Significance
To get more insights into the disease mechanism of T-cell acute lymphoblastic leukemia (T-ALL), particularly in an adult group, we addressed the genomic landscape in 130 patients, including 61 cases of adult T-ALL. A number of new genetic aberrations were identified using integrated transcriptome and genomic analysis. Distinct T-ALL subgroups were defined according to the interplay among different genetic abnormalities and gene transcription patterns. Characterization of genomic features of T-ALL is valuable not only for a better understanding of leukemogenesis, but also for patient stratification and tailored therapy.
Keywords: T-ALL, transcriptome, fusion gene, ZBTB16-ABL1, gene mutation
Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is a clonal malignancy of immature T cells. Recently, the next-generation sequencing approach has allowed systematic identification of molecular features in pediatric T-ALL. Here, by performing RNA-sequencing and other genomewide analysis, we investigated the genomic landscape in 61 adult and 69 pediatric T-ALL cases. Thirty-six distinct gene fusion transcripts were identified, with SET-NUP214 being highly related to adult cases. Among 18 previously unknown fusions, ZBTB16-ABL1, TRA-SALL2, and involvement of NKX2-1 were recurrent events. ZBTB16-ABL1 functioned as a leukemogenic driver and responded to the effect of tyrosine kinase inhibitors. Among 48 genes with mutation rates >3%, 6 were newly found in T-ALL. An aberrantly overexpressed short mRNA transcript of the SLC17A9 gene was revealed in most cases with overexpressed TAL1, which predicted a poor prognosis in the adult group. Up-regulation of HOXA, MEF2C, and LYL1 was often present in adult cases, while TAL1 overexpression was detected mainly in the pediatric group. Although most gene fusions were mutually exclusive, they coexisted with gene mutations. These genetic abnormalities were correlated with deregulated gene expression markers in three subgroups. This study may further enrich the current knowledge of T-ALL molecular pathogenesis.
T-cell acute lymphoblastic leukemia (T-ALL) is a clonal malignancy of immature T cells, accounting for 15% of pediatric ALL and 25% of adult ALL. Currently, the 5-y survival rate of pediatric T-ALL has reached more than 80% (1). In adult T-ALL, while significant therapeutic progress has been made in advanced hematology/oncology centers with a 5-y survival rate of over 60% (2), there are still challenges to improve the clinical prognosis in many cases. A better understanding of T-ALL in an adult group may allow more rational disease stratification and precision therapy. Over the past three decades, many genetic abnormalities have been found in T-ALL (3–5). Aberrant expression of genes such as LMO1, LMO2, TAL1, TLX1, TLX3, and other transcription factors (TFs) can be either due to chromosomal rearrangements juxtaposing T-cell receptor (TCR) loci to these genes or due to the recently described somatic mutations in enhancer regions recruiting relevant TFs such as MYB (6, 7). These abnormalities can be detected in 40–50% T-ALL (8). On the other hand, fusion genes are also common (20–30% in T-ALL), generating overexpression of mRNAs with ORFs for wild-type protein (such as TAL1 in STIL-TAL1) (9, 10) or transcripts containing fusions between two truncated ORFs such as SET-NUP214 (11). A number of gene abnormalities in pathways regulating differentiation, proliferation, self-renewal, and survival of T-cell precursors are also found in high frequencies, such as mutations of NOTCH1, JAK-STAT, PI3K-AKT, or RAS-MAPK pathway genes and CDKN2A/2B deletions (2, 5, 12–15).
In recent years, next-generation sequencing techniques, including whole-genome sequencing, whole-exome sequencing (WES), and RNA sequencing (RNA-seq), have extended the list of genetic abnormalities in T-ALL to epigenetic factors and translation/RNA stability pathways (16–21). Of note, a subgroup of T-ALL with a characteristic immunophenotype (CD3+/CD1a−/CD8−/CD5weak) has been designated as early T-cell precursor ALL (ETP-ALL), with special gene mutation patterns (16, 22). The integrated analysis of gene alterations in two recent series of pediatric T-ALL provided an enlarged view of the genomic landscape in this heterogenous disease (20, 23). However, complex interplay of gene fusions, sequence abnormalities, and transcriptional expression profiles, especially in adult cases, needs to be further addressed to refine the current model of T-ALL leukemogenesis and to reveal potential new biomarkers and therapeutic targets.
In this study, RNA-seq, WES, and other genomic analyses were performed in 61 adult and 69 pediatric T-ALL cases. A number of previously undescribed gene fusions, mutations, aberrant transcripts, and gene expression patterns were identified. Specifically, we also addressed the leukemogenic potential of the recurrent fusion gene ZBTB16-ABL1.
Results
Clinical Characteristics of Patients.
Consecutive samples were enrolled in ChiCTR-ONRC-14004968 (Shanghai Institute of Hematology) and ChiCTR-ONC-14005003 (Shanghai Children’s Medical Center) trials. The clinical and hematological information is listed in Table 1 and Dataset S1. In the pediatric group, a complete remission (CR) rate of 92.8% (64/69) and the 3-y overall survival (OS) rate of 72.8% (95% CI 67.3–78.3) were achieved. In contrast, in adults, the CR rate was 75.4% (46/61) while the 3-y OS rate was 29.0% (95% CI 22.4–35.6) (SI Appendix, Fig. S1), revealing an inferior prognosis and the need for further improved treatment strategy.
Table 1.
Clinical characteristics of 130 patients with T-ALL
| Characteristics | Adult (n = 61), n (%) | Child (n = 69), n (%) | P value |
| Gender, male/female | 42/19 | 53/16 | 0.307 |
| Age, y, median (range) | 34 (18–62) | 11 (1–17) | |
| WBC count | 0.004 | ||
| ≥100 × 109/L | 15 (24.6) | 34 (49.3) | |
| <100 × 109/L | 46 (75.4) | 35 (50.7) | |
| Immunophenotype | |||
| ETP | 17 (27.9) | 7 (10.1) | 0.009 |
| Pro | 6 (9.8) | 3 (4.3) | 0.219 |
| Pre | 13 (21.3) | 21 (30.4) | 0.238 |
| Cortical | 18 (29.5) | 28 (40.6) | 0.188 |
| Medullary | 7 (11.5) | 10 (14.5) | 0.611 |
Overview of Gene Fusion Transcripts.
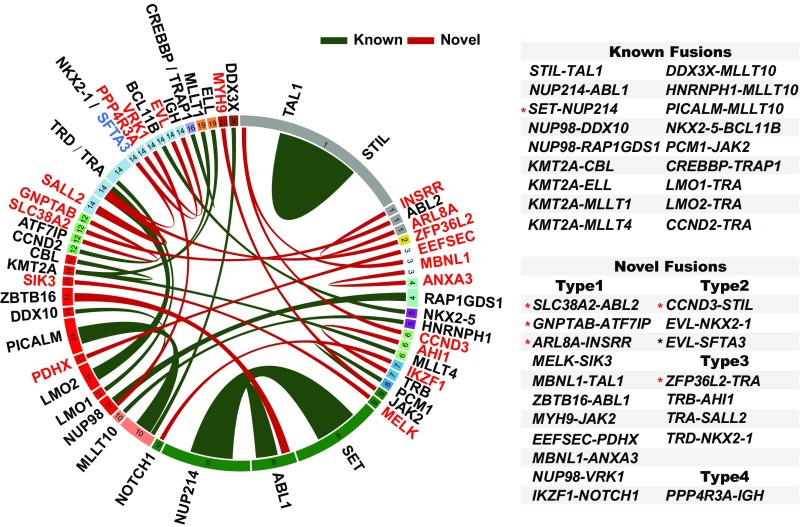
Based on RNA-seq data and reverse transcription PCR (RT-PCR) confirmation, 69 cases (53.08%) harbored a fusion gene. Thirty-six different fusions were found, including 18 known fusions covering common types such as STIL-TAL1, SET-NUP214, and NUP214-ABL1 (24–26), and 18 previously unknown fusions, of which five were identified in adults (Fig. 1, SI Appendix, Fig. S2, and Datasets S2 and S3). Although similar frequencies of gene fusions were found in the two groups (50.82% in adults vs. 55.07% in children), the types of fusions seemed less diverse in adults than in children (16 vs. 26) (Dataset S2). Of note, newly discovered ZBTB16-ABL1 and TRA-SALL2 fusions, and the involvement of NKX2-1, were recurrent events (Dataset S3). Four types of gene abnormalities were identified among previously unknown fusions. In ZBTB16-ABL1, the N-terminal moiety of ZBTB16 was fused to the body region of ABL1, defining a typical chimeric ORF (type 1). The type 2 fusion was represented by case C47 where the 5′ untranslated region (UTR) of EVL, a gene highly expressed in T-ALL cells found in this work (SI Appendix, Fig. S3A), was fused to the entire ORF of homeobox gene NKX2-1. This fusion led to an aberrantly high expression of NKX2-1 (SI Appendix, Fig. S3B). In the same case, an EVL-SFTA3 fusion was also detected, probably caused by a splicing between EVL and SFTA3, the latter being located downstream from NKX2-1 (Fig. 1). Type 3 fusion was characterized by genes fused to TCR-alpha, -beta, or -delta (TRA/TRB/TRD) loci, leading to overexpression of SALL2 [a TF deregulated in various cancers (27)] and NKX2-1 (SI Appendix, Fig. S3C). Although the initiation ATG codon of SALL2 in cases C22 and C38 was deleted in the fusion mRNA, an alternative initiation ATG codon was found at amino acid 63. The major functional domains of SALL2 could thus be maintained. One case (C60) was found to carry on a type 4 fusion, with a short transcript of PPP4R3A fused to IGH. Since PPP4R3A is a known tumor suppressor gene (28), the disruption of its ORF could cause gene inactivation. It is worth pointing out that in most cases (92.8%, 64/69), gene fusions were mutually exclusive.
Fig. 1.
Circos plot of the genomic landscape of gene fusions discovered by RNA-seq. Ribbon widths are proportional to the frequency of a fusion event. Chromosomes are individually colored and are arranged clockwise from chromosome 1 to X, starting with chromosome 1 at 12 o’clock. The newly found genes involved in T-ALL fusions are marked in red. Previously unknown fusion gene partners are connected by red ribbon. All previously identified gene fusions are shown as green ribbons. The detailed classification of novel fusions is described in the text. The black asterisk indicates that in addition to a fusion between 5′ UTR of EVL and NKX2-1, a second fusion transcript, EVL-SFTA3, was found probably as an accompanying event in case C47. The red asterisk in known fusions indicates the fusion involved mainly in adult patients. The red asterisk in novel fusions indicates the fusion discovered in adult patients.
Leukemogenic Power of the Recurrent Fusion Gene ZBTB16-ABL1.
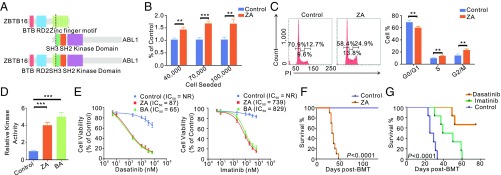
ZBTB16 (also known as promyelocytic leukemia zinc finger PLZF) contains one BTB domain and nine zinc fingers. The ZBTB16-ABL1 chimeric protein maintained the BTB and two or three zinc fingers (Fig. 2A). The ZBTB16-ABL1 occurred in two young children (case C11, 1 y old, accompanied by NOTCH1 and ZEB2 mutations and case C23, 2 y old, with mutations of PTEN, MYCN, and PIK3CD; Figs. 3 and 4). Both cases died within 1 y after diagnosis. When the proliferation rates of Jurkat cells transfected with ZBTB16-ABL1 or vehicle were compared, a stronger stimulatory effect on cellular proliferation by ZBTB16-ABL1 was observed (Fig. 2B). Moreover, ZBTB16-ABL1 promoted cell-cycle progression and DNA replication (Fig. 2C). We then examined protein tyrosine kinase (PTK) activity of ZBTB16-ABL1. The in vitro PTK activity of ZBTB16-ABL1 was fourfold that of ABL1, comparable to that of BCR-ABL1, which was fivefold the wild-type protein (Fig. 2D). Both fusion kinases had similar sensitivity to PTK inhibitors including imatinib and dasatinib (Fig. 2E). In retrovirus-mediated bone marrow (BM) transplantation experiments, all 10 mice carrying ZBTB16-ABL1 developed chronic myeloid leukemia-like myeloproliferative disease and quickly succumbed after transplantation (Fig. 2F and SI Appendix, Fig. S4). Massive elevation of maturing myeloid cells in BM, peripheral blood, and spleen was revealed. BM examination confirmed a significant increase in Mac-1+Gr-1+ myeloid elements (SI Appendix, Fig. S4). The ZBTB16-ABL1 BMT mice treated with imatinib or dasatinib showed significantly prolonged survival (Fig. 2G).
Fig. 2.
Molecular and functional characterization of the ZBTB16 (PLZF)-ABL1 fusion gene. (A) Structural and functional domains of wild-type ZBTB16, ABL1, and the representative fusion protein. Dashed lines indicated breakpoints of the wild-type proteins. (B) Cell proliferation of Jurkat cells transfected with ZBTB16-ABL1 (ZA) or vehicle (Control) seeded at a different density per well. (C) Cell-cycle distribution of Jurkat cells transfected with ZBTB16-ABL1 (ZA) or vehicle (Control). (D) Immunoprecipitated ABL1, BCR-ABL1 (BA), or ZBTB16-ABL1 (ZA) proteins were assayed for tyrosine kinase activity. The kinase activity of BA or ZA protein was compared with the activity of ABL1. (E) Viability of Jurkat cells transfected with ZBTB16-ABL1 (ZA) or vehicle (Control) upon exposure to dasatinib or imatinib. NR denotes “not reached.” (F) Kaplan–Meier survival analysis of ZBTB16-ABL1 (ZA) mice (n = 10) and control mice (n = 10). (G) Kaplan–Meier survival curves of control mice with ZBTB16-ABL1 (ZA) fusion (n = 6), imatinib (50 mg/kg) treated mice with ZA (n = 6), and dasatinib (5 mg/kg) treated mice with ZA (n = 6). Survival curves were compared by two-sided log-rank test. **P < 0.01; ***P < 0.001. Data are presented as mean ± SD from three independent experiments. Statistical significance was determined using two-sided Student’s t test.
Fig. 3.
Seven functional categories (C1–C7) of gene mutations in 130 T-ALL patients. Vertically, genes are ordered by functional categories and gene mutation rates are shown after gene names within each category. Newly found genes mutated in T-ALL are indicated by red color. Mutation events of all relevant genes in every case are summarized by a green label in each category to facilitate an integrated view of pathway abnormality. In C2, genetic abnormalities in JAK-STAT, RAS, and PI3K pathways are also summarized under relevant gene mutation events. Horizontally, samples are ordered into three groups (G1–G3) according to transcriptome-based clustering so that gene mutation patterns can be compared with gene expression markers (Fig. 4). Gene mutations with significantly different frequencies among G1, G2, and G3 groups are marked by a red star next to the gene names. Note: mutations of KMT2A, KMT2B, KMT2C, KMT2D, and KMT2E were collectively found in 23 cases (17.69%). RPL4, RPL5, RPL10, RPL13, and RPL22 were found in six cases (4.62%).
Fig. 4.
Integrated phenotypic and molecular architecture of leukemic cells from 61 adult and 69 pediatric T-ALL patients. (Top) Age, gender, and immunophenotype for each sample within each of the three unique gene expression groups: TLX1/3/HOXA (G1), ETP/LYL1/HOXA (G2), and TAL1/LMO1 (G3). The second vertical panel presents aberrant gene overexpression known to be associated with chromosome rearrangements and/or fusion transcripts. White labels on red rectangle are used to indicate cases with chromosomal translocations possibly involving TLX3 [t (1, 5) (p11;q35)] and LMO2 [t (11, 14) (p13;q11)] revealed by karyotype analysis (circles; Dataset S1), with amplification of TAL1, NKX2-1, or LYL1 genes revealed by SNP array (triangles) or with insertion of the enhancer regions of TAL1, LMO1, or LMO2 genes (plus signs), respectively. The third vertical panel (heatmap) shows the abnormal expression pattern of a group of genes potentially involved in early T-cell differentiation and hematopoietic regulation. Gene names with an aberrant expression pattern revealed in this work are in red. In the fourth vertical panel, aberrant transcript and fusion genes are presented. (Bottom) CDKN2A/2B deletions and mutations from C1 to C4 categories.
Identification of Distinct Gene Expression Groups and Aberrant RNA Splicing/Transcripts.
Unsupervised clustering methods were applied to the classification of gene expression from T-ALL cases, and three distinct subgroups were identified by using a set of the 1,484 most differentially expressed genes (SI Appendix, Figs. S5 and S6). The well-known expression marker genes in T-ALL, such as TAL1, LMO1, LYL1, HOXA, TLX1, and TLX3, showed distinct clustering patterns. TLX1/TLX3 overexpressions were found in one subgroup (G1), and LYL1 expression was enriched mainly in another subgroup (G2), whereas TAL1 and LMO1 overexpressions were clustered in the third subgroup (G3). HOXA overexpression was found in both G1 and G2. We also searched for aberrant mRNA splicing products or transcripts. An unusual transcript of SLC17A9, encoding a transmembrane transporter for adenoside triphosphate (ATP) and other small molecules, was highly expressed in 58 of 130 cases. Different from well-annotated SLC17A9 transcripts containing 13 exons, this short transcript had only exon 9–13, with a 5′ UTR from intron 8 and a putative initiation ATG codon at the 314th amino acid in exon 9, leading to an ORF of 123 amino acids and a truncation of the major facilitator superfamily (MFS) domain (SI Appendix, Fig. S7). Karyotype analysis revealed chromosomal translocations in one case with TLX3 overexpression and in two cases with LMO2 up-regulation. Among cases with overexpressed LMO1/2, TAL1/2, NKX2-1, and LYL1, amplification of TAL1, NKX2-1, or LYL1 was detected by single-nucleotide polymorphism (SNP) array in four cases, whereas insertion of an enhancer region in TAL1, LMO1, or LMO2 was found in eight other cases (Fig. 4).
Gene Mutation Profiling.
RNA-seq data-based mutation detection was used in all 130 cases according to a recently published highly stringent procedure (29, 30). In total, 119 genes were found mutated at least twice (Datasets S4 and S5). Attention was given to 48 genes with mutation rates over 3% (4/130 cases), including 6 newly identified mutated genes [CELSR3, PAK4, MINK1, NR4A1, BOD1L1, and VCP (Fig. 3 and SI Appendix, Fig. S8)], two of them (PAK4 and BOD1L1) being mainly involved in adults. Abnormalities of NOTCH1, FBXW7, PHF6, JAK3, PTEN, and JAK1 exhibited high mutation rates (74.6–10%). In addition, some gene families had multiple members affected by mutations, such as the histone-lysine methyltransferase family (KMT) and the ribosomal protein family (RPL) (Fig. 3). Mutated genes (>3%) were functionally divided into seven categories (C1–C7) (3): NOTCH1 pathway, signaling pathways, epigenetic factors, TFs, cell-cycle regulators, translation, and RNA stability-associated molecules, as well as others (Fig. 3). In the great majority (95.7%) of cases with gene fusions, at least one of the C1–C7 mutations coexisted, whereas FBXW7 and DNMT3A mutations occurred more frequently in cases without fusions (Dataset S6). Of note, the frequencies of mutations in adults were much higher than those in children (P = 0.001, SI Appendix, Fig. S9A and Dataset S7), especially among C2 and C3 genes. A significant correlation between the age and the number of gene mutations was found (R2 = 0.1147, P = 0.001; SI Appendix, Fig. S9B). The status of CDKN2A/2B was investigated in 115 cases, and CDKN2A and CDKN2B deletions were found in 75 and 64 cases, respectively (Fig. 4).
Correlation Among Gene Fusions/Mutations/Aberrantly Overexpressed Transcripts and Distinct Gene Expression Subgroups.
A close correlation was found among gene fusions/mutation profiles/aberrant transcripts and gene expression patterns, enabling a synthetic view of genomic abnormalities in T-ALL (Fig. 4 and SI Appendix, Table S1). In the G1 subgroup [28 cases (21.5%), 14 adults and 14 children], 21 cases bearing overexpressed TLX1/TLX3 shared a similar transcriptome profile to 7 cases with high expression of HOXA family genes. The G2 subgroup (37 cases, 28.5%), including 91.7% of our ETP cases, was composed mainly of adults (31/37 cases). One feature of G2 was a close association with NUP family genes containing fusions (15/37 cases). An array of core TFs for early hematopoiesis (HOXA, LYL1, MEF2C, SPI1, RARA, ELK3, BLNK, STAP1, ZBTB46, BTK, and NFKBIE) was also highly expressed in G2. The fact that MEF2C is overexpressed in 30/37 (81.8%) cases (SI Appendix, Fig. S10) suggested a common disease mechanism (31). In addition, ETP cases had higher mutation rates of epigenetic factors, IDH2, DNMT3A, and EZH2 in particular, than in other T-ALL cases (Dataset S8). Notably, all 20 cases with STIL-TAL1 and 4 cases with LMO1/2-TRA were clustered in G3 [65 cases (50%), 17 adults and 48 children], as were cases with variant fusions involving TAL1 or STIL. Several genes related to STIL-TAL1 (TAL1, RUNX1, MYB, ETS1, and BCL11B) were found overexpressed (Fig. 4), while up-regulated SIX6 was detected in 53 of 65 cases (81.5%). Interestingly, the aberrantly overexpressed SLC17A9 short transcript was grouped in G3 (55/65 cases), contributing to its unique signature. Scrutiny of gene abnormalities further divided G3 into two subgroups (G3a, 20 cases including 17 children; G3b, 45 cases comprising 14 adults and 31 children). The NOTCH1 (20 vs. 86.67%, P < 0.001) and FBXW7 (10 vs. 53.33%, P = 0.002) mutations were found significantly lower in G3a than in G3b (Dataset S9). Compared with G1/G2, G3 displayed lower gene mutation rates of JAK-STAT (4.62 vs. 46.15%, P < 0.001) and RAS (6.15 vs. 32.31%, P < 0.001) pathways, but a higher mutation frequency of the PI3K pathway (21.54 vs. 3.08%, P = 0.003) and the PTEN gene (18.46 vs. 1.54%, P = 0.003).
Genetic Characteristics of T-ALL Patients According to Immunophenotypes.
In ETP cases, the genetic feature was characterized by more SET-NUP214 fusions, overexpression of HOXA/MEF2C/LYL1, and high frequencies of mutations of RAS pathway/epigenetic factors. The genetic characteristics of early precursor (pro) cohorts were similar to those of ETP. However, among groups of precursor (pre), cortical, and medullary T-ALL, the frequencies of STIL-TAL1 and aberrantly overexpressed SLC17A9 short transcript were much higher (SI Appendix, Table S2). In addition, genes with specific overexpression patterns can be assigned to pro, pre, cortical, medullary, and ETP immunophenotypic subgroups (SI Appendix, Table S3).
Prognosis Analyses Related to the Genetic Features of Adult and Pediatric T-ALL Patients.
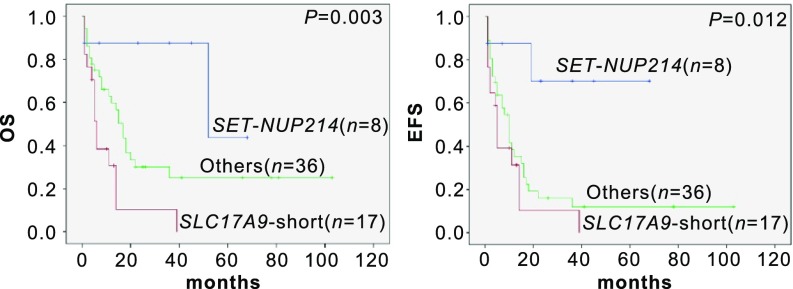
We compared the genomic landscapes between adult and pediatric T-ALL patients (SI Appendix, Table S4). In adults, cases with the aberrantly overexpressed short SLC17A9 transcript exhibited very poor outcome, with 3-y OS and 3-y EFS rates of 10.3% (95% CI 1.0–19.6) and 10.5% (95% CI 1.1–19.9), respectively. Conversely, eight cases with SET-NUP214 showed relatively good prognosis, with a 3-y OS rate of 87.5% (95% CI 75.8–99.2) and a 3-y EFS rate of 70.0% (95% CI 51.8–88.2) (Fig. 5). In the pediatric group, only Children's Oncology Group (COG) risk classification could differentiate three prognostic groups, while no individual genetic markers alone were found to clearly stratify the clinical outcome (SI Appendix, Fig. S11).
Fig. 5.
Kaplan–Meier survival curves of adult T-ALL patients with an aberrantly overexpressed SLC17A9 short transcript or SET-NUP214 fusion.
Discussion
In this study, by performing genomic landscape analysis, we identified a series of previously unknown fusion genes, gene mutations, and aberrant mRNA transcripts in 130 Chinese T-ALL cases. These findings were made through scrutiny of the molecular data, and it is possible that some features could be ascribed to the ethnic genetic background of T-ALL from the Chinese population. The genomic information of T-ALL in adults was also compared with that of pediatric cases. Taken as a whole, each of the 61 adult and 69 child T-ALL cases in our series harbored at least one major genetic lesion, including gene fusions, mutations of C1–C7 genes, and aberrant expression of genes key to leukemogenesis. This dataset further enriched our understanding of T-ALL and may be translated into useful clinical stratification markers or drug targets. Indeed, molecular markers predicted three distinct risk groups in our adult T-ALL cases.
Among 18 newly discovered gene fusions, ZBTB16-ABL1, TRA-SALL2 fusions, and involvement of NKX2-1 were recurrent events. It is well known that ZBTB16 functions as a hematopoietic regulator and is a fusion partner to retinoic acid receptor alpha (RARA) in a subset of acute promyelocytic leukemia (APL) (32). Here we found that ZBTB16-ABL1 protein retained the same portion of ZBTB16 as in ZBTB16-RARA, which could result in disturbed hematopoiesis. More importantly, ZBTB16-ABL1 also had SH3, SH2, and the tyrosine kinase domain of ABL1 and possessed constitutive PTK activity with strong transforming power (33). The fact that leukemia mice bearing ZBTB16-ABL1 responded to the therapeutic effect of PTK inhibitors suggested a potential clinical application of the drugs in relevant cases. In the future, patient-derived xenograft animal models will be used to further investigate the leukemogenesis and drug response.
An interesting finding of this work is that the number of gene mutations per case was higher in adult than in child T-ALL. The gene fusions/mutations and transcriptome features in our T-ALL series clearly showed an interplay of essential genetic abnormalities. Fusion genes rarely coexisted in the same case, suggesting that they play driver roles. However, fusions were often accompanied by at least one of the C1–C7 mutations, suggesting possible cooperative effects. Another dimension of genomic abnormalities lies at the level of transcription. Among 62 cases without fusions, 41 (66.1%) harbored deregulated expression of TAL1/2, LYL1, LMO1/2, TLX1/TLX3, HOXA, and NKX2-1 genes, at least in part due to intergenic structural aberrations or enhancer mutations. With regard to transcriptome analyses, SPI1 overexpression is noteworthy, since it serves as a regulator for MEF2C expression in normal lymphoid development and is highly expressed in prethymic progenitors (34). Recently, SPI1 was found to form a fusion gene with STMN1 or TCF7 (35). Although no such fusion was identified in our cohort, simultaneous high expression of SPI1 and MEF2C in a subset of our cases might be essential in leukemogenesis. In addition, most cases harboring STIL-TAL1/TAL1 overexpression presented an aberrantly overexpressed short SLC17A9 transcript with truncation of the MFS functional domain, and overexpression of this transcript predicted a poor outcome in adult patients. Further investigation is warranted to explore the role of this aberrantly overexpressed transcript in T-ALL pathogenesis. Moreover, larger cohort studies are needed to validate the prognostic significance of the overexpression of SLC17A9 short transcript and the SET-NUP214 fusion in adult T-ALL.
Materials and Methods
Patients.
The study cohort was composed of 61 adults and 69 pediatric patients who were followed from 2007 to 2016. The patients' clinical study was approved by the ethical boards of the participating centers (Institutes of Hematology in Shanghai, Soochow, and Fuzhou). All patients gave informed consent for treatment and cryopreservation of BM and peripheral blood samples according to the Declaration of Helsinki. Details of treatment protocols are available in SI Appendix, SI Materials and Methods.
RNA-Seq, WES Data, and CDKN2A/2B Analyses.
RNA-seq was performed according to a previously described method (36). WES was performed in 36 patients, 16 individuals having their own normal control samples (blood samples in CR). Reading pairs were aligned to the human reference genome hg19. Procedures of reading pairs alignment, mutation calling from RNA-seq or WES data, and the gene expression/pathway analysis are listed in SI Appendix, SI Materials and Methods. RT-PCR was used to confirm the newly identified fusion genes. A SNP array was performed to detect the intergenic rearrangements. Detection of CDKN2A/2B deletion is described in SI Appendix, SI Materials and Methods.
Functional Study of the ZBTB16-ABL1 Fusion Gene.
The wild-type ZBTB16 and ABL1 cDNA clones were kindly provided by OriGene. Procedures for plasmid construction, proliferation assay, cell-cycle analysis, PTK assay, and the drug inhibition test are described in SI Appendix, SI Materials and Methods. All animal experiments were approved by the Animal Care and Use Committee of Shanghai Jiao Tong University School of Medicine. Detailed information is available in SI Appendix, SI Materials and Methods.
Data Availability.
The datasets have been deposited in the Chinese Leukemia Genotype-Phenotype Archive (bioinfo.rjh.com.cn/cga) under accession no. CGAS00000000002.
Supplementary Material
Acknowledgments
This work was supported by the Chinese National Key Basic Research Project 973 (Grant 2013CB966800); the Chinese Ministry of Health (Grant 201202003); the Mega-Projects of Scientific Research for the 12th Five-Year Plan (2013ZX09303302); the National Natural Science Foundation of China (Grants 81470311, 81670137, 81570122, 81670147); the National Key Research and Development Program (Grant 2016YFC0902800); the Samuel Waxman Cancer Research Foundation; and the Center for High Performance Computing at Shanghai Jiao Tong University.
Footnotes
The authors declare no conflict of interest.
Data deposition: The datasets reported in this paper have been deposited in the Chinese Leukemia Genotype-Phenotype Archive, bioinfo.rjh.com.cn/cga (accession no. CGAS00000000002).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717125115/-/DCSupplemental.
References
- 1.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 2.Trinquand A, et al. Toward a NOTCH1/FBXW7/RAS/PTEN-based oncogenetic risk classification of adult T-cell acute lymphoblastic leukemia: A group for research in adult acute lymphoblastic leukemia study. J Clin Oncol. 2013;31:4333–4342. doi: 10.1200/JCO.2012.48.5292. [DOI] [PubMed] [Google Scholar]
- 3.Girardi T, Vicente C, Cools J, De Keersmaecker K. The genetics and molecular biology of T-ALL. Blood. 2017;129:1113–1123. doi: 10.1182/blood-2016-10-706465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iacobucci I, Mullighan CG. Genetic basis of acute lymphoblastic leukemia. J Clin Oncol. 2017;35:975–983. doi: 10.1200/JCO.2016.70.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vicente C, et al. Targeted sequencing identifies associations between IL7R-JAK mutations and epigenetic modulators in T-cell acute lymphoblastic leukemia. Haematologica. 2015;100:1301–1310. doi: 10.3324/haematol.2015.130179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahman S, et al. Activation of the LMO2 oncogene through a somatically acquired neomorphic promoter in T-cell acute lymphoblastic leukemia. Blood. 2017;129:3221–3226. doi: 10.1182/blood-2016-09-742148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mansour MR, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrando AA, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 9.Chen Q, et al. Coding sequences of the tal-1 gene are disrupted by chromosome translocation in human T cell leukemia. J Exp Med. 1990;172:1403–1408. doi: 10.1084/jem.172.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aplan PD, et al. Involvement of the putative hematopoietic transcription factor SCL in T-cell acute lymphoblastic leukemia. Blood. 1992;79:1327–1333. [PubMed] [Google Scholar]
- 11.Van Vlierberghe P, et al. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood. 2008;111:4668–4680. doi: 10.1182/blood-2007-09-111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez-Martin M, Ferrando A. The NOTCH1-MYC highway toward T-cell acute lymphoblastic leukemia. Blood. 2017;129:1124–1133. doi: 10.1182/blood-2016-09-692582. [DOI] [PubMed] [Google Scholar]
- 13.Zenatti PP, et al. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat Genet. 2011;43:932–939. doi: 10.1038/ng.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nosaka K, et al. Increasing methylation of the CDKN2A gene is associated with the progression of adult T-cell leukemia. Cancer Res. 2000;60:1043–1048. [PubMed] [Google Scholar]
- 15.Karrman K, Johansson B. Pediatric T-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2017;56:89–116. doi: 10.1002/gcc.22416. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann M, et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood. 2013;121:4749–4752. doi: 10.1182/blood-2012-11-465138. [DOI] [PubMed] [Google Scholar]
- 18.De Keersmaecker K, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet. 2013;45:186–190. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S, et al. Whole-genome non-coding sequence analysis in T-cell acute lymphoblastic leukemia identifies oncogene enhancer mutations. Blood. 2017;129:3264–3268. doi: 10.1182/blood-2017-03-771162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49:1211–1218. doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neumann M, et al. Mutational spectrum of adult T-ALL. Oncotarget. 2015;6:2754–2766. doi: 10.18632/oncotarget.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coustan-Smith E, et al. Early T-cell precursor leukaemia: A subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10:147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seki M, et al. Recurrent SPI1 (PU.1) fusions in high-risk pediatric T cell acute lymphoblastic leukemia. Nat Genet. 2017;49:1274–1281. doi: 10.1038/ng.3900. [DOI] [PubMed] [Google Scholar]
- 24.Janssen JW, Ludwig WD, Sterry W, Bartram CR. SIL-TAL1 deletion in T-cell acute lymphoblastic leukemia. Leukemia. 1993;7:1204–1210. [PubMed] [Google Scholar]
- 25.von Lindern M, Breems D, van Baal S, Adriaansen H, Grosveld G. Characterization of the translocation breakpoint sequences of two DEK-CAN fusion genes present in t(6;9) acute myeloid leukemia and a SET-CAN fusion gene found in a case of acute undifferentiated leukemia. Genes Chromosomes Cancer. 1992;5:227–234. doi: 10.1002/gcc.2870050309. [DOI] [PubMed] [Google Scholar]
- 26.Graux C, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36:1084–1089. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- 27.Hermosilla VE, et al. Developmental SALL2 transcription factor: A new player in cancer. Carcinogenesis. 2017;38:680–690. doi: 10.1093/carcin/bgx036. [DOI] [PubMed] [Google Scholar]
- 28.Byun HJ, Kim BR, Yoo R, Park SY, Rho SB. sMEK1 enhances gemcitabine anti-cancer activity through inhibition of phosphorylation of Akt/mTOR. Apoptosis. 2012;17:1095–1103. doi: 10.1007/s10495-012-0751-0. [DOI] [PubMed] [Google Scholar]
- 29.Meyer JA, et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet. 2013;45:290–294. doi: 10.1038/ng.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lilljebjörn H, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790. doi: 10.1038/ncomms11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Vlierberghe P, et al. Prognostic relevance of integrated genetic profiling in adult T-cell acute lymphoblastic leukemia. Blood. 2013;122:74–82. doi: 10.1182/blood-2013-03-491092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, et al. Fusion between a novel Krüppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 1993;12:1161–1167. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 34.Nutt SL, Metcalf D, D’Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med. 2005;201:221–231. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seki M, et al. Recurrent SPI1 (PU.1) fusions in high-risk pediatric T cell acute lymphoblastic leukemia. Nat Genet. 2017;49:1274–1281. doi: 10.1038/ng.3900. [DOI] [PubMed] [Google Scholar]
- 36.Liu YF, et al. Genomic profiling of adult and pediatric B-cell acute lymphoblastic leukemia. EBioMedicine. 2016;8:173–183. doi: 10.1016/j.ebiom.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets have been deposited in the Chinese Leukemia Genotype-Phenotype Archive (bioinfo.rjh.com.cn/cga) under accession no. CGAS00000000002.