Significance
Understanding how microbes adapt to changing conditions is fundamental to biological science and engineering. For example, adaptation is a key driver of antimicrobial resistance, and adaptive laboratory evolution has become a key tool in biotechnology. Here, we present a comprehensive genetic and fluxomic analysis of 10 adaptively evolved Escherichia coli phosphoglucose isomerase (pgi) knockout strains. The loss of PGI, a key enzyme in glycolysis, results in massive redirection of carbon catabolic flux and reduction in growth rate. Adaptive evolution results in a 3.6-fold increase in growth rate, enabled by key mutations and metabolic flux rewiring. These include global transcriptional regulators, cofactor transhydrogenases, and the phosphotransferase system component crr. Overcoming key bottlenecks, rather than a broad metabolic response, is the dominant mechanism of adaptation.
Keywords: adaptive evolution, Escherichia coli, metabolism, gene knockout, transhydrogenase
Abstract
Unraveling the mechanisms of microbial adaptive evolution following genetic or environmental challenges is of fundamental interest in biological science and engineering. When the challenge is the loss of a metabolic enzyme, adaptive responses can also shed significant insight into metabolic robustness, regulation, and areas of kinetic limitation. In this study, whole-genome sequencing and high-resolution 13C-metabolic flux analysis were performed on 10 adaptively evolved pgi knockouts of Escherichia coli. Pgi catalyzes the first reaction in glycolysis, and its loss results in major physiological and carbon catabolism pathway changes, including an 80% reduction in growth rate. Following adaptive laboratory evolution (ALE), the knockouts increase their growth rate by up to 3.6-fold. Through combined genomic–fluxomic analysis, we characterized the mutations and resulting metabolic fluxes that enabled this fitness recovery. Large increases in pyridine cofactor transhydrogenase flux, correcting imbalanced production of NADPH and NADH, were enabled by direct mutations to the transhydrogenase genes sthA and pntAB. The phosphotransferase system component crr was also found to be frequently mutated, which corresponded to elevated flux from pyruvate to phosphoenolpyruvate. The overall energy metabolism was found to be strikingly robust, and what have been previously described as latently activated Entner–Doudoroff and glyoxylate shunt pathways are shown here to represent no real increases in absolute flux relative to the wild type. These results indicate that the dominant mechanism of adaptation was to relieve the rate-limiting steps in cofactor metabolism and substrate uptake and to modulate global transcriptional regulation from stress response to catabolism.
In the study of microbial metabolism, understanding responses to genetic perturbation and adaptive evolution is fundamental. Mutations in metabolic enzymes force a rewiring of flux in the cell, the nature of which can inform our understanding of alternative pathways, kinetics, and regulation (1, 2). Adaptive laboratory evolution (ALE) is a powerful approach by which a microbe is cultured continuously for many generations, typically achieving improved fitness (e.g., faster growth rate) through natural selection. The final mutants are then sequenced and phenotypically characterized (3, 4), with the identification of causal genetic mutations and mechanistic insights enabled by replicate experiments and detailed “omics” analysis (5). Often used to study adaptations to environmental conditions like varied substrates (3, 5–8) or the presence of toxic chemicals (9–12), ALE has also been previously applied to study the adaptive responses to genetic perturbations such as the loss of major metabolic enzymes (13–15). These studies provide a valuable dimension for both evolutionary and metabolic research, as new metabolic phenotypes are evolved subject to significant and unnatural constraints. The metabolic response to knockouts before and after adaptive evolution has been an area of significant theorizing and in silico model development (15–17).
Phosphoglucose isomerase (pgi) knockouts of Escherichia coli are of significant interest in metabolic engineering and have been the subject of many investigations (1). Pgi catalyzes the first reaction in glycolysis, the conversion of glucose 6-phosphate (G6P) to fructose 6-phosphate (F6P), which in the wild type during aerobic growth on glucose catabolizes ∼70% of glucose (18, 19). Its loss results in a correspondingly severe growth impairment (70–80% lower growth rate) (13, 20) as the oxidative pentose phosphate pathway (oxPPP) and Entner–Doudoroff (ED) pathway must compensate. Several studies have used 13C-metabolic flux analysis (13C-MFA) to characterize Δpgi, frequently describing the activation of normally latent (i.e., nonutilized) pathways and a redox imbalance caused by overproduction of NADPH in the pentose phosphate pathway (2, 21–24). The major flux, redox, and growth rate changes caused by loss of pgi make it a rich target for ALE experiments (13, 14). Previously, Charusanti et al. (14) adaptively evolved 10 strains in replicate experiments over 50 d of continuous culture in glucose minimal media, reporting significant growth recovery of 3.6-fold. However, no underlying intracellular fluxes have been reported for these strains or any similarly large-scale ALE study of genetic mutants.
To gain fundamental insight into the mechanisms and outcomes of adaptive evolution, both the mutations and the selected-upon phenotype (here, metabolism) must be measured. In this study, we applied high-resolution 13C-MFA and next-generation sequencing to the 10 evolved Δpgi strains and the parental strain reported previously (14). Novel mutations were identified, and comparisons to recently reported wild-type ALE studies (5, 6) helped to identify mutations unique to Δpgi. Together with complimentary fluxomic information, a detailed picture of how Δpgi metabolically adapts to achieve faster growth is attained. Areas of convergence and divergence on the genetic and fluxomic levels highlight the large number of genetic solutions possible for achieving similar metabolic phenotypes as well as some differences in metabolic optima. In several cases, specific causal mutation–flux relationships were identified.
Results
Recovery of Growth Fitness in Δpgi ALE Strains Is Associated with Unique Mutations.
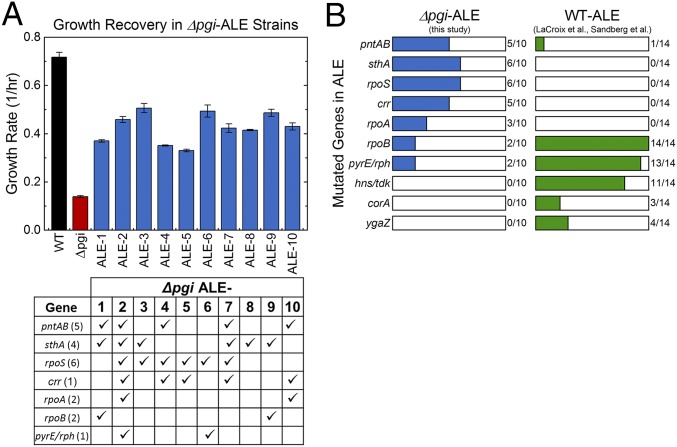
In E. coli K-12 MG1655, the knockout of pgi results in a reduction in growth rate of ∼80% compared with the wild type, from 0.72 h−1 to 0.14 h−1. Following ALE, a significant fraction of this growth rate can be recovered (46–71% of the wild-type growth rate, Fig. 1A). These 2.4–3.6-fold increases in growth rate are quite large compared with, for example, similar ALE experiments with wild-type E. coli, which reported 1.6-fold increases in growth rate (5).
Fig. 1.
Growth rate recovery in evolved Δpgi strains is supported by unique mutations. (A) The growth rate is severely reduced in Δpgi strain relative to wild type (WT). This is significantly, but not completely, recovered through ALE (growth rate, mean ± SEM, n ≥ 3). (B) The frequency of mutations in 10 Δpgi-ALE strains is compared with those previously reported for 14 WT-ALE strains of E. coli (5, 6). The profile of mutations is quite distinct for Δpgi-ALE and WT-ALE strains. The distribution of mutations from the seven most frequently affected genes in the 10 Δpgi-ALE is also shown in A, Bottom, with the number of unique mutations per gene noted.
To assess the genetic basis of the large increases in growth rate, whole-genome sequencing was performed and sequences were mapped to the E. coli reference to identify mutations in 10 independent ALE experiments. Recent advances in sequencing allowed for improved determination of mutations in clones isolated from final populations, particularly of insertion sequences (IS elements), compared with a previous effort that utilized both a microarray hybridization-based method and an earlier Illumina short-read technology protocol (14). In all, 52 unique mutations were identified across the 10 ALE strains, spanning 34 different genetic regions (29 structural mutations—i.e., within an ORF—and 5 intergenic regions). The complete mutation table is in Supporting Information. A key advantage and reason for using replicate ALE experiments is to use mutation frequency to help differentiate causal mutations from genetic “hitch-hikers” that do not affect fitness (5). Fig. 1B lists the genes that were mutated in ≥2 of the 10 Δpgi ALE strains as well as the genes frequently mutated in reported wild-type K-12 MG1655 ALE studies performed in similar conditions (5, 6). The two sets are striking in their lack of overlap; mutations that occur in almost every reported wild-type ALE experiment, in rpoB, pyrE/rph, and hns/tdk, occur rarely or not at all in Δpgi strains. Instead, the Δpgi ALE strains have a high frequency of mutations in the pyridine nucleotide transhydrogenases pntAB and sthA, the transcription factor rpoS, and the phosphotransferase system (PTS) sugar transport system component crr. The distribution of these mutations across the 10 Δpgi ALE strains is also in Fig. 1A, showing that while some strains had many of these common mutations (ALE-2 had six out of the top seven), others had fewer (ALE-8 had only 1/7). This likely reflects less common but equally effective adaptive routes. It is worthwhile to mention that some genes or genetic regions had many unique mutations in parallel evolutions (e.g., six for rpoS and five for pntAB). In contrast, the IS element insertion in crr was identical in five different strains. Both patterns clearly demonstrate evidence for causality. It was previously demonstrated that the combination of rpoS and sthA mutations are causal for increased growth in Δpgi and exhibit positive epistasis (14). The difference in mutation profiles demonstrate that there are unique selective pressures in Δpgi, which result in unique adaptive responses. To further investigate how these mutations enabled the large increases in growth rates from the initial perturbed Δpgi state, we next characterized the carbon metabolism of each strain using 13C-MFA.
Activation of Latent Pathways, or Not?
To characterize the rewiring of central carbon metabolism in the parental and ALE Δpgi strains, high-resolution 13C-MFA was performed. The analysis consisted of two parallel labeling experiments with [1,2-13C] and [1,6-13C]glucose [an experimental design previously identified as providing optimal flux estimate precision (25)], and the simultaneous fitting of labeling from proteinogenic amino acids, the ribose moiety of RNA, and glucose moiety of glycogen (26) to estimate fluxes. For the wild type, data from parallel labeling experiments previously reported were refitted (19). The full network model, the measured isotopomer distributions, and the estimated metabolic fluxes are in Supporting Information.
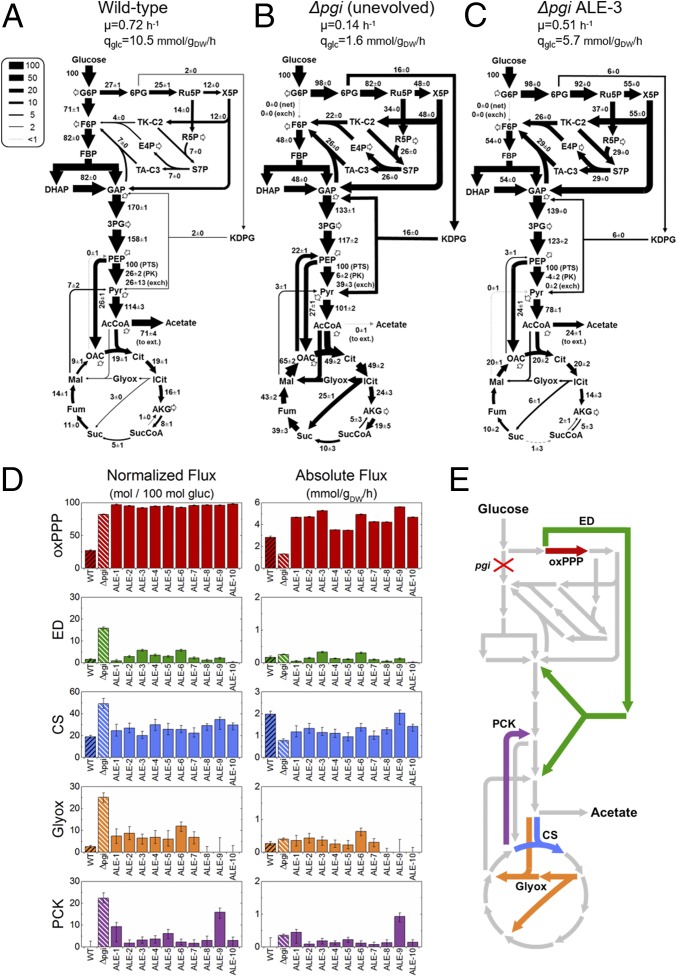
In Fig. 2 A–C, the estimated intracellular fluxes of the wild-type, Δpgi parental strain, and Δpgi ALE-3 are summarized. ALE-3 was the fastest growing Δpgi strain (0.51 h−1) and had an intracellular flux distribution typical of most of the ALE strains. The growth rates and glucose uptake rates for each strain are noted, and the fluxes shown are normalized to 100 units of glucose uptake. In all Δpgi strains, the forward and reverse fluxes of the PGI reaction were estimated to be zero, thus confirming the pgi knockout. The unevolved Δpgi (Fig. 2B) was found to utilize reactions and pathways that carry minimal flux in the wild type (Fig. 2A), including the ED pathway, glyoxylate shunt, and phosphoenolpyruvate carboxykinase (PCK) reaction [oxaloacetate to phosphoenolpyruvate (PEP)]. These flux changes have been noted in previous studies and have been described as “latent pathway activation” (2, 22–24, 27). After adaptive evolution, ALE-3 (Fig. 2C) and the other ALE strains significantly reduced the usage of these pathways; for example, the glyoxylate shunt flux was reduced from 25 to 6, and the PCK reaction from 22 to 3 in ALE-3. This “rerepression” following adaptive evolution has also been previously observed and been the focus of various speculations and computational analyses (13, 16). The previous terminology implies the presence of a transient regulatory response, activated in response to the stress caused by the knockout and then repressed during evolution to facilitate faster growth.
Fig. 2.
13C-MFA reveals large flux redistributions but ambiguous activation of latent pathways. Intracellular flux distributions, normalized to 100 units of glucose uptake, are shown for the wild type (A), unevolved Δpgi (B), and a representative evolved Δpgi strain ALE-3 (C). (D) Selected fluxes are shown in normalized flux units (as in A–C) as well as absolute flux units (mmol·gDW−1·h−1), in the context of central metabolic pathways (E). The error bars in D reflect the 95% confidence intervals of flux estimates.
When interpreting metabolic fluxes, it is important to consider both normalized fluxes (e.g., relative to glucose uptake) and absolute fluxes (mmol·gDW−1·h−1), as each provide complimentary information. Several key fluxes are shown in both units in Fig. 2D, corresponding to the pathway map shown in Fig. 2E. The glucose uptake rates used to calculate absolute fluxes are in Fig. S1. In Fig. 2 B and D, we can see that although the oxPPP is the dominant route of glucose catabolism in Δpgi, the absolute flux is reduced by roughly half in the unevolved strain relative to the wild type. It has been previously reported that G6P accumulates in Δpgi and that G6PDH (encoded zwf), the first step in the oxPPP, is likely rate limiting for growth due to allosteric inhibition caused by an elevated NADPH/NADP+ ratio (23). This limitation is overcome in the evolved strains, where oxPPP fluxes are increased by three- to fourfold, to rates higher even than in the WT (by up to twofold). Intriguingly, the highest absolute flux was observed in ALE-9, which was the only evolved strain with a mutation in zwf (Supporting Information).
In cases of “activated latent” pathways, the absolute fluxes provide an especially illuminating perspective. There was no statistically significant increase in the absolute ED, glyoxylate shunt, or PCK fluxes in Δpgi compared with the wild type. Instead, a similar low level of absolute flux was maintained, which only appeared much larger in relative terms due to the dramatic reductions in absolute glucose uptake caused by the pgi knockout. Very little change was observed in the ALE strains for these latent pathway fluxes, with the lone exception of an elevated PCK flux in ALE-9. These results challenge the notion that these latent pathways are “activated” in a regulatory sense that increases their absolute flux capacity. Perhaps more likely is that these enzymes are expressed at low levels in the wild type, and this is maintained in the Δpgi strain, where due to the perturbation in glycolysis the same small rates of flux play a larger relative role. The rerepression in the ALE strains, then, is instead simply the recovery of faster glucose uptake rate (Fig. S1).
Energy Metabolism Is Not Significantly Affected by Adaptive Evolution in Δpgi.
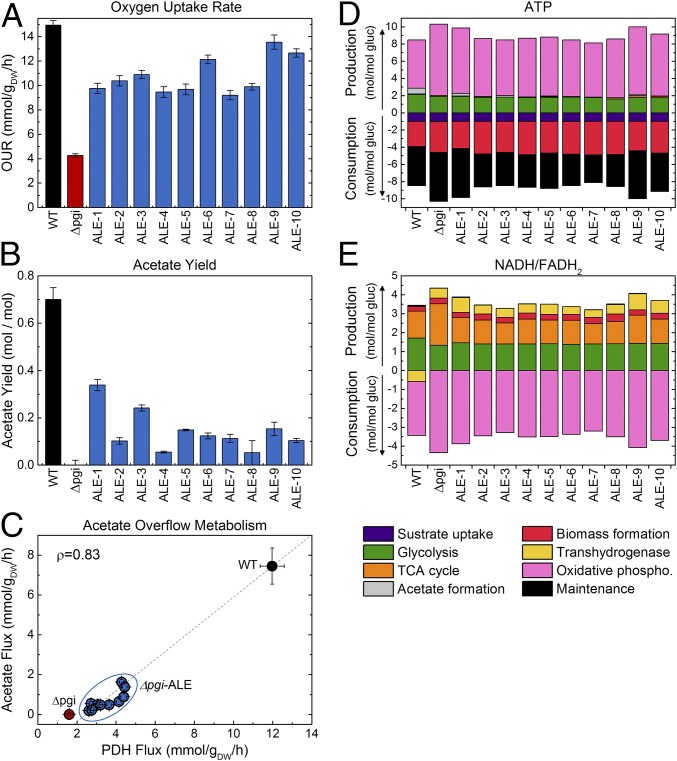
The measured metabolic fluxes can also inform a broader analysis of energy metabolism in these strains. In Fig. 3 A and B, oxygen uptake rates and acetate yields are shown. In the unevolved Δpgi, the oxygen uptake is reduced to 4.3 mmol·gDW−1·h−1, down from 15 mmol·gDW−1·h−1 in the wild type, corresponding to the overall slowed metabolism and growth rate. The unevolved Δpgi does not produce acetate, as the citrate synthase (CS) flux can easily accommodate all of the flux from acetyl-CoA at less than half of its wild-type rate (Fig. 2D). In the ALE strains, oxygen uptake recovers to 62–91% of the wild-type flux and some strains produce acetate. This may represent a limit in TCA cycle or oxidative phosphorylation capacity that these strains encounter, above which excess glycolytic flux is diverted to acetate production. This acetate overflow effect is shown in Fig. 3C, which shows that the absolute flux through the pyruvate (Pyr) dehydrogenase (acetyl-CoA generation) strongly correlates with the acetate secretion flux in all strains. Fig. 3 D and E show the normalized cofactor balances for ATP and NADH/FADH2 (the electron carriers used in oxidative phosphorylation for ATP production), with contributions to production and consumption by the various pathways and cell functions. One noticeable difference in the unevolved Δpgi strain is an increased contribution of the TCA cycle and oxidative phosphorylation to energy metabolism, leading to a slightly higher overall ATP yield. The ALE strains mostly reverted to normalized levels of total cofactor production and consumption that were very similar to the wild type. Overall, the profile of energy metabolism is remarkably conserved in these strains. Perhaps the most significant difference between the wild-type and Δpgi strains in cofactor metabolism is in the reversed role of transhydrogenase (Fig. 3D).
Fig. 3.
Energy metabolism is mostly unaffected by adaptive evolution in Δpgi. Oxygen uptake rates (OURs) (A) and acetate yields (B) are shown for the wild type, unevolved Δpgi, and 10 Δpgi-ALE strains. OUR was estimated by 13C-MFA, and the error bars indicate SDs of the estimates. The acetate overflow effect is represented in C, which shows the absolute acetate production flux plotted against the absolute Pyr dehydrogenase (PDH) flux in absolute units. Error bars represent 95% confidence intervals. The two fluxes strongly correlate (Pearson correlation coefficient of 0.83). Both the oxygen uptake and acetate secretion phenotypes directly impact cellular energy metabolism, summarized in more detail through ATP (D) and NADH/FADH2 (E) balances. Overall, energy metabolism was not significantly altered following adaptive evolution.
Transhydrogenase Genes Are Mutated in Many but Not All Δpgi ALE Strains.
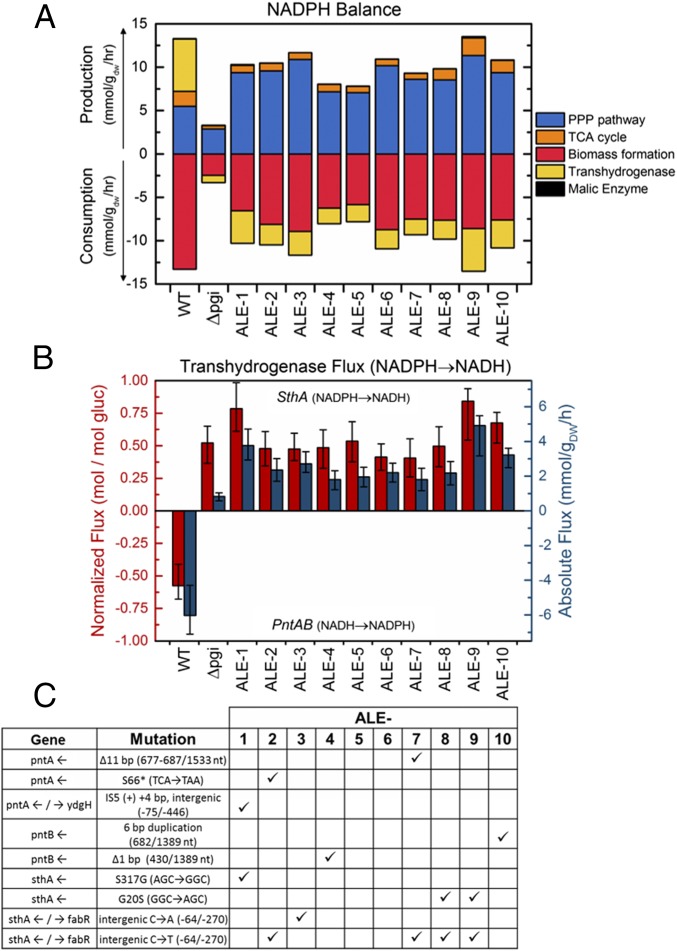
E. coli is able to interconvert reduced cofactors with two pyridine nucleotide transhydrogenases, the membrane-bound PntAB, which primarily converts NADH to NADPH, and the soluble form SthA (also referred to as UdhA), which primarily converts NADPH to NADH (28). In the wild type, excess NADH produced in glycolysis and the TCA cycle is used to produce approximately half of the needed NADPH through the transhydrogenase (Figs. 3D and 4A and Fig. S2). In Δpgi, the redirection of glucose flux in upper central carbon metabolism, away from glycolysis and into the oxPPP, results in a significant excess of NADPH generation. Some of this is utilized by elevated biosynthesis flux (i.e., biomass yield) (Fig. S1), but most of the imbalance must be corrected by a reversal of the transhydrogenase to convert NADPH to NADH.
Fig. 4.
Reversal of transhydrogenase flux corresponds to genetic mutations in many but not all Δpgi ALE strains. The function of the pyridine nucleotide transhydrogenases changes dramatically in Δpgi strains. In A, the pathway contributions to the NADPH production and consumption are shown in absolute units. In the wild type, excess NADH is used to produce approximately half of NADPH needed for biomass synthesis. In Δpgi strains, elevated oxPPP pathway flux creates an excess of NADPH, leading to a reversal of net transhydrogenase flux. This flux is shown in normalized and absolute units in B. The three transhydrogenase enzymes pntA, pntB, and sthA were frequently mutated in the ALE strains (C). Check marks reflect the presence of the described mutations in specific ALE strain. At least one transhydrogenase mutation occurred in 8 out of 10 strains.
The absolute rates of NADPH production and consumption are shown in Fig. 4A. Here again is shown the impact of the forcing of flux through the oxPPP, which generates a large excess of NADPH and necessitates the reversal of the transhydrogenase. This transhydrogenase flux is shown in Fig. 4B in both absolute and relative flux units, with the 95% confidence intervals calculated from 13C-MFA. In the Δpgi strains (both unevolved and evolved), the normalized flux does not change significantly (also Fig. S2), reflecting that other parts of central carbon metabolism were not rewired to relieve the cofactor imbalance. Instead, the absolute transhydrogenase fluxes increase significantly in the ALE strains. To gain insight into how these flux increases were achieved, the mutations directly related to the regions of the transhydrogenase genes in the ALE strains were compared with the flux changes (Fig. 4C). As noted above (Fig. 1), pntA, pntB, and sthA were some of the most frequently mutated genes in this study. Eight of the 10 ALE strains had at least one transhydrogenase mutation, with five having two. Based on the nature of the mutations, they presumably increase SthA activity, reduce PntAB activity, or both. The pntAB mutations were diverse, including deletions and a duplication, as well as an upstream IS element insertion. Three of the four mutations in the coding regions likely result in truncated, nonfunctional proteins: ALE-2 has a nonsense mutation in pntA, and ALE-4 and ALE-7 have major truncations in pntB and pntA, respectively (14). The sthA mutations were SNPs, including a commonly mutated site (five strains) 64 bp upstream. The exact effect of the upstream mutations on transcriptional regulation is uncertain, but we hypothesize that they increase sthA and reduce pntAB expression. Despite the high frequency of these mutations, two strains (ALE-5 and ALE-6) achieved the increased flux with no observed mutations directly in the transhydrogenase genes, raising questions about other possible mechanisms for cofactor rebalancing. Both ALE-5 and ALE-6 possess mutations in genes that directly affect transcription levels (e.g., lrp and rpoA). In fact, pntAB has been identified as a regulatory target of Lrp (29).
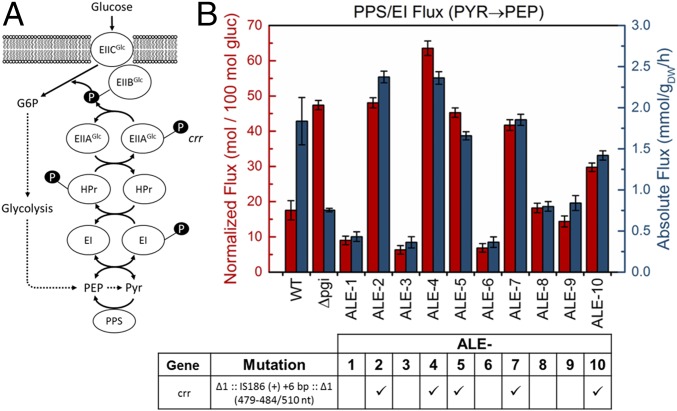
Mutations in PTS Component crr Are Associated with Increased Back Flux from Pyr to PEP.
Another frequently occurring, and more unexpected, mutation was an IS element insertion in crr in 5 of 10 ALE strains (Fig. 5). Crr encodes EIIAGlc, the cytosolic subunit of enzyme II (EII) in the PTS glucose transport system. The PTS system is the primary mode of glucose uptake in E. coli and links the uptake and phosphorylation of glucose (by EII) to the glycolytic conversion of PEP to Pyr (by EI, linked by the intermediary phosphotransferase HPr) (30, 31) (Fig. 5A). Previous work has shown that EI of the PTS sugar transport system is reversible in vivo (32) and that in the wild type 10% of PEP is produced from Pyr in the reverse (i.e., gluconeogenic) direction by this mechanism. Previous work has also shown that in a Δcrr strain, this flux is increased by more than twofold due to perturbation of the PTS equilibrium as well as some possible activation of PPS, the gluconeogenic enzyme also able to facilitate the conversion of Pyr to PEP. Given the prevalence of the crr mutation and global metabolic perturbations in Δpgi, we hypothesized that the PPS/EI flux (Pyr to PEP) would be altered in these ALE strains.
Fig. 5.
Elevated back flux from PYR to PEP corresponds with a frequently occurring mutation in crr. (A) The PTS glucose transport system couples the uptake and phosphorylation of glucose to the glycolytic conversion of PEP to Pyr. The terminal phosphotransferase EI has been previously shown to be reversible and capable of converting Pyr to PEP. The back flux from Pyr to PEP was quantified using [U-13C]alanine tracer experiments, shown here in both normalized and absolute flux units (B), with error bars reflecting SDs of the estimate. This flux was significantly elevated in Δpgi relative to the wild type in normalized units but decreased in absolute units. In the ALE strains, the back flux strongly correlated with the presence of a specific and frequent insertion element mutation in crr in 5 of 10 strains. Strains containing this mutation had highly elevated back fluxes.
The PPS/EI flux was measured using an [U-13C]alanine tracer approach developed recently (32) (see Materials and Methods and Fig. S3). As shown in Fig. 5B, the extent of this flux varied widely among the ALE strains, and its magnitude corresponded strongly with the presence of the crr mutation. Expressed as normalized flux (relative to 100 units of glucose uptake), the PPS/EI flux was significantly elevated in the unevolved Δpgi (from 18 in the wild type to 47) and was reduced subsequently in strains lacking the crr mutation but maintained at a high level in the strains with the mutated crr. In absolute terms, the flux was significantly reduced in all Δpgi strains except for those with the crr mutation, where the flux was more similar to the wild type. Without further analysis of the activity of PPS in these strains, it is difficult to deduce the exact mechanism of these changes, but they are consistent with the previous observations in Δcrr (32). This result strongly supports a genetic–metabolic flux relationship between the crr IS element mutation and elevated PPS/EI flux. The high frequency of this identical mutation indicates a strong selective pressure for this mutation, but the exact mechanism of the fitness benefit is unclear. In addition to its direct role in glucose uptake, crr is also involved in signaling the global regulator Crp (33), which controls the transcription of over 100 genes (34).
Discussion
In this work, we have explored how recovery of growth rate in Δpgi is enabled by unique genetic mutations and significant metabolic rewiring. In this system, fitness recovery was driven by global transcriptional regulation (i.e., sigma factors and other RNA polymerase components) and relief of a rate-limiting step at the cofactor transhydrogenase. This led to increased absolute flux through the oxidative pentose phosphate pathway and a corresponding recovery of faster glucose uptake and growth rates. The usage of latent pathways including the ED pathway, glyoxylate shunt, and PCK reaction were shown to represent no increase in absolute flux relative to the wild type and in absolute terms did not appreciably change after adaptive evolution. The availability of these pathways, expressed at low levels in the wild type, may offer flexibility when facing changing conditions. However, they do not appear to be “activated” as part of a general stress response in this case. These insights and the dataset presented here should help advance predictive metabolic modeling (35–37). Overall, these results add to our understanding of adaptive evolution by elucidating how challenges to specific cellular subsystems—that is, central carbon metabolism and glycolysis—are overcome. Future application of this approach to other significant, growth-limiting metabolic perturbations may similarly illuminate associated kinetic and regulatory limitations as well as reveal useful solutions to ameliorate them. Paired with the appropriate measurements (as shown here with fluxomics), such studies are likely to uncover detailed mechanisms of adaptation.
Comparing the mutations and phenotypes of wild-type and Δpgi-derived ALE strains deepens our mechanistic understanding of adaptive evolution by providing insight into the context-dependent selective forces and impacts of various mutations. For example, RNA polymerase components are commonly mutated in ALE, and in experiments started with wild type, mutations to rpoB and rpoC dominate (Fig. 1) (3, 5, 6, 38, 39). These have been shown to contribute to a rebalancing of the proteome to promote growth (40), corresponding to broad and proportional intracellular metabolic flux increases (i.e., no changes in normalized flux distributions) (41). In contrast, ALE experiments started with Δpgi-acquired frequent mutations in the stress response-associated sigma factor rpoS. This would seem to indicate the presence of a unique maladaptive rpoS-mediated stress response in Δpgi. However, rpoB mutations were also seen in 2 of 10 Δpgi ALE experiments here but never co-occurring with rpoS mutations. Further work is needed to deconvolute the role of each in adapting the global transcriptome and whether the two mutations would have additive benefits in Δpgi. Across both the wild type and Δpgi studies, mutations to more than two RNA polymerase subunits (rpo) genes are rarely observed, possibly pointing to overlapping mechanisms where the selected-for mutations depend on the state of the cell [in this case, perturbed (Δpgi) versus a wild-type state]. Other mutations seen more when starting with a wild-type strain, such as in pyrE/rph, are likely relevant based on the overall growth state (the evolved strains started with wild type are significantly faster when evolved under the same conditions); thus, they may never be selected for in a “crippled” starting strain such as the Δpgi starting strain (42). However, such mutations and a convergence of mutations may be seen if evolved for more extended lengths of time under the same conditions (43).
Other unique mutations in Δpgi were in the transhydrogenases and in crr. With regards to the former, a recent report of an ALE study of an oxPPP mutant (i.e., reduced NADPH production) found a high frequency of mutations in pntAB that led to increased activity (44). Given the importance of cofactors to the formation of metabolic products, the reported mutations from these two studies may provide valuable new candidates for rationally manipulating transhydrogenase activity in metabolic engineering (45). Lastly, the mechanism of fitness enhancement of the crr IS element mutation identified here requires further study. One possibility is that the reduced glycolytic flux in Δpgi could be sensed (46) by the PTS system—for example, via perturbation of the PEP/Pyr concentration ratio—and result in feedback inhibition of glucose uptake. Another is that the accumulated G6P (23) could induce a maladaptive regulatory response via CRP activation by P ∼ EIIBGlc. In these scenarios, a crr mutation may help to decouple feedback inhibition or limit the harmful regulatory effect. Ultimately, it will be desirable to confirm the identities and interactions of causal mutations by reproducing them synthetically. Previous work (14, 21) confirmed causality and epistasis for sthA and rpoS mutations but did not fully recapitulate the observed growth phenotypes of the evolved clones themselves. This likely points to a complex landscape that will become more feasible to explore as high-throughput genome engineering methods mature and many strains can be tested efficiently.
Materials and Methods
DNA Sequencing.
Sequences were obtained using Illumina MiSeq. The breseq pipeline (47) was used to map sequenced reads and identify mutations relative to the E. coli K-12 MG1655 genome.
Tracer Experiments.
For 13C-tracer experiments, strains were cultured aerobically in batch culture in M9 minimal medium at 37 °C in minibioreactors with 10 mL working volume (20). For the quantification of the Pyr to PEP flux, [U-13C]alanine tracer experiments were performed (32).
Gas Chromatography–Mass Spectrometry.
Gas chromatography–mass spectrometry (GC-MS) analysis was performed on an Agilent 5977A mass spectrometer to measure labeling of proteinogenic amino acids (48, 49), glucose (derived from glycogen), and ribose (from RNA) (26, 50, 51). Mass isotopomer distributions (MIDs) were obtained by integration and corrected for natural isotope abundances.
13C-MFA.
13C-MFA calculations were performed using the Metran software. For integrated analysis of parallel labeling experiments, the datasets were fitted simultaneously to a single flux model (18).
Data Availability.
All data generated or analyzed during this study are included in this published article (and its Supporting Information files).
Supplementary Material
Acknowledgments
This work was supported by NSF Grant MCB-1616332. C.P.L. and J.E.G. were also supported by the University of Delaware Graduate Fellows Award. A.M.F. and B.O.P. were supported under Novo Nordisk Foundation Center for Biosustainability Grant NNF10CC1016517.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716056115/-/DCSupplemental.
References
- 1.Long CP, Antoniewicz MR. Metabolic flux analysis of Escherichia coli knockouts: Lessons from the Keio collection and future outlook. Curr Opin Biotechnol. 2014;28:127–133. doi: 10.1016/j.copbio.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishii N, et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316:593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- 3.Herring CD, et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- 4.Dragosits M, Mattanovich D. Adaptive laboratory evolution—Principles and applications for biotechnology. Microb Cell Fact. 2013;12:64. doi: 10.1186/1475-2859-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LaCroix RA, et al. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol. 2015;81:17–30. doi: 10.1128/AEM.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandberg TE, et al. Evolution of E. coli on [U-13C]glucose reveals a negligible isotopic influence on metabolism and physiology. PLoS One. 2016;11:e0151130. doi: 10.1371/journal.pone.0151130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee DH, Palsson BO. Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a nonnative carbon source, L-1,2-propanediol. Appl Environ Microbiol. 2010;76:4158–4168. doi: 10.1128/AEM.00373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad TM, et al. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 2009;10:R118. doi: 10.1186/gb-2009-10-10-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atsumi S, et al. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol Syst Biol. 2010;6:449. doi: 10.1038/msb.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horinouchi T, et al. Transcriptome analysis of parallel-evolved Escherichia coli strains under ethanol stress. BMC Genomics. 2010;11:579. doi: 10.1186/1471-2164-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mundhada H, et al. Increased production of L-serine in Escherichia coli through adaptive laboratory evolution. Metab Eng. 2017;39:141–150. doi: 10.1016/j.ymben.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Reyes LH, Almario MP, Winkler J, Orozco MM, Kao KC. Visualizing evolution in real time to determine the molecular mechanisms of n-butanol tolerance in Escherichia coli. Metab Eng. 2012;14:579–590. doi: 10.1016/j.ymben.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Fong SS, Nanchen A, Palsson BO, Sauer U. Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. J Biol Chem. 2006;281:8024–8033. doi: 10.1074/jbc.M510016200. [DOI] [PubMed] [Google Scholar]
- 14.Charusanti P, et al. Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene. PLoS Genet. 2010;6:e1001186. doi: 10.1371/journal.pgen.1001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong SS, Palsson BO. Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nat Genet. 2004;36:1056–1058. doi: 10.1038/ng1432. [DOI] [PubMed] [Google Scholar]
- 16.Cornelius SP, Lee JS, Motter AE. Dispensability of Escherichia coli’s latent pathways. Proc Natl Acad Sci USA. 2011;108:3124–3129. doi: 10.1073/pnas.1009772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Reed JL. RELATCH: Relative optimality in metabolic networks explains robust metabolic and regulatory responses to perturbations. Genome Biol. 2012;13:R78. doi: 10.1186/gb-2012-13-9-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leighty RW, Antoniewicz MR. COMPLETE-MFA: Complementary parallel labeling experiments technique for metabolic flux analysis. Metab Eng. 2013;20:49–55. doi: 10.1016/j.ymben.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Crown SB, Long CP, Antoniewicz MR. Integrated 13C-metabolic flux analysis of 14 parallel labeling experiments in Escherichia coli. Metab Eng. 2015;28:151–158. doi: 10.1016/j.ymben.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long CP, Gonzalez JE, Sandoval NR, Antoniewicz MR. Characterization of physiological responses to 22 gene knockouts in Escherichia coli central carbon metabolism. Metab Eng. 2016;37:102–113. doi: 10.1016/j.ymben.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canonaco F, et al. Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiol Lett. 2001;204:247–252. doi: 10.1111/j.1574-6968.2001.tb10892.x. [DOI] [PubMed] [Google Scholar]
- 22.Usui Y, et al. Investigating the effects of perturbations to pgi and eno gene expression on central carbon metabolism in Escherichia coli using (13)C metabolic flux analysis. Microb Cell Fact. 2012;11:87. doi: 10.1186/1475-2859-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toya Y, et al. 13C-metabolic flux analysis for batch culture of Escherichia coli and its Pyk and Pgi gene knockout mutants based on mass isotopomer distribution of intracellular metabolites. Biotechnol Prog. 2010;26:975–992. doi: 10.1002/btpr.420. [DOI] [PubMed] [Google Scholar]
- 24.Fischer E, Sauer U. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur J Biochem. 2003;270:880–891. doi: 10.1046/j.1432-1033.2003.03448.x. [DOI] [PubMed] [Google Scholar]
- 25.Crown SB, Long CP, Antoniewicz MR. Optimal tracers for parallel labeling experiments and (13)C metabolic flux analysis: A new precision and synergy scoring system. Metab Eng. 2016;38:10–18. doi: 10.1016/j.ymben.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long CP, Au J, Gonzalez JE, Antoniewicz MR. (13)C metabolic flux analysis of microbial and mammalian systems is enhanced with GC-MS measurements of glycogen and RNA labeling. Metab Eng. 2016;38:65–72. doi: 10.1016/j.ymben.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hua Q, Yang C, Baba T, Mori H, Shimizu K. Responses of the central metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts. J Bacteriol. 2003;185:7053–7067. doi: 10.1128/JB.185.24.7053-7067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sauer U, Canonaco F, Heri S, Perrenoud A, Fischer E. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J Biol Chem. 2004;279:6613–6619. doi: 10.1074/jbc.M311657200. [DOI] [PubMed] [Google Scholar]
- 29.Cho B-K, Barrett CL, Knight EM, Park YS, Palsson BO. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc Natl Acad Sci USA. 2008;105:19462–19467. doi: 10.1073/pnas.0807227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deutscher J, et al. The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: Regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev. 2014;78:231–256. doi: 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Escalante A, Salinas Cervantes A, Gosset G, Bolívar F. Current knowledge of the Escherichia coli phosphoenolpyruvate-carbohydrate phosphotransferase system: Peculiarities of regulation and impact on growth and product formation. Appl Microbiol Biotechnol. 2012;94:1483–1494. doi: 10.1007/s00253-012-4101-5. [DOI] [PubMed] [Google Scholar]
- 32.Long CP, Au J, Sandoval NR, Gebreselassie NA, Antoniewicz MR. Enzyme I facilitates reverse flux from pyruvate to phosphoenolpyruvate in Escherichia coli. Nat Commun. 2017;8:14316. doi: 10.1038/ncomms14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng D, Constantinidou C, Hobman JL, Minchin SD. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lerman JA, et al. In silico method for modelling metabolism and gene product expression at genome scale. Nat Commun. 2012;3:929. doi: 10.1038/ncomms1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khodayari A, Zomorrodi AR, Liao JC, Maranas CD. A kinetic model of Escherichia coli core metabolism satisfying multiple sets of mutant flux data. Metab Eng. 2014;25:50–62. doi: 10.1016/j.ymben.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 37.Chowdhury A, Khodayari A, Maranas CD. Improving prediction fidelity of cellular metabolism with kinetic descriptions. Curr Opin Biotechnol. 2015;36:57–64. doi: 10.1016/j.copbio.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 38.Sandberg TE, Lloyd CJ, Palsson BO, Feist AM. Laboratory evolution to alternating substrate environments yields distinct phenotypic and genetic adaptive strategies. Appl Environ Microbiol. 2017;83:e00410-17. doi: 10.1128/AEM.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandberg TE, et al. Evolution of Escherichia coli to 42 °C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol Biol Evol. 2014;31:2647–2662. doi: 10.1093/molbev/msu209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Utrilla J, et al. Global rebalancing of cellular resources by pleiotropic point mutations illustrates a multi-scale mechanism of adaptive evolution. Cell Syst. 2016;2:260–271. doi: 10.1016/j.cels.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long CP, Gonzalez JE, Feist AM, Palsson BO, Antoniewicz MR. Fast growth phenotype of E. coli K-12 from adaptive laboratory evolution does not require intracellular flux rewiring. Metab Eng. 2017;44:100–107. doi: 10.1016/j.ymben.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen KF. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiser MJ, Ribeck N, Lenski RE. Long-term dynamics of adaptation in asexual populations. Science. 2013;342:1364–1367. doi: 10.1126/science.1243357. [DOI] [PubMed] [Google Scholar]
- 44.Chou HH, Marx CJ, Sauer U. Transhydrogenase promotes the robustness and evolvability of E. coli deficient in NADPH production. PLoS Genet. 2015;11:e1005007. doi: 10.1371/journal.pgen.1005007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jan J, Martinez I, Wang Y, Bennett GN, San KY. Metabolic engineering and transhydrogenase effects on NADPH availability in Escherichia coli. Biotechnol Prog. 2013;29:1124–1130. doi: 10.1002/btpr.1765. [DOI] [PubMed] [Google Scholar]
- 46.Kochanowski K, et al. Functioning of a metabolic flux sensor in Escherichia coli. Proc Natl Acad Sci USA. 2013;110:1130–1135. doi: 10.1073/pnas.1202582110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez JE, Long CP, Antoniewicz MR. Comprehensive analysis of glucose and xylose metabolism in Escherichia coli under aerobic and anaerobic conditions by (13)C metabolic flux analysis. Metab Eng. 2017;39:9–18. doi: 10.1016/j.ymben.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Long CP, Antoniewicz MR. Quantifying biomass composition by gas chromatography/mass spectrometry. Anal Chem. 2014;86:9423–9427. doi: 10.1021/ac502734e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cordova LT, Antoniewicz MR. (13)C metabolic flux analysis of the extremely thermophilic, fast growing, xylose-utilizing Geobacillus strain LC300. Metab Eng. 2016;33:148–157. doi: 10.1016/j.ymben.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 51.McConnell BO, Antoniewicz MR. Measuring the composition and stable-isotope labeling of algal biomass carbohydrates via gas chromatography/mass spectrometry. Anal Chem. 2016;88:4624–4628. doi: 10.1021/acs.analchem.6b00779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supporting Information files).