Significance
We have investigated how the directed cell migration toward stiffer extracellular matrix (ECM) is guided by filopodial mechanosensing of the surrounding ECM stiffness. As filopodia bind to the surrounding 3D ECM fibers, local force and displacement are created near the filopodium tip in response to the contraction of filopodia. We have looked into the time rate of changes to the force and the relative displacement between each binding site of ECM fiber (or fluorescent microbeads) and that of the filopodium tip. To quantify the local ECM stiffness sensed by the cell, we have aggregated these effects using two approaches of continuum and discrete mechanics and formulated two different equations for the effective rigidities of local ECM perceived by the cell.
Keywords: filopodia, mechanosensing, ECM, durotaxis, computational modeling
Abstract
Filopodia have a key role in sensing both chemical and mechanical cues in surrounding extracellular matrix (ECM). However, quantitative understanding is still missing in the filopodial mechanosensing of local ECM stiffness, resulting from dynamic interactions between filopodia and the surrounding 3D ECM fibers. Here we present a method for characterizing the stiffness of ECM that is sensed by filopodia based on the theory of elasticity and discrete ECM fiber. We have applied this method to a filopodial mechanosensing model for predicting directed cell migration toward stiffer ECM. This model provides us with a distribution of force and displacement as well as their time rate of changes near the tip of a filopodium when it is bound to the surrounding ECM fibers. Aggregating these effects in each local region of 3D ECM, we express the local ECM stiffness sensed by the cell and explain polarity in the cellular durotaxis mechanism.
Cellular mechanosensing plays a crucial role in tumor growth and cancer invasion since cancer cells invade and metastasize more aggressively for stiffer surrounding extracellular matrix (ECM) (1). For example, the promotion of the malignant progression of breast tumors is related to an elevated level of mechanosensing proteins in the stiffest ECM (2). Cellular mechanosensing in the surrounding ECM also influences many cellular functions such as cell spreading (3, 4), cell migration (5), and stem cell differentiation (6, 7). During cell migration, cells sense the direction and magnitude of complex cues and exhibit multifaceted responses, such as chemotaxis, haptotaxis, and durotaxis responses in the 3D extracellular environment. Durotaxis can be described as a form of cell migration in which the cell is guided by an ECM-rigidity gradient (5) and is thought to be critical to epithelial-to-mesenchymal transition (8) and cancer metastasis (9). Examining how cells sense the stiffness and how they preferentially polarize toward stiffer direction within the surrounding ECM will provide valuable insights into the intricacies of tumor growth and cancer invasion.
It has been found that the formation of long and abundant filopodia is a characteristic of invasive cancer cells (10, 11). Filopodia are finger-like actin-rich plasma membrane protrusions that are linked to enhancement of directed cell migration (12). Filopodia play two critical roles in interacting with the ECM: (i) As sensors they probe the environment (12) and detect chemotactic (13), durotactic (14), and haptotactic (15) cues, and (ii) as actuators they bind to ECM fibers and generate traction forces with actin–myosin machinery (16, 17). In particular, filopodial mechanosensing occurs during the retractile or contractile phase in which the retrograde flow of F-actin is driven by myosin II contractile motion (18). Accordingly, ECM-rigidity–regulated traction fluctuation in focal adhesions (19) was demonstrated in neuronal growth cones. Distinct behaviors of filopodia dynamics at hard and soft ECMs were explained using a stochastic model based on a motor-clutch mechanism at the filopodial shaft (20). The model predicts that the retrograde flow rate of F-actin in the filopodial shaft increases and filopodial traction force decreases with increasing the stiffness of the substrate.
Local ECM rigidity can dynamically vary over time during tumor growth and cancer invasion, since cancer cells cleave the ECM by secreting a group of matrix metalloproteinases (MMPs) to degrade the surrounding ECM. Thus, durotaxis requires filopodia to continuously sample and measure temporal variations of local ECM stiffness (19).
Previously, many modeling approaches have provided insights into the mechanism of durotaxis in cell migration. Such approaches include a simple cell model by considering the contraction of F-actin bundles containing myosin motors mediated by the movement of adhesions (21), an agent-based model with deformable cells and ECM fibers to explain matrix remodeling and durotaxis (22), a 3D numerical model of single-cell migration incorporating the mechanosensing process of the cell as the main mechanism regulating its movement (23), a 3D finite-element model of cell migration by extending pseudopodia as sensors on a 2D substrate (24), and a hybrid cellular Potts and finite-element computational model of collective cell migration with mechanical cell–substrate coupling (25). While providing many new insights, these previous models simplified cellular mechanosensing by considering the migration (i) only on 2D substrates or (ii) within a 3D material but modeled as a continuum without considering the discrete nature of ECM fibers. Consequently, those prior models needed to invoke certain assumptions about the detailed interactions between filopodia and the matrix fibers regarding how these interactions enable stiffness sensing.
A migrating cell probes the stiffness of ECM through filopodia interactions with the surrounding 3D network of ECM fibers. Here we present two methods for characterizing and measuring the stiffness of ECM that is sensed by filopodia; one is based on the theory of elasticity (26), and the other is derived from the elastic energy of discrete ECM fibers. In the existing literature, there is a gap between the ECM stiffness as a mechanical property and that of durotaxis sensed by filopodia penetrating into the 3D ECM. In continuum mechanics, local stiffness is direction dependent, and therefore it is a tensor. In contrast, a migrating cell does not probe the whole tensor quantity, but senses an effective stiffness by extending and contracting or retracting its filopodia in the 3D mesh of matrix filaments of the ECM. The cellular durotaxis mechanism hinges on this “sensed” stiffness through filopodia–ECM interactions. The objective of the present work is to better analyze the stiffness sensed by a cell during durotaxis based on 3D filopodia–ECM interactions. A detailed computational model has recently been made available for predicting dynamics of filopodia interacting with a 3D ECM fiber network (16). In this model each filopodium experiences dynamic changes of forces due to binding and rupture of integrin–ligand clusters, as the filopodium cycles through extension and contraction phases. Here we obtained an aggregate force-displacement relationship in each local region of 3D ECM. Furthermore, we have conducted experiments of filopodia–ECM interactions, using fluorescent beads for observing ECM deformation. Integrating theoretical model and experimental data, we have obtained a formula for estimating the effective total force at the tip of each filopodium and displacements of the surrounding ECM fibers, from which the local sensed stiffness is derived. Using this sensed stiffness, the direction of stiffer ECM is determined, and the polarity of cell migration is explained as the mechanism to follow the direction of sensed stiffer ECM.
This report on a 3D durotactic migration model predicts the guided cell migration based on filopodial mechanosensing of local effective stiffness of cross-linked ECM fibers that are sensed and degraded by the cell.
Results
Filopodia Dynamics and Discrete ECM Mechanics.
The directed cell migration toward stiffer ECM is a multifaceted process where at least four dynamic mechanisms are coupled: (i) intracellular dynamics, (ii) dynamics of filopodia penetrating into 3D ECM, (iii) mechanics of the discrete ECM fiber network, and (iv) reaction–diffusion mass transfer over ECM (SI Appendix, Method S1 and Figs. S1–S4). Among others, dynamic behaviors of filopodia and their interactions with discrete ECM fibers (collagen type 1) are complex, but can be predicted with a computational model based on a few assumptions and observations: (i) The discrete ECM fiber network is composed of viscoelastic ECM fibers [diameter of 41 nm (27) and modulus of 1 MPa (16)] and cross-links, and (ii) filopodia penetration dynamics into 3D ECM consist of four different phases of actions: an outgrowing phase driven by protrusive actin polymerization, a retractile or contractile phase due to motor activity of actin–myosin assemblies along the shaft of filopodia, and a tugging phase due to the attachment of a filopodial tip to nearby ECM fibers (16). Of particular importance is the tugging phase, where each filopodium is tethered to the ECM with multiple focal complexes (FCs) (Fig. 1A) (28). Depending on the strength and spatiotemporal properties of the FC formation, the bond of FCs at the filopodial tip either ruptures or results in the generation of a significant traction force. This phase plays a critical role in coordinating and switching among the diverse phases of filopodia actions, leading to either success or failure of cell migration depending on local ECM conditions.
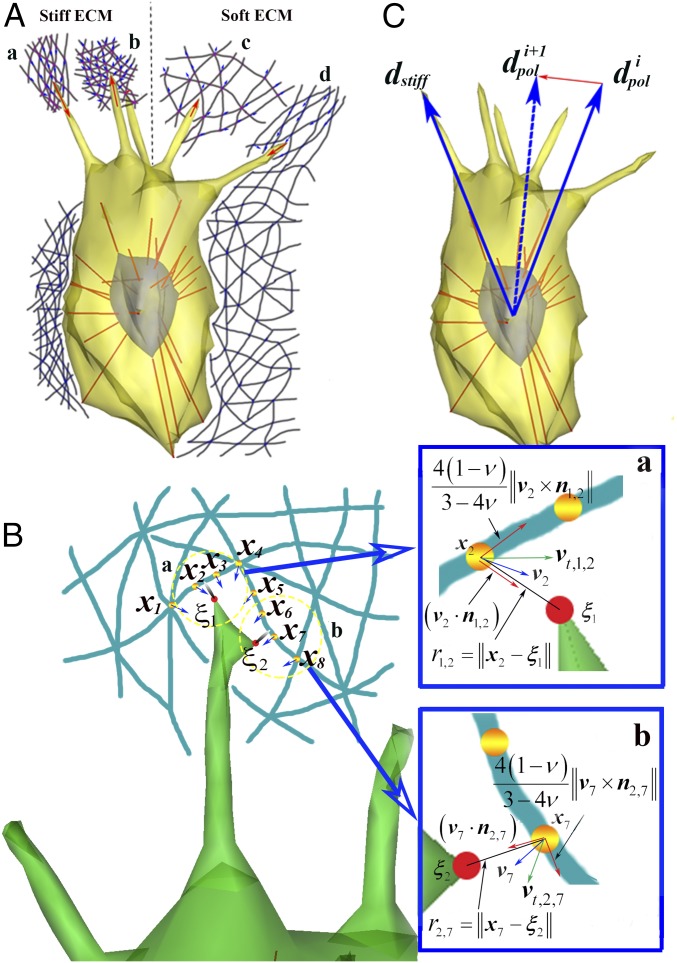
Fig. 1.
Mechanosensing at the filopodial tip. (A) Schematic of directed cell migration toward stiffer local ECM among (a) tensioned stiff ECM, (b) relaxed stiff ECM, (c) relaxed soft ECM, and (d) tensioned soft ECM. (B) A magnified view of A, a showing mathematical descriptions of mechanosensing at the filopodial tip. is a coordinate of the mth ECM fiber node, and is a coordinate of the kth FC at the filopodial tip. Note and directly interact with and respectively. Blue-colored Insets a and b show magnified views of a and b in B, respectively. For example, in Inset a, a blue arrow indicates a velocity vector () of displacement at the second ECM fiber node, and a total speed (; green arrow) has two orthogonal components of an axial speed () and a transverse speed (). (C) A schematic showing temporal variation in the direction of cellular polarity from to toward the direction of stiffer local ECM (). Here superscripts i and i + 1 indicate time points of t and t + Δt, respectively.
Mechanosensing at the Filopodial Tip.
We aim to obtain the local ECM stiffness sensed by a cell through filopodial interactions with ECM fibers. The filopodia simulation described above produces time profiles of ECM fiber movements at their nodes and the force between the nodes and the filopodial tip (Fig. 1). We introduce two expressions for calculating the effective local stiffness and from these data, respectively. First, using a mathematical theory of elasticity (SI Appendix, Method S2) (26), we can find that is proportional to the rate of traction force with time at a filopodial tip (29) and inversely proportional to a total speed of displacement () at the node on ECM fibers. The total speed is defined as a vector summation of the two velocity components at the node, and is given by
| [1] |
where is Poisson’s ratio [taken as 0.5 (30)], v is a velocity vector of displacement at the node on ECM fibers, is a unit vector in the direction of is defined as an axial speed of displacement along the parallel direction of and is defined as a transverse speed of displacement along the orthogonal direction of (Fig. 1B, Inset). Thereby, the equation for calculating can be expressed as
| [2] |
Second, using the elastic energy of discrete ECM fibers (SI Appendix, Method S3), we can find that is also proportional to at a filopodial tip (29) and inversely proportional to a total speed of displacement of the fiber () at the node on the ECM fiber, and the equation for calculating can be expressed as
| [3] |
where is defined as a vector summation of the two velocity components at the fiber and is given by
| [4] |
where and are velocity vectors of two end nodes at the fiber, and and are lengths of unstressed and stressed fiber, respectively. is a unit vector in the direction of fiber orientation.
As shown in Fig. 1B, there are a finite number of neighboring nodes (M) in the vicinity of a filopodial tip which has a finite number of FCs (K). Vectors and respectively, indicate a coordinate and velocity vector of displacement at the mth node of ECM fibers, which are obtained through the simulation of filopodia dynamics and discrete ECM mechanics at time points of t. Using these in Eq. 1 we can obtain the mth total speed where is a unit vector in the direction of and is a coordinate of the kth FC (Fig. 1B). The effective local stiffness can be given by aggregating these data of all of the nodes of ECM fibers within the pore size of ECM which associate with all of the FCs on the filopodial tip. Using a least-squares estimate (SI Appendix, Method S2), it is given by
| [5] |
where is the rate of traction force with time at the kth FC on the filopodial tip, and Similarly, using a least-squares estimate (SI Appendix, Method S3), the effective local stiffness of ECM fibers, is given by
| [6] |
Note that the effective local stiffness in Eq. 5 and the effective local stiffness of ECM fibers in Eq. 6 are based on the force and displacement data that the cell experiences by probing around the ECM fibers with the filopodia. We call and the effective cell-probed stiffnesses (CPSs), using approaches of continuum and discrete fiber in the present work, respectively.
Based on the effective CPS at the filopodial tip, the cell determines the direction of migration (or polarization direction). Polarization direction is important to redefine both leading and trailing edges of the cell. For example, when filopodial locations are changed from the leading edge to the trailing edge as polarization direction is updated, those filopodia have high probabilities of retraction and disappearance. Thus, as filopodia are located within the angle between the direction of filopodial protrusion and polarization direction is less than 45°, lifespans of filopodia are long. In addition, polarization direction also affects the lamellipodium protrusion at the leading edge of the cell. According to ref. 23, the direction of cell polarization, denoted by unit vector is steered in relation to the direction of the stiffest ECM denoted a unit vector pointing in the direction of a maximum of or among multiple filopodia at every time point of t (Fig. 1C). The dynamic transition of polarization direction is expressed as (23)
| [7] |
where is the response time (0.1 min−1) for reorientation of I is an identity matrix, represents tensor product and represents the projection linear transformation.
Characterization of CPS.
First, we aim to simulate mechanosensing tests for the two ECM fiber network models (SI Appendix, Figs. S5–S7) to characterize and compare and Second, we aim to simulate the mechanosensing of a cell interacting with the ECM of varying stiffness (SI Appendix, Figs. S8 and S9). The detailed explanations are found in SI Appendix, Text S1.
Experimental Observation of Filopodial Mechanosensing.
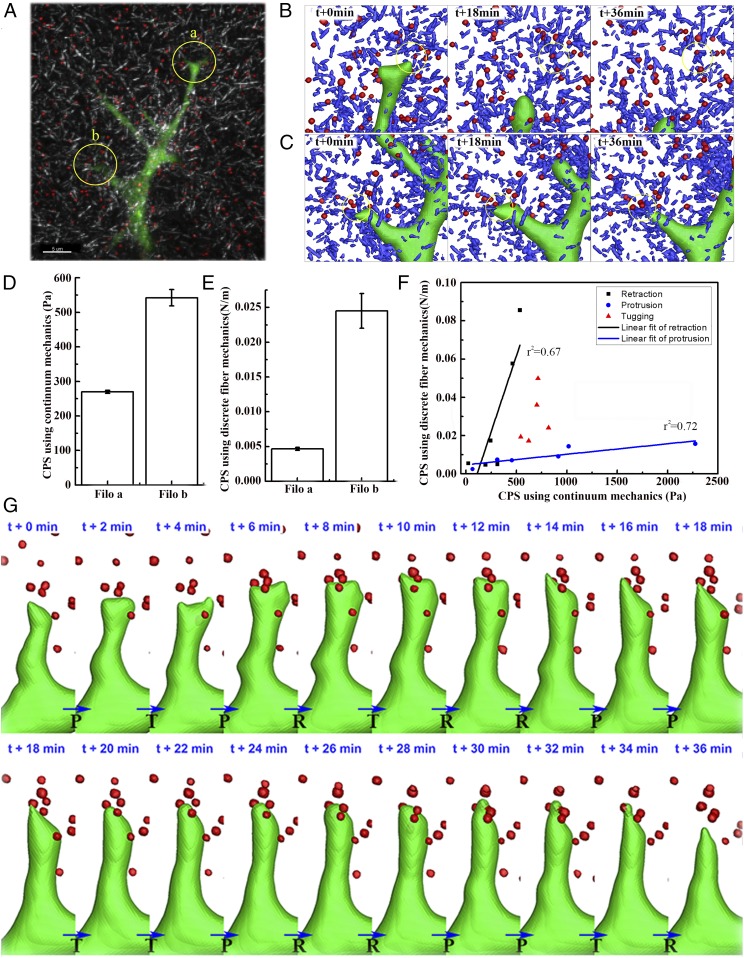
Following successful characterizations of CPS through simulations of mechanosensing tests for soft and stiff ECM models, experiments of filopodia dynamics in the collagen I gel were performed to justify the application of Eqs. 5 and 6 to measuring the local ECM stiffness sensed by filopodia. We have observed an interesting phenomenon which reveals an independent motion of each filopodium; as shown in Fig. 2A, one top filopodium a was retracted, but the other lower filopodium b was protruded (Movie S1). We hypothesized that these distinct motions of filopodia in ECM resulted from the difference of local ECM stiffness sensed by individual filopodium. In addition, it has been reported that the retraction of filopodia is due to zero or weak FC forces at the filopodial tip (31). Thereby, for the comparison of the measured CPS between these two locations of filopodia, each individual filopodium and its surrounding ECM fibers and beads were reconstructed to 3D objects with a time interval of 2 min for 36 min, respectively (Fig. 2 B and C). From these reconstructions, we can note the relaxation of both fibers and beads at filopodium a (beads displace away from the filopodial tip) (Movie S2). In turn, filopodium b mechanically interacts with fibers, which results in fiber densification and remodeling (Movie S3). We observed that beads were bound or embedded into the cross-linked ECM fibers (Fig. 2 B and C), so that each CPS using continuum or discrete fiber mechanics was calculated by aggregating the force and displacement data of beads (Fig. 2 D and E). As a result, both CPSs at filopodium b were found to be higher than those at filopodium a. Furthermore, we compared time-varying force, time rate of change to force, and two CPSs using continuum mechanics and discrete fiber mechanics at filopodium b (SI Appendix, Fig. S10) and found two linear fits of retraction and protrusion (Fig. 2F). These results also imply switching cellular polarity toward stiffer ECM after the competition between filopodia a and b. Note that the determination for the filopodial motion mode of protrusion (P), retraction (R), or transition (T) at every time point was based on forward, backward, or zero motions of filopodium, respectively (Fig. 2G). We found that the simulated data (SI Appendix, Fig. S9C) reproduced the experimental data in a similar trend that the slope of linear fit at the retraction is steeper than that at the protrusion (Fig. 2F and another example shown in SI Appendix, Fig. S11).
Fig. 2.
Experimental measurements of filopodial mechanosensing local ECM stiffness. (A) Three-dimensional confocal images showing filopodia protrusive and retractile motions in GFP-transfected HUVECs, collagen type 1 fiber network (gray), and fluorescent beads (red). (Scale bar: 5 µm.) Three-dimensional shade plots in B and C indicate 3D reconstructions of filopodia (green), surrounding collagen fibers (blue), and beads (red) at time points of 0 min, 18 min, and 36 min. These two kinds of shade plots in B and C are reconstructed from confocal images of two distinct filopodia a and b (yellow circles) in A, respectively. (D and E) Bar graphs showing average CPS using approaches of (D) continuum mechanics and (E) discrete ECM fiber mechanics and SE of means (n = 3) at filopodia a and b. Note that two kinds of CPS in D and E are calculated using Eqs. 5 and 6, respectively. (F) Linear regressions of protrusive (r2 = 0.72) and retractile phases (r2 = 0.67) between calculated two CPSs of the local ECM at filopodium b in D and E, respectively. (G) Series of still shots with time interval of 2 min for 36 min. P, R, and T indicate motion modes of protrusion, retraction, and transition, respectively.
Effect of Separation Distance in Directed Cell Migration Toward Stiffer ECM.
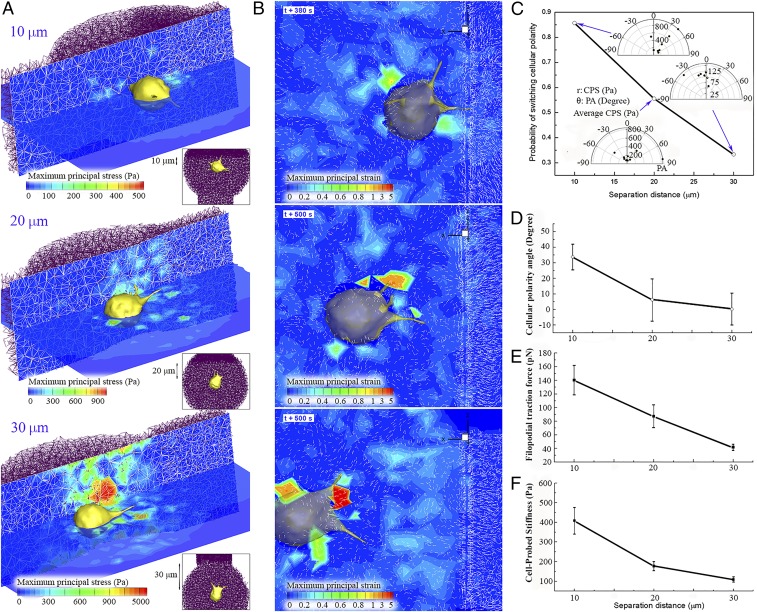
First, three series of simulations were performed and compared to investigate the effect of directed cell migration as a function of separation distance from the centroid of the cell to the sharp interface between stiff and soft ECMs. Computational models of soft and stiff ECM fiber network are constructed with two different pore sizes of 3.0 µm and 1.5 µm, respectively, and identical single collagen fiber diameter of 41 nm. To characterize and compare bulk moduli and pore sizes of these ECM segments, simulations of mechanical stretching tests for each ECM fiber segment were performed, and bulk moduli and pore sizes of soft and stiff ECMs were found to be 2,558 Pa and 4,999 Pa and 3.12 ± 0.57 µm and 1.81 ± 0.34 µm, respectively (SI Appendix, Fig. S12). The cell was initially placed at distances of 10 µm, 20 µm, and 30 µm from the boundary of stiffer ECM, respectively, and the volume exclusion effect of the cell was considered to prevent the ECM fiber from penetrating through cellular and filopodial membranes (SI Appendix, Fig. S13). As a result, ECM fibers are densified and aligned along the surface of the cellular membrane. Note that cellular polarity is initially set to be parallel to the boundary of stiffer ECM; that is, the cellular polarity angle (PA) is zero. Here PA is defined as an angle between a unit vector tangential to the boundary of stiffer ECM (Fig. 3A, Upper direction) and a cellular polarity direction (blue arrow in Fig. 3A). Fig. 4 A and B shows selected examples of contour distributions of maximum principal stress and strain in the domain of ECM as a function of separation distance from the boundary of stiffer ECM to the cell. Significantly higher values of stress are shown at the leading edge of the cell whose cellular polarity directs toward stiffer ECM than those at the trailing edge of the cell. In addition, colored displacement and maximum principal stress contours show that higher stresses (yellow and red) are concentrated near the cell and filopodia and that these stresses rapidly decay out of the cell and filopodia (SI Appendix, Fig. S14). Based on elasticity theory, we find that the strains decay with 1/r2 (SI Appendix, Eqs. S4-15 and S4-16) (32). Contours of maximum principal stress and strain fields in discrete ECM mechanics are drawn by using (i) mathematical theory of elasticity and (ii) discrete deformation gradient methods, respectively (SI Appendix, Method S4 and Figs. S15 and S16).
Fig. 3.
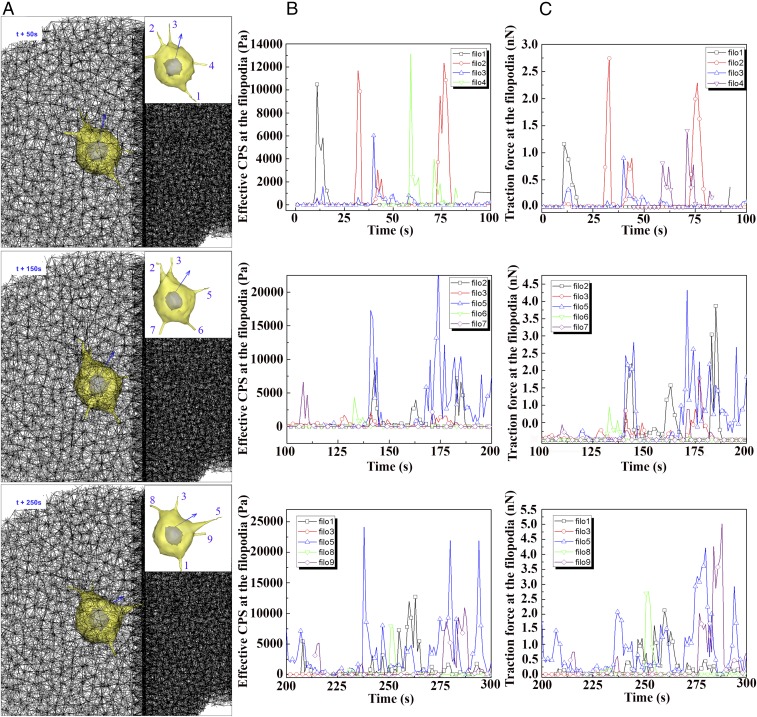
Time evolution of cellular polarity, traction force, and modulus during directed cell migration toward stiffer ECM. (A) Selected still shots of simulated cell migration toward stiffer ECM at three time points of 50 s, 150 s, and 250 s. Each Inset shows cellular and filopodial morphologies, and the direction of cellular polarity (blue arrow). Filopodia are identified with numbers for the analysis of time-varying CPS and traction force in B and C. Here diameter, length, and number of filopodia are 0.6 µm, 3 µm, and 5, respectively. The three graphs in B and C show time-varying CPS and traction force at identified filopodial tips in A at three time ranges, 0∼100 s, 100∼200 s, and 200∼300 s, respectively.
Fig. 4.
(A) Selected examples of maximum principal stress contour slices along both the migrating cell and the domain of ECM as a function of separation distance from the surface of stiffer ECM in which the cell migrates from three different separation distances of 10 µm, 20 µm, and 30 µm. White meshes on the vertical slice indicate ECM fiber network. Each Inset in A also indicates corresponding separation distance from the surface of stiffer ECM to the centroid of the cell. Here diameter, length, and number of filopodia in three cases are 0.6 µm, 3 µm, and 5, respectively. (B) Corresponding contour plots of maximum principal strain in the domain of ECM. White vectors in the plot indicate displacement vectors at the ECM fiber nodes. (C) Probability of directed cell migration toward to stiffer ECM fiber network as a function of separation distance. Three scatter plots in polar coordinates of r and θ represent distributions of time-averaged CPS at the filopodia (r) and time-averaged PA of the cell (θ). Note that PAs of 0°, 90°, and −90° indicate initial direction of cellular polarity and directions of stiff and soft ECM, respectively. (D–F) Variations of (D) time-averaged cellular PA, (E) traction force, and (F) CPS at the filopodial tip under three different separation distances of 10 µm, 20 µm, and 30 µm. All error bars in graphs indicate SE of mean.
Furthermore, comparing the probability of switching cellular polarity toward stiffer ECM with diverse separation distances to the boundary of stiffer ECM, we find that this probability decays as the separation distance from the surface of stiffer ECM increases (Fig. 4C). It is determined as the occurrence of polarity switching when the time-averaged cellular PA becomes positive. Accordingly, the probability is evaluated by counting the number of cells directed toward stiffer ECM: seven of eight, five of nine, and two of seven for the separation distances of 10 µm, 20 µm, and 30 µm, respectively (see scatter plots in Fig. 4C). Also, time-averaged cellular PAs are found to decrease with increasing separation distances of 10 µm, 20 µm, and 30 µm (33.7 ± 8.4°, 6.2 ± 13.6°, and 0.2 ± 10.3°, respectively) (Fig. 4D). Interestingly, both time-averaged traction forces and effective CPS at filopodial tips are also found to be highest in the case of the shortest separation distance, 10 µm (∼140 pN and 400 Pa) (Fig. 4 E and F).
Time Evolution of Cellular Polarity, Traction Force, and Cell-Probed Stiffness.
Comparing the series of simulated data of ECM fiber and filopodia provides insights into the time evolution of the cellular polarity, CPS, and traction force profiles during the directed cell migration toward stiffer ECM. Among the three cases of separation distances simulated (Fig. 4 A and B), the separation distance of 10 µm is chosen to analyze dynamic behaviors of filopodia since the time-averaged cellular PA is the highest. Here, the response time for switching the cellular polarity toward stiffer ECM tends to be shorter. As shown in Fig. 3A, three selected plots of ECM fiber network at time points of 50 s, 150 s, and 250 s show how fast the cellular polarity (blue arrow on the nuclear membrane) is switched toward stiffer ECM; PAs at 50 s, 150 s, and 250 s are 11.2°, 30.9°, and 57.3°, respectively (SI Appendix, Fig. S17). As filopodia mechanosense the highest value of CPS in the surrounding ECM, both the cellular polarity and the leading edge of the cell are rapidly switched toward stiffer ECM. Fig. 3 B and C shows temporal variations of CPS (Fig. 3B) and traction force (Fig. 3C) at the filopodial tips in three time ranges of 0–100 s (Top), 100–200 s (Middle), and 200–300 s (Bottom), respectively.
First, at the time range from 0 s to 100 s, cellular PA changes from 0° to 16.7° (SI Appendix, Fig. S17) since both the highest peak values of CPS and traction force are found to be 13 kPa and 0.8 nN at “filo 4” (Fig. 3 B and C), which is located on the trailing edge of the cell, at time 59 s. Although filo 4 is retracted later, it turns out to be instrumental in switching cellular polarity toward stiffer ECM since the duration of high CPS is longer at filo 4 than at any other filopodia over the time range from 0 s to 100 s.
Second, at the time range from 100 s to 200 s, cellular PA is further changed from 16.7° to 43.7° (SI Appendix, Fig. S17). At this time range, the most significant filopodia for switching the cellular polarity is filo 5 since the highest values of both CPS and traction force are recorded as 5∼20 kPa and 1∼4 nN, respectively. Interestingly, filo 5 started mechanosensing the boundary of stiffer ECM at the time point of 170 s, which results in highest values of both CPS and traction force (Movie S4).
Finally, at the time range from 200 s to 300 s, cellular polarity is completely switched toward stiffer ECM as cellular PA is changed from 43.7° to 66.0° (SI Appendix, Fig. S17). Thereby, the leading edge of the cell is also switched so that filo 5 and filo 9 are located on the leading edge of the cell. At this time range, filo 5 is the most significant filopodium among any filopodia since it further penetrates into stiffer ECM and gains substantial traction force ranging from 1 nN to 4.5 nN. Additionally, at filo 9, the highest values of CPS and traction force are observed at the time point of 284 s.
Three Simulated Examples of Durotaxis.
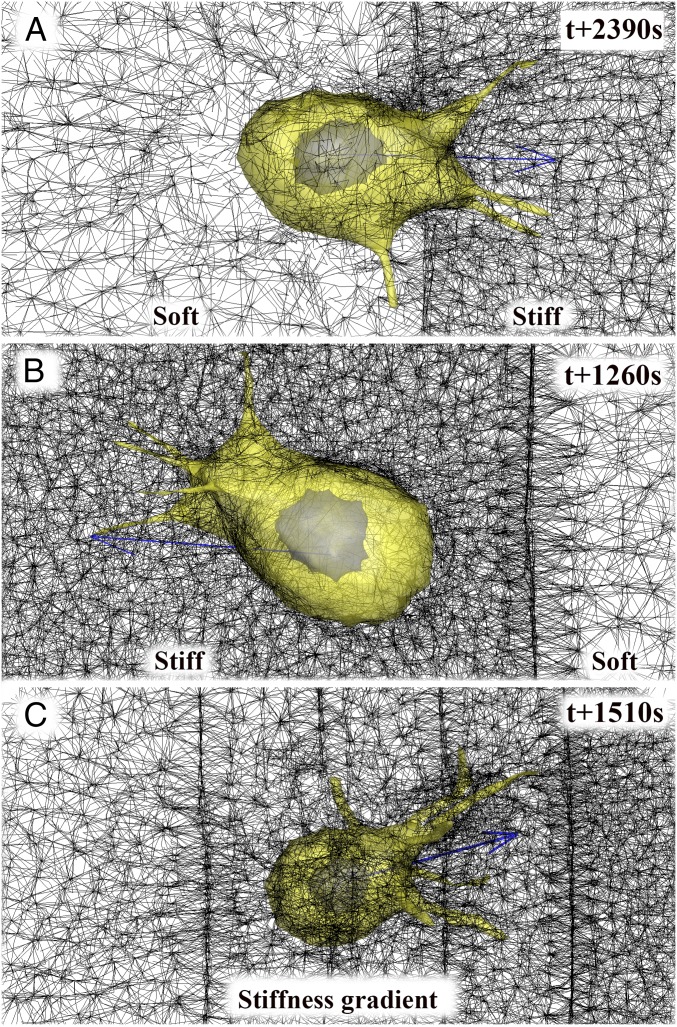
To test our computational model further, three cases of simulations on durotaxis were performed: (i) cell migration from soft to stiff ECM (Fig. 5A), (ii) cell migration from stiff to soft ECM (Fig. 5B), and (iii) cell migration along the stiffness gradient (Fig. 5C). Similar to previous experimental studies of durotaxis (5), we find that the cell model crosses the sharp interface when migrating from the soft to the stiff ECM (Movie S5), but the cell model steers away from the sharp interface when migration is from the stiff to the soft ECM (Movie S6). To confirm our filopodial mechanosensing using CPS methods, we further considered and generated a sharp stiffness transition at the interface with a gradual change of pore size from the softest ECM (3.0 µm) to the stiffest ECM (1.5 µm) and simulated cell migration along the stiffness gradient. As a result, we find that the cell model migrates toward stiffer ECM as cellular polarity directs toward stiffer ECM (Movie S7).
Fig. 5.
Three examples of durotaxis. (A–C) Still shots of (A) simulated cell crossing the sharp interface at the time point of 2,390 s when the cell starts migrating from the soft ECM, (B) simulated cell steering away from the sharp interface at the time point of 1,260 s when the cell starts migrating from the stiff ECM, and (C) simulated cell migration along the stiffness gradient at the time point of 1,510 s.
Cancer Cell with Numerous Filopodia.
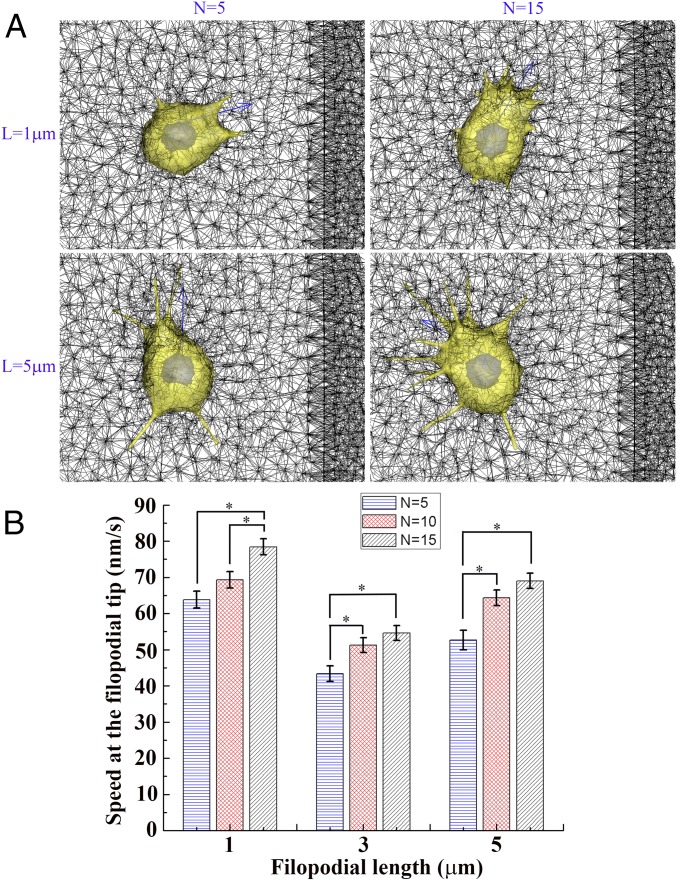
Abundant filopodia in cancer cells have been known to be a critical characteristic of aggressive cancer cells which invade into tissues. In addition, increased filopodia formation has been shown to promote migration (33), and increased motility is highly related with increased fascin up-regulation in the filopodia that bundles actin filaments to protrude (34). Thereby, to quantify the invasiveness of cancer cells with long (or short) and abundant filopodia, we further simulated cancer cell invasion into stiffer ECM to investigate how numbers and lengths of filopodia affect filopodia penetration dynamics into ECM and cell motility. Fig. 6 and SI Appendix, Fig. S18 show the simulations for filopodia numbers (N) of 5, 10, and 15 and the unstressed lengths of filopodia (L) of 1 µm, 3 µm, and 5 µm. Here initial cancer cell locations were identically set to be 20 µm from the boundary of stiffer ECM to quantitatively compare time-averaged speeds of filopodial tips models with various filopodial numbers and lengths. As a result, we find that, for all of the filopodial lengths of 1 µm, 3 µm, and 5 µm, the time-averaged speed at the filopodial tip is higher as the number of filopodia is increased. Probability distributions of the speed at filopodial tips show peak shifts toward higher speeds as filopodial number is increased more for all of the filopodial lengths of 1 µm, 3 µm, and 5 µm (SI Appendix, Fig. S19). However, in cases of filopodial lengths of 3 µm and 5 µm, the speed at the filopodial tip is significantly increased when N is varied from 5 to 10, but the speed at the filopodial tip is slightly increased when N is varied from 10 to 15 (Fig. 6B and SI Appendix, Fig. S19). Additionally, a graph of the average speed at the filopodial tip vs. three different lengths of filopodia with the same number of filopodia shows a biphasic distribution with a minimum value at the unstressed filopodial length of 3 µm, and similar distributions are also found under all three identical numbers of filopodia (Fig. 6B). Our simulations of cancer cell models indicate that speeds at filopodial tips are increased more as the filopodial length is either increased from 3 µm to 5 µm or decreased from 3 µm to 1 µm. However, interestingly, the cancer cell model with short and abundant filopodia (L = 1 µm, n = 15) turns out to be the most aggressive since its average speed at the tip of filopodia is the highest among all cases of cancer cell simulations (Movie S8).
Fig. 6.
Cancer cell simulations with numerous and long (or short) filopodia. (A) Selected plots of simulation for the directed cancer cell migration toward stiffer ECM by tuning unstressed filopodial lengths (L) of 1 µm and 5 µm (rows) and filopodial numbers (N) of 5 and 15 (columns) at the time point of 500 s. Blue arrow indicates the direction of cellular polarity. Pore sizes of soft (Left) and stiff (Right) are 3 µm and 1 µm, respectively. (B) Bar graphs showing time-averaged speeds and SE of means at the tip of filopodium for nine cases under three different unstressed filopodial lengths of 1 µm, 3 µm, and 5 µm and three different filopodial numbers of 5, 10, and 15 [*P < 0.05, n = 85, 155, and 122 (L = 1 µm, and n = 5, 10, and 15); n = 72, 79, and 83 (L = 3 µm, and n = 5, 10, and 15); and n = 45, 97, and 116 (L = 5 µm, and n = 5, 10, and 15), one-way ANOVA, posthoc Tukey’s test].
Discussion
In this report, we introduce a method of elucidating the local stiffness of ECM sensed by a cell. The method is twofold. One is to compute time series data of force and displacement that a cell experiences through filopodial interactions with surrounding discrete ECM fibers (SI Appendix, Method S1). The other is an elasticity-theoretic model relating force to displacement: two kinds of parametric relationships consisting of (i) Young’s modulus and Poisson’s ratio derived from elasticity theory (SI Appendix, Method S2) and (ii) stiffness of ECM fiber derived from the elastic energy of the discrete ECM fiber (SI Appendix, Method S3). The force-displacement data are then synthesized and reduced to a single scalar quantity delineating the effective CPS of local ECM fibers, and The CPS is not merely the physical stiffness of the ECM fiber network, but the one that is experienced by the cell through the characteristic filopodia movements in interacting with ECM fibers. The importance of ECM stiffness has been addressed by many authors, yet its real meaning in conjunction with the migratory cell behaviors has seldom been addressed. Thus, the presented method provides the foundation of stiffness probed by the cell and, using this CPS, polarity in cell migration is explained.
First, the presented method was characterized through a series of simulations for mechanosensing tests in soft and stiff ECM models (SI Appendix, Fig. S7), and two distinct linear regions were found in their hysteresis curves of and One is a region of force buildup (or protrusion of filopodia), and the other is a region of force dropdown (or retraction of filopodia) (SI Appendix, Figs. S5E and S6E). The slope of linear fit at the force drop-off was found to be steeper than that at the force buildup. However, it should be noted that a linear correlation at the force buildup region becomes poor (r2 = 0.18) when the density of the fiber network is very low (average pore size ∼5 µm), which indicates a distinct difference between continuum and discrete approaches (SI Appendix, Fig. S20). Second, we have applied this method to the filopodial mechanosensing model for the computation of the directed cell migration toward stiffer ECM and also identified two distinct linear regions at filopodial protrusive and retractile phases, which indicates that the slope of linear fit at the retractile phase was steeper than that at the protrusive phase (SI Appendix, Fig. S9C). Finally, experiments of filopodia dynamics in the collagen I gel were performed to justify the application of this method to measuring the local ECM stiffness sensed by filopodia. We have observed that values of both CPSs at a protrusive filopodium are higher than those at a retractile filopodium, which implies switching cellular polarity toward stiffer ECM after the mechanosensing competition among filopodia. The experimental results are in good agreement with the computational results of filopodia dynamics in two characteristic regions where the slope of the linear fit at the retractile phase is steeper than that at the protrusive phase (Fig. 2F and SI Appendix, Fig. S11E).
As for the application to experimental measurement of both effective CPSs, is suitable for gaining the intuition of area elasticity using unstructured (or scattered) data of fluorescent beads which are bound to ECM fibers. On the other hand, is suggested for gaining the effective ECM fiber stiffness of the ECM fiber network consisting of structured data of the ECM fiber network, which includes the identification of ECM fiber alignment and connectivity between fibers and cross-links. However, this additionally requires accurate segmentations of ECM fiber data from 3D confocal images. Our current experimental results are limited in this aspect because slice thickness (500 nm) is about five times larger than the diameter of a single collagen fibril (∼100 nm), which resulted in locally disconnected fibers with a high-aspect noncircular cross-sectional area in the z stack. Our simulations have demonstrated the application of the effective CPS using discrete fiber information, ECM fiber alignment, and connectivity between fibers and cross-links to switching cellular polarity toward stiffer ECM. Therefore, in the case of high remodeling of ECM fibers and a high-strain regime, the discrete approach of the effective CPS seems to be better than the continuum approach, and its further characterization, experimental observation, and improved 3D fiber reconstructions will be necessary in the future.
The presented method and its computational results are consistent with experimental observations reported recently (16). Cell migration experiments using human umbilical vein cells (HUVECs) transfected with cytosolic GFP were performed in the microfluidic device for the validation of the model, and it was found that speeds of both tip and root of filopodia increase as the pore size of the collagen gel is increased in experiments. The simulated speeds of both tip and root of filopodia also showed a good correlation to the experiments (16). It has been reported that, as the ECM pore size increases, a discrete ECM model shows a better agreement with experimental data, particularly for long-range displacements, than continuous models (35). The presented method is also based on the discrete ECM model consisting of a network of ECM fibers. The experimental results show that the buildup of traction forces is faster for higher substrate stiffness; is proportional to the stiffness (up to 80 nN/μm) (29, 36). This can be explained with the proposed computational model. For example, faster buildup of traction force can be found when a filopodium tugs more along a single ECM fiber which is grounded to the ECM fiber network (SI Appendix, Fig. S21). Effective stiffness of this ECM fiber is increased more as the filopodium tugging time is longer, which results from the nonlinear behavior of the single ECM fiber (SI Appendix, Fig. S21B): As the filopodium crawls more on the fiber, the unstressed length of the single ECM fiber is reduced more, and the stiffness of the ECM fiber is inversely proportional to its unstressed length [ where and are sectional area and time-varying unstressed length of single ECM fiber, respectively] (16). Thereby, the buildup of traction forces is faster as effective stiffness of the ECM fiber increases more rapidly.
We have established a detailed computational model to predict dynamics of filopodia interacting with a 3D ECM fiber network (Materials and Methods). Our computational model allows for the calculation of time-varying cellular polarity, as well as time-varying traction force and effective CPS at the tips of filopodia when they are bound to their surrounding ECM fibers. Through monitoring this traction force and effective CPS at tips of filopodia, we show that both the traction force and the sensed effective CPS at its tip take the highest peaks when a particular filopodium starts interacting with stiffer ECM fibers. After repeating multiple simulations for three different cases, our model also allows for the prediction of probabilities for switching the cellular polarity toward stiffer ECM as a function of separation distance from the boundary between soft and stiff ECMs to the cell in the soft ECM. As a result, we show that this probability decays as the separation distance increases. Additionally, the computational model includes reaction–diffusion mass transfers of six biochemical concentrations related to proteolysis and degradation in ECM to incorporate chemical interactions of 3D ECM with filopodia and cellular membranes (SI Appendix, Method S1). We visualize the distributions of biochemical concentrations such as MMP-2 and TIMP-2 (SI Appendix, Fig. S22 A and B). In particular, this computational model enables us to investigate variations of traction forces and CPS in the surrounding ECM in the cases of overexpressed and deficient MT1-MMP secretion rates (SI Appendix, Fig. S22 C and D). For the justification of the model, further experimental measurements of effective CPS in those cases will be necessary in the future.
We note that our model of “filopodia dynamics in the ECM fiber network” is complex, depending on many factors, including the strength and spatiotemporal properties of the FC formation in switching the diverse phases of filopodial dynamics, the sliding rate of nonmuscle myosin II, the filopodial protrusive force due to actin polymerization force, and membrane tension. Among them, a key sensitive factor is a stalled myosin sliding rate related to filopodial mechanosensing with ECM fibers. A recent experimental work revealed that the actin flow by the myosin motor is fed back by surrounding ECM rigidity (20). In addition, the maximum number of clustered integrins per node on the filopodial membrane is set to be 100, but as filopodia are located at the leading edge of the cell, those filopodia recruit and transport abundant integrins by myosin-X, forming integrin clustering at those tips to gain strong traction force (12). Therefore, the direction of the future filopodia dynamic model will be to improve the feedback mechanism of filopodial mechanosensing with ECM fibers and integrin clustering dynamics on the filopodial membrane. This method may fail if effects of integrin transmembrane, cytoplasmic domains, and the integrin “inside-out activation” process are neglected (37).
Finally, the goal of the present model is aimed at understanding how the filopodial mechanosensing model plays an important role in the directed cell migration toward stiffer ECM. The ultimate goal of the future cell model will be the development of a collective cell migration in 3D ECM with a gradient of stiffness. A recent interesting study has shown that collective cell migration on the elastic substrate more preferentially directs toward stiffer substrate than single-cell migration on the identical substrate, and the speed of cell migration tends to be faster as cells are located on the substrate with a steeper gradient of stiffness (38). Like their observation in the single-cell migration, we observe that single-cell models with various conditions of filopodial lengths, numbers, and diameters occasionally tend to migrate in opposite directions. Therefore, it is of great interest to capture an emergent behavior, which is defined as a functional response emerging from a complex multicellular system that cannot be observed in a single cellular system,† of collective cell migration in 3D ECM in our upcoming model. Furthermore, using the filopodial mechanosensing model, the future model can be extended to more complex models of vascular network formation and cancer metastasis, including a collective migration mediated by both cell–cell and cell–ECM adhesions to understand other functions of filopodia in sensing neighboring cells in 3D ECM.
Conclusions
There have been vague notions of stiffness in the literature of cell–ECM interactions due to the complexity of the ECM fiber network. It has been poorly understood in conjunction with the detection of local ECM stiffness. This present work manifested local stiffness in terms of time-series data of force and displacement across the fiber network in the vicinity of each filopodium. Stiffness depends on both force and displacement and two unique formulas are justified with numerous simulations which reproduce experimental observations. These formulations reflect filopodial protrusive and retractile activities interacting with surrounding ECM fibers which cause time series of position and force information. The computational model also reproduces two cases of experimental observations on durotaxis: (i) The cell steers away from the sharp interface when the cell starts migrating from the stiff ECM and (ii) the cell crosses the sharp interface when the cell starts migrating from the soft ECM. The computational model was then applied to quantifying the invasiveness of cancer cells with long (or short) and abundant filopodia. Specifically, we have found that the cancer cell model with short and abundant filopodia turns out to be the most aggressive. Thus, we feel strongly that our computational model has the potential to be a critical tool for the determination of invasiveness of cancer cells in the ECM microenvironment.
Materials and Methods
Cell Culture.
GFP-expressing HUVECs (Angio-Proteomie) were cultured in EGM2-MV growth medium (Lonza) and used in our experiments at passage 6. Cells were grown in T25 flasks (Thermo Fisher Scientific) until 90% confluent. Then they were removed from the flask surface by incubation with trypsin/EDTA (Gibco) for 2 min, centrifuged, and then resuspended with the collagen I solution, before seeding into the 3D microfluidic devices.
Preparation of Microfluidic Devices and Filopodia Imaging.
Preparation of 3D microfluidic devices, followed by cell seeding, was performed as previously described in detail (39). In brief, microfluidic devices were prepared using polydimethylsiloxane (PDMS) and bonded to a glass coverslip. Microfluidic channels were coated with poly-D-lysine (PDL) (Sigma). Before injection of the gel solution into the devices, fluorescent beads (red) of 200 nm diameter (Sigma) were mixed with the solution at a dilution of 1:500 from the original stock. Following that, the central gel channel, in which filopodia dynamics were later imaged, was filled with the collagen I solution, at a density of 2.5 mg/mL. Devices were then incubated for 30 min at 37 °C to polymerize. The concentration of NaOH in the collagen I solution was varied to allow accurate control of pH 7.0 during polymerization of the solution. Following polymerization of the gel, HUVECs were seeded in the bottom channel by pipetting 40 μL of cell suspension at 2 × 106 cells/mL. Following 24 h of incubation at 37 °C, the medium was replaced with fresh EGM2-MV medium supplemented with 50 ng/mL VEGF (Peprotech) and 250 nM S1P (Sigma). In the following 2–3 d, cells were left to sprout into the collagen gel. During sprouting, single filopodia were imaged using a confocal laser-scanning microscope (FV-1200; Olympus) at 60× magnification. Protrusive and retractile filopodia of the GFP-expressing HUVECs (green), fluorescent 200-nm beads (red), and collagen fibers (confocal reflectance microscopy) were simultaneously imaged with a time interval of 2 min for 36 min, while keeping cells humidified at 37 °C and 5% CO2.
Three-Dimensional Reconstructions of Confocal Images.
Confocal 3D image stacks were visualized using a 3D image analysis tool (IMARIS; Bitplane). To extract 3D objects of cells, fluorescent beads, and ECM fibers from 3D confocal images, the contour surface method was used as the image segmentation with options of the surface area level (smooth), the absolute intensity (thresholding), and the number of voxels (filter type) over time. Then, each of the 3D objects at every time frame was exported with a file format of virtual reality modeling language (VRML2). Afterward, all VRML2 files were converted to a format of stereolithography (STL), using a 3D digitization tool (MeshLab). Surfaces of cells, fluorescent beads, and ECM fibers were processed with triangular meshes.
Calculations of CPS from 3D Reconstructions.
All STL files of cells, fluorescence beads, and ECM fibers at every time interval were integrated using a commercial tool for the visual data analysis (Tecplot 360). Afterward, multiple zones of data were grouped with zones per group of three (data specifications) and time interval of 2 min (solution time). For the calculation of CPS using the continuum or discrete ECM fiber approach, three to four beads were selected with the following conditions: (i) Beads were bound to ECM fibers and (ii) located within a distance of 1 µm from the filopodial tip. Positions of beads were extracted with an option of “discrete points” at every time frame and used to calculate two datasets of rate of force by time and velocities of beads. Finally, calculations of CPS were processed using these two datasets.
Simulation of Mechanical Stretching Test for ECM Model.
For the simulation of stretching ECM fiber network models, two ECM stretching models with pore sizes of 1.5 μm and 3.0 μm, respectively, and fiber diameters of 41 nm were built with cube structures with equilateral length of 20 μm as shown in SI Appendix, Fig. S12 A and B. As for boundary conditions for the stretching simulations, time-varying stretching speed conditions were imposed at both top and bottom sides of each ECM model. Each ECM model was strained up to 0.25 and then relaxed to investigate its viscoelastic behavior. Bulk moduli of soft and stiff ECM models were calculated with linear fits of strain–stress curves.
Simulation of Mechanosensing Test for ECM Model.
For simulations of mechanosensing tests for ECM fiber models, two ECM models with pore sizes of 1.5 μm and 3.0 μm, respectively, and fiber diameters of 41 nm were built with cube structures with equilateral length of 20 μm. The mechanosensing tests were simulated for a physical time of 30 s and consist of two time zones (one is passive strain for 0–15 s and the other is active strain for 15–30 s). As for boundary conditions for the ECM model, constant stretching speed conditions were imposed at both top and bottom sides of each ECM model up to strain of 0.3 for 15 s (passive strain). Afterward, zero velocity conditions were imposed to maintain the strain of the ECM model at 0.3 for another 15 s. Simultaneously, a single fiber in the center of the ECM model was chosen to stretch up to three levels of fiber strain, 0.1, 0.3, and 0.5, where unstressed fiber length is 1 μm (active strain). As for the boundary condition for the fiber, a time-varying stretching speed condition (sine function) was imposed for 15 s.
Simulation of Cell Invasion Dynamics into 3D ECM.
The detailed equations that govern each of these dynamical processes are summarized in SI Appendix, Method S1, and the list of simulation parameters is summarized in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
This material is based upon work supported by the National Science Foundation, Science and Technology Center on Emergent Behaviors in Integrated Cellular Systems under Grant CBET-0939511 (to M.-C.K., R.D.K., and H.H.A.). This research was also supported by the National Research Foundation Singapore through the Singapore–MIT Alliance for Research and Technology’s BioSystem and Micromechanics Interdisciplinary Research Group research program (to Y.R.S., R.D.K., and H.H.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
†Raman R, Bashir R, DeStefano L, Kamm RD (2016) Module 4: Emergent Behavior. Emergent Behaviors of Integrated Cellular Systems (EBICS) Annual Retreat. August 3 and 4, 2016, St. Charles, IL.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717230115/-/DCSupplemental.
References
- 1.Low BC, et al. YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett. 2014;588:2663–2670. doi: 10.1016/j.febslet.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Rubashkin MG, et al. Force engages vinculin and promotes tumor progression by enhancing PI3K activation of phosphatidylinositol (3,4,5)-triphosphate. Cancer Res. 2014;74:4597–4611. doi: 10.1158/0008-5472.CAN-13-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engler A, et al. Substrate compliance versus ligand density in cell on gel responses. Biophys J. 2004;86:617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Fibroblast adaptation and stiffness matching to soft elastic substrates. Biophys J. 2007;93:4453–4461. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engler AJ, et al. Myotubes differentiate optimally on substrates with tissue-like stiffness: Pathological implications for soft or stiff microenvironments. J Cell Biol. 2004;166:877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 8.de Rooij J, Kerstens A, Danuser G, Schwartz MA, Waterman-Storer CM. Integrin-dependent actomyosin contraction regulates epithelial cell scattering. J Cell Biol. 2005;171:153–164. doi: 10.1083/jcb.200506152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Kovbasnjuk O, et al. The glycosphingolipid globotriaosylceramide in the metastatic transformation of colon cancer. Proc Natl Acad Sci USA. 2005;102:19087–19092. doi: 10.1073/pnas.0506474102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vignjevic D, et al. Fascin, a novel target of beta-catenin-TCF signaling, is expressed at the invasive front of human colon cancer. Cancer Res. 2007;67:6844–6853. doi: 10.1158/0008-5472.CAN-07-0929. [DOI] [PubMed] [Google Scholar]
- 12.Mattila PK, Lappalainen P. Filopodia: Molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 13.Allen WE, Zicha D, Ridley AJ, Jones GE. A role for Cdc42 in macrophage chemotaxis. J Cell Biol. 1998;141:1147–1157. doi: 10.1083/jcb.141.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong S, Guo WH, Wang YL. Fibroblasts probe substrate rigidity with filopodia extensions before occupying an area. Proc Natl Acad Sci USA. 2014;111:17176–17181. doi: 10.1073/pnas.1412285111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson HE, et al. F-actin bundles direct the initiation and orientation of lamellipodia through adhesion-based signaling. J Cell Biol. 2015;208:443–455. doi: 10.1083/jcb.201406102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim MC, Whisler J, Silberberg YR, Kamm RD, Asada HH. Cell invasion dynamics into a three dimensional extracellular matrix fibre network. PLoS Comput Biol. 2015;11:e1004535. doi: 10.1371/journal.pcbi.1004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacquemet G, Hamidi H, Ivaska J. Filopodia in cell adhesion, 3D migration and cancer cell invasion. Curr Opin Cell Biol. 2015;36:23–31. doi: 10.1016/j.ceb.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Lin CH, Espreafico EM, Mooseker MS, Forscher P. Myosin drives retrograde F-actin flow in neuronal growth cones. Neuron. 1996;16:769–782. doi: 10.1016/s0896-6273(00)80097-5. [DOI] [PubMed] [Google Scholar]
- 19.Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 2012;151:1513–1527. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan CE, Odde DJ. Traction dynamics of filopodia on compliant substrates. Science. 2008;322:1687–1691. doi: 10.1126/science.1163595. [DOI] [PubMed] [Google Scholar]
- 21.Harland B, Walcott S, Sun SX. Adhesion dynamics and durotaxis in migrating cells. Phys Biol. 2011;8:015011. doi: 10.1088/1478-3975/8/1/015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinhardt JW, Krakauer DA, Gooch KJ. Complex matrix remodeling and durotaxis can emerge from simple rules for cell-matrix interaction in agent-based models. J Biomech Eng. 2013;135:71003. doi: 10.1115/1.4024463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borau C, Kamm RD, García-Aznar JM. Mechano-sensing and cell migration: A 3D model approach. Phys Biol. 2011;8:066008. doi: 10.1088/1478-3975/8/6/066008. [DOI] [PubMed] [Google Scholar]
- 24.Allena R. Cell migration with multiple pseudopodia: Temporal and spatial sensing models. Bull Math Biol. 2013;75:288–316. doi: 10.1007/s11538-012-9806-1. [DOI] [PubMed] [Google Scholar]
- 25.van Oers RFM, Rens EG, LaValley DJ, Reinhart-King CA, Merks RM. Mechanical cell-matrix feedback explains pairwise and collective endothelial cell behavior in vitro. PLoS Comput Biol. 2014;10:e1003774. doi: 10.1371/journal.pcbi.1003774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sokolnikoff IS. Mathematical Theory of Elasticity. Krieger; Malabar, FL: 1983. pp. 336–337. [Google Scholar]
- 27.Yamamura N, Sudo R, Ikeda M, Tanishita K. Effects of the mechanical properties of collagen gel on the in vitro formation of microvessel networks by endothelial cells. Tissue Eng. 2007;13:1443–1453. doi: 10.1089/ten.2006.0333. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann B, Schäfer C. Filopodial focal complexes direct adhesion and force generation towards filopodia outgrowth. Cell Adhes Migr. 2010;4:190–193. doi: 10.4161/cam.4.2.10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trichet L, et al. Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness. Proc Natl Acad Sci USA. 2012;109:6933–6938. doi: 10.1073/pnas.1117810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grant CA, Brockwell DJ, Radford SE, Thomson NH. Tuning the elastic modulus of hydrated collagen fibrils. Biophys J. 2009;97:2985–2992. doi: 10.1016/j.bpj.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bornschlögl T, et al. Filopodial retraction force is generated by cortical actin dynamics and controlled by reversible tethering at the tip. Proc Natl Acad Sci USA. 2013;110:18928–18933. doi: 10.1073/pnas.1316572110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Abhilash AS, Chen CS, Wells RG, Shenoy VB. Long-range force transmission in fibrous matrices enabled by tension-driven alignment of fibers. Biophys J. 2014;107:2592–2603. doi: 10.1016/j.bpj.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, et al. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat Cell Biol. 2004;6:523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- 34.Arjonen A, Kaukonen R, Ivaska J. Filopodia and adhesion in cancer cell motility. Cell Adhes Migr. 2011;5:421–430. doi: 10.4161/cam.5.5.17723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall MS, et al. Fibrous nonlinear elasticity enables positive mechanical feedback between cells and ECMs. Proc Natl Acad Sci USA. 2016;113:14043–14048. doi: 10.1073/pnas.1613058113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fouchard J, Mitrossilis D, Asnacios A. Acto-myosin based response to stiffness and rigidity sensing. Cell Adhes Migr. 2011;5:16–19. doi: 10.4161/cam.5.1.13281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wegener KL, Campbell ID. Transmembrane and cytoplasmic domains in integrin activation and protein-protein interactions (review) Mol Membr Biol. 2008;25:376–387. doi: 10.1080/09687680802269886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sunyer R, et al. Collective cell durotaxis emerges from long-range intercellular force transmission. Science. 2016;353:1157–1161. doi: 10.1126/science.aaf7119. [DOI] [PubMed] [Google Scholar]
- 39.Shin Y, et al. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nat Protoc. 2012;7:1247–1259. doi: 10.1038/nprot.2012.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.