Significance
Contributions of rare variants to common and complex traits such as type 2 diabetes (T2D) are difficult to measure. This paper describes our results from deep whole-genome analysis of large Mexican-American pedigrees to understand the role of rare-sequence variations in T2D and related traits through enriched allele counts in pedigrees. Our study design was well-powered to detect association of rare variants if rare variants with large effects collectively accounted for large portions of risk variability, but our results did not identify such variants in this sample. We further quantified the contributions of common and rare variants in gene expression profiles and concluded that rare expression quantitative trait loci explain a substantive, but minor, portion of expression heritability.
Keywords: genetics, sequencing, type 2 diabetes, eQTL, rare variants
Abstract
A major challenge in evaluating the contribution of rare variants to complex disease is identifying enough copies of the rare alleles to permit informative statistical analysis. To investigate the contribution of rare variants to the risk of type 2 diabetes (T2D) and related traits, we performed deep whole-genome analysis of 1,034 members of 20 large Mexican-American families with high prevalence of T2D. If rare variants of large effect accounted for much of the diabetes risk in these families, our experiment was powered to detect association. Using gene expression data on 21,677 transcripts for 643 pedigree members, we identified evidence for large-effect rare-variant cis-expression quantitative trait loci that could not be detected in population studies, validating our approach. However, we did not identify any rare variants of large effect associated with T2D, or the related traits of fasting glucose and insulin, suggesting that large-effect rare variants account for only a modest fraction of the genetic risk of these traits in this sample of families. Reliable identification of large-effect rare variants will require larger samples of extended pedigrees or different study designs that further enrich for such variants.
Type 2 diabetes (T2D) is a common complex disease affecting >340 million individuals worldwide. Genomewide association studies (GWASs) have identified ∼88 common loci contributing to T2D (1). The role of rare variants in T2D is largely unknown, because large samples are required to have high power for the rarest variants and, until recently, strategies for genotyping rare variants in large samples have been prohibitively expensive. Rare variants typically have recent origins, and may therefore have large deleterious effects that have not yet been removed from the population by natural selection. If many large-effect rare variants underlie T2D, they could jointly explain a large fraction of trait heritability and their discovery could accelerate the transition from genetic association signals to biological understanding (2, 3).
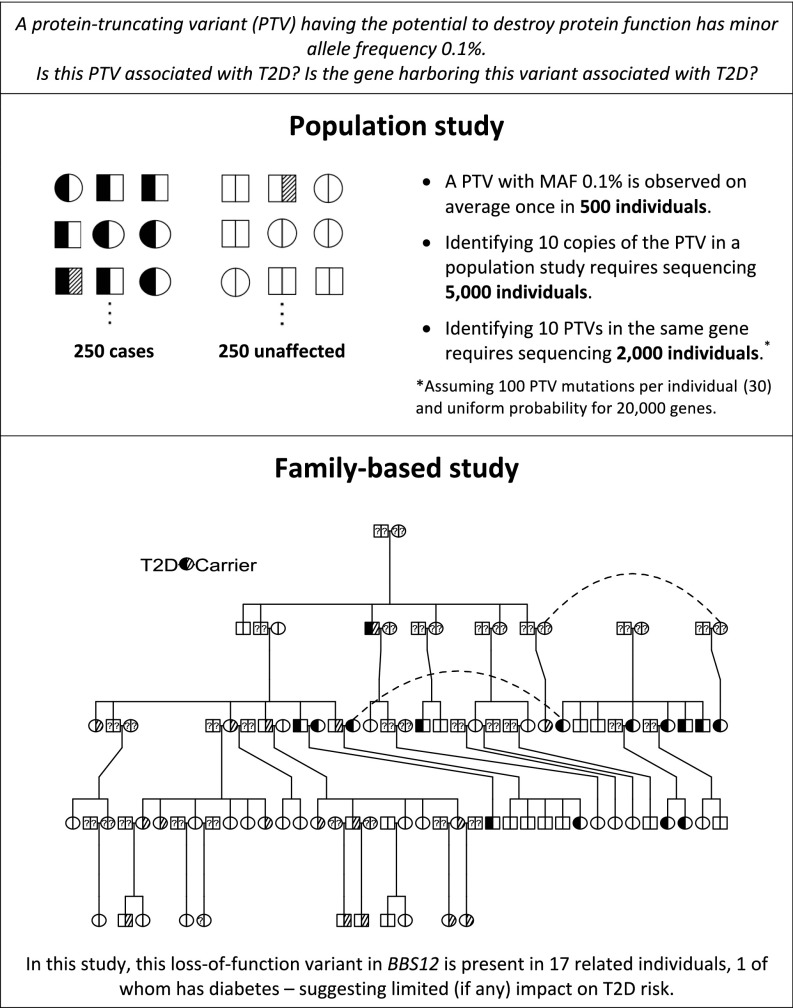
Although we can now discover and genotype rare genetic variants in large study cohorts, the majority of these variants will be present in only a few individuals—in population-based genetic studies, >50% of variants are seen in a single individual—making it difficult to establish evidence of association. Increased association power can be achieved by increasing the number of copies of each rare allele—for example, by sequencing very large numbers of unrelated individuals (4)—but even these studies have little power to detect association with variants with minor allele frequency (MAF) <0.1%. Here we describe an alternate strategy for testing rare variants, with a focus on private, family-specific variants, combining the classical genetic approach of large, well-characterized families with modern whole-genome sequencing technology. The rationale for the experiment is to increase allele counts for private variants by tracking Mendelian segregation among related individuals within pedigrees. By chance, some private variants will segregate to multiple related individuals, providing a sufficient number of observed alleles to allow association testing, which would be nearly impossible in even large studies of unrelated samples (Fig. 1).
Fig. 1.
Large pedigrees are a valuable tool for investigating the role of rare variants in complex disease.
Results
To determine the extent to which private and rare variants contribute to T2D and related quantitative phenotypes, we examined 20 large Mexican-American pedigrees drawn from the San Antonio Family Heart Study (5, 6) and San Antonio Family Diabetes/Gallbladder Study (7, 8). Pedigrees contained 22 to 86 individuals distributed across 3 to 5 generations, for a total of 1,034 individuals; 305 (∼30%) had T2D (Table 1). In addition to T2D, we tested diabetes-related quantitative traits reflecting glycemic control (fasting/2-h glucose and insulin levels) for association in the 729 nondiabetic individuals and lipid traits (total cholesterol, HDL, LDL, and triglycerides) in all samples. The high prevalence of T2D in these families is consistent with the possible segregation of large-effect, private risk variants, making them ideally suited for this experimental study design.
Table 1.
Sample distributions and phenotype statistics at the most recent examination
| Family | T2D cases | Unaffected |
| No. of individuals sequenced (% female) | 186 (60.8) | 400 (59.6) |
| No. of individuals imputed (% female) | 119 (55.4) | 329 (58.0) |
| Age, y | 62.9 ± 12.7 | 46.8 ± 15.7 |
| BMI, kg/m2 | 32.0 ± 7.23 | 31.5 ± 7.28 |
| Fasting glucose, mmol/L | 9.29 ± 4.08 | 5.71 ± 2.18 |
| Fasting insulin, mU/L | 29.3 ± 40.9 | 14.7 ± 13.3 |
| No. of individuals with expression data | 215 | 416 |
Mean ± SDs.
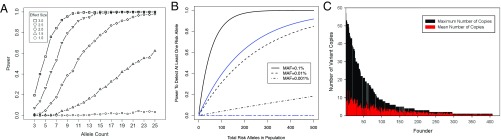
Power to detect the effect of a single rare variant on disease risk is a function of pedigree size, pedigree structure, and the effect size of the variant. Together, these determine the number of copies that can be observed for each private variant. In our 20 Mexican-American pedigrees, the 413 founders have varying numbers of descendants and potential transmitted copies for a private variant (Fig. 2C); >40 founders can transmit ≥25 copies of the rare variants they carry. Using gene-dropping simulation and averaging over all contributing founders, there is probability 16, 4.5, and 1.3% of capturing ≥5, ≥10, and ≥15 copies of any variant present only in a single founder, respectively; the average number of copies is 2.5.
Fig. 2.
Enrichment of allele counts within pedigrees and the effect on analysis power. (A) Power to detect private risk variants conditional on the number of observed allele counts. Effect sizes are expressed in SD units for normalized traits. (B) Power to detect at least one of N private risk alleles with an effect size of 2 phenotype SDs in our pedigree samples (black) and in 1,034 unrelated samples (blue). Blue curves for MAF 0.01% and MAF 0.001% are shown overlapped in one line at power 0. (C) Distribution of the maximum possible and expected numbers of minor alleles for 413 pedigree founders, where maximum numbers are the numbers of all descendent haploids and expected numbers are averaged over 1,000 gene-drop simulations.
In our study, a T2D variant with 80% penetrance and observed ≥25 times within a single pedigree had 50% power of detection at genomewide significance (α = 5 × 10−8) (SI Appendix, Fig. S1A). Although power to detect a single private variant is low, this study had 60% power to detect at least one such variant if at least 500 variants with MAF 0.1% existed in the population (SI Appendix, Fig. S1B) for T2D and 100% power for quantitative traits (Fig. 2B). The existence of large numbers of rare variants with large effect is compatible with current understanding of complex diseases, for which only a minority of heritability is typically explained by common variants (9–11). For example, given the 30% prevalence of type 2 diabetes, if fully penetrant rare variants with MAF ∼0.1% explain >20% of diabetes cases, at least 60 such variants must exist in the population; if causal variants have frequency 0.01%, at least 600 must exist in the population.
We had greater power to detect variants influencing quantitative traits, even though for analysis of these traits we excluded individuals with T2D. For example, we had 80% power to detect a rare variant that modifies a quantitative trait by 2.0 SDs provided it was transmitted to 16 individuals. Supposing that variants modifying traits with an effect size of 2.0 SDs have MAF ∼0.1% and jointly account for 33% of the heritability of a quantitative trait, there must be at least 400 such variants in the population. If most causal variants have lower frequency, then there must be even more of them. In any situation where variants with frequency <0.01% and effect sizes of ≥2.0 SDs jointly explain >33% of the heritability of a diabetes-relevant quantitative trait, our pedigrees provided ∼80% power to detect genomewide significant association (α = 5 × 10−8) with at least one of these variants. In contrast, sequencing a similar number of unrelated samples would be a hopeless strategy—any variants sampled would be present in only one or two individuals, and power would be <0.001% (Fig. 2B).
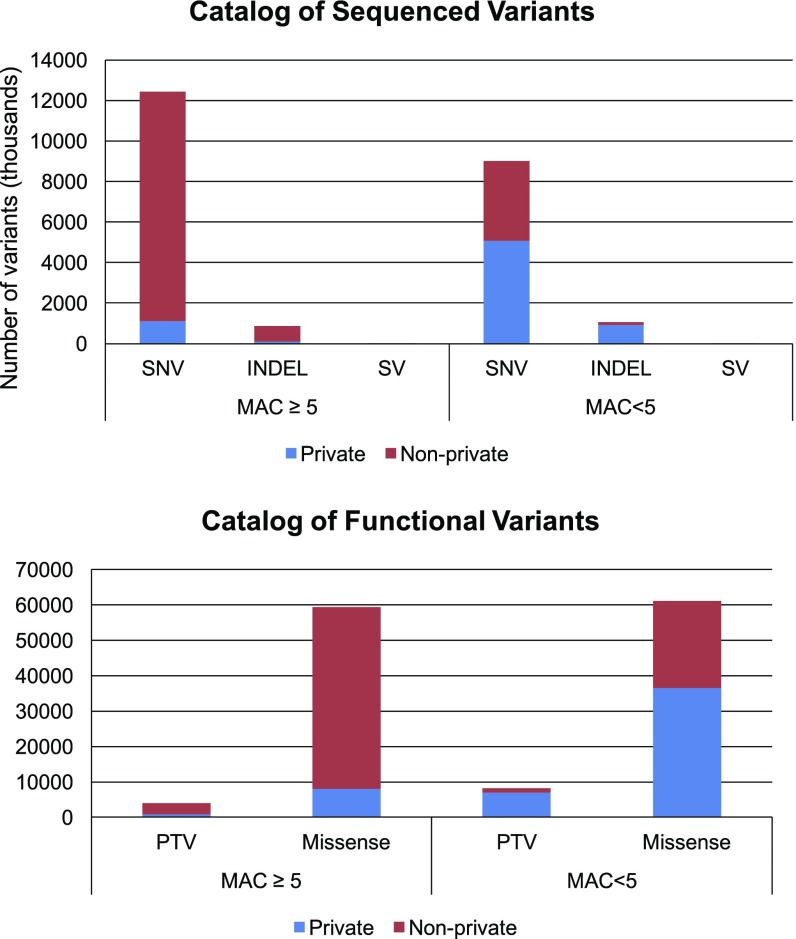
We strategically sequenced 586 individuals from the 20 pedigrees at >40× coverage using Complete Genomics services. Sequenced individuals were specifically chosen to maximize the capture of genetic variation in each pedigree and, by sequencing of parent–offspring pairs, to facilitate estimation of haplotypes. Sequencing identified 23.4 million (M) variants: 21.6M single-nucleotide variants (SNVs) and 1.9M more complex genetic variants including insertions, deletions, and copy-number variants (Fig. 3). As expected, most variants were rare: 15.1M had maximum-likelihood estimation (MLE) MAF <1% by SOLAR-estimated MAF; 7.2M are private, family-specific variants that enter our pedigrees through a single founder and do not appear in the 1000 Genomes Project data (12).
Fig. 3.
Catalog of variants identified by whole-genome sequencing. MAC, minor allele count; SV, structural variation.
We genotyped 448 additional pedigree members using Illumina HumanHap550v3, Human1M-Duov3, Human1Mv1, and Human660W-Quad_v1 GWAS arrays. SNVs not present in one platform were imputed and a comprehensive set of 1 million SNVs was defined. These data allow us to track haplotypes through each family and identify additional carriers of variants identified in the sequenced samples (13). We evaluated the accuracy of the genotypes (sequenced or imputed) by comparing our genotypes with rare variants genotyped using the Illumina HumanExome-12 v1 exome array. For variants with MLE MAF <1%, nonreference genotypes called by sequencing and by haplotype imputation were accurate 99.9 and 96.7% of the time, respectively. Many novel, private variants were transmitted to multiple descendants; 514K such variants were transmitted to >10 individuals. We observed 1.74M variants inherited from a single founder having enriched allele counts with ≥5 copies in pedigree members; these variants are likely to be singletons in the same number of samples of unrelated individuals.
Analysis of 1,000 simulated null phenotypes shows that a P value of 7.1 × 10−8 is required to achieve genomewide significance in this experiment (versus ∼1 × 10−9 using Bonferroni adjustment) (SI Appendix). This reflects the large linkage disequilibrium blocks observed in the Mexican-American pedigrees and the restricted number of segregating founder haplotypes.
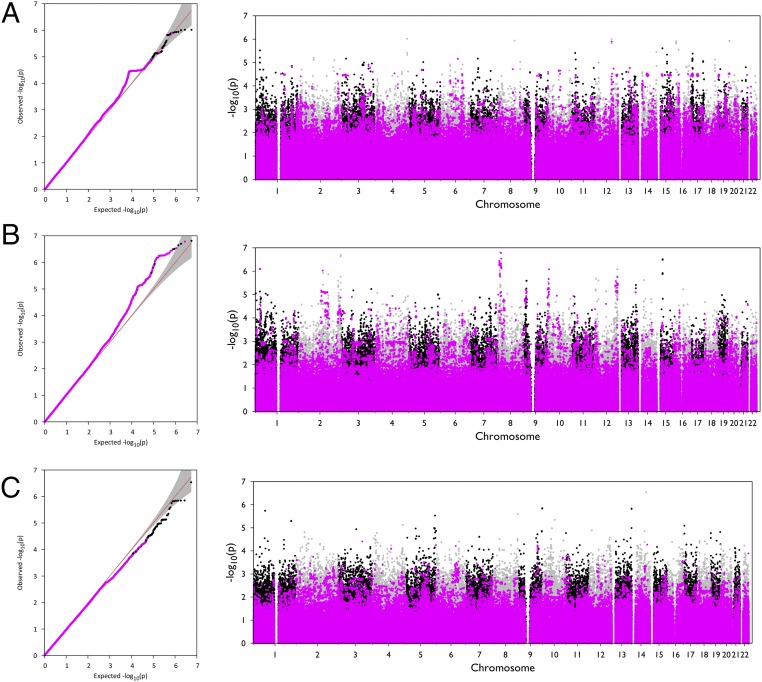
We did not observe significant evidence of association between individual rare variants and T2D, glucose, or insulin levels (Fig. 4). These results suggest that large-effect rare variants (those with near-complete penetrance for T2D or with an effect size >2 SDs for quantitative traits) are very unlikely to explain ≥20% of T2D risk or ≥33% heritability of quantitative traits in this sample; as noted previously, situations where this occurs would require large numbers of such variants and, in that case, we expect to detect a few. In the analyses of additional quantitative traits, we reidentified several previously known common variants associated with lipid traits but did not observe significant signals from individual rare variants (SI Appendix, section 4.2).
Fig. 4.
Single-variant association results for type 2 diabetes and glycemic traits. QQ and Manhattan plots for (A) T2D, (B) fasting glucose (adjusted for BMI), and (C) fasting insulin (adjusted for BMI). Only variants with MAF ≤1% in the 1000 Genomes phase I dataset are plotted. Variants that are only seen in one pedigree (that would be private in an unrelated sample) are highlighted in purple. The “step” in the T2D QQ plot is due to a group of variants shared by a nuclear family in one pedigree in which five members have T2D. No variant achieved a P value exceeding the experimentwide significance threshold of 7.1 × 10−8 for any of these three traits.
We carried out gene-based analyses that grouped functional rare variants within each gene (Methods). Using each of four grouping strategies, test statistics fit the null hypothesis and no gene reached exomewide significance (α = 2.5 × 10−6) for T2D. We observed exomewide significant association between the CYP3A4 gene and fasting glucose levels (P = 9.2 × 10−7) and between the OR2T11 gene and 2-h insulin levels (P = 1.9 × 10−6). We also observed that the LDLR gene is associated with LDL cholesterol levels (P = 8.3 × 10−7). We investigated evidence of rare variants with large effect sizes enriched in these gene-based results but did not find evidence of such variants. More details about gene-based results are provided in SI Appendix, section 4.3. We next examined single-variant and gene-level association results in regions linked to our traits by our prior linkage results. A linkage peak was considered significant if present with a logarithm of the odds (LOD) score above 3, and we set the respective boundaries by the peak LOD value minus 1 unit. We also investigated regions identified by GWAS as harboring trait-associated common genetic variants, regions harboring genes implicated in monogenic forms of diabetes, and single-gene disorders that affect fasting blood glucose and insulin levels. Each of these more focused analyses offered us the opportunity to prioritize strong signals that did not reach genomewide significance. Again, we did not observe association with T2D, fasting insulin, or fasting glucose even with appropriately relaxed stringency.
To allow investigation of rare-variant effects over a wider range of traits, we took advantage of array-based lymphocyte gene expression available for 643 individuals in 17 of the 20 pedigrees (14). cis-eQTL (expression quantitative trait locus) analysis of 21,677 transcripts identified 4,307 independent variant–expression associations at familywise error rate (FWER) <5% (α = 7.0 × 10−6); 3,144 expression traits had at least one associated variant. The average effect size across all 4,307 cis-eQTLs was 0.81 SD unit but, as expected, varied dramatically according to variant MAF: The 785 associated variants with MLE MAF <1% had an average effect size of 2.0 SD units, and the 3,522 associated variants with MLE MAF >1% had an average effect size of 0.55 SD unit. We observed 92 instances in which both rare and common eQTLs contributed to the same expression trait. Recently, the Genotype-Tissue Expression Consortium reported rare variants with large expression effects in genes with outlier expression levels in multitissue samples (15), while we have power to assess overall effects of rare variations over a wider spectrum of expression-level changes with the pedigrees.
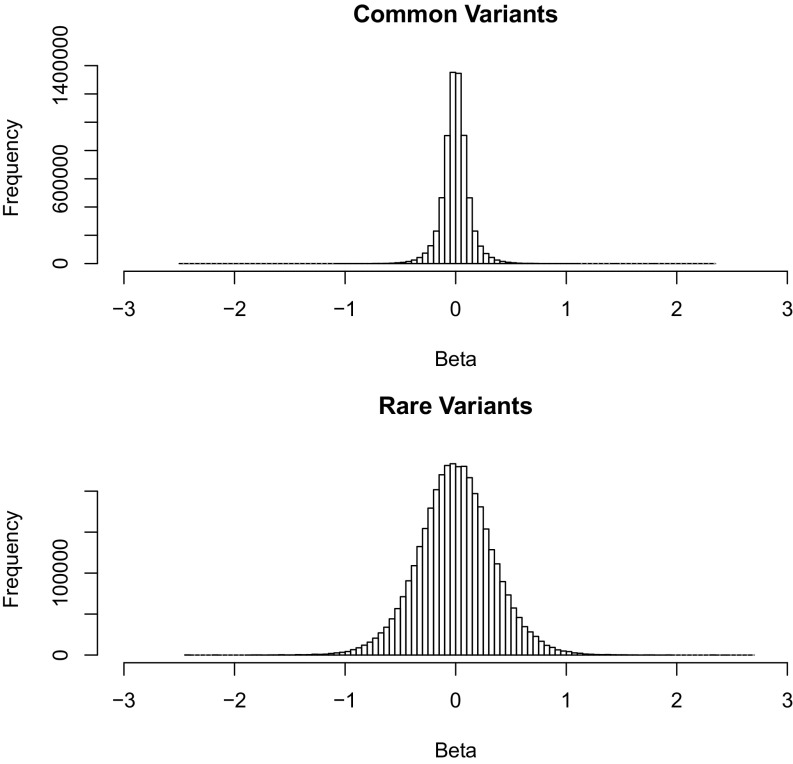
To formally test whether rare eQTLs have larger average effect sizes than common eQTLs, we compared the full distributions of standardized quantitative trait effect sizes regardless of whether a variant was significantly associated with expression traits (Fig. 5). We reasoned that evaluating the full distribution of rare-variant effect sizes would avoid the winner’s curse (16), given the asymptotic unbiasedness of the effect size estimates, and would help evaluate whether, overall, there is evidence that rare-variant effect sizes are larger in magnitude (and, thus, have higher variance) than those for common variants. The observed variance of effects estimated for rare variants is 5.65 times greater than that observed for common variants, suggesting that there are rare variants with substantially larger effects overall. After correcting for the estimated sampling error, which is greater for rare variants, the ratio of effect size variance of rare and common variants was 4.18. This is remarkably consistent with the ratio of effect sizes observed for statistically significant rare and common eQTLs (2.0 SDs compared with 0.55 SD), despite the fact that the winner’s curse results in inflated estimated effect sizes when a statistical threshold is applied. Finally, we randomly sampled from these empirical effect sizes and overall minor-allele frequency spectrum to estimate that as much as 25% of genetic variation in quantitative gene expression in these families may be due to rare variants with MLE MAF <1%. Overall, these results suggest that an average rare eQTL has a substantially greater biological effect than an average common eQTL—although we cannot rule out an unexpected artifact (such as an unmodeled population structure) that would increase rare-variant effect size variance beyond what we expected based on sampling error.
Fig. 5.
Distribution of estimated effect sizes (betas) of minor alleles on quantitative gene expression for common (n = 43,517,300) and rare (n = 927,244,054) variants.
Many rare eQTLs were undetected in this study because causal variants were not present in the 413 founders or were present in a founder but in too few of their descendants. Based on the numbers of detected associations, the allele frequency spectrum, and statistical power, we estimate that ∼23,000 common eQTLs with effect sizes of ∼0.5 SD unit are required to explain our observation of 3,522 detected common eQTLs with an average effect size of 0.55 SD unit. If we assume that rare variants have an average effect size of 1 SD unit (twofold higher than that of common variants), the detection of 765 rare eQTLs suggests that a total of ∼220,000 true rare eQTLs exist. With a larger true effect size of 2.0 SD units (fourfold that of common variants), ∼20,000 true rare eQTLs would be required to explain our 765 observed rare eQTLs. Overall, with ∼20,000 common eQTLs with effect sizes averaging 0.5 SD unit and 20,000 to 200,000 rare eQTLs with effect sizes averaging 1 to 2 SD units, rare variants would explain 5 to 20% of eQTL heritability. This estimate is smaller than the 25% observed in the simulation experiment due to the restriction to the distribution of observed significant effects. Taken together, our results suggest the existence of very large numbers of rare eQTLs with larger biological effects than those of common variants but a minority contribution to overall expression heritability.
Discussion
Genetic association studies have identified >88 common T2D-associated loci, most with small biological effect sizes (1, 17, 18). It has been hypothesized that many rare variants of large effect may exist and, that taken together, such variants could explain a considerable proportion of the variance in T2D risk (19). This hypothesis has not been well-tested before, because of the difficulty and cost involved in assessing very rare variants in large samples, while recently Fuchsberger et al. (4) showed that common-variant GWAS signals are not the results of clustered rare-variant signals residing on common haplotypes. Here, using a combination of deep whole-genome sequencing and analysis of large families, we designed an experiment specifically powered to identify variants with effect sizes >2.0 SDs and population frequency <0.01%. In models where these variants cumulatively explain ∼33% of the variation in risk for a diabetes-related trait, our experiment would have identified at least one such variant for each trait examined. We did not identify any rare variants associated with T2D, glycemic, or lipid traits, suggesting that large-effect, extremely rare variants are unlikely to explain a large portion of the variability in type 2 diabetes risk in this sample of pedigrees.
Our results are sensitive to stochastic effects. Most founder lineages are simply not large enough to identify private functional variants, because there is a limit on the number of copies of rare-variant alleles that can be transmitted. Thus, we expect an experiment such as ours to miss most such rare variants. However, our experiment will sample many copies (≥15) of a proportion of the variants that would be private in similar-sized samples of unrelated individuals. If larger numbers of rare, large-effect, T2D-associated variants were to exist, we would be uniquely well-placed to detect these. Some evidence for the likely importance of rare variants in quantitative phenotypic variation was observed for available gene expression data. For this larger set of phenotypes relatively close to gene action, rare variants exhibited demonstrably larger biological effects sizes and are estimated to account for as much as 25% of observed transcript-level genetic variance in these pedigrees.
Our analyses show that large families can be used to identify many copies of rare variants—which we expect will be especially important for genetic studies outside coding regions, where burden-based tests aggregating the effects of many variants remain challenging because of a lack of annotation strategies. Our results suggest that while rare variants might be plentiful enough to help understand causality and may be biologically important for specific individuals/lineages, they are unlikely to account for much heritability in diabetes and related traits in this sample. Our analyses further suggest that the identification of robust associations between variants private to single large families and diabetes-related traits will require larger numbers of extended pedigrees and/or different study designs that further increase the probability of functional rare-variant segregation. Alternative strategies that maximize the number of observed rare-variant alleles include focusing on population “isolates,” as recently illustrated by the identification of a variant with an increased allele frequency only in this specific population predisposing to type 2 diabetes in Greenland (20). Such isolates represent extended kindreds with large lineages.
Methods
We selected 1,034 individuals from 20 pedigrees who are part of the San Antonio Family Heart Study (SAFHS) (2, 5) and San Antonio Family Diabetes/Gallbladder Study (SAFDGS) projects (7, 8). Written informed consent was obtained from all participants. This study was approved by the Institutional Review Boards of the University of Texas Health Science Center at San Antonio and the University of Texas Rio Grande Valley. We then selected 600 samples to be sequenced to gain maximal genetic information about the remaining samples in the pedigrees using ExomePick software (ExomePicks, https://genome.sph.umich.edu/wiki/ExomePicks); EPACTS (including EMMAX), https://genome.sph.umich.edu/wiki/EPACTS; Famrvtest, https://genome.sph.umich.edu/wiki/Famrvtest; GotCloud, https://genome.sph.umich.edu/wiki/GotCloud) (21). Whole-genome sequencing for 600 samples was done by Complete Genomics (CGI). After stringent sample-level quality control, we analyzed 586 individuals with sequence data. Variant calls generated by the CGI pipeline were filtered based on multisample statistics using support vector machine filtering of the GotCloud pipeline (22). Merlin (13) was used to obtain sequence-scale genotype information for the remaining GWAS samples using sequenced family members. Variants were grouped into several functional categories using five prediction algorithms (LRT, Mutation Tester, PolyPhen2-HumDiv, PolyPhen2-HumVar, SIFT) assisted by extensive external information (23–26). We used EMMAX (27) to generate empirical kinship coefficients between samples to account for known and hidden family structures. Details on study design and data generation are described in SI Appendix, section 1.
We analyzed T2D-related metabolic traits: fasting glucose, fasting insulin, 2-h glucose, 2-h insulin, LDL cholesterol, HDL cholesterol, and triglyceride levels. Trait values were measured at up to five examinations. Regressions were performed at each examination adjusting for covariates as appropriate, producing examination-specific residuals. The examination-specific residuals were then averaged over multiple measurements and an inverse-normal transformation was applied to averaged residuals. Covariates were chosen to align with strategies taken by consortia participating in the metaanalysis of GWASs of the given traits, as well as the T2D-GENES and GoT2D consortia’s trait transformation strategy (4) and included age, age2, sex, and BMI (body mass index). T2D samples were excluded from glycemic trait analyses, and cholesterol levels were preadjusted by a fixed amount per lipid medication status.
Two different variance component models, SOLAR (28) and Famrvtest (29), were used for association analyses with the empirical kinship coefficients. More details on each of the analysis steps are described in SI Appendix, Methods. All software tools used in this project are publicly available.
To estimate overall contributions of common and rare variants to overall expression levels, we used the number of common and rare eQTLs from our association results together with the externally supplied allele frequency spectrum. Since sample allele frequencies in these pedigrees have a lower bound of 1/the number of founder chromosomes (1/816 = 0.12%), we simulated each possible founder allele count and used the allele frequency spectrum from 2,000 unrelated Mexican-American samples to obtain more accurate power estimates.
We restricted gene-based rare variant tests to variants with MLE MAF <1% by maximum-likelihood MAF estimation, and applied four different variant masks based on functional annotations: (i) protein-truncating variants (PTVs) only, (ii) PTVs + missense variants, (iii) PTVs + variants predicted to be deleterious by five different functional prediction algorithms, and (iv) PTVs + variants predicted to be deleterious by at least one functional prediction algorithm.
All data used in this paper are publicly available through the database of Genotypes and Phenotypes (accession no. phs000462.v2.p1).
Supplementary Material
Acknowledgments
We warmly thank the participants of the SAFHS and SAFDGS for their contribution, enthusiasm, and cooperation. This study is part of the Type 2 Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples (T2D-GENES) Consortium, funded by the European Commission (HEALTH-F4-2007-201413), Wellcome Trust (090367, 090532, 098381), Medical Research Council (G0601261), and NIH/NIDDK (RC2-DK08839, DK105535, DK085524, DK085545, DK085584, DK085501, DK098032, DK078616, DK085526). The whole-genome sequencing was done commercially by Complete Genomics, Inc. Additional genetic and phenotypic data were provided by the San Antonio Family Heart Study and San Antonio Family Diabetes/Gallbladder Study, which are supported by NIH Grants R01 HL0113323, P01 HL045222, R01 DK047482, and R01 DK053889. SAFHS gene expression data were generated through a donation from the Azar and Shepperd families. J.G.W. was supported by U54GM115428 from the National Institute of General Medical Sciences. S.C., S.L., J.K., J. Lee, and T.P. were supported by the Bio-Synergy Research Project (2013M3A9C4078158) of the Ministry of Science, ICT and Future Planning through the National Research Foundation of Korea, and Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health and Welfare (HI15C2165, HI16C2037). A.K.M. was supported by American Diabetes Association Mentor-Based Postdoctoral Fellowship #7-12-MN-02. M.I.M. is a Wellcome Trust Senior Investigator. The research was supported by the National Institute for Health Research (NIHR), Oxford Biomedical Research Centre. The views expressed are those of the authors and not necessarily those of the National Health Service, NIHR, or Department of Health, United Kingdom.
Footnotes
Conflict of interest statement: S.E.L., J. Laramie, and R.G.T. were employees of Complete Genomics during this study. T.M.T. is an employee of Regeneron Pharmaceuticals. D.A. is an employee of Vertex Pharmaceuticals.
This article is a PNAS Direct Submission. X.Z. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705859115/-/DCSupplemental.
References
- 1.Mohlke KL, Boehnke M. Recent advances in understanding the genetic architecture of type 2 diabetes. Hum Mol Genet. 2015;24:R85–R92. doi: 10.1093/hmg/ddv264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 3.Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuchsberger C, et al. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell BD, et al. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 1996;94:2159–2170. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- 6.MacCluer JW, et al. Genetics of atherosclerosis risk factors in Mexican Americans. Nutr Rev. 1999;57:S59–S65. doi: 10.1111/j.1753-4887.1999.tb01790.x. [DOI] [PubMed] [Google Scholar]
- 7.Hunt KJ, et al. Genome-wide linkage analyses of type 2 diabetes in Mexican Americans: The San Antonio Family Diabetes/Gallbladder Study. Diabetes. 2005;54:2655–2662. doi: 10.2337/diabetes.54.9.2655. [DOI] [PubMed] [Google Scholar]
- 8.Puppala S, et al. A genomewide search finds major susceptibility loci for gallbladder disease on chromosome 1 in Mexican Americans. Am J Hum Genet. 2006;78:377–392. doi: 10.1086/500274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 10.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locke AE, et al. LifeLines Cohort Study ADIPOGen Consortium AGEN-BMI Working Group CARDIOGRAMplusC4D Consortium CKDGen Consortium GLGC ICBP MAGIC Investigators MuTHER Consortium MIGen Consortium PAGE Consortium ReproGen Consortium GENIE Consortium International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auton A, et al. 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—Rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 14.Göring HH, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 15.Li X, et al. 2016. The impact of rare variation on gene expression across tissues. bioRxiv:10.1101/074443.
- 16.Zöllner S, Pritchard JK. Overcoming the winner’s curse: Estimating penetrance parameters from case-control data. Am J Hum Genet. 2007;80:605–615. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prasad RB, Groop L. Genetics of type 2 diabetes—Pitfalls and possibilities. Genes (Basel) 2015;6:87–123. doi: 10.3390/genes6010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris AP, et al. Wellcome Trust Case Control Consortium Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators Genetic Investigation of ANthropometric Traits (GIANT) Consortium Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium South Asian Type 2 Diabetes (SAT2D) Consortium DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 20.Moltke I, et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature. 2014;512:190–193. doi: 10.1038/nature13425. [DOI] [PubMed] [Google Scholar]
- 21.Sidore C, et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat Genet. 2015;47:1272–1281. doi: 10.1038/ng.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jun G, Wing MK, Abecasis GR, Kang HM. An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data. Genome Res. 2015;25:918–925. doi: 10.1101/gr.176552.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 25.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 27.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blangero J, et al. A kernel of truth: Statistical advances in polygenic variance component models for complex human pedigrees. Adv Genet. 2013;81:1–31. doi: 10.1016/B978-0-12-407677-8.00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng S, et al. Methods for association analysis and meta-analysis of rare variants in families. Genet Epidemiol. 2015;39:227–238. doi: 10.1002/gepi.21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.