Significance
All life uses the same 20 amino acids, but only 7–13 early amino acids seem to be indispensable to build functional proteins. Thus, what triggered the introduction of the additional amino acids? Employing quantum chemical calculations and biochemical experiments, we find that the additional amino acids have become systematically “softer” (Ralph G. Pearson) over time, more redox-reactive, and more capable of protecting cells from destruction by oxygen free radicals. Hence, it appears that molecular oxygen forced life to incorporate novel amino acids with augmented redox properties into the genetic code. The present study provides a plausible scenario for a more than 80-y-old problem of fundamental biochemistry: Why these 20 amino acids?
Keywords: amino acids, genetic code, molecular oxygen, origin of life, redox reactivity
Abstract
All extant life employs the same 20 amino acids for protein biosynthesis. Studies on the number of amino acids necessary to produce a foldable and catalytically active polypeptide have shown that a basis set of 7–13 amino acids is sufficient to build major structural elements of modern proteins. Hence, the reasons for the evolutionary selection of the current 20 amino acids out of a much larger available pool have remained elusive. Here, we have analyzed the quantum chemistry of all proteinogenic and various prebiotic amino acids. We find that the energetic HOMO–LUMO gap, a correlate of chemical reactivity, becomes incrementally closer in modern amino acids, reaching the level of specialized redox cofactors in the late amino acids tryptophan and selenocysteine. We show that the arising prediction of a higher reactivity of the more recently added amino acids is correct as regards various free radicals, particularly oxygen-derived peroxyl radicals. Moreover, we demonstrate an immediate survival benefit conferred by the enhanced redox reactivity of the modern amino acids tyrosine and tryptophan in oxidatively stressed cells. Our data indicate that in demanding building blocks with more versatile redox chemistry, biospheric molecular oxygen triggered the selective fixation of the last amino acids in the genetic code. Thus, functional rather than structural amino acid properties were decisive during the finalization of the universal genetic code.
The genetic code embodies the selection and assignment of amino acids (AAs) to be universally employed for protein synthesis. Many aspects of this selection have remained unsettled despite intense intellectual efforts, such that the evolution of the genetic code has occasionally been termed the “universal enigma” of biology (1). Specifically, the choice of the proteinogenic AAs out of a much larger prebiotic and metabolic pool has remained hardly understood, even if a core triad of attributes (size, charge, and hydrophobicity) probably played a major role, especially in the beginning (2). At the same time, theoretical and statistical investigations have clearly suggested that the choice of AAs to be encoded by the standard genetic code was nonrandom and adaptive (2–4).
Attractive candidates to trigger the introduction of new AAs into the genetic code are properties that would enable an improved folding of proteins or higher protein stability (2, 5). Surprisingly, though, investigations on the minimum number of different AA types required to build a foldable polypeptide have concluded that, in general, this number is only about 7–13 (5–9) and may be as low as 3 (10). In various examples of engineered proteins with a reduced AA repertoire, function was maintained (5, 8, 11). Hence, protein folding presently does not provide a ready explanation for the modern expansion of the genetic code beyond the 14-AA stage, and, thus, into the “codon capture” phase of genetic code evolution (12).
Interestingly, the last AAs may have been added substantially later than hitherto assumed. Structural analyses of their aminoacyl-tRNA synthetases indicate that the fixation of tyrosine (Y) and tryptophan (W) in the genetic code only occurred at the time of speciation of the first bacterial lineages (13). Hence, these aminoacyl-tRNA synthetases may not have been present in LUCA, the last universal common ancestor, but rather would have been distributed throughout life at a later time by lateral gene transfer (14). Similarly, the capacity to distinguish the late AA methionine from its genetic code neighbor isoleucine has probably developed only after the radiation of life (15). These observations call for selective factors behind the addition of methionine, Y, and W to the genetic code that have only become relevant to life rather lately.
Quantum chemical properties have not been considered as factors potentially significant for the evolutionary choice of AAs, even though they form the basis of chemical reactivity. For example, out of a list of 566 AA properties assembled in a scholarly database (16), none was referring to characters such as orbital energies or excited states. Hence, we have analyzed the quantum chemistry of the 20 proteinogenic AAs, of numerous abiotic AAs found in meteorites, and of various modern reference molecules. We find that the genetically encoded AAs have become systematically “softer” during early evolution, and we trace this distinctive shift toward higher chemical reactivity of the proteinogenic AAs to the advent of molecular oxygen in the habitat of the early cells.
Results and Discussion
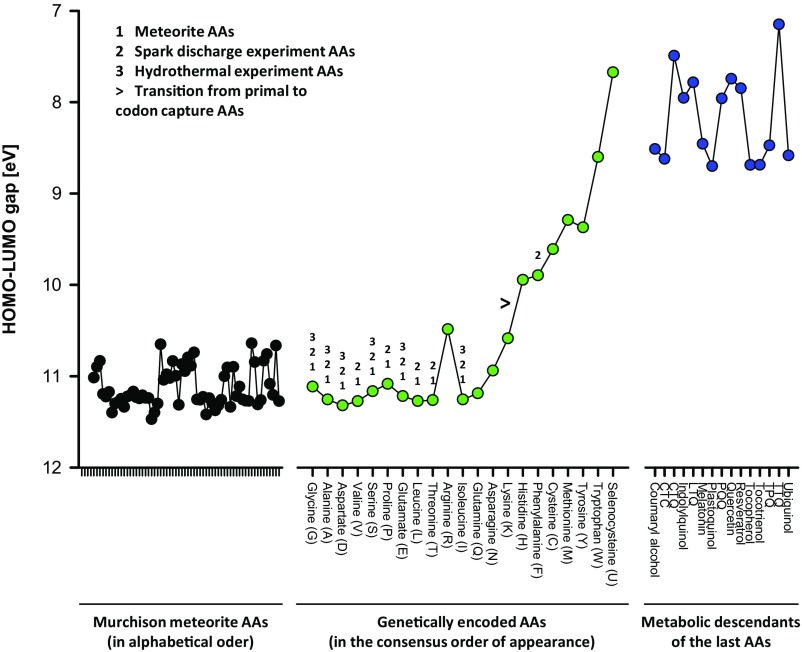
Analyzing the orbital energies of the 20 canonical, proteinogenic AAs and selenocysteine we have found that the energetic distance between the highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO), the HOMO–LUMO gap, exhibited a notable pattern related to the temporal emergence of the AAs in the standard genetic code (Fig. 1): Late additions to the genetic code (5, 17) have substantially smaller gaps than all early additions, such as the encoded prebiotic AAs found in the Murchison meteorite (18) or recovered from the Urey–Miller experiment (19). Emanating from frontier molecular orbital theory (20), the HOMO–LUMO gap is one of the most widely applicable theoretical predictors of chemical reactivity, particularly reflecting the kinetic stability of a compound toward reactions involving electron transfer or rearrangement (21, 22).
Fig. 1.
AA HOMO–LUMO gaps. HOMO–LUMO gaps of 62 Murchison meteorite AAs, 21 genetically encoded AAs, and various metabolic descendants of the shikimate pathway (listed in Table S1) were calculated using semiempirical methods (AM1). The 21 proteinogenic AAs are plotted in the consensus order of their evolutionary appearance according to Trifonov (17), which is in agreement with many single-factor assumptions (e.g., the early introduction of prebiotic AAs, the late introduction of single-codon AAs, or the late introduction of AAs with unusual aminoacyl-tRNA synthetases). Prebiotic AAs identified in cell-free systems (5) and codon capture AAs (12) are indicated. The results of corresponding calculations using a different basis set (PM6) and an ab initio algorithm [Hartree–Fock 6–31+G(d)] are shown in Figs. S1 and S2, respectively. Individual differences notwithstanding, they recapitulate the general principle.
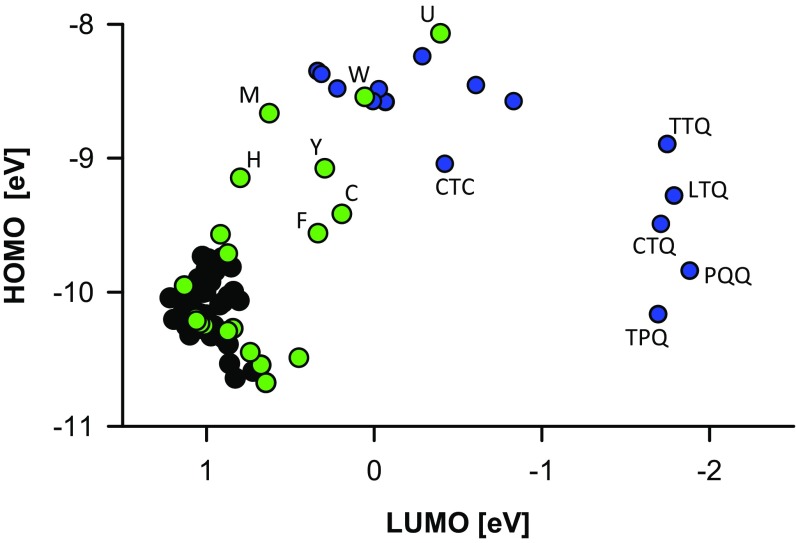
What type of chemical reactivity may explain the evolutionary trend toward smaller HOMO–LUMO gaps? We sought to find reasonable cues in the modern metabolic descendants of the last standard AAs, Y and W, as those descendants might in part represent refinements of the originally selected advantage. Thus, a representative panel of metabolites originating from the shikimate pathway was assembled (Fig. S3). Surprisingly, a rather clear common denominator emerged: The primary function attributable to essentially all examined structures is to catalyze or undergo redox reactions (justified in Table S2). For instance, the role of plastoquinol, ubiquinol, tocopherol, or the quinoid cofactors as committed two-electron and one-electron redox shuttles and redox catalysts is beyond doubt. In terms of their HOMO–LUMO gaps, though, these compounds are quite similar to the last standard AA, W, and the 21st AA, selenocysteine, even if the quinoid cofactors reach absolute orbital energy levels that are unattained by the proteinogenic AAs (Fig. 2). Hence, could the latest AAs indeed be redox cofactors that were integrated into protein structure?
Fig. 2.
AA frontier orbital energies. Individual frontier orbital energies are shown of the same compounds as analyzed in Fig. 1. In this representation, small HOMO–LUMO gaps appear in the upper right edge of the graph. All 62 Murchison meteorite AAs and the first 14 genetically encoded AAs cluster in the lower left edge of the graph. Colors and abbreviations are used as in Fig. 1.
Reviewing the biosynthetic origins of the metabolites of Fig. S3, we noted that all of them involved molecular oxygen (O2) in at least one step (Table S2). In contrast, all steps of the shikimate pathway, including W and Y biosynthesis, proceed anaerobically. In a framework of Darwinian evolution applied to molecules, W and Y would thus precisely occupy the position of random and initially useless “variants” of the primordial shikimate pathway (toward phenylalanine), whose permanent presence (and introduction into the genetic code) would have only been selected for after the appearance of oxygen in the biosphere.
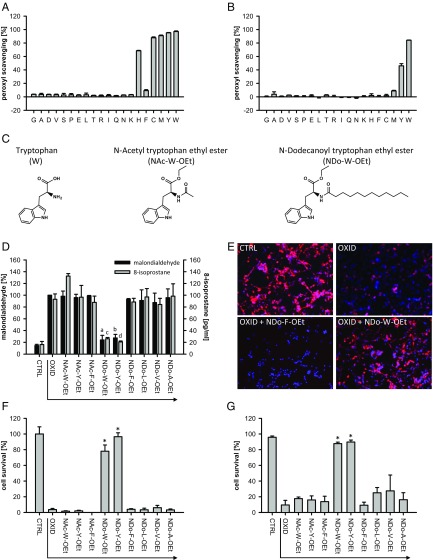
With this insight, we were able to experimentally test the conclusion emerging from HOMO–LUMO gap analysis that the redox reactivity of the AAs increases with their evolutionary position in the genetic code: We investigated their reactivity toward peroxyl radicals, which are species that must have risen substantially after the cellular emergence of oxygen. The results indicate that, in fact, late AAs are much more reactive toward peroxyl radicals than early AAs (Fig. 3A), with the latest additions W and Y being the most effective (Fig. 3B). Notably, reactivity under the employed, plausible conditions (aqueous solvent, 1 mM AA, 37 °C) (23) just begins with histidine, the first of the six presumed codon capture AAs: These AAs have probably required a redefinition of already assigned codons (instead of filling up vacancies) (12), particularly calling for selective mechanisms behind their introduction. We have also tested the reactivity of the proteinogenic AAs toward various other radicals. All AAs showing substantial reactivity were newer additions to the genetic code (Fig. S4). However, not all of the late AAs showed reactivity, as a small HOMO–LUMO gap only indicates basic kinetic accessibility of a compound toward electron-rearranging reactants (20); unfavorable transition state energies or other factors may still preclude reactivity, as exemplified in Fig. 3A on the inert AA phenylalanine with its high radicalization enthalpy (Table S3).
Fig. 3.
Redox reactivity of AAs. (A and B) Effect of the 20 standard AAs in a peroxyl radical scavenging assay. The relative activity of the AAs was tested at ratios 1:3 (A) and 1:2,000 (B) of AA versus radical initiator (n = 3). All AAs below a certain HOMO–LUMO threshold (∼10 eV) demonstrated activity under the conditions in A, except phenylalanine, whose inertia might be related to its unusually high radicalization enthalpy (Table S3). W and Y were the most effective AAs at high initiator/scavenger ratios (B). (C) Chemical structures of W, NAc-W-OEt, and NDo-W-OEt. (D) Inhibition of lipid peroxidation by acetylated or dodecanoylated AA ethyl esters (100 µM). Ferrous iron-induced lipid peroxidation was monitored by measuring the formation of malondialdehyde (F1 = 379, df = 1; F2 = 31, df = 12; aP < 0.001, bP < 0.001 versus “OXID” by post hoc test, n = 3 for compounds, n = 9 for controls) and 8-isoprostane [F1 = 35, df = 1; F2 = 11, df = 12; cP = 0.002, dP = 0.001 versus “OXID” by post hoc test, n = 2 for compounds, n = 4 for controls (all from duplicates)]. (E) Fluorescence microscopic images of the effect of NDo-W-OEt and NDo-F-OEt (10 µM) on the survival of neurons treated with 100 µM tert-butyl hydroperoxide (tBuOOH). Cells were immunostained for microtubule-associated protein 2 (red) indicative of neuronal survival and counterstained with the chromatin marker dye Hoechst 33342 (blue). (F) Quantification of cell survival experiments conducted as in E. Only lipophilic W and Y derivatives elicited cytoprotective activity against peroxide toxicity (F1 = 946, df = 1; F2 = 133, df = 9; *P < 0.001 versus “OXID” by post hoc test, n = 3 for compounds, n = 6 for controls). (G) Survival of fibroblasts treated with 50 µM tBuOOH in the presence of different AA derivatives at 10 µM concentration (F1 = 511, df = 1; F2 = 48, df = 12; *P < 0.001 versus “OXID” by post hoc test, n = 3 for compounds, n = 9 for controls).
The biosynthetic selection of a molecule for a specific property does not necessarily imply its introduction into the genetic code. In general, such an introduction indicates the deployment of a molecule to a multitude of macromolecular sites rather than a small number of catalytic centers. In the latter case, the noncovalent binding of a soluble new cofactor might usually be the preferred solution (24). The specific case of selenocysteine demonstrates that an intermediate number of sites (∼35 in humans) can suffice to trigger an incomplete integration into the genetic code, whereas a full integration of this AA was not achieved (25). Thus, which topological site of the primordial cell might have demanded the addition of a new, redox-active AA (such as W or Y) as a constituent of proteins in substantial amounts, to fulfill a function that high concentrations of the same AA in soluble form would not have attained? We argue that AA introduction into proteins residing in the lipid bilayer was crucial, for two reasons. First, in many modern cells the lipid membrane is the prime topological site of the cell to be collectively attacked and destroyed by peroxyl radicals in a process called lipid peroxidation, a free radical chain reaction (26, 27). Notably, the toxic effects of high oxygen concentrations are thought to be mediated by such free radical chain reactions (28). However, free AAs are excluded from the lipid bilayer, even those AAs usually classified as lipophilic. Second, many of the assumedly refined metabolic descendants of W and Y that are found ubiquitously primarily differ from their precursors in terms of their sizeable lipid anchors, for example ubiquinol, tocopherol, or plastoquinol (Fig. S3). Their phenolic head groups, however, appear relatively retained, indicating that there was no need for major refinement beyond the original phenolic stage in the three cited examples. Hence, we hypothesized that integration into transmembrane proteins was decisive for the eventual introduction of W and Y into the genetic code.
To explore this idea we have employed differentially lipophilic AA derivatives, as represented by acetylated and dodecanoylated tryptophan ethyl ester shown in Fig. 3C. These AA–lipid hybrid molecules were then investigated under “modern” conditions, in classic lipid peroxidation experiments and cellular survival assays under oxidative stress. Exposure of native lipid membranes to Fe2+ (10 µM, recycled by 200 µM ascorbate), an abundant component of primordial, low-oxygen, and rising-oxygen habitats (29), led to a pronounced induction of lipid peroxidation, as measured by the formation of malondialdehyde and 8-isoprostane (Fig. 3D). Lipophilic derivatives of Y and W (NDo-Y-OEt and NDo-W-OEt) indeed accumulated in membranes (Table S4) and significantly attenuated these radical-mediated reactions, whereas aqueous W and Y derivatives had no effect. To verify this biochemical activity in whole cells, living neurons and fibroblasts were directly exposed to an organic peroxide initiating cellular lipid peroxidation. Fluorescence microscopic images (Fig. 3E) and the corresponding quantifications (Fig. 3F) demonstrate that supplying cells with lipophilized Y or W prevented peroxide-induced neuronal death, a phenomenon recently discussed in relation to membrane protein oxidation in neurological disease (30). A similarly robust effect was seen in primary fibroblasts (Fig. 3G).
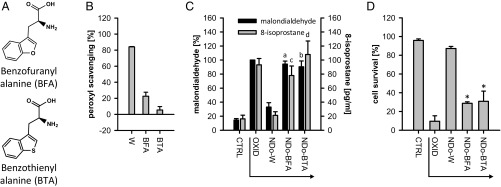
We sought to experimentally probe the precision of nature’s choice of W as the postulated last redox cofactor to be introduced into the standard genetic code. Hence, we investigated the two most plausible alternatives to W in classical terms (size and shape, charge, and hydrophobicity), which have not been introduced into the code, namely benzofuranyl alanine (BFA) and benzothienyl alanine (BTA). Was the evolutionary choice of W specific for its redox reactivity? As shown in Fig. 4, BFA and BTA were found to be much less reactive toward peroxyl radicals (Fig. 4B), and their N-dodecanoyl derivatives were essentially unable to prevent lipid peroxidation (Fig. 4C) or cell death (Fig. 4D), despite the compelling similarity of their side chains to W’s side chain in classical terms (Table S5). In what is a rerun of the situation with phenylalanine, BFA’s and BTA’s relative inertia was unrelated to their HOMO–LUMO gaps but attributable to their very unfavorable radicalization enthalpies (Table S3), which reasserts the role of a small HOMO–LUMO gap as a necessary but insufficient condition for reactivity.
Fig. 4.
Specificity of W. (A) Chemical structures of BFA and BTA. (B) W, BFA, and BTA as scavengers of peroxyl radicals under conditions as in Fig. 3B (n = 3). (C) Inhibition of lipid peroxidation by NDo-W, NDo-BFA, and NDo-BTA under conditions as in Fig. 3D [malondialdehyde: F1 = 379, df = 1; F2 = 31, df = 12; aP < 0.001, bP < 0.001 versus “NDo-W” by post hoc test, n = 3 for compounds, n = 9 for controls; 8-isoprostane: F1 = 35, df = 1; F2 = 11, df = 12; cP = 0.014, dP = 0.001 versus “NDo-W” by post hoc test, n = 2 for compounds, n = 4 for controls (all from duplicates)]. (D) Survival of tBuOOH-treated fibroblasts in the presence of NDo-W, NDo-BFA, and NDo-BTA under conditions as in Fig. 3G (F1 = 511, df = 1; F2 = 48, df = 12; *P < 0.001 versus “NDo-W” by post hoc test, n = 3 for compounds, n = 9 for controls).
The stepwise increases of oxygen in the biosphere have frequently been cited to represent major, fundamental transitions in the history of life (31–34). However, relatively few examples of specific biochemical changes in response to oxygenation have been presented beyond the expectable consequence of directly O2-dependent metabolic transformations (32). Here, we present theoretical, metabolic, and experimental evidence that the genetic code itself has been changed in answer to early local oxygenation. Rising oxygen concentrations forced primordial life to deploy increasingly soft molecules [Pearson’s absolute chemical hardness η is defined (21) as double of a molecule’s HOMO–LUMO gap], to be able to react flexibly to the new challenges imposed by reactive oxygen species that would otherwise disintegrate expensive cellular components designed only for an anoxic habitat (35, 36). With the new AAs, chains of redox reactions starting on protein surfaces could be developed, which would deflect and detoxify radicals in a similar way as present-day organisms (30, 37–39), culminating in the complex one-electron transfer cascades based on W and Y that characterize many modern, oxygen-adapted enzymes (38, 39). For instance, about half of all oxygen-dependent oxidoreductases possess chains of W and Y with three or more members which are stereochemically arranged in such a way as to enable reparative single-electron transfer from the outside (38). Avenues of W and Y seem to play a particularly important role in the control of reactivity of high-potential metal centers in enzymatic oxidation reactions. In case of a prolonged lack of substrate, these activated metal centers are reductively disarmed through chains of Y and W to avoid the looming self-destruction of the enzyme (38).
Intriguingly, a similar strategy of soft AA enrichment was pursued by animal life during the more recent gain in biospheric molecular oxygen about 550 million y ago, at the outset of the Phanerozoic (31, 34). Here, various animal phyla and fungal clades independently changed their mitochondrial genetic code to massively accumulate the soft AA methionine for the purpose of autoprotection (37, 40). Only animal phyla with high metabolic rates and thus high free radical load changed their genetic code assignment (40, 41), and a direct proportionality of aerobic metabolic rate and methionine accumulation (in respiratory chain complex I) has been noted in contemporary animals (41). After all, methionine has long been suspected to represent a genuinely antioxidant AA as it has neither been found to function in enzymatic catalytic cycles nor to be structurally indispensable for its characteristic thioether side chain (37).
Materials and Methods
Reagents.
Natural AAs were from Sigma-Aldrich, and N-acetylated natural AA derivatives were from Bachem. N-dodecanoylated natural AA derivatives were synthesized from free amine precursors as described (42). Enantiopure L-(benzofuran-3-yl)alanine (BFA) and L-(benzo[b]thiophen-3-yl)alanine (BTA) were obtained through chemoenzymatic synthesis starting from benzo[b]thiophene and 1-(2-hydroxyphenyl)ethanone as detailed before (43). N-dodecanoylated BFA and BTA were synthesized as described in SI Materials and Methods.
Reagents and solvents were purchased from Sigma-Aldrich if not otherwise stated. In particular, this supplier provided ascorbic acid, 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), 2,2′-azobis(2-methylpropionamidine) (APPH), tert-butyl hydroperoxide, 2,2-diphenyl-1-picrylhydrazyl (DPPH), fluorescein, galvinoxyl, Hoechst 33342, iron(II) sulfate, potassium peroxodisulfate, and 7,7,8,8-tetracyanoquinodimethane (TCNQ).
Compound abbreviations denote the following: NAc-W-OEt is N-acetyl tryptophan ethyl ester, NAc-Y-OEt is N-acetyl tyrosine ethyl ester, NAc-F-OEt is N-acetyl phenylalanine ethyl ester, NDo-W-OEt is N-dodecanoyl tryptophan ethyl ester, NDo-Y-OEt is N-dodecanoyl tyrosine ethyl ester, NDo-F-OEt is N-dodecanoyl phenylalanine ethyl ester, NDo-L-OEt is N-dodecanoyl leucine ethyl ester, NDo-V-OEt is N-dodecanoyl valine ethyl ester, NDo-A-OEt is N-dodecanoyl alanine ethyl ester, NDo-W is N-dodecanoyl tryptophan, NDo-BFA is N-dodecanoyl benzofuranyl alanine, and NDo-BTA is N-dodecanoyl benzothienyl alanine.
Chemical Calculations.
Quantum-chemical properties of Murchison meteorite AAs, proteinogenic AAs, modern metabolic descendants of the aromatic AAs, and synthetic tryptophan analogs were calculated using the academic software packages MOPAC (Molecular Orbital Package, releases 2003 and 2009) (44), embedded in the commercial software suites Chem3D 9.0 and ChemBio3D 13.0, respectively, from CambridgeSoft (PerkinElmer), and GAMESS (US) (General Atomic Molecular and Electronic Structure System, release of January 12, 2009) (45), embedded in ChemBio3D 13.0. Chemical structures were assembled, geometry-optimized by energy minimization using a molecular mechanics algorithm (a modified Allinger’s MM2 as implemented in Chem3D and ChemBio3D 13.0), and manually inspected for plausibility. Afterward, quantum chemical calculations of electronic energy levels were performed using three different methods.
-
i)
Semiempirical calculations using the MOPAC 2003 AM1 Hamiltonian, via the according MOPAC interface in Chem3D 9.0.
-
ii)
Semiempirical calculations using the MOPAC 2009 PM6 Hamiltonian, via the according MOPAC interface in ChemBio3D 13.0. Compared with AM1, the PM6 basis set provides a parameterization of more chemical elements and a modified treatment of core–core interactions (46).
-
iii)
Ab initio calculations based on the Hartree–Fock method [6-31+G(d) basis set] as implemented in GAMESS (US), via the according GAMESS interface in ChemBio3D 13.0.
Heats of formation and radicalization enthalpies of formation were generally calculated by method i. To this end, the standard enthalpy of formation of each structure was calculated before and after the abstraction of a hydrogen radical. The difference of these enthalpies was defined as radicalization enthalpy. If several chemically plausible sites of potential hydrogen radical abstraction were present in a given molecule, all were explored, and the site yielding the lowest radicalization enthalpy was considered further. Dipole moments of the binuclear AA side chains were also calculated using method i.
The geometric properties, molecular area, and solvent-excluded volume (in water) as well as molecular hydrophobicity values (logP) were estimated with the ChemPropPro tool set implemented in ChemBio3D 13.0.
UV-visible absorption spectra of W, BFA, and BTA were measured in a spectrophotometer (Beckman Coulter). Compounds were dissolved at 1 mM in PBS, pH 7.4.
Chemical and Biochemical Experiments.
Details on the conducted radical assays (peroxyl, TCNQ, galvinoxyl, DPPH, and ABTS radical scavenging) and lipid peroxidation measurements (formation of malondialdehyde and 8-iso prostaglandin F2α) can be found in SI Materials and Methods.
Cell Culture.
Primary cerebellar neurons were prepared from P2 Sprague-Dawley rats and cultivated in Neurobasal medium under a 2% oxygen atmosphere as also present in mammalian brain parenchyma. All other parameters were set as described (30). Analysis of neuronal survival was done by immunocytochemical staining for the neuronal marker protein anti-microtubule-associated protein 2 (MAP-2) following standard protocols (30). Animal procedures were performed in accordance with federal law and the institutional guidelines of the Central Animal Facility of the University of Mainz.
Human lung fibroblasts (type IMR-90; repository number I90) (47) were obtained from the Coriell Institute for Medical Research. The Coriell Institute adheres to Department of Health and Human Services (DHHS) regulations for the protection of human subjects, including informed consent, and distributes samples after submission and review by the Coriell Institutional Review Board of a statement of research intent declaring compliance with DHHS regulations. IMR-90 cells were cultured in DMEM supplemented with 10% heat-inactivated FCS, 1 mM pyruvate, and 1× antibiotic–antimycotic mix (all from Invitrogen). Cells were grown in 100-mm dishes at 37 °C in a humidified atmosphere containing ambient oxygen and 5% CO2. For the survival experiments, fibroblasts at 80% confluency were seeded into 96-well plates in 0.1 mL medium. After 24 h, the cells were pretreated with the relevant AA derivatives (10 µM) for 5 h, before adding tert-butyl hydroperoxide (50 µM). To assess cell survival, the cells were stained with Hoechst 33342 (1 µg/mL) and evaluated by bright-field and fluorescence microscopy.
Statistical Analysis.
All data are presented as mean ± SD. Two-way ANOVA was used for all statistical evaluations provided in this work. In general, factor 1 was the presence of a prooxidant or cytotoxin, and factor 2 was the presence of a potential antioxidant or cytoprotector. The indicated levels of significance in the figures (P values) result from Holm–Sidak’s multiple comparisons procedure executed after the two-way ANOVA under the condition that overall significance (of P = 0.001 or less) was reached in the two-way ANOVA independently for factor 1 and factor 2. Cited n values refer to independent biological or biochemical experiments; technical replicates are referred to as duplicates, triplicates, etc.
Supplementary Material
Acknowledgments
We thank C. Behl, U.-G. Maier, and G. Unden for their support. M.I.T. and F.-D.I. thank the Romanian National Authority for Scientific Research, CNCS - UEFISCDI, for financial sponsorship (Project PN-II-ID-PCE-2011-3-0775).
Footnotes
Conflict of interest statement: Some of the chemical compounds used in this work have been patented by the Max Planck Society, naming author B.M. as one of the inventors (EP 1113795 B1).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717100115/-/DCSupplemental.
References
- 1.Koonin EV, Novozhilov AS. Origin and evolution of the genetic code: The universal enigma. IUBMB Life. 2009;61:99–111. doi: 10.1002/iub.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu Y, Freeland SJ. A quantitative investigation of the chemical space surrounding amino acid alphabet formation. J Theor Biol. 2008;250:349–361. doi: 10.1016/j.jtbi.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Sengupta S, Higgs PG. Pathways of genetic code evolution in ancient and modern organisms. J Mol Evol. 2015;80:229–243. doi: 10.1007/s00239-015-9686-8. [DOI] [PubMed] [Google Scholar]
- 4.Ilardo M, Meringer M, Freeland S, Rasulev B, Cleaves HJ., 2nd Extraordinarily adaptive properties of the genetically encoded amino acids. Sci Rep. 2015;5:9414. doi: 10.1038/srep09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longo LM, Blaber M. Protein design at the interface of the pre-biotic and biotic worlds. Arch Biochem Biophys. 2012;526:16–21. doi: 10.1016/j.abb.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Riddle DS, et al. Functional rapidly folding proteins from simplified amino acid sequences. Nat Struct Biol. 1997;4:805–809. doi: 10.1038/nsb1097-805. [DOI] [PubMed] [Google Scholar]
- 7.Murphy LR, Wallqvist A, Levy RM. Simplified amino acid alphabets for protein fold recognition and implications for folding. Protein Eng. 2000;13:149–152. doi: 10.1093/protein/13.3.149. [DOI] [PubMed] [Google Scholar]
- 8.Akanuma S, Kigawa T, Yokoyama S. Combinatorial mutagenesis to restrict amino acid usage in an enzyme to a reduced set. Proc Natl Acad Sci USA. 2002;99:13549–13553. doi: 10.1073/pnas.222243999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan K, Wang W. What is the minimum number of letters required to fold a protein? J Mol Biol. 2003;328:921–926. doi: 10.1016/s0022-2836(03)00324-3. [DOI] [PubMed] [Google Scholar]
- 10.Davidson AR, Lumb KJ, Sauer RT. Cooperatively folded proteins in random sequence libraries. Nat Struct Biol. 1995;2:856–864. doi: 10.1038/nsb1095-856. [DOI] [PubMed] [Google Scholar]
- 11.Walter KU, Vamvaca K, Hilvert D. An active enzyme constructed from a 9-amino acid alphabet. J Biol Chem. 2005;280:37742–37746. doi: 10.1074/jbc.M507210200. [DOI] [PubMed] [Google Scholar]
- 12.Trifonov EN. The triplet code from first principles. J Biomol Struct Dyn. 2004;22:1–11. doi: 10.1080/07391102.2004.10506975. [DOI] [PubMed] [Google Scholar]
- 13.Yang XL, et al. Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains. Proc Natl Acad Sci USA. 2003;100:15376–15380. doi: 10.1073/pnas.2136794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukai T, Reynolds NM, Crnković A, Söll D. Bioinformatic analysis reveals archaeal tRNA-Tyr and tRNA-Trp identities in bacteria. Life (Basel) 2017;7:8. doi: 10.3390/life7010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones TE, Ribas de Pouplana L, Alexander RW. Evidence for late resolution of the aux codon box in evolution. J Biol Chem. 2013;288:19625–19632. doi: 10.1074/jbc.M112.449249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawashima S, et al. AAindex: Amino acid index database, progress report 2008. Nucleic Acids Res. 2008;36:D202–D205. doi: 10.1093/nar/gkm998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trifonov EN. The origin of the genetic code and of the earliest oligopeptides. Res Microbiol. 2009;160:481–486. doi: 10.1016/j.resmic.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Cronin JR, Pizzarello S. Amino acids in meteorites. Adv Space Res. 1983;3:5–18. doi: 10.1016/0273-1177(83)90036-4. [DOI] [PubMed] [Google Scholar]
- 19.Miller SL. A production of amino acids under possible primitive earth conditions. Science. 1953;117:528–529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- 20.Fukui K. Role of frontier orbitals in chemical reactions. Science. 1982;218:747–754. doi: 10.1126/science.218.4574.747. [DOI] [PubMed] [Google Scholar]
- 21.Pearson RG. Absolute electronegativity and hardness correlated with molecular orbital theory. Proc Natl Acad Sci USA. 1986;83:8440–8441. doi: 10.1073/pnas.83.22.8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aihara J. Reduced HOMO−LUMO gap as an index of kinetic stability for polycyclic aromatic hydrocarbons. J Phys Chem A. 1999;103:7487–7495. [Google Scholar]
- 23.Hren MT, Tice MM, Chamberlain CP. Oxygen and hydrogen isotope evidence for a temperate climate 3.42 billion years ago. Nature. 2009;462:205–208. doi: 10.1038/nature08518. [DOI] [PubMed] [Google Scholar]
- 24.Szathmáry E. Coding coenzyme handles: A hypothesis for the origin of the genetic code. Proc Natl Acad Sci USA. 1993;90:9916–9920. doi: 10.1073/pnas.90.21.9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutteridge JM. Lipid peroxidation and antioxidants as biomarkers of tissue damage. Clin Chem. 1995;41:1819–1828. [PubMed] [Google Scholar]
- 27.Yin H, Xu L, Porter NA. Free radical lipid peroxidation: Mechanisms and analysis. Chem Rev. 2011;111:5944–5972. doi: 10.1021/cr200084z. [DOI] [PubMed] [Google Scholar]
- 28.Fridovich I. Oxygen toxicity: A radical explanation. J Exp Biol. 1998;201:1203–1209. doi: 10.1242/jeb.201.8.1203. [DOI] [PubMed] [Google Scholar]
- 29.Ilbert M, Bonnefoy V. Insight into the evolution of the iron oxidation pathways. Biochim Biophys Acta. 2013;1827:161–175. doi: 10.1016/j.bbabio.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Hajieva P, Bayatti N, Granold M, Behl C, Moosmann B. Membrane protein oxidation determines neuronal degeneration. J Neurochem. 2015;133:352–367. doi: 10.1111/jnc.12987. [DOI] [PubMed] [Google Scholar]
- 31.Holland HD. The oxygenation of the atmosphere and oceans. Philos Trans R Soc Lond B Biol Sci. 2006;361:903–915. doi: 10.1098/rstb.2006.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raymond J, Segrè D. The effect of oxygen on biochemical networks and the evolution of complex life. Science. 2006;311:1764–1767. doi: 10.1126/science.1118439. [DOI] [PubMed] [Google Scholar]
- 33.Crowe SA, et al. Atmospheric oxygenation three billion years ago. Nature. 2013;501:535–538. doi: 10.1038/nature12426. [DOI] [PubMed] [Google Scholar]
- 34.Lyons TW, Reinhard CT, Planavsky NJ. The rise of oxygen in Earth’s early ocean and atmosphere. Nature. 2014;506:307–315. doi: 10.1038/nature13068. [DOI] [PubMed] [Google Scholar]
- 35.Martin W, Russell MJ. On the origins of cells: A hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos Trans R Soc Lond B Biol Sci. 2003;358:59–83, discussion 83–85. doi: 10.1098/rstb.2002.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiss MC, et al. The physiology and habitat of the last universal common ancestor. Nat Microbiol. 2016;1:16116. doi: 10.1038/nmicrobiol.2016.116. [DOI] [PubMed] [Google Scholar]
- 37.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci USA. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gray HB, Winkler JR. Hole hopping through tyrosine/tryptophan chains protects proteins from oxidative damage. Proc Natl Acad Sci USA. 2015;112:10920–10925. doi: 10.1073/pnas.1512704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aubert C, Vos MH, Mathis P, Eker AP, Brettel K. Intraprotein radical transfer during photoactivation of DNA photolyase. Nature. 2000;405:586–590. doi: 10.1038/35014644. [DOI] [PubMed] [Google Scholar]
- 40.Bender A, Hajieva P, Moosmann B. Adaptive antioxidant methionine accumulation in respiratory chain complexes explains the use of a deviant genetic code in mitochondria. Proc Natl Acad Sci USA. 2008;105:16496–16501. doi: 10.1073/pnas.0802779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schindeldecker M, Moosmann B. Protein-borne methionine residues as structural antioxidants in mitochondria. Amino Acids. 2015;47:1421–1432. doi: 10.1007/s00726-015-1955-8. [DOI] [PubMed] [Google Scholar]
- 42.Fincher TK, Yoo SD, Player MR, Sowell JW, Sr, Michniak BB. In vitro evaluation of a series of N-dodecanoyl-L-amino acid methyl esters as dermal penetration enhancers. J Pharm Sci. 1996;85:920–923. doi: 10.1021/js9600787. [DOI] [PubMed] [Google Scholar]
- 43.Podea PV, Tosa MI, Paizs C, Irimie FD. Chemoenzymatic preparation of enantiopure L-benzofuranyl- and L-benzo[b]thiophenyl alanines. Tetrahedron Asymmetry. 2008;19:500–511. [Google Scholar]
- 44.Stewart JJ. 2008 MOPAC2009 (Stewart Computational Chemistry, Colorado Springs, CO). Available at OpenMOPAC.net. Accessed May 13, 2016.
- 45.Schmidt MW, et al. General atomic and molecular electronic structure system. J Comput Chem. 1993;14:1347–1363. [Google Scholar]
- 46.Stewart JJ. Optimization of parameters for semiempirical methods V: Modification of NDDO approximations and application to 70 elements. J Mol Model. 2007;13:1173–1213. doi: 10.1007/s00894-007-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nichols WW, et al. Characterization of a new human diploid cell strain, IMR-90. Science. 1977;196:60–63. doi: 10.1126/science.841339. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto Y, Niki E, Kamiya Y. Oxidation of lipids: III. Oxidation of methyl linoleate in solution. Lipids. 1982;17:870–877. doi: 10.1007/BF02534581. [DOI] [PubMed] [Google Scholar]
- 49.Betigeri S, Thakur A, Raghavan K. Use of 2,2′-azobis(2-amidinopropane) dihydrochloride as a reagent tool for evaluation of oxidative stability of drugs. Pharm Res. 2005;22:310–317. doi: 10.1007/s11095-004-1199-x. [DOI] [PubMed] [Google Scholar]
- 50.Cao G, Alessio HM, Cutler RG. Oxygen-radical absorbance capacity assay for antioxidants. Free Radic Biol Med. 1993;14:303–311. doi: 10.1016/0891-5849(93)90027-r. [DOI] [PubMed] [Google Scholar]
- 51.Melby LR, et al. Substituted quinodimethans. II. Anion-radical derivatives and complexes of 7,7,8,8-tetracyanoquinodimethan. J Am Chem Soc. 1962;84:3374–3387. [Google Scholar]
- 52.Grilj J, Zonca C, Daku LM, Vauthey E. Photophysics of the galvinoxyl free radical revisited. Phys Chem Chem Phys. 2012;14:6352–6358. doi: 10.1039/c2cp23577c. [DOI] [PubMed] [Google Scholar]
- 53.Litwinienko G, Ingold KU. Abnormal solvent effects on hydrogen atom abstractions. 1. The reactions of phenols with 2,2-diphenyl-1-picrylhydrazyl (dpph*) in alcohols. J Org Chem. 2003;68:3433–3438. doi: 10.1021/jo026917t. [DOI] [PubMed] [Google Scholar]
- 54.Re R, et al. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic Biol Med. 1999;26:1231–1237. doi: 10.1016/s0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
- 55.Moosmann B, Behl C. The antioxidant neuroprotective effects of estrogens and phenolic compounds are independent from their estrogenic properties. Proc Natl Acad Sci USA. 1999;96:8867–8872. doi: 10.1073/pnas.96.16.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hajieva P, Mocko JB, Moosmann B, Behl C. Novel imine antioxidants at low nanomolar concentrations protect dopaminergic cells from oxidative neurotoxicity. J Neurochem. 2009;110:118–132. doi: 10.1111/j.1471-4159.2009.06114.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.