Abstract
Seed germination paves the way for the dormant embryo to establish itself as a new plant marking the first critical step in postembryonic plant growth and development. Germination starts with the uptake of water (imbibition), followed by induction of transcription, translation, energy metabolism, and cell division processes. Although small RNAs have been implicated in many developmental processes, their role during seed germination stages and conditions remained elusive. Here we show that seed germination conditions, like imbibition and temperature, dynamically regulate the expression of many developmentally important miRNAs and their targets. We have identified 58 miRNAs belonging to 30 different families at different seed germination conditions. Amongst these, 15 miRNAs and their targets were significantly differentially expressed in Arabidopsis seeds in dry and 12 h, 24 h and 48 h of imbibition. Interestingly, differential expression of miR390, which targets trans-acting siRNA locus (TAS3) derived transcripts, resulted in alteration of tasiR-ARF mediated regulation of expression of target AUXIN RESPONSE FACTORs (ARF2/3/4). Our results suggest that the dynamic expression of several miRNAs, their targets, and a crosstalk between miRNA and ta-siRNA pathways contribute to the regulation of seed germination in Arabidopsis thaliana.
Introduction
Seed germination process, which marks the transition from seed to seedling stage, plays a crucial role in the complete life cycle of higher plants. Dormant seeds germinate when the environmental conditions, such as temperature, water or humidity are favourable1. The contrasting physiological event of seed germination is dormancy, which is regarded as the temporary failure or block of a viable seed to complete germination under seemingly unfavourable conditions and is an adaptive feature for optimizing the timing of germination2. Seed germination starts with the uptake of water (imbibition) by the quiescent dry seed and is considered as complete when radicle emerges out from the seed coat. A complex co-ordination of different molecular, physiological and environmental factors govern the dynamic and triphasic process of seed germination2,3.
Small non-coding RNAs (of 19–24 nucleotides length) play diverse roles in growth, development, morphogenesis, and stress responses in both plants and animals4–8. There are two major classes of small non-coding RNAs - short interfering RNAs (siRNAs) and microRNAs (miRNAs) that regulate their target genes by binding to the complementary sequences and generally by making a cleavage in the target mRNA9,10 or by blocking their translation10–12. Recently, trans-acting small interfering RNAs (ta-siRNA) have also been implicated in plant development (Axtell, 2013). ta-siRNAs belong to a plant-specific class of endogenous small RNAs, whose biogenesis requires an initial process of specific miRNA-mediated cleavage of their precursors2,13. The functions of miRNAs are also regulated by several hormones and stresses14–16. Recently, it has been shown that proteins involved in small RNA biogenesis such as DICER LIKE1 (DCL1) plays significant role in embryogenesis and seed development in Arabidopsis1. This indicates involvement of small RNAs in seed development. Mutation in ARF10, which is the target of miR160, results in developmental defects in seeds through induced expression of ABA responsive genes15. miR159 regulates floral development, fertility and seed germination by targeting and negatively regulating MYB transcript level16; miR417 negatively regulate seed germination under salt stress condition and over expression of miR402 enhances the seed germination under stress conditions in Arabidopsis thaliana17,18.
Previous study reported 115 known miRNAs and 167 novel miRNAs in maize seeds imbibed for 24 h, which is very early stages of seed germination19. They identified 24 conserved miRNA families in both dry and imbibed maize seeds through deep sequencing. Few novel and known miRNAs and some of their targets were validated20. Deep sequencing of small RNAs from rice seed embryos identified 59 known and 230 novel miRNAs differentially regulated in the early stages of seed germination21. Moreover, in eudicot (Nelumbo nucifera) seed germination, 145 known and 78 novel miRNAs were identified22. These reports indicate the association of more number of small RNAs in the dynamic seed germination procedure. However, little is known about the condition specific regulation of small RNAs and their targets, which are potentially important contributors to the early stages of Arabidopsis seed germination.
In this study, we have identified both conserved and non-conserved small RNAs in both dry and imbibed Arabidopsis seeds under stratified (4 °C) and non-stratified conditions. We used miRNA- microarray approach to identify miRNAs functionally relevant to early stages of seed germination. We validated the expression of 15 such miRNAs and their targets at 12 h, 24 h and 48 h imbibition at 4 °C and room temperatures. Besides this, the expression level of miR390 was significantly high at both 24 h/RT and 24 h/4 °C conditions, which is required for processing of TAS3 transcripts23. miR390 is required for the processing of TAS3 transcript into functional tasiR-ARF that targets and negatively regulate the ARF family members ARF2, ARF3, and ARF414,24. The correlation of altered expression of miR390 and ARF2/3/4 indicate that a crosstalk of both miRNAs, ta-siRNAs, and their targets contributes to the dynamic seed germination process.
Results
Expression of miRNAs are differentially regulated in seeds under germination conditions
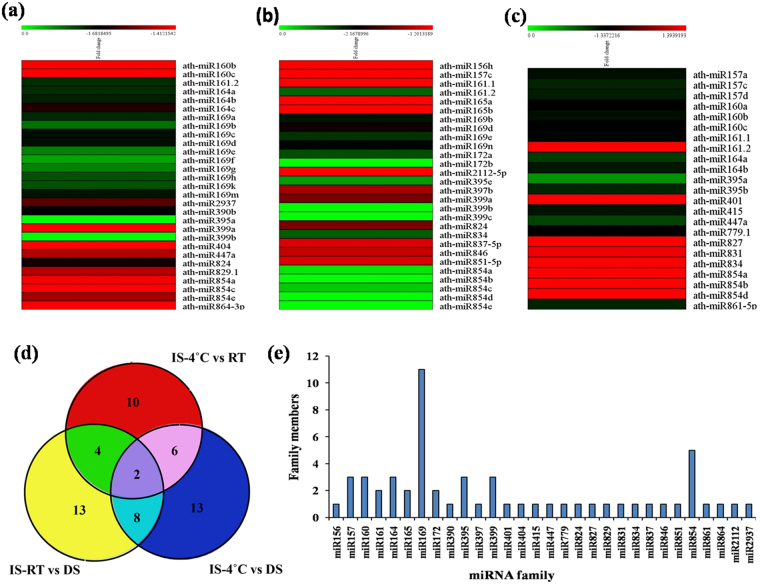
In order to identify miRNAs that are involved in seed germination process in Arabidopsis, we used miRNA microarray approach. Microarray was performed using total RNA from imbibed seeds (IS) at room temperature (RT), imbibed seeds at 4 °C and dry seeds (DS), and a three way comparison was made to identify differently expressed miRNAs. We identified a total of 58 miRNAs differently expressed in (i) IS-4 °C vs. DS (Fig. 1a), (ii) IS-RT vs. DS (Fig. 1b) and lastly, (iii) IS-4 °C vs. IS-RT (Fig. 1c). We found that specific miRNAs were upregulated and down regulated in each of the three cases. The results identified 29 miRNA genes in IS- 4 °C vs. DS (Fig. 1a), 28 miRNA genes in IS-RT vs. DS (Fig. 1b) and 23 miRNA genes in IS-4 °C vs. IS-RT (Fig. 1c). Among the 29 miRNAs in IS- 4 °C vs. DS (Fig. 1a), only two miRNAs were upregulated and rest were downregulated. In case of 28 miRNAs in IS-RT vs. DS (Fig. 1b), five miRNAs were upregulated and rest were downregulated, whereas among 23 miRNAs in IS-4 °C vs. IS-RT (Fig. 1c), only eight miRNA genes were upregulated and rest were downregulated.
Figure 1.
Microarray based expression patterns of miRNAs at different conditions of seed germination in Arabidopsis thaliana. (a) Heat map at cold imbibitions (4 °C) vs. dry seeds (DS). (b) Heat map at room temperature (RT) vs. dry seed (DS). (c) Heat map at cold imbibitions (4 °C) vs. room temperature (RT) [For generating heat map we used MeV(Multiple Experiment Viewer) (http://mev.tm4.org/)]. The bars represent the scale of expression levels of the miRNAs. During microarray, we have pooled all the cold imbibed (IS-4 °C) and room temp imbibed (IS-RT) total RNAs separately, and the microarray was performed with two biological replicates for each individual samples. (d) The Venn diagram represents the comparison of the known miRNAs between three different conditions (DS, IS-4 °C and IS-RT) used in the microarray experiment. (e) The graph represents the miRNA families and their respective family members that were detected in the microarray analysis.
Through Venn diagram representation (Fig. 1d), we observed that 10 miRNA genes were common in both IS-4 °C vs DS and IS-RT vs. DS and they are miR161.2; miR169b, d, e; miR399a, b; miR824 and miR854a, c, e. Similarly, 8 miRNA genes were common in IS-4 °C vs. DS and IS-4 °C vs. RT and they are miR160b, c; miR161.2; miR164a, miR395a; miR447a and miR854a; while 6 miRNA genes were common in IS-RT vs. DS and 4 °C vs. RT and they are miR157c; miR161.2; miR834 and miR854a, b, d. Only two miRNAs, miR161.2 and miR854a, were common in all three cases here. Analysis of the data showed that amongst the differently expressed miRNAs, most abundant families were miR169 and miR854, with 11 and 5 family members, respectively (Fig. 1e). Among these miRNAs, eighteen precursor miRNA families were selected based on their p-values (≤0.05) and fold change values (≥2.0) across the three data sets for validation. Among these eighteen precursor miRNA, miR157a, miR157c, miR157d consist of same mature sequence; so we took only one mature sequence of miR157 (Fig. 2f) among the three. Also miR399b and miR399c contain the same mature sequence in Arabidopsis, therefore we had chosen only one among them (Fig. 3e). The other miRNAs for validation were miR165/166 (Fig. 2a), miR172a (Fig. 2b), miR390b (Fig. 2c), miR160a (Fig. 2d), miR156h (Fig. 2e), miR164a (Fig. 3a), miR169b (Fig. 3b), miR161.1 (Fig. 3c), miR399a (Fig. 3d), miR824 (Fig. 3f), miR834 (Fig. 3g), miR854 (Fig. 3h) and miR2112-5p (Fig. 3i).
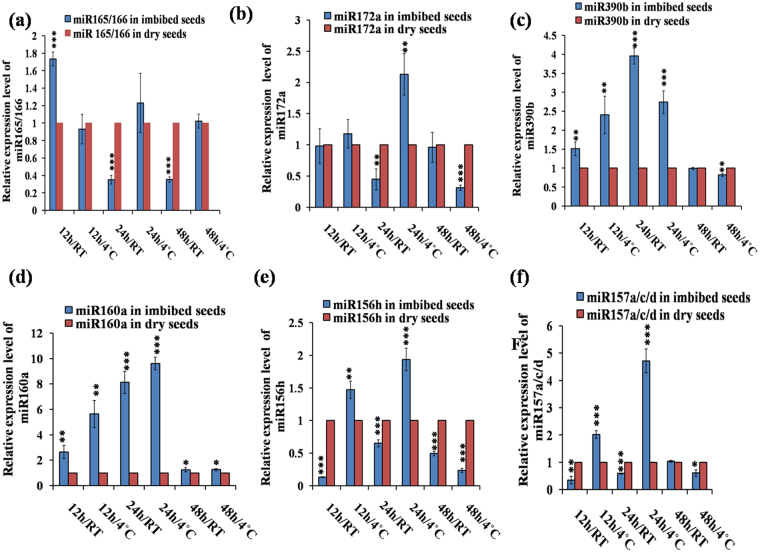
Figure 2.
Quantitative RT-PCR based validation of expression of mature miRNAs in imbibed seeds, in comparison to dry seeds, in Arabidopsis thaliana. (The expression profiles obtained from qRT-PCR slightly varied with the microarray based miRNA expressions, probably due to the pulling of total RNAs during microarray). The qRT-PCR validation of miRNAs were done at six different germination stages as 12 h/RT, 12 h/4 °C, 24 h/RT, 24 h/4 °C, 48 h/RT and 48 h/4 °C each compared to dry seed. The expression values presented here were the means of three biological replicates ± standard deviation (SD). The Arabidopsis ACTIN7 was used for each samples as an endogenous control. (a) ath-miR165/166; (b) ath-miR172a; (c) ath-miR390b; (d) ath-miR160a; (e) ath-miR156h; (f) ath-miR157a/or, c/or, d. In Arabidopsis, miR157a, c and d have the same mature sequence. Asterisks indicate significant statistical differences: ***P < 0.001, **P < 0.01, *P < 0.05 (One-way ANOVA).
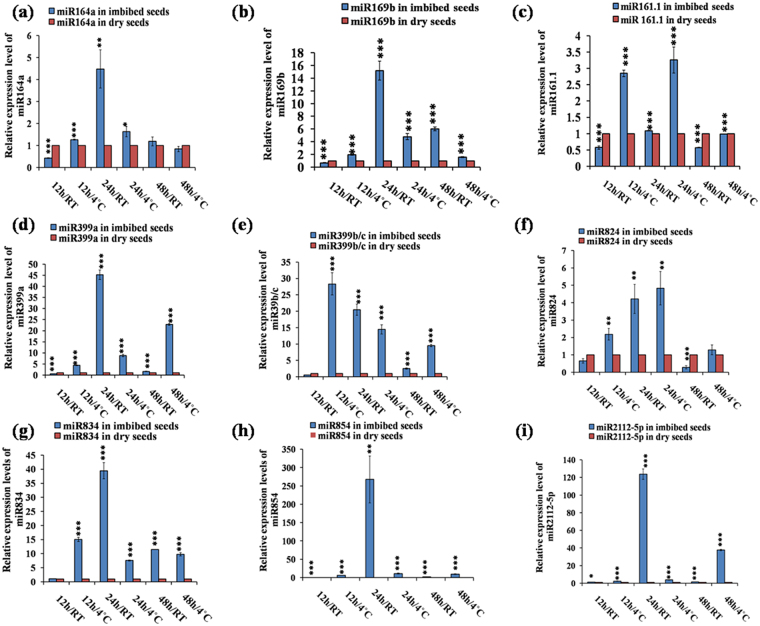
Figure 3.
Quantitative RT-PCR based validation of expression of mature miRNAs in imbibed seeds, in comparison to dry seeds, in Arabidopsis thaliana. The qRT-PCR validation of miRNAs were done at six different germination conditions as 12 h/RT, 12 h/4 °C, 24 h/RT, 24 h/4 °C, 48 h/RT and 48 h/4 °C each compared to dry seed. The expression values presented here were the means of three biological replicates ± SD. The Arabidopsis ACTIN7 was used for each sample as an endogenous control. (a) ath-miR164a; (b) ath-miR169b; (c) ath-miR161.1; (d) ath-miR399a; (e) ath-miR399b/or, c (in Arabidopsis, miR399b and c contain the same mature sequences); (f) ath-miR824; (g) ath-miR834; (h) ath-miR854; (i) ath-miR2112-5p.Asterisks indicate significant statistical differences: ***P < 0.001, **P < 0.01, *P < 0.05 (One-way ANOVA).
Validation of expression reflects dynamic regulation of miRNAs during seed germination
The different time points used for validation of miRNAs were 0 h (DS), 12 h/RT, 12 h/4 °C, 24 h/RT, 24 h/4 °C, 48 h/RT and 48 h/4 °C. The highest expression of miR165/166 was observed in case of 12 h/RT (Fig. 2a), which drastically changed in case of 12 h/4 °C. The lowest expressions were observed in 24 h/RT and 48 h/RT (Fig. 2a). However, it showed high expression at 24 h/4 °C and nearly equal expression to DS at 48 h/4 °C (Fig. 2a). We observed highest expression level of miR172a (Fig. 2b) during 24 h/4 °C and then 12 h/4 °C compare to DS (Fig. 2b), and the level reduced at the later stage of germination at 48 h/4 °C (Fig. 2b). Interestingly, its expression reduced at RT. We observed high level of expression of miR390 (Fig. 2c) compared to DS in almost all the conditions that we used. We found higher expression of miR390 (Fig. 2c) in both 24 h/RT and 24 h/4 °C conditions, and this expression level decreases at later stages of germination like 48 h (RT/4 °C) (Fig. 2c). The highest expression of miR160a was observed at 24 h/4 °C (Fig. 2d), then gradually in decreasing order at 24 h/RT, 12 h/4 °C, 12 h/RT compared to DS. We observed the lowest and almost same expression in case of 48 h/RT and 48 h/4 °C (Fig. 2d). Similar expression patterns were observed in case of both miR156h (Fig. 2e) and miR157a/c or, d (Fig. 2f). The miR157 families a, c and d constitute the same mature miRNA sequences. In both of the cases, we observed highest level of expression at 24 h/4 °C (2 fold in miR156 and ~5 fold in miR157). During cold imbibition, these miRNAs showed higher expressions compared to RT (Fig. 2e,f). Both the miRNAs showed reduced expression pattern with increasing time. We found maximum expression of miR164a (~4.5 fold) at 24 h/RT (Fig. 3a) and lowest expression at 12 h/RT (Fig. 3a). Also during 24 h cold imbibed condition, miR164a was showing upregulation compare to DS (Fig. 3a). Mainly, during very early cold imbibition stages (12 h, 24 h) miR164a showed high expression (Fig. 3a) but it gradually decreases at the later stages (48 h). Except 12 h/RT, in other germination conditions we observed upregulation of miR169b (Fig. 3b). The maximum expression of this miRNA was observed at 24 h/RT (Fig. 3b) like miR164a (Fig. 3a). The highest expression of miR161.1 was observed at 24 h/4 °C and then 12 h/4 °C (Fig. 3c), and the lower expressions were observed at 48 h/RT and 12 h/RT, which were almost same (Fig. 3c). At 48 h/4 °C, almost similar expression of miR169b was observed in both dry and imbibed seeds. Interestingly, in spite of single nucleotide difference in miR399a and miR399b/c, we observed a significant variation in their expression levels at different germination stages. Highest expression of miR399a (Fig. 3d) was at 24 h/RT and then 48 h/4 °C, whereas highest expression of miR399b/c (Fig. 3e) was at 12 h/4 °C and then 24 h/RT. But expression of both of the miRNAs was less at 12 h/RT and 48 h/RT compared to DS. Interestingly, all the miR399 (a, b and c) were induced in case of cold imbibition (4 °C) rather than RT. This indicates that cold imbibition plays significant role in miR399 expression during seed germination. In this study, we found higher expression of miR824 (Fig. 3f) in almost every stages, especially in cold imbibed conditions. We observed the highest expression of this miRNA at 24 h/4 °C (Fig. 3f). Although, miR834 was highly up regulated in every stages of germination compared to DS (Fig. 3g) and the maximum expression level (~40 fold) was observed at 24 h/RT (Fig. 3g). In case of miR854 (Fig. 3h) and miR2112-5p (Fig. 3i), highest expression of ~275 fold and ~130 fold respectively were observed at 24 h/RT. Overall, the expression pattern of most mature miRNAs were dynamically and differentially regulated at different time points under various germination conditions. Since miRNAs negatively regulate their targets, these results indicated possible differential regulation of their targets as well, which we further tested.
Differential expression of miRNAs negatively correlates the expression of target transcripts during seed germination conditions
The targets of the validated miRNAs were identified through “psRNATarget” tool25. Our prediction indicated more than one targets for each miRNA under standard settings of prediction tool. Some of the targets were novel and not indicated earlier. For experimental validation of miRNA-target expression correlation, we chose the targets for each miRNA considering its respective expectation value of 0.5 to 2.5 range. In cases where more than two significant targets were identified, all of them were used in the validation experiments. Target gene specific primers were designed in the region flanking to miRNA binding site. The list of selected targets and target gene specific primers used in this study were provided in (supplementary table S2 and S3).We analysed the expression of total 27 targets of 15 differentially expressed (dE) miRNAs using fast SYBR green fluorescent based qRT-PCR.
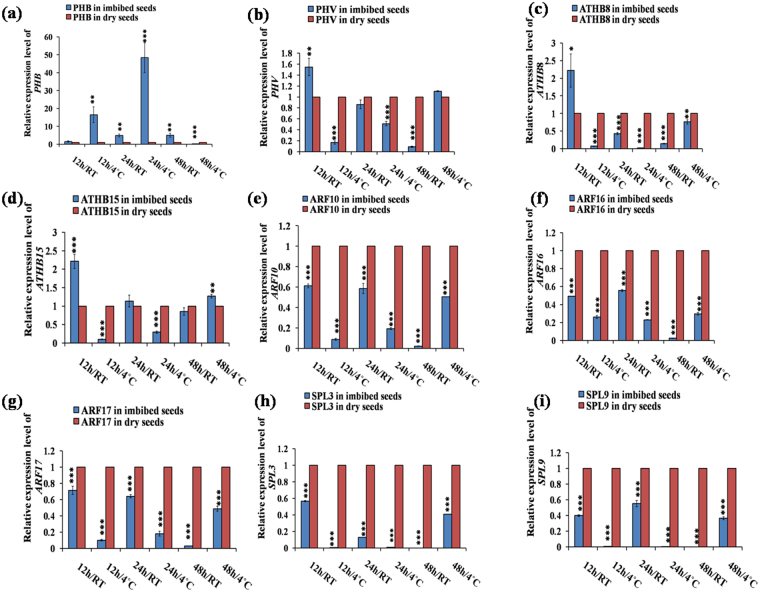
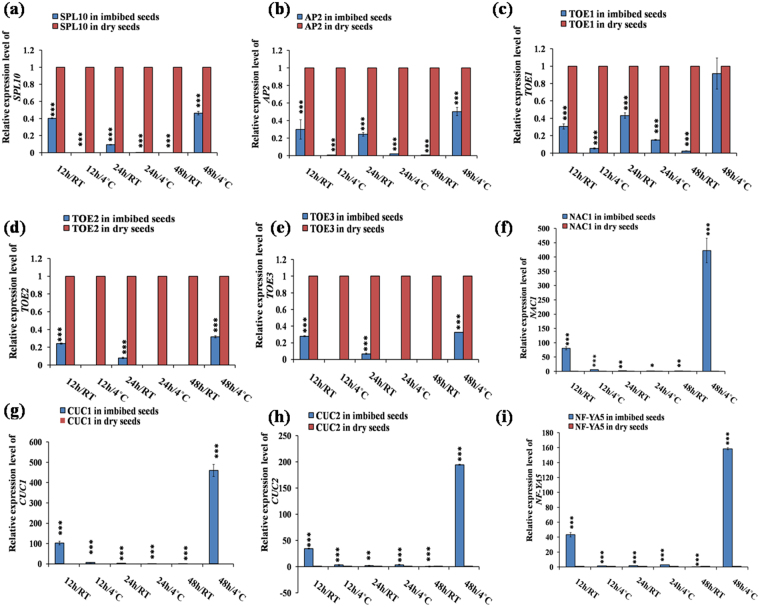
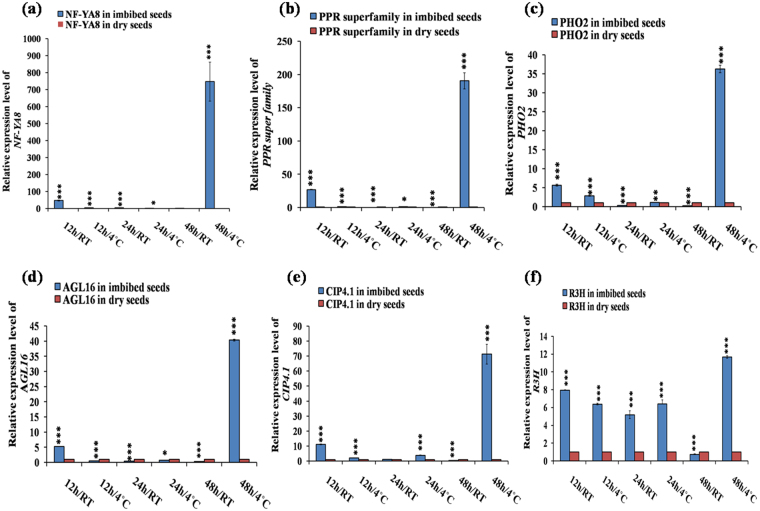
The expression of the targets was determined using the same germination conditions as those for the miRNAs, and in most of the cases we obtained opposite expression of the targets compare to their corresponding miRNAs. This inverse correlation partially validated the miRNA targets, since miRNAs mostly post-transcriptionally cleave their respective target gene transcripts. However, the inverse relationship of expression pattern between miRNAs and their targets were limited in some cases during specific stages of germination, which might be because of condition induced transcriptional regulation. We validated the expression of PHB (PHABULOSA) (Fig. 4a), PHV (PHAVOLUTA) (Fig. 4b), ATHB8 (Arabidopsis thaliana HOMEOBOX GENE8) (Fig. 4c) and ATHB15 (Arabidopsis thaliana HOMEOBOX GENE15) (Fig. 4d), which are targets of miR165/166. In cold imbibition (4 °C) condition at 12 h, miR165 expression was less compared to DS, where as PHB (Fig. 4a) expression was high. At 48 h/4 °C, miR165 expression was upregulated compared to DS but the expression of PHB was severely decreased in 48 h/4 °C (Fig. 4a). We found maximum expression of PHB in 24 h/4 °C. The expression of PHB was slightly different as compare to PHV, ATHB8 and ATHB15, since both PHV, ATHB8 and ATHB15 showed high expressions at 12 h/RT whereas PHV expression was reduced (Fig. 4a–d). The expressions of ARF10 (Fig. 4e), ARF 16 (Fig. 4f) and ARF 17 (Fig. 4g) as the target of miR160 were highly downregulated in all germination stages compare to DS. We analysed the expression of SPL3 (SQUAMOSA PROMOTER BINDING PROTEIN LIKE3) (Fig. 4h), SPL9 (Fig. 4i) and SPL10 (Fig. 5a), which are the targets of both miR156/157. We observed low transcript level of the target genes SPL3, 9 and 10 in all the germination conditions as compared to DS. However, the transcript level started to increase in the later stage of germination at 48 h/4 °C. We validated the expression of 4 targets of miR172, namely AP2 (APETALA2), TOE1 (TARGET OF EARLY ACTIVATION TAGGED1), TOE2, TOE3 out of 6. We observed highest expression of the targets at 48 h/4 °C (Fig. 5b–e). Comparatively, high expression of these targets was observed in case of RT, rather than cold imbibition (4 °C), which inversely correlates with increased expression of miR172 under cold imbibitions. We validated expression of three different targets of miR164, such as NAC1 (NO APICAL MERISTEM1) (Fig. 5f), CUC1 (CUP SHAPED COTYLEDON1) (Fig. 5g) and CUC2 (Fig. 5h). The maximum transcript level of these targets was observed in 48 h/4 °C and followed by expression in 12 h/RT. In these conditions, the expression level of NAC1 was ~430 fold, whereas CUC1 and CUC2 expression was induced by ~450 and ~190 folds respectively. In other conditions, we found low expression of these targets. For miR169, we validated the expression of two targets - NF-YA5 (NUCLEAR FACTOR Y, SUBUNIT A5) and NF-YA8. Like the targets of miR164a, we mostly observed upregulation of expression of the targets of miR169b in 48 h/4 °C and 12 h/RT (Figs 5i, 6a), and highest transcript level was observed in 48 h/4 °C (Figs 5i, 6a). The significantly high transcript level at 48 h/4 °C in case of NF-YA5 was 160 fold, and for NF-YA8 it was ~750 fold. We validated expression of miR161 target PPR (PENTATRICOPEPTIDE REPEAT) super family (Fig. 6b). The highest expression of the target was observed at 48 h/4 °C (Fig. 6b), and the transcript level was observed as ~190 fold. The second upregulation of this target was observed at 12 h/RT (~30 fold). In other cases were observed either same or downregulation of the targets as compared to DS. PHO2 (PHOSPHATE 2) expression correlation was validated as the target of miR399 (Fig. 6c). The highest transcript level of the target was observed at 48 h/4 °C (~37 fold). The second upregulation was observed at 12 h/RT (~6 fold) and then 12 h/4 °C (~2.5 fold). Expression in other conditions showed either downregulation or almost similar transcript level w.r.t. DS (Fig. 6c). We validated the expression correlation AGL16 (AGAMOUS-like 16) (Fig. 6d), CIP4.1(COP1-interacting protein4.1) (Fig. 6e) and R3H (Fig. 6f) as the targets of miR824, miR834 and miR854 respectively. The maximum target transcript level of AGL16 was observed at 48 h/4 °C (~41 fold) and then 12 h/RT (~5 fold). In other cases, the downregulation of the targets were observed. The highest transcript level for CIP4.1 and R3H were observed at 48 h/4 °C (Fig. 6e,f). It was ~70 fold for CIP4.1 and ~12 fold for R3H. We observed upregulation of the target w.r.t. DS in almost all conditions except 48 h/RT.
Figure 4.
Quantitative RT-PCR based validation of different target genes in imbibed seeds in comparison to dry seeds in Arabidopsis thaliana. The qRT-PCR validation of miRNAs and the targets were done at six different germination stages as 12 h/RT, 12 h/4 °C, 24 h/RT, 24 h/4 °C, 48 h/RT and 48 h/4 °C each compared to dry seed. The expression values presented here were the means of three biological replicates ± SD. The Arabidopsis ACTIN7 was used for each sample as an endogenous control.(a–d) targets of miR165/166; (a) PHB; (b) PHV; (c) ATHB8; (d) ATHB15. (e–g) targets of miR160; (e) ARF10; (f) ARF16; (g) ARF17. (h,i) targets of miR156/or, miR157; (h) SPL3; (i) SPL9. Asterisks indicate significant statistical differences, ***P < 0.001, **P < 0.01, *P < 0.05 (One-way ANOVA).
Figure 5.
Quantitative RT-PCR based validation of various predicted target genes in imbibed vs dry seeds in Arabidopsis. The target genes were selected through psRNATarget tool based on their expectation values. The qRT-PCR based validation of the targets were done at six different germination stages, like 12 h/RT, 12 h/4 °C, 24 h/RT, 24 h/4 °C, 48 h/RT and 48 h/4 °C, each compared to dry seed. The expression values presented here were the means of three biological replicates ± SD. The Arabidopsis ACTIN7 was used as an endogenous control for each sample. (a) SPL10- target of miR156/or, miR157. (b–e) targets of miR172; (b) AP2; (c) TOE1; (d) TOE2; (e) TOE3. (f–h) targets of miR164; (f) NAC1; (g) CUC1; (h) CUC2; (i) NF-YA5- target of miR169. Asterisks indicate significant statistical differences; ***P < 0.001, **P < 0.01, *P < 0.05 (One-way ANOVA).
Figure 6.
Quantitative RT-PCR based validation of various predicted target genes in imbibed vs dry seeds in Arabidopsis. The target genes were selected through psRNA Target tool based on their expectation values. The qRT-PCR validation of the targets were done at six different germination stages as 12 h/RT, 12 h/4 °C, 24 h/RT, 24 h/4 °C, 48 h/RT and 48 h/4 °C each compared to dry seed. The expression values presented here were the means of three biological replicates ± SD. The Arabidopsis ACTIN7 was used for each sample as an endogenous control. Expression of (a) NF-YA8-target of miR169; (b) PPR superfamily- target of miR161.1; (c) PHO2- target of miR399; (d) AGL16- target of miR824; (e) CIP4.1 or CIP4- target of miR834; (f) R3H- target of miR854. Asterisks indicate significant statistical differences; ***P < 0.001, **P < 0.01, *P < 0.05 (One-way ANOVA).
Expression pattern of miR390b in seeds correlates with the expression of tasiR-ARF target ARF2/3/4, and indicates its role in seed germination
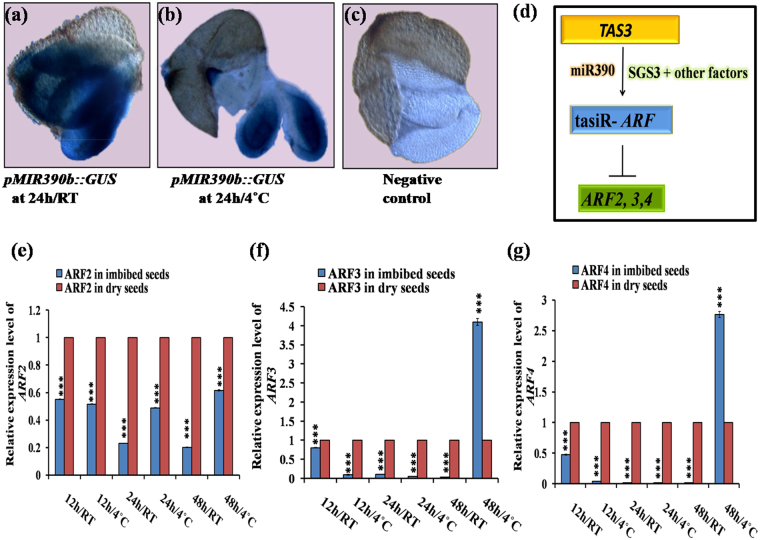
Since miR390 is required for TAS3 transcript and ta-siRNA production, we chose to chracterise it further to understand possible involvement of miRNA-tasiR-ARF module in seed germination. We have found maximum expression of miR390b at 24 h/RT following 24 h/4 °C through qRT PCR assay (Fig. 2c). To verify the expression at tissue level, we performed histochemical GUS assay with seeds of pMIR390b::GUS homozygous line along with the WT (Col-0) seeds negative control. We observed high level of expressions of miR390b at 24 h/RT (Fig. 7a) and then 24 h/4 °C (Fig. 7b) of germinating seeds, where as Col-0 as a negative control, showing no expressions (Fig. 7c). Since miR390 is essential for maturation of TAS3 and regulate the production of functional tasiR-ARF, its expression should regulate transcript level of tasiR-ARF targets ARF2, ARF3 and ARF4 through different feedback mechanisms24. We observed that the transcript levels of targets ARF2/3/4 were low at the early stages of germination and became high in later stage at 48 h/4 °C (Fig. 7e, f and g). However, interestingly, at 48 h/RT, the expression of the targets ARF2/3/4 was not high. Expression of ARF3 (Fig. 7f) and ARF4 (Fig. 7g) transcript level increased significantly at 48 h/4 °C (for ARF3 it was 4 fold, and for ARF4 it was ~2.75 fold).
Figure 7.
Spatial expression of pMIR390b::GUS in germinating seeds, function of miR390, and expression of ARF2, ARF3, and ARF4 during seed germination. (a) GUS expression of pMIR390b::GUS at 24 h/RT imbibed condition; (b) GUS expression of pMIR390b::GUS at 24 h/4 °C imbibed condition; (c) Negative control of GUS assay at 24 h/4 °C imbibed Col-0 seed. 24 h/RT-imbibed seeds were showing the higher expression of miR390b compare to 24 h/4 °C-imbibed seeds, which is similar to the expression pattern of the qRT-PCR based validation result of miR390b; (d)The model depicting the role of miR390 inthe biogenesis of tasiR-ARFs and regulation of ARF2/3/4 (According to Marin et al., 2010). (e–g) qRT-PCR based expression pattern of ARF2/3/4, which are the targets of tasiR-ARF; (e) transcript level of target ARF2; (f) transcript level of targetARF3; (g) transcript level of target ARF4.
In the previous study, it was reported that first 24 h (Phase-II of triphasic seed germination events) of imbibition is very critical for seed germination, because of maximum cellular repairments, RNA transcription and metabolism resumptions occur in this phase26–28. Our results indicate potential role of miR390-tasiR-ARF module in seed germination process.
Discussion
In higher plants, seed germination is one of the most vital phase transition from seed to seedling stage2. Although, small RNAs have been implicated in various aspects of plant development, the regulatory role of small RNAs in seed germination is less explored area till date. However, few recent reports indicated involvement of miRNAs in different plant species such as rice, maize and other monocot but in Arabidopsis, whether and how small RNAs might be regulating dynamic process of seed germination is largely unknown to us. Our present study focused on the identification and characterization of miRNAs mediated gene regulation in early stages of seed germination in Arabidopsis thaliana.
miRNAs and their targets are dynamically regulated at different conditions during germination
We have identified 58 miRNA precursors (pre-miRNAs) belonging to 30 miRNA families to be differently expressed in comparative study of three different conditions – (1) IS-4 °C vs. DS (Fig. 1a), (2) IS-RT vs. DS (Fig. 1b) and (3) IS-4 °C vs. IS-RT (Fig. 1c) during germination. Among these, 15 miRNA precursors belonging to 14 families were of P ≤ 0.05 and fold change ≥ 2.0 and considered to be significant. Since mature miRNAs are the main functional molecule that regulates their targets, we observed their dynamic regulation using SL-qRT-PCR at three different time points (12 h, 24 h and 48 h) of germination conditions. Expression of miR399a and miR399b/c were analyzed independently, since their mature sequence differed.
We observed significant upregulaion of miR165/166 in 12 h/RT and downregulation at 24 h/RT and 48 h/RT (Fig. 2a). The expression of target PHB was downregulated at 12 h/RT (Fig. 4a), where as it was upregulated at 24 h/RT and 48 h/RT (Fig. 4a), indicating their post-transcriptional regulation by miR165/166 in these conditions. Additionally, ATHB15 was also slightly upregulated in 24 h/RT (Fig. 4d). Previous reports implicated miR165/166 module in leaf, shoot, root vascular patterning (Zhou et al., 2007) and also in seed germination in maize20 and rice21. Previous reports indicated role of miR172 in regulation of vegetative to reproductive phase change and cold stress induced response affecting root growth29,30. We observed that miR172a was significantly upregulated in 24 h/4 °C (Fig. 2b) and downregulated in 24 h/RT and 48 h/4 °C (Fig. 2b), whereas its target AP2 (Fig. 5b), TOE1 (Fig. 5c), TOE2 (Fig. 5d) and TOE3 (Fig. 5e) were significantly downregulated at 24 h/4 °C. This inverse correlation of expression of miR172a and its targets indicate their post-transcriptional regulation by miRNAs. Our results indicate possible role of miR172-target AP2/TOEs module in seed germination process. miR160 is involved in auxin signalling pathway during various plant growth and developmental processes14,15,31 by negatively regulating its target transcription factors ARF10, ARF16 and ARF17 via hormonal crosstalk32. We observed the significant upregulation of miR160 during all of the germination conditions we used and found maximum upregulation at 24 h/4 °C followed by 24 h/RT (Fig. 2d). This consistent upregulation of miR160 led to the consistent downregulation of its targets ARF10/16/17 (Fig. 4e, f, g) during all germination conditions. This indicates the potential significant role of miR160 during early stages of seed germination irrespective of their stratification status. The downregulation of ARFs by miR160 during imbibition indicates possible auxin-ABA crosstalk during germination since ABA became downregulated in over expressing miR160 plants15 and ARFs are known to be involved in auxin signalling. Earlier findings also indicate the role of miR160 in seed germination in rice21 and Nelumbo nucifera22. According to the previous report, miR156 and its closely related miR15733 are the principal regulators of transition from juvenile to adult phase. They are also involved in shoot development, floral induction, initiation of leaf etc34 by negatively regulating their target SPLs. Among the ten SPLs in Arabidopsis, we chose SPL3 (Fig. 4h), SPL9 (Fig. 4i) and SPL10 (Fig. 5a), since they are the targets of both miR156 and miR157. We observed significant upregulation of these two miRNAs at 24 h/4 °C, followed by 12 h/4 °C (Fig. 2e,f); and the significant downregulation of their targets in the same above said germination conditions. This inverse correlation of miR156/157 and its target SPLs indicate their post-transcriptional regulation. Earlier studies also indicated the role of miR156 in the dynamic seed germination process of maize20 and Nelumbo nucifera22. miR164 plays a significant role in formation of proper organ boundaries35, floral patterning36, leaf morphogenesis37 and lateral root development38 by negative regulation of its target NAC1, CUC1 and CUC2. We observed the maximum significant upregulation of miR164a at 24 h/RT (Fig. 3a), followed by 24 h/4 °C (Fig. 3a), which was further followed by 12 h/4 °C (Fig. 3a). Here we also observed the inverse correlation of miRNA–target. We found maximum significant upregulation of the targets at 48 h/4 °C, followed by 12 h/RT (Fig. 5f–h). The inverse correlation of target- miRNA indicates the post-transcriptional regulation by miRNAs. In the previous report, there is the indication of the involvement of miR164 in seed germination in maize20. The miR169 family is the largest miRNA family in Arabidopsis and is encoded by 14 members39; however, only a few members have been annotated with specific functions. The miR169 targets members of the Arabidopsis NF-YA gene family40. NF-Y encodes a CCAAT-binding transcription factor, which participates in transcriptional regulation of a large number of genes41. In Arabidopsis, there are 10 genes coding for the AtNF-YA subunit42. It was reported that over expression of NF-YA5 caused hypersensitivity to ABA during seed germination43,44. We observed the maximum significant upregulation of miR169b at 24 h/RT (Fig. 3b), followed by 48 h/RT (Fig. 3b); and we observed maximum significant downregulation in the above said germination conditions and maximum upregulation of the targets NF-YA5 and NF-YA8 at 48 h/4 °C, followed by 12 h/RT (Figs 5i, 6a). This inverse correlation of target-miRNA indicates the post-transcriptional regulation of miRNA. The miR161 is a non conserved miRNA, since it is represented by single genes rather than multigene families45. The miR161 locus is unusual as it encodes overlapping miRNAs (miR161.1 and miR161.2) from a single precursor sequence45. miR161.1 targets PPR superfamily through negative regulation, which has a major impact on evolutionary background46. We observed the maximum significant expression of miR161.1 during 24 h/4 °C (Fig. 3c), followed by 12 h/4 °C (Fig. 3c); and we observed the maximum significant downregulation of the target PPR superfamily at 24 h/4 °C (Fig. 6b), then 12 h/4 °C (Fig. 6b). This inverse correlation of miR161.1 and its target PPR superfamily indicates the post- transcriptional regulation of miRNA. Our result indicates the role of miR161.1 –target PPR superfamily module in seed germination process. The Arabidopsis genome encodes six miR399 genes (miR399a -f), which are all induced by Phosphorus starvation to different extents47. miR399 is involved in orthophosphate (Pi) deficiency signalling pathway targets PHOSPHATE2 (PHO2) gene encoding E2 enzyme that negatively regulates phosphate uptake and root-to-shoot allocation47,48. The miR399a and miR399b/c (miR399b and miR399c have same mature sequence in Arabidopsis) has single nucleotide difference in the 13th position from 5’ end. Throughout the germination stages miR399a and miR399b/c expression levels were upregulated in comparison to DS and the target PHO2 expression level was downregulated, except 48 h/4 °C (Fig. 6c). Previous reports also indicated the expression of miR399 in seeds of maize20 and Nelumbo nucifera22. Availability of phytate and orthophosphate (Pi) in germinating seeds may regulate miR399 expression, which is known to be regulated by Pi availability, during germination47. miR824 is Brassicaceae-specific miRNA9,49,50. miR824 has function in rosette and cauline leaves, shoots, inflorescence and roots50,51 by negative regulation of its target AGAMOUS-LIKE16 (AGL16), which encodes a MADS box transcription factor49,52. We observed the maximum significant upregulation of miR824 during 24 h/4 °C (Fig. 3f), followed by 24 h/RT (Fig. 3f), which is further followed by 12 h/4 °C (Fig. 3f). We observed the significant downregulation of the target also. At 48 h/4 °C, followed by 12 h/RT (Fig. 6d), we observed the significant upregulation of the target. The inverse correlation of miR824 and its target indicates their post-transcriptional regulation by miRNA. Our results indicate the role of miR824-target AGL16 module in seed germination process. miR834 is a non-conserved miRNA. The predicted targets of miR834 are DEMETER-LIKE 2 (DML2) and COP1-INTERACTING PROTEIN1 (CIP1). In this study we have validated CIP4.1(AT4G00930.1). CIP1was the first reported interacting protein for CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) of Arabidopsis53. CIP1 is a positive regulator of abscissic acid (ABA) response53. CIP4, a homologue of CIP4.1, is CIP4 is COP1 interactive partner and acts downstream of COP154. We observed the significant upregulation of miR834 in all of the germination conditions that we used irrespective of stratification status; and found maximum significant upregulation during 24 h/RT (Fig. 3g), followed by 12 h/4 °C (Fig. 3g). We found maximum significant downregulation of target CIP4.1 during 48 h/RT, followed by 12 h/4 °C (Fig. 6e). The inverse correlation of miRNA-target also indicates the post-transcriptional regulation by miRNA and the miR834-target CIP4.1 module in the dynamic seed germination process. miR854 is a conserved and stress responsive55 miRNA. Plants and animals share miRNAs of the miR854 family, suggesting a common origin of these miRNAs as regulators of basal transcriptional mechanisms56. Recently, it was shown that miR854 regulated the rhizome development and the essential oil biosynthesis in ginger57. We observed significant downregulation of miR854 expression in most of the germination conditions, except 24 h/RT (Fig. 3h), we observed the inverse correlation of miRNA and its target R3H (Fig. 6f), which indicates the post-transcriptional regulation by miRNA and miR854-R3H module in seed germination. miR2112- 5p is a non-conserved miRNA. We observed the maximum significant expression of this miRNA during 24 h/RT (Fig. 3i), followed by 48 h/4 °C (Fig. 3i). According to psRNATarget tool, we found two targets of miR2112-5p. One is ergosterol biosynthesis ERG4/ERG24 family and another is pentatricopeptide repeat PPR superfamily protein. miR2112- 5p targets both of the target proteins through translational inhibition.
We observed upregulation of most of the miRNAs in 24 h imbibition, which indicates their role in seed germination during imbibition onwards in Arabidopsis thaliana. Some of the miRNAs showed the minor discrepancies between microarray data and the qRT-PCR results. The reason behind that is, during microarray we had pulled the RNAs of both RT and 4 °C imbibed seeds individually. Another reason could be the differences in the specificity, sensitivity and algorithm used between the two techniques. In most of the cases we found higher accumulation of the targets during 48 h and 12 h of germination conditions. In some conditions we didn’t observe the inverse correlation between miRNAs and their targets, this indicates seed germination condition induced transcriptional regulation of these targets, besides their post-transcriptional regulation by miRNAs. This further implies that both transcriptional and post-transcriptional regulation of miRNA targets play important role in seed germination process.
Expression pattern of miR390 and downstream ARF2/3/4 indicates potential role of miRNA-ta-siRNA crosstalk in seed germination process
Spatial expression pattern of pMIR390b::GUS in embryonic root, cotyledon and endosperm of germinating seeds (Fig. 7a,b) indicates its potential role in seed germination. We observed that the induced expression of miR390b (Fig. 2c) correlates downregulation of tasiR-ARF targets ARF2, ARF3 and ARF4 in early stages of seed germination (Fig. 7e,f and g). Upregulation of miR390 should enhance the production of tasiR-ARF leading to the down regulation of its target ARF2/3/4, which we observed in most of the germination conditions we used. This result indicates the role of miR390-tasiR-ARF mediated post-transcriptional regulation and a crosstalk of two classes of small RNAs (miRNA and ta-siRNA) in seed germination. Although we cannot rule out additional involvement of other ta-siRNAs. Thus, our study indicates that the miR390 – tasiR-ARF - ARF2/3/4 module and crosstalk of miRNA and ta-siRNA pathways (Fig. 7d) contributes to the regulation of the dynamic process of seed germination in Arabidopsis, besides role of other miRNAs and their targets.
Methods
Plant material and growth conditions
Arabidopsis thaliana ecotype Col-0 was used throughout the study. Seeds were surface sterilized and germinated on half-strength Murashige and Skoog (MS) medium (HiMedia) supplemented with 1% sucrose and 0.8% agar58. The plates were kept at 4 °C in dark for 3 days for stratification and then transferred to normal growth conditions. Plants were grown in a controlled environment at 22 ± 2 °C, under 16:8 h light (~120 µmol m−2 s−1)/dark photoperiodic cycle. The pMIR390b::GUS was described earlier23; Col-0 seeds were obtained from ABRC.
Total RNA extraction and cDNA synthesis
Total RNA was extracted from germinating wild type Col-0 Arabidopsis seeds at 0 (dry seeds), 12, 24 and 48 h after imbibition both at room temperature (RT) and 4 °C, using slightly modified guanidine hydrochloride method59. The integrity, quantity and quality of the RNAs were assessed by Bio Analyser 2100 (Agilent Technologies), Nanodrop spectrophotometer (Thermo Fisher Scientific) and also in 1.2% TAE agarose gel (Supplementary Fig. S1a) and in MOPS-formaldehyde gel (Supplementary Fig. S1b). The total RNA was further treated with DNase-I (Thermo Fisher Scientific, USA) and purified with lithium chloride (Sigma). First-strand cDNA synthesis was performed using 500ng of purified RNA with Super Script III reverse transcriptase (Invitrogen, USA) as per manufacturer’s instructions.
miRNA microarray analysis
For miRNA microarray, total RNA was isolated, and the quality of RNA was tested as described above. Approximately 250 ng of total RNA was used from each sample in two biological replicates for setting up in-vitro transcription reaction (Affymetrix, USA). The RNA purification, fragmentation and hybridization reactions were done using Arabidopsis miRNA chip miRNA v1.0 array (Affymetrix, USA). Washing and scanning were performed as suggested in Affymetrix Gene Chip total RNA procedure. Results obtained after scanning were analyzed using Gene spring GX software v11.5. The processed raw signal intensities were subjected to normalization using the same software. Expression of differentially expressed and selected miRNA genes in microarray were further validated by real time stem-loop quantitative RT-PCR (qRT-PCR).
Stem-loop and regular quantitative RT- PCR (qRT- PCR)
The expression level of mature miRNAs was analysed by stem-loop qRT-PCR, as described earlier60–62. SuperScript III reverse transcriptase (Invitrogen, USA) was used according to the manufacturer’s instructions and as described earlier60. The primer sequences used for stemloop and regular qRT-PCR were provided in (Supplementary Table S1 and S2). The transcript levels were normalized using ACTIN7 (ACT7) as an endogenous control.
β-glucuronidase (GUS) histochemical assay
During germination, the highest expression of miR390b was observed at 24 h germination condition after imbibition both at room temperature and 4 °C. To check this in vitro, we imbibed pMIR390b::GUS transgenic homozygous seeds 24 h in room temperature and 4 °C. After that, the germinating seeds were transferred to GUS staining buffer and staining was performed as described earlier63. The GUS expression was checked under stereomicroscope.
Statistical analysis
For statistical analysis, we performed one way ANOVA to determine significant differences among the samples. Differences were taken as significant when P < 0.05.
Electronic supplementary material
Acknowledgements
SSD thanks Department of Science and Technology (DST, India) for the Woman Scientist-A (WOS-A) fellowship (grant no. WOS-A/LS-1276/2014) and National Institute of Plant Genome Research (NIPGR) for short-term fellowship. SSD also thanks International Centre for Genetic Engineering and Biotechnology (ICGEB) for instrument and basic lab facilities during WOS-A fellowship. AKS and MM acknowledge Central Instrument Facility (CIF) and financial support from NIPGR. Fellowship to SY was provided by University Grant Commission (UGC, India) and fellowship to AS and VG was provided by Council of Scientific and Industrial Research (CSIR, India). We also thank M. Karthikeyan for help in preliminary stage of the work.
Author Contributions
S.S.D. performed the experiments and wrote the manuscript. S.Y., A.S. and V.G. made partial contribution to resource development and experiments. A.K.S., A.K.N., P.K., M.M. and N.S.M. conceptualised and designed the experiments and revised the manuscript. All authors read and approved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18823-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Willmann MR, Mehalick AJ, Packer RL, Jenik PD. MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol. 2011;155:1871–1884. doi: 10.1104/pp.110.171355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das SS, Karmakar P, Nandi AK, Sanan-Mishra N. Small RNA mediated regulation of seed germination. Front Plant Sci. 2015;6:828. doi: 10.3389/fpls.2015.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weitbrecht K, Muller K, Leubner-Metzger G. First off the mark: early seed germination. J Exp Bot. 2011;62:3289–3309. doi: 10.1093/jxb/err030. [DOI] [PubMed] [Google Scholar]
- 4.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Chen X. Small RNAs in development—insights from plants. Curr. Opin. Genet.Dev. 2012;22:361–367. doi: 10.1016/j.gde.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanan-Mishra N, Mukherjee SK. A peep into the plant miRNA world. Open Plant Journal. 2007;1:1–9. doi: 10.2174/1874294700701010001. [DOI] [Google Scholar]
- 8.Sanan-Mishra N, Varanasi SP, Mukherjee SK. Micro-regulators of auxin action. Plant Cell report. 2013;32:733–740. doi: 10.1007/s00299-013-1425-2. [DOI] [PubMed] [Google Scholar]
- 9.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poethig, R. S. et al. The function of RNAi in plant development. Cold. Spring Harb. Symp. Quant. Biol. (2006). [DOI] [PubMed]
- 13.Allen E, Xie Z, Gustafson AM, Carrington J. C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu PP, et al. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007;52:133–146. doi: 10.1111/j.1365-313X.2007.03218.x. [DOI] [PubMed] [Google Scholar]
- 16.Reyes JL, Chua NH. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007;49:592–606. doi: 10.1111/j.1365-313X.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- 17.Jung HJ, Kang H. Expression and functional analyses of microRNA417 in Arabidopsis thaliana under stress conditions. Plant Physiol Biochem. 2007;45:805–811. doi: 10.1016/j.plaphy.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 18.Kim JY, Kwak KJ, Jung HJ, Lee HJ, Kang H. MicroRNA402 affects seed germination of Arabidopsis thaliana under stress conditions via targeting DEMETER-LIKE Protein3 mRNA. Plant Cell Physiol. 2010;51:1079–1083. doi: 10.1093/pcp/pcq072. [DOI] [PubMed] [Google Scholar]
- 19.Wang, L., Liu, H., Li, D. & Chen, H. Identification and characterization of maize microRNAs involved in the very early stage of seed germination BMC Genomics (2011). [DOI] [PMC free article] [PubMed]
- 20.Li D, et al. Deep sequencing of maize small RNAs reveals a diverse set of microRNA in dry and imbibed seeds. PLoS One. 2013;8:e55107. doi: 10.1371/journal.pone.0055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He D, Wang Q, Wang K, Yang P. Genome-Wide Dissection of the MicroRNA Expression Profile in Rice Embryo during Early Stages of Seed Germination. PLoS One. 2015;10:e0145424. doi: 10.1371/journal.pone.0145424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu J, Jin J, Qian Q, Huang K, Ding Y. Small RNA and degradome profiling reveals miRNA regulation in the seed germination of ancient eudicot Nelumbo nucifera. BMC Genomics. 2016;17:684. doi: 10.1186/s12864-016-3032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montgomery TA, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 24.Marin E, et al. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell. 2010;22:1104–1117. doi: 10.1105/tpc.109.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai X, Zhao P. X. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Research. 2011;39:W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han C, He D, Li M, Yang P. In-depth proteomic analysis of rice embryo reveals its important roles in seed germination. Plant Cell Physiol. 2014;55:1826–1847. doi: 10.1093/pcp/pcu114. [DOI] [PubMed] [Google Scholar]
- 27.Galland M, et al. Dynamic proteomics emphasizes the importance of selective mRNA translation and protein turnover during Arabidopsis seed germination. Mol Cell Proteomics. 2014;13:252–268. doi: 10.1074/mcp.M113.032227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holdsworth MJ, Finch-Savage WE, Grappin P, Job D. Post-genomics dissection of seed dormancy and germination. Trends Plant Sci. 2008;13:7–13. doi: 10.1016/j.tplants.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Wu G, et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao L, Kim Y, Dinh TT, Chen X. miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J. 2007;51:840–849. doi: 10.1111/j.1365-313X.2007.03181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang JW, et al. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell. 2005;17:2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naya L, et al. Cleavage of a non-conserved target by a specific miR156 isoform in root apexes of Medicago truncatula. Plant Signal Behav. 2010;5:328–331. doi: 10.4161/psb.5.3.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu M, et al. Developmental Functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Genes in Arabidopsis thaliana. PLoS Genet. 2016;12:e1006263. doi: 10.1371/journal.pgen.1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mallory AC, Dugas DV, Bartel DP, Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol. 2004;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 36.Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development. 2007;134:1051–1060. doi: 10.1242/dev.02817. [DOI] [PubMed] [Google Scholar]
- 37.Nikovics K, et al. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18:2929–2945. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo HS, Xie Q, Fei JF, Chua NH. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu MY, et al. Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana. Journal of Experimental Botany. 2013 doi: 10.1093/jxb/ert353. [DOI] [PubMed] [Google Scholar]
- 40.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 41.Zhao M, Ding H, Zhu JK, Zhang F, Li WX. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 2011;190:906–915. doi: 10.1111/j.1469-8137.2011.03647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorin C, et al. A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytologist. 2014;202:1197–1211. doi: 10.1111/nph.12735. [DOI] [PubMed] [Google Scholar]
- 43.Li, W. X. et al. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. The Plant Cell, 2238–2251 (2008). [DOI] [PMC free article] [PubMed]
- 44.Mu J, Tan H, Hong S, Liang Y, Zuo J. Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol Plant. 2013;6:188–201. doi: 10.1093/mp/sss061. [DOI] [PubMed] [Google Scholar]
- 45.Allen E, et al. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- 46.Barkan A, Small I. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol. 2014;65:415–442. doi: 10.1146/annurev-arplant-050213-040159. [DOI] [PubMed] [Google Scholar]
- 47.Kuo, H. F. & Chiou, T. J. The Role of MicroRNAs in Phosphorus Deficiency Signaling. Plant Physiology156 (2011). [DOI] [PMC free article] [PubMed]
- 48.Hackenberg M, Shi BJ, Gustafson P, Langridge P. Characterization of phosphorus-regulated miR399 and miR827 and their isomirs in barley under phosphorus-sufficient and phosphorus-deficient conditions. BMC Plant Biology. 2013;13:1471–2229. doi: 10.1186/1471-2229-13-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fahlgren N, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kutter C, et al. MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell. 2007;19:2417–2429. doi: 10.1105/tpc.107.050377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alvarez-Buylla ER, et al. MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant Journal. 2000;24:457–466. doi: 10.1046/j.1365-313x.2000.00891.x. [DOI] [PubMed] [Google Scholar]
- 52.de Meaux J, Hu JY, Tartler U, Goebel U. Structurally different alleles of the ath-MIR824 microRNA precursor are maintained at high frequency in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2008;105:8994–8999. doi: 10.1073/pnas.0803218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ren C, Zhu X, Zhang P, Gong Q. Arabidopsis COP1-interacting protein 1 is a positive regulator of ABA response. Biochemical and Biophysical Research Communications. 2016;477:847–853. doi: 10.1016/j.bbrc.2016.06.147. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto, Y. Y., Deng, X. & Matsui, M. CIP4, a New COP1 Target, Is a Nucleus-Localized Positive Regulator of Arabidopsis Photomorphogenesis. Plant Cell13, 399–411(2001). [DOI] [PMC free article] [PubMed]
- 55.Srivastava S, Srivastava AK, Suprasanna P. S.F., D. S. & Identification and profiling of arsenic stress-induced microRNAs in Brassica juncea. Journal of Experimental Botany. 2012;64:303–315. doi: 10.1093/jxb/ers333. [DOI] [PubMed] [Google Scholar]
- 56.Arteaga-Vazquez M, Caballero-Perez J, Vielle-Calzada JP. A family of microRNAs present in plants and animals. Plant Cell. 2006;18:3355–3369. doi: 10.1105/tpc.106.044420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh, N., Srivastava, S. & Sharma, A. Identification and analysis of miRNAs and their targets in ginger using bioinformatics approach Gene, 570–576 (2015). [DOI] [PubMed]
- 58.Singh S, Singh A, Roy S, Sarkar AK. SWP1 negatively regulates lateral root initiation and elongation in Arabidopsis. Plant Signal Behav. 2012;7:1522–1525. doi: 10.4161/psb.22099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh G, Kumar S, Singh P. A Quick Method to Isolate RNA From Wheat and Other Carbohydrate-RichSeeds. Plant Mol. Bio. Rep. 2003;21:93a–93f. doi: 10.1007/BF02773401. [DOI] [Google Scholar]
- 60.Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gautam, V., Singh, A., Singh, S. & Sarkar, A. K. An Efficient LCM-Based Method for Tissue Specific Expression Analysis of Genes and miRNAs. Scientific Reports6, 10.1038/srep21577 (2016). [DOI] [PMC free article] [PubMed]
- 62.Singh A, Singh S, Panigrahi KC, Reski R, Sarkar AK. Balanced activity of microRNA166/165 and its target transcripts from the class III homeodomain-leucine zipper family regulates root growth in Arabidopsis thaliana. Plant Cell Rep. 2014;33:945–953. doi: 10.1007/s00299-014-1573-z. [DOI] [PubMed] [Google Scholar]
- 63.Rai V, et al. Iron Availability Affects Phosphate Deficiency-Mediated Responses, and Evidences of Cross Talk with Auxin and Zinc in Arabidopsis. Plant Cell Physiol. 2015;56(6):1107–1123. doi: 10.1093/pcp/pcv035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.