Abstract
The cyclic expression of pituitary gonadotropin-releasing hormone receptors (GnRHRs) may be an important checkpoint for leptin regulatory signals. Gonadotrope Lepr-null mice have reduced GnRHR levels, suggesting these receptors may be leptin targets. To determine if leptin stimulated GnRHR directly, primary pituitary cultures or pieces were exposed to 1 to 100 nM leptin. Leptin increased GnRHR protein levels and the percentages of gonadotropes that bound biotinylated analogs of gonadotropin-releasing hormone (bio-GnRH) but had no effect on Gnrhr messenger RNA (mRNA). An in silico analysis revealed three consensus Musashi (MSI) binding elements (MBEs) for this translational control protein in the 3′ untranslated region (UTR) of Gnrhr mRNA. Several experiments determined that these Gnrhr mRNA MBE were active: (1) RNA electrophoretic mobility shift assay analyses showed that MSI1 specifically bound Gnrhr mRNA 3′-UTR; (2) RNA immunoprecipitation of pituitary fractions with MSI1 antibody pulled down a complex enriched in endogenous MSI protein and endogenous Gnrhr mRNA; and (3) fluorescence reporter assays showed that MSI1 repressed translation of the reporter coupled to the Gnrhr 3′-UTR. In vitro, leptin stimulation of pituitary pieces reduced Msi1 mRNA in female pituitaries, and leptin stimulation of pituitary cultures reduced MSI1 proteins selectively in gonadotropes identified by binding to bio-GnRH. These findings show that leptin’s direct stimulatory actions on gonadotrope GnRHR correlate with a direct inhibition of expression of the posttranscriptional regulator MSI1. We also show MSI1 interaction with the 3′-UTR of Gnrhr mRNA. These findings now open the door to future studies of leptin-modulated posttranscriptional pathways.
This study provides strong evidence that leptin is a modulator of GnRHR expression directly at the level of the gonadotrope and implicates the RNA binding protein Musashi as a potential mediator.
Pituitary gonadotropes in females are extremely dynamic. These cells are remodeled every cycle to support a preovulatory luteinizing hormone (LH) surge and a postovulatory rise in follicle-stimulating hormone (FSH) (1). For decades, researchers studied mechanisms underlying this remodeling, focusing on hormonal regulators and modulators that facilitate optimal gonadotrope function. The earliest studies identified the cyclic production of gonadotropin-releasing hormone receptors (GnRHRs) as a critical rate-limiting step (2, 3). With the use of radioreceptor assays, these studies showed that gonadotropes undergo an increase in receptor numbers early in diestrus I (metestrus) to reach a peak in late diestrus or the morning of proestrus. The increase in GnRHR precedes the increase in stores of gonadotropins needed for the surge activity (1). Just before the LH surge, GnRHR numbers fall precipitously, to remain low throughout the remaining stages of the cycle, rendering gonadotropes less responsive to gonadatropin-releasing hormone (GnRH). Cytochemical studies from our laboratory used biotinylated analogs of GnRH (bio-GnRH) to detect receptor binding in living cells. They revealed that this increase in GnRHR is mediated through an increase in overall percentages of gonadotropes that express binding sites for bio-GnRH (4, 5).
The cyclicity of gonadotrope remodeling (which regulates pituitary responses to GnRH pulses) thus forms one of several regulatory steps by which reproduction can be slowed or delayed in response to environmental influences. For example, reproductive activity is permitted when nutrition is adequate. If energy stores are low, signals from the metabolome inform the brain, which directs the slowing of metabolism and reproduction. The adipokine leptin is a critically important metabolic informant (6–13) because serum levels of leptin reflect the adequacy of fat stores as well as nutritional status. It is well established that a rise in leptin stimulates metabolism and permits reproductive cyclicity only when energy stores are adequate. However, leptin receptors are expressed in many tissues and cell types throughout the body; the significance of each set of target sites for leptin action in the brain, pituitary, and gonads is still being explored. Pituitary gonadotropes are known to be leptin receptive (14). However, the molecular mechanisms underlying the action of leptin on gonadotrope function are not well understood.
Leptin plays an important permissive role in regulating the populations of hypothalamic neurons that release GnRH (15–25). Two laboratories have reported that restoration of leptin receptors (LEPRs) in the brain partially or completely restored fertility in mice lacking LEPR (Lepr-null) (26–28). In contrast, selective restoration of LEPR in GnRH-target cells in the pituitary did not restore fertility, although FSH levels were elevated (29). Thus, the role of leptin in directly regulating gonadotropes that respond to GnRH is somewhat controversial. Leptin has been shown to directly stimulate gonadotrope LH secretion (30, 31). Our recent dual-labeling cytochemical studies revealed a cyclic expression of LEPR in female gonadotropes (14), which correlates well with the timing of the rise in serum leptin (8). We have also shown that a 24-hour fast that results in reduced serum leptin also results in reduced pituitary LH stores and reduced percentages of bio-GnRH–bound cells in rats (9). As little as 100 pM of leptin treatment of 1 hour restores LH stores in fasting-depleted gonadotropes in vitro (9). Collectively, this evidence points to the potential for direct regulatory influences of leptin on gonadotrope stores and responses to GnRH.
Recently, we determined that leptin signaling is critical for optimal gonadotrope function by selectively deleting the signaling domain of LEPR (containing the Janus kinase–binding site) in gonadotropes (14). Gonadotrope Lepr-null (mutant) females showed a substantial delay in time to their first litter and had 50% fewer pups/litter. Mutant diestrous females also had reduced serum levels of LH (by 40%) and FSH (by 70%). The reduction in FSH was correlated with reduced expression of activin messenger RNA (mRNA). In addition, bio-GnRH binding was reduced in Lepr-null gonadotropes coexpressing LH or FSH. These specific reductions correlated well with reductions in GnRHR proteins detected by immunolabeling (14). Thus, the ablation of the signaling domain of LEPR in gonadotropes clearly impaired or slowed reproduction. Loss of LEPR signaling was also shown to affect expression of two target gene products (activin β subunit mRNA and GnRHR proteins) that are known to be critical for optimal expression and secretion of gonadotropins. Inhibin α subunit mRNA levels were unaffected by the loss of LEPR in gonadotropes.
The current study builds upon our previous findings in several ways. First, it addresses the question of whether leptin stimulates the expression of GnRHR directly by evaluating the responses of pituitary cells to leptin in vitro. This study led to the second group of studies based on our discovery that leptin stimulated an increase in GnRHR proteins and GnRH binding, but not an increase in Gnrhr mRNA levels. An in silico analysis of Gnrhr mRNA untranslated regions (UTRs) identified three consensus-binding sites for the mRNA translational regulator Musashi (MSI) in the 3′-UTR of Gnrhr mRNA. Additional studies were performed to determine if these MSI-binding elements (MBEs) were active binding sites for MSI1 and whether leptin could regulate expression of MSI1 in pituitary gonadotropes. Collectively, our findings show that leptin directly stimulates GnRHR because it inhibits endogenous MSI1 expression specifically in gonadotropes. We also present RNA immunoprecipitation (IP) findings showing that endogenous MSI1 binds endogenous Gnrhr mRNA and that MSI1 represses Gnrhr 3′-UTR–controlled translation in an independent cellular system. This repression correlates well with the higher expression of MSI1 in Lepr-null gonadotropes that show GnRHR protein deficiency. Collectively, these studies suggest a potential posttranscriptional pathway for leptin regulation of GnRHR protein expression. We discuss how these findings open the door to future studies to fully define this pathway and how a posttranscriptional regulatory pathway might work to support gonadotropes’ role as metabolic sensors.
Materials and Methods
Animal care
The Institutional Animal Care and Use Committee approved all animal care protocols. The 129P2 variant of the FVB strain was used to avoid the confounding influence of retinal degeneration in adult FVB/NJ mice. After weaning, experimental and control mice were housed four to five mice/cage. Stages of the estrous cycle were detected by vaginal smears, as reported previously (8, 14). Females were used after they exhibited two normal estrous cycles. Mice were exposed to a 14-hour light/10-hour dark cycle, given food and water ad libitum, and fed the standard rodent chow (Rodent diet 8640; Harlan).
Production of gonadotrope Lepr-null mutant mice
Mice in which Lepr-exon 17 was deleted in gonadotropes were produced as described in our recent study (14). The Cre-recombinase was driven by LH-β (Lh-β cre) and was passed via the females because of known expression of this transgene in the testes. Our previous studies have validated this line by organ genotyping and a complete study of cyclicity and breeding. All mice in this study were 3 to 5 months of age.
Pituitary cell dispersion and leptin stimulation
The cell dispersion, culture, and leptin stimulation protocols have been described in previous studies (14). A subset of experiments used pituitary pieces stimulated with leptin (1 to 100 nM) for 3 hours followed by extraction of proteins or mRNA as described previously (32–34). We detected biotinylated analogs of D-Lys6-GnRH (bio-GnRH) in cultures grown for 24 hours with protocols described previously (9, 14).
Quantitative real-time polymerase chain reaction
Pituitary RNA was isolated with the Maxwell 16 LEV simplyRNA Tissue kit (AS1280; Promega). For quantitative real-time polymerase chain reaction (qRT-PCR), complementary DNA samples and primers were added to Power SYBR Green PCR Master Mix (4367659; Applied Biosystems) and all reactions performed with the QuantStudio 12K Flex system (Applied Biosystems, Life Technologies) with the following protocol in three stages. Incubation/denaturation stage: 50°C for 2 minutes and 95°C for 10 minutes; polymerase chain reaction (PCR) amplification stage (40 cycles): 95°C for 15 seconds, 55°C for 15 seconds, and 72°C for 1 minute; and melt curve stage: 95°C for 15 seconds (ramp rate = 1.6°/s), 60°C for 1 second (ramp rate = 1.6°/s), and 95°C for 15 seconds (ramp rate = 0.5°/s). Samples were normalized to cyclophilin expression, and relative expression values were determined by the QuantStudio 12K Flex software, version 1.0, using the ΔΔ cycle threshold method.
Immunocytochemistry
Dispersed pituitary cells from wild-type mice were stimulated for 3 hours with leptin followed by exposure to bio-GnRH. The cells were fixed, and the bio-GnRH was identified cytochemically by avidin-biotin complex and nickel intensified diaminobenzidine (gray-black reaction). The cells were then dual-immunolabeled for MSI1 (Table 1) as detected by an orange-amber diaminobenzidine reaction product. Cell counts were focused on the cells labeled for bio-GnRH, which were scored positive or negative for MSI1 content. At least 100 GnRH-bound cells were analyzed for each group.
Table 1.
Antibodies Used
| Peptide/Protein Target | Antigen Sequence (if Known) | Name of Antibody | Manufacturer, Catalog No., and/or Name of Individual Providing the Antibody | Species Raised in; Monoclonal or Polyclonal | Dilution Used | RRID |
|---|---|---|---|---|---|---|
| LH | β subunit | Bovine LH-β | JG Pierce | Rabbit; polyclonal | 1:150,000 | AB_2716794 |
| Musashi 1 | Musashi1 isoform | Anti-Musashi1 | Abcam, ab2168 | Rabbit; polyclonal | 1:1000–1:5000 | AB_2042574 |
| GAPDH | GAPDH | GAPDH (14C10) | Cell Signaling, 2118 | Rabbit; monoclonal | 1:1000 | AB_561053 |
| Musashi 1 | Musashi 1 (N terminal) | Anti-Musashi1 | Abcam, ab52865 | Rabbit; monoclonal | 5 μg per sample (IP) | AB_881168 |
Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; RRID, Research Resource Identifier.
Immunolabeling focused on MSI1 in this study because our ongoing analysis showed that only MSI1 (and not MSI2) was changed in the female mice bearing Lepr-null gonadotropes. Because this was a new immunolabeling protocol for the laboratory, we performed a complete set of method, sensitivity, and specificity controls for the MSI1 immunolabeling (Supplemental Fig. 1 (11MB, pptx) ). We tested three sets of cells known to differentially express MSI1; these thus serve as biological controls. These included SH-SY5Y neuroblastoma tumor cells, which are known to produce MSI1 by Western analysis, and NIH 3T3 cells, which produce very little (if any) MSI1 based on Western analysis. The third set of cells was normal pituitary cells from wild-type mice grown in culture for 1 day.
Our tests of antibody efficiency (sensitivity) showed optimal labeling at 1:5000 of the anti-MSI1 antibody (30 minutes, 37°C) detected by 1:100 biotinylated goat antirabbit immunoglobulin G (IgG) and streptavidin horseradish peroxidase (each step, 30 minutes at room temperature). No labeling was seen in cultures with a 1:15,000 dilution of anti-MSI1. Labeling in the two cell lines was seen only after fixation in 4% paraformaldehyde, whereas anterior pituitary cells were successfully labeled after either paraformaldehyde or 1% glutaraldehyde fixation.
GnRHR enzyme immunoassay
Following dissection, whole pituitaries were immediately placed in radioimmunoprecipitation assay buffer (Sigma Aldrich, R0278) with 10 μL/mL protease inhibitor cocktail (Fisher Scientific, PI78425) on ice. Pituitaries were homogenized and incubated at 4°C overnight. The extracts were then centrifuged at 4°C at 14,000 rpm for 20 minutes. Supernatant was removed and stored at −20°C. Whole pituitary protein extracts were assayed for GnRHR protein content using an enzyme-linked immunosorbent assay (MyBiosource.com, MBS9311067).
Electrophoretic mobility shift assay detection of MSI binding to Gnrhr mRNA 3′-UTR
Glutathione S-transferase (GST) fusion proteins were in vitro transcribed/translated using TNT SP6-coupled Reticulocyte Lysate System (Promega) from pXen1 (for GST), or pXen plasmids encoding wild-type Msi1 or RNA-binding defective Msi1 (Msi1-bm), which have been described previously (35). The entire 191-bp GnRHR 3′-UTR (RefSeq Accession NM_010323), which contains three consensus MBEs was PCR amplified from murine whole pituitary poly[A]+ RNA preparations after 3′ RNA ligation to an oligonucleotide primer P1 (36) using primers to add a 5′ EcoR1 and a 3′ BamH1 and subcloned into EcoR1/BamH1 digested pGEM4z (Promega). Quikchange (Agilent) site-directed mutagenesis was used to sequentially disrupt all three MBEs (ATAGA to ATccA; GTTAGA to GTTccA; and GTTAGA to GTTccA, 5′ to 3′ respectively). A 5′ biotin-labeled RNA oligonucleotide probe was synthesized by Integrated DNA Technologies corresponding to the last 90 nucleotides of the murine GnRHR 3′-UTR, which contains two of the consensus MBEs. An 80-fmol portion of labeled probe was incubated with 1 μL of reticulocyte lysate in binding buffer (50 mM Tris pH 7.5, 20 mM KCl, 150 mM NaCl, 2 mM EGTA, 0.05% NP-40, 6 mM DTT, 8U RNase OUT) (37) in a final volume of 20 μL. For competition assays, unlabeled competitor mRNA was transcribed in vitro and the indicated fold molar excess of unlabeled mRNA added to the binding reaction. The binding reaction was incubated at room temperature for 20 minutes and then 0.5 μL of 200 mg/mL heparin was added (to reduce nonspecific binding) and incubated for a further 20 minutes. A 5-μL volume of the binding reaction was run on a 6% DNA retardation gel (Invitrogen) and transferred to Biodyne B membranes (Pierce) according to the manufacturer’s instructions. After ultraviolet crosslinking, biotinylated RNA was detected using Chemiluminescent Nucleic Acid Detection Module (Pierce) and an AlphaInnotech ChemiImager as previously described (35).
RNA IP of endogenous MSI1-Gnrhr mRNA complexes
To confirm Gnrhr binding by MSI1 in the mouse, the Millipore Magna-RIP kit (Millipore Sigma, 17-700) was used to immunoprecipitate MSI1 protein. Briefly, six pituitaries from adult control (FVB) mixed-cycle females were pooled, washed, and lysed in a single tube. The sample was then equally split for (1) IP by MSI1 antibody (Abcam, ab52865) or (2) IP with a control rabbit IgG. Magnetic beads were preincubated with antibody, washed, and then incubated with the samples overnight. The protein was degraded, and RNA was precipitated and analyzed by qRT-PCR for Gnrhr mRNA content. Results are expressed as fold-enrichment compared with rabbit IgG control and are compared with our housekeeping transcript cyclophilin. MSI1 IP was confirmed by western blot (data not shown). The IP experiments were performed twice, and each qRT-PCR run was repeated.
Luciferase mRNA translation reporter assays
The 191-bp murine GnRHR 3′-UTR was synthesized as a geneblock fragment (IDT) with a 5′ Xho1 site and 3′ Sal1 site and cloned into Xho1/Sal1 digested pmiRGLO (Promega) dual firefly luciferase (FLuc) RNA reporter and control Renilla Luciferase plasmid. The resultant clone placed the GnRHR 3′-UTR downstream of the FLuc open reading frame and was designated pmiRGLO GnRHR UTR. NIH3T3 cells (ATCC) were cotransfected with the pmiRGLO GnRHR UTR plasmid along with either MSI1-AA-eGFP, Msi1-bm-eGFP, or eGFP control plasmids as described previously (38, 39). The MSI1-AA protein is mutated in two sites of regulatory phosphorylation that are necessary for derepression and translation of target mRNAs (40). Consequently, the MSI1-AA exerts only repression. Expression of the MSI1-AA-eGFP, MSI-bm-eGFP, and eGFP proteins was confirmed by fluorescence microscopy. Luciferase activity was determined in quadruplicate after 24 hours, using the Dual-Luciferase Reporter Assay System (Promega) and Turner Biosystems luminometer (Promega) according to the supplier's protocol. Data are expressed as relative luciferase activity in arbitrary units. All experiments were repeated on three to eight separate occasions.
Statistics
At least five animals were used for each test, and in vitro tests were repeated three times. Cell counts and assay values were analyzed with Prism statistical software with analysis of variance (ANOVA) followed by Tukey or Bonferroni post hoc tests, as described previously (14, 33, 34). When two groups were compared, the Student t test was run. A post hoc power analysis was done with counts of GnRHR-bearing cells in the pituitary culture. This analysis showed that a 1-nM treatment with leptin will stimulate an increase in percentages of biotinylated-GnRH–labeled cells from 16.6% (controls) to 24% (standard deviation, 3.6%). This increase has 99% statistical power with five replicates in a two-tailed, 0.05-level t test. If we compare results following stimulation with 1 and 100 nM leptin, the increase in number of GnRH-labeled cells (from 24% to 28%; standard deviation, 3.5%) will have 81% power with five replicates in a two-tailed 0.05-level t test.
Results
Leptin stimulates GnRHR protein levels
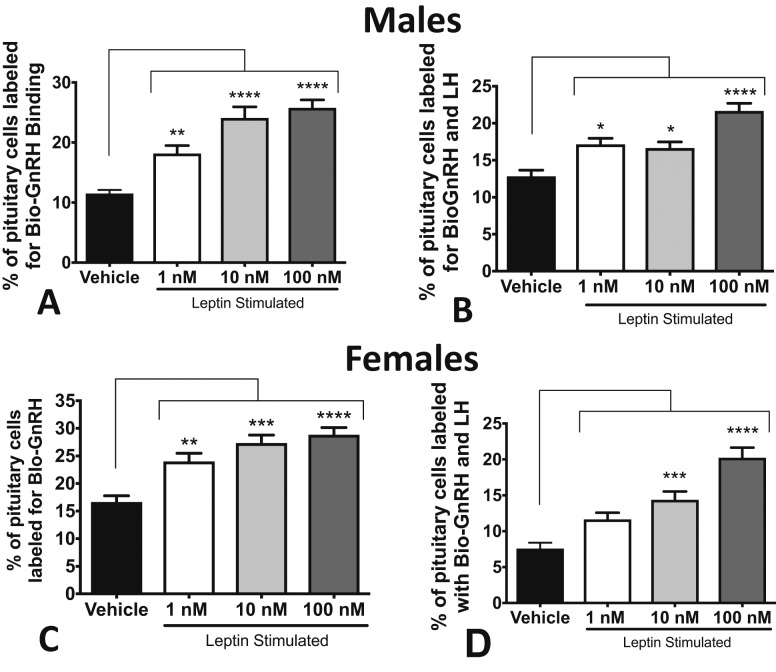
The first set of studies was performed on 1-day cultures or pituitary pieces (from wild-type males or diestrous females) to determine if leptin directly stimulates GnRHR levels. Specifically, we determined (1) if leptin stimulates an increase in GnRH binding by living gonadotropes and (2) if leptin stimulation increases total pituitary GnRHR protein levels. Figure 1A–1C shows that leptin stimulation for 3 hours resulted in a dose-dependent increase in cells that expressed GnRHR as identified by bio-GnRH binding in males and females. Analysis of dual labeling also showed a dose-dependent increase in cells expressing both LH and GnRHR (Fig. 1B–1D). The changes in dual labeling are illustrated in micrographs in Fig. 2A and 2B.
Figure 1.
Leptin stimulates GnRHr expression in wild-type pituitaries in vitro. (A) Dispersed pituitary cells from male mice were stimulated with 1, 10, or 100 nM leptin. All concentrations of leptin stimulated an increase in the percentages of living pituitary cells that bound 1 nM biotinylated GnRH. (B) All concentrations of leptin stimulated a substantial increase in cells dual-labeled for bio-GnRH and LH. Stimulation with 100 nM resulted in the highest percentages of dual-labeled cells (compared with 1 and 10 nM, P < 0.005). (C) All concentrations of leptin stimulated an increase in the percentage of living diestrous female pituitary cells that bound biotinylated GnRH when compared with vehicle, with 1 nM being significantly lower than 10 nM (P < 0.05) and 100 nM (P < 0.005). (D) Both 10 and 100 nM leptin stimulated an increase in the percentage of diestrous pituitary cells dual labeled for LH and bio-GnRH, and 100 nM stimulated significantly more dual-labeled cells than 1 nM (P < 0.0001) and 10 nM (P < 0.005). *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.0001.
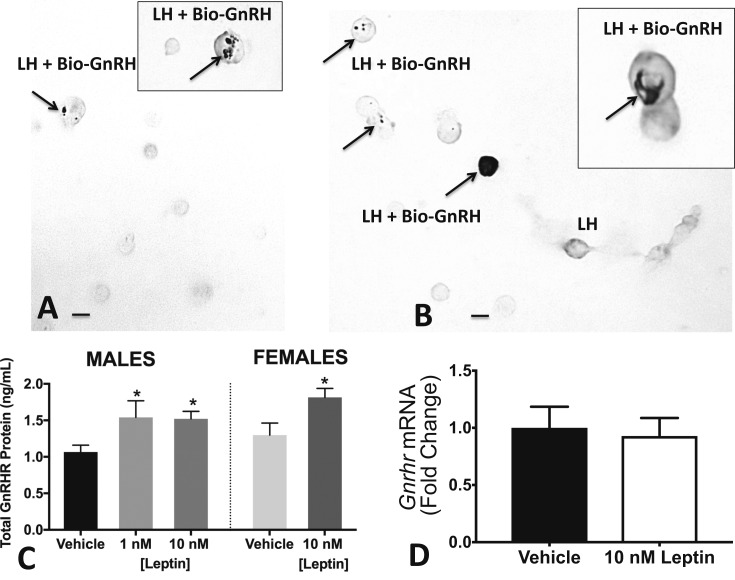
Figure 2.
Leptin stimulates bio-GnRH binding and GnRHr protein expression, but not Gnrhr mRNA in vitro. (A) Representative field showing dual labeling for bio-GnRH (black patches, arrows) and gray label for LH. Vehicle control field shows only one dual-labeled cell. (B) Multiple dual-labeled cells in field treated with 1 nM leptin for 3 hours. Arrows point to the cells with black labeling for bio-GnRH. Inset, the increased density of labeling in some of the cells. This field also contains LH cells that do not express bio-GnRH (labeled LH). Bar = 10 μm. (C) GnRHR proteins were assayed following 1 and 10 nM leptin stimulation of wild-type male or female pituitary pieces. Both treatments significantly increased GnRHR proteins. (D) Leptin stimulation of wild-type female pituitary pieces for 3 hours had no effect on expression of Gnrhr mRNA levels. *ANOVA, Fisher least significant difference test, P < 0.05. **P < 0.005; ***P < 0.0005; ****P < 0.0001.
When dual-labeled LH cells were analyzed further, diestrous females showed a significant increase in the proportion of LH cells that bound bio-GnRH following leptin stimulation (vehicle: 58% ± 3% of LH cells; 10 nM LEP: 70% ± 4%; or 100 nM LEP: 80% ± 2%; ANOVA, P = 0.0004). In control males, a higher percentage of LH cells expressed bio-GnRH binding in vehicle-treated cultures (78% ± 4%) and only 100 nM leptin resulted in a significant increase in percentages of LH cells with GnRHRs to 88% ± 2% (P = 0.03).
Parallel studies of leptin stimulation of pituitary pieces were used to obtain protein fractions that were assayed for GnRHR by enzyme immunoassay. Figure 2C shows a representative experiment that involved cells from both male and female mice. Leptin stimulated substantial increases in GnRHR proteins in both groups (Fig. 2C), which correlated well with the cytochemical studies of bio-GnRH binding to living cells. Additional experiments also showed that 1 and 100 nM leptin stimulated GnRHR in females (data not shown). We also extracted RNA from additional pituitaries stimulated with vehicle or leptin and performed RT-PCR for Gnrhr mRNA. Figure 2D shows that 10 nM leptin did not affect levels of Gnrhr mRNA, which complements results from our previous studies that showed no changes in Gnrhr mRNA in the gonadotrope Lepr-null mice (14).
In silico analysis of the Gnrhr mRNA 3′-UTR and evidence for MSI-dependent translational control
The data from this and our previous study (14) showed that GnRHR proteins were reduced in gonadotrope Lepr-null pituitaries. The current data now show that GnRHR proteins are increased following direct leptin stimulation in vitro (Fig. 2). However, Gnrhr mRNA levels did not change either in gonadotrope Lepr-null mutant pituitaries (14) or following in vitro stimulation with 10 nM leptin (Fig. 2). Collectively, these findings led us to hypothesize that the Gnrhr mRNA may be subject to translational control. Therefore, we subjected the Gnrhr mRNA to in silico analyses and determined that the 45-nucleotide murine Gnrhr mRNA 5′-UTR contains no oligopyrimidine tracts indicative of mTOR-mediated control. However, the 3′-UTR of murine Gnrhr mRNA contained three consensus MBEs of sequence (G/A)U1-3U (41). Figure 3A shows the map of murine Gnrhr mRNA 3′-UTR showing the MBE distribution.
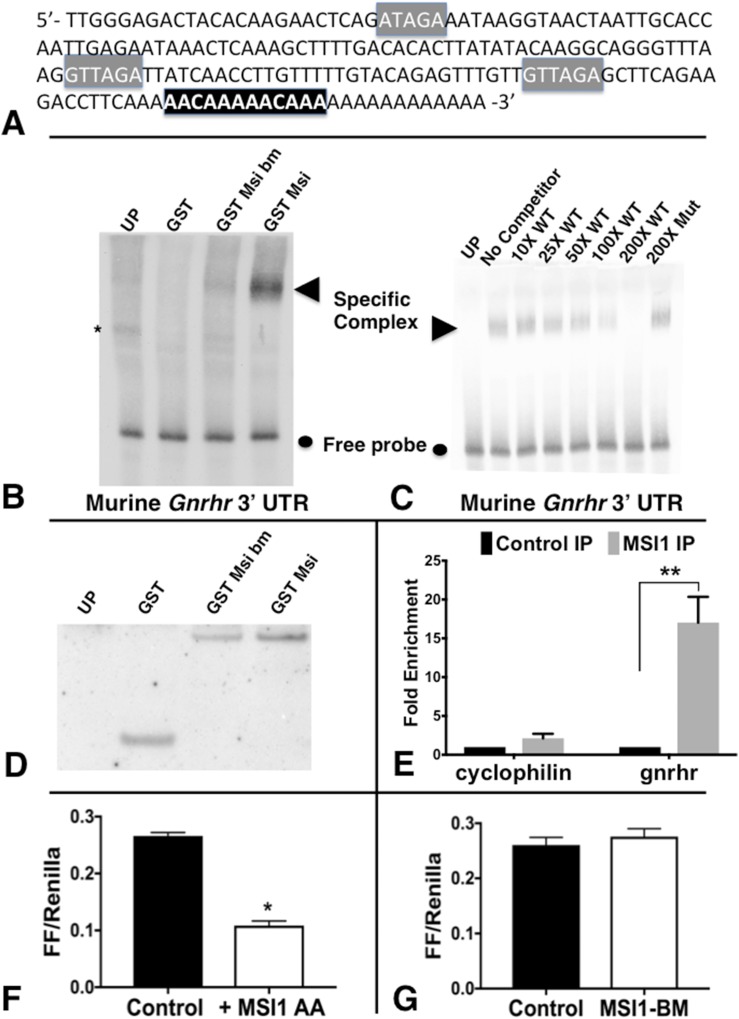
Figure 3.
EMSA and FLuc assay data showing that Musashi1 interacts directly with the mGnrhr 3′-UTR. (A) Sequence-verified mGnrhr 3′-UTR. Gray, consensus MBEs; black, two adjacent AACAAA polyadenylation signals. (B) RNA EMSA using biotinylated murine GnRHR 3′-UTR incubated with UP, or GST, GST Msi bm, or GST-Msi. Musashi1-specific complex formation is indicated by an arrowhead; free probe is marked by a circle; a nonspecific complex is indicated by an asterisk. (C) RNA EMSA similar to that in panel B, except MSI binding to Gnrhr 3′-UTR was challenged with increasing molar excess (10 to 200×) WT or 200× MBE Mut unlabeled Gnrhr 3′-UTR. Disruption of MBEs in the mutant Gnrhr 3′-UTR attenuated competition for MSI binding to the biotinylated probe. (D) UP or reticulocytes expressing GST, GST fused to either GST Msi or Msi-bm used in (B) and (C) were processed for western blotting to confirm that each protein was expressed to comparable levels. (E) Gnrhr mRNA is significantly enriched with MSI1 immunoprecipitation. The data represent two experiments analyzed with quantitative PCR. Numbers are the average fold-enrichment as analyzed from qRT-PCR. Student t test, control IP vs MSI1 IP for each target mRNA. (F, G) FLuc assay run on NIH3T3 cells that do not normally produce MSI1. These cells were cotransfected with a construct including the Gnrhr 3′-UTR attached to FLuc RNA reporter and EGFP plasmids containing vehicle or a mutant form of MSI that cannot be phosphorylated on two regulatory sites (MSI-AA) or the MSI-bm. The FLuc values were normalized to expression of a control Renilla Luciferase expressed from the same plasmid (FF/Renilla). (F) Average from three representative experiments showing that MSI-AA suppressed translation 2.7-fold (D: P = 0.0002). (G) The values from 2 experiments in which the MSI-bm mutant, which is disrupted for RNA binding, had no effect on translation of Gnrhr-3′-UTR (E: P = 0.5) Student t test with Welch correction. *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.0001. GST, lysates expressing GST alone; GST Msi, wild-type Musashi1; GST Msi bm, RNA-binding mutant Musashi1; GST-Msi, Musashi1; Msi-bm, RNA-binding mutant Msi; Mut, mutant; UP, unprogrammed rabbit reticulocyte lysate; WT, wild-type.
Electrophoretic mobility shift assay (EMSA) RNA-binding studies were then run to determine if MSI1 interacted directly with the mGnrhr 3′-UTR. Figure 3B shows results with the use of biotinylated murine Gnrhr 3′-UTR incubated with unprogrammed rabbit reticulocyte lysate, the GST moiety alone, or GST fused to either an RNA-binding mutant MSI1 (GST MSI1-bm) or wild-type MSI1 (GST MSI1). Although the GST MSI1–expressing lysate was able to form a specific complex, neither the GST MSI1-bm or GST moiety alone was able to do so. We conclude that MSI1 interacts directly with the Gnrhr 3′-UTR. To determine if the MBEs in the Gnrhr 3′-UTR were necessary for MSI1 binding, a competition experiment was performed (Fig. 3C). Increasing molar excess of unlabeled wild-type Gnrhr 3′-UTR progressively competed for binding to MSI1, with complete ablation of specific binding seen with 200-fold molar excess. The MSI1 interaction specifically required the MBEs as 200-fold molar excess of an MBE-disrupted Gnrhr 3′-UTR showed no attenuation of MSI1 interaction with the biotinylated Gnrhr 3′-UTR probe. Indeed, the level of complex formation in the presence of 200-fold excess MBE-disrupted Gnrhr 3′-UTR was indistinguishable from complex formation seen in the absence of competitor RNA (no competitor). Equivalent levels of GST and the GST fusion proteins, GST MSI1-bm and GST MSI1, were expressed in the reticulocyte lysates used for the EMSA reactions (Fig. 3D). We conclude that MSI1 binds specifically to the Gnrhr 3′-UTR via one or more of the three identified MBEs.
Next we confirmed MSI1-Gnrhr association by immunoprecipitating a complex containing endogenous MSI1 proteins that was also enriched in endogenous Gnrhr mRNA (Fig. 3E). Pituitaries from six females were lysed and split for IP with an anti-MSI1 antibody or control IgG (rabbit IgG). We found that the MSI1 antibody precipitates Gnrhr 17-fold as compared with the control (IgG) samples (average of two repeats). Our housekeeping gene cyclophilin was only enriched twofold in the MSI1 samples compared with control samples.
To determine if the binding of MSI1 to the GnRHR 3′-UTR exerted translational control, we used a luciferase reporter assay in NIH3T3 cells. For this assay, the FLuc coding region was placed under the control of the GnRHR 3′-UTR. Whereas NIH3T3 cells do not express MSI1 (Supplemental Fig. 1C (11MB, pptx) ), they are capable of reconstituting MSI1-dependent repression of target mRNAs upon transfection of ectopic MSI1 (41). The MSI1-AA protein is mutated in two sites of regulatory phosphorylation that are necessary for derepression and translation of target mRNAs (40). Consequently, the MSI1-AA exerts only repression (38). In contrast with cells cotransfected with vehicle or a mutant MSI that is RNA-binding defective where FLuc was efficiently expressed (Fig. 3G), translation of the FLuc reporter was repressed significantly (by 63%) in cells that were transfected with MSI1-AA (Fig. 3F). Taken together, our data from this independent system indicate that MSI1 binds to MBEs within the murine GnRHR 3′-UTR and exert translational repression. Of note, the longer human GnRHR 3′-UTR (NM_001012763) has 19 putative MBEs and the shorter one (NM_000406) has 29 putative MBEs, suggesting that MSI may be a conserved regulator of Gnrhr mRNA translation.
Leptin regulation of expression of pituitary Msi1 mRNA
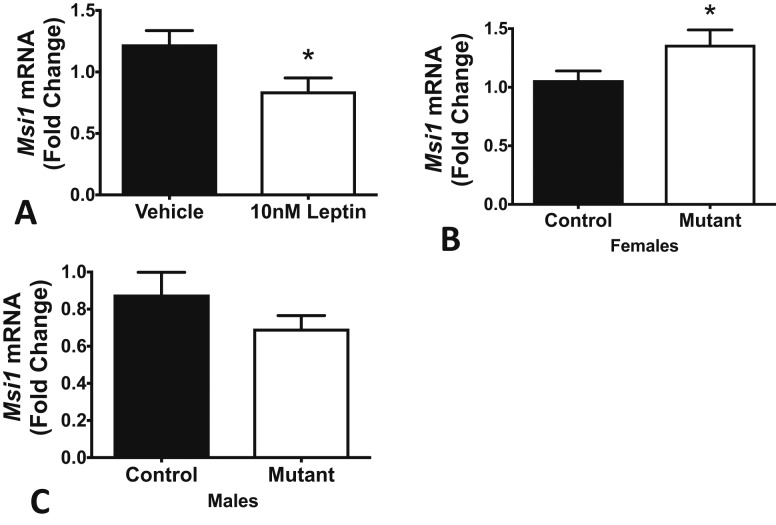
The foregoing showed that leptin regulated GnRHR proteins, and MSI1 bound Gnrhr mRNA and repressed translation of a reporter linked to the Gnrhr mRNA 3′-UTR. This supports the hypothesis that the MBEs in the Gnrhr 3′-UTR are active repressor sites. The next two sets of studies were designed to determine if leptin interacted in this pathway by altering expression of MSI in the pituitary and, most importantly, in pituitary gonadotropes. Figure 4 shows the results from qRT-PCR assays of mRNA from pituitary pieces from proestrous female mice, which were stimulated for 3 hours with 10 nM leptin. Leptin reduced Msi1 mRNA levels (Fig. 4A). In parallel studies, we assayed Msi1 mRNA levels in control and gonadotrope Lepr-exon 17 null mutant pituitaries. Figure 4B shows that mutant proestrous females exhibit an increase in pituitary Msi1 mRNA levels. Taken together, these results indicate that leptin stimulation causes a rapid reduction in Msi1 mRNA expression. Conversely, in the absence of leptin signaling, levels of Msi1 mRNA increase. Notably, mutant males showed no substantial change in Msi1 mRNA (Fig. 4C).
Figure 4.
Leptin regulates Msi1 mRNA levels in females. (A) Leptin stimulation of pituitary pieces for 3 hours significantly reduces levels of Msi1 (whole pit mRNA, extracted and measured by RT-PCR). (B) Gonadotrope Lepr-null females have significantly higher levels of Msi1 mRNA compared with littermate controls. (C) In contrast, mutant gonadotrope Lepr-null males exhibit no changes in Msi1 mRNA. *Significantly different from vehicle or wild-type control, Student t test, P < 0.05. **P < 0.005; ***P < 0.0005; ****P < 0.0001.
Leptin regulation of expression of MSI proteins in pituitary gonadotropes
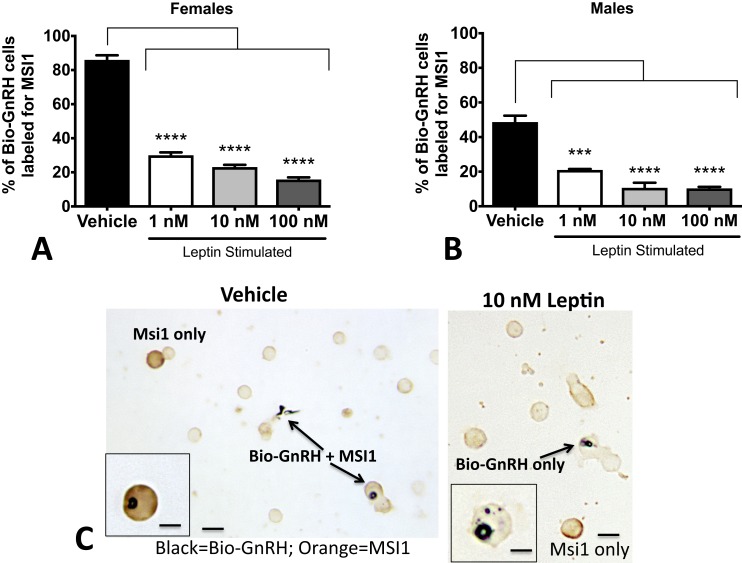
Because these changes in pituitary MSI1 reflect the net result of changes in all pituitary cells, we used a cytochemical approach to determine changes specific to gonadotropes. We analyzed the same cell populations stimulated with leptin (analyzed in Fig. 1). However, instead of dual labeling with LH, we substituted anti-MSI1. We then counted the number of cells labeled for bio-GnRH that also contained MSI1. The results shown in Fig. 5A–5B indicate that leptin caused a substantial reduction in the percentages of GnRH target cells with orange MSI labeling. Images taken of double-immunolabeled diestrous female pituitary cells can be seen in Fig. 5C. Note that vehicle control dual-labeled cells are intensely labeled black for bio-GnRH and orange for MSI1. However, in the field treated with 10 nM leptin for 3 hours, the bio-GnRH cells are devoid of orange labeling for MSI1. Yet, as shown in Fig. 2, the black label for bio-GnRH is very strong in the presence of leptin, which reflects its stimulatory effect.
Figure 5.
Leptin stimulation of MSI1 in gonadotropes. Monolayer cultures of pituitary cells were stimulated with leptin for 3 hours and then exposed to biotinylated GnRH for the last 10 minutes of culture. After fixation, cells were dual labeled for bio-GnRH and MSI1. MSI proteins are found (A) in 84% of bio-GnRH bound gonadotropes in females and (B) in ∼50% of bio-GnRH cells in males. After 3 hours of leptin stimulation, there is a dramatic loss in expression of MSI1 proteins in both males and females. ANOVA followed by Bonferroni multiple comparisons test. (C) A sample photomicrograph demonstrates the dual-labeled cells containing black label for bio-GnRH and orange label for MSI1 (left). A field treated with 10 nM leptin and the loss of orange labeling for MSI1 in the biotinylated GnRH-labeled cells (right). The label for bio-GnRH is stronger, as would be expected following leptin stimulation. Low magnification bar = 20 μm; high magnification bars = 10 μm. *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.0001.
Discussion
The requirement for dynamic, cyclic changes in GnRHR in female gonadotropes makes this gene product a prime target for regulators that modulate the reproductive system, including the adipokine leptin. Our earliest reports of this cyclic expression in females showed that cells that bound bio-GnRH increased in number, reaching a peak on the morning of proestrus (4). Our more recent reports have shown that percentages of cells with bio-GnRH binding are significantly reduced in rat gonadotropes that lack leptin signals after fasting (9) and in mice that have gonadotrope leptin receptors ablated (14). Thus, GnRHR proteins or GnRH-binding sites appear to be a target for metabolic signaling. Whereas these findings are compelling, we also recognize that GnRHR levels are regulated by many factors, including by GnRH itself (42, 43). Thus, the first objective of this study was to determine if leptin regulated GnRHR levels at the level of the pituitary.
In the present report, we tagged living gonadotropes with bio-GnRH and detected leptin-mediated changes in GnRH-bound cells from both males and females. We also showed that leptin stimulates a dose-dependent increase in GnRHR proteins in gonadotropes (Figs. 1 and 2), especially in females, which matches that normally seen in the diestrous to proestrous transition (4, 5). Also, as reported previously, gonadotropes from males and diestrous females have different expression levels of GnRHR, with higher basal levels of bio-GnRH binding in males (70% of LH cells) than in diestrous females (58% of LH cells). Lower levels of leptin were needed to bring the GnRH binding by diestrous female LH cells up to levels seen in the male. This differential sensitivity to leptin correlates well with our report that there are sex differences in the responses to the ablation of LEPR in gonadotropes (14). Males have reduced GnRHR, but this reduction did not alter fertility. In contrast, the reduction in GnRHR in females is more severe, resulting in subfertility (14) and, in the case of more complete ablation of leptin signaling via conditional LEPR-exon 1 deletion, infertility (44).
We also examined the direct effect of leptin on expression of Gnrhr mRNA and showed no changes in levels of this mRNA. This correlated well with our recent studies of the Lepr-null gonadotropes, which also showed no changes in Gnrhr mRNA when compared with control animals. These findings opened the door to a series of studies designed to test the hypothesis that leptin acts through candidates in a posttranscriptional pathway. The studies included an in silico analysis, which showed evidence for potential microRNA and MSI-binding sites in the 3′-UTR of Gnrhr mRNA. Focusing on MSI, we now report that MSI1 binds to the Gnrhr 3′-UTR through one or more consensus MBEs. This present report begins the characterization of MSI1 binding by showing evidence that it is active and modulated by leptin. At this time, we do not exclude additional posttranscriptional regulation of GnRHR levels involving other RNA-binding proteins and/or microRNAs. Future studies will be necessary to address contributions from such additional regulators.
The two MSI RNA-binding protein family members, Msi1 and Msi2, regulate both physiological and pathological stem/progenitor cell self-renewal (45–57). The MSI1 and MSI2 proteins each contain two highly conserved N-terminal RNA recognition motifs, and recent crosslinking and functional studies indicate that they may regulate overlapping mRNA targets (58, 59). Indeed, Msi1 and Msi2 have been shown to effectively substitute for each other in the control of Xenopus oocyte maturation, mammalian neuronal stem cell self-renewal, intestinal stem cell quiescence, and colorectal cancer (58–62). MSI functions to oppose cell maturation in multiple cell types through repression of target mRNA translation (45, 46, 57). In the present studies, Msi2 mRNA levels are unchanged in the female gonadotrope Lepr-null mutants and are not affected by leptin stimulation (data not shown), which is why we have focused on MSI1 here.
In the context of gonadotrope cyclic remodeling, we hypothesized that MSI1 may act to repress translation of the Gnrhr mRNA early in the cycle to prevent inappropriate gonadotrope responses to GnRH pulses. Whereas gonadotropes lacking GnRHR might not be considered “immature” per se, they are clearly functionally repressed during certain stages of the cycle. To begin to test this hypothesis, this phase of the study addressed the key questions: does MSI1 directly bind the Gnrhr mRNA? and are the MBEs active repressor sites? With respect to the first question, we tested binding of MSI1 to wild-type and mutated Gnrhr mRNA in vitro by RNA EMSA and determined that MSI1 does specifically interact with the MBEs in the Gnrhr mRNA. Interaction is competed by the wild-type, but not by the MBE mutant Gnrhr 3′-UTR.
Additional support for endogenous MSI1 binding to endogenous Gnrhr mRNA in the pituitary was then seen in the findings from the RNA IP experiments that pulled down complexes enriched in endogenous MSI1 protein and endogenous Gnrhr mRNA from female mouse pituitary fractions. The Gnrhr mRNA was significantly enriched compared with control (normal rabbit IgG) IP samples, providing strong evidence for MSI1-Gnrhr binding in the mouse pituitary.
Finally, to test activity of the MBEs, we used FLuc RNA translation reporter assays in an independent cellular system that does not normally express MSI1 but can reconstitute MSI-dependent repression upon ectopic MSI expression. We determined that transfection of MSI1 specifically represses the translation of the FLuc reporter driven by Gnrhr 3′-UTR, but a mutant MSI (which is attenuated for RNA binding) does not. Whereas these studies are compelling, they do not provide proof of translational regulation in the pituitary per se. Future studies are planned that address the need to directly assess MSI1 effects on translation of Gnrhr mRNA by polysomal profiling. These studies are beyond the purview of this report, but will be necessary to prove that MSI1 represses translation in the pituitary.
Collectively, the data on MSI1 binding and activity strongly support the hypothesis that this regulatory protein can interact with Gnrhr mRNA as a repressor. Ongoing studies of its expression throughout the estrous cycle are designed to determine if there are cyclic changes in gonadotropes that correlate with this regulatory role.
Our overall hypothesis also states that leptin may permit reproduction and cyclicity by regulating MSI1 function and derepressing the MSI1 target Gnrhr mRNA early in the cycle, thus allowing the translation of GnRHR protein. However, does leptin regulate MSI1 expression and function in gonadotropes? In the present report, we provide data showing that leptin exposure of pituitary pieces and pituitary cells in culture results in changes overall in MSI1 expression as well as changes selective to gonadotropes.
Specifically, leptin caused a substantial reduction in expression of Msi1 mRNA in pituitary pieces from wild-type proestrous female mice. This finding correlates well with the assays showing the higher Msi1 mRNA levels in female mutants lacking leptin signals. Interestingly, we noted a sex-specific difference in the effect of leptin upon expression of Msi1 mRNA in which the male mutants showed no changes in Msi1 mRNA. As discussed previously, our in vitro studies show that higher levels of leptin are needed to affect GnRHR levels in the male, suggesting that the female may be more sensitive to leptin for cyclic regulation of GnRHR. These findings also correlate well with the fact that ablation of LEPR in gonadotropes affects female fertility but not male fertility (14). This further highlights the need for future studies of normal cycling females to determine if there is cyclic regulation and/or expression of MSI1 in gonadotropes in a pattern that correlates with its proposed regulatory function.
The last group of studies for this report focused on detection of leptin-mediated changes in gonadotropes, with the recognition that the aforementioned changes may include results from leptin regulation of multiple target cells in the anterior pituitary. With the use of dual affinity cytochemistry and immunocytochemistry, we detected a selective reduction in MSI1 in gonadotropes that were both defined and identified by their binding to bio-GnRH. The results were striking because the leptin stimulation brought out increased numbers of black GnRHR-labeled cells that were emptied of the orange label for MSI1 proteins (Fig. 5). Future studies are needed to determine if this lack of MSI1 labeling reflects degradation of MSI1 during the 3-hour leptin treatment or if it reflects a change in conformation of MSI1 resulting from regulatory phosphorylation that masks it for antibody recognition. These selective studies of gonadotropes show that leptin regulates MSI1 expression specifically in gonadotropes. Although these results are compelling and support the hypothesis, future studies of purified populations of gonadotropes from cycling female mice will be needed to prove that the leptin regulation of MSI1 actually stimulates an increase in GnRHR in this population.
Our findings may thus have uncovered a unique role for MSI in control of adult gonadotrope physiology. Rather than only functioning in stem/progenitor cells, MSI proteins may also exert homeostatic control of differentiated secretory cell lineages. Alternatively, MSI1 may function to maintain gonadotrope progenitor cells, providing a population that can support the turnover in the gonadotrope cell population. Or, MSI1 may serve to alter gonadotrope metabolism. Ongoing studies of selective gonadotrope Msi1 knockouts may provide information about its role in this specific cell population. In addition, although Msi1 and Msi2 are generally considered to function redundantly, it will also be important to characterize MSI2 function (if any) in the adult pituitary in future studies.
To summarize, this report presents evidence that leptin regulates both GnRH binding of, and the expression and function of, an mRNA regulatory protein, MSI1. Our evidence thus far shows that MSI1 binds Gnrhr 3′-UTR and represses the translation of the FLuc reporter attached to Gnrhr 3′-UTR. Its interaction with endogenous pituitary Gnrhr mRNA is validated by RNA IP studies, which pulled down complexes enriched in endogenous MSI1 protein and Gnrhr mRNA from female pituitaries.
These findings open the door to future studies that will use RNA-sequencing and polysomal profiling in purified gonadotropes to identify and characterize the underlying mechanisms by which leptin exerts mRNA translational regulation of gonadotrope function. Thus, this is the first in a series of planned studies to elucidate the timing of the leptin-mediated changes that permit cyclicity and optimize reproduction. We hypothesize that the transcription and translation of Gnrhr early in the cycle represents an important checkpoint that allows a metabolic signal to intervene in case of nutritional deprivation.
Acknowledgments
Financial Support: This work was funded by National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development Grants R01HD059056 (to G.V.C.) and R01HD087057 (to G.V.C. and A.M.-J.); National Institute of General Medical Sciences Grants P20 GM103425 and P30GM11070 (to Dr. Edgar Garcia-Rill), and a University of Arkansas for Medical Sciences Development Enhancement Award (to G.V.C. and A.M.M.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ANOVA
- analysis of variance
- bio-GnRH
- biotinylated analogs of gonadotropin-releasing hormone
- EMSA
- electrophoretic mobility shift assay
- FLuc
- firefly luciferase
- FSH
- follicle-stimulating hormone
- GnRH
- gonadotropin-releasing hormone
- GnRHR
- gonadotropin-releasing hormone receptor
- GST
- glutathione S-transferase
- IgG
- immunoglobulin G
- IP
- immunoprecipitation
- LEPR
- leptin receptor
- LH
- luteinizing hormone
- MBE
- Musashi binding element
- mRNA
- messenger RNA
- MSI
- Musashi
- PCR
- polymerase chain reaction
- qRT-PCR
- quantitative real-time polymerase chain reaction
- UTR
- untranslated region.
References
- 1.Childs GV. Gonadotropes and lactotropes. In: Neill J, Knobil E, eds. Physiology of Reproduction. New York, NY: Elsevier Press; 2006:1483–1579. [Google Scholar]
- 2.Clayton RN, Solano AR, Garcia-Vela A, Dufau ML, Catt KJ. Regulation of pituitary receptors for gonadotropin-releasing hormone during the rat estrous cycle. Endocrinology. 1980;107(3):699–706. [DOI] [PubMed] [Google Scholar]
- 3.Savoy-Moore RT, Schwartz NB, Duncan JA, Marshall JC. Pituitary gonadotropin-releasing hormone receptors during the rat estrous cycle. Science. 1980;209(4459):942–944. [DOI] [PubMed] [Google Scholar]
- 4.Lloyd JM, Childs GV. Changes in the number of GnRH-receptive cells during the rat estrous cycle: biphasic effects of estradiol. Neuroendocrinology. 1988;48(2):138–146. [DOI] [PubMed] [Google Scholar]
- 5.Childs GV, Unabia G, Miller BT. Cytochemical detection of gonadotropin-releasing hormone-binding sites on rat pituitary cells with luteinizing hormone, follicle-stimulating hormone, and growth hormone antigens during diestrous up-regulation. Endocrinology. 1994;134(4):1943–1951. [DOI] [PubMed] [Google Scholar]
- 6.Vamvini MT, Aronis KN, Chamberland JP, Mantzoros CS. Energy deprivation alters in a leptin- and cortisol-independent manner circulating levels of activin A and follistatin but not myostatin in healthy males. J Clin Endocrinol Metab. 2011;96(11):3416–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mantzoros CS, Magkos F, Brinkoetter M, Sienkiewicz E, Dardeno TA, Kim SY, Hamnvik OP, Koniaris A. Leptin in human physiology and pathophysiology. Am J Physiol Endocrinol Metab. 2011;301(4):E567–E584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akhter N, Crane C, Childs GV. Pituitary leptin-a paracrine regulator of gonadotropes: a review. Open Neuroendocrinol J. 2011;4(1):25–42. [Google Scholar]
- 9.Crane C, Akhter N, Johnson BW, Iruthayanathan M, Syed F, Kudo A, Zhou YH, Childs GV. Fasting and glucose effects on pituitary leptin expression: is leptin a local signal for nutrient status? J Histochem Cytochem. 2007;55(10):1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blüher S, Mantzoros CS. Leptin in reproduction. Curr Opin Endocrinol Diabetes Obes. 2007;14(6):458–464. [DOI] [PubMed] [Google Scholar]
- 11.Schneider JE, Buckley CA, Blum RM, Zhou D, Szymanski L, Day DE, Bartness TJ. Metabolic signals, hormones and neuropeptides involved in control of energy balance and reproductive success in hamsters. Eur J Neurosci. 2002;16(3):377–379. [DOI] [PubMed] [Google Scholar]
- 12.Ahima RS, Dushay J, Flier SN, Prabakaran D, Flier JS. Leptin accelerates the onset of puberty in normal female mice. J Clin Invest. 1997;99(3):391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barash IA, Cheung CC, Weigle DS, Ren H, Kabigting EB, Kuijper JL, Clifton DK, Steiner RA. Leptin is a metabolic signal to the reproductive system. Endocrinology. 1996;137(7):3144–3147. [DOI] [PubMed] [Google Scholar]
- 14.Akhter N, CarlLee T, Syed MM, Odle AK, Cozart MA, Haney AC, Allensworth-James ML, Beneš H, Childs GV. Selective deletion of leptin receptors in gonadotropes reveals activin and GnRH-binding sites as leptin targets in support of fertility. Endocrinology. 2014;155(10):4027–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahima RS, Prabakaran D, Mantzoros C, Qu D, Lowell B, Maratos-Flier E, Flier JS. Role of leptin in the neuroendocrine response to fasting. Nature. 1996;382(6588):250–252. [DOI] [PubMed] [Google Scholar]
- 16.Elias CF, Lee C, Kelly J, Aschkenasi C, Ahima RS, Couceyro PR, Kuhar MJ, Saper CB, Elmquist JK. Leptin activates hypothalamic CART neurons projecting to the spinal cord. Neuron. 1998;21(6):1375–1385. [DOI] [PubMed] [Google Scholar]
- 17.Finn PD, Cunningham MJ, Pau KY, Spies HG, Clifton DK, Steiner RA. The stimulatory effect of leptin on the neuroendocrine reproductive axis of the monkey. Endocrinology. 1998;139(11):4652–4662. [DOI] [PubMed] [Google Scholar]
- 18.Casanueva FF, Dieguez C. Neuroendocrine regulation and actions of leptin. Front Neuroendocrinol. 1999;20(4):317–363. [DOI] [PubMed] [Google Scholar]
- 19.Korner J, Savontaus E, Chua SC Jr, Leibel RL, Wardlaw SL. Leptin regulation of Agrp and Npy mRNA in the rat hypothalamus. J Neuroendocrinol. 2001;13(11):959–966. [DOI] [PubMed] [Google Scholar]
- 20.Balthasar N, Coppari R, McMinn J, Liu SM, Lee CE, Tang V, Kenny CD, McGovern RA, Chua SC Jr, Elmquist JK, Lowell BB. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron. 2004;42(6):983–991. [DOI] [PubMed] [Google Scholar]
- 21.Bouret SG, Draper SJ, Simerly RB. Trophic action of leptin on hypothalamic neurons that regulate feeding. Science. 2004;304(5667):108–110. [DOI] [PubMed] [Google Scholar]
- 22.Elmquist JK, Flier JS. Neuroscience. The fat-brain axis enters a new dimension. Science. 2004;304(5667):63–64. [DOI] [PubMed] [Google Scholar]
- 23.Morrison CD, Morton GJ, Niswender KD, Gelling RW, Schwartz MW. Leptin inhibits hypothalamic Npy and Agrp gene expression via a mechanism that requires phosphatidylinositol 3-OH-kinase signaling. Am J Physiol Endocrinol Metab. 2005;289(6):E1051–E1057. [DOI] [PubMed] [Google Scholar]
- 24.Lehman MN, Coolen LM, Goodman RL. Minireview: kisspeptin/neurokinin B/dynorphin (KNDy) cells of the arcuate nucleus: a central node in the control of gonadotropin-releasing hormone secretion. Endocrinology. 2010;151(8):3479–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehman MN, Merkley CM, Coolen LM, Goodman RL. Anatomy of the kisspeptin neural network in mammals. Brain Res. 2010;1364:90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donato J Jr, Silva RJ, Sita LV, Lee S, Lee C, Lacchini S, Bittencourt JC, Franci CR, Canteras NS, Elias CF. The ventral premammillary nucleus links fasting-induced changes in leptin levels and coordinated luteinizing hormone secretion. J Neurosci. 2009;29(16):5240–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Luca C, Kowalski TJ, Zhang Y, Elmquist JK, Lee C, Kilimann MW, Ludwig T, Liu SM, Chua SC Jr. Complete rescue of obesity, diabetes, and infertility in db/db mice by neuron-specific LEPR-B transgenes. J Clin Invest. 2005;115(12):3484–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elias CF, Purohit D. Leptin signaling and circuits in puberty and fertility. Cell Mol Life Sci. 2013;70(5):841–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allen SJ, Garcia-Galiano D, Borges BC, Burger LL, Boehm U, Elias CF. Leptin receptor null mice with reexpression of LepR in GnRHR expressing cells display elevated FSH levels but remain in a prepubertal state. Am J Physiol Regul Integr Comp Physiol. 2016;310(11):R1258–R1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu WH, Kimura M, Walczewska A, Karanth S, McCann SM. Role of leptin in hypothalamic-pituitary function. Proc Natl Acad Sci USA. 1997;94(3):1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu WH, Walczewska A, Karanth S, McCann SM. Nitric oxide mediates leptin-induced luteinizing hormone-releasing hormone (LHRH) and LHRH and leptin-induced LH release from the pituitary gland. Endocrinology. 1997;138(11):5055–5058. [DOI] [PubMed] [Google Scholar]
- 32.Odle AK, Allensworth-James M, Haney A, Akhter N, Syed M, Childs GV. Adipocyte versus somatotrope leptin: regulation of metabolic functions in the mouse. Endocrinology. 2016;157(4):1443–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Odle AK, Allensworth-James ML, Akhter N, Syed M, Haney AC, MacNicol M, MacNicol AM, Childs GV. A sex-dependent, tropic role for leptin in the somatotrope as a regulator of POU1F1 and POU1F1-dependent hormones. Endocrinology. 2016;157(10):3958–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allensworth-James ML, Odle A, Haney A, Childs G. Sex differences in somatotrope dependency on leptin receptors in young mice: ablation of LEPR causes severe growth hormone deficiency and abdominal obesity in males. Endocrinology. 2015;156(9):3253–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charlesworth A, Wilczynska A, Thampi P, Cox LL, MacNicol AM. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006;25(12):2792–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rassa JC, Wilson GM, Brewer GA, Parks GD. Spacing constraints on reinitiation of paramyxovirus transcription: the gene end U tract acts as a spacer to separate gene end from gene start sites. Virology. 2000;274(2):438–449. [DOI] [PubMed] [Google Scholar]
- 37.Okabe M, Imai T, Kurusu M, Hiromi Y, Okano H. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 2001;411(6833):94–98. [DOI] [PubMed] [Google Scholar]
- 38.MacNicol AM, Hardy LL, Spencer HJ, MacNicol MC. Neural stem and progenitor cell fate transition requires regulation of Musashi1 function. BMC Dev Biol. 2015;15(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacNicol MC, Cragle CE, MacNicol AM. Context-dependent regulation of Musashi-mediated mRNA translation and cell cycle regulation. Cell Cycle. 2011;10(1):39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arumugam K, MacNicol MC, Wang Y, Cragle CE, Tackett AJ, Hardy LL, MacNicol AM. Ringo/cyclin-dependent kinase and mitogen-activated protein kinase signaling pathways regulate the activity of the cell fate determinant Musashi to promote cell cycle re-entry in Xenopus oocytes. J Biol Chem. 2012;287(13):10639–10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, Nakafuku M, Okano H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21(12):3888–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaiser UB, Sabbagh E, Katzenellenbogen RA, Conn PM, Chin WW. A mechanism for the differential regulation of gonadotropin subunit gene expression by gonadotropin-releasing hormone. Proc Natl Acad Sci USA. 1995;92(26):12280–12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaiser UB, Jakubowiak A, Steinberger A, Chin WW. Differential effects of gonadotropin-releasing hormone (GnRH) pulse frequency on gonadotropin subunit and GnRH receptor messenger ribonucleic acid levels in vitro. Endocrinology. 1997;138(3):1224–1231. [DOI] [PubMed] [Google Scholar]
- 44.Odle AK, Akhter N, Syed MM, Allensworth-James ML, Beneš H, Melgar Castillo AI, MacNicol MC, MacNicol AM, Childs GV. Hypothesis and Theory: Leptin regulation of gonadotrope gonadotropin-releasing hormone receptors as a metabolic checkpoint and gateway to reproductive competence. Front Endocrinol. In press. doi: 10.3389/fendo.2017.00367. [DOI] [PMC free article] [PubMed]
- 45.MacNicol AM, Wilczynska A, MacNicol MC. Function and regulation of the mammalian Musashi mRNA translational regulator. Biochem Soc Trans. 2008;36(Pt 3):528–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res. 2005;306(2):349–356. [DOI] [PubMed] [Google Scholar]
- 47.Glazer RI, Vo DT, Penalva LO. Musashi1: an RBP with versatile functions in normal and cancer stem cells. Front Biosci. 2012;17(1):54–64. [DOI] [PubMed] [Google Scholar]
- 48.Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100(25):15178–15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, Lento WE, Zhao C, Lagoo A, Gerrard G, Foroni L, Goldman J, Goh H, Kim SH, Kim DW, Chuah C, Oehler VG, Radich JP, Jordan CT, Reya T. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466(7307):765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanemura Y, Mori K, Sakakibara S, Fujikawa H, Hayashi H, Nakano A, Matsumoto T, Tamura K, Imai T, Ohnishi T, Fushiki S, Nakamura Y, Yamasaki M, Okano H, Arita N. Musashi1, an evolutionarily conserved neural RNA-binding protein, is a versatile marker of human glioma cells in determining their cellular origin, malignancy, and proliferative activity. Differentiation. 2001;68(2-3):141–152. [DOI] [PubMed] [Google Scholar]
- 51.Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S, Morgan K, Tam W, Paktinat M, Okabe R, Gozo M, Einhorn W, Lane SW, Scholl C, Fröhling S, Fleming M, Ebert BL, Gilliland DG, Jaenisch R, Daley GQ. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16(8):903–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, Downey RJ, Manova-Todorova K, Brogi E, Massagué J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17(7):867–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, Ramalingam S, Bishnupuri KS, Natarajan G, Anant S, Houchen CW. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134(5):1448–1458. [DOI] [PubMed] [Google Scholar]
- 54.Toda M, Iizuka Y, Yu W, Imai T, Ikeda E, Yoshida K, Kawase T, Kawakami Y, Okano H, Uyemura K. Expression of the neural RNA-binding protein Musashi1 in human gliomas. Glia. 2001;34(1):1–7. [DOI] [PubMed] [Google Scholar]
- 55.Wang XY, Penalva LO, Yuan H, Linnoila RI, Lu J, Okano H, Glazer RI. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol Cancer. 2010;9(1):221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang XY, Yu H, Linnoila RI, Li L, Li D, Mo B, Okano H, Penalva LO, Glazer RI. Musashi1 as a potential therapeutic target and diagnostic marker for lung cancer. Oncotarget. 2013;4(5):739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fox RG, Park FD, Koechlein CS, Kritzik M, Reya T. Musashi signaling in stem cells and cancer. Annu Rev Cell Dev Biol. 2015;31(1):249–267. [DOI] [PubMed] [Google Scholar]
- 58.Li N, Yousefi M, Nakauka-Ddamba A, Li F, Vandivier L, Parada K, Woo DH, Wang S, Naqvi AS, Rao S, Tobias J, Cedeno RJ, Minuesa G, y K, Barlowe TS, Valvezan A, Shankar S, Deering RP, Klein PS, Jensen ST, Kharas MG, Gregory BD, Yu Z, Lengner CJ. The Msi family of RNA-binding proteins function redundantly as intestinal oncoproteins. Cell Reports. 2015;13(11):2440–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.MacNicol MC, Cragle CE, McDaniel FK, Hardy LL, Wang Y, Arumugam K, Rahmatallah Y, Glazko GV, Wilczynska A, Childs GV, Zhou D, MacNicol AM. Evasion of regulatory phosphorylation by an alternatively spliced isoform of Musashi2. Sci Rep. 2017;7(1):11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arumugam K, Wang Y, Hardy LL, MacNicol MC, MacNicol AM. Enforcing temporal control of maternal mRNA translation during oocyte cell-cycle progression. EMBO J. 2010;29(2):387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakakibara S, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, Ueda S, Uchiyama Y, Noda T, Okano H. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci USA. 2002;99(23):15194–15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yousefi M, Li N, Nakauka-Ddamba A, Wang S, Davidow K, Schoenberger J, Yu Z, Jensen ST, Kharas MG, Lengner CJ. Msi RNA-binding proteins control reserve intestinal stem cell quiescence. J Cell Biol. 2016;215(3):401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]