation media as described previously.45 For siRNA transfections, 2.5 x 104 cells per well were cultured in the 24-well dish. After 24hrs, cells were transfected with smartpool siRNA (from Dharmacon) against each gene using Optifect reagent as per the manufacturer protocol. On the following day, differentiation of OP9-K cells into adipocytes was initiated by replacing the media with 500 µL of differentiation media. After 48hrs of differentiation induction, differentiated cells were stained with adipored to assess lipid accumulation using fluorescent spectroscopy. For quantitative-PCR, total RNA was isolated from differentiated OP9-K cells and cDNA was synthesized using TaqMan® Fast Cells-to-CT™ Kit (Applied Biosystems) according to manufacturer's instructions. Quantitative real-time PCR analysis was performed on a TaqMan ABI Prism 7900 Sequence Detector System (Applied Biosystems). Expression results were analysed relative to GAPDH mRNA content in the same sample.
Supplementary Material
Acknowledgements
This study was funded by the United Kingdom’s Medical Research Council through grants MC_UU_12015/1, MC_PC_13046, MC_PC_13048 and MR/L00002/1. This work was supported by the MRC Metabolic Diseases Unit (MC_UU_12012/5) and the Cambridge NIHR Biomedical Research Centre and EU/EFPIA Innovative Medicines Initiative Joint Undertaking (EMIF grant: 115372). EPIC-InterAct Study funding: funding for the InterAct project was provided by the EU FP6 programme (grant number LSHM_CT_2006_037197). This work was funded, in part, through an EFSD Rising Star award to R.A.S. supported by Novo Nordisk. D. B. S. is supported by the Wellcome Trust grant n. 107064. M. McC. is a Wellcome Trust Senior Investigator and is supported by the following grants from the Wellcome Trust: 090532 and 098381. M. vdB. is supported by a Novo Nordisk postdoctoral fellowship run in partnership with the University of Oxford. I. B. is supported by the Wellcome Trust grant WT098051. S. O. R. acknowleges funding from the Wellcome Trust (Wellcome Trust Senior Investigator Award : 095515/Z/11/Z and Wellcome Trust Strategic Award: 100574/Z/12/Z).
We are grateful for OP9-K cells kindly shared by Alan Kopin’s laboratory, Tufts Medical Center, Boston, USA. The authors gratefully acknowledge the help of the MRC Epidemiology Unit Support Teams, including Field Teams, Laboratory Team and Data Management Team, and of the staff of the Wellcome Trust Clinical Research Facility.
EPIC-InterAct consortium authors
Claudia Langenberg(1), Robert A Scott(1), Stephen J Sharp(1), Nita G Forouhi(1), Nicola D Kerrison(1), Matt Sims(1), Debora ME Lucarelli(1), Inês Barroso(2,3), Panos Deloukas(2), Mark I McCarthy(5,6), Larraitz Arriola(12,13,14), Beverley Balkau(15,16), Aurelio Barricarte(14,17,18), Heiner Boeing(19), Paul W Franks(20,21), Carlos Gonzalez(22), Sara Grioni(23), Rudolf Kaaks(24), Timothy J Key(25), Carmen Navarro(14,26,27), Peter M Nilsson(28), Kim Overvad(29,30), Domenico Palli(31), Salvatore Panico(32), J. Ramón Quirós(33), Olov Rolandsson(21), Carlotta Sacerdote(34,35), Elena Salamanca-Fernández(14,36,37), Nadia Slimani(38), Anne Tjonneland(39), Rosario Tumino(40), Annemieke MW Spijkerman(41), Daphne L van der A(41), Yvonne T van der Schouw(42), Elio Riboli(43), Nicholas J Wareham(1).
EPIC-InterAct consortium affiliations: (12) Public Health Division of Gipuzkoa, San Sebastian, Spain, (13) Instituto BIO-Donostia, Basque Government, San Sebastian, Spain, (14) CIBER Epidemiología y Salud Pública (CIBERESP), Spain, (15) Inserm, CESP, U1018, Villejuif, France, (16) Univ Paris-Sud, UMRS 1018, Villejuif, France, (17) Navarre Public Health Institute (ISPN), Pamplona, Spain, (18) Navarra Institute for Health Research (IdiSNA) Pamplona, Spain, (19) German Institute of Human Nutrition Potsdam-Rehbruecke, Germany, (20) Lund University, Malmö, Sweden, (21) Umeå University, Umeå, Sweden, (22) Catalan Institute of Oncology (ICO), Barcelona, Spain, (23) Epidemiology and Prevention Unit, Milan, Italy, (24) German Cancer Research Centre (DKFZ), Heidelberg, Germany, (25) Nuffield Department of Population Health, University of Oxford, Oxford, United Kingdom, (26) Department of Epidemiology, Murcia Regional Health Council, IMIB-Arrixaca, Murcia, Spain, (27) Unit of Preventive Medicine and Public Health, School of Medicine, University of Murcia, Murcia, Spain, (28) Department of Clinical Sciences, Lund University, Skane University Hospital, Malmo, Sweden, (29) Department of Public Health, Section for Epidemiology, Aarhus University, Aarhus, Denmark, (30) Aalborg University Hospital, Aalborg, Denmark, (31) Cancer Research and Prevention Institute (ISPO), Florence, Italy, (32) Dipartimento di Medicina Clinica e Chirurgia, Federico II University, Naples, Italy, (33) Public Health Directorate, Asturias, Spain, (34) Unit of Cancer Epidemiology, Citta' della Salute e della Scienza Hospital-University of Turin and Center for Cancer Prevention (CPO), Torino, Italy, (35) Human Genetics Foundation (HuGeF), Torino, Italy, (36) Andalusian School of Public Health, Granada, Spain, (37) Instituto de Investigación Biosanitaria de Granada (Granada.ibs), Granada, Spain, (38) International Agency for Research on Cancer, Lyon, France, (39) Danish Cancer Society Research Center, Copenhagen, Denmark, (40) ASP Ragusa, Ragusa, Italy, (41) National Institute for Public Health and the Environment (RIVM), Bilthoven, The Netherlands, (42) University Medical Center Utrecht, Utrecht, the Netherlands, (43) School of Public Health, Imperial College London, London, United Kingdom.
Cambridge FPLD1 consortium authors
Robert Semple(2), Claire Adams(2), Anna Stears(44), Stella George(45), Mark Walker(46), Ellie Gurnell(44), Deirdre Maguire(47), Rasha Mukhtar(48), Sath Nag(48), Amanda Adler(44), Maarten Soeters(49), Ken Laji(50), Alistair Watt(51), Simon Aylwin(52), Andrew Johnson(53), Gerry Rayman(54), Fahmy Hanna(55), Sian Ellard(56), Richard Ross(57), Kristina Blaslov(58), Lea Smirčić Duvnjak(58), Stephen O’Rahilly(2), David B. Savage(2).
Cambridge FPLD1 consortium affiliations: (44) Wolfson Diabetes and Endocrine Clinic, Institute of Metabolic Science, Addenbrooke's Hospital, Cambridge University Hospitals NHS Foundation Trust, Cambridge, United Kingdom, (45) East and North Herts NHS Trust, Lister Hospital, Herts, United Kingdom, (46) Institute of Cellular Medicine (Diabetes), Newcastle University Medical School, Newcastle upon Tyne, United Kingdom, (47) Harrogate and District Hospital, Harrogate, United Kingdom, (48) James Cook University Hospital, Middlesborough, United Kingdom, (49) Department of Endocrinology & Metabolism, Internal Medicine, Academic Medical Center, Amsterdam, The Netherlands, (50) St Richard's Hopsital, Spitalfield Lane, Chichester, United Kingdom, (51) North Devon District Hospital, Raleigh Park, Barnstaple, United Kingdom, (52) King's College Hospital, London, United Kingdom, (53) Department of Diabetes and Endocrinology, Southmead Hospital, Bristol, United Kingdom, (54) Ipswich Hospital, Ipswich, United Kingdom, (55) University Hospital of North Midlands NHS Trust, Royal Stoke University Hospital, Stoke-on-Trent, United Kingdom, (56) Royal Devon and Exeter NHS Foundation Trust, Exeter, United Kingdom, (57) The University of Sheffield, The Medical School, Sheffield, United Kingdom, (58) Vuk Vrhovac University Clinic for Diabetes, Endocrinology and Metabolic diseases, Zagreb, Croatia.
Footnotes
Data Access Statement
This research has been conducted using the UK Biobank resource. Data on glycemic traits were contributed by the MAGIC consortium investigators. Associations with type 2 diabetes were obtained from the DIAGRAM (DIAbetes Genetics Replication And Meta-analysis) consortium investigators. Data on coronary heart disease / myocardial infarction have been contributed by the CARDIoGRAMplusC4D consortium investigators. Data on body mass index, waist, hip, waist-to-hip ratio were contributed by the GIANT consortium investigators. Data about triglycerides and high-density lipoprotein cholesterol were contributed by the Global Lipids Genetics Consortium investigators. We are very grateful to the GENESIS consortium for provision of summary statistic results for clamp- or insulin suppression test-based insulin resistance. The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about. Raw exome sequence data from FPLD1 individuals and family members is available from the European Genome-Phenome Archive (https://www.ebi.ac.uk/ega/home, see full accession codes in Supplementary Table 7). Understanding Society: The UK Household Longitudinal Study is led by the Institute for Social and Economic Research at the University of Essex and funded by the Economic and Social Research Council. The survey was conducted by NatCen and the genome-wide scan data were analysed and deposited by the Wellcome Trust Sanger Institute. Information on how to access the data can be found at https://www.understandingsociety.ac.uk/. Genome-wide genotyping data of 5,296 unrelated women from UKHLS is publically available through the European Genome-phenome Archive (Dataset Accession: EGAD00010000891).
Data download:
MAGIC consortium (http://www.magicinvestigators.org/)
GLGC consortium (http://csg.sph.umich.edu//abecasis/public/lipids2013/)
GIANT (https://www.broadinstitute.org/collaboration/giant/)
DIAGRAM consortium (http://diagram-consortium.org/)
CARDIoGRAMplusC4D (http://www.cardiogramplusc4d.org/)
Exome Aggregation Consortium (http://exac.broadinstitute.org/about)
European Genome-Phenome Archive (https://www.ebi.ac.uk/ega/home)
Study websites:
Fenland (http://www.mrc-epid.cam.ac.uk/research/studies/fenland/)
EPIC-Norfolk (http://www.srl.cam.ac.uk/epic/)
EPIC-InterAct (http://www.inter-act.eu/)
UK Biobank (http://www.ukbiobank.ac.uk/)
Understanding Society: The UK Household Longitudinal Study (https://www.understandingsociety.ac.uk/)
Author Contributions
Concept and design: L. A. Lotta, I. Barroso, N. J. Wareham, D. B. Savage, C. Langenberg, S. O’Rahilly, R. A. Scott. Generation, acquisition, analysis, or interpretation of data: all authors. Drafting of the manuscript: L. A. Lotta, I. Barroso, N. J. Wareham, D. B. Savage, C. Langenberg, S. O’Rahilly, R. A. Scott. Critical review of the manuscript for important intellectual content and approval of the final version of the manuscript: all authors.
Competing Financial Interests Statement
The authors report no conflict of interest relative to this study.
References
- 1.Samuel VT, Shulman GI. The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. The Journal of clinical investigation. 2016;126:12–22. doi: 10.1172/JCI77812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ginsberg HN. Insulin resistance and cardiovascular disease. The Journal of clinical investigation. 2000;106:453–458. doi: 10.1172/JCI10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shulman GI. Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. The New England journal of medicine. 2014;371:1131–1141. doi: 10.1056/NEJMra1011035. [DOI] [PubMed] [Google Scholar]
- 4.Lillioja S, Bogardus C. Obesity and insulin resistance: lessons learned from the Pima Indians. Diabetes/metabolism reviews. 1988;4:517–540. doi: 10.1002/dmr.5610040508. [DOI] [PubMed] [Google Scholar]
- 5.Perry RJ, Samuel VT, Petersen KF, Shulman GI. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature. 2014;510:84–91. doi: 10.1038/nature13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arner P. The adipocyte in insulin resistance: key molecules and the impact of the thiazolidinediones. Trends in endocrinology and metabolism: TEM. 2003;14:137–145. doi: 10.1016/s1043-2760(03)00024-9. [DOI] [PubMed] [Google Scholar]
- 7.Friedman JM. Obesity in the new millennium. Nature. 2000;404:632–634. doi: 10.1038/35007504. [DOI] [PubMed] [Google Scholar]
- 8.Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nature reviews. Molecular cell biology. 2008;9:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman JM. A war on obesity, not the obese. Science. 2003;299:856–858. doi: 10.1126/science.1079856. [DOI] [PubMed] [Google Scholar]
- 10.Hardy OT, Czech MP, Corvera S. What causes the insulin resistance underlying obesity? Current opinion in endocrinology, diabetes, and obesity. 2012;19:81–87. doi: 10.1097/MED.0b013e3283514e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefan N, Haring HU, Hu FB, Schulze MB. Metabolically healthy obesity: epidemiology, mechanisms, and clinical implications. Lancet Diabetes Endocrinol. 2013;1:152–162. doi: 10.1016/S2213-8587(13)70062-7. [DOI] [PubMed] [Google Scholar]
- 12.Robbins AL, Savage DB. The genetics of lipid storage and human lipodystrophies. Trends in molecular medicine. 2015;21:433–438. doi: 10.1016/j.molmed.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Semple RK, Savage DB, Cochran EK, Gorden P, O'Rahilly S. Genetic syndromes of severe insulin resistance. Endocrine reviews. 2011;32:498–514. doi: 10.1210/er.2010-0020. [DOI] [PubMed] [Google Scholar]
- 14.Samuel VT, Petersen KF, Shulman GI. Lipid-induced insulin resistance: unravelling the mechanism. Lancet. 2010;375:2267–2277. doi: 10.1016/S0140-6736(10)60408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danforth E., Jr Failure of adipocyte differentiation causes type II diabetes mellitus? Nature genetics. 2000;26:13. doi: 10.1038/79111. [DOI] [PubMed] [Google Scholar]
- 16.Unger RH. Lipid overload and overflow: metabolic trauma and the metabolic syndrome. Trends in endocrinology and metabolism: TEM. 2003;14:398–403. doi: 10.1016/j.tem.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Virtue S, Vidal-Puig A. Adipose tissue expandability, lipotoxicity and the Metabolic Syndrome--an allostatic perspective. Biochimica et biophysica acta. 2010;1801:338–349. doi: 10.1016/j.bbalip.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Shulman GI. Cellular mechanisms of insulin resistance. The Journal of clinical investigation. 2000;106:171–176. doi: 10.1172/JCI10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karpe F, Pinnick KE. Biology of upper-body and lower-body adipose tissue--link to whole-body phenotypes. Nature reviews. Endocrinology. 2015;11:90–100. doi: 10.1038/nrendo.2014.185. [DOI] [PubMed] [Google Scholar]
- 20.Robbins DC, et al. The effect of diet on thermogenesis in acquired lipodystrophy. Metabolism: clinical and experimental. 1979;28:908–916. doi: 10.1016/0026-0495(79)90090-8. [DOI] [PubMed] [Google Scholar]
- 21.Shimomura I, Hammer RE, Ikemoto S, Brown MS, Goldstein JL. Leptin reverses insulin resistance and diabetes mellitus in mice with congenital lipodystrophy. Nature. 1999;401:73–76. doi: 10.1038/43448. [DOI] [PubMed] [Google Scholar]
- 22.Oral EA, et al. Leptin-replacement therapy for lipodystrophy. The New England journal of medicine. 2002;346:570–578. doi: 10.1056/NEJMoa012437. [DOI] [PubMed] [Google Scholar]
- 23.Gavrilova O, et al. Surgical implantation of adipose tissue reverses diabetes in lipoatrophic mice. The Journal of clinical investigation. 2000;105:271–278. doi: 10.1172/JCI7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JY, et al. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. The Journal of clinical investigation. 2007;117:2621–2637. doi: 10.1172/JCI31021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray SL, et al. Leptin deficiency unmasks the deleterious effects of impaired peroxisome proliferator-activated receptor gamma function (P465L PPARgamma) in mice. Diabetes. 2006;55:2669–2677. doi: 10.2337/db06-0389. [DOI] [PubMed] [Google Scholar]
- 26.Medina-Gomez G, et al. PPAR gamma 2 prevents lipotoxicity by controlling adipose tissue expandability and peripheral lipid metabolism. PLoS genetics. 2007;3:e64. doi: 10.1371/journal.pgen.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knowles JW, et al. Identification and validation of N-acetyltransferase 2 as an insulin sensitivity gene. The Journal of clinical investigation. 2016;126:403. doi: 10.1172/JCI85921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott RA, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nature genetics. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manning AK, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nature genetics. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salazar MR, et al. Comparison of the abilities of the plasma triglyceride/high-density lipoprotein cholesterol ratio and the metabolic syndrome to identify insulin resistance. Diabetes & vascular disease research. 2013;10:346–352. doi: 10.1177/1479164113479809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott RA, et al. Common genetic variants highlight the role of insulin resistance and body fat distribution in type 2 diabetes, independent of obesity. Diabetes. 2014;63:4378–4387. doi: 10.2337/db14-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nature genetics. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prokopenko I, et al. A central role for GRB10 in regulation of islet function in man. PLoS genetics. 2014;10:e1004235. doi: 10.1371/journal.pgen.1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nature genetics. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikpay M, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nature genetics. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozaki K, et al. Mutational analysis of human lipoprotein lipase by carboxy-terminal truncation. Journal of lipid research. 1993;34:1765–1772. [PubMed] [Google Scholar]
- 38.Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. The New England journal of medicine. 2016 doi: 10.1056/NEJMoa1507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mailly F, et al. A common variant in the gene for lipoprotein lipase (Asp9-->Asn). Functional implications and prevalence in normal and hyperlipidemic subjects. Arteriosclerosis, thrombosis, and vascular biology. 1995;15:468–478. doi: 10.1161/01.atv.15.4.468. [DOI] [PubMed] [Google Scholar]
- 40.Avila M, et al. Clinical reappraisal of SHORT syndrome with PIK3R1 mutations: towards recommendation for molecular testing and management. Clinical genetics. 2015 doi: 10.1111/cge.12688. [DOI] [PubMed] [Google Scholar]
- 41.Thauvin-Robinet C, et al. PIK3R1 mutations cause syndromic insulin resistance with lipoatrophy. American journal of human genetics. 2013;93:141–149. doi: 10.1016/j.ajhg.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chudasama KK, et al. SHORT syndrome with partial lipodystrophy due to impaired phosphatidylinositol 3 kinase signaling. American journal of human genetics. 2013;93:150–157. doi: 10.1016/j.ajhg.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dyment DA, et al. Mutations in PIK3R1 cause SHORT syndrome. American journal of human genetics. 2013;93:158–166. doi: 10.1016/j.ajhg.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pers TH, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nature communications. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lane JM, Doyle JR, Fortin JP, Kopin AS, Ordovas JM. Development of an OP9 derived cell line as a robust model to rapidly study adipocyte differentiation. PloS one. 2014;9:e112123. doi: 10.1371/journal.pone.0112123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yaghootkar H, et al. Genetic evidence for a normal-weight "metabolically obese" phenotype linking insulin resistance, hypertension, coronary artery disease, and type 2 diabetes. Diabetes. 2014;63:4369–4377. doi: 10.2337/db14-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu Y, et al. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nature communications. 2016;7:10495. doi: 10.1038/ncomms10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kilpelainen TO, et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nature genetics. 2011;43:753–760. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yaghootkar H, et al. Genetic evidence for a link between favorable adiposity and lower risk of type 2 diabetes, hypertension and heart disease. Diabetes. 2016 doi: 10.2337/db15-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biggs ML, et al. Association between adiposity in midlife and older age and risk of diabetes in older adults. JAMA : the journal of the American Medical Association. 2010;303:2504–2512. doi: 10.1001/jama.2010.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pischon T, et al. General and abdominal adiposity and risk of death in Europe. The New England journal of medicine. 2008;359:2105–2120. doi: 10.1056/NEJMoa0801891. [DOI] [PubMed] [Google Scholar]
- 52.Vague J. The degree of masculine differentiation of obesities: a factor determining predisposition to diabetes, atherosclerosis, gout, and uric calculous disease. The American journal of clinical nutrition. 1956;4:20–34. doi: 10.1093/ajcn/4.1.20. [DOI] [PubMed] [Google Scholar]
- 53.Smith U. Abdominal obesity: a marker of ectopic fat accumulation. The Journal of clinical investigation. 2015;125:1790–1792. doi: 10.1172/JCI81507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Despres JP, Lemieux I. Abdominal obesity and metabolic syndrome. Nature. 2006;444:881–887. doi: 10.1038/nature05488. [DOI] [PubMed] [Google Scholar]
- 55.Dahlman I, et al. Numerous Genes in Loci Associated With Body Fat Distribution Are Linked to Adipose Function. Diabetes. 2016;65:433–437. doi: 10.2337/db15-0828. [DOI] [PubMed] [Google Scholar]
- 56.Ryden M, Andersson DP, Bergstrom IB, Arner P. Adipose tissue and metabolic alterations: regional differences in fat cell size and number matter, but differently: a cross-sectional study. The Journal of clinical endocrinology and metabolism. 2014;99:E1870–1876. doi: 10.1210/jc.2014-1526. [DOI] [PubMed] [Google Scholar]
- 57.Pinnick KE, et al. Distinct developmental profile of lower-body adipose tissue defines resistance against obesity-associated metabolic complications. Diabetes. 2014;63:3785–3797. doi: 10.2337/db14-0385. [DOI] [PubMed] [Google Scholar]
- 58.Baughman BM, Pattenden SG, Norris JL, James LI, Frye SV. The L3MBTL3 Methyl-Lysine Reader Domain Functions As a Dimer. ACS chemical biology. 2016;11:722–728. doi: 10.1021/acschembio.5b00632. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Randall JC, et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS genetics. 2013;9:e1003500. doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paternoster L, et al. Adult height variants affect birth length and growth rate in children. Human molecular genetics. 2011;20:4069–4075. doi: 10.1093/hmg/ddr309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wood AR, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nature genetics. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta GD, et al. A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell. 2015;163:1484–1499. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singaraja RR, et al. Identification of four novel genes contributing to familial elevated plasma HDL cholesterol in humans. Journal of lipid research. 2014;55:1693–1701. doi: 10.1194/jlr.M048710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chasman DI, et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS genetics. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DeFronzo RA, et al. Pioglitazone for diabetes prevention in impaired glucose tolerance. The New England journal of medicine. 2011;364:1104–1115. doi: 10.1056/NEJMoa1010949. [DOI] [PubMed] [Google Scholar]
- 66.DREAM (Diabetes REduction Assessment with ramipril and rosiglitazone Medication) Trial Investigators. Effect of rosiglitazone on the frequency of diabetes in patients with impaired glucose tolerance or impaired fasting glucose: a randomised controlled trial. Lancet. 2006;368:1096–1105. doi: 10.1016/S0140-6736(06)69420-8. [DOI] [PubMed] [Google Scholar]
- 67.Lincoff AM, Wolski K, Nicholls SJ, Nissen SE. Pioglitazone and risk of cardiovascular events in patients with type 2 diabetes mellitus: a meta-analysis of randomized trials. JAMA : the journal of the American Medical Association. 2007;298:1180–1188. doi: 10.1001/jama.298.10.1180. [DOI] [PubMed] [Google Scholar]
- 68.Kernan WN, et al. Pioglitazone after Ischemic Stroke or Transient Ischemic Attack. The New England journal of medicine. 2016 doi: 10.1056/NEJMoa1506930. [DOI] [PubMed] [Google Scholar]
- 69.Nissen SE, Wolski K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. The New England journal of medicine. 2007;356:2457–2471. doi: 10.1056/NEJMoa072761. [DOI] [PubMed] [Google Scholar]
- 70.Food and Drug Administration. Guidance for Industry. Diabetes Mellitus — Evaluating Cardiovascular Risk in New Antidiabetic Therapies to Treat Type 2 Diabetes. 2008 URL: http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm071627.pdf.
- 71.Swerdlow DI, et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet. 2015;385:351–361. doi: 10.1016/S0140-6736(14)61183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dewey FE, et al. Inactivating Variants in ANGPTL4 and Risk of Coronary Artery Disease. The New England journal of medicine. 2016 doi: 10.1056/NEJMoa1510926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Do R, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jorgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjaerg-Hansen A. Loss:of-function mutations in APOC3 and risk of ischemic vascular disease. The New England journal of medicine. 2014;371:32–41. doi: 10.1056/NEJMoa1308027. [DOI] [PubMed] [Google Scholar]
- 75.TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. The New England journal of medicine. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Myocardial Infarction Genetics Consortium Investigators. Inactivating mutations in NPC1L1 and protection from coronary heart disease. The New England journal of medicine. 2014;371:2072–2082. doi: 10.1056/NEJMoa1405386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moltke I, et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature. 2014;512:190–193. doi: 10.1038/nature13425. [DOI] [PubMed] [Google Scholar]
- 78.Day N, et al. EPIC-Norfolk: study design and characteristics of the cohort. European Prospective Investigation of Cancer. British journal of cancer. 1999;80(Suppl 1):95–103. [PubMed] [Google Scholar]
- 79.Riboli E, et al. European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public health nutrition. 2002;5:1113–1124. doi: 10.1079/PHN2002394. [DOI] [PubMed] [Google Scholar]
- 80.InterAct Consortium et al. Design and cohort description of the InterAct Project: an examination of the interaction of genetic and lifestyle factors on the incidence of type 2 diabetes in the EPIC Study. Diabetologia. 2011;54:2272–2282. doi: 10.1007/s00125-011-2182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Collins R. What makes UK Biobank special? Lancet. 2012;379:1173–1174. doi: 10.1016/S0140-6736(12)60404-8. [DOI] [PubMed] [Google Scholar]
- 82.Lyssenko V, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. The Journal of clinical investigation. 2007;117:2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aschard H, Vilhjalmsson BJ, Joshi AD, Price AL, Kraft P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. American journal of human genetics. 2015;96:329–339. doi: 10.1016/j.ajhg.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Day FR, Loh PR, Scott RA, Ong KK, Perry JR. A Robust Example of Collider Bias in a Genetic Association Study. American journal of human genetics. 2016;98:392–393. doi: 10.1016/j.ajhg.2015.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genetic epidemiology. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lawrence RD. Types of human diabetes. British medical journal. 1951;1:373–375. doi: 10.1136/bmj.1.4703.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Herbst KL, et al. Kobberling type of familial partial lipodystrophy: an underrecognized syndrome. Diabetes care. 2003;26:1819–1824. doi: 10.2337/diacare.26.6.1819. [DOI] [PubMed] [Google Scholar]
- 88.Payne F, et al. Hypomorphism in human NSMCE2 linked to primordial dwarfism and insulin resistance. The Journal of clinical investigation. 2014;124:4028–4038. doi: 10.1172/JCI73264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.UK10K Consortium. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nature methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 92.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature genetics. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 93.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic acids research. 2012;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johnson AD, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic acids research. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang X, et al. Synthesis of 53 tissue and cell line expression QTL datasets reveals master eQTLs. BMC genomics. 2014;15:532. doi: 10.1186/1471-2164-15-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Buil A, et al. Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nature genetics. 2015;47:88–91. doi: 10.1038/ng.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nica AC, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS genetics. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.