Significance
Identifying the drivers of the interindividual diversity of the human immune system is crucial to understand their consequences on immune-mediated diseases. By examining the transcriptional responses of 1,000 individuals to various microbial challenges, we show that age and sex influence the expression of many immune-related genes, but their effects are overall moderate, whereas genetic factors affect a smaller gene set but with a stronger effect. We identify numerous genetic variants that affect transcriptional variation on infection, many of which are associated with autoimmune or inflammatory disorders. These results enable additional exploration of the role of regulatory variants in the pathogenesis of immune-related diseases and improve our understanding of the respective effects of age, sex, and genetics on immune response variation.
Keywords: human immune variation, gene expression, genetics, sex, age
Abstract
The contribution of host genetic and nongenetic factors to immunological differences in humans remains largely undefined. Here, we generated bacterial-, fungal-, and viral-induced immune transcriptional profiles in an age- and sex-balanced cohort of 1,000 healthy individuals and searched for the determinants of immune response variation. We found that age and sex affected the transcriptional response of most immune-related genes, with age effects being more stimulus-specific relative to sex effects, which were largely shared across conditions. Although specific cell populations mediated the effects of age and sex on gene expression, including CD8+ T cells for age and CD4+ T cells and monocytes for sex, we detected a direct effect of these intrinsic factors for the majority of immune genes. The mapping of expression quantitative trait loci (eQTLs) revealed that genetic factors had a stronger effect on immune gene regulation than age and sex, yet they affected a smaller number of genes. Importantly, we identified numerous genetic variants that manifested their regulatory effects exclusively on immune stimulation, including a Candida albicans-specific master regulator at the CR1 locus. These response eQTLs were enriched in disease-associated variants, particularly for autoimmune and inflammatory disorders, indicating that differences in disease risk may result from regulatory variants exerting their effects only in the presence of immune stress. Together, this study quantifies the respective effects of age, sex, genetics, and cellular heterogeneity on the interindividual variability of immune responses and constitutes a valuable resource for further exploration in the context of different infection risks or disease outcomes.
Unraveling the contributions of host and environmental factors to interindividual variability in immune responses is crucial to understand immune pathology (1). Immunological research has largely neglected the concept of interindividual heterogeneity, but there is now growing biomedical interest in studies of the variation of the immune response and its determinants in healthy populations (2)—a strategy known as systems or population immunology (1, 3, 4). Recent cohort-based studies have shed light on how host genetic and nongenetic factors, including environmental variables (e.g., annual seasonality, nutrition, latent infections) and variation of the commensal microbiota, drive the plasticity of immune responses. For example, intrinsic factors, such as age and sex, have an impact on cellular and molecular phenotypes, such as immune cell and protein levels (5–12), and genetic variants also account for a significant fraction of the observed variation of these immune traits (5, 6, 8, 13–16).
In terms of gene expression, immune responses vary markedly between individuals and populations (17–22), but the extent of this variation and its drivers are only beginning to be clarified (1, 3, 23). Recent whole-blood studies have shown that age and sex strongly affect gene expression in the basal state (12, 24, 25). Likewise, genetic variation is an important source of variability in gene expression (20, 26–28). The mapping of expression quantitative trait loci (eQTLs; genetic variants that affect gene expression variation) has become an important approach in translational medicine (29), as regulatory variants are increasingly recognized as contributing to complex disease risk (22, 23, 26–28, 30, 31). eQTLs are particularly informative in studies of immune phenotypes, in which interactions between genetic and environmental factors, such as infection, may be required for phenotypic manifestations (23, 32). In this context, thousands of eQTLs that only appear after immune challenge (i.e., response eQTLs) have been identified over the last years (17–19, 21, 22, 33, 34), establishing putative functional links between expression phenotypes and organismal traits, such as immunity to infection (23, 26, 32). Furthermore, recent data suggest that immune-related response eQTLs play an important role in the genetic architecture of human diseases (35).
Despite the major contribution of systems immunology studies to the increased comprehension of human immune system variation (4), important questions remain to be systematically explored. The investigation of how intrinsic factors impact gene expression variation on infection is missing, yet this is critical to understand the observed inequalities among individuals of different ages and sexes in immune responses and disease risk (36, 37). Furthermore, most studies have focused on isolated cell types treated with single agonists and have not quantified jointly the influence of the genetic and nongenetic drivers of gene expression variation on immune stimulation or infection in a multicellular environment.
In this study, we adopted an integrative approach, combining genetic, transcriptomic, and cytometric data. We generated 7,000 immune transcriptional profiles for whole-blood samples, after stimulation with a wide range of microbes, from 1,000 healthy individuals of European ancestry stratified by age (20–69 y old, 200 per decade) and sex (500 women, 500 men). This balanced experimental design (Fig. S1) provided a unique opportunity to delineate the respective effects of age, sex, and genetic factors and of inherent variation in immune cell populations on the interindividual variability of immune responses to infection. In doing so, our study lays the foundations for future precision medicine clinical strategies that may stratify patient groups based on age, sex, or genetic background.
Results
Distinctive Transcriptional Responses to Bacterial, Fungal, and Viral Challenges.
We stimulated whole blood with three bacteria, Escherichia coli, Staphylococcus aureus, and Bacillus Calmette–Guérin (BCG); a fungus, Candida albicans; a live virus, influenza A virus (IAV); and a superantigen, staphylococcal enterotoxin B (SEB) (Fig. S1). To limit the burden of multiple testing, we quantified the expression of 560 immune-related genes before and after immune stimulation in the 1,000-donor cohort using NanoString hybridization arrays, which produce highly reproducible transcriptional data (38). Furthermore, we measured the proportions of eight major immune cell types (i.e., neutrophils, CD19+ B cells, CD4+ T cells, CD8+ T cells, CD4+CD8+ T cells, CD4−CD8− T cells, natural killer (NK) cells, and CD14+ monocytes) in all individuals by standardized flow cytometry.
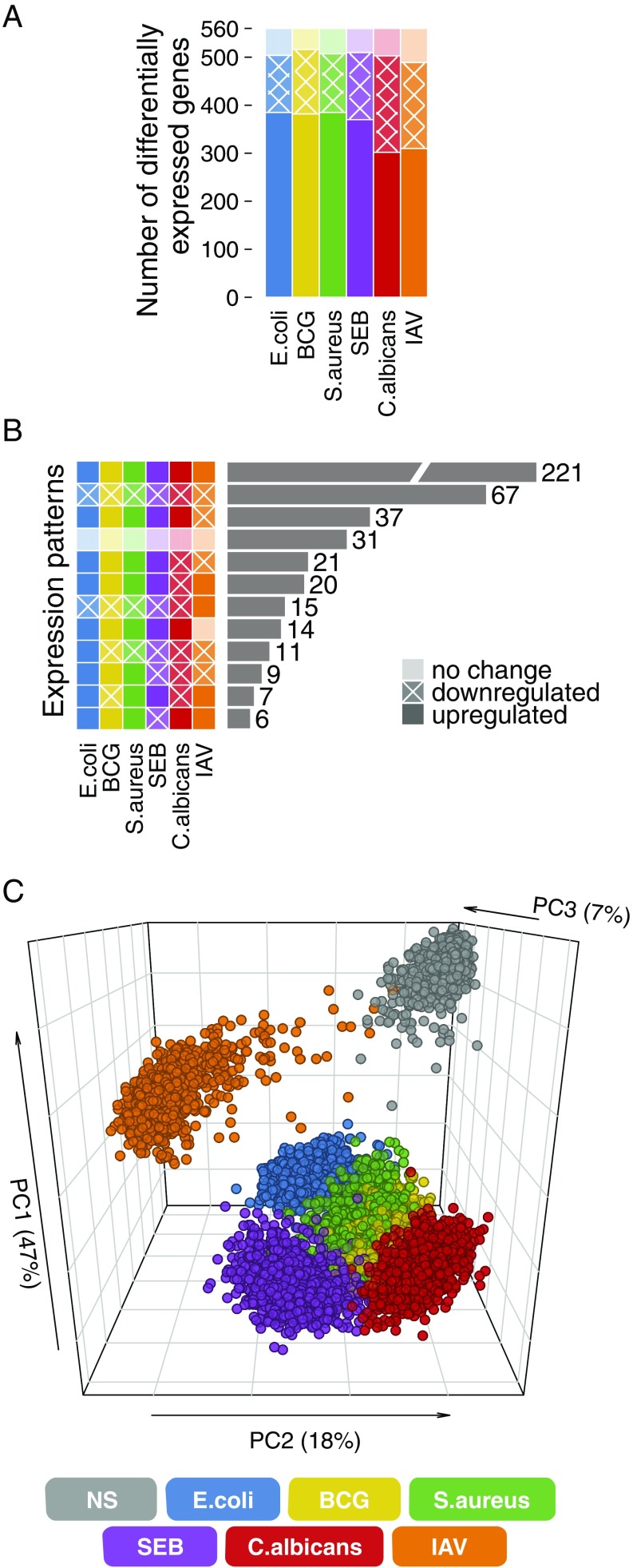
Immune stimulation altered the expression of 87–92% of the genes tested (Dataset S1), with most genes being up-regulated [paired t test; false discovery rate (FDR) < 0.01] (Fig. 1A and Dataset S2). The direction of expression changes was globally shared across stimuli, with IAV inducing the most distinctive response (Fig. 1B). Principal component analysis confirmed these observations (Fig. 1C); principal component 1 (PC1) was driven mainly by genes induced by all stimuli other than IAV, reflecting a bacterial signature, whereas PC2 was driven by IAV-induced genes, reflecting a strong type 1 IFN signature (Fig. S2 A and B). PC2 further distinguished C. albicans-induced genes from those stimulated by E. coli and SEB, whereas PC3 separated the genes induced by E. coli from those stimulated by C. albicans and SEB (Fig. 1C and Fig. S2C). Thus, all stimuli, except BCG and S. aureus, triggered distinguishable transcriptional responses, the largest differences being observed between viral and bacterial/fungal challenges.
Fig. 1.
Distinct transcriptional responses to bacterial, viral, and fungal infections. (A) Number of genes presenting differential expression on immune stimulation. (B) Number of genes presenting common patterns of expression changes across stimulation conditions. Only expression patterns common to at least five genes are presented. (C) Principal component analysis of immune gene expression profiles in the nonstimulated state and on immune simulation. NS, nonstimulated control.
Widespread Effects of Age and Sex on Gene Expression Variation on Immune Stimulation.
We investigated the effect of age on transcriptional responses to immune activation. For each gene, in each condition, we regressed its expression on age, while adjusting for sex, blood cell composition, and technical variables. The expression of 473 genes was linearly correlated with age in at least one condition (FDR < 0.01) (Dataset S3 and Table S1), with 267 being affected by age only on immune stimulation, highlighting the importance of examining induced transcriptional profiles. Unlike previous reports of increased gene expression variance with age in different species (39–41), we found no statistical support for age-dependent changes in expression variance in our age-balanced cohort (Breusch–Pagan test; FDR < 0.01).
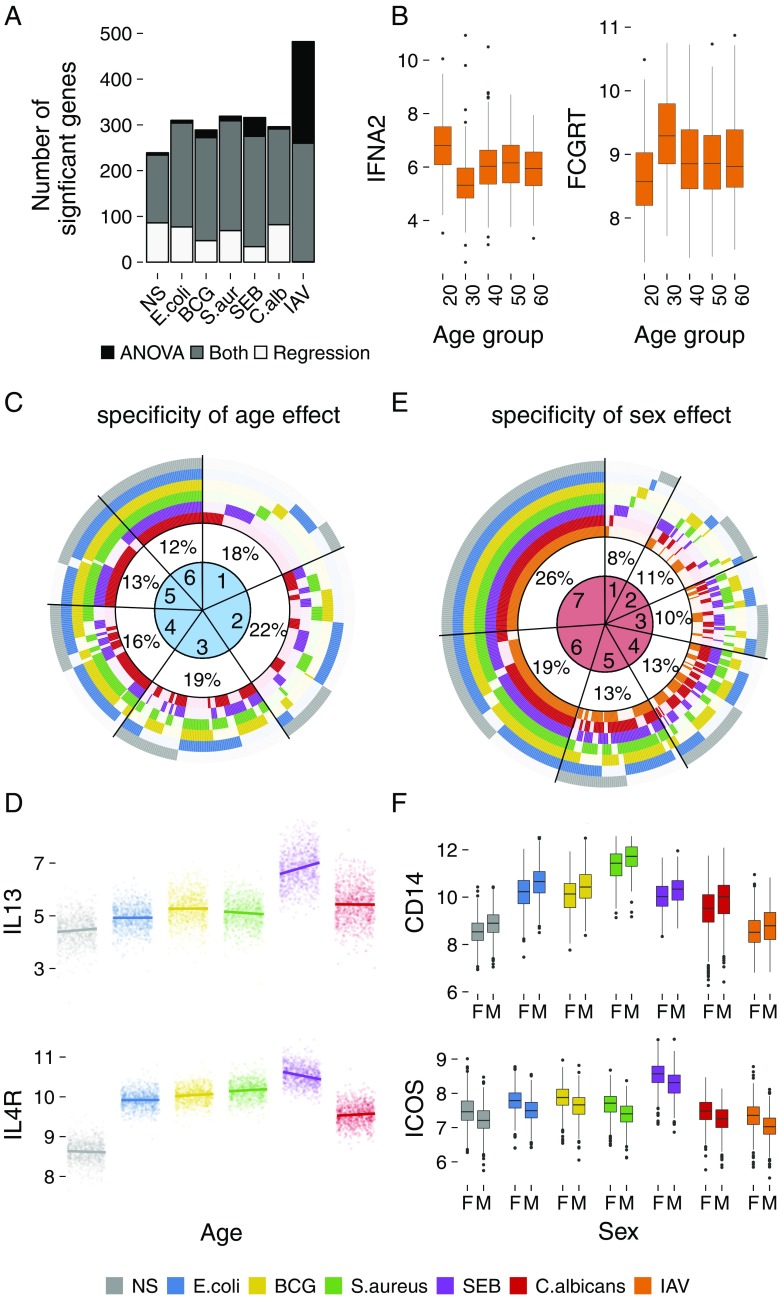
To test for nonlinear associations between gene expression and age, we used ANOVA and stratified the cohort into five age groups. The results obtained were similar to those for the regression analysis for all conditions, except for IAV (Fig. 2A). The 20- to 29-y-old age group displayed a response to IAV that was different from all other groups, this difference being most marked relative to the 30- to 39-y-old age group (Fig. 2B). The relevance of the age groups selected was confirmed by testing alternative age stratification strategies (SI Materials and Methods and Fig. S3). We found 87 and 119 genes displaying increased and decreased expression, respectively, in the 20- to 29-y-old age group compared with 30- to 39-y-old individuals (Tukey Honest Significant Differences test; P < 0.05) (Fig. 2B and Dataset S4). Genes with increased expression were enriched in functions relating to innate immune responses as annotated in the innateDB database (P = 6.5 × 10−3; e.g., type 1 IFNs), whereas genes with decreased expression included genes with known roles in antibody-associated responses (e.g., FCGRT, CR1). Interestingly, the detected age differences echo a recent study that reported similar gene expression differences in H1N1 vaccine recipients under the age of 35 y old (42).
Fig. 2.
Effects of age and sex on the variation of gene expression. (A) Number of genes presenting age-dependent expression changes, as detected by linear regression and ANOVA, in the absence of stimulation and after stimulation. (B) Expression patterns of IFNA2 and FCGRT in response to IAV stimulation across five decades of life. (C) Specificity of age effects on gene expression across conditions. Numbers in the circle sectors (1–6) denote the numbers of stimuli for which the expression of the corresponding genes was age-dependent. The IAV condition was not considered, as it presented a nonlinear association with age. (D) Age-specific (20–69 y) expression of IL13 and IL4R for each stimuli. A significant age association was observed in response to SEB stimulation. (E) Specificity of the effect of sex on gene expression across conditions. Numbers in the circle sectors (1–7) denote the numbers of stimuli for which the expression of the corresponding genes was sex-dependent. (F) Expression differences between men and women for CD14 and ICOS, common to all conditions. Gene expression is presented as normalized gene counts. The legend with color-coding applies to C–F. F, female; M, male; NS, nonstimulated control.
We then explored the stimulus specificity of age effects for each of the 467 genes presenting age-dependent expression (FDR < 0.01, except for IAV because of the nonlinear effect). We found that the effect of age was often stimulus-specific, with 40% of genes presenting age-dependent expression in only one or two conditions (Fig. 2C and Dataset S3). An example is shown for SEB stimulation, which displayed increased IL-13 expression and a parallel decrease in IL-4R (IL-13 receptor) expression as a function of age (Fig. 2D).
As age is associated with increased pathogen infection, in particular with CMV that is known to have a broad influence on immune variation (5), we tested whether CMV seropositivity could explain the detected age-dependent expression changes. When comparing the proportions of variance explained by age in two linear regression models with and without CMV as an independent variable, no significant differences were detected between the models (Fig. S4). Although we cannot rule out that infection with other pathogens could mediate, at least partially, some age-specific effects, our results indicate that infection with CMV does not explain the widespread effects of age on microbial-induced gene expression.
Next, we investigated the influence of sex on immune response variation by regressing gene expression on sex, while adjusting for age, blood cell heterogeneity, and technical variables. We found 509 genes with expression that was sex-dependent in at least one condition (FDR < 0.01) (Dataset S3 and Table S1), with 181 being affected by sex only after stimulation. More genes displayed higher expression in women than in men across all conditions, this difference being significant for BCG, S. aureus, SEB, and C. albicans (test for one proportion; P < 0.01). Furthermore, 33 genes displayed differences in expression variance between the sexes in at least one condition (Levene test; FDR < 0.01) (Dataset S5). When assessing the stimulus specificity of sex effects, we found that these were often shared across stimuli; 45% of genes presented sex-specific differences in expression in more than six conditions (Fig. 2E and Dataset S3). Two key examples are CD14 (encoding the LPS coreceptor), which was more strongly expressed in men, and ICOS (encoding CD278, which down-regulates T-cell activation), which was more strongly expressed in women, for all stimuli (Fig. 2F).
Together, our findings suggest an important contribution of age and sex to immune response variation, with the effects of age on transcriptional variability being highly dependent on the infectious stimulus used and those of sex being largely shared across microbial challenges.
Unraveling the Direct and Indirect Effects of Age and Sex on Gene Expression.
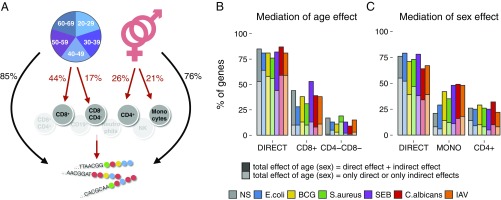
As both age and sex influence blood cell composition (43), we next investigated whether their effects on gene expression are mediated by the eight major cell populations studied using structural equation modeling (44). For each gene with an age-related expression pattern, we built a model with eight indirect effects (one for each cell type) and one direct effect of age, while adjusting for sex, the genetic factors identified below, and technical variables (Fig. S5A). An analogous model was built for sex (Fig. S5B). The outcome of these models is such that, for any given gene, direct and/or indirect effects of age and sex may be observed simultaneously.
We found that, in the absence of stimulation, the expression of 85% of genes was directly affected by age (Fig. 3 A and B and Dataset S6) and that this was the only (i.e., total) effect observed for 53% of genes (Fig. 3B). The expressions of 44 and 17% of genes were influenced by age through a decrease with age in the proportions of CD8+ and CD4−CD8− T cells, respectively (the mediation of the other cell types was not statistically significant). These indirect, cell-mediated effects were often coupled with direct effects of age (Fig. 3B). Thus, only for a very small number of genes could the total effect of age on expression be explained by cellular composition. On immune stimulation, age affected expression through the same cell populations, but the CD8+ T-cell mediation was generally less marked than in the absence of stimulation (Fig. 3B).
Fig. 3.
Structural equation models. (A) Mediation of the effects of age and sex on gene expression by blood cell composition in the nonstimulated control. (B) Fraction of genes displaying expression directly or indirectly affected by age across stimulation conditions. (C) Fraction of genes displaying expression directly or indirectly affected by sex across conditions. NS, nonstimulated control.
Similarly, the expression of 76% of genes was directly affected by sex in the absence of stimulation (Fig. 3 A and C and Dataset S7), this being the only effect observed for 55% of genes (Fig. 3C). The expressions of 26 and 21% of genes were indirectly affected by sex because of a decrease in the proportion of CD4+ T cells in men and a decrease in monocytes in women, respectively. After stimulation, sex effects were also mediated by CD4+ T cells and monocytes, and the mediation effect of monocytes was observed for a larger proportion of genes (29–49%) compared with nonstimulated cells (21%), reflecting that many of our stimuli activate monocytes.
Overall, although specific cell populations can mediate the effects of age and sex on immune-related gene expression, our analyses detected a direct effect of these intrinsic factors for the majority of immune genes affected, suggesting that age and sex effects are often mediated by mechanisms other than those affecting blood cell heterogeneity.
Mapping the Genetic Basis of Transcriptional Responses to Microbial Challenges.
To investigate the contribution of genetic factors to immune gene expression variation, we mapped eQTLs through tests for associations between genome-wide SNPs (minor allele frequency >0.05) and 560 expression traits. Individuals were genotyped using the HumanOmniExpress and the HumanExome BeadChips that, after imputation using the 1,000 Genomes reference panel (45), yielded a final dataset of 5,265,361 SNPs. We used a linear mixed model to account for possible relatedness and population structure in our cohort, which was further adjusted for age, sex, blood cell heterogeneity, and technical variables.
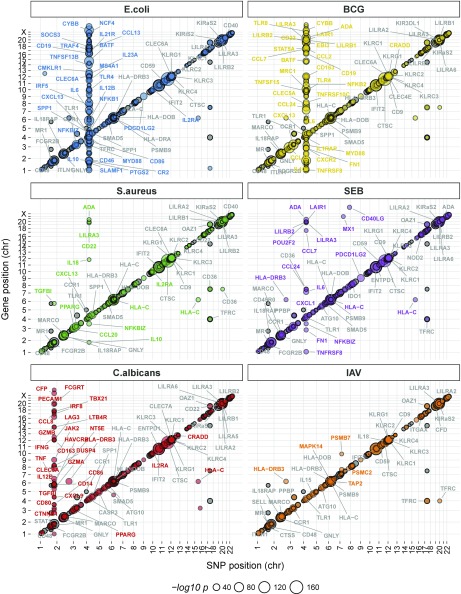
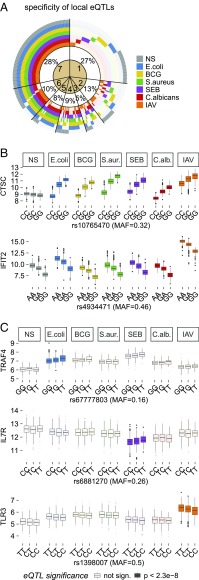
We first mapped local, likely cis-acting eQTLs (within 1 Mb of each gene) and detected 239 genes presenting an eQTL in at least one condition (P < 2.3 × 10−8, Bonferroni-adjusted P < 0.05) (Fig. 4, Datasets S8–S14, and Table S2). A large proportion of eQTLs was either detected in all conditions (28%) or specific to a single condition (27%) (Fig. 5A) as illustrated by the shared eQTLs at CTSC and IFIT2 and the condition-specific eQTLs at TRAF4, IL7R, and TLR3 (Fig. 5 B and C). Notably, the latter cases represent response eQTLs, of which a total of 104 were identified, indicating gene–environment (G × E) interactions.
Fig. 4.
Local and trans-genetic factors associated with gene expression variation. The genomic position of the regulatory SNP is presented on the x axis, whereas that of the gene for which expression variation is associated with the regulatory variant is presented on the y axis. The numbers along the x and y axes are the chromosome numbers. Circles on the diagonals represent genes with expression patterns regulated by local eQTLs, whereas off-diagonal circles correspond to genes with expression patterns regulated in trans. On each panel, the eQTLs detected in the absence of stimulation were plotted first and were then overlaid with eQTLs detected in the corresponding stimulation conditions. Labels are shown only for genes regulated by local eQTLs with a P value <10−32. For trans-eQTLs regulating multiple genes, only the top 30 most significant genes are highlighted. Colored labels indicate genes with significant eQTLs only after stimulation; gray labels indicate the genes with significant eQTLs in both the presence and absence of stimulation. The exact position and rs numbers of both local and trans-associated SNPs are provided in Datasets S8–S21.
Fig. 5.
Stimulus specificity of immune response eQTLs. (A) Specificity of local eQTLs across stimulation conditions. Numbers in the circle sectors (1–7) denote the numbers of stimuli for which the expression of the corresponding genes was associated with a nearby genetic variant. (B) Cases of CTSC and IFIT2 presenting local eQTLs across all seven conditions. The eQTL effect at CTSC differed between nonstimulated and stimulated conditions. (C) Cases of TRAF4, IL7R, and TLR3 presenting local response eQTLs specific to E. coli, SEB, and IAV stimulations, respectively, highlighting G × E interactions. Gene expression is presented as normalized gene counts. MAF, minor allele frequency; NS, nonstimulated control.
We then searched for master regulatory regions by mapping trans-eQTLs (i.e., variants regulating the expression of distant genes or gene networks; >1 Mb away from the transcriptional start/end site). We further verified that the detected trans-effects of each eQTL did not result from heterogeneity in immune cell subpopulations (SI Materials and Methods). In the absence of stimulation, we identified four trans-eQTLs (P < 1.7 × 10−11, Bonferroni-corrected P < 0.05), the strongest of which was located in the FCGR3A/HSPA6 region and regulated GBP5, STAT1, FCGR1A, GBP1, and IRF1. We found that trans-regulation had a stronger effect after stimulation, particularly with E. coli, BCG, C. albicans, and SEB (Fig. 4, Datasets S15–S21, and Table S2). The strongest trans-eQTL was detected for the TLR1/6/10 locus, which regulated 105 genes after stimulation with E. coli, 80 genes after stimulation with BCG, 7 genes after stimulation with S. aureus, and 13 genes after stimulation with SEB. On E. coli stimulation, individuals with the TT genotype (rs4833095, T-allele frequency =0.79) displayed lower expression for many inflammatory response genes (e.g., IL1B, IL6, IL12B) and higher expression for regulatory response genes (e.g., TGFB1, TGFBI, IL1RAP) (Dataset S22). This variant is in strong linkage disequilibrium (r2 = 0.89) in Europeans with an SNP detected as a trans-eQTL after the stimulation of monocytes with Pam3CSK4 (19).
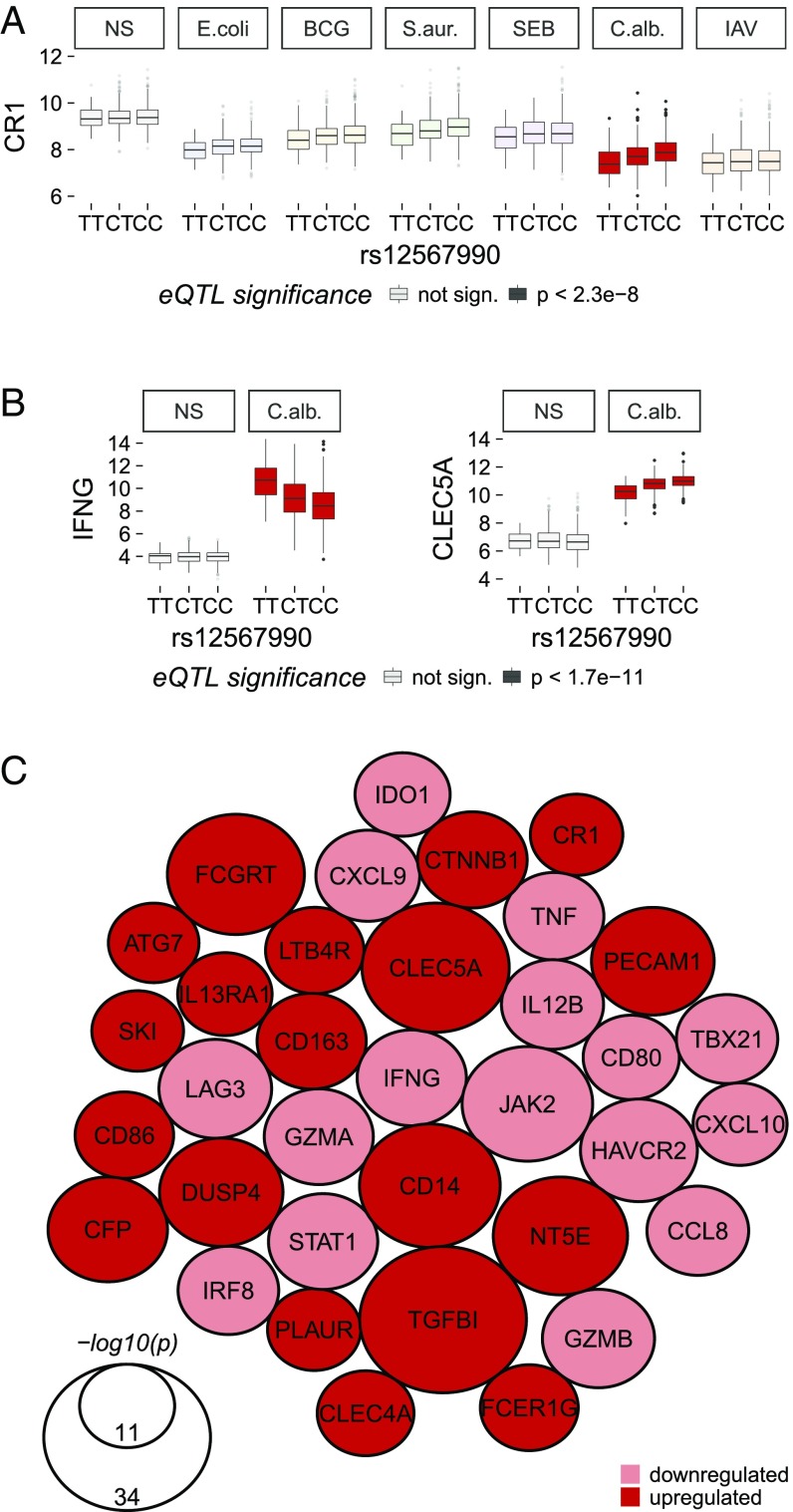
We also identified a strong trans-eQTL located near the CR1 locus, which regulated 34 genes specifically on C. albicans stimulation (Figs. 4 and 6). Individuals with the CC genotype (rs12567990, C-allele frequency =0.81) (Fig. 6C) had lower levels of expression for 16 genes, including genes involved in the IFNγ-related response (IFNG, STAT1, JAK2, CXCL10), and higher expression of 18 genes, including several encoding cell surface proteins (CLECL5A, FCGRT, CD14, IL-13RA1) and signaling molecules (e.g., DUSP4).
Fig. 6.
Stimulus-specific trans-acting eQTL at the CR1 locus. (A) Local eQTL at CR1 acting specifically in response to C. albicans stimulation. (B) rs12567990 was significantly associated, in trans, with the expression of IFNG and CLEC5A only after C. albicans stimulation. Gene expression is presented as normalized gene counts. (C) Network of the 34 genes significantly trans-regulated by the CR1 locus (P < 1.7 × 10−11). The size of the nodes is proportional to −log10(p) of the association between rs12567990 and gene expression. Colors indicate the direction of the change in expression associated with the C allele (frequency = 0.81). NS, nonstimulated control.
As previous studies have reported that genetic variants can impact gene expression at the steady state in an age- or sex-specific manner (46, 47), we explored how these intrinsic factors affect eQTLs on immune stimulation. We included in the linear mixed model an interaction term, separately for sex (SEX × SNP) and age (AGE × SNP), and screened all variants within 1 Mb of each gene. We detected only one significant AGE × SNP interaction affecting the expression of SPP1 after stimulation with E. coli (rs28628889; P = 1.3 × 10−8, Bonferroni-corrected P < 0.05) (Fig. S6A) and no significant SEX × SNP interaction. Our analysis had sufficient power to detect loci displaying strong interactions with sex and age but limited power to detect intermediate and weak interactions (Fig. S6 B and C), suggesting that there are no major differences in the genetic control of immune gene expression between subjects of different ages and sexes.
Collectively, our eQTL analyses revealed that a substantial fraction of the interindividual variation in gene expression can be attributed to local or trans-acting genetic factors, many of which manifest their effects only in the presence of an immunological challenge.
Immune Response eQTLs Are Enriched in Disease Risk Variants.
Because a functional role of regulatory variants in disease risk is increasingly recognized (22, 23, 26, 27, 30, 31, 35), we examined the extent to which the detected eQTLs were enriched in genome-wide association study hits (Dataset S23 and Table S3). We found that local and trans-eQTLs in the nonstimulated and response states were enriched in disease-associated variants with respect to random sets of SNPs sampled from all variants tested in local eQTL and trans-eQTL mapping, respectively (resampling P < 0.05). For example, in the absence of stimulation, we confirmed that NOD2 genetic variation, which has been associated with leprosy (48, 49), affects mRNA levels for this gene (50). We also found that a TNFSF15-TNFSF8 variant, contributing to inflammatory bowel disease risk (51), strongly affects TNFSF8 mRNA levels (P = 3.7 × 10−15), while having only a moderate effect on TNFSF15 (P = 6.5 × 10−4), identifying TNFSF8 as the probable causal gene.
Remarkably, the observed enrichment in disease-causing variants was stronger for response eQTLs than for eQTLs in the absence of stimulation (4.0–7.3× vs. 3.5×, respectively) (Table S3). Several response eQTLs at IRF5, ITGAM, and IKZF1 have been associated with systemic lupus erythematous, the pathogenesis of which is linked to an aberrant regulation of innate and adaptive immune response genes (52). Likewise, three loci associated with self-reported allergy and other immune diseases (51, 53) were response eQTLs (Dataset S23), controlling the expression of SMAD3, PTGER4, and IKZF3 after stimulation with BCG, C. albicans, and IAV, respectively. The strong trans-eQTL that we detected at CR1 (rs12567990) on C. albicans stimulation (Fig. 6) has also been associated with interindividual variability in biomarkers of inflammation (erythrocyte sedimentation rate) (54).
Overall, our results indicate that the identification of the genetic factors controlling transcriptional responses to microbial challenges can shed light on the etiology of organismal traits related to infection, inflammation, and autoimmunity.
Quantifying the Impact of Genetic and Nongenetic Factors on Immune Response Variation.
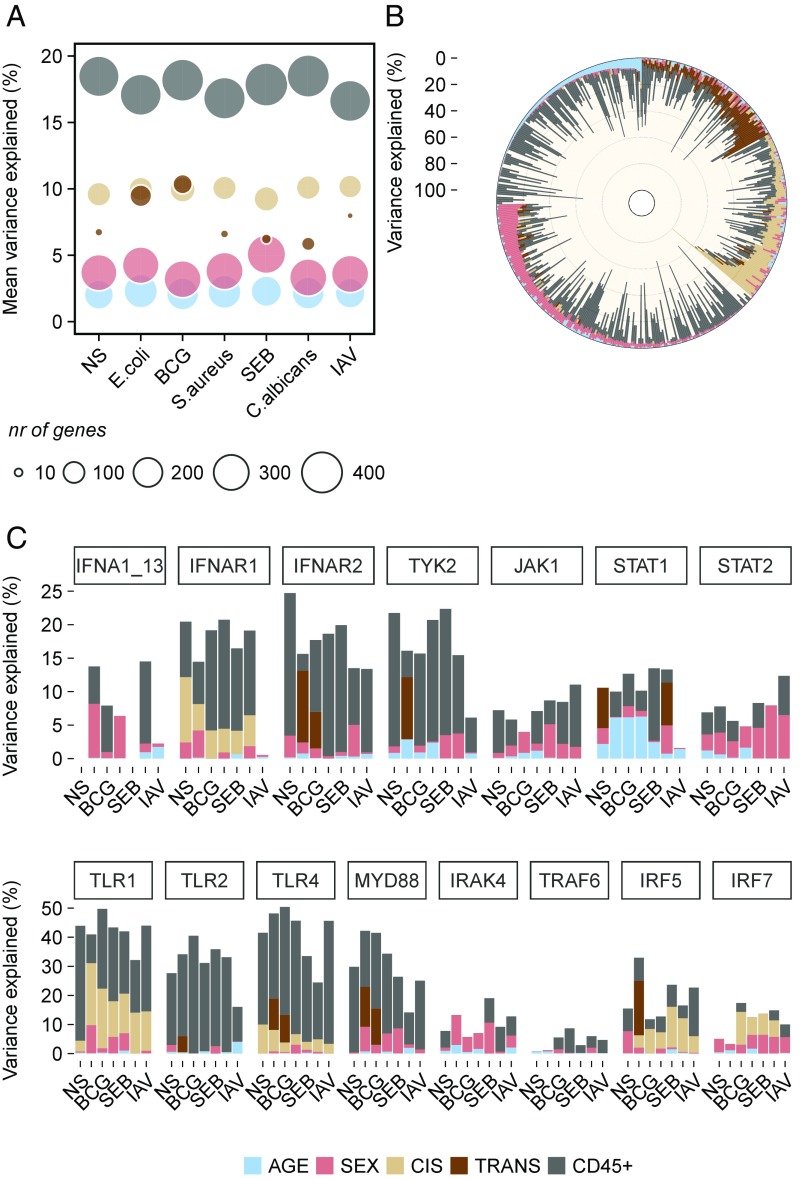
Finally, we quantified the respective impacts of age, sex, blood cell heterogeneity, and genetics on transcriptional response variation. The largest proportion of gene expression variance was explained by heterogeneity in blood cell composition both in number of genes affected (>400 per condition) and in mean proportion of the variance explained (∼18% across genes and conditions) (Fig. 7A and Dataset S24). Age and sex also contributed to the expression variance of a large number of genes (>200 and >300 per condition, respectively), but their individual influences were weak (∼2 and ∼4%, respectively). Conversely, genetic factors affected the expression of a smaller gene set (∼140 per condition for local eQTLs and <40 for trans-eQTLs), but their influence was stronger than those of age or sex: ∼10 and ∼7% for local and trans-eQTLs, respectively. The contribution of trans-genetic factors was stronger for E. coli and BCG than for the other stimuli, affecting ∼100 genes and explaining ∼10% of their expression variance (Fig. 7A). The genetic component was generally the strongest contributory factor other than heterogeneity in blood cell composition for genes with expression variation associated with genetic factors (Fig. 7B and Fig. S7).
Fig. 7.
Expression variance explained by age, sex, genetics, and blood cell composition. (A) Mean variance, across genes, explained by age, sex, genetics, and the proportions of CD45+ cell populations in the absence of stimulation and in the six stimulation conditions. The sizes of the circles correspond to the number of genes affected by each factor in each condition. (B) Proportion of the expression variance explained by age, sex, genetics, and proportions of CD45+ cells for all genes expressed in response to E. coli stimulation. (C) Proportion of the expression variance explained by the different intrinsic and heritable factors for the genes of the type 1 IFN and TLR-MyD88 pathways. The order of stimuli on the x axis is the same as on A. The legend with color-coding applies to A–C. NS, nonstimulated control.
Leaving aside these broad patterns, the respective contributions of nongenetic and genetic factors to immune response variation differed considerably at the individual gene level both within pathways and between stimuli as illustrated by the type 1 IFN and TLR-MyD88 pathways (Fig. 7C). Cellular composition had a stronger impact on expression variation for receptors, such as IFNAR1/2 and TLR1/2/4, than for downstream molecules, with the exception of TyK2 and MyD88. The impact of genetics also varied considerably between genes: the expression of IFNAR1 was under the control of a local eQTL, whereas that of IFNAR2 was regulated by a stimulus-specific trans-eQTL. The impact of genetics was particularly strong on TLR1 expression, where it accounted for up to 21% of the total variance. Age and sex generally had weak effects on gene expression, but several interesting differences were apparent, such as the impact of age on STAT1 and that of sex on STAT2 (Fig. 7C).
Together, our analyses indicate that the main source of immune response variation, at least at the transcriptional level, is whole-blood cellular heterogeneity. Age and sex influence the expression of a wide range of genes, but their effect sizes are moderate, whereas genetic factors acting locally or distantly have stronger effects but on a smaller number of immune genes.
Discussion
Using a systems immunology approach in a 1,000-individual healthy cohort specifically designed for the comprehension of the diversity of the human immune system (55), our study represents a systematic investigation of the respective contributions of age, sex, genetics, and cellular heterogeneity to variation in transcriptional responses to immune activation. We found that the variation of immune cell populations in whole blood was the main driver of interindividual differences in immune responses, accounting for ∼18% of the total variance in gene expression. The effects of age and sex were overall moderate (<5% of the total variance), consistent with reports based on steady-state expression (24, 25, 56), but they were widespread among immune genes and were generally not mediated by immune cell composition. We also found that age effects were more stimulus-specific compared with those of sex. Although future studies with increased power and the inclusion of donors more than 70 y of age may provide a more nuanced view of their respective effects, our results suggest that the microbial specificity of age effects may be driven by environmental exposures that change throughout life, whereas sex effects are more constant. The detected differences of age effects across stimuli echo recent studies, in which immune cell frequencies were found to be more similar between younger than older monozygotic twins (5) and older individuals presented more heterogeneous immunotypes (i.e., cell populations, cell signaling, and antibody responses) than younger donors in unrelated individuals with ages between 8 and 89 y old (11).
We show that genetic factors affect fewer genes than age or sex but that their effect sizes are stronger (∼10% of the total variance) (3, 57). Although the contribution of genetic factors to expression variance can reach even higher values for specific genes or pathways, our findings are in accordance with the moderate influence of genetics in shaping the variation of other immune traits, such as cell proportions (5–7, 15). We nevertheless detected local and trans-eQTLs for 43 and 42%, respectively, of the immune genes studied. About 100 genes presented an immune response eQTL, enabling the identification of G × E interactions in the context of infection.
We also identified master regulators of immune responses, including the trans-eQTL at TLR1/6/10, which we previously detected in monocytes (19). We now extend the description of this trans-effect to whole-blood responses to E. coli, BCG, and to a lesser extent, S. aureus and SEB, highlighting this locus as a major source of immune response variation to bacterial challenges in Europeans. Likewise, the detected trans-eQTL at the CR1 locus reveals a source of variation related to responses to C. albicans. CR1 is a receptor for the C3b and C4b split products, opsonins formed as a result of complement system activation. Both C. albicans and BCG trigger C3 and C4 cleavage, but only opsonized C. albicans engages CR1 (58). We found that CR1 variation regulated the strength of induced IFNγ responses, downstream signaling pathways (JAK2/STAT1), and subsequent chemokine induction (CXCL10/CXCL9). The trans-eQTL SNP also had a local effect on CR1 expression (Fig. 6A), but this effect could not account for all of the variance of the trans-effect, perhaps reflecting differential temporal regulation as previously observed for the IFNB1 gene (18). CR1 variants have been previously associated with differences in erythrocyte sedimentation rate (54, 59), which increases during fungal infections. This highlights the need for studies evaluating the clinical impact of CR1 variation on susceptibility to fungal infections.
Our age- and sex-stratified cohort allowed us to explore if interactions between these intrinsic variables and the numerous genetic factors identified affect immune responses. With the exception of an AGE × SNP interaction for E. coli-induced SPP1 expression, we found no interactions of age or sex with genetic factors, contrary to recent reports (46, 60). This discrepancy may stem from our focus on immune functions and suggests that the effects of genetic variants on immune gene expression are constant across age groups and between sexes. Consistent with this view, previously reported interactions between age, sex, and genetics did not affect immune genes (with the exception of NOD2) (46, 60) or were not identified in whole blood (57).
A nonnegligible fraction of immune response variance remains presently unexplained. Hence, the contribution of other determinants requires additional investigation, including the effects of environmental exposures, epigenetic modifications, interactions with the microbiota, or more complex genetic control (57). In this context, our quantification of the effects of genetic factors on immune response variance (i.e., ∼10% on average) should be considered as a conservative estimate, especially if one considers the many regulatory variants with small effect sizes that our analyses cannot detect.
Our study also presents some limitations. Gene expression variation was assessed at 560 immune genes, providing a partial view of the impact that nongenetic and genetic factors have on gene expression sensu lato. This choice was based on the robust, highly reproducible gene expression measurements generated by the NanoString arrays in whole blood, avoiding technical variability introduced by amplification steps (38). It was also our strategy to deliberately focus on the expression variation of only immune genes to limit the burden of multiple testing. The use of RNA sequencing will make it possible to extend these analyses to a wider array of genes and layers of transcriptional variability, such as potential differential isoform usage on infection. Furthermore, analyses of other immune system measurements after stimulation (proteins and metabolites after microbial challenges, antibody responses to vaccines, etc.) are necessary to provide a more comprehensive view of the different intermediate phenotypes that constitute the human immune system.
Despite these perceived limitations, the use of gene expression variation and its genetic determinants remains a powerful tool for systems immunology studies, as variants affecting gene expression, in particular after immune stimulation (35), are increasingly recognized as contributing to ultimate organismal phenotypes (22, 26, 27, 30, 31). This notion is strongly supported by the cases of G × E interactions detected in this study, as response eQTLs exhibited a stronger enrichment in risk variants for human diseases, such as autoimmune and inflammatory disorders, than eQTLs in the nonstimulated state.
Overall, this study provides an assessment of how intrinsic and genetic factors drive interindividual differences in transcriptional responses to bacterial, fungal, and viral challenges. Our dataset is freely accessible via a web-based browser (misage.pasteur.fr/), making it possible to query and visualize the contribution of these factors to immune response variation. It also constitutes a valuable resource for additional exploration in the context of different infection risks or disease outcomes in human populations.
Materials and Methods
The cohort consists of 1,000 healthy volunteers (500 men and 500 women) aged 20–69 y old equally distributed across five decades of life who were selected based on stringent inclusion and exclusion criteria (55). The study was approved by the Comité de Protection des Personnes—Ouest 6 and the Agence Nationale de Sécurité du Médicament and is sponsored by the Institut Pasteur (ID-RCB no. 2012-A00238-35). The study protocol was designed and conducted in accordance with the Declaration of Helsinki and good clinical practice as outlined in the International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH) Guidelines for Good Clinical Practice (https://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Efficacy/E6/E6_R1_Guideline.pdf), and all subjects gave informed consent. Stimulations were performed on 1 mL whole blood for 22 h using TruCulture tubes (61), and flow cytometry analyses were performed with an eight-color cytometry panel (62). Gene expression was performed using the Human Immunology v2 Gene Expression CodeSet, which contains 594 gene probes that encompass major immune pathways and functions, such as the TLR, Jak-STAT, and MAPK signaling pathways, cytokine–cytokine receptor interactions, apoptosis, and the complement and coagulation cascades. For each gene in each stimulated condition, a paired t test was used to compare expression levels in stimulated and nonstimulated states, controlling for FDR. Seven multiple regression models were built to estimate the effects of age and sex on gene expression. Structural equation modeling (44) was used to investigate the ways in which the different cell populations mediate the effects of age and sex on gene expression. DNA genotyping was performed using the HumanOmniExpress-24 BeadChip and the HumanExome-12 BeadChip (Illumina). After imputation using the 1,000 Genomes Project imputation reference panel (45), a final dataset of 5,265,361 SNPs was obtained. eQTLs mapping was performed with a linear mixed model implemented in GenABEL (63). Interaction effects between variables (genetics, sex, and age) on gene expression were estimated using ProbABEL v.0.4.5 (64).
Detailed information about the experimental methods and statistical analyses may be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We acknowledge Stephanie Thomas for managing the Milieu Intérieur Consortium. This work was supported by the French Government’s Investissement d’Avenir Program, Laboratoire d’Excellence “Milieu Intérieur” Grant ANR-10-LABX-69-01.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genotype data reported in this paper have been deposited in the European Genome-Phenome Archive (EGA; accession no. EGAS00001002460).
2M.L.A. and L.Q.-M. jointly directed this work.
4A complete list of the Milieu Interieur Consortium can be found in the Supporting Information.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714765115/-/DCSupplemental.
Contributor Information
Collaborators: Laurent Abel, Andres Alcover, Hugues Aschard, Kalle Aström, Philippe Bousso, Pierre Bruhns, Ana Cumano, Darragh Duffy, Caroline Demangel, Ludovic Deriano, James Di Santo, Françoise Dromer, Gérard Eberl, Jost Enninga, Jacques Fellay, Magnus Fontes, Antonio Freitas, Odile Gelpi, Ivo Gomperts-Boneca, Serge Hercberg, Olivier Lantz, Claude Leclerc, Hugo Mouquet, Etienne Patin, Sandra Pellegrini, Stanislas Pol, Antonio Raussel, Lars Rogge, Anavaj Sakuntabhai, Olivier Schwartz, Benno Schwikowski, Spencer Shorte, Vassili Soumelis, Frédéric Tangy, Eric Tartour, Antoine Toubert, Marie-Noëlle Ungeheuer, Lluis Quintana-Murci, and Matthew L. Albert
References
- 1.Brodin P, Davis MM. Human immune system variation. Nat Rev Immunol. 2017;17:21–29. doi: 10.1038/nri.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Jager PL, et al. ImmVar project: Insights and design considerations for future studies of “healthy” immune variation. Semin Immunol. 2015;27:51–57. doi: 10.1016/j.smim.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Liston A, Carr EJ, Linterman MA. Shaping variation in the human immune system. Trends Immunol. 2016;37:637–646. doi: 10.1016/j.it.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Davis MM, Tato CM, Furman D. Systems immunology: Just getting started. Nat Immunol. 2017;18:725–732. doi: 10.1038/ni.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodin P, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orrù V, et al. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr EJ, et al. The cellular composition of the human immune system is shaped by age and cohabitation. Nat Immunol. 2016;17:461–468. doi: 10.1038/ni.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ter Horst R, et al. Host and environmental factors influencing individual human cytokine responses. Cell. 2016;167:1111–1124.e13. doi: 10.1016/j.cell.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen-Orr SS, et al. Defective signaling in the JAK-STAT pathway tracks with chronic inflammation and cardiovascular risk in aging humans. Cell Syst. 2016;3:374–384.e4. doi: 10.1016/j.cels.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furman D, et al. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proc Natl Acad Sci USA. 2014;111:869–874. doi: 10.1073/pnas.1321060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaczorowski KJ, et al. Continuous immunotypes describe human immune variation and predict diverse responses. Proc Natl Acad Sci USA. 2017;114:E6097–E6106. doi: 10.1073/pnas.1705065114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blazkova J, et al. Multicenter systems analysis of human blood reveals immature neutrophils in males and during pregnancy. J Immunol. 2017;198:2479–2488. doi: 10.4049/jimmunol.1601855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, et al. A functional genomics approach to understand variation in cytokine production in humans. Cell. 2016;167:1099–1110.e14. doi: 10.1016/j.cell.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, et al. Inter-individual variability and genetic influences on cytokine responses to bacteria and fungi. Nat Med. 2016;22:952–960. doi: 10.1038/nm.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roederer M, et al. The genetic architecture of the human immune system: A bioresource for autoimmunity and disease pathogenesis. Cell. 2015;161:387–403. doi: 10.1016/j.cell.2015.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aguirre-Gamboa R, et al. Differential effects of environmental and genetic factors on T and B cell immune traits. Cell Rep. 2016;17:2474–2487. doi: 10.1016/j.celrep.2016.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee MN, et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairfax BP, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quach H, et al. Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell. 2016;167:643–656.e17. doi: 10.1016/j.cell.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raj T, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nedelec Y, et al. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell. 2016:167, 657–669.e21. doi: 10.1016/j.cell.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 22.Kim-Hellmuth S, et al. Genetic regulatory effects modified by immune activation contribute to autoimmune disease associations. Nat Commun. 2017;8:266. doi: 10.1038/s41467-017-00366-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fairfax BP, Knight JC. Genetics of gene expression in immunity to infection. Curr Opin Immunol. 2014;30:63–71. doi: 10.1016/j.coi.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peters MJ, et al. NABEC/UKBEC Consortium The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6:8570. doi: 10.1038/ncomms9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jansen R, et al. Sex differences in the human peripheral blood transcriptome. BMC Genomics. 2014;15:33. doi: 10.1186/1471-2164-15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 27.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raj T, et al. Common risk alleles for inflammatory diseases are targets of recent positive selection. Am J Hum Genet. 2013;92:517–529. doi: 10.1016/j.ajhg.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibson G, Powell JE, Marigorta UM. Expression quantitative trait locus analysis for translational medicine. Genome Med. 2015;7:60. doi: 10.1186/s13073-015-0186-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am J Hum Genet. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li YI, et al. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quach H, Quintana-Murci L. Living in an adaptive world: Genomic dissection of the genus Homo and its immune response. J Exp Med. 2017;214:877–894. doi: 10.1084/jem.20161942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barreiro LB, et al. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc Natl Acad Sci USA. 2012;109:1204–1209. doi: 10.1073/pnas.1115761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Çalışkan M, Baker SW, Gilad Y, Ober C. Host genetic variation influences gene expression response to rhinovirus infection. PLoS Genet. 2015;11:e1005111. doi: 10.1371/journal.pgen.1005111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alasoo K, et al. 2017. Shared genetic effects on chromatin and gene expression reveal widespread enhancer priming in immune response. bioRxiv:10.1101/102392.
- 36.Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 37.vom Steeg LG, Klein SL. SeXX matters in infectious disease pathogenesis. PLoS Pathog. 2016;12:e1005374. doi: 10.1371/journal.ppat.1005374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urrutia A, et al. Milieu Intérieur Consortium Standardized whole-blood transcriptional profiling enables the deconvolution of complex induced immune responses. Cell Rep. 2016;16:2777–2791. doi: 10.1016/j.celrep.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Viñuela A, Snoek LB, Riksen JA, Kammenga JE. Genome-wide gene expression regulation as a function of genotype and age in C. elegans. Genome Res. 2010;20:929–937. doi: 10.1101/gr.102160.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somel M, Khaitovich P, Bahn S, Pääbo S, Lachmann M. Gene expression becomes heterogeneous with age. Curr Biol. 2006;16:R359–R360. doi: 10.1016/j.cub.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 41.Bahar R, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 42.Sobolev O, et al. Adjuvanted influenza-H1N1 vaccination reveals lymphoid signatures of age-dependent early responses and of clinical adverse events. Nat Immunol. 2016;17:204–213. doi: 10.1038/ni.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melzer S, et al. Reference intervals for leukocyte subsets in adults: Results from a population-based study using 10-color flow cytometry. Cytometry B Clin Cytom. 2015;88:270–281. doi: 10.1002/cyto.b.21234. [DOI] [PubMed] [Google Scholar]
- 44.Gunzler D, Chen T, Wu P, Zhang H. Introduction to mediation analysis with structural equation modeling. Shanghai Jingshen Yixue. 2013;25:390–394. doi: 10.3969/j.issn.1002-0829.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao C, et al. Sex- and age-interacting eQTLs in human complex diseases. Hum Mol Genet. 2014;23:1947–1956. doi: 10.1093/hmg/ddt582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dimas AS, et al. MuTHER Consortium Sex-biased genetic effects on gene regulation in humans. Genome Res. 2012;22:2368–2375. doi: 10.1101/gr.134981.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang FR, et al. Genomewide association study of leprosy. N Engl J Med. 2009;361:2609–2618. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 49.Liu H, et al. Discovery of six new susceptibility loci and analysis of pleiotropic effects in leprosy. Nat Genet. 2015;47:267–271. doi: 10.1038/ng.3212. [DOI] [PubMed] [Google Scholar]
- 50.Naranbhai V, et al. Genomic modulators of gene expression in human neutrophils. Nat Commun. 2015;6:7545. doi: 10.1038/ncomms8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu JZ, et al. International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bentham J, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hinds DA, et al. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet. 2013;45:907–911. doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naitza S, et al. A genome-wide association scan on the levels of markers of inflammation in Sardinians reveals associations that underpin its complex regulation. PLoS Genet. 2012;8:e1002480. doi: 10.1371/journal.pgen.1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas S, et al. Milieu Intérieur Consortium The Milieu Intérieur study–An integrative approach for study of human immunological variance. Clin Immunol. 2015;157:277–293. doi: 10.1016/j.clim.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 56.Whiting CC, et al. Large-scale and comprehensive immune profiling and functional analysis of normal human aging. PLoS One. 2015;10:e0133627. doi: 10.1371/journal.pone.0133627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vinuela A, et al. 2016. Age-dependent changes in mean and variance of gene expression across tissues in a twin cohort. bioRxiv:10.1101/063883.
- 58.Kozel TR, Brown RR, Pfrommer GS. Activation and binding of C3 by Candida albicans. Infect Immun. 1987;55:1890–1894. doi: 10.1128/iai.55.8.1890-1894.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kullo IJ, et al. Complement receptor 1 gene variants are associated with erythrocyte sedimentation rate. Am J Hum Genet. 2011;89:131–138. doi: 10.1016/j.ajhg.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kukurba KR, et al. Impact of the X chromosome and sex on regulatory variation. Genome Res. 2016;26:768–777. doi: 10.1101/gr.197897.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duffy D, et al. Milieu Intérieur Consortium Functional analysis via standardized whole-blood stimulation systems defines the boundaries of a healthy immune response to complex stimuli. Immunity. 2014;40:436–450. doi: 10.1016/j.immuni.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 62.Hasan M, et al. Milieu Intérieur Consortium Semi-automated and standardized cytometric procedures for multi-panel and multi-parametric whole blood immunophenotyping. Clin Immunol. 2015;157:261–276. doi: 10.1016/j.clim.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 63.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: An R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 64.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.