Abstract
Hypersaline environments close to saturation harbor the highest density of virus-like particles reported for aquatic systems as well as low microbial diversity. Thus, they offer unique settings for studying virus–host interactions in nature. However, no viruses have been isolated so far infecting the two most abundant inhabitants of these systems (that is, the euryarchaeon Haloquadratum walsbyi and the bacteroidetes Salinibacter ruber). Here, using three different co-occurring strains, we have isolated eight viruses infecting the ubiquitous S. ruber that constitute three new different genera (named as ‘Holosalinivirus’, ‘Kryptosalinivirus’ and ‘Kairosalinivirus’) according to their genomic traits, different host range, virus–host interaction capabilities and abundances in natural systems worldwide. Furthermore, to get a more complete and comprehensive view of S. ruber virus assemblages in nature, a microcosm experiment was set with a mixture of S. ruber strains challenged with a brine virus concentrate, and changes of viral populations were monitored by viral metagenomics. Only viruses closely related to kairosalinivirus (strictly lytic and wide host range) were enriched, despite their low initial abundance in the natural sample. Metagenomic analyses of the mesocosms allowed the complete recovery of kairosalinivirus genomes using an ad hoc assembly strategy as common viral metagenomic assembly tools failed despite their abundance, which underlines the limitations of current approaches. The increase of this type of viruses was accompanied by an increase in the diversity of the group, as shown by contig recruitment. These results are consistent with a scenario in which host range, not only virus and host abundances, is a key factor in determining virus fate in nature.
Introduction
Hypersaline environments, in addition of their interest per se (that is, ~50% of continental waters are hypersaline (Shiklomanov, 1998)), represent a remarkable opportunity to study virus–host interactions in natural settings as they are mainly inhabited by a relatively low number of prokaryotic species and their viruses (Santos et al., 2012; Gomariz et al., 2015). In fact, they present the highest concentrations of virus-like particles (VLPs) reported so far for aquatic systems, with concentrations as high as 1010 VLP/ml of water (Santos et al., 2012). Virus abundance is well co-related to the number of cells, and they are frequently the main biological factor controlling their host’s populations as predation of prokaryotes is normally absent in waters over 25% of total salt concentration (Guixa-Boixareu et al., 1996).
Among the microbes inhabiting hypersaline environments, the extremely halophilic Salinibacter ruber (phylum Bacteroidetes) offers an excellent model for microdiversity studies. S. ruber is present in most hypersaline waters analyzed so far and harbors a very wide intraspecies genomic diversity, even for co-occurring strains (for example, M8 and M31 strains were simultaneously isolated and have 99.9% identical rRNA but harbor strain-specific genes concentrated in hypervariable regions; Pasić et al., 2009; Peña et al., 2010, 2005). In many hypersaline environments worldwide, S. ruber and relatives are the main bacteria, whereas the microbial community as a whole is dominated by archaea, often of the Haloquadratum assemblage (Ventosa et al., 2015). However, despite the relevance of these two microbial groups, none of their viruses has been brought into pure culture. Conversely, many of the haloviruses isolated so far infect hosts that are not abundant (Atanasova et al., 2012), which hampers the characterization of ecologically meaningful virus–host interactions in hypersaline environments.
Virus–host interactions in hypersaline and saline environments have been addressed by culture-independent techniques such as metagenomics and other newly developed techniques (Kamke et al., 2013; Martínez-García et al., 2014; Roux et al., 2016; Martínez-Hernández et al., 2017). The analysis of ‘environmental’ viral fosmid libraries and shotgun metagenomes has allowed the recovery of genomic sequences from viruses tentatively infecting S. ruber and relatives. These putative virus–host assignments have been based on GC content, similarity and di- and tetra-nucleotide frequency analyses (Santos et al., 2010; Garcia-Heredia et al., 2012). A microarray-based metatranscriptomic analysis suggested that Salinibacter-related viruses were indeed very active in crystallizer (that is, saltern ponds where NaCl precipitates) waters, which could partially explain why S. ruber is generally outnumbered by haloarchaea, although it is as halophilic as the most halophilic archaeon (Santos et al., 2011).
However, in terms of fine-scale characterization nothing beats isolation, and, here, we present the isolation of the first S. ruber viruses cultured so far. They have been obtained using as hosts three S. ruber strains that were co-isolated from the same crystallizer pond in 1999 (Antón et al., 2002). Thus, these hosts have shared their environment and have probably been exposed to similar virus assemblages. A total of eight viruses from different hypersaline waters were isolated and selected for an in-depth phenotypic, genomic and ecogenomic study combined with targeted viral metagenomics to get some insights into the S. ruber viral population dynamics. Our results show that bacteroidetes coexisting in hypersaline environments are exposed to a suite of viruses with markedly different strategies that likely have very different origins and evolutionary trajectories, underlining the complexity of virus–host interactions in nature.
Materials and methods
Isolation and culture of S. ruber viruses
The three S. ruber strains M1, M8 and M31T used in the species description of S. ruber (Antón et al., 2002) and co-isolated from brines of S’Avall solar salterns (Mallorca, Balearic Islands) were used as hosts for virus isolation in a plaque assay using 32 different hypersaline, natural viral assemblages (see Supplementary Material). Exponentially grown S. ruber cultures were mixed with 100 μl of non-diluted and 10-fold diluted filtered natural viral assemblages and then incubated for 30 min at room temperature without shaking. After the incubation, the cultures were mixed with 25% sea water (SW) soft agar, poured into 25% SW+0.2% yeast extract agar plates and incubated at 37 °C during 10–15 days. The formed plaques were resuspended into 25% SW+0.2% yeast extract liquid medium to collect the viruses. To obtain enough viruses for further experiments, liquid cultures of each resulting virus–host pair were prepared as described in Supplementary Material.
In order to determine the host range of each isolated virus, a collection of 61 S. ruber strains from different hypersaline samples (Supplementary Material) was used. The susceptible hosts were first detected with a spot test using direct, 10- and 100-fold dilutions over individual plates containing a strain culture and then the positive interactions were quantified using regular plaque assay as described above.
Transmission electron microscopy and pulsed-field gel electrophoresis (PFGE)
One milliliter from each viral culture was ultracentrifuged at 25 000 g (1 h 20 ºC), the viral pellet was washed with ammonium acetate 0.1 M and centrifuged at 25 000 g (1 h 20 ºC) twice and finally it was resuspended in 50 μl ammonium acetate 0.1 M. A 5 μl drop of purified viruses was placed onto a Formvar-coated carbon grid (Electron Microscopy Sciences, Hatfield, PA, USA), allowed to adsorb for 1 min and stained five consecutive times (10 s each) with 2% uranyl acetate. VLPs were observed in a Jeol JEM-2010 transmission electron microscope (JEOL Manufacturer, Tokyo, Japan) operating at 200 kV.
Fifty microliters from each viral concentrate were used for plug preparation in order to determine the genome size using pulsed-field gel electrophoresis as described in Santos et al., 2007.
DNA extraction, sequencing and assembly of viral genomes
Viruses were purified from 120 ml liquid cultures using centrifugation at 40 000 g for 20 min and the supernatant was concentrated by centrifugation using Amicon Ultra 100 K devices (Millipore, Billerica, MA, USA) into 250 μl of final volume in order to make agarose plugs. The virus plugs were then digested and their DNA extracted as described in Santos et al., 2010. Genome sequencing was performed by Life Sequencing at the Scientific Park at the University of Valencia using 454 sequencing technology on purified viral DNA. In order to achieve a proper assembly of the raw viral sequences, the ends of the viral genomes were sequenced after cloning in fosmids as described before (Boujelben et al., 2012). In addition, the assembled viral genomes were submitted to an in silico digestion that was confirmed by an in vitro digestion of viral genomes with BamHI and NdeI enzymes (see Supplementary Material).
DNA sequence analyses
Analyses of average nucleotide identity values were performed by using JSpecies (Richter and Rosselló-Móra, 2009) and BLASTn analysis with 90% cutoff was performed using Easyfig Phyton application (Sullivan et al., 2011). Sequence alignment was performed with MUSCLE as implemented in Geneious 8.1.6 with default parameters in order to compare the sequences from the isolates and the results were represented in a cladogram reconstructed using the UPGMA clustering method implemented in the bioinformatic package Geneious 8.1.6. To determine the tetranucleotide frequency of the isolates, the web-based analysis site Mobyle portal was used. These results were compared by principal component analysis, (IBM SPSS Statistics v.22, Armonk, NY, USA) with the tetranucleotide frequencies found in virus genomes cloned directly from natural samples (Garcia-Heredia et al., 2012), chromosomes and plasmids of S. ruber strains M1, M8 and M31, and Hqr. walsbyi strains C23 and DSM16790 genomes (Bolhuis et al., 2006; Dyall-Smith et al., 2011).
The genome annotation was performed using the DOE-JGI Microbial Annotation Pipeline (Grigoriev et al., 2011). A BLASTp was performed using standalone blast-2.2.30+ (Shiryev et al., 2007) using the isolated virus open reading frames (ORFs) as a query and as a subject. ORFs were considered homologous above 30% of amino-acid identity.
Terminase sequences from the new isolates were used to infer a phylogenetic tree using Geneious Tree Builder with default parameters (Geneious 6.1.7). Terminases from well-characterized viruses of different origins were also included in the analysis.
The presence of the sequenced viral genomes in halophilic environments was studied by performing a recruitment analysis using the following halophilic metaviromes and metagenomes as databases: Lake Tyrrell, Australia, PRJNA81851 (Emerson et al., 2013); Bras del Port, Alicante, Spain, GU735099–GU735367, GU735369–GU735406, HM030731–HM030733 (Santos et al., 2010) and PRJNA82917 (Garcia-Heredia et al., 2012); South Bay Salt Works, San Diego, CA, USA, PRJNA28457 (Rodriguez-Brito et al., 2010); E2 metavirome from Campos Salterns, Balearic Islands, Spain (unpublished). A standalone BLASTn analysis was performed with a cutoff of 70% coverage and an e-value ⩾10−1.
Identification of structural proteins
A liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed with a tryptic-digested viral protein concentration, and the results were mapped into the viral annotation with a BLASTp analysis using the hypothetical proteins determined by annotation of the eight viruses as a database (Geneious 6.1.7; Supplementary Material).
‘Targeted’ metavirome
A brine sample from crystallizer CR30 (Bras del Port, Santa Pola, Alicante, Spain) was taken in February 2014 (36% salt concentration and 5.21 × 108 VLP/ml). The sample was centrifuged at 30 000 g, 30 min at 20 °C and then filtered through 0.2 μm filters to eliminate cells. A total of 270 ml of the viral fraction was incubated with a 5 ml of S. ruber mix of concentrate cultures containing equal amount of nine strains (M1, M8, M31T, RM158, RM225 Mallorca (Spain) isolates and P13, P18, SP38, SP73 Alicante (Spain) isolates), final concentration 2.26 × 107 cells/ml, and with 30 ml of 2% yeast extract in SW 25%, for 9 days, at 37 °C and stirred gently two or three times per week to prevent cells from sinking. After the incubation, the viral fraction was separated as described above. Both the incubated and 700 ml of the natural viral fraction were then (separately) concentrated using tangential flow filtration through a Vivaflow filter cassette system with a molecular weight cutoff of 30 000 daltons. Then, they were concentrated using ultracentrifugation; a 5 μl sample was taken in order to compare viral morphology percentages by transmission electron microscopy (TEM), and the rest was used to extract DNA from agarose plugs as described above.
Sequencing of viral DNA extracted was performed using Illumina (FISABIO, Valencia, Spain) Mi-seq Nextera XT 300 × 2 bp paired-end run (at FISABIO, Valencia, Spain). Paired-end reads were joined using Fastq-join from the eatools suite (Aronesty, 2011). Reads were quality-assessed and trimmed using PRINSEQ software (Schmieder and Edwards, 2011). Reads shorter than 50 bp, and with quality lower than 20, were discarded. Nonpareil (Rodriguez-R and Konstantinidis, 2014) was used to estimate the coverage of the community on each metagenome data set with default parameters. De novo assemblies of trimmed reads were generated using IDBA assembler (Peng et al., 2010) with the ‘-pre_correction’ option resulting in two large contigs (over 20 kb) that were then merged as described in Supplementary Material. Functional annotation of predicted genes from virus metagenomes were carried out using JGI (IMG/M ER: http://img.jgi.doe.gov/mer) and MetaVir (Roux et al., 2011).
Nucleotide sequence accession number
The genomic sequences of the newly isolated S. ruber viruses have been deposited in the GenBank database under the accession numbers MF580955 to MF580962; Metaviromes are deposited under the BioProject ID PRJNA396958 and the SR-uncultured virus (SRUTV-1) under the accession number MF629150.
Results and discussion
Virus isolation and host range
Three strains of S. ruber (M31T, M8 and M1) were used as hosts for virus isolation from different brine viral assemblages. These strains were co-isolated in 1999 from the same crystallizer pond of Campos solar salterns (Mallorca, Balearic Islands, Spain), and were included in the species description (Antón et al., 2002). Each of them was challenged with a total of 32 hypersaline water samples as described above. Only some combinations with Bras del Port (Santa Pola, Alicante, Spain) and Campos samples (Table 1) yielded plaques on host lawn and were thus used for subsequent virus isolation. Viral isolates were labeled with the name of the host strain, followed by the sample of origin and the plaque identifier number. Transmission electron microscopy (TEM; Supplementary Figure S1) showed that all the selected viruses presented head–tail morphologies and, therefore, belonged to the order Caudovirales. The isolation of these new haloviruses expands the haloviriosphere considerably, given that only four viruses infecting the extremely halophilic bacteria Salicola sp. and Salisaeta sp. (Kukkaro and Bamford, 2009; Atanasova et al., 2012; Aalto et al., 2012) had been isolated previously, from a total of 110 (now 118) viruses infecting halophilic and extremely halophilic organisms (Atanasova et al., 2015a, 2015b).
Table 1. General characteristics of the isolated viruses.
| Saltern | Sample (salinity) | Host | PFU/ml sample | Selected viruses | Head/tail (nm) | Genome (bp) a | GC content (%) a | Group |
|---|---|---|---|---|---|---|---|---|
| Bras del Port (Alicante) | CC (23.2%) April 2009 | M8 | 2.0 × 101 | M8CC-19 | 51/111c | 53 812 | 53 | II |
| M31 | 1.0 × 101 | M31CC-1 | 60/118 | 53 808 | 53 | II | ||
| CR41 (34%) March 2011 | M31 | 1.0 × 102 | M31CR41-2 | 59/114 | 51 326 | 59.7 | III | |
| M31CR41-3 | 60/115 | 50 128 | 59.7 | III | ||||
| CR30 (34.2%) April 2009 | M8 | 5.4 × 104 | M8CR30-2 | 55/121 | 35 009 | 64.6 | I | |
| M8CR30-4 | 54/121 | 35 434 | 64.6 | I | ||||
| Campos (Mallorca) | CRM (22.9%) April 2011 | M8 | 3.0 × 101 | M8CRM-1 | 59/120 | 53 197 | 53.4 | II |
| M31 | 1.0 × 101 | M31CRM-1 | ND | b | ND | ND | ||
| EM (29%) April 2011 | M1 | 2.3 × 102 | M1EM-1 | 56/119 | 35 883 | 64.7 | I |
Abbreviation: ND, not determined.
As determined by sequencing.
The size of M31CRM-1 was similar to that of M8CRM-1 as determined by PFGE. This virus was lost during subsequent culturing.
Measured in positively stained samples.
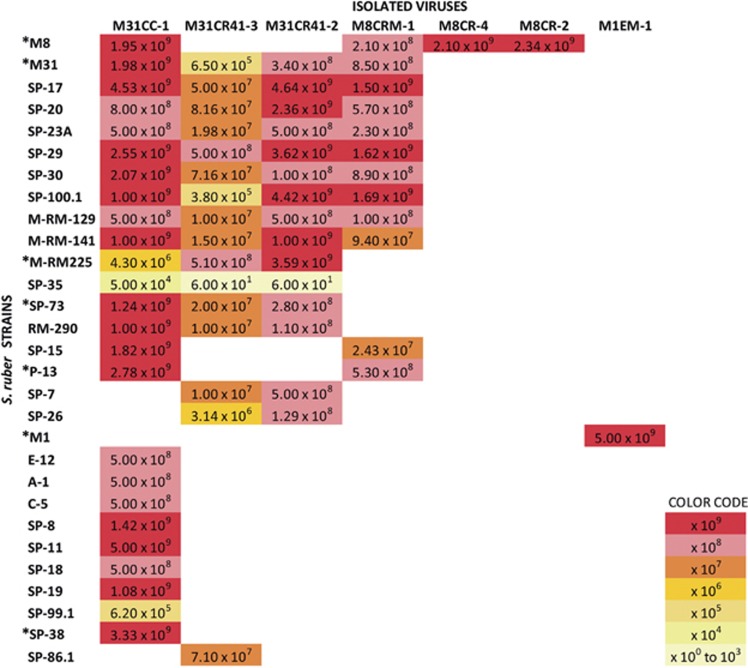
The three host strains, although very closely related based on their 16S rRNA gene similarity (above 99.8% Peña et al., 2005), displayed different susceptibilities to virus infection, with M1 as the most resistant strain, at least with the analyzed water samples (Table 1). The isolated viruses were challenged with a collection of 61 S. ruber strains isolated from salterns around the world, and previously characterized by a suit of genomic, phylogenetic and metabolomic tools (Peña et al., 2014). As shown in Figure 1, the analyzed viruses showed very different infection patterns that, as discussed below, can be explained according to the genomic differences among the viruses.
Figure 1.
Plaque-forming units (PFUs) per milliliter obtained when challenging the different S. ruber strains shown in the left (in vertical) with the newly isolated viruses (top horizontal names), colored according to the order of magnitude of the obtained PFU/ml. All the strains yielding plaques with the viruses were isolated from Mediterranean salterns except C-5 (Atlantic). Other strains analyzed that did not yield PFU with the assayed viruses were: Mediterranean (P-18*, ES-4, SP-3, SP-22, SP-23B, SP-24, SP-51, SP-57, SP-79, M-RM-30.2, M-RM-84, M-RM-101, M-RM-103, M-RM-117, M-RM-131, M-RM-174.1, M-RM272 and M-RM-158*), Atlantic (C-6, C-7, C-9, C-14, C-15, C-17, C-24, C-27 and C-29) and Peruvian (PR-1, PR-2, PR-3, PR-4 and PR-8). ‘M’ and ‘SP/P’ at the beginning of the name of the strains indicate that they were isolated from Mallorca or Santa Pola, respectively. Strains marked with asterisks (in the figure and the figure legend) were used in the targeted metavirome experiment.
At a global biogeographical scale (Figure 1), while host strains isolated from Mediterranean salterns showed varying levels of sensitivity to viral infection, Peruvian and Atlantic strains were resistant (with one exception) to all assayed viruses. These strains, isolated from three distant geographical areas separated by a distance of 10 000 km, showed quantitative differences in components generally associated with cell membranes (Rosselló-Móra et al., 2008). It is tempting to speculate that such differences are also involved in viral susceptibility of the analyzed strains. In addition, at a finer scale, we observed clear differences in virus sensitivity of Mediterranean S. ruber strains, as isolates from Bras del Port salterns presented a higher sensitivity to infection than those from Campos (70 and 30%, for 23 and 13 strains, respectively; z-value −2.3199). This could be because of the fact that most of the analyzed viruses were isolated from Bras del Port salterns (Table 1). Other isolation-based studies (Weitz et al., 2013) also suggest that ‘phages preferentially infect hosts from the same site more than host isolates from similar but distant sites’. Thus, our data show that, both at local and global scales, there is a clear biogeographical pattern in the interactions between S. ruber and its viruses in the analyzed virus–host pairs.
Overall genomic characterization
Pulsed-field gel electrophoresis indicated that viral genome sizes ranged from 35 to 53 kb, as further confirmed by sequencing (Table 1 and Supplementary Table S1). The GC content of the viral genomes ranged from 53 to 64.7% (Table 1), whereas the host strains had GC contents of ~66% (Peña et al., 2005). With some exceptions (see, for instance, Bath et al., 2006), the GC difference between phages and their microbial hosts is normally ~4% (Willner et al., 2009; Santos et al., 2010). Thus, the GC content of phages M8CC-19, M8CRM-1 and M31CC-1 is considerably lower than what could be expected for S. ruber viruses (Santos et al., 2010). It is remarkable that the analyzed lowest GC viruses had a wide host range and were isolated from medium salinity ponds (for example, 22.9–23.2%), where S. ruber is not expected to be as abundant as in close-to-saturation ponds (Antón et al., 2000; Gomariz et al., 2015). Therefore, it is possible that S. ruber is not the main natural host for these three low GC viruses, as discussed further below.
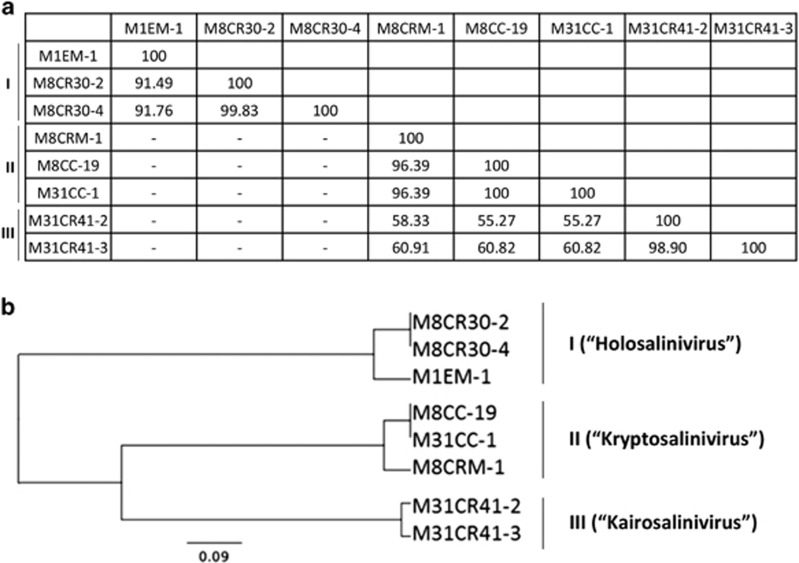
The assembly of the newly sequenced genomes was checked by restriction analysis of the purified viral genomes as shown in Supplementary Figure S2. Whole-genome alignment indicated that isolated viruses clustered into three distinct groups (labeled as I, II and III) with low sequence similarity among them (Figure 2) and marked differences in GC content and genome sizes, as shown in Table 1.
Figure 2.
Overall genomic comparison of the newly isolated viruses. (a) Average nucleotide identity (ANIb) values obtained with JSpecies software, ‘−’ indicates no significant homology. (b) UPGMA clustering tree based on the MUSCLE analysis showing the three viral groups.
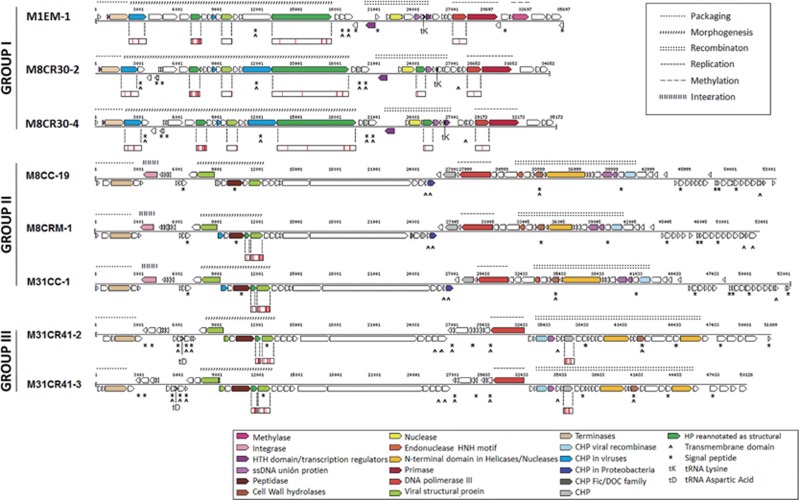
As in many caudoviral genomes (Krupovic et al., 2011), these eight viruses showed densely packed coding sequences (from 85.5 to 91.2% of the genome) and a modular distribution of protein functions (Figure 3). Overall, five categories of modules were found: DNA packaging, virion morphogenesis, genomic replication and recombination, and, in some of the genomes, modification of the DNA and lysogeny. However, the proportion of hypothetical proteins in the newly sequenced genomes was still very high (~83%). A proteomic approach, by means of LC-MS/MS (see the Material and methods section), was thus undertaken in order to identify new structural proteins and refine the in silico-based annotation (see Supplementary Table S1). Viruses within each of the groups in Figure 3 displayed identical peptide patterns (Figure 3 and Supplementary Table S1).
Figure 3.
Genomic organization of Salinibacter ruber viruses. The genome of each virus is represented with a line and ORFs are represented with arrows. Every ORF is colored according to its predicted function (see legend). ORFs with transmembrane segments or signal peptides are labeled with ^ and *, respectively. Proteins detected using LC-MS/MS are represented as small boxes below the corresponding ORF, with red bars marking the detected parts.
Overall, the nucleotide similarity among the eight viral genomes was low, although they displayed some common characteristics, in addition to the above-mentioned modular structure. All the genomes harbored genes coding for terminases (a hallmark of caudovirales and closely related herpes viruses (Casjens, 2011)) and DNA polymerases. They also presented similarities to proteins from environmental ‘halophages’ previously described by a metagenomic approach (Garcia-Heredia et al., 2012), most specifically to the ‘environmental halophages’ eHP25, that was loosely hypothesized to infect the Nanohaloarchaeota based on tetranucleotide frequency and codon usage analyses. The hits with eHP25 proteins were generally partial and frequently corresponded (see Supplementary Table S1) with S. ruber virus structural proteins, as identified by proteomics. Thus, it is likely that these hits correspond to structural domains important in hypersaline environments, as also shown by metagenomic data (see below). In addition, all the viral genomes have some orthologs in their host genomes (Supplementary Table S1), either chromosomal or plasmidic, as further indication of the ‘high level of recombination among viruses, cells and mobile genetic elements in hypersaline environments’ pointed out by Atanasova et al. (2016).
As TEM images could not unambiguously identify the newly isolated viruses as members of Myoviridae or Siphoviridae, a phylogenetic tree using large subunit terminase sequences (Supplementary Figure S3) was built to clarify this issue (Sullivan et al., 2009; Huang et al., 2012). Terminase phylogeny identified S. ruber viruses as belonging to the Siphoviridae family, in good agreement with VIRFAM analyses (http://biodev.extra.cea.fr/virfam/, Lopes et al. Automated detection and classification of head-to-tail connection proteins in bacteriophages (submitted)) that use head-neck-tail module genes to assign viruses to families.
The eight newly isolated S. ruber constitute three new viral genera
According to the recommendations of the International Committee of Taxonomy of Virus (Adriaenssens and Brister, 2017), the newly isolated S. ruber viruses can be classified into three new genera (that is, they share over 50% nucleotide identity, have similar genome sizes and GC content, and carry the same tRNAs, if any). These three genera also meet the characteristics found in Adriaenssens et al., (2015) (for example, a group of phages sharing at least 40% of their genes, combined with other characteristics like morphology and genome size and organization, among others, Supplementary Figure S4).
The main characteristics of each of the three groups are described below:
Group I (35 kb): narrow host range viruses
This group is constituted by viruses M1EM-1, M8CR30-2 and M8CR30-4. The two viruses infecting M8 are very closely related but not identical (99.8% nucleotide identity, Supplementary Figure S5), which means that, in fact, we have picked two strains from the ‘same’ virus (Adriaenssens and Brister, 2017). The M1 virus is more distant, although still in the same genus and closely related to the rest (~91.5% nucleotide identity); in addition, this is the only isolated virus carrying a methylase. Remarkably, these three viruses, despite their relatedness, have been isolated from different salterns and using different hosts. In addition to most of their gene content (Supplementary Table S1), they share the same gene distribution (Figure 3) with a packaging module that carries a terminase large subunit and a small protein with a DNA-binding domain that could indeed be a terminase small subunit (Gao and Rao, 2011; Garcia-Heredia et al., 2012). Adjacent in the genome, the morphogenesis module is constituted by at least four structural proteins, sharing the same LC-MS/MS pattern. Finally, several proteins with DNA-binding domains and nuclease activity as well as a DNA polymerase III-sliding clamp subunit and a protein with a primase domain constitute a genome recombination and replication module in which a tRNA for lysine is found. According to Bailly-Bechet et al. (2007), tRNAs present in phages tend to correspond to codons that are simultaneously highly used by the phage genes but are rare in the host genomes. This could be the case for the viruses in group I, as the codon (AAA) corresponding to the viral tRNA represents ~50% of the viral lysine codons and only 20% of the host’s.
One likely explanation of the observed host specificity differences among the group I viruses is the presence in S. ruber M1 genome (González-Torres et al., manuscript in preparation) of a CRISPR-Cas system (Makarova et al., 2015), which carries a spacer that is 100% identical to the regions (that is, the protospacers) spanning from genome nucleotide positions 24 881–24 922 and 25 200–25 241 in M8CR30-2 and M8CR30-4 genomes, respectively, and could therefore may be acting as a host defense mechanism against these viruses. This spacer, according to its relative position within the CRISPR cassette, corresponds to one of the most ancient incorporations within the extant spacers, and its conservation indicates the presence of recent selective pressure. In contrast, a previous study of virus–host dynamics in an acid mine drainage system (Andersson and Banfield, 2008) demonstrated that only the most recently acquired CRISPR spacers matched coexisting viruses and showed that viruses rapidly recombined to evade CRISPR targeting. Thus, the spacer incorporation in the S. ruber M1 CRISPR-cas system would have changed its susceptibility and subsequently selected for new variants of virulent phages, as a new example of the host–virus arms race.
Group II (53 kb): lysogenic potential and wide host range
The three viruses in this group were isolated from ponds of medium salinity (~23% total salts) and have wide host ranges among the analyzed S. ruber strains. Viruses M8CC-19 and M31CC-1 are almost identical and were recovered from the same Santa Pola sample using two different hosts (Table 1). The third virus, M8CRM-1, was isolated from a Mallorca saltern 400 km away from Santa Pola’s but which still shares ~96.3% nucleotide identity with M8CC-19 and M31CC-1 (Supplementary Figure S5). They have very similar gene content and distribution (Figure 3) and present an ORF coding for an integrase. Therefore, the possibility exists that these are temperate viruses. In addition, viruses in this group are the only S. ruber viruses described here that lack tRNAs, which is in agreement with previous observations showing that virulent phages contain more tRNAs than temperate ones (Bailly-Bechet et al., 2007).
All the viruses in group II also have an ORF coding for a signal peptide containing protein with more than 40% amino-acid identity with cell wall hydrolases from different members of Bacteroidetes, such as Rhodothermus marinus, Salisaeta longa and S. ruber. Cell wall hydrolases (pfam07486) have been implicated in cell wall hydrolysis, and thus it seems reasonable to speculate that these cellular enzymes have been incorporated into the viral genomes to help virion releasing after infection.
The main difference among the viruses in this group is the presence of several ORFs coding for hypothetical proteins and thus their involvement in infectiveness and host range cannot be ascertained. In addition, the most dissimilar genome region between M8CRM-1 and the other viruses in the group encodes a large protein of unknown function, as also observed for viruses in group III (see below). M8CRM-1 also codes for a conserved hypothetical protein not present in M8CC-19 and M31CC-1 that is related to Fic/Doc family, which includes proteins involved in the cell division regulation and in the preservation of integrated viruses by killing the cells that lose their prophage (Komano et al., 1991; Lehnherr et al., 1993).
Group III (50 kb): the ‘same’ wide-range virus twice
Viruses M31CR41-2 and M31CR41-3 were isolated using brine from crystallizer CR41 that only yielded infection of S. ruber M31T. These two viruses are very closely related (98.9% nt identity) and have almost identical host range (albeit with different infectivity). Most differences between the two genomes (Supplementary Figure S5) are located in the 3′ end of a gene coding for a large hypothetical protein containing a sugar-binding domain in M31CR41-3. They share 27.9% of the genes, such as the cell wall hydrolase, with group II viruses (Supplementary Table S1), although the synteny is interrupted by the inversion of several genomic regions (see Supplementary Figure S4). In addition, viruses in group III lack integrases, and thus are likely strictly lytic. They harbor a tRNA for aspartic acid corresponding to the codon GAC right next to the morphogenesis module. In this case, the presence of tRNA in the viral genome can be explained by the higher proportion of aspartic acid in the viral proteome (8.2% among the totality of proteins and 8.9% in the structural proteins) compared to the host’s (7.0%), although in both cases the codon usage is rather similar.
Overall viruses in group I share few genetic similarities with groups II and III, although they may share host strains and thus can be assumed to have been in direct genetic contact with each other (Hatfull, 2015). In addition, they display different types of infection networks (Weitz et al., 2013), suggesting a one-to-one structure for viruses in group I and a nested-like network for groups II and III (Figure 1). On the basis of these differences we proposed the names ‘Holosalinivirus’ (holós, ‘completely’ in Greek), ‘Kryptosalinivirus’ (krypto, ‘hide’ in Greek) and ‘Kairosalinivirus’ (kairós, ‘the right moment’ in Greek), for groups I, II and III, respectively.
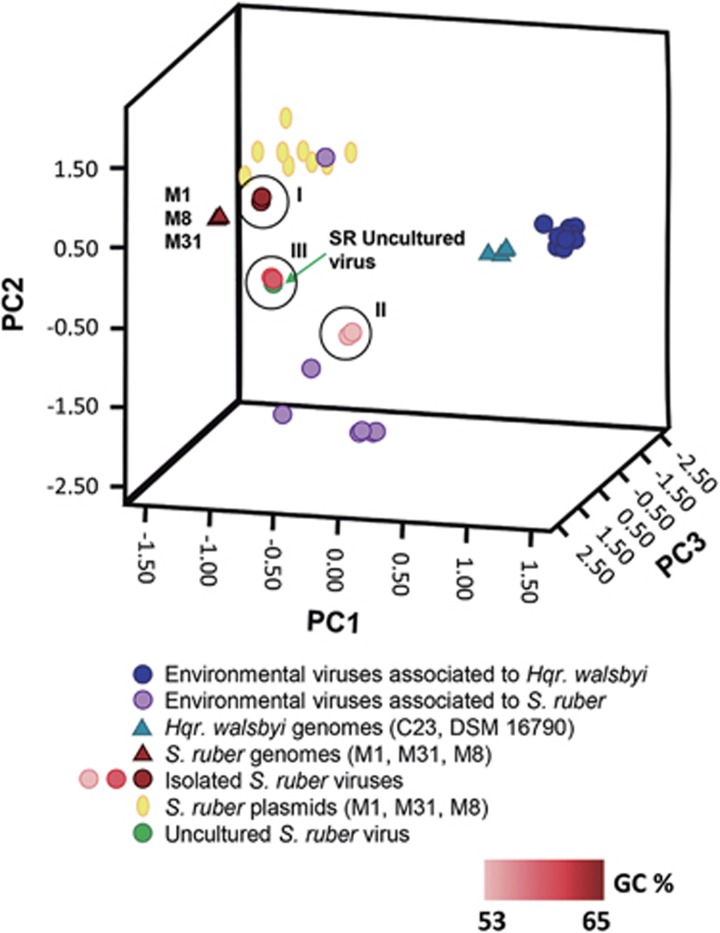
In previous studies (Santos et al., 2010; Garcia-Heredia et al., 2012), di- and tetranucleotide frequency analyses were used to tentatively assign hosts to the viruses detected in metaviromes from hypersaline environments. This allowed for the identification of some ‘environmental halophages’ putatively infecting S. ruber. Here, we have widened the analysis by including the eight newly isolated S. ruber viruses (Figure 4). As expected, viruses in each of the three groups cluster together and are separated from the rest. Holosalinivirus and kairosalinivirus are located very close to their hosts’s genomes and within the same space as S. ruber ‘environmental halophages’ (orange circles in the figure) while the putatively temperate kryptosalinivirus still falls relatively close to the S. ruber strains M8 and M31T but apart from the rest. Virulent phages contain more significant compositional differences with respect to their hosts’ genomes than temperate viruses (Bailly-Bechet et al., 2007). Therefore, one would expect that kryptosalinivirus would cluster closer to their host genomes, unless S. ruber M8 and M31T and close relatives are not their preferred hosts in nature, a hypothesis that is also consistent with the wide host range of these viruses. In addition, the exposure of these viruses to different bacterial genomes may have a wider impact on their own evolution than on that of narrower range viruses (Sullivan et al., 2005). As pointed out by Hatfull (2015), ‘the % GC composition along with associated codon usage biases do not necessarily reflect that of a known host but reflect the variety of hosts encountered in their evolutionary past’.
Figure 4.
principal component analysis of the tetranucleotide frequencies in S. ruber strains and their isolated and environmental viruses as well as S. ruber and Hqr. walsbyi strains and environmental viruses. The viruses isolated in this work are colored in increasing shades of red, according to their GC content.
Abundance and distribution of S. ruber viruses in the environment
In order to assess the abundance and distribution of the newly isolated and the ‘environmental’ S. ruber viruses in different systems, a comparison was performed with metaviromes from hypersaline environments using the viral genomes as references. The output of the analysis can be represented as the percentage of metavirome recruited by the viral genome (Supplementary Figure S6) or as the distribution of these recruited sequences along the viral genome (Figure 5). A high recruitment of a given virus in a metavirome can be due either to the high abundance of the virus or to the very frequent presence of some of its ORFs in the viral metagenome; in order to distinguish between the two situations, the coverage along the whole viral genome has to be considered.
Figure 5.
Presence of S. ruber virus groups I and III in metaviromes from different origins. BLASTn searches of the following metavirome reads against S. ruber viral genomes: Lake Tyrrell (Australia), siteA (2010); Campos salterns (Spain), E2 pond (2014); Bras del Port salterns (Spain), CR30 pond (2010); San Diego salterns (CA, USA), High Salt 111605 (2005). Viruses from group II are not represented as their presence is below the detection limit.
The normalized results (Supplementary Figure S6) of the fragment recruitment analysis showed equivalent representation of both isolated and ‘environmental’ viruses in the metaviromes analyzed, with the exception of ‘environmental virus’ eHP7, with S. ruber as the putative host, which displayed a very high recruitment in a San Diego pond. This viral genome was obtained from a Santa Pola sample by directly cloning ‘environmental’ viral DNA in fosmids (Garcia-Heredia et al., 2012). In general, both for ‘environmental’ and isolated viruses, the recruitments were higher in the metaviromes from the salterns where they were isolated, albeit with some exceptions, like the above-mentioned eHP7 and the presence in San Diego salterns of phages from group I. However, this effect is most likely because of the presence of genes highly conserved in S. ruber viruses in these salterns, as shown by the contig recruitment plots (data not shown). Overall, our results indicate that most of the newly isolated viruses of S. ruber represent relevant populations in nature.
The recruitment plots of the three new viral groups isolated in this work (Figure 5) offered a distinct distribution pattern, in good agreement with the features discussed above. Holosalinivirus, regardless their place of isolation, rendered a higher recruitment with metaviromes from Mallorca (unpublished), Santa Pola (Santos et al., 2010), and high-salt ponds from San Diego salterns (Rodriguez-Brito et al., 2010). Furthermore, this recruitment was considerable even along the viral genomes, indicating that these viruses, or very close relatives, were present in these three environments. Kryptosalinivirus, all isolated from medium salinity ponds, showed little recruitment in the analyzed halophilic metaviromes but were the only viral genomes recruiting from a medium salinity pond in Santa Pola (data not shown). Finally, kairosalinivirus had a modest recruitment only in Mediterranean salterns. In viral assemblages from high-salt ponds (from 20% salts to saturation), only a pulsed-field gel electrophoresis band of ~37 kb has been observed in most of the samples analyzed so far (Santos et al., 2012). This is in agreement with the highest recruitment of holosalinivirus that harbor 35 kb genomes. Most likely, the differences in recruitment between holosalinivirus and kairosalinivirus are related to their different rates of evolution due to exposure to different hosts. Group I, according to our data, has a very narrow host range and, therefore, fewer chances of infection (unless, obviously, the host is very abundant). These viruses seem to have a specific niche in the environment that allowed them to prevail for long periods of time (note the dates of metavirome analysis in Figure 5) and to spread to distant places with little genome variation. Conversely, kairosalinivirus can develop a more complex network of host interactions, ‘traveling’ through different hosts with different efficiency and experiencing more chances of evolution. The switching of hosts could eventually lead to an increase of virus diversity in the system and allow them to escape host resistance mechanisms more efficiently than narrow host range viruses. Therefore, we anticipated that, provided the appropriate hosts, viruses in this group might become abundant in the environment with a high degree of intragroup diversity.
Testing the hypothesis: S. ruber-targeted metavirome
To gain further insight into the issues of abundance and diversity of the S. ruber viral population discussed above, the natural viral assemblage from a crystallizer pond (CR-30) was enriched in S. ruber viruses using a protocol that is frequently used before virus isolation. In brief, a viral concentrate from crystallizer CR30 was incubated with a mixture of 9 S. ruber strains for ~200 h. These strains were chosen based on their response to virus infection (Figure 1, strains labeled with asterisks) in order to cover a wide range of susceptibilities. The increase in the number of S. ruber hosts (not necessarily previously present in the sample) provides their viruses with an increased chance to interact with them and multiply, thus enriching the viral assemblage in S. ruber viruses. Changes in the viral assemblages through incubation were monitored by TEM that showed a decrease (33%) of spindle-shaped viruses and an increase of 30% in head-tailed viruses. This change in the community is compatible with an increase in S. ruber viruses since, while head-tailed morphologies have been found in viruses infecting bacteria and archaea, spindle-shaped viruses are typical of Archaea (Pietilä et al., 2014).
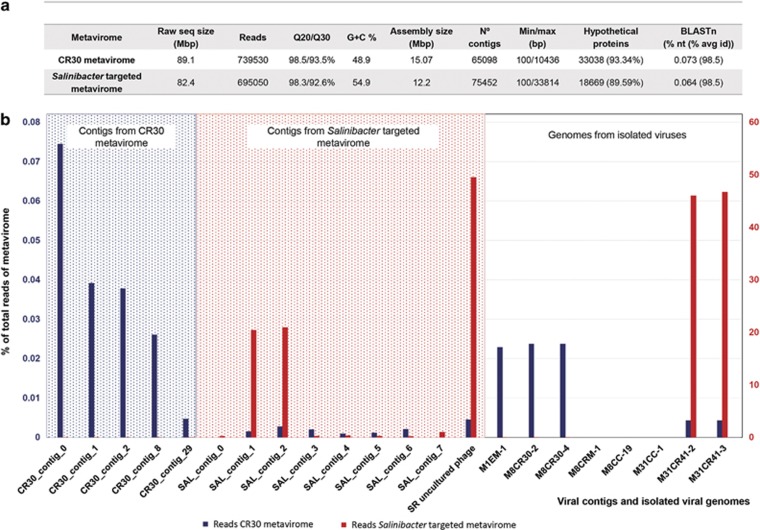
In addition, the original crystallizer metavirome and the ‘targeted’ metavirome (that is, the metagenome of the viral community generated after incubation with S. ruber strains) were sequenced and analyzed. Comparison of these metaviromes (see general aspects of both metaviromes in Figure 6) with the genomes of the newly isolated S. ruber viruses indicated a decrease in holosalinivirus after the incubation and a remarkable increase (100-fold) in group kairosalinivirus, while kryptosalinivirus remained below the level of detection (Figure 6). This increase was not due to the presence of a set of conserved genes but to the actual production of these viruses during the incubation with their hosts, as shown by the fragment recruitment plots in Figure 7. Furthermore, the largest contigs retrieved from both metaviromes after assembly were very different among them (Figure 6). In other words, most assembled viruses in the natural CR30 virome were not detected in the targeted virome, whereas some of the enriched viruses obtained in the targeted virome were already present in the natural virome, albeit at very low concentration.
Figure 6.
Characteristics of CR30 and S. ruber-targeted metaviromes. (a) Overall traits of both metaviromes. BLASTn: percentage of sequences shared between metaviromes at 98.5% identity. (b) Abundance of different viral genomes (either >5 kb metagenomics contigs or genomes from isolated S. ruber viruses) in the CR30 and targeted metaviromes. Abundances in CR30 and the targeted metavirome are represented, respectively, as blue (left scale, 0–0.08%) and red (right scale, 0–60%) bars. Left panel: abundance of contigs retrieved from CR30 metavirome; notice that these contigs are under the detection limit in the targeted metavirome. Central panel: abundance of contigs retrieved from the targeted metavirome; each of these contigs is present in low amounts in CR30 metavirome and makes up almost 20% of the targeted metavirome. The last bar represents the SR-uncultured virus genome obtained after extending SAL_contig_1 and SAL_contig_2. Right panel: percentage of reads recruited in the metaviromes by the isolated S. ruber viruses described in this work.
Figure 7.
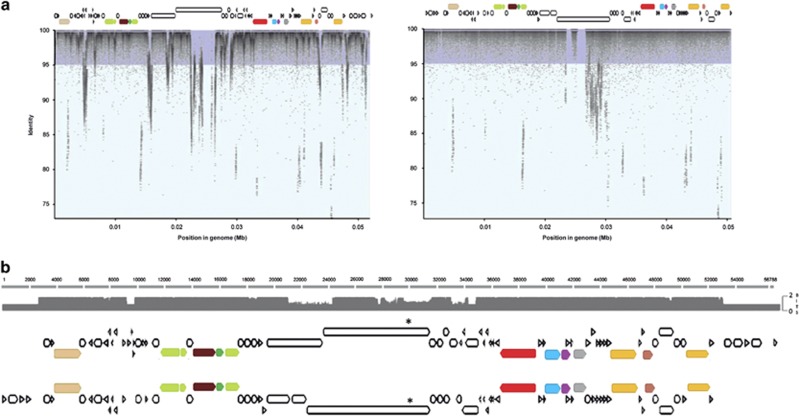
Presence of group III S. ruber viruses in the targeted metavirome. (a) BLASTn search of S. ruber-targeted metavirome reads against S. ruber viral genomes (left: M31CR41-2, right: uncultured S. ruber viral genome retrieved from the targeted metavirome). The figures represent the reads recruited by genome position (x axis) and identity (y axis). (b) Alignment between M31CR41-2 (top) and the uncultured viral genome (bottom). The asterisk shows the position of the sugar-binding domain mentioned in the text.
A detailed analysis of the targeted metavirome showed that the two largest contigs (SAL_contig_1 and SAL_contig_2 in Figure 6) matched, respectively, the 5′ and 3′ ends of M31CR41-2 and could thus correspond to ‘the same’ viral genome. This was confirmed by extension of the contig ends that allowed for the recovery of a complete viral genome (labeled as SR-uncultured virus, see Supplementary Figure S7) that accounted for 49.5% of the targeted metavirome reads and had a similarity of ~90% with group III virus. As expected, this SR-uncultured virus was very close to type III viruses in the principal component analysis plot shown in Figure 4. Most differences between SR-uncultured virus and the kairosalinivirus isolates were found again in the gene coding for a large protein containing a sugar-binding domain (Figure 7). Although this is a hypothetical protein, it could be involved in the adsorption machinery of the virus, which according to Chaturongakul and Ounjai, 2014 is ‘the most rapidly evolving part of the tailed phage genomes’.
The recruitment plots of SR-uncultured virus and kairosalinivirus with the targeted metavirome (Figure 7) presented low coverage regions that corresponded to the previously described metagenomic islands (Garcia-Heredia et al., 2012; Mizuno et al., 2014) that include the above-mentioned large protein region containing sugar-binding domains. Indeed, the viruses generated after S. ruber incubation constituted a heterogeneous assemblage of closely related genotypes sharing conserved regions (that is, the contigs SAL_contig_1 and SAL_contig_2) but harboring specific sequences in the island region, hence the different levels of coverage between these two types of genomic regions. This genomic microdiversity poses a challenge to metagenomics analysis as illustrated by the fact that the standard assembly protocol retrieved two large contigs which were indeed part of the same viral genome. This is likely one of the main drawbacks of metagenomic analyses of viral communities (Martínez-García et al., 2014). Finally, although some authors anticipated that metagenomic islands are characteristic of narrow host range viruses (Mizuno et al., 2014), it does not seem to be the case for S. ruber viruses, at least considering host range at the strain level.
Conclusions
In this study, virus isolation has allowed us to pinpoint for the first time viruses infecting coexisting strains of the cosmopolitan extremely halophilic bacteroidetes S. ruber. A combination of cultivation and metagenomic approaches indicates that these new viral isolates are abundant in natural environments and represents a gradient of different ecological strategies from low-abundance specialists to bloomers that can be easily enriched in the presence of the host. They may present different host range and different evolutionary trajectories and interactions (that is, lytic and lysogenic) with different subsets of the S. ruber assemblage in nature. Our results suggest a complex network of infections in which viruses with different levels of relatedness travel at different paces through different subsets of their host populations, interacting with them in different ways. Low-abundance wide host-range viruses could be amplified by traveling through their different hosts in a sort of chain reaction that in turn increases the chances of interactions with different subsets of hosts, as a side-effect of viral infection of the most abundant hosts. This introduces a factor into virus–host interactions, which could be largely independent of the host growth rate and its adaptation to the environment.
Acknowledgments
We warmly thank the classical language expert Fernando Nicolás for his help with the genus names, as well as Elina Roine, Evelien Adriaenssens, Francisco Martínez and Ana B Martín-Cuadrado for helpful discussion and the critical reading of the manuscript. The help of the owners of the salterns Campos and Bras del Port is greatly appreciated. We thank Karen Neller for her professional English editing. This research was supported by the Spanish Ministry of Economy projects CLG2015_66686-C3-1 (to JA) and CLG2015_66686-C3-3 (to RRM), which were also supported with European Regional Development Fund (FEDER) funds.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Aalto AP, Bitto D, Ravantti JJ, Bamford DH, Huiskonen JT, Oksanen HM. (2012). Snapshot of virus evolution in hypersaline environments from the characterization of a membrane-containing Salisaeta icosahedral phage 1. Proc Natl Acad Sci USA 109: 7079–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adriaenssens EM, Brister JR. (2017). How to name and classify your phage: an informal guide. bioRxiv 9: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adriaenssens EM, Edwards R, Nash JHE, Mahadevan P, Seto D, Ackermann HW et al. (2015). Integration of genomic and proteomic analyses in the classification of the Siphoviridae family. Virology 477: 144–154. [DOI] [PubMed] [Google Scholar]
- Andersson AF, Banfield JF. (2008). Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320: 1047–1050. [DOI] [PubMed] [Google Scholar]
- Antón J, Oren A, Benlloch S, Rodríguez-Valera F, Amann R, Rosselló-Mora R. (2002). Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int J Syst Evol Microbiol 52: 485–491. [DOI] [PubMed] [Google Scholar]
- Antón J, Rosselló-mora R, Amann R, Antón J. (2000). Extremely halophilic bacteria in crystallizer ponds from solar salterns. Appl Environ Microbiol 66: 3052–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronesty E. (2011). ea-utils: Command-line tools for processing biological sequencing data. Available from: http://code.google.com/p/ea-utils (Durham, NC).
- Atanasova NS, Bamford DH, Oksanen HM. (2016). Virus-host interplay in high salt environments. Environ Microbiol Rep 8: 431–444. [DOI] [PubMed] [Google Scholar]
- Atanasova NS, Demina TA, Buivydas A, Bamford DH, Oksanen HM. (2015. a). Archaeal viruses multiply: Temporal screening in a solar saltern. Viruses 7: 1902–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasova NS, Oksanen HM, Bamford DH. (2015. b). Haloviruses of archaea, bacteria, and eukaryotes. Curr Opin Microbiol 25: 40–48. [DOI] [PubMed] [Google Scholar]
- Atanasova NS, Roine E, Oren A, Bamford DH, Oksanen HM. (2012). Global network of specific virus-host interactions in hypersaline environments. Environ Microbiol 14: 426–440. [DOI] [PubMed] [Google Scholar]
- Bailly-Bechet M, Vergassola M, Rocha E. (2007). Causes for the intriguing presence of tRNAs in phages. Genome Res 17: 1486–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath C, Cukalac T, Porter K, Dyall-Smith ML. (2006). His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group Salterprovirus. Virology 350: 228–239. [DOI] [PubMed] [Google Scholar]
- Bolhuis H, Palm P, Wende A, Falb M, Rampp M, Rodriguez-Valera F et al. (2006). The genome of the square archaeon Haloquadratum walsbyi : life at the limits of water activity. BMC Genomics 7: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boujelben I, Yarza P, Almansa C, Maalej S, Antón J, Santos F. (2012). Virioplankton community structure in Tunisian solar salterns. Appl Environ Microbiol 78: 7429–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens SR. (2011). The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Microbiol 9: 647–657. [DOI] [PubMed] [Google Scholar]
- Chaturongakul S, Ounjai P. (2014). Phage-host interplay: examples from tailed phages and Gram-negative bacterial pathogens. Front Microbiol 5: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall-Smith ML, Pfeiffer F, Klee K, Palm P, Gross K, Schuster SC et al. (2011). Haloquadratum walsbyi: limited diversity in a global pond. PLoS ONE 6: e20968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JB, Thomas BC, Andrade K, Heidelberg KB, Banfield JF. (2013). New approaches indicate constant viral diversity despite shifts in assemblage structure in an Australian hypersaline lake. Appl Environ Microbiol 79: 6755–6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Rao VB. (2011). Specificity of interactions among the DNA-packaging machine components of T4-related bacteriophages. J Biol Chem 286: 3944–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Heredia I, Martin-Cuadrado A-B, Mojica FJM, Santos F, Mira A, Antón J et al. (2012). Reconstructing viral genomes from the environment using fosmid clones: the case of haloviruses. PLoS One 7: e33802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomariz M, Martínez-García M, Santos F, Rodriguez F, Capella-Gutiérrez S, Gabaldón T et al. (2015). From community approaches to single-cell genomics: the discovery of ubiquitous hyperhalophilic Bacteroidetes generalists. ISME J 9: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D et al. (2011). The genome portal of the department of energy joint genome institute. Nucleic Acids Res 40: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guixa-Boixareu N, Calderón-Paz JI, Heldal M, Bratbak G, Pedrós-Alió C. (1996). Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat Microb Ecol 11: 215–227. [Google Scholar]
- Hatfull GF. (2015). Dark matter of the biosphere: the amazing world of bacteriophage diversity. J Virol 89: 8107–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Wang K, Jiao N, Chen F. (2012). Genome sequences of siphoviruses infecting marine Synechococcus unveil a diverse cyanophage group and extensive phage-host genetic exchanges. Environ Microbiol 14: 540–558. [DOI] [PubMed] [Google Scholar]
- Kamke J, Sczyrba A, Ivanova N, Schwientek P, Rinke C, Mavromatis K et al. (2013). Single-cell genomics reveals complex carbohydrate degradation patterns in poribacterial symbionts of marine sponges. ISME J 7: 2287–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano T, Utsumi R, Kawamukai M. (1991). Functional analysis of the fic gene involved in regulation of cell division. Res Microbiol 142: 269–277. [DOI] [PubMed] [Google Scholar]
- Krupovic M, Prangishvili D, Hendrix RW, Bamford DH. (2011). Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere. Microbiol Mol Biol Rev 75: 610–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukkaro P, Bamford DH. (2009). Virus-host interactions in environments with a wide range of ionic strengths. Environ Microbiol Rep 1: 71–77. [DOI] [PubMed] [Google Scholar]
- Lehnherr H, Maguin E, Jafri S, Yarmolinsky MB. (1993). Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J Mol Biol 233: 414–428. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ et al. (2015). An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol 13: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García M, Santos F, Moreno-Paz M, Parro V, Antón J. (2014). Unveiling viral–host interactions within the ‘microbial dark matter’. Nat Commun 5: 1–8. [DOI] [PubMed] [Google Scholar]
- Martínez-Hernández F, Fornas O, Lluesma Gomez M, Bolduc B, de la Cruz Peña MJ, Martinez JM et al. (2017). Single-virus genomics reveals hidden cosmopolitan and abundant viruses. Nat Commun 8: 15892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno CM, Ghai R, Rodriguez-Valera F. (2014). Evidence for metaviromic islands in marine phages. Front Microbiol 5: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasić L, Rodriguez-Mueller B, Martin-Cuadrado A-B, Mira A, Rohwer F, Rodriguez-Valera F. (2009). Metagenomic islands of hyperhalophiles: the case of Salinibacter ruber. BMC Genomics 10: 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Leung H, Yiu SM, Chin F. (2010). IDBA - A practical iterative De Bruijn graph De Novo assembler. In: Research in Computational Molecular Biology, pp 426–440.
- Peña A, Gomariz M, Lucio M, González-Torres P, Huertas-Cepa J, Martínez-García M et al. (2014) Salinibacter ruber: the never ending microdiversity?. In: Papke T, Oren A, Ventosa A (eds). Genetics and Genomics of Halophiles. Caister Academic Press: Norfolk, UK, pp 37–53. [Google Scholar]
- Peña A, Teeling H, Huerta-Cepas J, Santos F, Yarza P, Brito-Echeverría J et al. (2010). Fine-scale evolution: genomic, phenotypic and ecological differentiation in two coexisting Salinibacter ruber strains. ISME J 4: 882–895. [DOI] [PubMed] [Google Scholar]
- Peña A, Valens M, Santos F, Buczolits S, Antón J, Kämpfer P et al. (2005). Intraspecific comparative analysis of the species Salinibacter ruber. Extremophiles 9: 151–161. [DOI] [PubMed] [Google Scholar]
- Pietilä MK, Demina TA, Atanasova NS, Oksanen HM, Bamford DH. (2014). Archaeal viruses and bacteriophages: comparisons and contrasts. Trends Microbiol 22: 334–344. [DOI] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106: 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M et al. (2010). Viral and microbial community dynamics in four aquatic environments. ISME J 4: 739–751. [DOI] [PubMed] [Google Scholar]
- Rodriguez-R LM, Konstantinidis KT. (2014). Estimating coverage in metagenomic data sets and why it matters. ISME J 8: 2349–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosselló-Mora R, Lucio M, Peña A, Brito-Echeverría J, López-López A, Valens-Vadell M et al. (2008). Metabolic evidence for biogeographic isolation of the extremophilic bacterium Salinibacter ruber. ISME J 2: 242–253. [DOI] [PubMed] [Google Scholar]
- Roux S, Brum JR, Dutilh BE, Sunagawa S, Duhaime MB, Loy A et al. (2016). Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537: 689–693. [DOI] [PubMed] [Google Scholar]
- Roux S, Faubladier M, Mahul A, Paulhe N, Bernard A, Debroas D et al. (2011). Metavir: A web server dedicated to virome analysis. Bioinformatics 27: 3074–3075. [DOI] [PubMed] [Google Scholar]
- Santos F, Meyerdierks A, Peña A, Rosselló-Mora R, Amann R, Antón J. (2007). Metagenomic approach to the study of halophages: the environmental halophage 1. Environ Microbiol 9: 1711–1723. [DOI] [PubMed] [Google Scholar]
- Santos F, Moreno-Paz M, Meseguer I, López C, Rosselló-Mora R, Parro V et al. (2011). Metatranscriptomic analysis of extremely halophilic viral communities. ISME J 5: 1621–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Yarza P, Parro V, Briones C, Antón J. (2010). The metavirome of a hypersaline environment. Environ Microbiol 12: 2965–2976. [DOI] [PubMed] [Google Scholar]
- Santos F, Yarza P, Parro V, Meseguer I, Rosselló-Móra R, Antón J. (2012). Culture-independent approaches for studying viruses from hypersaline environments. Appl Environ Microbiol 78: 1635–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiklomanov IA. (1998). World Water Resources: A New Appraisal and Assessment for the 21st Century. International Hydrological Programme report. Paris UNESCO. [Google Scholar]
- Shiryev SA, Papadopoulos JS, Schäffer AA, Agarwala R. (2007). Improved BLAST searches using longer words for protein seeding. Bioinformatics 23: 2949–2951. [DOI] [PubMed] [Google Scholar]
- Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. (2005). Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol 3: e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MB, Krastins B, Hughes JL, Kelly L, Chase M, Sarracino D et al. (2009). The genome and structural proteome of an ocean siphovirus: a new window into the cyanobacterial ‘mobilome’. Environ Microbiol 11: 2935–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MJ, Petty NK, Beatson SA. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27: 1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventosa A, de la Haba RR, Sánchez-Porro C, Papke RT. (2015). Microbial diversity of hypersaline environments: a metagenomic approach. Curr Opin Microbiol 25: 80–87. [DOI] [PubMed] [Google Scholar]
- Weitz JS, Poisot T, Meyer JR, Flores CO, Valverde S, Sullivan MB et al. (2013). Phage-bacteria infection networks. Trends Microbiol 21: 82–91. [DOI] [PubMed] [Google Scholar]
- Willner D, Thurber RV, Rohwer F. (2009). Metagenomic signatures of 86 microbial and viral metagenomes. Environ Microbiol 11: 1752–1766. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.