Abstract
Dermatophytes are the group of filamentous fungi infecting keratinized structures such as skin, hair, and nails. Knowledge about genes and molecular mechanisms responsible for pathogenicity, as well as other biological properties of Microsporum canis is still relatively poor. The qRT-PCR is a reliable technique for quantifying gene expression across various biological processes, and choosing a set of suitable reference genes to normalize the expression data is a crucial step of this technique. We investigated the suitability of nine candidate reference genes: β-act, β-tub, adp-rf, ef1-α, sdha, rpl2, mbp1, psm1, and rGTPa for gene expression analysis in the dermatophyte M. canis in response to different carbon sources, phosphate levels, and pH shifts - factors that are extremely important and necessary for growth of dermatophyte in the host tissue. The transcription stability of these genes was evaluated using NormFinder, geNorm, BestKeeper, and RefFinder software. Regarding expression stability, mbp1, β-act, and sdha were the most stable housekeeping genes which we recommend for future qRT-PCR studies on M. canis strains. To the best of our knowledge this is the first study on selection and validation of reference genes for qRT-PCR data normalization in M. canis growth in culture media which promote adhesion-inducing conditions.
Introduction
Microsporum canis is a member of dermatophytes – a group of pathogenic fungi able to invade keratinized structures, leading to infection of skin, hair, and nails. This species is distributed worldwide and in many areas such as Central and Southern Europe, Middle East, North Africa, South America, and China it is the most prevalent dermatophyte1–11 responsible for tinea corporis and tinea capitis in humans and animals12. The disease mainly affects preadolescents, the elderly, as well as immunocompromised individuals, including AIDS patients and transplant recipients13–16, and manifests by severe scalp itching, hair loss as well as skin scaling, especially around hair shafts17,18. Eradication of the disease is difficult because human asymptomatic carriers are common, moreover pets such as cats, dogs, and rabbits may transmit M. canis to humans1,19–21. Molecular mechanisms of interaction between dermatophytes and host cells are still poorly understood. Therefore, there is an explicit need for identification of factors involved in dermatophyte pathogenicity, and the logical starting point is identification of genes which expression increases or attenuates in response to different environmental stimuli typical for the stage of invasion. Analysis of expression profiles of such genes may enable to study more thoroughly virulence factors and mechanisms of resistance against common antifungal drugs. M. canis genome sequencing22 improved functional genomic studies to identify factors involved in dermatophyte pathogenicity. Dermatophyte adherence and secretion of enzymes are the key factors in colonization of the host tissues, which may be regulated in response to different carbon sources, phosphate levels, and ambient pH shifts23. Gene expression profiling is an effective means of studying response to different environmental stimuli and a necessary component in identifying genes and regulatory mechanisms associated with biological processes in any organism. Choosing a proper reference gene remains one of the golden rules to increase the credibility of qRT-PCR data interpretation23. Due to the limited knowledge on such genes suitable for expression analysis in dermatophytes24 we investigated the transcription level of a group of nine candidate housekeeping genes: β-act (β-actin), β-tub (β-tubulin), adp-rf (ADP ribosylation factor), ef1-α (elongation factor 1-alpha), sdha (succinate dehydrogenase complex flavoprotein subunit A), rpl2 (ribosomal protein L2), mbp1 (multiubiquitin chain binding protein 1), psm1 (mitotic cohesion complex subunit Psm1), rGTPa (rho GTPase activating-protein 5) (Table 1) as reference genes for selected clinical strain of M. canis grown under different adhesion-inducing environmental stimuli typical for the stage of host infection, such as: different carbon sources (glucose, keratin, keratin with soy protein, elastin, collagen, colloidal chitin, keratinocyte free medium), low-Pi environments, pH shifts. The candidate reference genes were selected from among endogenous controls in some species of fungi as well as in other organisms22,24 and to the best of our knowledge this is the first such complex search in case of dermatophyte species. The stability of each candidate reference gene was evaluated by algorithms: geNorm module of qbase + (Biogazelle), NormFinder, BestKeeper, and RefFinder web-based comprehensive tool.
Table 1.
Microsporum canis candidate reference genes used for qRT-PCR.
| Gene symbol/accession no. | Gene name | Primers (5′-3′) forward reverse | Length bp | Tm [°C] | Ct range | Efficiency (%) | R2 |
|---|---|---|---|---|---|---|---|
|
β-act
XM_002845542 |
β-actin | CTCCTGAGGCTCTCTTCC GTAGTACCGCCGGACATG |
142 | 60.5 | 15.08–18.70 | 110 | 0.99936 |
|
β-tub
XM_002848601 |
β-tubulin | AAGAGTTCCCAGACCGTATG TGTTGTACAAGGCCTCATTG |
159 | 60.5 | 17.86–22.57 | 109 | 0.99975 |
|
adp-rf
XM_002848521 |
ADP ribosylation factor | GAATTCTCATGGTCGGTCTC AACGTTGAATCCGATGGTG |
104 | 60.5 | 15.93–20.24 | 100 | 0.99855 |
|
ef1-α
XM_002850842 |
elongation factor 1-alpha | CCTAAGTTCGTCAAGTCTGG CTTCTCGACAGCCTTGATG |
159 | 60.5 | 16.43–22.41 | 104 | 0.99798 |
|
sdha
XM_002843730 |
succinate dehydrogenase complex flavoprotein subunit A | TCTAGGAAACATGCACAAGG TTCGATAACACTCTGAGGGG |
127 | 60.5 | 16.05–19.80 | 103 | 0.99895 |
|
rpl2
XM_002844911 |
ribosomal protein L2 | GATCTATATTCACGGCTCGC ATGATGTTCTTCACGACACC |
109 | 60.5 | 19.40–25.26 | 107 | 0.99912 |
|
mbp1
XM_002843632 |
multiubiquitin chain binding protein 1 | AGTCCTAGTTACCTTGACCG CGGTGTTTAAGTGCTAGATAGG |
123 | 60.5 | 18.73–22.18 | 99 | 0.99924 |
|
psm1
XM_002844171 |
mitotic cohesion complex subunit Psm1 | AGCGTACCTGGATATTGAAG GGATAGCGAATAACAGAGCC |
149 | 60.5 | 22.84–26.65 | 104 | 0.99980 |
|
rGTPa
XM_002843982 |
rho GTPase activating-protein 5 | GACTCCCTCTGGCATATTTG ATCGGTTGCTTTCTCCTTC |
160 | 60.5 | 16.44–18.97 | 104 | 0.99980 |
Results
Standard curve, PCR efficiency, and product specificity
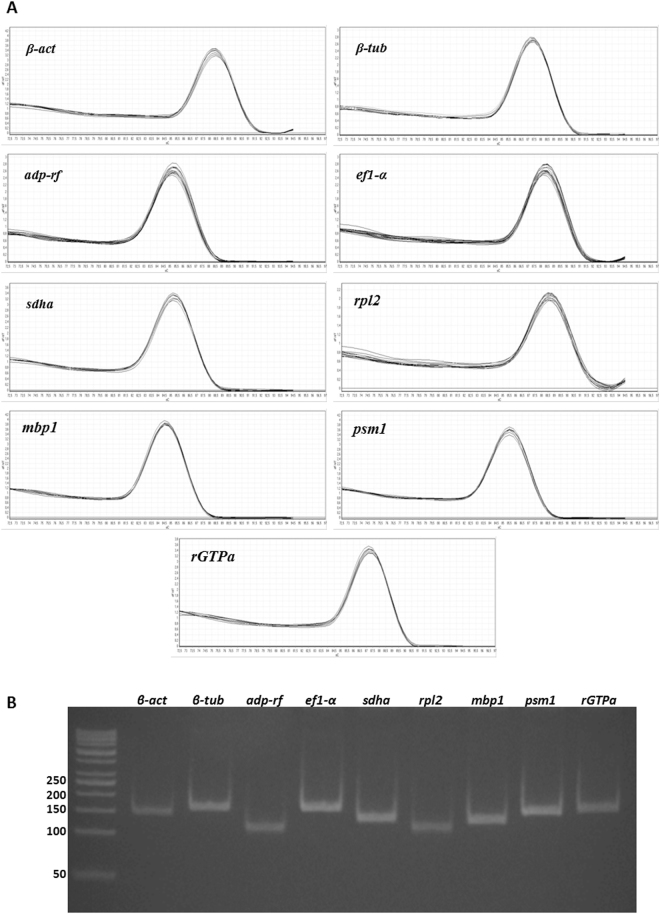
The cDNA samples were obtained using total RNA isolated from three independent repetitions of M. canis cultures subjected to 30 environmental conditions (Supplementary Table S1). The expression of nine reference gene candidates (Table 1) was evaluated in control (M. canis cultivated in MM-Cove medium) and experimental conditions (Table S1). The raw Ct values (Supplementary Table S1) were used to calculate the mean Ct for each amplicon in each sample. The candidate reference genes exhibited Ct values ranging from 15.08–26.65 (Fig. 1, Table 1). The calculated efficiencies for the candidate reference genes, shown in Table 1, were between 99–110%. The efficiency curves for nine analyzed candidate genes were found to have linear correlation coefficients (R2) ranging from 0.997–0.999. Melt peak analysis demonstrated a single homogenous peak for all primer sets (Fig. 2A). Polyacrylamide gel electrophoresis analysis of the amplified products for all primer sets revealed single bands of the expected size (Table 1, Fig. 2B).
Figure 1.
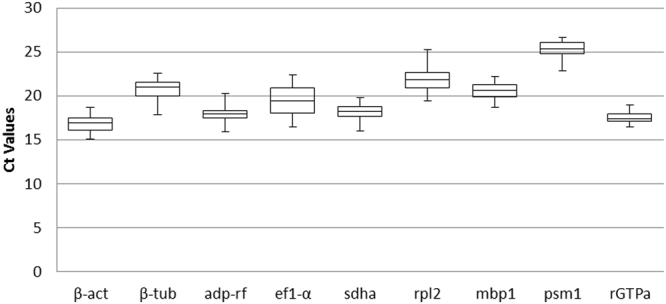
Expression level of nine reference genes in M. canis. The mean Ct values for all experimental conditions of each candidate reference gene are shown as box plot representations. The box indicates the 25th and 75th percentiles, the line across the box represents the median and whisker caps the maximum and minimum values.
Figure 2.
Melting curves of the nine M. canis candidate reference genes show single peaks (A). 8% polyacrylamide gel electrophoresis indicated the amplification of a single product of the expected size for nine reference genes (B).
Analysis of candidate reference gene expression using geNorm, NormFinder, BestKeeper, and RefFinder
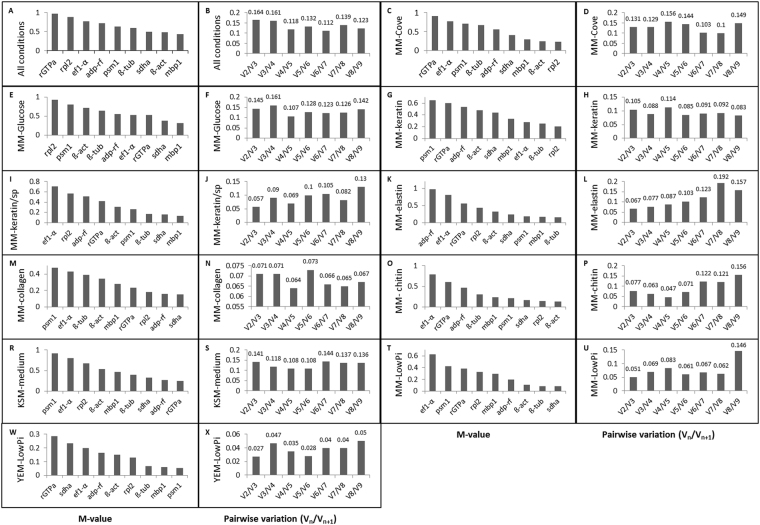
The GeNorm module of qbase + (Biogazelle) ranks the reference genes based on the stability value (M-value). The lower M-value corresponds to the more stable gene. In order to examine the minimal number of genes required for reliable normalization, the V-value for all gene pairs was calculated (Vn/Vn+1) between two subsequent normalization factors (NFn and NFn+1) with the cut-off value set at 0.15. Analyses were conducted using eleven groups of samples: A and B – all experimental conditions; C and D - growth in MM-Cove medium (control medium); E and F - growth in MM-Cove medium with glucose; G and H - growth in MM-Cove medium with keratin; I and J - growth in MM-Cove medium with keratin/soy protein; K and L - growth in MM-Cove medium with elastin; M and N - growth in MM-Cove medium with collagen; O and P - growth in MM-Cove medium with colloidal chitin; R and S - growth in KSM medium; T and U - growth in MM-LowPi medium; W and X - growth in YEM-LowPi medium. mbp1, β-act, sdha, and β-tub genes were the most stable genes under all tested conditions with mbp1 ranked as the best reference gene (M-value = 0.438). On the other hand rGTPa was the least stable gene (M-value = 0.968) in all analyzed samples (Fig. 3A). The pairwise variation (Vn/Vn+1) indcated that the use of four reference genes was reliable for normalization (V4/5 = 0.118) (Fig. 3B). The stability analysis of nine candidate reference genes of M. canis cultivated on control medium (MM-Cove) showed that rpl2 and β-act were the most stable ones (Fig. 3C), and the use of two reference genes was adequate to achieve credible data normalization (V2/3 = 0.131) (Fig. 3D). Given that mbp1 and sdha genes presented the most stable expression level in the presence of glucose in a culture medium (Fig. 3E), we estimated according to pairwise variation (Vn/Vn+1) that the use of two reference gene gave reliable results for qRT-PCR data normalization (V2/3 = 0.145) (Fig. 3F). The stability analysis with geNorm module showed that rpl2 and β-tub genes were the most stable when keratin was the main substrate during M. canis cultivation (Fig. 3G), and according to the pairwise variation (Vn/Vn+1) the use of these two reference genes was sufficient for the process of proper normalization (V2/3 = 0.105) (Fig. 3H). In the presence of keratin and soy protein in cultivation medium, mbp1, and sdha reference genes of M. canis reached the lowest M-value (Fig. 3I), and the use of two reference genes was suitable to achieve the best normalization data (V2/3 = 0.057) (Fig. 3J). The stability analysis of M. canis candidate reference genes expressed in MM-Cove medium supplemented with elastin showed that β-tub and mbp1 genes were the most stable (Fig. 3K), while analysis of pairwise variation (Vn/Vn+1) revealed that the use of two reference genes was the best combination for normalization results (V2/3 = 0.067) (Fig. 3L). sdha and adp-rf were the most stable candidate reference genes of M. canis in the presence of collagen in a culture medium (Fig. 3M). Pairwise variation (Vn/Vn+1) demonstrated that the use of these two reference genes was reliable for accurate normalization (V2/3 = 0.071), moreover addition of one more gene (rpl2) resulted in variation of such normalization factor (V3/4 = 0.071) (Fig. 3N). β-act and rpl2 reference genes were the most stable (Fig. 3O) when M. canis was cultivated on MM-Cove medium supplemented with colloidal chitin. According to (Vn/Vn+1) analysis the use of two genes was credible for normalization (V2/3 = 0.077) (Fig. 3P). rGTPa and adp-rf genes were ranked by geNorm as the best reference genes in Keratinocyte-SFM medium (KSM) containing L-glutamine (Fig. 3R), as the use of these genes resulted in accurate normalization (V2/3 = 0.141) (Fig. 3S). Sdha and β-tub were found to be the most stable genes in the presence of low concentration of organic Pi in MM-Cove medium at different pH (Fig. 3T) while psm1 and mbp1 were the most stable genes in the presence of low concentration of organic Pi in YEM- medium at different pH (Fig. 3W). In both cases, the use of two reference genes combination provided the best normalization (V2/3 = 0.051, and V2/3 = 0.027, respectively) (Figs. 3U, 3x). Summarizing, our analysis using geNorm module algorithms showed that mbp1, β-act, sdha, and β-tub with an M-value of 0.438, 0.478, 0.496, and 0.593, respectively (Table 2) were overall the most stable candidates for normalization of M. canis target gene expression across all experimental conditions. However, different sample sets required individual, most stable reference gene combinations (Fig. 3), what emphasizes the importance of such reference genes identification before qRT-PCR analysis.
Figure 3.
Gene expression stability (M-value) and pairwise variation (Vn/Vn+1) of the Microsporum canis candidate reference genes analyzed by geNorm.
Table 2.
Ranking of M. canis candidate reference genes ordered by their expression stability, determined by geNorm, NormFinder, BestKeeper, and RefFinder for all experimental conditions.
| Rank | geNorm | NormFinder | BestKeeper | RefFinder | ||||
|---|---|---|---|---|---|---|---|---|
| Gene | Expression Stability (M) | Gene | Stability Value (p) | Gene | Coefficient of Correlation (r) | Gene | Geomean of ranking values | |
| 1 | mbp1 | 0.438 | sdha | 0.081 | β-act | 0.898 | mbp1 | 1.178 |
| 2 | β-act | 0.478 | mbp1 | 0.102 | mbp1 | 0.891 | sdha | 2.060 |
| 3 | sdha | 0.496 | β-act | 0.184 | sdha | 0.876 | β-act | 2.449 |
| 4 | β-tub | 0.593 | β-tub | 0.244 | β-tub | 0.821 | rGTPa | 3.807 |
| 5 | psm1 | 0.636 | psm1 | 0.327 | ef1-α | 0.802 | adp-rf | 4.162 |
| 6 | adp-rf | 0.721 | adp-rf | 0.334 | psm1 | 0.761 | psm1 | 4.427 |
| 7 | ef1-α | 0.774 | ef1-α | 0.381 | adp-rf | 0.716 | β-tub | 6.735 |
| 8 | rpl2 | 0.886 | rpl2 | 0.413 | rpl2 | 0.691 | ef1-α | 8.239 |
| 9 | rGTPa | 0.968 | rGTPa | 0.464 | rGTPa | 0.492 | rpl2 | 8.739 |
NormFinder analysis showed that sdha had the lowest stability value SV = 0.081 and demonstrated that when the samples were subjected to different environmental stimuli the best combination of two reference genes was sdha and mbp1 with SV = 0.070 (Table 3). NormFinder demonstrated that mbp1 was the most stable gene during M. canis growth in MM-Cove medium supplemented, respectively, with glucose (SV = 0.053), keratin (SV = 0.063), keratin with soy protein (SV = 0.188), elastin (SV = 0.040), collagen (SV = 0.096), and colloidal chitin (SV = 0.106). sdha was the most stable gene during M. canis growth in MM-Cove medium (SV = 0.096), KSM medium (SV = 0.224), and MM-Cove medium with low concentration of organic Pi at different pH shifts (SV = 0.093). In case of M. canis growth in YEM medium with low concentration of organic Pi and at different pH, NormFinder analysis demonstrated that β-tub was the most stable reference gene (SV = 0.254).
Table 3.
Ranking of M. canis candidate reference genes determined by NormFinder algorithm.
| Rank | All conditions | MM-Cove growth | Glucose growth | Keratin growth | Keratin/soy protein growth | Elastin growth | Collagen growth | Colloidal chitin growth | KSM Medium growth | MM-LowPi medium growth | YEM-LowPi medium growth | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gene | SV | gene | SV | gene | SV | gene | SV | gene | SV | gene | SV | gene | SV | gene | SV | gene | SV | gene | SV | gene | SV | |
| 1 | sdha | 0.081 | sdha | 0.096 | mbp1 | 0.053 | mbp1 | 0.063 | mbp1 | 0.188 | mbp1 | 0.040 | mbp1 | 0.096 | mbp1 | 0.106 | sdha | 0.224 | sdha | 0.093 | β-tub | 0.254 |
| 2 | mbp1 | 0.102 | mbp1 | 0.096 | sdha | 0.057 | sdha | 0.117 | β-act | 0.250 | sdha | 0.100 | sdha | 0.113 | β-act | 0.280 | mbp1 | 0.269 | β-act | 0.182 | psm1 | 0.319 |
| 3 | β-act | 0.184 | rpl2 | 0.323 | ef1-α | 0.207 | rpl2 | 0.173 | sdha | 0.272 | β-act | 0.141 | β-act | 0.230 | sdha | 0.315 | β-act | 0.359 | β-tub | 0.212 | ef1-α | 0.325 |
| 4 | β-tub | 0.244 | β-act | 0.345 | adp-rf | 0.248 | adp-rf | 0.210 | β-tub | 0.414 | β-tub | 0.175 | β-tub | 0.252 | rpl2 | 0.334 | ef1-α | 0.553 | mbp1 | 0.248 | mbp1 | 0.326 |
| 5 | psm1 | 0.327 | adp-rf | 0.420 | β-act | 0.252 | β-act | 0.220 | adp-rf | 0.440 | psm1 | 0.176 | ef1-α | 0.310 | adp-rf | 0.363 | adp-rf | 0.557 | rpl2 | 0.299 | sdha | 0.337 |
| 6 | adp-rf | 0.334 | psm1 | 0.543 | β-tub | 0.303 | β-tub | 0.235 | rGTPa | 0.567 | rpl2 | 0.254 | psm1 | 0.314 | psm1 | 0.427 | β-tub | 0.610 | psm1 | 0.369 | rpl2 | 0.428 |
| 7 | ef1-α | 0.381 | β-tub | 0.562 | rpl2 | 0.305 | ef1-α | 0.277 | psm1 | 0.593 | adp-rf | 0.400 | rpl2 | 0.324 | β-tub | 0.468 | psm1 | 0.645 | adp-rf | 0.375 | β-act | 0.431 |
| 8 | rpl2 | 0.413 | ef1-α | 0.587 | psm1 | 0.377 | psm1 | 0.302 | ef1-α | 0.664 | ef1-α | 0.448 | adp-rf | 0.337 | rGTPa | 0.542 | rGTPa | 0.912 | rGTPa | 0.492 | adp-rf | 0.437 |
| 9 | rGTPa | 0.464 | rGTPa | 0.921 | rGTPa | 0.392 | rGTPa | 0.390 | rpl2 | 0.767 | rGTPa | 0.474 | rGTPa | 0.374 | ef1-α | 0.737 | rpl2 | 1.151 | ef1-α | 0.547 | rGTPa | 0.583 |
| best combination | sdha, mbp1 | 0.070 | a | — | a | — | a | — | a | — | a | — | a | — | a | — | a | — | a | — | a | — |
The Excel-based BestKeeper algorithm25 was used to evaluate the expression stability of reference genes based on the coefficient of variance (CV) and the standard deviation (SD) of the average Ct values. The descriptive statistics for the nine candidate genes is given in Table 4. Among nine evaluated candidates, seven genes exhibited a recommended standard deviation value [0.5 < SD[±Ct] ≤ 1.00]. Data analysis using pairwise correlation and regression analysis assessed the inter-gene relations, which eliminated ef1-α and rpl2 genes as showing highest standard deviation (SD = 1.21, and 1.04, respectively) and variation (CV = 6.25, and 4.75, respectively). The lowest variation was observed for the rGTPa gene (CV = 2.74). Moreover, rGTPa gene demonstrated a weak correlation to BestKeeper index (BI) as compared to other candidates (r = 0.492). Therefore, ef1-α, rpl2, and rGTPa gene were excluded from further normalization. The analysis of the remaining six genes (β-act, β-tub, adp-rf, mbp1, and psm1) showed a significant correlation of 0.716 < r < 0.898 between their expression levels and BI (p < 0.001). Ranking of the four tightly correlated genes on the basis of variation from the most stable to the least stable one was as follows: β-act, mbp1, sdha, and β-tub. Among these four genes β-act, and mbp1 were the most highly correlated 0.898 < r < 0.891 to the BI (Table 4).
Table 4.
Descriptive statistical analysis of candidate reference genes using BestKeeper.
| Gene | β-act | β-tub | adp-rf | ef1-α | Sdha | rpl2 | mbp1 | psm1 | rGTPa |
|---|---|---|---|---|---|---|---|---|---|
| n | 90 | 90 | 90 | 90 | 90 | 90 | 90 | 90 | 90 |
| GM [Ct] | 16.73 | 20.75 | 17.86 | 19.37 | 18.15 | 21.79 | 20.58 | 25.25 | 17.48 |
| AM [Ct] | 16.75 | 20.78 | 17.88 | 19.43 | 18.17 | 21.83 | 20.60 | 25.27 | 17.49 |
| Min [Ct] | 15.08 | 18.87 | 15.93 | 16.43 | 16.05 | 19.41 | 18.74 | 22.84 | 16.44 |
| Max [Ct] | 18.70 | 22.57 | 20.25 | 22.41 | 19.81 | 25.27 | 22.19 | 26.66 | 18.98 |
| SD [±Ct] | 0.72 | 0.83 | 0.60 | 1.21 | 0.70 | 1.04 | 0.66 | 0.71 | 0.48 |
| CV [% Ct] | 4.30 | 3.99 | 3.37 | 6.25 | 3.83 | 4.75 | 3.22 | 2.79 | 2.74 |
| Min [x-fold] | −3.12 | −7.39 | −3,79 | −7.66 | −4.28 | −5.20 | −3.60 | −5.31 | −2.06 |
| Max [x-fold] | 3.94 | 3.53 | 5.26 | 8.23 | 3.16 | 11.17 | 3.05 | 2.65 | 2.82 |
| SD [±x-fold] | 1.65 | 1.78 | 1.52 | 2.32 | 1.62 | 2.05 | 1.58 | 1.63 | 1.39 |
| BI Index [r] | 0.898 | 0.821 | 0.716 | 0.802 | 0.876 | 0.691 | 0.891 | 0.761 | 0.492 |
| p-value | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.006 |
Finally, RefFinder which is a comprehensive web-based tool that integrates geNorm, NormFinder and BestKeeper was applied to generate a comprehensive final ranking of the candidate reference genes. As shown in Table 2, mbp1, sdha, and β-act were ranked as the top stable genes under all experimental conditions.
Stability of mbp1, β-act and sdha reference genes in M. canis strains
We compared the data of mbp1, β-act, and sdha reference genes expression in M. canis CBS 113480 and M. canis 23/10 strain (isolated from cat) with those obtained for M. canis 267/10 strain, which was used in the stability evaluation. The M. canis CBS, M. canis 23/10, and M. canis 267/10 strain were cultivated at 28 °C for 48 h in MM-Cove medium (control medium), and MM-Cove medium supplemented with keratin. Degradation of keratin as well as other proteins releases amino acids, metabolism of which leads to secretion of ammonia what changes pH from acidic to alkaline, after 48 hours22, which is extremely important for growth of dermatophytes in the host tissue. Therefore, to check stability of the selected reference genes in other M. canis strains it is reasonable to choose keratin as a carbon source, as it builds stratum corneum of mammalia. We observed slight variation in the Ct values (Fig. 4) but mbp1, β-act, and sdha genes expression was not significantly different under the experimental conditions (pmbp1 = 0.91; pβ-act = 0.46; psdha = 0.93, ANOVA) (Fig. 4). These results confirmed the stability of mbp1, β-act, and sdha reference gene expression in different M. canis strains, suggesting that these genes are sufficient for effective normalization of qRT-PCR data.
Figure 4.
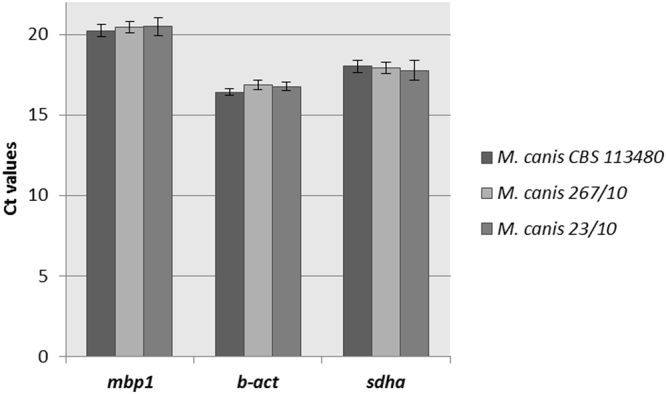
Stability of mbp1, β-act and sdha reference genes expression, evaluated in three M. canis strains cultured on MM-Cove medium (control medium) and MM-Cove medium supplemented with keratin. Gene expression levels are represented by average Ct values. mbp1 (p = 0.91, ANOVA), β-act (p = 0.46, ANOVA) and sdha (p = 0.93, ANOVA) gene expression level was not significantly different across analyzed culture conditions. Error bars indicate standard error.
Validation of selected M. canis reference genes
To validate the selected reference genes for qRT-PCR normalization, the relative expression profiles of the MEP3 gene were analyzed. MEP3 is known as the gene encoding M. canis keratynolityc metalloprotease (43.5 kDa) expressed in the presence of keratin26. The validation analysis was conducted using three different reference gene variants selected by GeNorm, NormFinder, BestKeeper, and RefFinder in M. canis 267/10 strain cultivated at 28 °C for 48 h in MM-Cove medium (control medium), MM-Cove medium supplemented with keratin, and MM-Cove medium supplemented with keratin and soy protein which increases proteolytic activity. These variants were as follows: variant A - two least stable reference genes (rpl2 and rGTP); variant B - three most stable reference genes (mbp1, β-act, sdha); variant C - all candidate reference genes. Relative expression was calculated using the 2−ΔΔCt method. The statistical significance between control and treatment conditions was analysed by one-way ANOVA test. The best result indicating an increase of MEP3 transcript of M. canis 267/10 growing in MM-Cove medium supplemented with keratin and keratin/soy protein in relation to control medium was found when the three reference genes (variant B) were used, as revealed by all four algorithms (Fig. 5).
Figure 5.
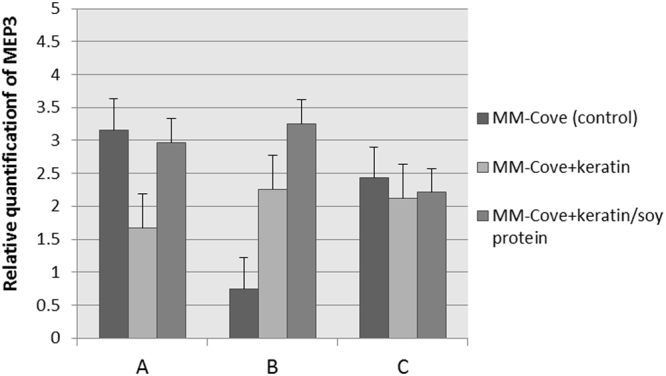
Relative quantification of MEP3 gene expression in control, keratin and keratin/soy protein samples using different reference gene variants: A (p = 0.67, ANOVA) - two least stable reference genes rpl2 and rGTP; B (p = 0.72, ANOVA) – three most stable reference genes mbp1, β-act and sdha; C (p = 0.55, ANOVA) – all candidate reference genes. Error bars indicate standard error.
Discussion
qRT-PCR technique is a frequently used platform to quantify transcript abundance. However its heavily dependent on the stability of the internal reference genes used to normalize measurements of transcription level. Normalizer genes are defined as those with a stable expression under previously defined conditions, thus appropriate to quantify gene expression level of specific targets. In agreement with our results, increasing numbers of studies demonstrated that no single reference gene is stably expressed in all cell types and under different experimental conditions27. Therefore, the expression stability of candidate reference genes needs to be verified before each qRT-PCR experiment.
In the present study, we used four different algoritmhs such as geNorm, NormFinder, BestKeeper, and RefFinder to evaluate nine candidate reference genes for their potential use as internal controls for expression analysis of target genes of M. canis in culture media supplemented with different carbon sources, inorganic phosphate, and at optimal pH what promotes adhesion-inducing conditions. To our knowledge, this is the first identification and validation of mbp1, β-act, and sdha as the most suitable and stably expressed reference genes of M. canis tested among the larger set consisting of nine candidates (Table 2). Transcription factor 1 (mbp1) involved in transition of cell cycle from G1 to S phase was classified in our study as one of the three most stable genes (Table 2). This is the unique use of this gene as there have been no reports on validation of mpb1 as a reference in other fungi transcription studies so far.
β-act, one of the three major proteins of cytoskeleton and sdha gene that codes for a subunit of succinate dehydrogenase and is important in cellular respiration were also found in our study to be suitable reference genes for expression analyses in M. canis growing under different adhesion inducing conditions. β-act as well as 18 S and gapdh were some of the most commonly used housekeeping genes in qRT-PCR analysis28. β-act, performed as highly reliable reference gene when evaluated in fungi such as Trichophyton rubrum24, Aspergillus niger29, Candida albicans30 but also in other eukariots such as: barley (Hordeum vulgare L.)31, tung tree (Vernicia fordii Hemsl.)32, crop Musa33 or humans (prostate cancer studies)34. However, it has to be pointed out that there were also reports in which β-act was found to be unstable e.g. in Saccharomyces cerevisiae35, Candida glabrata36, Benjaminiella poitrasii37, Poria cocos38, Monopteus albus39 or cell culture models infected with influenza viruses40. Sdha gene was found to be suitable housekeeping gene for expression analyses in bovine tuberculosis41, rat tissue under toxicological conditions42, human glioma43, stress response of the athlete horses cells44, neutrophils, and untreated total blood leukocytes45. Again, some other studies have demonstrated that the stability of sdha gene expression was low in Candida glabrata36, rat vocal fold model of mucosal injury46 or different tissues of yak fetuses47. So, despite the fact that β-act and sdha genes were often used as housekeeping genes, a number of studies have provided evidence that their transcription level may vary between different organisms, cell types, developmental stages or experimental conditions27 and for that reason the selection of the most appropriate internal controls for each experimental model used for qRT-PCR is a crucial prerequisite for reliable gene expression analysis. In this study several optimal sets of reference genes that are suitable for qRT-PCR data normalization in M. canis were identified (Table 3, Fig. 3). These results indicate that the stability of reference gene expression in M. canis needs to be investigated for each experimental condition, what confirms the thesis that there is no universal reference gene (Fig. 3, Table 3).
In order to validate the reliability of the selected reference genes, relative quantification of MEP3 gene known to be induced in the presence of keratin was performed. The MEP3 up-regulation was detectable in M. canis 267/10 growing in MM-Cove medium supplemented with keratin and also with keratin/soy protein only when the three reference genes: mbp1, β-act, and sdha previously selected as most stable internal controls were used in combination. The results clearly proved that normalization using inappropriately chosen housekeeping genes could lead to erroneous conclusions (Fig. 5).
In conclusion, our study was the first attempt to evaluate and validate M. canis reference genes. These findings will allow further analysis of M. canis gene expression under different adhesion-inducing environmental stimuli, with improved accuracy and reliability, and may also provide a starting point for selection of candidate genes for gene expression analysis in other related species.
Materials and Methods
Selection of candidate genes and primer design
Nine candidate reference genes (β-act, β-tub, adp-rf, ef1-α, sdha, rpl2, mbp1, psm1, and rGTPa) were selected based on the NCBI database (http://www.ncbi.nlm.nih.gov) in some fungal species and other organisms (Table 1). Primers were designed based on nucleotide sequences deposited in GenBank (Table 1) and theoretically evaluated using Primer3 software48. Default program settings were selected except for following categories: “monovalent cations concentration” which was set at 50 mM, “divalent cations concentration” set at 3 mM, “dNTP concentration” set at 1.2 mM, and “annealing oligo concentration” set at 250 mM. All PCR products were within 80–150 bp range and one of the primers from each pair was anchored within exon-exon junction. Each primer pair underwent experimental evaluation and was accepted if all following conditions were true: (1) product PCR reaction using cDNA as a template was specific, (2) reaction using genomic DNA as a template gave no product, and (3) the efficiency of a real time PCR reaction was between 90–110% (Table 1).
Fungal strain and growth conditions
For all experiments Microsporum canis 267/10 strain was used, which is a clinical isolate from tinea capitis of a 54 years old woman. Microsporum canis CBS 11348 and Microsporum canis 23/10, a clinical isolate from cat, were used in evaluation of the most stable reference genes, selected by geNorm, NormFinder, BestKeeper, and RefFinder algorithms. Standard mycological identification based on the phenotypic features was performed in the specialized hospital laboratory and confirmed in our laboratory by PCR-RFLP analysis targeting ITS1–5.8S-ITS2 followed by sequencing49. Conidia from M. canis strain (approximately 107 cells/ml) were isolated as described previously50 and incubated separately for 24 h, 48 h, and 72 h at 28 °C with agitation in a liquid minimal medium (MM-Cove)51 (control medium) supplemented with 70 mM sodium nitrate at pH 5.0, and, respectively, with different carbon sources such as: glucose (55 mM), 0.5% (w/v) keratin, 0.5%/1% (w/v) keratin/soy protein, 0.25% (w/v) elastin, 0.25% (w/v) collagen, or 1% (w/v) colloidal chitin, as well as in Keratinocyte serum- free medium (1X) (Thermo Scientific). The M. canis conidia were also inoculated into Low-Pi minimal liquid medium (MM), and yeast extract medium (YEM) pH 5.0, 8.0, 10.0 and incubated at 37 °C for 17 h at 200 rpm. Final concentrations of Pi in low-Pi cultures was 200 μmol in case of MM-medium and 700 μmol in YEM medium52.
RNA extraction, cDNA synthesis and qRT-PCR
Total RNA was extracted from M. canis cells using RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions, with addition of DNase I to eliminate potential DNA contamination. Quantity and purity of the RNA was assessed by NanoPhotometerTM Pearl Version 1.0 (IMPLEN). Only RNA samples with A260/A280 ratio between 1.9 and 2.1, and A260/A230 ratio higher than 2.0 were used in the analysis. RNA integrity was further assessed by 1% denaturing agarose gel electrophoresis. Two micrograms of total RNA were reverse transcribed into cDNA to a final volume of 40 µl using RevertAid Transcriptase (Thermo Scientific) according to the manufacturer’s instructions. qRT-PCR was performed in RotorGene Q System (Qiagen). The reaction mixture (20 µl) contained 10 µl of SsoAdvanced Universal SYBR®Green Supermix (2X), 1 μl of each primer (500 nM), 5 μl of diluted cDNA (1:40) and 4 μl of nuclease-free water. Amplifications were performed using the following cycling profile: an initial activation step (95 °C for 1 min) followed by 40 cycles of denaturation at 95 °C for 20 s, annealing at 60.5 °C for 20 s, and extension at 72 °C for 15 s. For melting curve analysis, a dissociation step cycle (72 °C for 10 s, and then 0.5 °C for 10 s until 95 °C) was added. All qRT-PCR experiments were performed in three biological and three technical replicas. Amplification efficiency (E) and correlation coefficient (R2) were calculated using the Rotor-Gene Q Series Software Version 2.3.1. (Qiagen) by standard curve method with 4-fold serial dilutions (Table 1).
Data analysis
The expression stability of the 9 reference genes was evaluated by geNorm module of qbase + Version 3.1 (Biogazelle), NormFinder, BestKeeper and RefFinder algorithms in 90 samples (three biological replicas and 30 different conditions, Supplementary Table S1) under different experimental conditions. The geNorm module was used to compute expression stability values for all reference targets. As an input for analysis, Ct values exported directly from the Rotor-Gene Q Series Software Version 2.3.1. (Qiagen) were used. The candidate reference genes were ranked according to the expression stability M value, which is the average pairwise variation of a particular gene with all other reference genes. The gene showing the lower M-value is assigned to be expressed in a more stable fashion, while the one with the higher M-value has the less stable expression53. This algorithm was used to rank the optimal number of reference genes for each experimental condition. The geNorm module determines the pairwise variation Vn/Vn+1 between two subsequent normalization factors NFn and NFn+1 in order to rank the minimum number of reference genes for normalization with a cut-off value of 0.15. NormFinder is a Visual Basic application software which takes into account intra- and inter-group variations and calculates a stability M-value. The genes with lower M-values indicate low inter- and intra-group variations and are considered to have greater stability54. The BestKeeper algorithm is usually performed by assessing the calculation of standard deviation SD (±Ct) and correlation coefficients of variance CV (%Ct) for all reference genes in all samples. All stably expressed reference genes are combined into the BestKeeper index using the geometric mean of each candidate reference gene’s Ct value25. The webbased tool RefFinder (http://leonxie.esy.es/RefFinder/) was used in order to combine the results and rank the candidate genes. The lowest rank indicates the most stably expressed gene.
Electronic supplementary material
Acknowledgements
This work was supported by grant 2014/13/B/NZ7/02307 and by grant 2016/23/D/NZ7/03964 from National Science Centre, Poland. The experiments were partly performed using equipment of the Laboratory of Microscopic Imaging and Specialized Biological Techniques, Faculty of Biology and Environmental Protection, University of Łódź, Poland. The authors would like to thank Dr Marek Gadzalski for help in selecting candidate reference genes and primers design.
Author Contributions
A.C. conceived and conducted the experiments, analyzed the data and interpreted the results, wrote the manuscript. P.S. conceived and wrote the manuscript. Both authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-19680-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ginter-Hanselmayer G, Weger W, Ilkit M, Smolle J. Epidemiology of tinea capitis in Europe: current state and changing patterns. Mycoses. 2007;50:6–13. doi: 10.1111/j.1439-0507.2007.01424.x. [DOI] [PubMed] [Google Scholar]
- 2.Abanmi A, et al. Characteristics of superficial fungal infections in the Riyadh region of Saudi Arabia. Int. J. Dermatol. 2008;47:229–235. doi: 10.1111/j.1365-4632.2008.03563.x. [DOI] [PubMed] [Google Scholar]
- 3.Fuller LC. Changing face of tinea capitis in Europe. Curr. Opin. Infect. Dis. 2009;22:115–118. doi: 10.1097/QCO.0b013e3283293d9b. [DOI] [PubMed] [Google Scholar]
- 4.Costa M, et al. Epidemiology and etiology of dermatophytosis in Goiânia, GO, Brazil. Rev. Soc. Bras. Med. Trop. 2002;35(1):19–22. doi: 10.1590/S0037-86822002000100004. [DOI] [PubMed] [Google Scholar]
- 5.Li C, Liu W. Epidemiology of tinea capitis among children in China in recent years: a retrospective analysis. Chinese J. Mycol. 2011;6:77–82. [Google Scholar]
- 6.Ali-Shtayeh MS, Yaish S, Jamous RM, Arda H, Husein EI. Updating the epidemiology of dermatophyte infections in Palestine with special reference to concomitant dermatophytosis. J. Mycol. Med. 2015;25:116–122. doi: 10.1016/j.mycmed.2015.02.046. [DOI] [PubMed] [Google Scholar]
- 7.Razzaq Adel AA, Sultan AO, Basmiah AM, Aftab A, Nabel N. Prevalence of tinea capitis in southern Kuwait. Mycoses. 2007;50:317–320. doi: 10.1111/j.1439-0507.2007.01336.x. [DOI] [PubMed] [Google Scholar]
- 8.Panasiti V, et al. Epidemiology of dermatophytic infections in Rome, Italy: a retrospective study from 2002 to 2004. Med. Mycol. 2007;45:57–60. doi: 10.1080/13693780601028683. [DOI] [PubMed] [Google Scholar]
- 9.Hryncewicz-Gwóźdź A, Ploter-Niezgoda E, Maj J. Grzybica stóp, rąk i paznokci - epidemiologia, objawy, leczenie. Mikologia Lekarska. 2005;12(1):57–62. [Google Scholar]
- 10.del Boz J, Crespo V, Rivas-Ruiz F, de Troya M. A 30-year survey of pediatric tinea capitis in southern Spain. J. Eur. Acad. Dermatol. Venerol. 2011;25(2):170–174. doi: 10.1111/j.1468-3083.2010.03733.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhu M, et al. Tinea capitis in Southeastern China: a 16-year survey. Mycopathologia. 2010;169(4):235–239. doi: 10.1007/s11046-009-9260-2. [DOI] [PubMed] [Google Scholar]
- 12.Lakshmipathy DT, Kannabiran K. Review on dermatomycosis: pathogenesis and treatment. Nat. Science. 2010;2:726–731. [Google Scholar]
- 13.Berg JC, Hamacher KL, Roberts GD. Pseudomycetoma caused by Microsporum canis in an immunosuppressed patient: a case report and review of the literature. J. Cutan. Pathol. 2007;34:431–434. doi: 10.1111/j.1600-0560.2006.00628.x. [DOI] [PubMed] [Google Scholar]
- 14.Bournerias I, et al. Unusual Microsporum canis infections in adult HIV patients. J. Am. Acad. Dermatol. 1996;35:808–810. doi: 10.1016/S0190-9622(96)90089-4. [DOI] [PubMed] [Google Scholar]
- 15.Bennassar A, Grimalt R. Management of tinea capitis in childhood. Clin. Cosmet. Investig. Dermatol. 2010;3:89–98. doi: 10.2147/ccid.s7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King D, et al. Primary invasive cutaneous Microsporum canis infections in immunocompromised patients. J. Clin. Microbiol. 1996;34:460–462. doi: 10.1128/jcm.34.2.460-462.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel GA, Schwartz RA. Tinea capitis: still an unsolved problem? Mycoses. 2011;54:183–188. doi: 10.1111/j.1439-0507.2009.01819.x. [DOI] [PubMed] [Google Scholar]
- 18.Brillowska-Dabrowska A, Michalek E, Saunte DM, Nielsen SS, Arendrup MC. PCR test for Microsporum canis identification. Med. Mycol. 2013;51:576–579. doi: 10.3109/13693786.2012.755741. [DOI] [PubMed] [Google Scholar]
- 19.Cafarchia C, Romito D, Capelli G, Guillot J, Otranto D. Isolation of Microsporum canis from the hair coat of pet dogs and cats belonging to owners diagnosed with M. canis tinea corporis. Vet. Dermatol. 2006;17:327–331. doi: 10.1111/j.1365-3164.2006.00533.x. [DOI] [PubMed] [Google Scholar]
- 20.Kraemer A, Mueller RS, Werckenthin C, Straubinger RK, Hein J. Dermatophytes in pet Guinea pigs and rabbits. Vet. Microbiol. 2012;157(1–2):208–213. doi: 10.1016/j.vetmic.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Hermoso de Mendoza M, et al. A zoonotic ringworm outbreak caused by a dysgenic strain of Microsporum canis from stray cats. Rev. Soc. Bras. Med. Trop. 2010;27(2):62–65. doi: 10.1016/j.riam.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 22.Martinez DA, et al. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. mBio. 2012;3(5):e00259–12. doi: 10.1128/mBio.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan H, et al. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 2010;399:257–261. doi: 10.1016/j.ab.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Jacob TR, et al. rpb2 is reliable reference gene for quantitative gene expression analysis in the dermatophyte Trichophyton rubrum. Med. Mycol. 2012;50:368–377. doi: 10.3109/13693786.2011.616230. [DOI] [PubMed] [Google Scholar]
- 25.Pfaffl MW, et al. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 26.Brouta F, et al. Secreted metalloprotease gene family of Microsporum canis. 2002;70:5676–5683. doi: 10.1128/IAI.70.10.5676-5683.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozera B, Rapacz M. Reference genes in real-time PCR. J. Appl. Genetics. 2013;54:391–406. doi: 10.1007/s13353-013-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Jonge HJ, et al. Evidence based selection of housekeeping genes. PLoS ONE. 2007;2:e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bohle K, et al. Selection of reference genes for normalization of specific gebe quantification data of Aspergillus niger. J. Biotechnol. 2007;132:353–358. doi: 10.1016/j.jbiotec.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Nailis H, Coenye T, Van Nieuwerburgh F, Deforce D, Nelis HJ. Development and evaluation of different normalization strategies for gene expression studies in Candida albicans biofilms by real-time PCR. BMC Mol. Biol. 2006;7:25. doi: 10.1186/1471-2199-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rapacz M, Stępień A, Skorupa K. Internal standards for quantitative RT-PCR studies of gene expression under drought treatment in barley (Hordeum vulgare L.): the effects of developmental stage and leaf age. Acta Physiol. Plant. 2012;34:1723–1733. doi: 10.1007/s11738-012-0967-1. [DOI] [Google Scholar]
- 32.Han X, et al. Selection of Reliable Reference Genes for Gene Expression Studies Using Real-Time PCR in Tung Tree during Seed Development. PLoS One. 2012;7(8):e43084. doi: 10.1371/journal.pone.0043084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Podevin N, Krauss A, Henry I, Swennen R, Remy S. Selection and validation of reference genes for quantitative RT-PCR expression studies of the non-model crop Musa. Mol. Breed. 2012;30:1237–1252. doi: 10.1007/s11032-012-9711-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mori R, Wang Q, Danenberg KD, Pinski JK, Danenberg PV. Both β-actin and GAPDH are useful reference genes for normalization of quantitative RT-PCR in human FFPE tissue samples of prostate cancer. Prostate. 2008;68:1555–1560. doi: 10.1002/pros.20815. [DOI] [PubMed] [Google Scholar]
- 35.Teste MA, et al. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Sacharomyces cerevisiae. BMC Mol. Biol. 2009;10:99. doi: 10.1186/1471-2199-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li QQ, Skinner J, Bennet J. Evaluation of reference genes for real-time quantitative PCR studies in Candida glabrata following azole treatment. BMC Mol. Biol. 2012;13:22. doi: 10.1186/1471-2199-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pathan EK, Ghormade V, Deshpande MV. Selection of reference genes for quantitative real-time RT-PCR assays in different morphological forms of dimorphic zygomycetous fungus Benjaminiella poitrasii. PLoS One. 2017;12(6):e0179454. doi: 10.1371/journal.pone.0179454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, et al. Selection and validation of reference genes for normalization of quantitative real-time reverse transcription PCR analysis in Poria cocos(Schw.) Wolf (Fuling) Chin. Med. 2016;11:8. doi: 10.1186/s13020-016-0079-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu Q, Guo W, Gao Y, Tang R, Li D. Reference gene selection for real-time RT-PCR normalization in rice field eel (Monopterus albus) during gonad development. Fish Physiol. Biochem. 2014;40:1721–1730. doi: 10.1007/s10695-014-9962-3. [DOI] [PubMed] [Google Scholar]
- 40.Kuchipudi SV, et al. 18S rRNA is a reliable normalisation gene for real time PCR based on influenza virus infected cells. Virol. J. 2012;9:230. doi: 10.1186/1743-422X-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim A, Bovin S. Evaluation of Reference Genes for Differential Gene Expression Study of Bovine Tuberculosis. Adv. Zoology and Botany. 2017;5(2):17–24. [Google Scholar]
- 42.Svingen T, Letting H, Hadrup N, Hass U, Vinggaard AM. Selection of reference genes for quantitative RT-PCR (RT-qPCR) analysis of rat tissues under physiological and toxicological conditions. PeerJ. 2015;24:e855. doi: 10.7717/peerj.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kreth S, et al. Identification of valid endogenous control genes for determining gene expression in human glioma. Neuro-Oncology. 2010;12:570–579. doi: 10.1093/neuonc/nop072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cappelli K, et al. Exercise induced stress in horses: Selection of the most stable reference genes for quantitative RT-PCR normalization. BMC Mol. Biol. 2008;19:49. doi: 10.1186/1471-2199-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ledderose C, Heyn J, Limbeck E, Kreth S. Selection of reliable reference genes for quantitative real-time PCR in human T cells and neutrophils. BMC Res. Notes. 2011;20:427. doi: 10.1186/1756-0500-4-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang Z, Ling C, Yamashita M, Welham NV. Microarray-driven validation of reference genes for quantitative real-time polymerase chain reaction in a rat vocal fold model of mucosal injury. Anal. Biochem. 2010;406:214–221. doi: 10.1016/j.ab.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li M, et al. Identification of Optimal Reference Genesfor Examination of Gene Expression in Different Tissues of Fetal Yaks. Czech J. Anim. Sci. 2017;10:426–434. doi: 10.17221/75/2016-CJAS. [DOI] [Google Scholar]
- 48.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23(10):1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 49.Dobrowolska A, Stączek P, Kaszuba A, Kozłowska M. PCR-RFLP analysis of the dermatophytes isolated from patients in Central Poland. J. Dermatol. Sci. 2006;42(1):71–74. doi: 10.1016/j.jdermsci.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Dobrowolska A, Stączek P. Development of transformation system for Trichophyton rubrum by electroporation of germinated conidia. Curr. Genet. 2009;55(5):537–542. doi: 10.1007/s00294-009-0264-8. [DOI] [PubMed] [Google Scholar]
- 51.Cove DJ. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta. 1966;113:51–56. doi: 10.1016/S0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- 52.Trevisan GL, et al. Transcription of Aspergillus nidulans pacC is modulated by alternative RNA splicing of palB. FEBS Lett. 2011;585:3442–3445. doi: 10.1016/j.febslet.2011.09.037. [DOI] [PubMed] [Google Scholar]
- 53.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):research0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.