Abstract
Cucumber mosaic virus (CMV) infection could induce mosaic symptoms on a wide-range of host plants. However, there is still limited information regarding the molecular mechanism underlying the development of the symptoms. In this study, the coat protein (CP) was confirmed as the symptom determinant by exchanging the CP between a chlorosis inducing CMV-M strain and a green-mosaic inducing CMV-Q strain. A yeast two-hybrid analysis and bimolecular fluorescence complementation revealed that the chloroplast ferredoxin I (Fd I) protein interacted with the CP of CMV-M both in vitro and in vivo, but not with the CP of CMV-Q. The severity of chlorosis was directly related to the expression of Fd1, that was down-regulated in CMV-M but not in CMV-Q. Moreover, the silencing of Fd I induced chlorosis symptoms that were similar to those elicited by CMV-M. Subsequent analyses indicated that the CP of CMV-M interacted with the precursor of Fd I in the cytoplasm and disrupted the transport of Fd I into chloroplasts, leading to the suppression of Fd I functions during a viral infection. Collectively, our findings accentuate that the interaction between the CP of CMV and Fd I is the primary determinant for the induction of chlorosis in tobacco.
Introduction
Plant viruses generally induce mosaic symptoms on infected hosts. The chloroplast is a common target of plant viruses, and are closely related to the expression of mosaic symptoms1,2. Viruses affect chloroplasts via several ways1,3; they can repress the transcription of chloroplast-related genes4–7 and inhibit the translation of chloroplast -related proteins8–11, resulting in deformation and disruption chloroplast12–14. Viral factors also affect the function of some chloroplast proteins3,15–17. For example, the disease-specific protein of Rice stripe virus reportedly recruits the chloroplast oxygen-evolving complex protein, PsbP, into the cytoplasm by interacting with it14. Whereas, the HC-Pro proteinase of Potato virus Y was observed to interact with the chloroplast division-related factor NtMinD to inhibit the formation of NtMinD homodimers, thereby interfering with chloroplast division18. Furthermore, the P3 protein encoded by Shallot yellow stripe virus was revealed to directly disrupt ribulose-1,5-bisphosphate carboxylase/oxygenase activities in the infected plant host19.
Cucumber mosaic virus (CMV) is a member of the genus Cucumovirus in the family Bromoviridae20. This virus can infect more than 1200 plant species, causing significant economic losses in diverse crops of agricultural importance21. The CMV genome consists of three positive-sense RNA strands, encoding five proteins22. The 1a and 2a proteins are encoded by RNA1 and RNA2, respectively, and are obligatory for viral replication22,23. The 2b protein is encoded by a subgenomic RNA (RNA4A) from RNA2, and functions as a viral suppressor of RNA silencing24. RNA3 is dicistronic: The protein encoded by the 5′ open reading frame (ORF) is the designated movement protein, while the CP encoded by the 3′ ORF is translated from a subgenomic RNA4 synthesized de novo from RNA3 minus-strand progeny22,25,26.
The CMV strains are broadly divided into two subgroups (subgroup I and II) based on the nucleotide sequence homology26. CMV strains in the same subgroup, but not between the subgroups, share a high degree of sequence similarity22,26. Another feature that distinguishes CMV strains of the subgroup I and II is the severity of the symptom phenotype26. For example, CMV-Q belonging to subgroup I induce mild green mosaic phenotype27. By contrast, CMV strains M and Price 6 belonging to subgroup II induces strong chlorotic phenotype28. Analysis of pseudorecombinants revealed that RNA3 being the genetic determinant for the induction of symptom phenotype29,30. Finer mutational analysis later identified that amino acid residue located at position 129 affects symptom expression31–33. A more recent study showed that chloroplast damage is closely related to the onset of mosaic symptoms in Nicotiana tabacum cv. Samsun plants infected with CMV13. However, it remains unclear how the CP of CMV interferes with host chloroplasts to induce a given symptom phenotype.
In this study, the CP of CMV was confirmed to be the determinant for mosaic symptom phenotype in tobacco plants. Additionally, a chloroplast protein, ferredoxin I (Fd I), was found to interact with the CP of CMV both in vitro and in vivo, and was directly associated with the development of chlorosis symptoms. Further analyses revealed that the interaction destabilized Fd I and disrupted its import into chloroplasts, resulting in the development of chlorosis symptoms on tobacco leaves.
Results
Viral CP is the determinant of mosaic symptoms in the host
We used infectious CMV cDNA clones to investigate whether the viral CP regulated the development of mosaic symptoms. We exchanged the CP-encoding regions between two CMV strains that induced different symptoms. The M strain of CMV (CMV-M), from subgroup I, induced severe chlorosis symptoms on Nicotiana tabacum cv. Samsun plants, while the Q strain of CMV (CMV-Q), from subgroup II, caused no symptoms or mild green mosaic symptoms (Fig. 1A). When the CP-coding regions of CMV-M was replaced with that of CMV-Q, the recombinant virus, CMV-MQCP, did not cause any obvious symptoms at 14 days post-inoculation (dpi). However, infected plants started to exhibit mild green mosaic symptoms at 28 dpi (Fig. 1A). Plants inoculated with the reciprocal recombinant virus CMV-QMCP developed chlorosis symptoms at 14 dpi, and the symptoms were more severe by 28 dpi (Fig. 1A). The accumulation of CP in the top systemic leaves was assessed with an anti-CP monoclonal antibody. The level of virus accumulation differed among plants infected by CMV-M, CMV-Q, and their recombinant viruses (Fig. 1B). These results confirmed previous observations13,31–33 that the CP is determinant for mosaic phenotype.
Figure 1.
Symptoms induced by two CMV strains at 14 and 28 days post-inoculation (dpi). (A) The M strain of CMV (CMV-M) and recombinant virus, CMV-QMCP, induced chlorosis symptoms on Nicotiana tabacum cv. Samsun plants. The Q strain of CMV (CMV-Q) did not induce symptoms on tobacco plants, while an infection by the recombinant virus, CMV-MQCP, resulted in a lack of symptoms at 14 dpi and mild green mosaic symptoms at 28 dpi. (B) The accumulation of CP in the top systemic leaves were detected by western blot analysis with an anti-CP monoclonal antibody. Plant actin was used as the control.
Chloroplast ferredoxin I interacts with the CP of CMV-M, but not the CP of CMV-Q
A yeast two-hybrid (YTH) assay was used to detect host proteins that interact with the CP of CMV-M. The CP-encoding ORF was inserted into pGBKT7 to produce the pGBK-MCP as a bait plasmid. A cDNA library for N. tabacum leaves was constructed using the pGADT7-Rec vector. Approximately 1.08 × 107 independent cDNA library clones were obtained and used to transform yeast AH109 cells along with pGBK-MCP. A BLAST analysis against NCBI database sequences revealed that three clones had sequences that were almost identical to that of the gene encoding Fd I (GenBank accession No. AY552781), with only five nucleotides at the 5′ end of the gene being different.
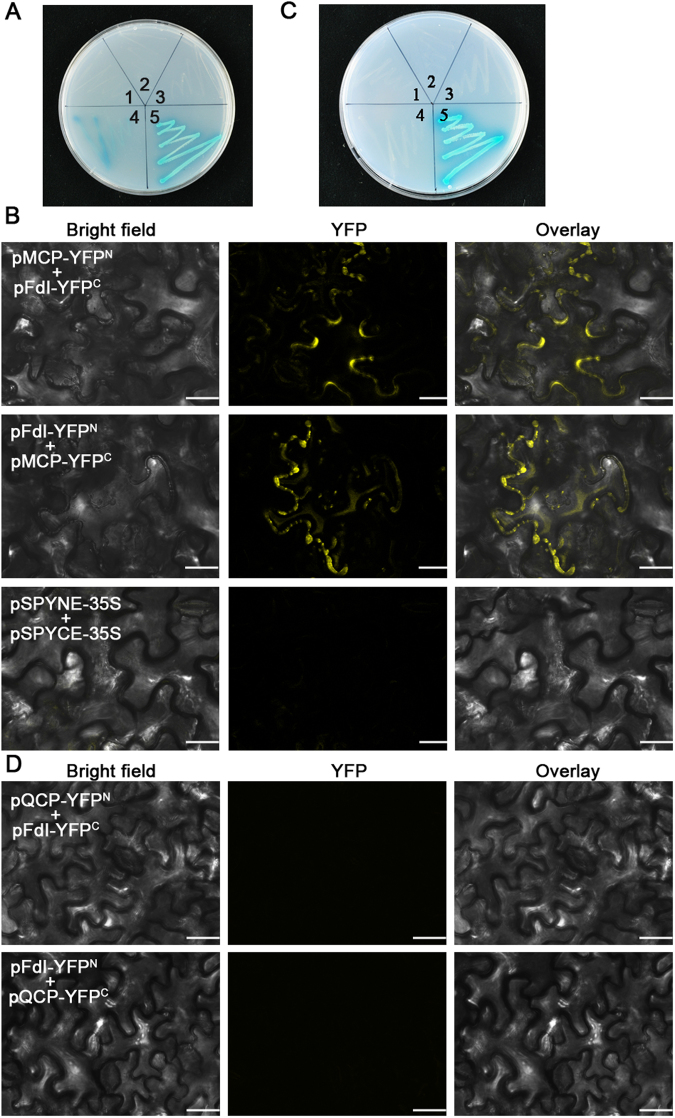
The full-length Fd I cDNA sequence was cloned and inserted into pGADT7 to generate the pGAD-FdI plasmid, which was used to co-transform cells along with pGBK-MCP. The pGADT7-RecT/pGBKT7-53 plasmid combination was used as the positive control, while pGADT7-RecT/pGBKT7-Lam, pGADT7/pGBK-MCP, and pGAD-FdI/pGBKT7 served as negative controls. The transformants were cultured on SD/−Ade/−His/−Leu/−Trp medium and further verified by growing on SD/−Ade/−His/−Leu/−Trp/X-α-Gal medium. The AH109 cells transformed with pGAD-FdI/pGBK-MCP and pGADT7-RecT/pGBKT7-53 grew well on SD/−Ade/−His/−Leu/−Trp/X-α-Gal medium, while transformants harbouring pGADT7-RecT/pGBKT7-Lam, pGADT7/pGBK-MCP, or pGAD-FdI/pGBKT7 did not (Fig. 2A). These results indicated that Fd I interacts with the CP of CMV-M in vitro.
Figure 2.
Interaction between ferredoxin I (Fd I) and the CP. (A) A yeast two-hybrid system was used to analyse the interaction between Fd I and the CP of CMV-M. Transformed Saccharomyces cerevisiae AH109 cells were cultured on SD/−Ade/−His/−Leu/−Trp/X-α-Gal medium. 1, pGADT7-RecT/pGBKT7-Lam (negative control); 2, pGADT7/pGBK-MCP; 3, pGAD-FdI/pGBKT7; 4, pGAD-FdI/pGBK-MCP; 5, pGADT7-RecT/pGBKT7-53 (positive control). (B) Bimolecular fluorescence complementation (BiFC) analysis to verify the interaction between Fd I and the CP of CMV-M in Nicotiana benthamiana epidermal cells. Two combinations of BiFC plasmids (pMCP-YFPN/pFdI-YFPC and pFdI-YFPN/pMCP-YFPC) were inserted into plant cells with Agrobacterium tumefaciens, and YFP fluorescence was detected at 3 days post-agroinfiltration. The pSPYNE-35S/pSPYCE-35S plasmids were used as controls. Scale bars = 25 µm. (C) Yeast two-hybrid system to test the interaction between Fd I and the CP of CMV-Q. 1, pGADT7-RecT/pGBKT7-Lam (negative control); 2, pGADT7/pGBK-QCP; 3, pGAD-FdI/pGBKT7; 4, pGAD-FdI/pGBK-QCP; 5, pGADT7-RecT/pGBKT7-53 (positive control). (D) BiFC to verify the interaction between Fd I and the CP of CMV-Q in N. benthamiana epidermal cells.
Bimolecular fluorescence complementation (BiFC) enables the direct visualization of complex protein interactions under natural conditions34–36. Plasmids for the production of the CP of CMV-M fused to the N-terminal of yellow fluorescent protein (YFP) and Fd I fused to the C-terminal of YFP or the reverse (pMCP-YFPN/pFdI-YFPC and pFdI-YFPN/pMCP-YFPC) were inserted into Agrobacterium tumefaciens strain EHA105 cells, which were infiltrated into Nicotiana benthamiana epidermal cells. The pSPYNE-35S/pSPYCE-35S plasmids served as the negative control. After 2–3 days, cytoplasmic YFP signals were observed in plants carrying pMCP-YFPN/pFdI-YFPC or pFdI-YFPN/pMCP-YFPC, but not in control plants (Fig. 2B). These results indicated that the CP of CMV-M interacts with Fd I in plant cells.
To investigate whether Fd I contributes to the variability in the expression of symptoms, we analyzed the interaction between Fd I and the CP of CMV-Q. The YTH and BiFC results indicated that Fd I was unable to interact with the CP of CMV-Q in yeast and plant cells (Fig. 2C and D). These findings implied that a specific interaction between Fd I and the CP of CMV-M affects the development of chlorosis symptoms.
Ferredoxin I expression was affected by CMV-M infection, but not by CMV-Q infection
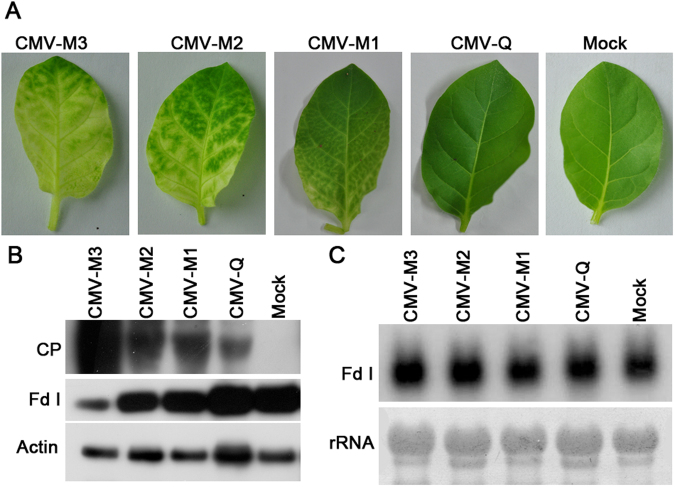
To verify the roles of Fd I during symptom development, we monitored its abundance after viral infection. We collected the following three sets of leaf samples from CMV-M-infected N. tabacum: the first set of systemic leaves (CMV-M1) with vein-clearing symptoms, the second set of systemic leaves (CMV-M2) with mild chlorosis symptoms, and the third set consisted of asymptomatic leaves from CMV-Q–infected tobacco plants (Fig. 3A). The CP and Fd I contents were evaluated by western blot analysis with an anti-CP monoclonal antibody and an anti-Fd I polyclonal antibody, respectively. Plant actin was used as the internal control. The CP abundance increased as the chlorosis symptoms became more severe (CMV-M1 < CMV-M2 < CMV-M3). However, the abundance of Fd I clearly decreased during CMV-M infection, especially in leaves with severe chlorosis symptoms (CMV-M3). There was no clear decrease in Fd I contents in CMV-Q–infected samples relative to the level in mock-inoculated samples (Fig. 3B), suggesting that stability of Fd I was affected by CMV-M, but not by CMV-Q.
Figure 3.
Detection of Fd I levels following an infection by CMV. (A) Leaves exhibiting severe chlorosis symptoms, mild chlorosis symptoms, or vein-clearing symptoms from CMV-M–infected Nicotiana tabacum cv. Samsun plants, and asymptomatic leaves from tobacco plants infected by CMV-Q and mock-inoculated plants. (B) Western blots using anti-CP monoclonal and anti-Fd I polyclonal antibodies to analyse CMV CP and Fd I in the abovementioned samples, with actin as a control. (C) Northern blot conducted with 10 µg total RNA to examine the accumulation of Fd I transcription using a specific probe corresponding to position 75–435 of Fd I mRNA. The loading control comprised 25 S rRNA.
Viral infection has been shown to affect the expression levels of chloroplast-related genes4,5,37–39. A northern blot analysis was conducted to determine whether the decreased Fd I levels were associated with decreased Fd I mRNA content. Approximately 10 µg total RNA from each of the above-mentioned samples was subjected to agarose gel electrophoresis. The Fd I mRNA contents were similar in all five samples (Fig. 3C), implying that Fd I transcription was unaffected by the viral infection. Thus, the decrease in the abundance of Fd I may be due to its interaction with the CP of CMV-M.
Decreased ferredoxin I levels induced chlorosis symptoms
To determine whether the down-regulation of Fd I is directly related to the development of chlorosis symptoms, Fd I was silenced using a Tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) system40. At 21 dpi, chlorosis symptoms were observed on N. benthamiana plants injected with TRV-FdI, similar to those induced by CMV-M infection (Fig. 4A). No obvious symptoms were detected on TRV-infected plants (Fig. 4A). A western blot analysis confirmed that the Fd I abundance had decreased in TRV-FdI–injected plants (Fig. 4B), which was likely directly related to the development of chlorosis symptoms.
Figure 4.
Symptoms induced by eliminating Fd I production. (A) Chlorosis symptoms on plants in which Fd I was silenced with a TRV-based virus-induced gene silencing system. The symptoms were similar to those induced by CMV-M on Nicotiana benthamiana plants. Additionally, there were no obvious symptoms on plants carrying the empty TRV vector. (B) Accumulation of Fd I in mock-, TRV-, and TRV-FdI–inoculated samples, as determined by western blot analysis, with plant actin as a control.
Viral infection inhibited transport of ferredoxin I into chloroplasts
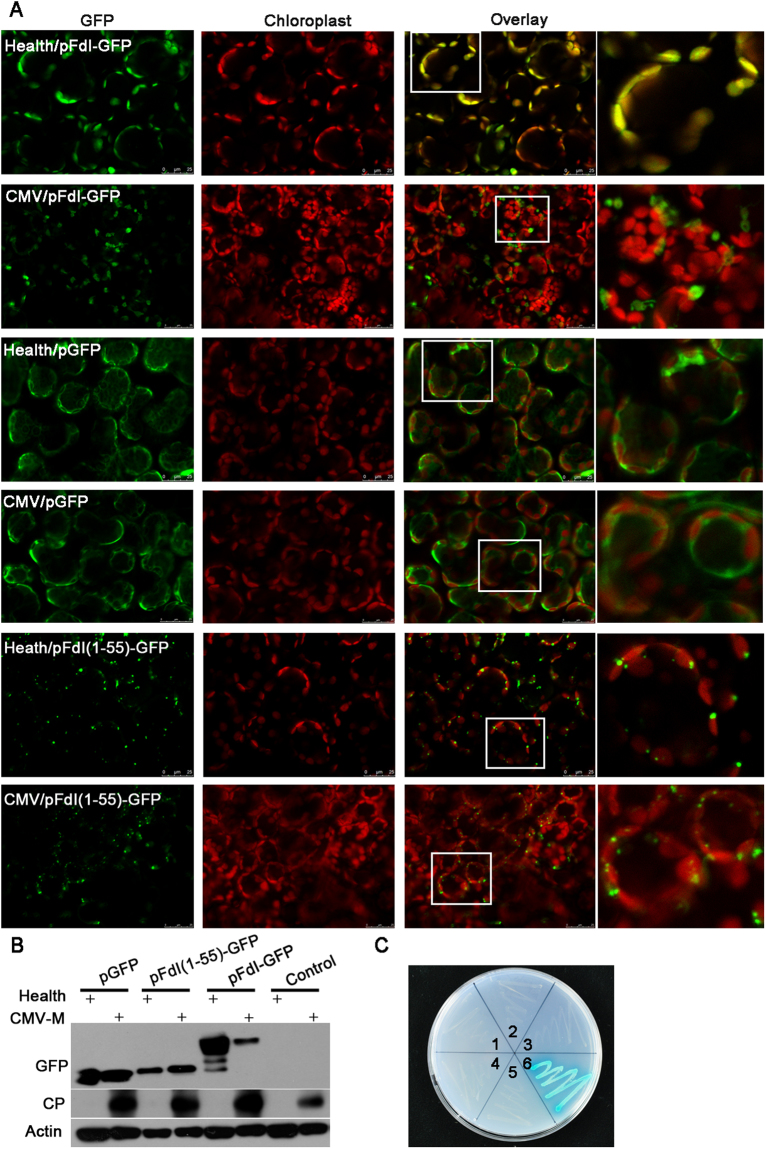
Fd I is encoded by the nuclear genome. The precursor of Fd I is produced in the cytoplasm and then imported into chloroplasts41. To investigate whether the transport of Fd I is influenced by the viral infection, the Fd I ORF was inserted into the pGFP vector, which contains the ORF of the gene encoding the green fluorescent protein (GFP). The pFdI-GFP plasmid was infiltrated into both healthy and CMV-M-infected N. benthamiana plants with A. tumefaciens. The empty pGFP vector was used as the control. When pFdI-GFP was transferred into healthy plants, the fluorescence of the Fd I-GFP fusion protein was observed in chloroplasts. However, when pFdI-GFP was injected into the chlorotic leaves of CMV-M–infected plants, most of the Fd I-GFP fusion protein was observed in the cytoplasm instead of chloroplasts (Fig. 5A). The localization of GFP alone was not affected by CMV-M infection, as GFP fluorescence was observed in the cytoplasm in both healthy and CMV-M–infected plants (Fig. 5A). We also investigated the subcellular localization of the CP of CMV-M, and observed that CP-GFP fusion protein was localized in the cytoplasm (Supplementary Fig. S1). These findings indicated that the CP of CMV-M interfered with the import of Fd I into chloroplasts.
Figure 5.
Subcellular production and localization of Fd I in CMV-M–infected or healthy Nicotiana benthamiana plants. (A) The Fd I-GFP fusion protein mainly localized in the chloroplasts and cytoplasm of healthy and CMV-M–infected plants, respectively. Whereas, GFP alone was mainly localized in the cytoplasm in healthy and CMV-M–infected hosts. Residues 1–55 at the N-terminal of Fd I formed a transit peptide targeting GFP into chloroplasts in both healthy and CMV-M–infected plants. Scale bars = 25 µm. (B) Western blot analysis with an anti-GFP antibody to detect GFP alone (pGFP), GFP fused to Fd I (pFdI-GFP), and GFP fused to the Fd I transit peptide [pFdI(1–55)-GFP]. (C) In yeast cells, the CP of CMV-M did not interact with the Fd I transit peptide or mature Fd I. 1, pGADT7-RecT/pGBKT7-Lam (negative control); 2, pGAD-FdI (1–55)/pGBKT7; 3, pGAD-FdI (56–145)/pGBKT7; 4, pGAD-FdI (1–55)/pGBK-MCP; 5, pGAD-FdI (56–145)/pGBK-MCP; 6, pGADT7-RecT/pGBKT7-53 (positive control).
The import of Fd I into chloroplasts is mediated by a transit peptide (TP) that is removed inside chloroplasts to release the mature form of Fd I42. To determine whether the function of the Fd I TP is inhibited by the infection of CMV-M, we first identified the TP by generating constructs for the production of fusion proteins in which different parts of the Fd I N-terminal were fused to GFP. When GFP was linked to residues 1–55 of Fd I [pFdI(1–55)-GFP], GFP fluorescence was detected in chloroplasts (Fig. 5A), indicating that residues 1–55 at the N-terminus of Fd I were sufficient to target the protein to chloroplasts. When the pFdI(1–55)-GFP plasmid was infiltrated into healthy or CMV-M–infected plants, the Fd I TP-GFP fusion proteins was localized in the chloroplasts (Fig. 5A), confirming that the function of the Fd I TP was unaffected by CMV-M.
Western blots were used to detect GFP or GFP-tagged fusion proteins in healthy and CMV-M–infected N. benthamiana plants after the A. tumefaciens-mediated infiltration. The accumulation of the Fd I-GFP fusion proteins was much lower in CMV-M–infected N. benthamiana plants than in healthy plants, while the accumulation of GFP alone or the Fd I TP-GFP fusion proteins were similar in CMV-M–infected and healthy N. benthamiana plants. These results confirmed that the Fd I production was influenced by CMV-M.
To investigate whether CMV-M interacts with the Fd I TP, individual constructs containing sequences encoding either the Fd I TP (residues 1–55) or mature Fd I (residues 56–145) were cloned into pGADT7, generating the pGAD-FdI (1–55) and pGAD-FdI (56–145) plasmids, respectively. When these plasmids were used in a YTH assay with pGBK-MCP, the CP of CMV-M did not interact with the Fd I TP or the mature form of Fd I (Fig. 5C). This explained why CMV-M only interfered with the function and stability of the Fd I precursor, and not the Fd I TP.
Discussion
In this study, we revealed that the chloroplast ptrotein, Fd I, interacted with the CP of CMV-M but not with that of CMV-Q. We also observed that the interaction influenced Fd I expresion and function to induce the formation of mosaic symptoms. The chloroplast is oftern targted by plant viruses and usually undergoes structural and functional changes during viral infection1–3. The CMV infection could induce mosaic symptoms, with the CP as an important determinant31–33. However, no chloroplast proteins had previously been confirmed to be associate with the CP. In this study, we confirmed that the chloroplasts protein Fd I can interact with the CP of CMV-M, but not the CP of CMV-Q. Subsequent analyses indicated that Fd I was associated with the expression of chlorotic symptoms, as the Fd I abundance decreased following an infection by CMV-M, but not by CMV-Q. Additionally, the chlorosis symptoms induced by the silencing of Fd I were the same as those resulting from an infection by CMV-M.
An earlier study concluded that Fd I interacts with the CP of Tomato mosaic virus to regulate chlorosis symptom development, although how the interaction induces the formation of the symptoms was not determined43. Researchers speculated that Fd I influences the transport of the viral CP into chloroplasts because the accumulation of the CP of Tobacco mosaic virus in chloroplasts increased when Fd I was silenced44. However, the results of our analyses suggest that Fd I has different functions during viral infections. We observed that the CMV-M infection interfered with the import of Fd I into chloroplasts, as most of the Fd I remained in the cytoplasm. Moreover, the CP of CMV-M localized in the cytoplasm, where it interacted with Fd I. Furthermore, the CP of CMV-M interacted with the precursor of Fd I synthesized in the cytoplasm, but not with the Fd I TP or the mature form of Fd I localized in chloroplasts. Thus, we concluded that the CP of CMV-M specifically interacts with Fd I to influence its function.
We also observed that Fd I levels decreased in response to an infection by CMV-M. Researchers previously suggested that the CP may influence the chloroplast-to-nucleus retrograde signals to repress the transcription of chloroplast-related genes13. However, our northern blot analyses indicated that the Fd I transcript levels were unaffected by the CMV infection. These results imply that the decrease in Fd I levels was related to the interaction with the CP of CMV-M. Consequently, we speculate that Fd I may fail to fold properly because of the interaction, thereby preventing it from being efficiently imported by chloroplasts. The accumulation of unfolded or misfolded proteins will inhibit normal cellular functions. Thus, the structurally unstable or abnormal Fd I may be selectively degraded by cell proteases machineries45–48.
Because Fd I is an indispensable part of photosystem I and donates electrons to various pathways41, a decrease in Fd I content alters the distribution of electrons49 (Fig. 6). The disrupted electrons transport in the plant host due to an infection by CMV stimulates the accumulation of H2O2 in chloroplasts50. The considerable oxidative stress imposed by H2O2 might be toxic to chloroplasts44,51. Moreover, many enzymes receive electrons from Fd I, implying a decrease in Fd I content affects the metabolic activities occurring in the chloroplast (Fig. 6), including the biosynthesis of chlorophyll and phytochrome41. Therefore, the abnormal production and translocation of Fd I resulting from an infection by CMV-M is responsible for the formation of chlorosis symptoms along with disrupted chloroplast structure and function51. However, the detailed mechanism by which Fd I is repressed via its interaction with the CP requires further research.
Figure 6.
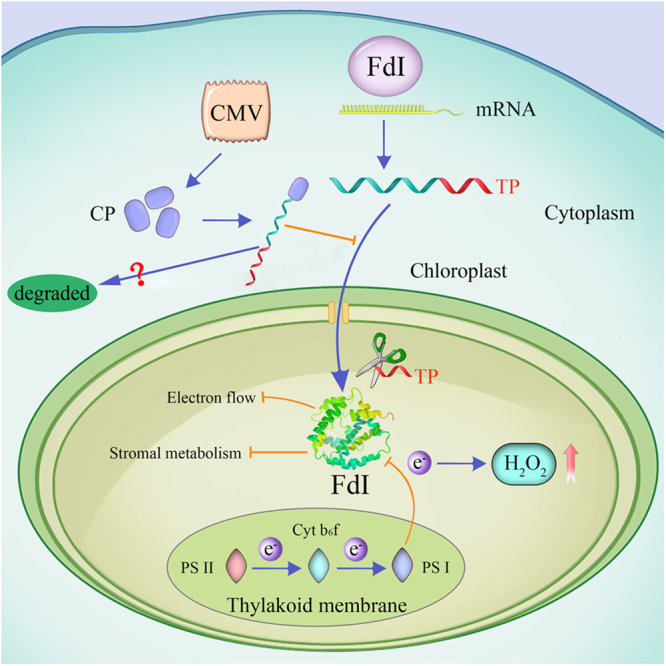
Model for Fd I functions during viral infections. After it is synthesized in the cytoplasm, the Fd I precursor is imported into chloroplasts because of the transit peptide (TP), which is removed inside the chloroplasts. Our data indicate that the CP of CMV interacts with the Fd I precursor to inhibit the import process. Decreases in Fd I production following an infection by CMV will need to be further verified. Because Fd I localizes on the stromal side of chloroplasts and transfers electrons from photosystem I (PS I) to various pathways, decreases in Fd I abundance will disrupt the electrons transport chain and influence Fd I-dependent metabolic activities. This will ultimately lead to an increase in H2O2 levels and inhibited chloroplast function.
In addition to the CP, the 2b protein of CMV is also involved in the development of mosaic symptoms26. An infection by a 2b protein-deficient CMV-M mutant induces relatively weak chlorosis symptoms and a low viral accumulation in N. tabacum plants52. This suggests the 2b protein can enhance the accumulation of viruses by inhibiting several steps of the RNA silencing process53, but does not directly induce the development of mosaic symptoms13,52,54. By exchanging the CP between CMV-M and CMV-Q, we confirmed that the CP is a determinant of mosaic symptom development. Furthermore, the amino acid residue at position 129 of the CP influences the formation of symptoms on host plants. Modifying this amino acid was suggested to affect the three-dimensional structure of the CP55, viral assembly32, or viral movement56,57, but the effects on CP functions have not been fully characterized. Moreover, whether this amino acid participates in the interaction between the CP of CMV-M and Fd I will need to be investigated further.
In summary, the results presented herein indicate the CP of CMV-M interacts with Fd I and disrupts its synthesis and function, leading to the development of chlorosis symptoms on tobacco plants.
Materials and Methods
Plasmid construction
All vectors used in this study are listed in Table 1. Details of the primers used for cloning are provided in Supplementary Information Table S1.
Table 1.
Details regarding the plasmids used in this study.
| Plasmids | Primer | Instruction |
|---|---|---|
| pM3QCP | M-5UTR-F/M-5UTR-R, QCP-F/QCP-R, M-3UTR-F/M-3UTR-R | CP mutants and based on viral infectious cDNA clones. |
| pQ3MCP | Q-5UTR-F/M-5UTR-R, MCP-F/MCP-R, Q-3UTR-F/Q-3UTR-R | |
| pGBK-MCP | BD-MCP-F/BD-MCP-R | For YTH work and based on vector pGBKT7 and pGADT7 (Clontech). |
| PGBK-QCP | BD-QCP-F/BD-QCP-R | |
| pGAD-FdI | AD-FdI-F/AD-FdI-R | |
| pGAD-FdI(1–55) | AD-FdI-F/AD-FdI-55R | |
| pGAD-FdI(56–145) | AD-FdI-56F/AD-FdI-R | |
| pMCP-YFPN | Bi-MCP-F/Bi-MCP-R | For BiFC assay and based on vector pSPYNE-35S and pSPYCE-35S36. |
| pMCP-YFPC | ||
| pQCP-YFPN | Bi-QCP-F/Bi-QCP-R | |
| pQCP-YFPC | ||
| pFdI-YFPN | Bi-FdI-F/Bi-FdI-R | |
| pFdI-YFPC | ||
| pFdI-GFP | Sub-FdI-F/Sub-FdI-R | For subcellular localization assay and based on modified vector pCAMBIA1300. |
| pFdI(1–55)-GFP | Sub-FdI-F/Sub-FdI-55R | |
| pTRV-FdI | TRV-FdI-F/TRV-FdI-R | For VIGS assay and based on TRV mediated VIGS system40. |
Plant growth and viral inoculation
Nicotiana tabacum cv. Samsun and N. benthamiana plants were grown in a glasshouse at 24 °C under a 16-h light/8-h dark photoperiod. The infectious CMV-M cDNA clones (i.e., pM1, pM2, and pM3) under the control of the T7 promoter were constructed in our laboratory. The infectious CMV-Q clones (i.e., pQ1, pQ2, and pQ3) were a gift from the Shouwei Ding Laboratory (University of California, Riverside, CA, USA)58. The CP-encoding ORF was exchanged between CMV-M and CMV-Q, resulting in two plasmids (i.e., pM3QCP and pQ3MCP). Transcripts for the viral infectious clones pM1/pM2/pM3 (CMV-M), pM1/pM2/pM3QCP (CMV-MQCP), pQ1/pQ2/pQ3 (CMV-Q), and pQ1/pQ2/pQ3MCP (CMV-QMCP) were obtained using a mMESSAGE mMACHINE® Kit (Ambion), and were used to inoculate the top two fully expanded leaves of N. tabacum plants at the four-leaf stage.
Yeast two-hybrid screening
A cDNA library for N. tabacum leaves was constructed using the Matchmaker™ Library Construction & Screening Kit (Clontech) according to the manufacturer’s instructions. The library was screened with a bait plasmid in co-transformed Saccharomyces cerevisiae strain AH109 cells. The yeast cells were then spread on synthetic dropout (SD) medium lacking adenine, histidine, leucine, and tryptophan (SD/−Ade/−His/−Leu/−Trp) to identify putative interacting proteins. To verify protein interactions, clones growing well on SD/−Ade/−His/−Leu/−Trp medium were used to inoculate into the same medium supplemented with X-α-Gal (SD/−Ade/−His/−Leu/−Trp/X-α-Gal). Plasmids were extracted from positive clones and sequenced for further functional analyses.
Bimolecular fluorescence complementation analysis
We used the pSPYNE-35S and pSPYCE-35S BiFC vectors, which enabled the expressio of the N- and C-termini of YFP (YFPN and YFPC, respectively)36. Using a double-digestion method, the ORFs of the CP genes from CMV-M and CMV-Q were cloned into pSPYNE-35S and pSPYCE-35S to produce the pMCP-YFPN, pMCP-YFPC, pQCP-YFPN, and pQCP-YFPC plasmids. The Fd I ORF was also cloned into pSPYNE-35S and pSPYCE-35S, generating the pFdI-YFPN and pFdI-YFPC plasmids. The following plasmid pairs were used to transform A. tumefaciens strain EHA105 cells, which were grown in medium until the cultures reached the exponential growth phase: pMCP-YFPN/pFdI-YFPC, pFdI-YFPN/pMCP-YFPC, pQCP-YFPN/pFdI-YFPC, and pFdI-YFPN/pQCP-YFPC. We used needles to infiltrate the leaves of healthy N. benthamiana plants with the collected cultures. Leaves were collected after 48 or 72 h and observed under an SP5 confocal laser scanning microscope (Leica Microsystems). The excitation and emission wavelengths for YFP were 514 nm and 520–550 nm, respectively.
Subcellular localization in plant cells
To study the subcellular localization of the target protein, the pCAMBIA1300 transient expression vector was modified to carry the ORF of the gene encoding GFP (pGFP). The Fd I ORF and an fragments encoding the N-terminal (corresponding to residues 1–55) were inserted into the pGFP vector to generate the pFdI-GFP and pFdI(1–55)-GFP plasmids, respectively. These constructs were incorporated into healthy and CMV-M–infected N. benthamiana plants using A. tumefaciens strain EHA105. To locate the GFP-tagged fusion proteins, GFP fluorescence and the auto-fluorescence of chloroplasts were induced by a laser (488 nm), and captured at emission wavelengths of 500–530 nm and 650–700 nm, respectively.
Virus-induced gene silencing
The VIGS system is commonly used to study the function of plant genes40. We applied a TRV-based VIGS system, which used modified infected clones to transfer the DNA fragments that targeted homologous genes40. A sequence corresponding to nucleotide position 63–433 of tobacco Fd I mRNA was amplified and inserted into the pTRV2 vector using restriction enzyme digestions to produce the pTRV-FdI plasmid. This plasmid was then inserted into A. tumefaciens strain EHA105 cells together with pTRV1 (TRV-FdI). The empty pTRV1/pTRV2 vector were used as the control (TRV). Cultures were then injected into N. benthamiana plants at the five-leaf stage, with N. benthamiana plants infected with CMV-M served as positive controls. Plants were photographed using a digital camera (Nikon, Tokyo, Japan) at 21 dpi.
Western blot analyses
Plant leaves were immediately ground in liquid nitrogen and mixed with 2× loading buffer for sodium dodecyl sulfate polyacrylamide gel electrophoresis. Proteins were then transferred to a polyvinylidene difluoride (PVDF) membrane (GE Healthcare) using a semi-dry transfer method. The PVDF membrane was blocked with 5% milk for 1 h at room temperature. After an overnight incubation with the primary antibody at 4 °C, the membrane was treated with a horseradish peroxidase-labelled secondary antibody (Sigma–Aldrich). Signals were visualized by exposing the PVDF membrane to an x-ray film after adding Pierce™ ECL Western Blotting Substrate (Thermo Scientific). The virus was detected with an anti-CP monoclonal antibody, while GFP was detected with an anti-GFP monoclonal antibody (Cell Signaling Technology). Plant actin was used as the internal control, and was detected with a polyclonal antibody (Sigma–Aldrich).
Northern blot hybridization
Total RNA was extracted from plant tissue using the TRIzol reagent (Invitrogen). The extracted RNA was separated on a 1.2% agarose gel containing formaldehyde, transferred to a nylon N+-membrane (GE Healthcare), and cross-linked with ultraviolet light. The membrane was stained with a methylene blue solution (0.5 N NaAc, pH5.2 and 0.04% methylene blue) and rinsed with distilled water to produce clearly visible bands, which were used as the loading control. The DNA fragments corresponding to position 75–435 of the Fd I mRNA sequence were labeled with [α-32P]-dCTP using the Rediprime II DNA Labeling System (GE Healthcare). Signals were visualized on exposed film.
Electronic supplementary material
Acknowledgements
This study was supported by the National Key Research and Development Program (2016YFC1200605) and the Basic Scientific Research Foundation of the Chinese Academy of Inspection and Quarantine (2017JK035). We thank Pro. Guifen Li (Chinese Academy of Inspection and Quarantine) for providing us with the antisera against the CMV CP (monoclonal antibody).
Author Contributions
Y.Q., Y.W., X. L. and S.Z. planned and designed the study. Y.Q., Y.Z. C.W., and R.L. conducted the experiments. All authors interpreted the results. Y.Q., Y.W., X.L. and S.Z. wrote the manuscript, which all authors reviewed.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-19525-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yupin Wu, Email: wuyuping@caiqtest.com.
Xinshi Li, Email: lixinshi2017@126.com.
Shuifang Zhu, Email: zhusf@caiq.gov.cn.
References
- 1.Zhao, J., Zhang, X., Hong, Y. G. & Liu, Y. Chloroplast in plant-virus interaction. Front Microbiol7, 10.3389/fmicb.2016.01565 (2016). [DOI] [PMC free article] [PubMed]
- 2.Li Y, Cui H, Cui X, Wang A. The altered photosynthetic machinery during compatible virus infection. Curr Opin Virol. 2016;17:19–24. doi: 10.1016/j.coviro.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharyya, D. & Chakraborty, S. Chloroplast: The trojan horse in plant-virus interaction. Mol Plant Pathol 12533 (2017). [DOI] [PMC free article] [PubMed]
- 4.Shi B, et al. Identification and regulation of host genes related to Rice stripe virus symptom production. New Phytol. 2016;209:1106–1119. doi: 10.1111/nph.13699. [DOI] [PubMed] [Google Scholar]
- 5.Mochizuki T, Ogata Y, Hirata Y, Ohki ST. Quantitative transcriptional changes associated with chlorosis severity in mosaic leaves of tobacco plants infected with Cucumber mosaic virus. Mol Plant Pathol. 2014;15:242–254. doi: 10.1111/mpp.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babu M, Gagarinova AG, Brandle JE, Wang A. Association of the transcriptional response of soybean plants with Soybean mosaic virus systemic infection. J Gen Virol. 2008;89:1069–1080. doi: 10.1099/vir.0.83531-0. [DOI] [PubMed] [Google Scholar]
- 7.Dardick C. Comparative expression profiling of Nicotiana benthamiana leaves systemically infected with three fruit tree viruses. Mol Plant Microbe Interact. 2007;20:1004–1017. doi: 10.1094/MPMI-20-8-1004. [DOI] [PubMed] [Google Scholar]
- 8.Di Carli M, Benvenuto E, Donini M. Recent insights into plant-virus interactions through proteomic analysis. J Proteome Res. 2012;11:4765–4780. doi: 10.1021/pr300494e. [DOI] [PubMed] [Google Scholar]
- 9.Di Carli M, et al. Proteomic analysis of the plant-virus interaction in Cucumber mosaic virus (CMV) resistant transgenic tomato. J Proteome Res. 2010;9:5684–5697. doi: 10.1021/pr100487x. [DOI] [PubMed] [Google Scholar]
- 10.Diaz-Vivancos P, et al. Alteration in the chloroplastic metabolism leads to ROS accumulation in pea plants in response to Plum pox virus. J Exp Bot. 2008;59:2147–2160. doi: 10.1093/jxb/ern082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehto K, Tikkanen M, Hiriart JB, Paakkarinen V, Aro EM. Depletion of the photosystem II core complex in mature tobacco leaves infected by the flavum strain of Tobacco mosaic virus. Mol Plant Microbe Interact. 2003;16:1135–1144. doi: 10.1094/MPMI.2003.16.12.1135. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharyya D, et al. A geminivirus betasatellite damages the structural and functional integrity of chloroplasts leading to symptom formation and inhibition of photosynthesis. J Exp Bot. 2015;66:5881–5895. doi: 10.1093/jxb/erv299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mochizuki T, Yamazaki R, Wada T, Ohki ST. Coat protein mutations in an attenuated Cucumber mosaic virus encoding mutant 2b protein that lacks RNA silencing suppressor activity induces chlorosis with photosynthesis gene repression and chloroplast abnormalities in infected tobacco plants. Virology. 2014;456:292–299. doi: 10.1016/j.virol.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Kong L, Wu J, Lu L, Xu Y, Zhou X. Interaction between Rice stripe virus disease-specific protein and host PsbP enhances virus symptoms. Mol Plant. 2014;7:691–708. doi: 10.1093/mp/sst158. [DOI] [PubMed] [Google Scholar]
- 15.Balasubramaniam M, Kim BS, Hutchens-Williams HM, Loesch-Fries LS. The photosystem II oxygen-evolving complex protein PsbP interacts with the coat protein of Alfalfa mosaic virus and inhibits virus replication. Mol Plant Microbe Interact. 2014;27:1107–1118. doi: 10.1094/MPMI-02-14-0035-R. [DOI] [PubMed] [Google Scholar]
- 16.Gao L, et al. NIa-pro of Papaya ringspot virus interacts with papaya methionine sulfoxide reductase B1. Virology. 2012;434:78–87. doi: 10.1016/j.virol.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Y, Liu Z, Xu J, Zhou T, Fan Z. HC-Pro protein of Sugar cane mosaic virus interacts specifically with maize ferredoxin-5 in vitro and in planta. J Gen Virol. 2008;89:2046–2054. doi: 10.1099/vir.0.2008/001271-0. [DOI] [PubMed] [Google Scholar]
- 18.Jin Y, et al. The HC-pro protein of Potato virus Y interacts with NtMinD of tobacco. Mol Plant Microbe Interact. 2007;20:1505–1511. doi: 10.1094/MPMI-20-12-1505. [DOI] [PubMed] [Google Scholar]
- 19.Lin L, et al. Interaction between potyvirus P3 and ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) of host plants. Virus Genes. 2011;43:90–92. doi: 10.1007/s11262-011-0596-6. [DOI] [PubMed] [Google Scholar]
- 20.Van Regenmortel, M. H. V. et al. In Virus taxonomy: seventh report of the international committee on taxonomy of viruses. (2000).
- 21.Scholthof KBG, et al. Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol. 2011;12:938–954. doi: 10.1111/j.1364-3703.2011.00752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palukaitis P, Garcia-Arenal F. Cucumoviruses. Adv Virus Res. 2003;62:241–323. doi: 10.1016/S0065-3527(03)62005-1. [DOI] [PubMed] [Google Scholar]
- 23.Hayes RJ, Buck KW. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell. 1990;63:363–368. doi: 10.1016/0092-8674(90)90169-F. [DOI] [PubMed] [Google Scholar]
- 24.Ding SW, Anderson BJ, Haase HR, Symons RH. New overlapping gene encoded by the Cucumber mosaic virus genome. Virology. 1994;198:593–601. doi: 10.1006/viro.1994.1071. [DOI] [PubMed] [Google Scholar]
- 25.Boccard F, Baulcombe D. Mutational analysis of cis-acting sequences and gene function in RNA3 of Cucumber mosaic virus. Virology. 1993;193:563–578. doi: 10.1006/viro.1993.1165. [DOI] [PubMed] [Google Scholar]
- 26.Mochizuki T, Ohki ST. Cucumber mosaic virus: viral genes as virulence determinants. Mol Plant Pathol. 2012;13:217–225. doi: 10.1111/j.1364-3703.2011.00749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi BJ, Palukaitis P, Symons RH. Differential virulence by strains of Cucumber mosaic virus is mediated by the 2b gene. Mol Plant Microbe Interact. 2002;15:947–955. doi: 10.1094/MPMI.2002.15.9.947. [DOI] [PubMed] [Google Scholar]
- 28.Shintaku M. Coat protein gene sequences of two Cucumber mosaic virus strains reveal a single amino acid change correlating with chlorosis induction. J Gen Virol. 1991;72:2587–2589. doi: 10.1099/0022-1317-72-10-2587. [DOI] [PubMed] [Google Scholar]
- 29.Mossop DW, Francki RI. Association of RNA 3 with aphid transmission of Cucumber mosaic virus. Virology. 1977;81:177–181. doi: 10.1016/0042-6822(77)90071-X. [DOI] [PubMed] [Google Scholar]
- 30.Rao ALN, Francki RIB. Distribution of determinants for symptom production and host range on the 3 RNA components of Cucumber mosaic virus. J Gen Virol. 1982;61:197–205. doi: 10.1099/0022-1317-61-2-197. [DOI] [Google Scholar]
- 31.Shintaku MH, Zhang L, Palukaitis P. A single amino acid substitution in the coat protein of Cucumber mosaic virus induces chlorosis in tobacco. Plant Cell. 1992;4:751–757. doi: 10.1105/tpc.4.7.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki M, Kuwata S, Masuta C, Takanami Y. Point mutations in the coat protein of Cucumber mosaic virus affect symptom expression and virion accumulation in tobacco. J Gen Virol. 1995;76:1791–1799. doi: 10.1099/0022-1317-76-7-1791. [DOI] [PubMed] [Google Scholar]
- 33.Mochizuki T, Ohki ST. Single amino acid substitutions at residue 129 in the coat protein of Cucumber mosaic virus affect symptom expression and thylakoid structure. Arch Virol. 2011;156:881–886. doi: 10.1007/s00705-010-0910-y. [DOI] [PubMed] [Google Scholar]
- 34.Chaturvedi S, Seo JK, Rao AL. Functionality of host proteins in Cucumber mosaic virus replication: GAPDH is obligatory to promote interaction between replication-associated proteins. Virology. 2016;494:47–55. doi: 10.1016/j.virol.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Citovsky V, et al. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol. 2006;362:1120–1131. doi: 10.1016/j.jmb.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 36.Walter M, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant Journal. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 37.Vozarova Z, Zilova M, Subr Z. Differentially expressed genes in healthy and Plum pox virus-infected Nicotiana benthamiana plants. Acta Virologica. 2015;59:389–397. doi: 10.4149/av_2015_04_389. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Yang J, Bi HP, Zhang P. Why mosaic? Gene expression profiling of African cassava mosaic virus-infected cassava reveals the effect of chlorophyll degradation on symptom development. J Integr Plant Biol. 2014;56:122–132. doi: 10.1111/jipb.12133. [DOI] [PubMed] [Google Scholar]
- 39.Lu J, et al. Transcriptome analysis of Nicotiana tabacum infected by Cucumber mosaic virus during systemic symptom development. Plos One. 2012;7:e43447. doi: 10.1371/journal.pone.0043447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Schiff M, Marathe R. & Dinesh-Kumar, S. P. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to Tobacco mosaic virus. Plant Journal. 2002;30:415–429. doi: 10.1046/j.1365-313X.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- 41.Hanke G, Mulo P. Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell and Environment. 2013;36:1071–1084. doi: 10.1111/pce.12046. [DOI] [PubMed] [Google Scholar]
- 42.Jarvis P. Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 2008;179:257–285. doi: 10.1111/j.1469-8137.2008.02452.x. [DOI] [PubMed] [Google Scholar]
- 43.Sun XC, et al. In vitro binding and bimolecular fluorescence complementation assays suggest an interaction between Tomato mosaic virus coat protein and tobacco chloroplast ferredoxin I. Arch Virol. 2013;158:2611–2615. doi: 10.1007/s00705-013-1778-4. [DOI] [PubMed] [Google Scholar]
- 44.Ma Y, Zhou T, Hong Y, Fan Z, Li H. Decreased level of ferredoxin I in Tobacco mosaic virus-infected tobacco is associated with development of the mosaic symptom. Physiol Mol Plant Pathol. 2008;72:39–45. doi: 10.1016/j.pmpp.2008.05.004. [DOI] [Google Scholar]
- 45.Nishimura K, Kato Y, Sakamoto W. Chloroplast Proteases: updates on proteolysis within and across suborganellar compartments. Plant Physiology. 2016;171:2280–2293. doi: 10.1104/pp.16.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Wijk KJ. Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Annu Rev Plant Biol. 2015;66:75–111. doi: 10.1146/annurev-arplant-043014-115547. [DOI] [PubMed] [Google Scholar]
- 47.Yoshioka-Nishimura M, Yamamoto Y. Quality control of Photosystem II: the molecular basis for the action of FtsH protease and the dynamics of the thylakoid membranes. Journal of photochemistry and photobiology B: Biology. 2014;137:100–106. doi: 10.1016/j.jphotobiol.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 48.Ling QH, Huang WH, Baldwin A, Jarvis P. Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science. 2012;338:655–659. doi: 10.1126/science.1225053. [DOI] [PubMed] [Google Scholar]
- 49.Holtgrefe S, et al. Decreased content of leaf ferredoxin changes electron distribution and limits photosynthesis in transgenic potato plants. Plant Physiology. 2003;133:1768–1778. doi: 10.1104/pp.103.026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song X, et al. Effects of Cucumber mosaic virus infection on electron transport and antioxidant system in chloroplasts and mitochondria of cucumber and tomato leaves. Physiologia Plantarum. 2009;135:246–257. doi: 10.1111/j.1399-3054.2008.01189.x. [DOI] [PubMed] [Google Scholar]
- 51.Lei R, Jiang H, Hu F, Yan J, Zhu S. Chlorophyll fluorescence lifetime imaging provides new insight into the chlorosis induced by plant virus infection. Plant Cell Rep. 2017;36:327–341. doi: 10.1007/s00299-016-2083-y. [DOI] [PubMed] [Google Scholar]
- 52.Qiu Y, Zhang Y, Hu F, Zhu S. Characterization of siRNAs derived from Cucumber mosaic virus in infected tobacco plants. Arch Virol. 2017;162:2077–2082. doi: 10.1007/s00705-017-3335-z. [DOI] [PubMed] [Google Scholar]
- 53.Csorba T, Kontra L, Burgyan J. viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology. 2015;479:85–103. doi: 10.1016/j.virol.2015.02.028. [DOI] [PubMed] [Google Scholar]
- 54.Du Z, et al. The 2b protein and the C-terminus of the 2a protein of Cucumber mosaic virus subgroup I strains both play a role in viral RNA accumulation and induction of symptoms. Virology. 2008;380:363–370. doi: 10.1016/j.virol.2008.07.036. [DOI] [PubMed] [Google Scholar]
- 55.Gellert A, Salanki K, Naray-Szabo G, Balazs E. Homology modelling and protein structure based functional analysis of five cucumovirus coat proteins. J Mol Graph Model. 2006;24:319–327. doi: 10.1016/j.jmgm.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 56.Mochizuki T, Ohki ST. Amino acid 129 in the coat protein of Cucumber mosaic virus primarily determines invasion of the shoot apical meristem of tobacco plants. J Gen Plant Pathol. 2005;71:326–332. doi: 10.1007/s10327-005-0207-2. [DOI] [Google Scholar]
- 57.Wong SM, Thio SSC, Shintaku MH, Palukaitis P. The rate of cell-to-cell movement in squash of Cucumber mosaic virus is affected by sequences of the capsid protein. Mol Plant Microbe Interact. 1999;12:628–632. doi: 10.1094/MPMI.1999.12.7.628. [DOI] [Google Scholar]
- 58.Ding SW, et al. Efficient infection from cDNA clones of cucumber mosaic cucumovirus RNAs in a new plasmid vector. J Gen Virol. 1995;76:459–464. doi: 10.1099/0022-1317-76-2-459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.