Abstract
METTL20 is a seven-β-strand methyltransferase that is localised to the mitochondria and tri-methylates the electron transfer flavoprotein (ETF) β subunit (ETFB) at lysines 200 and 203. It has been shown that METTL20 decreases the ability of ETF to extract electrons from medium-chain acyl-coenzyme A (CoA) dehydrogenase (MCAD) and glutaryl-CoA dehydrogenase in vitro. METTL20-mediated methylation of ETFB influences the oxygen consumption rate in permeabilised mitochondria, suggesting that METTL20-mediated ETFB methylation may also play a regulatory role in mitochondrial metabolism. In this study, we generated Mettl20 knockout (KO) mice to uncover the in vivo functions of METTL20. The KO mice were viable, and a loss of ETFB methylation was confirmed. In vitro enzymatic assays revealed that mitochondrial ETF activity was higher in the KO mice than in wild-type mice, suggesting that the KO mice had higher β-oxidation capacity. Calorimetric analysis showed that the KO mice fed a ketogenic diet had higher oxygen consumption and heat production. A subsequent cold tolerance test conducted after 24 h of fasting indicated that the KO mice had a better ability to maintain their body temperature in cold environments. Thus, METTL20 regulates ETF activity and heat production through lysine methylation when β-oxidation is highly activated.
Introduction
Protein post-translational modifications (PTMs) are involved in diverse cellular processes such as gene expression, signal transduction, and intracellular interactions. Lysine methylation in histones has been extensively studied for more than a decade; it is known to play important roles in gene expression, replication, and genome stability1–3. Although hundreds of non-histone lysine methylation sites have been reported, the enzymes responsible for methylation of these lysines and the physiological functions of these PTMs remain largely unknown.
There are approximately 40 MTase genes that may participate in mitochondrial functioning because the encoded proteins contain mitochondria-targeting signal sequences4. Nonetheless, only a small number of these genes has been characterised so far. METTL20 was the first mitochondrial lysine MTase (KMT) to be discovered (by two independent groups)5,6. METTL20 specifically methylates the β subunit (ETFB) of electron transfer flavoprotein (ETF), which acts as a mobile carrier of electrons from several flavin adenine dinucleotide–containing dehydrogenases to ETF: quinone oxidoreductase. The two METTL20 methylation sites in ETFB, i.e. K200 and K203, are located in the proximity of the recognition loop responsible for the interaction between ETF and medium-chain acyl-coenzyme A (CoA) dehydrogenase (MCAD)7; it is believed to be involved in interactions with other ETF-dependent dehydrogenases. Accordingly, it has been shown that METTL20 decreases the ability of ETF to extract electrons from MCAD and glutaryl-CoA dehydrogenase in vitro6. Moreover, METTL20-mediated methylation of ETFB influences the oxygen consumption rate in permeabilised mitochondria that are supplied with palmitoyl-L-carnitine5, suggesting that METTL20-mediated ETFB methylation may play a regulatory role in mitochondrial metabolism.
Several approaches have been developed to probe lysine-methylated proteins specifically. One such approach involves antibodies specific to methylated lysine8,9. The second approach takes advantage of methylated lysine-binding domains. It has been reported that an engineered version of the triple malignant-brain-tumour domain region, i.e. L3MBTL1, can serve as a probe for the detection and enrichment of proteins containing mono- and di-methylated lysine residue(s)10,11. Another approach is to use cofactor analogues for labelling, detection, and pull-down via copper-catalysed azide-alkyne cycloaddition, also known as click chemistry, to enrich PTM targets. Synthetic co-factors for lysine methylation of several SET domain KMTs have been reported, for example, SUV39H212, G9a/GLP12,13, ESET14, Set7/913, and a seven-β-strand MTase, METTL1015.
In the present study, we performed a proteomic substrate screening for METTL20 in mitochondria of cultured cells. We found that ETFB is a major substrate; this result is consistent with other findings5,6. To explore the biological function of METTL20-mediated ETFB methylation, we created Mettl20 knockout (KO) mice and characterised phenotypes related to their metabolism.
Results
ETFB is a major substrate for METTL20 in the mitochondria, and its catalytic activity is regulated by lysine methylation
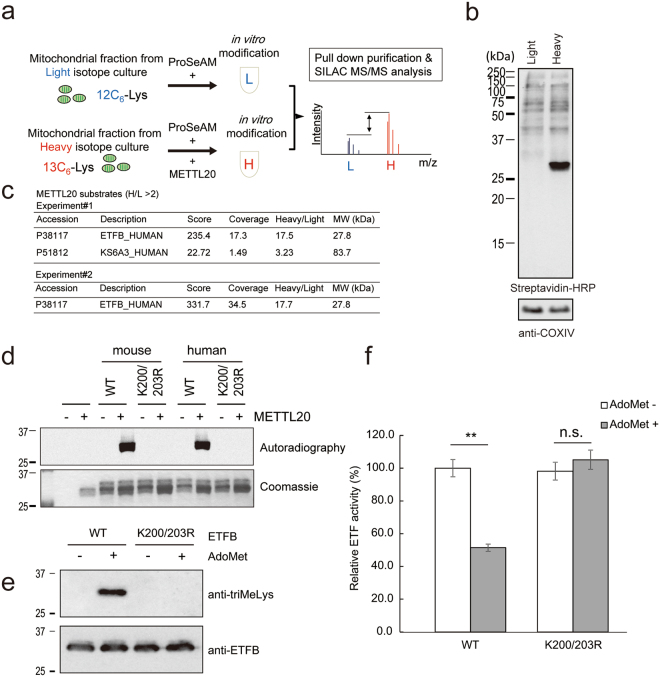
Recently, a mitochondrial MTase, METTL20, was identified as an ETFB MTase5,6. We also attempted proteomic identification of METTL20 substrates in mitochondria. A proteomic analysis using a synthetic S-adenosylmethionine (AdoMet) analogue, ProSeAM12,15, has been applied to identify the substrate (Fig. 1a). HEK293T cells were cultured in DMEM with 10% of dialyzed foetal calf serum (FCS) containing either light isotopic 12C6-Lys or heavy 13C6-Lys. Each mitochondrial fraction was incubated with ProSeAM12,13,15 in the absence or presence of His-METTL20. After biotinylation of ProSeAM-labelled proteins, a small part (5%) of the protein was subjected to western blotting with streptavidin conjugated with horseradish peroxidase (HRP; Fig. 1b). The samples were combined and mixed, and subsequent proteomic analysis revealed that ETFB was a major mitochondrial substrate for this enzyme (Fig. 1c).
Figure 1.
ETFB methylation is conserved in human and mouse, which regulates the catalytic activity. (a) A schematic drawing of screening for METTL20 substrates. Cells were cultured in either light isotope labeled Lys containing medium (L) or heavy isotope labeled Lys containing medium (H). The mitochondrial lysates were reacted with ProSeAM with (H) or without (L) His-METTL20. After the in vitro modification, biotin tags were introduced to the modified residues via click reaction. Then the two samples were mixed together, and biotinylated proteins were pull-down with Streptavidin beads. After Lys-C digestion, peptides were analyzed by LC-MS/MS. (b) Confirmation of ProSeAM labeling. After the ProSeAM mediated alkylation and biotinylation via click reaction, small aliquot of the protein samples were analyzed by SDS-PAGE followed by western blotting with Streptavidin-HRP and anti-COXIV antibody as a loading control. (c) METTL20 substrates identified in the screening. Summary of two independent experiments were shown. Note that only ETFB was the protein identified in both cases. (d) Recombinant ETF complex (ETFA WT/ ETFB WT or K200/203R) and His-METTL20 were incubated with 14C-labeled AdoMet for 2 h at 30 °C. The autoradiography was detected with the image analyzer BAS-5000. (e) 4 µg of the ETF complex were mixed with 4 µg of METTL20 in the presence or absence of AdoMet and incubated at 30 °C for 3 h. After the reaction, they were mixed on ice with 8 µg of His-MCAD, 50 µM octanoyl-CoA and 70 µM DCPIP. Initial rate of the reduction of DCPIP was calculated from OD600 measured at 30 °C for every 30 sec for 10 min with a microplate reader (SpectraMax 190, Molecular Devices). Average of three independent experiments, n = 3 for each experiments, mean ± SEM. Student’s T-test **p < 0.01. (f) Small aliquot of the ETF used in A were separated with SDS-PAGE, and their methylation were detected with anti-triMeLys antibodies (top) and anti-ETFB antibodies (bottom, as loading control).
We confirmed that ETFB was modified by ProSeAM (Supplementary Fig. S2a) and labelled with 14C-AdoMet (Supplementary Fig. S2b) in the presence of METTL20 in vitro. Because histones contain many methylated lysine residues, we also tested recombinant histone H3 (hereafter: H3) in an experiment and demonstrated that METTL20 can methylate both ETFB and H3 in vitro. It should be noted that because histones are nuclear proteins, they are possibly not a physiological target of METTL20, which is localised to mitochondria. Nevertheless, it is still possible that METTL20 has one or more additional mitochondrial substrates aside from ETFB. We also confirmed that the methylation site in ETFB is exactly the same as in other reports, i.e., K200 and K203 (Supplementary Fig. S2c–f)5,6. We then checked if this methylation is conserved between humans and mice. As shown in Fig. 1d, methylation of murine ETFB was abolished in K200/203R mutant, indicating that methylation of ETF is well conserved between these two species.
ETF functions as an electron transfer protein for acyl-CoA dehydrogenases. Given that the methylation of ETFB inhibits the catalytic activity5,6, we also checked the mouse ETF catalytic activity in vitro. The recombinant ETF complex was incubated with METTL20, with or without AdoMet for 3 h, after which the ETF complex was mixed with recombinant MCAD, octanoyl-CoA, and 2,6-Dichloroindophenol (DCPIP) as described previously16,17. The methylation status of ETFB after the methylation reaction was confirmed (Fig. 1e). As depicted in Fig. 1f, methylated ETF showed more reduced activity (51% ± 2%) than unmethylated ETF did (100% ± 5%). It can be inferred that either the KR mutant did not affect catalytic activity (98% ± 5%) or the methylation reaction did not influence its activity (105% ± 6%). These results confirmed that methylation of lysine in ETFB by METTL20 inhibits mouse ETF catalytic activity, in line with the results from other reports5,6,18.
Creation of Mettl20 KO mice by CRISPR/Cas9 genome editing
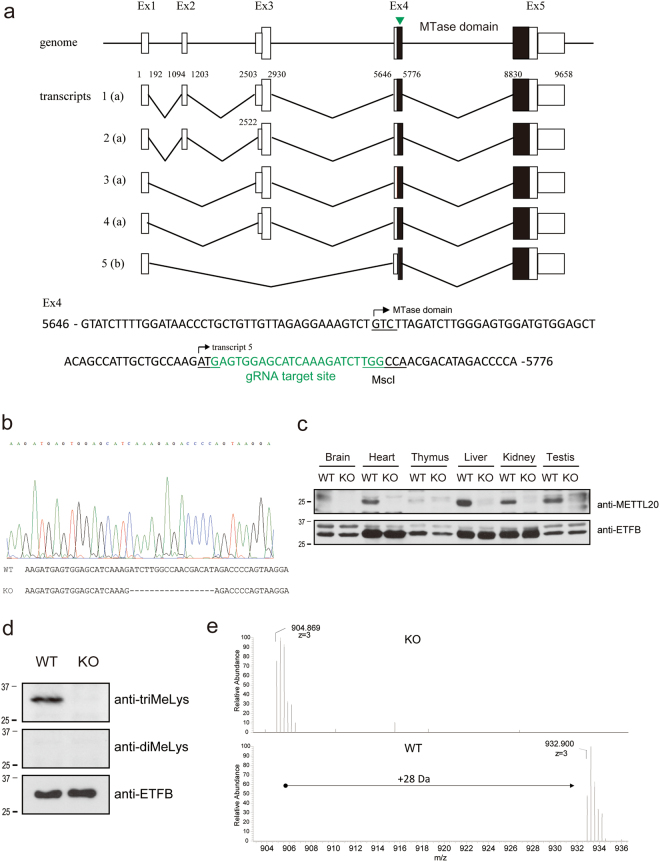
To understand the physiological function of METTL20 in vivo, Mettl20 KO mice were generated via the CRISPR/Cas9 genome editing technology19,20. Guide RNAs were designed against exon 4, which encodes the MTase domain of METTL20 (Fig. 2a). The guide RNA sequence contains the MscI site, which is convenient for genotyping. A Mettl20 KO mouse carries a 17 bp deletion, which results in a frameshift mutation in the MTase domain (Fig. 2b). Expression of mMETTL20 in several organs of WT and KO mice was examined by western blotting with an anti-METTL20 antibody. As shown in Fig. 2c, mMETTL20 protein expression levels were undetectable in all the tissues tested in KO mice but were the highest in the WT liver. Because proteins METTL20 and ETFB were highly expressed in the mouse liver, which is an important organ for fatty acid metabolism, we checked the ETFB methylation level in the liver of WT and KO mice. ETFB in the liver was immunoprecipitated with an anti-ETFB antibody, and methylation was detected with an anti-di-methyl lysine antibody (diMeLys) or anti-tri-methyl lysine antibody (triMeLys, Fig. 2d). Tri-methylation of ETFB was not detected in KO mice, suggesting that mMETTL20 was crucial for the tri-methylation. Of note, di-methylation of ETF was detected in neither WT nor KO mice. To verify this result, the ETFB immunoprecipitated from WT and Mettl20 KO liver samples was subjected to liquid chromatography-mass spectrometry (LC-MS). As shown in Fig. 2e, the K200 and K203-containing ETF peptide had an additional molecular weight (MW) of 28 Da in WT mice, in agreement with the MW observed after 2× trimethylation. In contrast, this additional MW was completely lost in KO mice. These observations implied that K200 and K203 were fully tri-methylated in ETFB of WT mice but remained completely unmethylated in Mettl20 KO mice.
Figure 2.
Generation of Mettl20 knockout mouse. (a) Schematics of mouse Mettl20 gene. Mettl20 has five transcripts. Transcript 1 to 4 encode same longer isoform (a) but possess different 5′ UTR, whereas transcript 5 encodes shorter isoform (b), which start translation in exon4. A guide RNA was designed against the Ex4. Small box; transcript including UTRs, large box; coding region, filled box; MTase domain, arrow head; gRNA target site. (b) DNA sequence of the Mettl20 KO mice. The KO mice lost 17 base pairs around the gRNA target site which resulted in a frame-shift mutation of Mettl20. (c) Protein expression of METTL20 in WT and KO mice. Several tissues were collected from WT and KO mice, and protein expression of mMETTL20 or ETFB were measured with a western blotting with anti-Mettl20 antibodies (top) or anti-ETFB antibodies (bottom). (d) Methylation of endogenous ETFB in the mice liver. Mitochondria were fractionated from WT or Mettl20 KO mice liver, and endogenous ETFB were immunoprecipitated with anti-ETFB antibodies. Precipitated proteins were separated with SDS-PAGE, and their methylation were detected with anti-triMeLys antibodies (top), anti-diMeLys antibodies (mixture of c4930 and c5123) (middle), and anti-ETFB antibodies (bottom, as loading control). (e) nanoLC-MS analysis of endogenous ETFB. Immunoprecipitated ETFB were separated with SDS-PAGE, digested with Asp-N, and methylation of peptides containing K200/203 were analyzed with nanoLC-MS (Q-Exactive, Thermo Fisher Scientific). Detected triply charged peptide ion were m/z = 904.869 (z = 3), which corresponds to un-modified EPRYATLPNIMKAKKKKIEVVKAG peptide in KO mice and m/z = 932.900 (z = 3), which corresponds to the peptide carrying 6 methyl groups in WT.
Mettl20 KO mice show increased ETF activity, oxygen consumption, and heat production when fed a ketogenic diet (KD) or when fasted
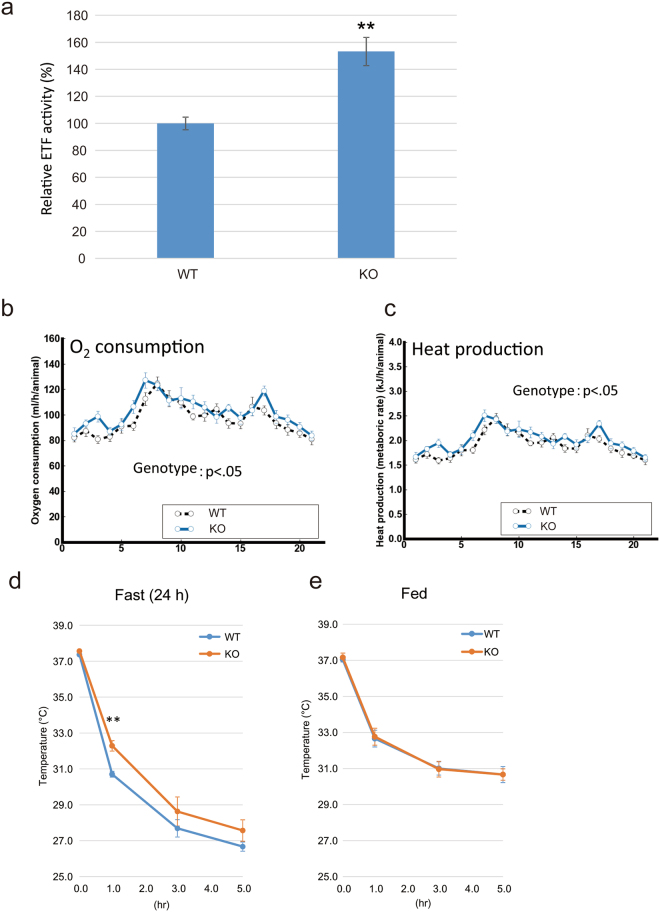
We measured ETF activity in Mettl20 KO mice as reported elsewhere17, with a slight modification. Briefly, the mitochondrial fractions from the WT or KO murine liver were sonicated, and the supernatant was mixed with recombinant MCAD, octanoyl-CoA, and DCPIP. As presented in Fig. 3a, the ETF activity in Mettl20 KO mouse mitochondria was ~1.5-fold greater than that in the WT. This result is consistent with that of in vitro experiments, suggesting that methylation of ETFB inhibited ETF activity (Fig. 1f) because ETFB in the WT mitochondria was fully methylated (Fig. 2d and e).
Figure 3.
The Mettl20 KO mice had higher O2 consumptions and heat production under KD and showed cold tolerance under fasting. (a) ETF activity in mitochondrial lysate. 300 µg of the mitochondrial fraction from mice liver were sonicated, and the supernatant was mixed with 3 µg recombinant MCAD, 80 µM octanoyl-CoA and 70 µM DCPIP. Initial rate of the reduction of DCPIP was calculated from OD600 measured at 30 °C for every 30 sec for 10 min with a microplate reader (SpectraMax 190, Molecular Devices). Average of three independent experiments, n >= 3 for each experiments, mean ± SEM. Student’s T-test **p < 0.01. (b) Oxygen consumption fed with KD. ANOVA, effect of genotype, F (1, 18) = 5.0746, P < 0.04; effect of time increment, F (20, 360) = 13.5184, P < 0.0001; interaction between genotype and time increment, F (20, 360) = 1.053, P > 0.39. Horizontal axis indicates time increment (h). Error bars represent the S.E.M. N = 10 male mice for each genotype at 12 weeks of age. (c) Heat production fed with KD. ANOVA, effect of genotype, F (1, 18) = 4.8976, P < 0.05; effect of time increment, F (20, 360) = 13.2229, P < 0.0001; interaction between genotype and time increment, F (20, 360) = 1.061, P > 0.38. Horizontal axis indicates time increment (h). Error bars represent the S.E.M. N = 10 male mice for each genotype at 12 weeks of age. (d) Cold tolerance test under fasting and (e), Fed. WT and Mettl20 KO mice were either fed on ND (e) or fasted for 24 h (d) before the experiment. Mice in the same cage per conditions/genotype were transferred to cold room (4 °C), and measured their core body temperatures after 1 h, 3 h and 5 h. n = 5, mean ± SEM. Student’s T-test **p < 0.01.
The growth of Mettl20 KO mice was normal and indistinguishable from that of the WT when these mice were fed a normal diet (ND) or KD. Additionally, the differences in the body weight and several parameters such as results of liver function testing (LDH, AST, ALT, and GGT), renal function testing (TP, ALB, UN, CRE), lipid metabolism analyses (T-Bil, HDL-C, LDL-C, T-CHO), glucose, IPGTT, and serum adipocytokine tests were statistically insignificantly different between WT and KO mice (data not shown).
To examine the possibility that Mettl20 KO mice were better at generating energy from fat, a calorimetric analysis was performed on the mice fed the ND or KD, because the KO mice showed increased ETF activity (Fig. 3a). Both strains of mice had similar food intake and motor activity throughout the experiments (data not shown). As shown in Fig. 3b and c, the Mettl20 KO mice fed the KD exhibited increased oxygen consumption and heat production (p < 0.05), whereas these parameters were unchanged when these mice were fed the ND (Supplementary Fig. S3).
Next, we checked heat production in a cold environment (Fig. 3d and e). The mice were either fed or fasted for 24 h before the experiment, given that fatty acid metabolism is known to be elevated during fasting. The cold tolerance test indicated that Mettl20 KO mice had a better ability to maintain their body temperature than WT mice did during fasting (Fig. 3d, KO: 32.3 °C ± 0.3 °C vs. WT: 30.7 °C ± 0.2 °C, p < 0.01 after a 1 h cold test). Again, no difference was observed during feeding (Fig. 3e). These results indicated that mMETTL20 is involved in the regulation of β-oxidation under ketogenic or fasting conditions, where fatty acid metabolism is known to be highly accelerated.
Metabolomic analysis of serum fatty acids in Mettl20 KO mice
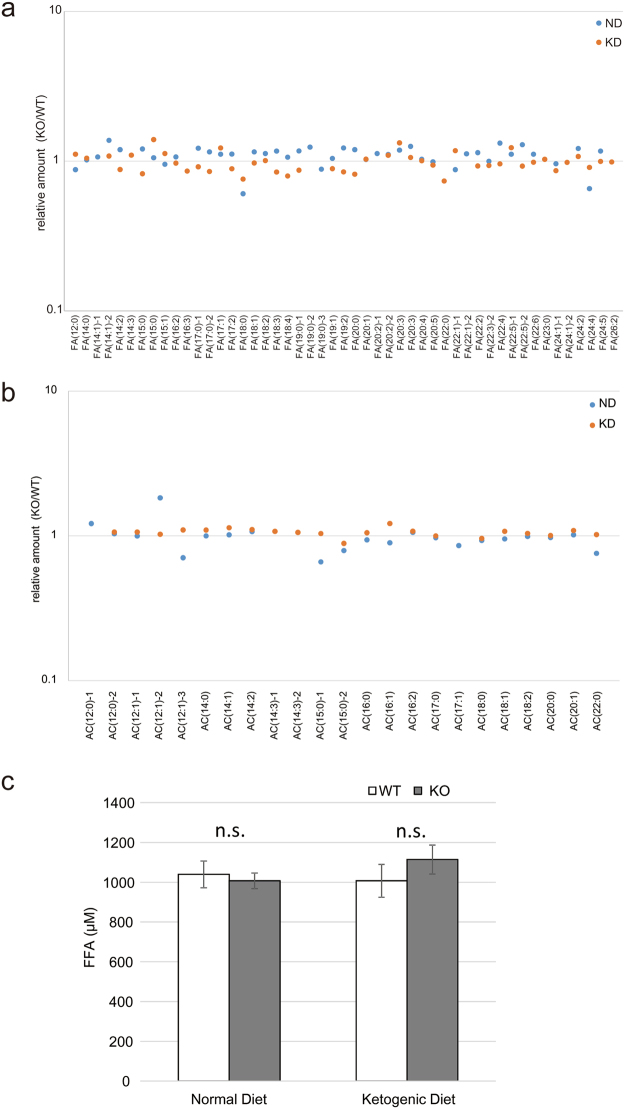
Because Mettl20 KO mice showed accelerated ETF activity and heat production during ketosis or fasting, it is possible that serum fatty acid contents are different between the WT and KO mice. To test this possibility, a metabolomic assay was performed to quantify fatty acids and acyl carnitines in WT or KO mouse serum samples. Overall, the levels of 177 metabolites detected in the assay were not significantly different between the two groups (<2-fold difference, data not shown). Moreover, levels of free fatty acids (FFAs) and acyl carnitine were similar between the two strains of mice (Fig. 4a and b). To confirm these results, we measured the total FFA concentration in serum (Fig. 4c). Again, there was no difference in the serum FFA concentration between the WT and KO mice.
Figure 4.
Metabolomic analysis of the Mettl20 KO mice fed with ND or KD. WT and Mettl20 KO male mice (n = 10 each condition) were fed with ND or KD from 4 wks old to 26 wks old. Then, mice were sacrificed and the serum were pooled in one tube per condition. Metabolites were extracted and analyzed with LC-TOF MS. (a) Serum fatty acids level. (b) Serum acyl carnitine level. All metabolomic data were collected as relative amount (KO/WT). (c) Blood samples were collected from the WT and Mettl20 KO male mice (n = 10 each condition) under ad libitum feeding. Concentration of serum free fatty acids (FFA) were measured. Data were presented as mean ± SEM, n = 10 per condition.
METTL20 is tolerant to loss-of-function (LoF) mutations
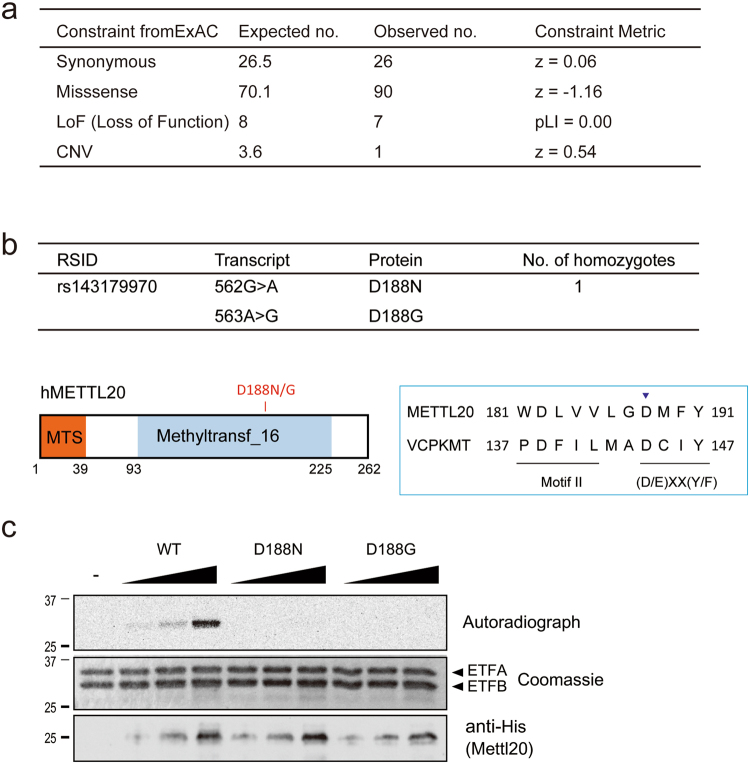
The METTL20/ETFBKMT homozygous loss-of-function mutation has been identified in an Icelandic population21, suggesting that this enzyme is not essential for viability of humans18. To further address the biological significance of genetic variants of METTL20 in the human population, we searched for genomic mutations within the exons of METTL20 in the ExAC Browser, which stores exome DNA sequence data for 60,706 individuals with diverse ancestries22. As shown in Fig. 5a, METTL20 was classified as a gene tolerant to LoF (if pLI = 0.00 or pLI < 0.01, then the gene is thought to be tolerant to LoF22). In addition to the frameshift mutation that was reported in the Icelandic population21, individuals exist that are homozygous for certain missense mutations in METTL20 (ref.22 and Supplementary Table S1). Among them, the c.562G > A mutation (rs143179970) induces the D188N substitution in Motif Post II of the METTL20 MTase domain (Fig. 5b), which is crucial for the catalytic activity of seven-β-strand MTases such as VCPKMT23. To evaluate the enzymatic activity of the corresponding mutant protein, recombinant METTL20 D188N and another mutant (D188G), which is also present as a heterozygous mutation in the ExAC database, were expressed in bacteria; an MTase activity assay was conducted in vitro (Fig. 5c). These mutant enzymes could not methylate ETFB, implying that the loss of catalytic activity by METTL20 does not cause an obvious disadvantage for survival of humans. These findings confirmed that METTL20 is a gene tolerant to LoF.
Figure 5.
Mettl20 is tolerant to loss of function (LoF) mutation. (a) Genomic variation within the exon of Mettl20 was screened with ExAC Browser. (b) Missense mutations in METTL20 MTase domain found in the ExAC. D188N was found as a homozygous mutation (top). Amino acid sequence of METTL20 and VCPKMT, a Methyltransferase Family 16 with hallmark (D/E)XX(Y/F) motif located downstream of Motif II (bottom). (c) Catalytic activity of METTL20 D188N and D188G mutants. Recombinant ETF complex and increased amount of His-METTL20 were incubated with 14C-labeled AdoMet for 2 h at 30 °C. Proteins were separated with SDS-PAGE and stained with Coomassie blue (middle, loading control), the autoradiography was detected with the image analyzer BAS-5000 (top). Loading amount of METTL20 was confirmed with a western blotting with anti-His antibody (bottom).
Discussion
Approximately 40 MTase genes may participate in mitochondrial functions4; however, only a few of these ‘mitochondrial’ MTase genes have been characterised so far. METTL20 has been identified as an ETFB MTase5,6. We also identified ETFB as a METTL20 substrate by a unique method that involves the synthetic co-factor ProSeAM12,15. During the screening, ETFB was found to be the sole candidate for METTL20 (Fig. 1c), suggesting that ETFB is a major target of METTL20 in mitochondrial lysates. In the in vitro experiments, METTL20 methylated ETFB at K200 and K203, and we could detect peptides carrying up to five methyl groups, but not fully methylated ones (Supplementary Fig. S2f). This observation is interesting because ETFB is fully methylated in the mouse liver (Fig. 2d,e). It is possible that other proteins or co-factors are required for full methylation of ETFB in vivo.
Given that methylation sites are located around the recognition loop, which is required for the interaction with MCAD, it is probable that methylation of mouse ETFB affects its catalytic activity (Fig. 1f), and this notion is consistent with the results from other reports5,6. In this study, we generated Mettl20 KO mice (Fig. 2) and demonstrated that methylation of ETF was lost in KO mice. The catalytic activity of ETF was higher in KO mice (Fig. 3a), suggesting that mMETTL20 has a regulatory role in fatty acid oxidation. Accordingly, Mettl20 KO mice also had a higher oxygen consumption rate and heat production when they consumed the KD (Fig. 3b and c). Furthermore, the KO mice showed greater cold tolerance than the WT did during fasting although there was no difference during ad libitum feeding, where glucose metabolism is known to be a primary energy source (Fig. 3d and e). From these results, we can conclude that β-oxidation activity is regulated by the methylation of ETFB by METTL20, when fatty acids serve as a primary energy source, i.e. β-oxidation is highly activated. It should be noted that serum fatty acid concentrations were not affected by the Mettl20 KO (Fig. 4), pointing to the existence of a compensatory mechanism ensuring lipid homeostasis. Further research is needed to reveal such a compensatory mechanism.
The fact that Mettl20 KO mice grew to be normal and undistinguishable from WT mice (data not shown) implies that this gene is not essential. To test whether this is true for humans, the ExAC database was searched for genomic variations in Mettl20 exons. The analysis revealed that METTL20 is classified as a gene tolerant to LoF (Fig. 5a). The identified homozygous missense mutation (D118N) completely eliminated the METTL20 catalytic activity (Fig. 5c). These results further indicate that METTL20-mediated protein methylation is not essential in humans but more likely modulates energy production during fasting or under other metabolic conditions where β-oxidation is up-regulated. One possible benefit of this regulation is that too much energy production during fasting is avoided. This mechanism may help animals survive long-term starvation. Besides, it is likely that methylation of ETFB dynamically changes according to the metabolic environment, which, in turn, regulates metabolism adaptability. In both cases, further experiments are needed to test these possibilities.
Materials and Methods
Animals
All the experiments involving mice were carried out according to protocols approved by the Animal Experiment Committee of the RIKEN Brain Science Institute. The animals were maintained in a 12 h light/dark cycle with ad libitum access to food and water. The temperature and humidity were maintained at 22–23 °C and 50–60%, respectively. Animal health was checked by the animal facility staff five times per week. The mice were subdivided into two groups and fed a standard chow (ND) containing 5% fat and 25% protein (CLEA Japan, Inc., CE-2) or a KD containing 91% fat and 9% protein (Harlan Teklad, TD96355) from the age of 4 weeks to 26 weeks.
Plasmids
Full-length cDNA corresponding to Mettl20 (NCBI ID: AK132096), Etfb (AK150293), Mcad (AK146444), METTL20 (NM_001135863), ETFA (NM_000126), and ETFB (NM_001985) were obtained either by PCR from a template plasmid, as a clone from the HeLa cDNA library, or as a clone from the Fantom3 clone collection24. The cDNAs of METTL20 and MCAD were cloned into the pET19b vector to generate N-terminally His-tagged constructs (pET19b-METTL20, pET19b-METTL20ΔMTS, and pET19b-MCADΔMTS). For tandem expression of the ETFA–ETFB complex, mouse Etfb cDNA was cloned at BamHI/NotI sites into MCS1, and human ETFA cDNA was cloned at NdeI/XhoI sites into MCS2 of the pETDuet-1 vector (resulting in pETDuet-1-His-ETFB/ETFA). In addition, the cDNAs for METTL20 and ETFB were cloned into the pcDNA3 vector with a C-terminal FLAG tag (pcDNA3-cFLAG) to generate mammalian expression constructs (pcDNA3-METTL20-cFLAG and pcDNA3-ETFB-cFLAG). The QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies, Inc., Santa Clara, CA USA) was used to generate mutant plasmids.
Antibodies
The following antibodies were used: an anti-ETFB antibody (N-16, Santa Cruz Biotechnology, USA, and PA5-22005, Thermo Fisher Scientific, USA), anti-α-tubulin antibody (clone B-5-1-2, Sigma-Aldrich, St. Louis, MO, USA), anti-FLAG M2 antibody (Sigma-Aldrich), anti-COXIV antibody (Cell Signaling Technology, USA), and anti-6xHis antibody (Wako). Monoclonal anti-di-methylated lysine antibodies (diMeLys) c4930 and c5123 were generated as described previously25,26 by means of a chemically lysine-di-methylated baculovirus as an antigen. Their specificity for lysine-di-methylation was confirmed by an ELISA as described elsewhere27 (Supplementary Fig. S1). A polyclonal rabbit anti-tri-methyl-lysine antibody (triMeLys) was purchased from Abcam, USA (#ab76118). To generate the anti-METTL20 antibody, 2 mg of recombinant His-METTL20ΔMTS was employed as an antigen to immunise rabbits. The antiserum was collected after three doses of immunisation. Specificity of the immunoreaction was confirmed with an ELISA. Prior to experiments, the antiserum was purified on a column with immobilised METTL20 (antigen).
Purification of recombinant proteins
BL21 (pLysS) strains were transformed with pET19b plasmids, and the bacteria were cultured in the 2xYT medium with ampicillin (100 µg/ml) and 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 18 h at 16 °C. The cells were pelleted and lysed with 1× PBS containing 0.5% of NP-40 by sonication with a Branson Sonifier (S-250D, Branson Ultrasonics Corp., CT, USA) for 5 min on ice. The lysates were centrifuged at 15,000× g for 10 min, and the supernatants were incubated with Ni-NTA Agarose (Qiagen, Valencia, CA, USA) for 1 h at 4 °C with gentle agitation. The agarose beads were washed five times with wash buffer (50 mM Tris-HCl pH 7.4, 25 mM imidazole) and then the bound proteins were eluted with elution buffer (50 mM Tris-HCl, pH 7.4, 250 mM imidazole). The purified proteins were dialyzed against 1× PBS containing 10% of glycerol, and their concentration was measured with the Bradford Protein Assay Kit (BioRad Laboratories, Hercules, CA, USA).
Stable isotopic labelling of cells and mitochondrial purification
For METTL20 target screening, HEK293T cells were cultured for at least six cell doublings in DMEM supplemented with 10% of dialysed FCS containing either a light (12C6) or stable heavy (13C6) isotope in lysine after processing with the SILAC labelling kit (Pierce, cat. # 89983). The mitochondrial fractions of each cell type were purified as described previously28. Briefly, cell pellets were resuspended in SEM buffer (10 mM 4-[2-hydroxyethyl]-1-piperazineethanesulfonic acid [HEPES]-KOH pH 7.5, 0.2 M mannitol, 50 mM sucrose, 10 mM KCl, 1 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride), and homogenised by means of the Dounce tissue grinder (2 ml, Pestle A, Fisher Scientific, USA). After that, the supernatants were centrifuged at 700× g for 10 min to separate nuclear pellets from the supernatants containing cytosolic materials and mitochondria. The supernatants were again centrifuged at 700× g to remove residual pellets, and then at 7,000× g for 10 min to separate the mitochondrial pellet from the cytosol-enriched supernatant. The mitochondrial pellets were next washed twice with SEM buffer, and the resulting mitochondrial fractions were stored at −80 °C.
ProSeAM labelling
Labelling of mitochondrial proteins with ProSeAM was carried out by a previously described procedure15, with a slight modification. Briefly, 10 ul of lysis buffer (50 mM Tris-HCl pH 8.0, 50 mM KCl, 10% glycerol, 0.1% of Tween 20) containing 150 μg of either light-isotope–labelled mitochondria (12C6-Mito) or heavy-isotope–labelled mitochondria (13C6-Mito) was incubated with 0.4 mM ProSeAM, with (13C-Mito) or without (12C-Mito) His-METTL20 (10 μg) (protein:enzyme at 15:1) in 50 ul of MTase reaction buffer (50 mM Tris-HCl pH 8.0) at 20 °C for 2 h. The reaction was stopped by adding four volumes of ice-cold acetone. The reaction tube was centrifuged at 15,000× g for 5 min, and the precipitate was washed once with ice-cold acetone. The pellet was dissolved in 58.5 µl of 1× PBS containing 0.2% of SDS, after which 15 µl of 5× click reaction buffer and 1.5 µl of 10 mM Azide-PEG4-Biotin (Click Chemistry Tools) were added; the reaction mixture was incubated for 60 min at room temperature. The click reaction was stopped with four volumes of ice-cold acetone. The pellet was dissolved in 75 µl of binding buffer (1× PBS, 0.1% of Tween-20, 2% of SDS, 20 mM dithiothreitol) and sonicated for 10 s. 12C-Mito and 13C-Mito samples were mixed in a tube; 450 µl of IP buffer (Tris Buffered Saline (TBS) with 0.1% Tween 20) containing 3 mg of Dynabeads M-280 Streptavidin (Life Technologies Japan Ltd., Minato-ku, Tokyo, Japan) was added into the tube, and it was incubated for 30 min at room temperature (the final SDS concentration in the reaction was 0.5%). The protein-bound beads were washed thrice with wash buffer (1× PBS, 0.1% of Tween 20, 0.5% of SDS) and twice with 100 mM ammonium bicarbonate buffer, and were analysed by western blotting or MS.
Quantitative tandem MS (MS/MS) analysis for target protein identification
Acetonitrile (1/10 volume) and dithiothreitol (20 mM) were added to protein-bound Dynabeads in 100 mM ammonium bicarbonate buffer, and the mixture was incubated for 30 min at 56 °C. Then, iodoacetamide was added, and the mixture was incubated for 30 min at 37 °C in the dark. Next, the protein samples were digested with 0.5 µg of Lys-C (Promega). The protein fragments were applied to a liquid chromatograph (EASY-nLC 1000; Thermo Fisher Scientific, Odense, Denmark) coupled to a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, Inc., San Jose, CA, USA), with a nanospray ion source in positive mode. The peptides derived from the protein fragments were separated on a NANO-HPLC C18 capillary column (0.075 mm inner diameter × 150 mm length, 3 mm particle size; Nikkyo Technos, Tokyo, Japan). Mobile phase A was composed of water with 0.1% of formic acid, and mobile phase B consisted of acetonitrile with 0.1% of formic acid. Two different slopes were used for gradient elution for 120 min at a flow rate of 300 nl/min: 0–30% of phase B during 100 min and 30–65% of phase B for 20 min. The mass spectrometer was operated in top-10 data-dependent scan mode. The operating parameters of the mass spectrometer were as follows: spray voltage, 2.3 kV; capillary temperature, 275 °C; mass-to-charge (m/z) ratio, 350–1800; normalised collision energy, 28%. Raw data were acquired via the Xcalibur software (Thermo Fisher Scientific). The MS and MS/MS data were subjected to searches in the Swiss-Prot database by means of Proteome Discoverer 1.4 (Thermo Fisher Scientific) in the MASCOT search engine software version No. 2.4.1 (Matrix Science, London, United Kingdom). The search parameters were as follows: enzyme, Lys-C; quantitation, SILAC K (+6); static modifications, carbamidomethyl (Cys); dynamic modifications, oxidation (Met); precursor mass tolerance, ± 6 ppm; fragment mass tolerance, ± 20 mDa; and maximum missed cleavages, 1. The proteins were considered identified when their false discovery rate was less than 1%. For substrate identification, proteins manifesting at least a 2-fold increase in levels in two independent experiments were defined as positive hit proteins.
An in vitro MTase assay
Recombinant histone H3.1 was purchased from NEB. One microgram of the substrate was incubated in 10 ul of 1× Reaction buffer (50 mM Tris-HCl pH 8.0) with His-METTL20 (1 µg) and 14C-labelled AdoMet (0.01 µCi, Perkin Elmer) from 2 h to overnight at 30 °C. The reaction was stopped by adding Laemmli SDS sample buffer. Proteins were resolved by SDS-PAGE in a 12.5% acrylamide gel. An imaging plate (Fuji-Film) was exposed to the dry gel for 48 h, and the autoradiograph was obtained on a BAS-5000 Image analyser (Fuji-Film).
An in vitro ETF activity assay
ETF enzymatic activity was determined by a previously described DCPIP dye assay16,17. The recombinant WT and mutant ETF complex were prepared with the pETDuet-1-His-ETFB/ETFA. Four micrograms of the ETF complex was mixed with 4 µg of METTL20 with or without AdoMet and incubated at 30 °C for 3 h in 20 ul of MTase reaction buffer. After that, 4 µg of the ETF complex was mixed on ice with 8 µg of His-MCAD, 50 µM octanoyl-CoA, and 70 µM DCPIP in potassium phosphate buffer (pH 7.6) containing 0.2 mM N-Ethylmaleimide (NEM). For the mitochondrial ETF activity assay, 300 µg of the mitochondrial fraction from the murine liver was sonicated, and the supernatant was mixed with 3 µg of recombinant MCAD, 80 µM octanoyl-CoA, and 70 µM DCPIP in potassium phosphate buffer (pH 7.6) containing 0.2 mM NEM. The initial rate of reduction of DCPIP was calculated from optical density at 600 nm measured at 30 °C every 30 s for 10 min on a microplate reader (SpectraMax 190, Molecular Devices). Data were obtained in triplicate for each biological sample.
Creation of Mettl20 KO mice
Guide RNAs that target exon 4 of mouse Mettl20 were screened with CRISPR design tools (http://crispr.mit.edu/). The T7 promoter sequence 5′-TTAATACGACTCACTATAGG-3′ and the 20-mer Mettl20 guide RNA sequence 5′-GAGTGGAGCATCAAAGATCT-3′ were cloned into the AflII site of the gRNA_Cloning Vector, a gift from George Church (Addgene #41824)19. Oligos Insert-F (5′-TTTCTTGGCTTTATATATCTTAATACGACTCACTATAGGAGTGGAGCATCAAAGATCT-3′) and Insert-R (5′-GACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAACAGATCTTTGATGCTCCACTC-3′) were annealed and extended to construct a 100-bp double-stranded DNA fragment using Phusion polymerase (New England Biolabs, Japan). The gRNA_Cloning Vector was linearised with AflII and the inserts were incorporated via Gibson assembly (NEB, Japan, E2611S). To prepare hCas9 mRNA and Mettl20 guide RNAs, an in vitro transcription reaction kit with T7 RNA polymerase (mMESSAGE mMACHINE T7 Ultra Kit, Life Technologies) was used; the transcribed RNAs were purified with the MEGAclear Kit (Life Technologies). The quality of the guide RNA was checked by an in vitro Cas9 cleavage assay with recombinant hCas9 nuclease (NEB) and a 1 kb PCR product around the guide RNA target site. The Cas9 mRNA and guide RNA were injected into fertilised eggs (mouse strain C57BL/6J), and the mutated Mettl20 gene versions were screened by PCR after MscI digestion, because the MscI site TGGCCA is located immediately after the guide RNA target site. One in 11 heterozygous (+/−) males carried the expected 17 bp deletion. The heterozygous mouse was crossed with a WT C57BL/6J mouse, and the male (+/−) and female (+/−) mice were crossed to obtain Mettl20 KO (−/−) mice and WT (+/+) mice.
Calorimetric analysis
We conducted this analysis according to the standard operating procedure for the indirect calorimetry test (IMPC_CAL_001, http://www.mousephenotype.org/impress/protocol/86) of the International Mouse Phenotyping Consortium29,30. Briefly, mice were acclimated to the test room (in a 12 h light–dark cycle; lights on at 8:00 h) and the calorimetry cage 24 h before testing. Into the calorimetry cage, a single mouse was placed; food and water were made available ad libitum during acclimation and calorimetric analysis. The measurements were started 5 h before lights were switched off for 21 h. Input and output levels of gases oxygen (O2) and carbon dioxide (CO2) were measured by an Oxymax indirect calorimetry system (Columbus Instruments, Columbus, OH). The changes in the CO2 and O2 levels between the input and output were used to compute O2 consumption (VO2; ml/h per animal) and CO2 production (VCO2; ml/h per animal).
The respiratory exchange ratio (RER), calorific value (CV), and heat production (KJ/h per animal) were computed from VCO2 and VO2 as follows: RER = VCO2/VO2, CV = 3.815 + (1.232 × RER); heat production (KJ/h per animal) = 4.184 × CV × VO2. Water intake was measured by the Food and Water Intake Monitor (FWI-16M, O’Hara Co., Ltd., Tokyo, Japan). Locomotor activity of the mice was quantified by the Infrared XY beam activity system for mice (IA-16M, O’Hara Co., Ltd.).
Cold tolerance test
This test was performed as described previously31, with a slight modification. WT and Mettl20 KO mice were either fed the ND or fasted for 24 h before the experiment. Mice in the same cage in terms of conditions and genotype were transferred to the cold room (4 °C), and their core body temperature was measured with a BAT-7001H Thermometer (Physitemp Instruments, Inc.) after 1, 3, and 5 h.
Metabolomic analysis
WT and Mettl20 KO male mice (n = 10 for each condition) were fed the ND containing 5% of fat and 25% of protein (CLEA Japan, Inc., CE-2) or the KD containing 91% fat and 9% protein (Harlan Teklad, TD96355) from the age of 4 weeks to 26 weeks. Twenty-six-week-old mice were killed, and the serum samples of mice from each group were pooled and kept at −80 °C until measurements. All LC-TOF MS analyses were conducted at Human Metabolome Technologies Inc., Tsuruoka, Japan. Briefly, 500 μl of pooled serum was mixed with 6 μM internal standards in 1.5 ml of the mixture of 1% formic acid and 99% acetonitrile. Samples were centrifuged at 2,300× g and 4 °C for 5 min, and the supernatants were collected and filtered through a solid-phase extraction column. Flow-through fractions were dried and dissolved in 100 μl of 50% 2-propanol. The samples were applied to a liquid chromatograph (Agilent 1200 series RRLC system SL, Agilent Technologies, Inc.) coupled to an Agilent LC/MSD TOF Mass Spectrometer (Agilent Technologies, Inc), with electrospray ionisation in both positive and negative modes. The samples were separated on an ODS column (2 mm inner diameter × 50 mm length, 2 μm particle size). Mobile phase A comprised water with 0.1% of formic acid, and mobile phase B comprised 65% of isopropanol, 30% of acetonitrile, 5% of H2O with 0.1% of formic acid, and 2 mM HCOONH4. Three slopes were used for gradient elution for 20 min at a flow rate of 300 μl/min: 0–1% of phase B for 0.5 min, 1–100% of phase B during 13.5 min, and then 100% of phase B for 20 min. The settings of the mass spectrometer were as follows: nebuliser pressure, 40 psi; dry gas flow, 10 l/min; dry gas temperature, 350 °C; capillary voltage, 3500 V; scan range, m/z 100–1700. The obtained data were subjected to searches in the HMT database by means of MasterHands ver.2.17.1.11 (ref.32) (Keio University). The overall data processing flow consisted of the following steps: noise filtering, baseline removal, migration time correction, peak detection, and integration of the peak area from a 0.02 m/z–wide slice of the electropherograms.
Fatty acid measurement
WT and Mettl20 KO male mice (n = 10 for each condition) were fed the ND or KD from the age of 4 weeks to 23 weeks. The blood samples were collected from the orbital sinus during ad libitum feeding. Concentrations of serum FFAs were analysed with a kit (Abcam, cat. # ab65341). The obtained data were presented as mean ± SEM, n = 10 per condition.
ExAC database analysis
The ExAC Browser was searched for the METTL20 gene mutation22 (constraint metrics: for synonymous and missense variants, a signed Z score calculating the deviation of observed counts from the expected number is indicated in ExAC analysis). Positive Z scores indicated increased constraint (intolerance to variation), and therefore the gene had fewer variants than expected. Negative Z scores were given to genes that had more variants than expected.
Regarding LoF, the closer pLI is to 1.0, the more LoF-intolerant the gene is expected to be. If pLI ≥ 0.9, then the set of genes is extremely LoF-intolerant. Among the 106 loss-of-function and missense variants in ExAC (as of September 19, 2017), a D188N mutant (RSID: rs143179970) was found to be a homozygous mutation (Supplementary Table S1).
Electronic supplementary material
Acknowledgements
We thank Drs Takeshi Inagaki and Juro Sakai for their valuable suggestions regarding metabolomic analyses in mice. This work was supported by the Mochida Memorial Foundation for Medical and Pharmaceutical Research, Naito Foundation, Inamori Grant Program, Takeda Science Foundation, Uehara Memorial Foundation, and a RIKEN internal research fund. This work was also supported by the Program for Development of New Functional Antibody Technologies of the New Energy and Industrial Technology Development Organization (NEDO) of Japan (for TH).
Author Contributions
T.S., T.Y., S.W., and Y.S. conceived and directed the study. T.S. conceived and conducted the experiments. T.F. performed the high-throughput phenotyping analysis in the W.T. and mutant mice. S.O., H.I., and T.H. generated anti-Kme2 monoclonal antibodies. S.B. carried out the ExAC database search. T.S. and N.D. performed M.S. analysis. T.S. and Y.S. wrote the manuscript with support and contributions from all the co-authors.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-19615-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tollervey JR, Lunyak VV. Epigenetics: judge, jury and executioner of stem cell fate. Epigenetics. 2012;7(8):823–40. doi: 10.4161/epi.21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai HC, Baylin SB. Cancer epigenetics: linking basic biology to clinical medicine. Cell Res. 2011;21(3):502–17. doi: 10.1038/cr.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13(5):343–57. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhein VF, Carroll J, Ding S, Fearnley IM, Walker JE. NDUFAF7 methylates arginine 85 in the NDUFS2 subunit of human complex I. J Biol Chem. 2013;288(46):33016–26. doi: 10.1074/jbc.M113.518803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhein VF, et al. Human METTL20 Methylates Lysine Residues Adjacent to the Recognition Loop of the Electron Transfer Flavoprotein in Mitochondria. J Biol Chem. 2014;289(35):24640–51. doi: 10.1074/jbc.M114.580464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malecki J, Ho AY, Moen A, Dahl HA, Falnes PO. Human METTL20 is a mitochondrial lysine methyltransferase that targets the beta subunit of electron transfer flavoprotein (ETFbeta) and modulates its activity. J Biol Chem. 2015;290(1):423–34. doi: 10.1074/jbc.M114.614115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toogood HS, et al. Extensive domain motion and electron transfer in the human electron transferring flavoprotein.medium chain Acyl-CoA dehydrogenase complex. J Biol Chem. 2004;279(31):32904–12. doi: 10.1074/jbc.M404884200. [DOI] [PubMed] [Google Scholar]
- 8.Cao XJ, Arnaudo AM, Garcia BA. Large-scale global identification of protein lysine methylation in vivo. Epigenetics. 2013;8(5):477–85. doi: 10.4161/epi.24547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo, A. et al. Immunoaffinity Enrichment and Mass Spectrometry Analysis of Protein Methylation. Mol Cell Proteomics, (2013). [DOI] [PMC free article] [PubMed]
- 10.Moore KE, et al. A general molecular affinity strategy for global detection and proteomic analysis of lysine methylation. Mol Cell. 2013;50(3):444–56. doi: 10.1016/j.molcel.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson SM, Moore KE, Green EM, Martin GM, Gozani O. Proteome-wide enrichment of proteins modified by lysine methylation. Nat Protoc. 2014;9(1):37–50. doi: 10.1038/nprot.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bothwell IR, et al. Se-adenosyl-L-selenomethionine cofactor analogue as a reporter of protein methylation. J Am Chem Soc. 2012;134(36):14905–12. doi: 10.1021/ja304782r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willnow S, Martin M, Luscher B, Weinhold E. A selenium-based click AdoMet analogue for versatile substrate labeling with wild-type protein methyltransferases. Chembiochem. 2012;13(8):1167–73. doi: 10.1002/cbic.201100781. [DOI] [PubMed] [Google Scholar]
- 14.Binda O, et al. A chemical method for labeling lysine methyltransferase substrates. Chembiochem. 2011;12(2):330–4. doi: 10.1002/cbic.201000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimazu T, Barjau J, Sohtome Y, Sodeoka M, Shinkai Y. Selenium-Based S-Adenosylmethionine Analog Reveals the Mammalian Seven-Beta-Strand Methyltransferase METTL10 to Be an EF1A1 Lysine Methyltransferase. PLoS One. 2014;9(8):e105394. doi: 10.1371/journal.pone.0105394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bross P, et al. A polymorphic variant in the human electron transfer flavoprotein alpha-chain (alpha-T171) displays decreased thermal stability and is overrepresented in very-long-chain acyl-CoA dehydrogenase-deficient patients with mild childhood presentation. Mol Genet Metab. 1999;67(2):138–47. doi: 10.1006/mgme.1999.2856. [DOI] [PubMed] [Google Scholar]
- 17.Amendt BA, Rhead WJ. The multiple acyl-coenzyme A dehydrogenation disorders, glutaric aciduria type II and ethylmalonic-adipic aciduria. Mitochondrial fatty acid oxidation, acyl-coenzyme A dehydrogenase, and electron transfer flavoprotein activities in fibroblasts. J Clin Invest. 1986;78(1):205–13. doi: 10.1172/JCI112553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falnes PO, Jakobsson ME, Davydova E, Ho A, Malecki J. Protein lysine methylation by seven-beta-strand methyltransferases. Biochem J. 2016;473(14):1995–2009. doi: 10.1042/BCJ20160117. [DOI] [PubMed] [Google Scholar]
- 19.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–8. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sulem P, et al. Identification of a large set of rare complete human knockouts. Nat Genet. 2015;47(5):448–52. doi: 10.1038/ng.3243. [DOI] [PubMed] [Google Scholar]
- 22.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kernstock S, et al. Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat Commun. 2012;3:1038. doi: 10.1038/ncomms2041. [DOI] [PubMed] [Google Scholar]
- 24.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 25.Iwabata H, Yoshida M, Komatsu Y. Proteomic analysis of organ-specific post-translational lysine-acetylation and -methylation in mice by use of anti-acetyllysine and -methyllysine mouse monoclonal antibodies. Proteomics. 2005;5(18):4653–64. doi: 10.1002/pmic.200500042. [DOI] [PubMed] [Google Scholar]
- 26.Hamakubo T, Kusano-Arai O, Iwanari H. Generation of antibodies against membrane proteins. Biochim Biophys Acta. 2014;1844(11):1920–1924. doi: 10.1016/j.bbapap.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Kimura H, Hayashi-Takanaka Y, Goto Y, Takizawa N, Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct Funct. 2008;33(1):61–73. doi: 10.1247/csf.07035. [DOI] [PubMed] [Google Scholar]
- 28.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA. 2006;103(27):10224–9. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koscielny, G. et al. The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res. 42(Database issue), D802–9 (2014). [DOI] [PMC free article] [PubMed]
- 30.Dickinson ME, et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537(7621):508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakakibara I, et al. Fasting-induced hypothermia and reduced energy production in mice lacking acetyl-CoA synthetase 2. Cell Metab. 2009;9(2):191–202. doi: 10.1016/j.cmet.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6(1):78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.