Abstract
Intravital microscopy (IVM) is an imaging tool that is capable of detecting subcellular signaling or metabolic events as they occur in tissues in the living animal. Imaging in highly scattering biological tissues, however, is challenging because of the attenuation of signal in images acquired at increasing depths. Depth-dependent signal attenuation is the major impediment to IVM, limiting the depth from which significant data can be obtained. Therefore, making quantitative measurements by IVM requires methods that use internal calibration, or alternatively, a completely different way of evaluating the signals. Here, we describe how ratiometric imaging of genetically encoded biosensor probes can be used to make quantitative measurements of changes in the activity of cell signaling pathways. Then, we describe how fluorescence lifetime imaging can be used for label-free measurements of the metabolic states of cells within the living animal.
Keywords: Intravital microscopy (IVM), Ratiometric imaging, Fluorescence lifetime imaging microscopy (FLIM), Intrinsic fluorescence, Biosensor probes
1. Introduction
Intravital microscopy (IVM) has become an essential tool for evaluating cellular physiology in the context of the living animal [1]. This method uses two-photon excitation (2PE) to gain access to the cellular environments within living tissues that are not typically accessible by conventional microscopy. The physics that underlies 2PE is described in detail elsewhere [2,3], but it is important to emphasize two key points here. First, the near infrared illumination used in IVM allows deeper penetration of the excitation light into highly scattering tissues. Second, because of the quadratic dependence of the 2PE process, the fluorescence emissions emanating from cellular sources are limited to the focal plane. This makes IVM uniquely capable of detecting subcellular signaling or metabolic events as they occur within a complex tissue in the living animal. However, depth-dependent signal attenuation remains a major limitation to intravital imaging, and is very dependent on the scattering characteristics of different biological tissues. Therefore, intravital imaging in a particular tissue is limited to depths where it is possible to obtain reasonable signal-to-noise.
The use of IVM to detect specific cellular events in intact organs presents some unique challenges. A primary challenge is maintaining the physiological welfare of the animal while on the microscope stage. Generally, the tissue of interest must be surgically exposed, requiring high standards of practice for anesthesia and delicate surgery. The anesthetized animal is placed on the microscope stage where its temperature must be maintained with a heating pad and continuously monitored. The surgically exposed organ is secured to a glass-bottomed dish on the environmentally controlled microscope stage to reduce motion artifacts caused by respiration and the beating of the heart. If necessary, the image collection is gated to the respiration to minimize the motion artifacts, and distortions in the collected images can be corrected in the digital image analysis [4,5].
Another substantial challenge for fluorescence imaging generally, and IVM in particular, is the quantitative analysis of the optical signals. When imaging in highly scattering biological tissues, both the excitation light and the emission signal are attenuated in a wavelength- and depth-dependent fashion. This reduces the effective excitation power at the focal plane, while creating a higher detection threshold [6,7]. The loss of signal prevents the direct comparison of intensities acquired at different depths in a tissue. The degradation of signal-to-noise limits how deep IVM can image into a tissue, and quantitative measurements require the use of probes that are internally calibrated, such as the biosensor probes. We will describe how ratiometric imaging of genetically encoded biosensor probes can be used in IVM to obtain quantitative measurements of changes in cell signaling pathways. Alternatively, IVM can exploit a completely different way of evaluating the signals to achieve quantitative measurements of cellular events. We will describe how fluorescence lifetime imaging can be used for label-free measurements of metabolic events within cells in the living animal.
2. Theory
2.1. Ratiometric imaging of biosensor probe activities
Ratiometric imaging of the genetically encoded fluorescent biosensor proteins provide internally calibrated measurements of cell signaling events [8,9]. Typically, these probes rely on Förster resonance energy transfer (FRET) between donor and acceptor fluorescent proteins (FPs), which are arranged in tandem within the reporter module of the biosensor. The reporter module is linked by a sensing unit that is designed to detect a specific cellular event, such as the activity of kinases, or the binding of small molecules or protein ligands [10,11]. Changes in the biosensor conformation that accompanies the targeted cellular event alter energy transfer within the reporter module. Because energy transfer quenches the donor signal while causing increased emission from the acceptor, the ratio of the donor and acceptor emissions provides a sensitive measure of the conformational change. Importantly, while the fluorescence intensity will vary with depth, provided the emission signals from the donors and the acceptors are similarly affected there will be no depth-dependent change in the ratio.
A limitation to ratiometric FRET measurements occurs when it becomes necessary to use two separate excitation wavelengths to acquire images of the donor and the acceptor fluorophores. Since IVM systems are usually equipped with a single infrared laser, switching to another wavelength requires re-tuning of the laser between acquisition of the two separate images. This may also involve realignment and a change in laser power, and this affects the speed at which images can be acquired, limiting the ability to measure dynamic changes in biosensor activity. Therefore, the ideal biosensors for IVM should need only a single 2PE wavelength to allow rapid acquisition of ratiometric measurements without retuning of the laser [12,13]. Here, we demonstrate an IVM method for making ratiometric measurements of FRET-based biosensor activity that requires only a single excitation wavelength. Additionally, we describe how some of the limitations to ratiometric measurements of fluorescence intensity can be overcome by measuring fluorescence lifetimes.
2.2. Fluorescence lifetime imaging microscopy (FLIM)
The fluorescence lifetime is the average time that a population of fluorophores spends in the excited state before returning to the ground state, and FLIM measures the duration of the excited state. Because the measurements are made in the time domain, they are unaffected by variations in probe concentration, changes in the excitation intensity, and, provided there is sufficient signal-to-noise, will be minimally affected by light scattering. The fluorescence lifetime is an intrinsic property of every fluorophore, and it carries information about events that affect the excited-state. For example, energy transfer is an excited-state process that quenches the donor fluorescence, and this shortens the donor lifetime. FLIM can quantify this change in lifetime, making it a powerful approach to measure FRET [14]. FLIM has been used for intravital measurements of calcium biosensor activity in the central nervous system of living mice [15]. This, however, requires the collection of sufficient numbers of photons to accurately assign lifetimes to the individual fluorescent species, which can be limiting for deep-tissue imaging.
2.3. Phasor plot analysis of lifetime distributions
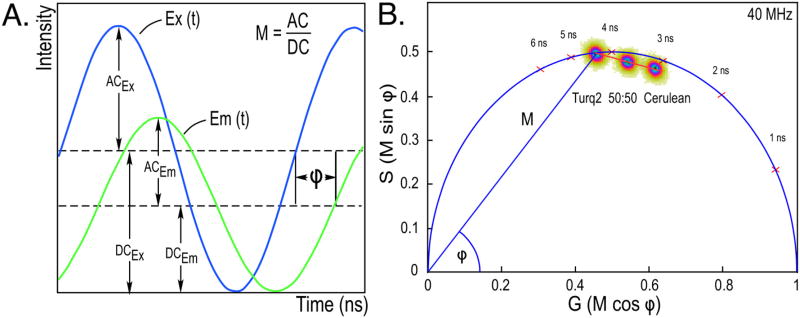
There is an alternative approach that does not require the assignment of lifetimes to individual fluorescent species. The method, known as the phasor plot, is used in frequency domain (FD) FLIM to provide a global representation of the distribution of lifetimes within a specimen. The mathematical theory that underpins FD lifetime measurements and the analysis by phasor plot are described elsewhere [16–18]. FD lifetime measurements are acquired using an excitation source that is modulated at high radio frequencies (usually between 10 to 400 MHz), which results in a modulated emission signal from the fluorophores. However, because of the persistence of the excited state, there is a phase delay (Φ) and a change in the modulation (M) of the emission signal relative to the corresponding excitation waveform (Fig. 1A). The fluorescence lifetime (τ) is determined directly from the Φ and the M of the emission signal for each excitation frequency.
Fig. 1.
FD FLIM measurements and analysis by phasor plot. (A) FD FLIM measures the phase delay (Φ) and the change in the modulation (M) of the emission signal (Em) relative to the corresponding excitation waveform (Ex). The fluorescence lifetime is determined from both the Φ and M of the emission signal for each excitation frequency. (B) The phasor plot analysis of the lifetime distributions for the cyan FPs mTurquoise2 and mCerulean. Images (256 × 256 pixels) were acquired of each FP, and the lifetime distribution for all image pixels is plotted relative to the universal semicircle, using color to indicate the incidence of lifetime values from blue (highest) to red to yellow (lowest). For fluorophores with a single-component decay, the lifetime distribution will fall directly on the semicircle, whereas the lifetime distributions for fluorophores with multi-exponential decays fall inside the semicircle (Section 2.3). The phasor analysis shows mTurquoise2 has a single component lifetime of 4.5 ns, whereas mCerulean has a multi-component decay with an average lifetime of 3.1 ns. The lifetime distribution for a 50:50 mixture of mTurquoise2 and mCerulean falls on a line directly between the distributions for the pure species. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The phasor plot represents the Φ and the M of the emission signal relative to the excitation waveform as the Fourier transforms G and S:
The frequency characteristics from each image pixel are mapped to the G and S coordinates using vectors with angles specified by the Φ and lengths determined by M (Fig. 1B). In this regard the phasor plot can be thought of as a histogram of the number of image pixels with the same phasor coordinates, thus providing a global view of the fluorescence decay at each pixel of an image. The distribution of lifetimes for a species with a short lifetime will fall to the right on the phasor plot, while those with longer lifetimes will fall to the left. For fluorophores with single-component decays, the relationship M = cos Φ describes a vector with an endpoint falling somewhere on a universal semicircle, irrespective of the modulation frequency or lifetime. Therefore, the lifetime distribution for fluorophores with single-component decays fall directly on the semicircle. In contrast, fluorophores with multi-component decays have phase and the modulation lifetimes that differ from one-another, and this causes the lifetime distribution to fall inside the semicircle [18].
This is illustrated by the phasor plot in Fig. 1B, which shows the lifetime distributions for two purified cyan FPs that have identical spectra but different lifetimes. The lifetime for all image pixels (65,536 pixels for a 256 × 256 pixel image) is plotted relative to the universal semicircle, using color to indicate the incidence of pixel lifetime values from blue (highest) to red to yellow (lowest). The lifetime distribution for purified mTurquoise2 falls on the semicircle, indicating a single component lifetime decay of 4.5 ns. In contrast, the lifetime distribution for purified mCerulean falls inside the universal semicircle, which indicates the presence of more than one lifetime component (Fig. 1B) [16]. Earlier chisquared analysis showed that mCerulean best fit a two-component decay, with fractional lifetimes of 4.8 ns and 2.1 ns, resulting in an intensity-weighted average lifetime of 3.1 ns [14]. Despite the identical emission profiles for these two FPs, their distinct lifetimes allow phasor analysis to be used to determine the relative contribution of each to a mixture. The location of the lifetime distribution for a 50:50 mixture of the two FPs falls on a straight line directly between the distributions for the different pure species (Fig. 1B) [18]. Together, this illustrates how the phasor plot analysis enables the direct visualization of complex lifetime decays, and allows the separation of distinct lifetime species in mixtures.
The phasor plot approach has been used to improve the sensitivity of biosensor measurements in cells in culture [19]. However, the most innovative application of phasor analysis is in the measurement of intrinsic cellular auto-fluorescence signals, which can be used as an index of cellular metabolism in intact tissues [20,21]. In the live animal, tissue auto-fluorescence arises principally from several biologically important metabolites, including the flavoproteins (flavin adenine dinucleotide, FAD) and the reduced form of nicotinamide adenine dinucleotide (NADH) [22,23]. The phasor analysis of these auto-fluorescence signals can be used to generate lifetime distribution “fingerprints” that identify the unique metabolic states of different cell-types within a tissue, providing a label-free imaging tool to study metabolism in living tissues [24]. Here, we describe how IVM of fluorescence lifetimes can map cell-specific metabolic signatures in the tissues of live animals.
3. Materials and methods
3.1. Plasmids encoding standards and a biosensor for IVM
The use of FRET standards allows the verification and optimization of the IVM system for biosensor measurements [12]. We developed a series of plasmids encoding validated standards ranging from high to low FRET efficiency (EFRET) that use the monomeric (m)Turquoise and mVenus FPs separated by linker peptides of increasing length. Turquoise is a brighter and more photo-stable variant of mCerulean that has the necessary spectral overlap with Venus for efficient energy transfer. Importantly, mTurquoise and mVenus have minimal 2PE crosstalk at 810 nm, permitting reasonably selective excitation of the donor [13]. Here, the FRET standards are used to verify biosensor measurements obtained with a modified version of the A kinase activity reporter (AKAR4.1) of protein kinase A (PKA), first in cells in culture, and then in cells in the liver of living mice. The plasmids encoding the mTurquoise-mVenus FRET standards and the AKAR4.1 biosensor are available through the National Institutes of Health O’Brien Center for Advanced Renal Microscopic Analysis at Indiana University School of Medicine. All plasmid inserts were confirmed by direct sequencing.
3.2. Cell culture and transfections
Human embryonic kidney (HEK)293 (American Type Cell Culture Collection (ATCC) CRL-1573) cells were maintained in monolayer culture and harvested at 80% confluence as recommended by ATCC. Fetal cardiomyocytes were isolated by enzymatic digestion from day 15 mouse embryo hearts as described earlier [12]. The cells were transfected using X-tremeGene HP DNA transfection reagent (Roche Diagnostics, Indianapolis, IN) following the manufacturer’s instructions. The cells were transferred to 4-chamber Lab-Tek II coverglass (Thermo Fisher Scientific, Waltham, MA), and maintained in 5% CO2-95% O2 at 37 °C.
3.3. Intravital ratiometric imaging of biosensor probes
All animal studies were approved by the Institutional Animal Care and Use Committee of Indiana University School of Medicine and conformed to the “Guide for the Care and Use of Laboratory Animals” published by the National Institutes of Health (NIH Publication No. 85-23, Revised 1996). IVM was conducted using an Olympus FV1000 system modified for multi-photon imaging and equipped with a heated 25× NA 1.05 water immersion objective as described earlier [25]. The laser was tuned to 810-nm and the emission signals were routed by spectral detection to the donor channel (454–494 nm) and acceptor channel (520–580 nm), and the signals were detected by photomultiplier tubes on the descanned detection pathway.
IVM of AKAR4.1 biosensor activity was conducted using 11 to 12-week-old C57BL/6 mice obtained from Jackson Laboratories (Bar Harbor, Maine). In vivo expression of AKAR4.1 was accomplished by tail vein injection of the Adenoviral (Ad)-CMV- AKAR4.1 vector (4.8 × 1010 viral particles in 200 microliters saline per mouse) as described earlier [12,13]. Seven days after Ad injection, mice were fasted for three hours, anesthetized with isoflurane and surgically prepared for intravital microscopy. A 2-cm ventral incision was placed just 1 cm below the rib cage to expose the liver, and a small section of PE50 tubing was secured in the abdominal cavity for intraperitoneal (IP) injection of glucagon on the stage of the microscope. The left lateral lobe of the liver was carefully secured to a glass bottom plate. The mouse was then placed ventral side down on a heated microscope stage and covered with a warming blanket [25]. After selecting a region of interest, image volumes from ten focal planes separated by one micron were collected before and for 15 min following the IP injection of 200 µg/kg glucagon.
3.4. FD FLIM measurements
The digital FD FLIM measurements were acquired using an Olympus FV1000 multiphoton microscope equipped with the FastFLIM data acquisition card and a two-detector unit (ISS Inc., Champaign, IL) connected to the fiber port of the microscope. The system is equipped with a Spectra Physics MaiTai laser and gallium arsenide detectors, and the collected signals were digitized to 12-bit resolution. Lifetime calibration is performed before each experiment using fluorescein (Sigma Aldrich #30181) dissolved in 1 mM NaOH (4 ns lifetime) [26]. Additional measurements are made of HPTS (8-hydroxypyrene-1,3,6-trisulfonic acid, Santa Cruz Biotechnology #291738) dissolved in pH 7.8 phosphate buffer (5.3 ns lifetime) [27] or Coumarin 6 (Sigma Aldrich #546283) dissolved in EtOH (2.5 ns lifetime) [28] to verify the calibration. The FD FLIM system uses the existing 80 MHz frequency of the titanium-sapphire laser, with additional measurements at 3 harmonics (160, 240, and 400 MHz). The VistaVision software (ISS Inc., Champaign, IL) records the time resolved photons in different cross frequency phase bins to generate the phase histogram. The information at higher harmonics is obtained by the digital frequency transform of the phase histogram. Each FLIM image was acquired by frame averaging 15 to 20 one-second scans to obtain an average of at least 100 photons per pixel. The VistaVision software was used to render the multi-frequency response curves that measure the phase delay and demodulation for each frequency, as well as the phasor plot to determine the distribution of lifetimes.
3.5. Measuring intrinsic fluorophore lifetime standards
Solutions of individual cellular fluorophores were used to determine the phasor lifetime positions for each intrinsic fluorescent species [20,21,29,30]. The intrinsic fluorophore standards were purchased from Sigma Aldrich (St. Louis, MO), and their concentrations in solutions were determined by UV–VIS spectrophotometry using the extinction coefficients supplied by the manufacturer or from the literature [31,32]. Retinol and all-trans retinoic acid (catalog #R7632 and #R2625) were prepared in DMSO at concentrations of 0.13 mM and 46 mM, respectively. NADH (catalog #N8129) was prepared at 45 mM in MOPS (3-(N-morpholino) propanesulfonic acid) buffer (pH 7.2). The protein-bound NADH was prepared by dilution of NADH in a solution of lactate dehydrogenase (LDH) to a final concentration of 4 mM. The LDH solution (catalog #L3916, 1000 units/mL) was prepared by 1:1 dilution by volume in MOPS buffer (pH 7.2), followed by brief centrifugation to remove precipitated protein. FAD (catalog #F6625) was prepared at 3.9 mM in water. The mixtures of NADH and FAD were prepared at 3 different molar ratios in MOPS buffer for a final total concentration of 4 mM. For lifetime measurements of the intrinsic fluorophore standards the laser was tuned to 740 nm excitation and 256 × 256 pixel images were acquired at a scan speed 12.5 µs/pixel. The emission signals were routed by a 515 nm long-pass beam splitter through the 470/100 nm or 525/39 nm band-pass filters to the detectors.
3.6. Intravital FLIM
Intravital FLIM was performed using the Olympus FV1000 multiphoton microscope described above equipped with a 60× NA 1.2 water-immersion objective. Animals were placed on the stage with the exposed intact kidney placed in a coverglass-bottomed dish (catalog #D5040P, Warner Instruments, Hamden, CT) bathed in isotonic saline as described earlier [33]. For FLIM measurements of cellular auto-fluorescence the laser was tuned to 800 nm excitation and 256 × 256 pixel images were acquired at a scan speed 12.5 µs/pixel. Typical laser power at the specimen plane was about 1 mW, and FLIM images were acquired by frame averaging 15–20 one-second scans to obtain an average of at least 100 photons per pixel. The emission signals were routed by a 560 nm long-pass beam splitter through the 470/100 nm or 630/92 nm band-pass filters to the detectors.
4. Results and discussion
4.1. Validating 2PE imaging of biosensor probe activities
The inherent challenges of measuring biosensor activities by IVM demand the use of standards to validate the method. This is accomplished by first measuring fluorescence from cells producing either the donor (mTurquoise) or the acceptor (mVenus) alone to determine the spectral bleedthrough (SBT) components resulting from excitation at 810 nm. The method described here uses a single excitation wavelength, so it is also necessary to estimate the fractional excitation of the acceptor (Venus) at 810 nm using cells that produce a mixture of mTurquoise and mVenus, and cells producing the low-FRET standard (Turquoise-TRAF-Venus). Together, these measurements determine the SBT corrections that are required to quantify the FRET efficiency (EFRET) of the standards (Fig. 2A) as described earlier [12,13].
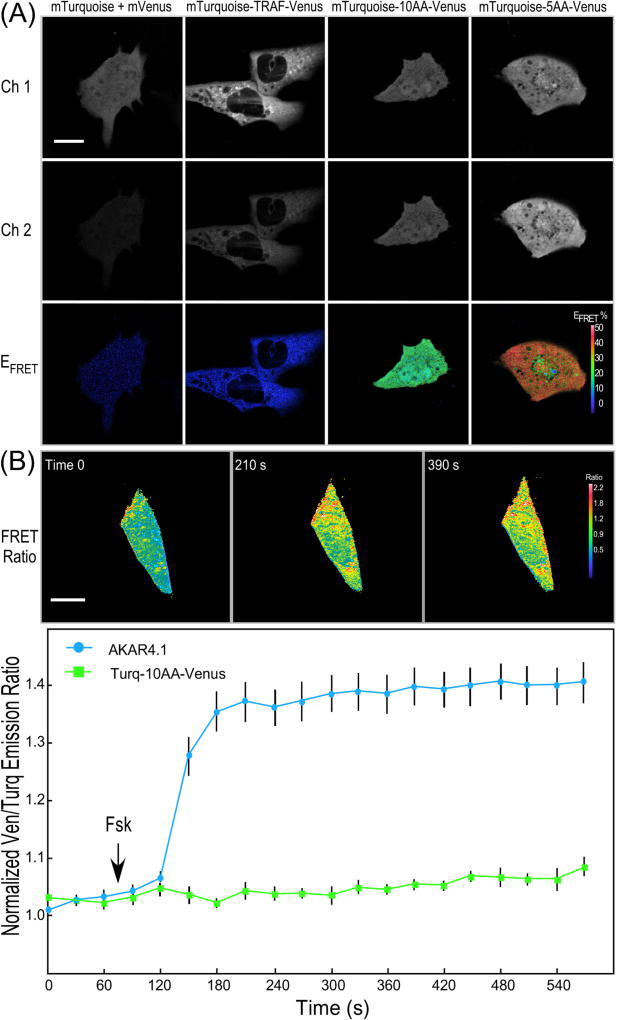
Fig. 2.
IVM ratiometric imaging of FRET standards and the AKAR4.1 biosensor. (A) Representative donor intensity (Ch. 1, 454–494 nm; scale bar is 10 µm), acceptor intensity (Ch. 2, 520–580 nm), and FRET efficiency (EFRET) was determined for HEK293 cells expressing the indicated FRET standard probes (scale bar is 10 µm and the look-up table indicates higher EFRET with warmer color). (B) Ratiometric measurements of the AKAR4.1 probe or the Turquoise-10AA-Venus FRET standard in living mouse cardiomyocytes. The cells were illuminated at 810 nm and the emission signals were simultaneously monitored in the donor (454–494 nm) and acceptor (520–580 nm) channels, and the FRET ratio was determined for a single cardiomyocyte at the indicated time points (scale bar is 10 µm and the look-up table indicates higher FRET ratio with warmer color). Ratiometric measurements show the rapid increase in the Ven/Turq ratio for the cells expressing AKAR4.1, but not the FRET standard over a 3-min time frame. The results for AKAR4.1 are from the 9 individual cells (±SE). [from Ref. [12]]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The FRET standards are then used to evaluate and optimize the IVM system for ratiometric detection of biosensor activity in transfected HEK293 cells. The cells expressing a 1:1 mixture of mTurquoise and mVenus were used as a negative control, while the cells producing the low EFRET (~5%) standard, mTurquoise-TRAF-mVenus, were used to assess the sensitivity of the imaging system for FRET detection (Fig. 2A). By contrast, the EFRET for cells producing the two high FRET standards, mTurquoise-5AA (amino acid)-Venus and mTurquoise-10AA-Venus were easily measured, and the two probes could be clearly distinguished from one-another. The EFRET determined from the cells expressing the highest EFRET standard, mTurquoise-5AA-Venus was about 45%, whereas measurements from cells expressing the mTurquoise-10AA-Venus standard produce EFRET of about 36% (Fig. 2A).
Once the system was validated with the FRET standards, the AKAR4.1 biosensor was used to monitor changes in intracellular PKA activities in response to Forskolin (Fsk). Freshly isolated primary cardiomyocytes were transfected with plasmids encoding either the AKAR4.1 or the mTurquoise-10AA-Venus FRET standard (Fig. 2B). The 2PE ratiometric imaging demonstrated that the addition of Fsk to cells expressing AKAR4.1 resulted in a rapid increase in emission ratio of Ven/Turq over a 3-min time frame. In contrast, the Ven/Turq emission ratio in cells expressing the mTurquoise- 10AA-Venus standard did not change with Fsk treatment (Fig. 2B) demonstrating the specificity of the biosensor for the detection of changes in intracellular PKA activity.
4.2. IVM measurements of biosensor activity from living animals
The in vitro measurements of the standards and the biosensor provides the basis for moving forward with measurements of cellular function in the living animal. A primary challenge is the delivery of the biosensor probe to the desired tissue and cell-type in living animal. This has been achieved using transgenic mouse animal models, but there can be problems with this approach relating to transgene silencing or recombination events that occur between the highly homologous sequences in the reporter module [34,35]. Viral transduction offers an alternative to the generation of transgenic animals, and enables the rapid expression of biosensor probes in living animals. Adenoviral transduction leads to robust, but transient, expression in certain tissues, including the liver [36].
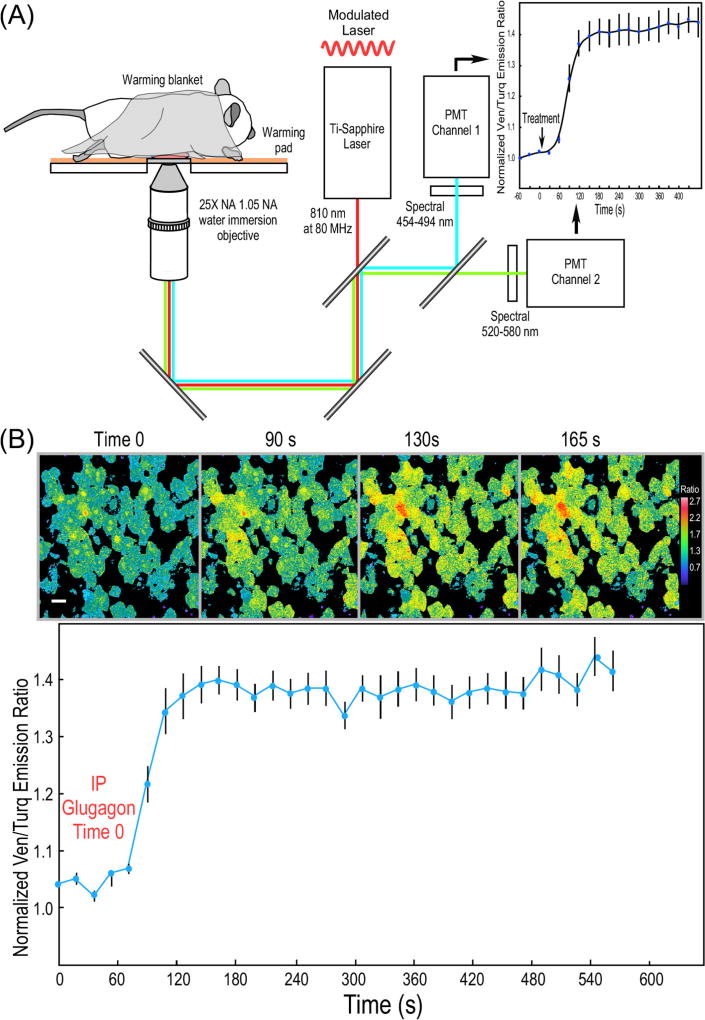
We used adenoviral transduction to achieve high level expression of the AKAR4.1 biosensor in the liver of mice. Seven days after injection with Ad AKAR4.1, the mice were fasted for 3 h, prepared for IVM, and ratiometric images were acquired with the IVM system illustrated in Fig. 3A. Baseline images were collected, and imaging was continued after IP injection of glucagon (200 µg/kg), a treatment that will rapidly activate the PKA pathway in the hepatocytes of fasted mice [37]. The IVM of the AKAR4.1 biosensor revealed a 1.4-fold change in the Ven/Turq emission ratio following the glucagon treatment, demonstrating a rapid and sustained activation of PKA, which is detected at the cellular level in the liver of the living mouse (Fig. 3B).
Fig. 3.
IVM ratiometric measurements of PKA activity in mouse liver. (A) Schematic diagram of the IVM system used for ratiometric measurements of biosensor probes. (B) IVM measurements of the response of the AKAR4.1 probe to glucagon in hepatocytes in the intact mouse liver. The AKAR4.1 biosensor was introduced into mice by tail vein injection of an adenoviral vector encoding AKAR4.1, resulting in extensive expression in the liver 7 days later. Mice were fasted for 3 h prior to imaging by IVM and 3D image volumes (10 planes spanning 10 mm) were collected over time prior to and following IP injection of glucagon (200 µg/kg). Ratio images from a single image plane in mouse liver were acquired at the indicated time points (scale bar is 10 µm and the look-up table indicates higher FRET ratio with warmer color). Each volume was then summed and background-corrected Ven/Turq ratios were determined for regions of interest from cells in the field. The results for AKAR4.1 are from the 10 individual cells (±SE). [from Ref. [12]]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The advantage of this ratiometric IVM method is that it requires only a single 2PE wavelength to monitor the activity of genetically encoded biosensor probes produced in the cells of living animals. The approach is limited, however, by the lack of cellular specificity in biosensor delivery, and by the possibility of inflammatory responses to Ad, which may limit its use in prolonged studies [36]. What is more, there have been reports that some biosensor probes have the potential to perturb the underlying cellular mechanism that they are designed to detect [38]. Therefore, there is a need for a label-free IVM method that can provide a fast, quantitative readout of the metabolic state of cells in the natural environment of the living tissue. Next, we describe how intravital FLIM can be used for label-free measurements of metabolic events within cells in the living animal.
4.3. FLIM measurements of intrinsic cellular fluorescence
Intravital FLIM can quantify the lifetime properties of the intrinsic fluorophores that contribute to cellular auto-fluorescence in living tissues. Several important cellular metabolites contribute to auto-fluorescence emissions, including NAD(P)H and FAD. NADPH cannot be easily distinguished from NADH, and the fluorescence lifetimes of NADPH and NADH in solution are identical. Therefore, the auto-fluorescence intensity will reflect both NADH and NADPH, and is typically denoted as NAD(P)H [22]. The emission wavelengths of these intrinsic fluorophores all strongly overlap, and their quantum yields depend on whether they are in a bound or free state [21–23]. This substantially limits the ability of intensity measurements to distinguish the individual intrinsic fluorescent species with certainty. In striking contrast the fluorescence lifetimes of the intrinsic auto-fluorescence can be distinguished by phasor analysis [20,21,29,30]. The phasor approach enables intravital FLIM to be used to map cell-specific metabolic signatures in the tissues of live animals without the need to determine the lifetimes for individual species.
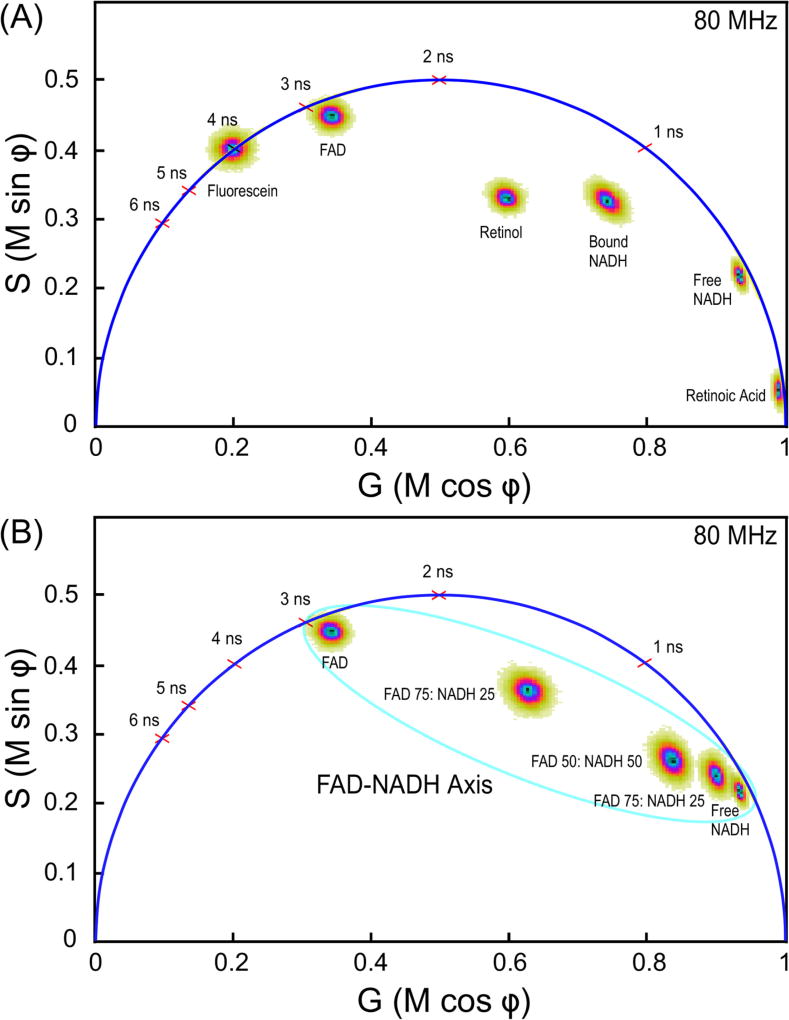
To demonstrate the separation of the distinct lifetimes for intrinsic fluorescent species by phasor plot, standard solutions of cellular fluorophores were prepared and lifetimes determined using 2PE FLIM (Fig. 4). The FLIM system was calibrated before each experiment using a solution of fluorescein [26], and verified with HPTS [27] or Coumarin 6 [28]. Measurements were then made from solutions of FAD, bound and free NADH, retinol, and retinoic acid. The phasor plot was used to represent the lifetime distributions for each individual fluorophore. The lifetime distribution for fluorophores with single-component decays, such as fluorescein, fall directly on a universal semicircle, whereas the lifetime distributions for fluorophores with multi-component decays fall inside the semicircle (Section 2.3, see Fig. 4A). Next, the indicated mixtures of NADH and FAD were measured, and the phasor plot confirmed that the lifetime distributions of the mixtures fall on a straight line between the pure species [18], which defines the FAD-NADH lifetime distribution axis (Fig. 4B). Together, this demonstrates how the phasor analysis of intrinsic lifetimes can separate the signals from cellular auto-fluorescent sources despite the strongly overlapping emissions.
Fig. 4.
FLIM measurements of different cellular fluorophore lifetimes. (A) The phasor plot analysis of the five different solutions of standard intrinsic auto-fluorescent species (Section 3.5). Fluorescein dissolved in 1 mM NaOH (4 ns lifetime) is used to calibrate the FLIM system before each experiment. The lifetime distribution for fluorophores with single-component decays, such as fluorescein, fall directly on a universal semicircle, whereas the lifetime distributions for fluorophores with multi-component decays fall inside the semicircle. (B) The phasor plot of the lifetime distributions for FAD, NADH, or the indicated different molar ratio mixtures of the two. The phasor plot illustrates the FAD-NADH axis where the lifetime distributions for the principle cellular auto-fluorescence species occurs.
4.4. FLIM in living animals
The combination of intravital FLIM and the phasor approach allows the direct visualization of complex lifetime decays emanating from individual cell-types in unlabeled tissues in the living animal. The distribution of intrinsic fluorescence lifetimes within the phasor plot is called the phasor fingerprint, and this represents all the contributions from the endogenous fluorophores. Importantly, the phasor fingerprint of intrinsic fluorescence can be acquired rapidly at relatively low laser power (about 1 mW for our system), and it provides a global overview of cellular metabolism without the need to identify the individual metabolites in the tissue [20,21].
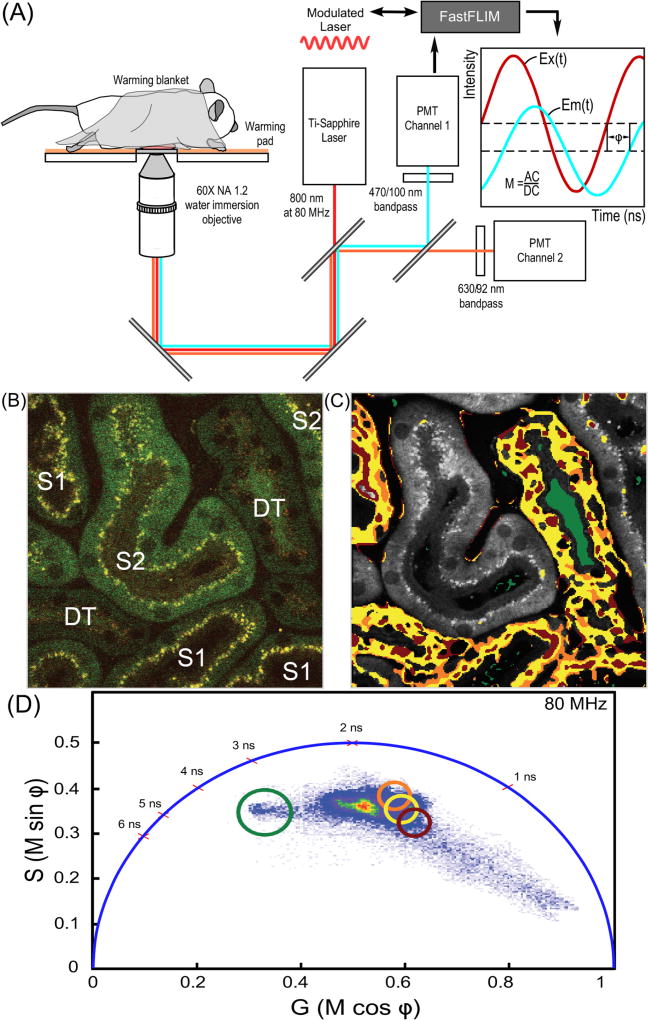
Here, intravital FLIM of the mouse kidney was accomplished using the imaging system illustrated in Fig. 5A. A 2PE fluorescence intensity image was first acquired from a region of the mouse kidney (Fig. 5B). The first (S1) and second (S2) segments of proximal tubules, as well as distal tubules (DT) can be identified in the intensity image based on morphology [33]. A FLIM image was then acquired from the same region, and the phasor plot is generated using the VistaVision software (ISS Inc., Champaign, IL). The software is used to create a FLIM image that is gated for specific lifetime distributions identified in the phasor plot, generating a color-coded phasor image. The color-coded phasor image in Fig. 5C was derived by highlighting the image pixels that correspond to the lifetimes within selected regions of interest (ROI) in the phasor fingerprint (Fig. 5D). The gated lifetime ROI in the phasor fingerprint are indicated by the red, yellow, orange, and green circles (Fig. 5D), and pixels that correspond to those gated lifetimes are highlighted in Fig. 5C with the color corresponding to each circle. The ROI in the phasor fingerprint were selected to illustrate the lifetime distributions most characteristic of the S1 segments of proximal tubules, and the DTs. These results revealed that S1 proximal tubules and DTs have very similar metabolic signatures despite the marked differences in their biological function and morphological appearance [39]. These intravital FLIM results also demonstrate that the lifetime distributions characteristic of the S1 and DTs differed substantially from that of the S2 segments, indicating different metabolic microenvironments in those two regions. Additionally, the phasor fingerprint identified the distinct lifetime signature for the non-cellular contents of the urinary lumen of the DT (Fig. 5C, green) that falls outside the FAD-NADH lifetime distribution axis.
Fig. 5.
IVM FLIM measurements in living animals. (A) Schematic diagram of the IVM system for FLIM measurements of intrinsic auto-fluorescence in the tissues of living animals. (B) The 2PE IVM intensity image collected from the mouse kidney showing the first (S1) and second (S2) segments of proximal tubules, and distal tubules (DT). (C) The color-coded phasor image projected on the grayscale 2PE intensity image. The pixels in the color-coded phasor image are highlighted with the same color as the circles used to define the gated lifetime ROI shown in panel (D). (D) The phasor fingerprint for the region of kidney shown in panel (C) with the gated lifetime ROI (red, yellow, orange, green circles) indicated. These lifetime ROI are characteristic of the S1 tubules and DTs, and indicate that the cellular metabolic signature in these regions differs from that of the S2 segments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
5. Conclusions
IVM provides some of the most relevant information available about cellular physiology and pathophysiology in the context of intact tissue in the living animal. This is critical, since cellular pathologies develop while integrated within the complex tissue environment in vivo. To examine the origins of pathologies it is necessary to study them in the setting of the full cellular organization, ideally in 3D and in real-time, inside the living animal [40]. However, imaging in highly scattering biological tissue presents substantial challenges to the quantitative analysis of the optical signals. Some of these challenges are overcome using ratiometric imaging of biosensor probes by IVM. Because these ratiometric probes are internally calibrated, they are less affected by depth-dependent signal attenuation than intensity-based imaging methods.
Measurements collected at greater depths, however, will suffer from signal degradation, and wavelength-dependent differences in scattering can introduce depth-dependent effects on ratiometric measurements collected in biological tissues [6,7]. We find that useful signal in both the blue (480 nm) and yellow (530 nm) channel diminish before there is an obvious wavelength-dependent effect on the emission ratio, but others demonstrated minor effects of depth on ratiometric FRET measurements [7]. Therefore, studies collected over a range of depths should be evaluated for systematic effects of depth on ratiometric measures.
The use of a single 2PE wavelength described here allows rapid measurements of the spatial and temporal characteristics of cell signaling, but it is particularly important to use standards to validate the IVM measurements of biosensor probes. A substantial limitation to the ratiometric approach, however, is the problem of delivering the biosensor probes to specific cell types in the living animal. Moreover, there is the potential that producing the exogenous biosensor probes in the target cells may perturb the underlying cellular mechanisms they are designed to detect [38].
Compared to ratiometric imaging methods, FLIM can provide a robust method for imaging in the complex tissue environments [41], particularly when it is applied to label-free measurements of cellular metabolism in the living animal. The analysis of intravital FLIM measurements of intrinsic auto-fluorescence by phasor plot allows the global representation of the distribution of lifetimes within a tissue, and can reflect differences in metabolism at the cellular level. Theoretically, it should be possible to identify and quantify the cellular contributions of individual intrinsic fluorophores using intravital FLIM, provided the integration times are sufficiently long to accumulate enough photon counts. However, this is not practical when imaging in the intravital setting, where acquisition time and laser power need to be carefully controlled. Nor is it necessary.
The phasor fingerprint approach described here provides a global representation of the auto-fluorescent signals coming from living tissue without the need to identify the individual sources. The application of the phasor fingerprint method can distinguish different metabolic signatures from cells in situ in complex living tissue. However, intravital FLIM typically uses relatively slow image acquisition (Section 3.6). While this can limit the ability of intravital FLIM to measure metabolic changes in dynamic systems, it is still possible to monitor changes in cellular metabolism that occur over time scales of minutes. A substantial strength of this method is that it enables a bias-free approach to characterize and monitor metabolism in vivo and offers the unique opportunity to uncover metabolic changes in living animals with subcellular resolution.
Acknowledgments
This research is supported by the National Institutes of Health O’Brien Center for Advanced Renal Microscopic Analysis (NIHNIDDK P30DK079312 to R.N.D.). Microscopy studies were conducted at the Indiana Center for Biological Microscopy, and the authors thank M. Kamocka for assistance in microscopy.
References
- 1.Pittet MJ, Weissleder R. Intravital imaging. Cell. 2011;147:983–991. doi: 10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 3.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat. Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 4.Presson RG, Jr, Brown MB, Fisher AJ, Sandoval RM, Dunn KW, Lorenz KS, Delp EJ, Salama P, Molitoris BA, Petrache I. Two-photon imaging within the murine thorax without respiratory and cardiac motion artifact. Am. J. Pathol. 2011;179:75–82. doi: 10.1016/j.ajpath.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn KW, Lorenz KS, Salama P, Delp EJ. IMART software for correction of motion artifacts in images collected in intravital microscopy. Intravital. 2014;3:e28210. doi: 10.4161/intv.28210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heim N, Garaschuk O, Friedrich MW, Mank M, Milos RI, Kovalchuk Y, Konnerth A, Griesbeck O. Improved calcium imaging in transgenic mice expressing a troponin C-based biosensor. Nat. Methods. 2007;4:127–129. doi: 10.1038/nmeth1009. [DOI] [PubMed] [Google Scholar]
- 7.Radbruch H, Bremer D, Mothes R, Gunther R, Rinnenthal JL, Pohlan J, Ulbricht C, Hauser AE, Niesner R. Intravital FRET: Probing Cellular and Tissue Function in Vivo. Int. J. Mol. Sci. 2015;16:11713–11727. doi: 10.3390/ijms160511713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyawaki A. Development of probes for cellular functions using fluorescent proteins and fluorescence resonance energy transfer. Annu. Rev. Biochem. 2011;80:357–373. doi: 10.1146/annurev-biochem-072909-094736. [DOI] [PubMed] [Google Scholar]
- 9.Kardash E, Bandemer J, Raz E. Imaging protein activity in live embryos using fluorescence resonance energy transfer biosensors. Nat. Protoc. 2011;6:1835–1846. doi: 10.1038/nprot.2011.395. [DOI] [PubMed] [Google Scholar]
- 10.DiPilato LM, Zhang J. Fluorescent protein-based biosensors: resolving spatiotemporal dynamics of signaling. Curr Opin Chem Biol. 2010;14:37–42. doi: 10.1016/j.cbpa.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X, Herbst-Robinson KJ, Zhang J. Visualizing dynamic activities of signaling enzymes using genetically encodable FRET-based biosensors from designs to applications. Methods Enzymol. 2012;504:317–340. doi: 10.1016/B978-0-12-391857-4.00016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tao W, Rubart M, Ryan J, Xiao X, Qiao C, Hato T, Davidson MW, Dunn KW, Day RN. A practical method for monitoring FRET-based biosensors in living animals using two-photon microscopy. Am. J. Physiol. Cell Physiol. 2015;309:C724–C735. doi: 10.1152/ajpcell.00182.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Day RN, Tao W, Dunn KW. A simple approach for measuring FRET in fluorescent biosensors using two-photon microscopy. Nat. Protoc. 2016;11:2066–2080. doi: 10.1038/nprot.2016.121. [DOI] [PubMed] [Google Scholar]
- 14.Day RN. Measuring protein interactions using Forster resonance energy transfer and fluorescence lifetime imaging microscopy. Methods. 2014;66:200–207. doi: 10.1016/j.ymeth.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rinnenthal JL, Bornchen C, Radbruch H, Andresen V, Mossakowski A, Siffrin V, Seelemann T, Spiecker H, Moll I, Herz J, et al. Parallelized TCSPC for dynamic intravital fluorescence lifetime imaging: quantifying neuronal dysfunction in neuroinflammation. PLoS One. 2013;8:e60100. doi: 10.1371/journal.pone.0060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jameson DM, Gratton E, Hall RD. The measurement and analysis of heterogeneous emissions by multifrequency phase and modulation fluorometry. Appl. Spectrosc. Rev. 1984;20:55–106. doi: 10.1146/annurev.bb.13.060184.000541. [DOI] [PubMed] [Google Scholar]
- 17.Redford GI, Clegg RM. Polar plot representation for frequency-domain analysis of fluorescence lifetimes. J. Fluoresc. 2005;15:805–815. doi: 10.1007/s10895-005-2990-8. [DOI] [PubMed] [Google Scholar]
- 18.Eichorst JP, Wen Teng K, Clegg RM. Polar plot representation of time-resolved fluorescence. Methods Mol. Biol. 2014;1076:97–112. doi: 10.1007/978-1-62703-649-8_6. [DOI] [PubMed] [Google Scholar]
- 19.Hinde E, Digman MA, Hahn KM, Gratton E. Millisecond spatiotemporal dynamics of FRET biosensors by the pair correlation function and the phasor approach to FLIM. Proc. Natl. Acad. Sci. USA. 2013;110:135–140. doi: 10.1073/pnas.1211882110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stringari C, Cinquin A, Cinquin O, Digman MA, Donovan PJ, Gratton E. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc. Natl. Acad. Sci. USA. 2011;108:13582–13587. doi: 10.1073/pnas.1108161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stringari C, Edwards RA, Pate KT, Waterman ML, Donovan PJ, Gratton E. Metabolic trajectory of cellular differentiation in small intestine by Phasor Fluorescence Lifetime Microscopy of NADH. Sci. Rep. 2012;2:568. doi: 10.1038/srep00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georgakoudi I, Quinn KP. Optical imaging using endogenous contrast to assess metabolic state. Annu. Rev. Biomed. Eng. 2012;14:351–367. doi: 10.1146/annurev-bioeng-071811-150108. [DOI] [PubMed] [Google Scholar]
- 23.Zipfel WR, Williams RM, Christie R, Nikitin AY, Hyman BT, Webb WW. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc. Natl. Acad. Sci. USA. 2003;100:7075–7080. doi: 10.1073/pnas.0832308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pate KT, Stringari C, Sprowl-Tanio S, Wang K, TeSlaa T, Hoverter NP, McQuade MM, Garner C, Digman MA, Teitell MA, et al. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J. 2014;33:1454–1473. doi: 10.15252/embj.201488598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryan JC, Dunn KW, Decker BS. Effects of chronic kidney disease on liver transport: quantitative intravital microscopy of fluorescein transport in the rat liver. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014;307:R1488–R1492. doi: 10.1152/ajpregu.00371.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magde D, Wong R, Seybold PG. Fluorescence quantum yields and their relation to lifetimes of rhodamine 6G and fluorescein in nine solvents: improved absolute standards for quantum yields. Photochem. Photobiol. 2002;75:327–334. doi: 10.1562/0031-8655(2002)075<0327:fqyatr>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 27.Ryder AG, Glynn TJ, Przyjalgowski MA, Szczupak BA. Compact violet diode laser-based fluorescent lifetime microscope. J. Fluoresc. 2002;12:177–180. [Google Scholar]
- 28.Kristoffersen AS, Erga SR, Hamre B, Frette O. Testing fluorescence lifetime standards using two-photon excitation and time-domain instrumentation: rhodamine B, coumarin 6 and lucifer yellow. J. Fluoresc. 2014;24:1015–1024. doi: 10.1007/s10895-014-1368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stringari C, Sierra R, Donovan PJ, Gratton E. Label-free separation of human embryonic stem cells and their differentiating progenies by phasor fluorescence lifetime microscopy. J. Biomed. Opt. 2012;17:046012. doi: 10.1117/1.JBO.17.4.046012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stringari C, Nourse JL, Flanagan LA, Gratton E. Phasor fluorescence lifetime microscopy of free and protein-bound NADH reveals neural stem cell differentiation potential. PLoS One. 2012;7:e48014. doi: 10.1371/journal.pone.0048014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaffer MW, Roy SS, Mukherjee S, Nohr D, Wolter M, Biesalski HK, Ong DE, Das SK. Qualitative and quantitative analysis of retinol, retinyl esters, tocopherols and selected carotenoids out of various internal organs form different species by HPLC. Anal. Methods. 2010;2:1320–1332. doi: 10.1039/c0ay00288g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munro AW, Noble MA. Fluorescence analysis of flavoproteins. In: Chapman SK, Reid GA, editors. Flavoprotein Protocols. Humana Press; Totowa, N.J.: 1999. pp. 25–48. [DOI] [PubMed] [Google Scholar]
- 33.Hato T, Winfree S, Kalakeche R, Dube S, Kumar R, Yoshimoto M, Plotkin Z, Dagher PC. The macrophage mediates the renoprotective effects of endotoxin preconditioning. J. Am. Soc. Nephrol. 2015;26:1347–1362. doi: 10.1681/ASN.2014060561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamioka Y, Sumiyama K, Mizuno R, Sakai Y, Hirata E, Kiyokawa E, Matsuda M. Live imaging of protein kinase activities in transgenic mice expressing FRET biosensors. Cell Struct. Funct. 2012;37:65–73. doi: 10.1247/csf.11045. [DOI] [PubMed] [Google Scholar]
- 35.Oldach L, Zhang J. Genetically encoded fluorescent biosensors for live-cell visualization of protein phosphorylation. Chem. Biol. 2014;21:186–197. doi: 10.1016/j.chembiol.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buchholz CJ, Friedel T, Buning H. Surface-engineered viral vectors for selective and cell type-specific gene delivery. Trends Biotechnol. 2015;33:777–790. doi: 10.1016/j.tibtech.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Miller RA, Chu Q, Xie J, Foretz M, Viollet B, Birnbaum MJ. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature. 2013;494:256–260. doi: 10.1038/nature11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lock JT, Parker I, Smith IF. A comparison of fluorescent Ca(2)(+) indicators for imaging local Ca(2)(+) signals in cultured cells. Cell Calcium. 2015;58:638–648. doi: 10.1016/j.ceca.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hato T, Winfree S, Day R, Sandoval RM, Molitoris BA, Yoder MC, Wiggins RC, Zheng Y, Dunn KW, Dagher PC. Two-photon intravital fluorescence lifetime imaging of the kidney reveals cell-type specific metabolic signatures. J. Am. Soc. Nephrol. 2017;28 doi: 10.1681/ASN.2016101153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Follain G, Mercier L, Osmani N, Harlepp S, Goetz JG. Seeing is believing: multi-scale spatio-temporal imaging towards in vivo cell biology. J. Cell. Sci. 2016 doi: 10.1242/jcs.189001. [DOI] [PubMed] [Google Scholar]
- 41.Yellen G, Mongeon R. Quantitative two-photon imaging of fluorescent biosensors. Curr. Opin. Chem. Biol. 2015;27:24–30. doi: 10.1016/j.cbpa.2015.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]