Abstract
Inherited platelet disorders are a heterogeneous group of rare diseases, caused by inherited defects in platelet production and/or function. Their genetic diagnosis would benefit clinical care, prognosis and preventative treatments. Until recently, this diagnosis has usually been performed via Sanger sequencing of a limited number of candidate genes. High-throughput sequencing is revolutionizing the genetic diagnosis of diseases, including bleeding disorders. We have designed a novel high-throughput sequencing platform to investigate the unknown molecular pathology in a cohort of 82 patients with inherited platelet disorders. Thirty-four (41.5%) patients presented with a phenotype strongly indicative of a particular type of platelet disorder. The other patients had clinical bleeding indicative of platelet dysfunction, but with no identifiable features. The high-throughput sequencing test enabled a molecular diagnosis in 70% of these patients. This sensitivity increased to 90% among patients suspected of having a defined platelet disorder. We found 57 different candidate variants in 28 genes, of which 70% had not previously been described. Following consensus guidelines, we qualified 68.4% and 26.3% of the candidate variants as being pathogenic and likely pathogenic, respectively. In addition to establishing definitive diagnoses of well-known inherited platelet disorders, high-throughput sequencing also identified rarer disorders such as sitosterolemia, filamin and actinin deficiencies, and G protein-coupled receptor defects. This included disease-causing variants in DIAPH1 (n=2) and RASGRP2 (n=3). Our study reinforces the feasibility of introducing high-throughput sequencing technology into the mainstream laboratory for the genetic diagnostic practice in inherited platelet disorders.
Introduction
Inherited platelet disorders (IPDs) are a heterogeneous group of rare diseases of variable clinical severity, usually characterized by mucocutaneous bleeding and excessive blood loss after trauma or surgery. Some IPDs are due to defects in genes that encode proteins that play critical roles in megakaryopoiesis and proplatelet production, leading to an inherited thrombocytopenia (IT). Alternatively, the molecular pathology may affect the development or maintenance of the platelet ultrastructure, the formation and cargo of granules, or platelet responses to agonists, which are known collectively as inherited platelet function disorders (IPFDs). Not infrequently, IPDs combine thrombocytopenia and impaired platelet function.1,2
Although phenotype-guided diagnosis is straightforward in some IPDs, including Bernard Soulier syndrome (BSS), Glanzmann Thrombasthenia (GT) and Hermansky-Pudlak syndrome (HPS), due to the severity of the bleeding and a readily available flow cytometry test, or to the syndromic nature of the disorder, most IPDs lack distinctive clinical and laboratory features. The diagnosis of this group remains a challenge even under expert analysis. Consequently, many IPDs remain underdiagnosed, despite the many diagnostic algorithms that have been proposed.3 Moreover, until recently, IPD diagnosis at the genetic level has been attained in fewer than half of patients, with the greatest success being seen in specialized centers.4,5
Genetic diagnosis for IPD families would facilitate better clinical care, prognosis and preventive treatments, which are especially important for clinically severe IPDs, or for those platelet syndromes that associate with an increasing risk of malignancy, and which require genetic counseling.6–8 While genotyping based on Sanger sequencing of candidate genes has been used successfully since the early 1990s for some IPDs,4,5 the approach is, at present, costly, time-consuming and not applicable to disorders whose phenotype-based diagnosis is not straightforward and for which there is no obvious candidate gene.6–8 The recent advent of high-throughput sequencing (HTS), either whole-genome sequencing (WGS), whole-exome sequencing (WES), and targeted sequencing of pre-specified genes, has begun to revolutionize the field of genetic diagnosis and is rapidly becoming established in clinical practice.9,10 With respect to bleeding disorders, HTS is emerging as a valuable tool for the molecular diagnosis of hemophilia and other rare coagulation disorders, and also for genotyping in IPDs.9,11 Thus, HTS is increasingly important as the first-line of the diagnostic investigation of these diseases.3,6,8,12,13
Herein, we report the design and implementation of a multigenic HTS platform for genetic analysis of IPD patients. The application of this novel approach has greatly aided our diagnostic process, resulting in a conclusive molecular diagnosis in the largest series of IPD patients investigated in the Iberian Peninsula.
Methods
Patient enrollment, blood sampling, platelet phenotyping and DNA isolation
We studied 92 unrelated patients with suspected IPD, enrolled prospectively within the aims of the multicenter “Functional and Molecular Characterization of Patients with Inherited Platelet Disorders” project, under the scientific sponsorship of the Spanish Society of Thrombosis and Haemostasis. Patients with known acquired disorders or clotting factor defects were excluded. Investigations abided by the Declaration of Helsinki and were approved by the Local Ethical Committees of the Instituto de Investigación Biomédica (IBSAL, Salamanca, Spain) and Hospital Universitario Reina Sofía (Murcia, Spain). All patients gave their written informed consent.
Clinical data of all patients were reviewed by the investigating team and their bleeding symptoms were reassessed and scored using the International Society on Thrombosis and Haemostasis Bleeding Assessment Tool (ISTH-BAT) questionnaire. Venous blood samples were obtained for platelet phenotyping4 and molecular studies (Online Supplementary Methods).
Custom HTS panel design, sequencing and data analysis
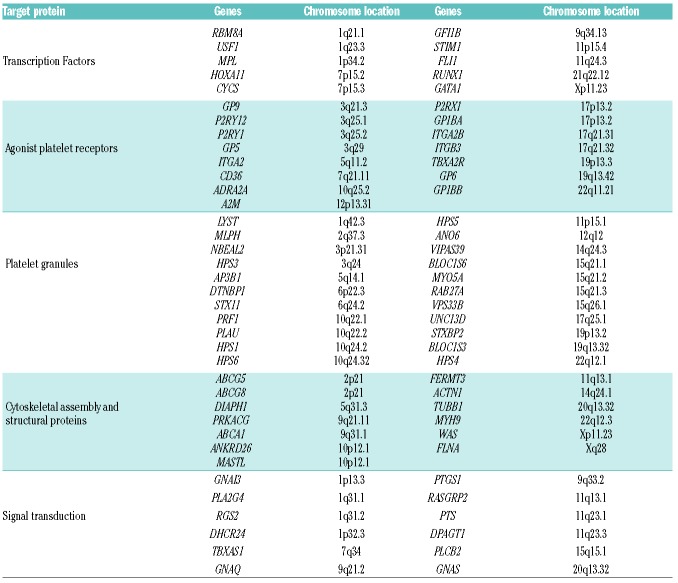
Using Design Studio (Illumina, San Diego, CA, USA), we designed a HTS platform with 1399 probes targeting 1106 regions, including exons, untranslated regions (UTRs) and flanking regions, of 72 genes that are associated with IPDs and/or are significant in platelet physiology (Table 1). Sequence data generated by HTS was mapped to the Reference Human Genome (hg19) with MiSeq integrated computer software (MiSeq Reporter, Illumina), which uses a Burrows-Wheeler-Aligner (BWA) and Genome Analysis Tool Kit (GATK) for variant-calling of single nucleotide variants (SNVs) and short insertions/deletions (InDels).14,15 Secondary data analysis, sequence alignment and variant detection was performed with Variant Studio Data Analysis and Integrative Genomics Viewer (IGV) (Broad Institute, Cambridge, MA, USA) software. The coverage per base was ≥100 reads. The first step consisted of a quality filter based on a Phred score >20, Quality >20 and Read coverage >30 at each position within the reads, to indicate high sequence quality. Data was then filtered according to the severity of the consequence, in order to prioritize variants leading to an amino acid change in the protein sequence (missense, nonsense, frameshift) and those in the splice site and UTRs. Apart from exceptional cases, synonymous and intronic variants were disregarded. Minor allelic frequencies (MAFs) were consulted in the Exome Variant Server, 1000 Genomes Browser and exome aggregation consortium (ExAC) databases; variants with a MAF of <0.05 were selected for further analysis. The other variants were searched for across sources, such as the dbSNP147, the Catalog of Somatic Mutations in Cancer (COSMIC), the National Center for Biotechnology Information (NCBI) ClinVar, the HGMD professional database, PubMed, Online Mendelian Inheritance in Man (OMIM), and locus-specific mutation databases in an attempt to identify variants known to cause IPDs (Online Supplementary Figure S1).16 Several in silico tools, Polymorphism Phenotyping v2 (PolyPhen-2), Sorting Intolerant From Tolerant (SIFT), Mutation Taster, MutationAssessor and Functional Analysis Through Hidden Markov Models (fathmm), were used to predict the functional effects and pathogenicity of the novel variants. We followed the guidelines of the American College of Medical Genetics and Genomics and Association for Molecular Pathology,17 to qualify each identified variant as a “pathogenic variant” (PV), “likely pathogenic variant” (LPV) or “variant of uncertain significance”(VUS; Online Supplementary Figure S2).
Table 1.
Genes included in the HTS platform for molecular screening of IPDs.
For more methodological details, see the Online Supplementary Methods.
Results
Patient characteristics
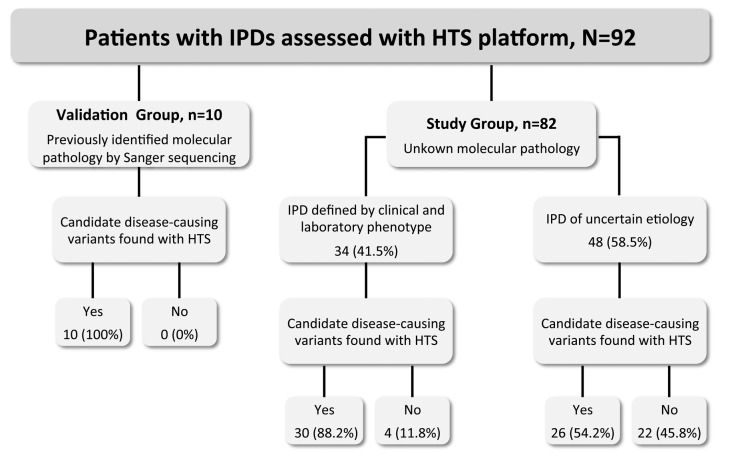
Patients were divided into 2 groups (Figure 1): i) a small validation group comprising 10 patients previously characterized at the functional and molecular level,4,18 harboring known pathogenic variants identified by Sanger sequencing (Online Supplementary Table S1), and ii) a larger study group of 82 patients who were enrolled in the project because of variable bleeding diathesis and abnormalities in the number or function of platelets, or both, but with unknown molecular pathology. Only 3 (3.7%) of these patients had undergone Sanger sequencing of candidate genes, and this had failed to identify candidate disease-causing variants.
Figure 1.
Classification of the 92 IPD patients sequenced with a novel HTS platform. Ninety-two unrelated patients with a suspicion of IPD were enrolled in the project “Functional and Molecular Characterization of Patients with Inherited Platelet Disorders”. Patients fell into 2 main groups: on the left, a validation group comprising 10 IPD patients harboring known pathogenic variants identified by Sanger sequencing (Online Supplementary Table S1), and on the right, a study group of 82 IPD patients with unknown molecular pathology. DNA from all patients was sequenced with an HTS platform targeting 72 genes (Table 1), as described in the Methods. The identified genetic variants were prioritized and assessed for pathogenicity, as stated in the Methods. IPD: inherited platelet disorder; HTS: high-throughput sequencing.
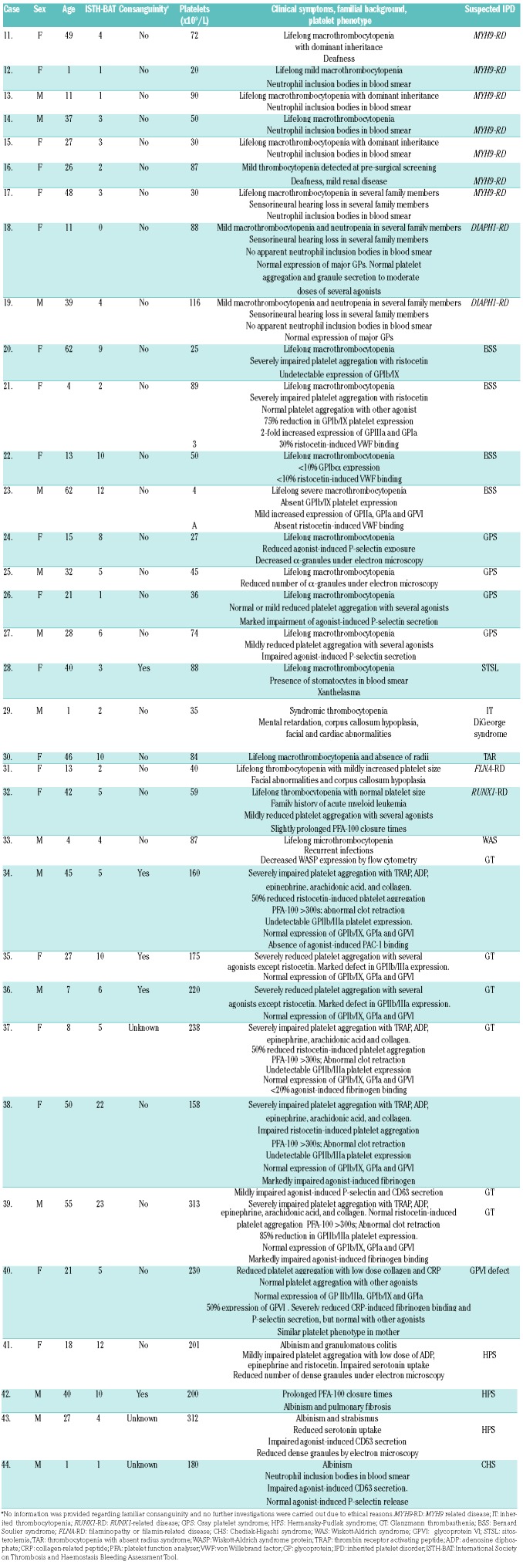
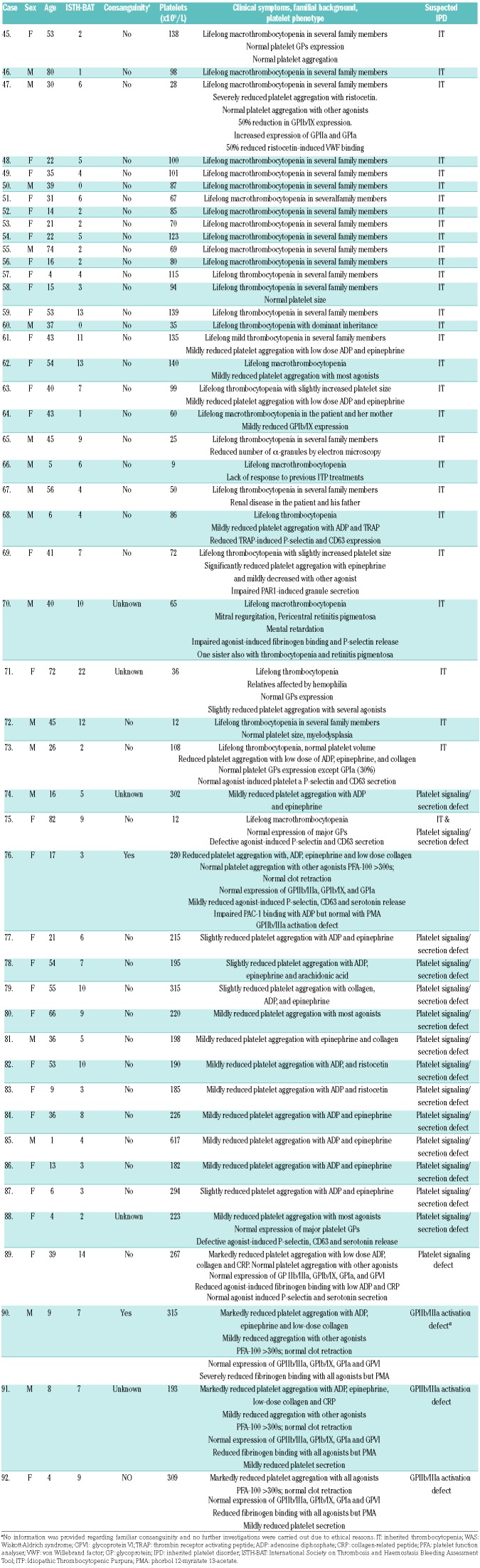
The major clinical and biological characteristics of the 82 patients in the study group are summarized in Table 2 and Table 3. The majority (62.2%) were female; median age was 29 (1–82) years; median bleeding score was 5 (0–23); median platelet count was 96 (4–617) ×109/L. Fifty-three cases (64.6%) presented with lifelong thrombocytopenia as the main inclusion criterion and the others had laboratory abnormalities consistent with an IPFD. Forty patients (50%) had a family history of bleeding, thrombocytopenia and/or hematological malignancies. In 34 cases (41.5%), clinical background and centralized laboratory assessment supported the diagnosis of a particular type of IT or IPFD: MYH9 related disease (MYH9-RD), n=7; DIAPH1 related disease (DIAPH1-RD), n=2; BSS, n=4; Gray platelet syndrome (GPS), n=4; sitosterolemia (STSL), n=1; DiGeorge syndrome, n=1; thrombocytopenia with absent radius syndrome (TAR), n=1; filaminopathy or filamin-related disease (FLNA-RD), n=1; RUNX1-related disease (RUNX1-RD), n=1; Wiskott-Aldrich syndrome (WAS), n=1; GT, n=6; glycoprotein VI (GPVI) signalling defect, n=1; HPS, n=3; Chediak-Higashi syndrome (CHS=1), n=1. The remaining 48 patients (58.5%) either had low platelet counts (n=30) and/or platelet function abnormalities (n=18) of uncertain etiology (Table 2 and Table 3).
Table 2.
General characteristics of 34 patients with a clinical and biological phenotype suggesting a particular type of IPD.
Table 3.
Clinical and biological characteristics of 48 patients with IPD of uncertain etiology.
General performance of the HTS assay and validation of the HTS multigenic platform
The 72 genes included in the panel (Table 1) were analyzed for all patients. We successfully sequenced 95.6% of the 1106 target regions at a minimum coverage depth of 100 for each nucleotide base-pair position of interest. The remaining 4.4% of regions could not be sequenced with adequate coverage using the NGS platform (Online Supplementary Table S2). In addition, the mean fraction of exonic bases covered at 20X and 50X was 0.991 and 0.987, respectively.
To validate the accuracy of our HTS platform for detecting causative variants within these genes, we assayed, in a blind manner, DNA from the 10 patients with previously ascertained pathogenic variants by Sanger sequencing. These included 12 SNV (9 missense variants and 3 nonsense changes) and 1 deletion within 8 genes (Online Supplementary Table S1).4,18 In each case the HTS test, and accompanying data analysis, identified the previously known genetic variant, thus demonstrating the high sensitivity of the platform. Reproducibility studies were also performed, in which 4 DNA samples from the validation groups (Cases 1, 4, 8 and 10) were assayed in triplicate in 3 separate runs. After applying the prioritization protocol, we found 100% concordance between runs in detecting the correspondent variants present in each DNA, demonstrating high intra-batch and inter-batch reproducibility of the platform.
General performance of the HTS multigenic platform in the IPD study group
The high sensitivity and reproducibility displayed by our HTS platform in the validation group prompted us to use it as the first genotyping method in the 82 patients of the study group. As stated above, this included 34 (41.5%) cases with a strong suspicion of a defined IPD, and 48 (58.5%) patients with phenotypes not suggestive of a particular type of IPD (Figure 1).
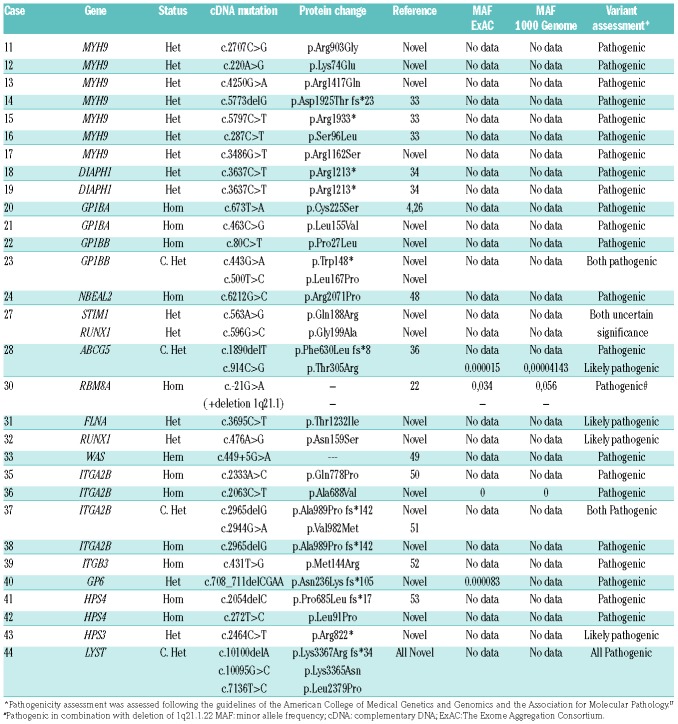
Overall, by applying the bioinformatic tools described in the Methods, there was a median of 169 (range: 128–230) sequence variations across the 72 genes in the 82 patients. We prioritized 62 of these candidate variants in 56 (68.3%) index cases (Table 4 and Table 5). This included 40 missense variants, 8 nonsense variants, 11 frameshift variants (deletion=11, Insertion-Deletion=1), 2 variants in the UTR region and 1 splice site variant within 29 genes. All these genetic changes were confirmed by Sanger sequencing and segregated into available family members. Variants were inherited in a homozygous/hemizygous manner in 14 patients, 4 cases were compound heterozygous defects and the other 38 were heterozygous. Forty-one (72%) variants had not been previously reported in public databases and are therefore novel variants. Assessment of variant pathogenicity, following consensus guidelines,17 led to 43 (69%) variants being classified as PV, 16 (26%) as LPV and 3 as VUS (5%) (Table 4 and Table 5).
Table 4.
Genetic variants identified with the HTS test in patients with suspicion of particular IPDs according to their clinical and biological phenotype.
Table 5.
Genetic variants identified with the HTS test in the cohort of patients with IPDs of uncertain etiology on the basis of clinical and biological phenotype.
As expected, the HTS platform was highly sensitive in detecting causal variants among patients with a strong suspicion of defined IPDs. In 30 out of 34 (88.2%) of these patients we identified 34 different candidate variants in 17 genes, which were identified in most cases as PVs (n=28), but in 4 cases as LPVs and in 2 as VUS (Table 4). These genetic findings gave rise to confirmation of diagnosis of BSS (n=4), MYH9-RD (n=7), TAR (n=1), GPS (n=1), FLNA-RD (n=1), RUNX1-RD, (n=1), GT (n=5), HPS (n=3), CHS (n=1), GPVI defect (n=1), and 2 rare cases of WAS and STSL that we have recently reported in detail (Online Supplementary Table S3). In 2 patients (Cases 18 and 19) with macrothrombocytopenia, mild neutropenia and familiar sensorineural hearing loss, the HTS identified the p.Arg1213* variant in DIAPH1. In these families, the p.Arg1213* variant displayed full penetrance with deafness, minor impact on platelet functional status, and a moderate effect on the platelet levels of DIAPH1 and Tubulin β1 (Online Supplementary Figure S3, S4, S5 and S6). Moreover, HTS failed to identify PV and LPV variants in 1 patient with what was previously thought to be a straightforward diagnosis of GT (Case 34) and in 3 cases with a suspicion of GPS (Cases 24–27; Table 2). Furthermore, in Case 27, who had suspected GPS, we found 2 novel missense variants in STIM1 and RUNX1 (Table 4). These variants, similarly to that of a variant in GNAS in Case 86 (Table 5), were classified as VUS on the basis of their identification only in the index case, since no relatives were available for screening, there are no previous reports in other patients, and no specific studies demonstrating a deleterious functional effect.17
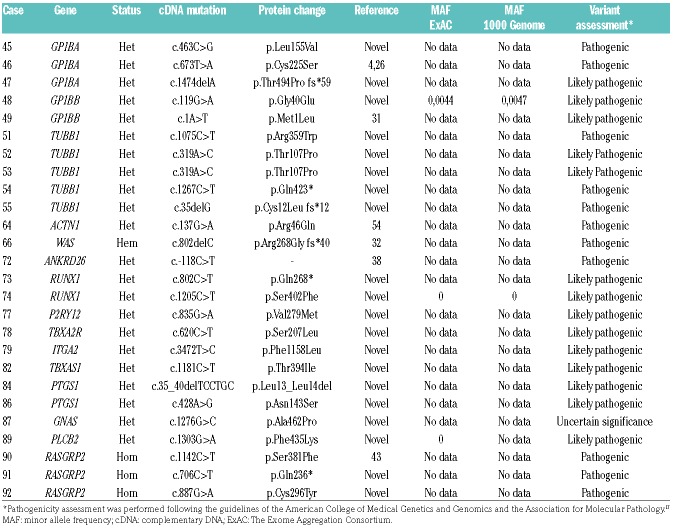
The overall sensitivity of the HTS platform was significantly lower (Z-score=3.0599; P=0.00222) among the subgroup of IPD of uncertain etiology. Herein, we identified 26 different variants located in 16 genes in 26 cases (54.2%). Most of them were classified as PV or LPVs (12 and 13, respectively) and one was identified as VUS. Accordingly, these patients were assigned a diagnosis of monoallelic BSS (n=5); tubulin β1-related thrombocytopenia (TUBB1-RT), n=5; RUNX1-RD, n=2; actinin-1 related thrombocytopenia (ACTN1-RT), n=1; type-2 thrombocytopenia (ANKRD26-RT), n=1; WAS, n=1. The other cases presumably had congenital defects in the gene encoding for ADP receptor P2Y12 (P2RY12)(n=1); thromboxane A2 receptor gene (TBXA2R) (n=1); Ca2+ DAG-regulated guanine nucleotide exchange factor I or CalDAG-GEFI (RAS-GRP2) (n=3) (Online Supplementary Table S3); G-protein α subunit or Gs-α (GNAS) (n=1); phospholipase β2 (PLCB2) (n=1); prostaglandin-endoperoxide synthase 1 (PTGS1) (n=2); thromboxane A synthase 1 (TBXAS1) (n=1); and integrin α2 (ITGA2)(n=1); (Table 5).
Discussion
In recent years, the identification of the underlying molecular pathology has become the cornerstone for establishing a conclusive diagnosis of IPDs, leading to better clinical care and follow-up of these patients.6,7 Until recently, Sanger sequencing of candidate genes and linkage studies have been the main tools for IPD genetic diagnosis, providing outstanding but limited results.4,5 In 2010, HTS emerged as the herald of the revolution in genetic diagnosis of human diseases, including IPDs.6–8,13
The study herein demonstrates the feasibility of an in-house designed HTS platform for rapid genetic characterization of patients with IPDs in a clinical setting. Indeed, 60% of patients in our study had no diagnostic features allowing for a straightforward selection of candidate genes and the use of Sanger sequencing. Further, for those patients who presented with a phenotype indicative of a particular type of disorder, most were associated with diseases that can be due to defects in several genes and/or in large genes, thus hampering Sanger sequencing (Table 4). We have already used a detailed phenotyping and Sanger sequencing approach for this group of IPD patients,4 and it compares negatively with our current HTS test both in terms of time and cost, although we did not undertake a full cost-benefit analysis.
The current version of this platform allows for multiplex analysis of coding and selected non-coding regions of 72 genes including those previously associated with IPDs (Table 1). However, it can be easily modified to include additional genes such as those recently identified: SLFN14, ETV6, and SCR.2,6,13 None of the patients in this IPD study had phenotypes consistent with these genes, but they are of interest for novel cases referred to our project. The ThromboGenomics platform does not currently include these genes, but patients with gain-of-function variants in SCR have been reported.19
We first performed a validation study of previously ascertained genetic variants in blinded samples, which confirmed the strong analytical sensitivity and reproducibility of the HTS platform, and the appropriateness of our variant-filtering strategy. We detected the 13 known variants in eight genes, including missense and nonsense variants and small deletions, from 10 patients (Online Supplementary Table S1).
Subsequently, we used the HTS multigenic platform for the first genetic analysis of a cohort of 82 patients prospectively enrolled from different hospitals in order to realize the aim of the collaborative project “Functional and Molecular Characterization of Patients with Inherited Platelet Disorders”. The clinical and platelet phenotypic presentation was highly variable among these patients, which is consistent with the widely recognized heterogeneity of IPDs. Most patients (65%) presented with thrombocytopenia as their main hematological feature. Many of them also displayed a variable degree of platelet dysfunction consistent with recent findings in other IT series.20 Not surprisingly, the majority of patients were women (62.2%), as more frequent bleeding complications in females facilitate their clinical identification. Moreover, most individuals (67%) were adults (aged >18 years) at the time of our centralized phenotypic evaluation and selection for molecular analysis. We have previously shown that, at least in our clinical setting, there is a significant delay between the time patients are suspected to have an IPD and the time confirmatory phenotypic diagnosis and molecular characterization are performed.4 The significance of this should not be underestimated, as it implies that many patients remain without a conclusive diagnosis for years, and thus are at risk of receiving inappropriate treatments. Multicentre collaboration, as supported by our project, and availability of HTS are expected to change this.
Overall, our HTS approach enabled a molecular diagnosis in 68.3% of the patients (Figure 1), which is a much higher success rate than we achieved using only candidate gene sequencing.4 Remarkably, this sensitivity increased to nearly 90% for patients presenting with a well-defined clinical and laboratory phenotype indicative of a particular type of IPD. These results resemble those recently reported for the ThromboGenomics HTS platform, which covers the molecular screening of 63 genes.11 However, among patients presenting with an unclear phenotype other than bleeding or low platelet count, i.e., IPDs of uncertain etiology, our HTS platform was only able to identify candidate variants in about 50% of patients. This value is higher than that reported by the ThromboGenomics consortium in the same category of patients,11 but similar to the sensitivity recently achieved by WES in a limited cohort of IT patients with unknown etiology.20 Several factors could potentially contribute to this difference. Our study recruited only patients with established IPDs, even though the etiology of about half of them could not be inferred from clinical and laboratory data. In contrast, the ThromboGenomics study enrolled, among “cases with a highly uncertain etiology”, patients with bleeding problems but normal platelet function tests, and a few patients who had experienced thrombotic events. In addition, the gene content in both platforms is different, with only 33 genes from our platform (Table 1) being present in the ThromboGenomics platform, which also included genes involved in coagulation disorders.11
The failure to identify candidate genetic defects in about 30% of cases may be due to intrinsic limitations of our HTS platform. First, the causative gene may not be present in the panel. Second, HTS methods can miss large deletions or duplications (>200bp), copy number variants involving >1000bp, or big structural chromosomal variants, translocations and aneuploidy, unless they have been specifically designed for such a purpose.21 Thus, for successful molecular diagnosis of certain cases, the HTS test must be combined with other molecular approaches such as comparative genomic hybridization (CGH) array, quantitative polymerase chain reaction (q-PCR), or multiplex ligation-dependent probe amplification (MLPA). For instance, in Case 30, who was diagnosed with TAR, the HTS test detected the uncommon rs139428292 single nucleotide polymorphism (SNP), inherited from the father (Online Supplementary Figure S7), but was insensitive to the pathogenic microdeletion in 1q21.1 which is associated with the disorder,22 and which was later detected by CGH-array analysis (Online Supplementary Figure S7). Standard analysis of HTS results also failed to identify candidate variants in Case 29, who had a clinical suspicion of DiGeorge syndrome (Table 2). However, massive parallel sequencing of CNVs analysis by HTS identified a RUNX1 deletion (Online Supplementary Figure S8), and hybridization in situ analysis revealed a 21q22 microdeletion (data not shown), resulting in RUNX1 haploinsufficiency.23 Moreover, our HTS platform also left a few target regions with insufficient coverage (<20×). This affected up to 21 genes, although none of them appeared to be related to the phenotypes of the corresponding patients (Online Supplementary Table S2). In Case 34, who had an unambiguous phenotype of GT but no HTS findings, no β3 messenger ribonucleic acid (mRNA) was detected, but Sanger sequencing of the candidate ITGB3 in the patient DNA also yielded negative results. It has been suggested that the few GT patients in whom no ITGA2B and ITGB3 variants are detected might benefit from whole genome analysis. This may unravel defects in regulatory elements and deep intronic regions that adversely affect the transcription or post-translational modifications and trafficking of αIIbβ3 integrin.24 In addition, for most IPDs of uncertain etiology and in a few suspected ones, such as our Cases 25–27 who had a suspicion of GPS, the lack of genetic variants in our study highlights that many genes which cause IPDs or pathogenic variants affecting noncoding regulatory regions of the genome remain unidentified. Large-scale HTS projects, such as the 100,000 Genomes project in the UK and other novel approaches to gene discovery6,7 will help to overcome this limitation.
In this series of patients, we found 57 different candidate variants in 28 genes, 70% of which were absent from the main reference databases, thus emphasizing the great heterogeneity of the molecular pathology underlying IPDs. Appropriate interpretation of the pathogenicity of candidate genetic variants found by HTS in IPDs remains a major challenge, especially for novel variants, even if present in well-established IPD genes. To prevent misinterpretation, the use of consensus guidelines is highly recommended,13,17 although there remains significant discordance between laboratories.25 Herein, following established guidelines,17 we classified 68.4% and 26.3% of the identified candidate variants as PV and LPV, respectively. In 2 cases (27 and 87) we found 3 (5.2%) novel variants affecting STIM1, RUNX1 and GNAS which qualified as VUS. Lack of segregation of novel candidate variants in the pedigrees is critical to prevent over-interpretation of pathogenicity. As an example, we disregarded novel variants in GP1BA (Case 45, c.1022C>G, p.Ser341Cys), FLNA (Case 63, c.5933 C>T, p.Thr1978Met), MASTL (Case 58, (c.836C>G; p.Pro279Arg) and NBEAL2 (Case 70, c.3424G>T, p.Gly1142Cys), since they were present in family members exhibiting no platelet defects.
The genetic findings of this study are of clinical and scientific relevance. We established a conclusive diagnosis of autosomal recessive severe IPDs or X-linked disorders in about 25% of the patients, which would have informed decisions regarding their clinical care. These included diagnosis of BSS (n=4), GT (n=6), HPS (n=3), CHS (n=1), GPS (n=1), TAR (n=1) and WAS (n=2). One BSS patient carried a missense p.Cys225Ser variant in GP1BA, which has been previously identified in other patients from the Iberian peninsula, thereby suggesting a common ancestry.4,26 Another patient carried the novel change p.Leu155Val, also in GP1BA, which resembles the previously reported Bolzano variant p.Ala156Val, as it associates with severe biallelic BSS and nearly asymptomatic monoallelic BSS.26 It is worth mentioning that heterozygous variants in the GP1BA and GP1BB genes were a common cause of dominant IT in our series of patients, lending further weight to the idea that this condition might be more common than previously recognized.26,27 Six GT patients were diagnosed in this study. HTS revealed no pathogenic variants in ITGA2B and ITGB3 in 1 of these (Case 34), although the patient presented with an obvious type I GT platelet phenotype and no detectable levels of ITGB3 mRNA by quantitative real-time (qRT)-PCR. This case, along with those few GT patients previously reported to have no detectable mutations in ITGA2B and ITGB3,24,28 might benefit from whole-genome analysis with the aim of identifying defects in regulatory elements and deep intronic regions that adversely affect the transcription or post-translational modifications and trafficking of αIIbβ3. HTS cannot fully replace clinical evaluation and platelet phenotyping as it may result in misdiagnosis, but molecular characterization should be a component of an integral protocol for diagnosis. A similar argument may be valid for the 3 patients presenting with a phenotype suggestive of GPS (Cases 25–27), in whom we found no candidate variants in NBEAL or GFI1B. This is in accordance with multiple unknown genotypic alterations that may underlie the GPS phenotype. The clinical value of patient care and an accessible HTS test is also well exemplified in the case of severe multi-system IPDs such as HPS (Cases 41–43), CHS (Case 44) and WAS (Cases 33 and 66), in which the gene affected, as in the cases of HPS, or the type of mutation, regarding CHS and WAS, is likely to predict phenotype.29–31 Early identification of patients with these potentially life-threatening IPDs, depending on the genotype, is critical to the successful application of hematopoietic stem cell transplantation, and possibly gene therapy in some cases. Remarkably, in this project 3 young children had their molecular diagnosis confirmed as having either CHS or WAS defects, and 1 of the WAS patients (Case 66) atypically presented with macrothrombocytopenia of uncertain phenotype.32
The importance of genotype in predicting a patientś clinical phenotype is also apparent in MYH9-RD.33 Herein, our HTS platform identified 7 pathogenic variants in MYH9 in 7 unrelated cases. Three were unreported variants affecting the head domain of the protein, the SH3/MDi region, and another novel variant affected the tail domain. Notably, 5 of these patients were young individuals (<30 years old) who presented with no extra-hematological manifestations, and who could therefore benefit from close follow-up.
Two notable pedigrees, Cases 18 and 19, were originally referred to us with a clinical suspicion MYH9-RD on the basis of mild macrothrombocytopenia and autosomal dominant sensorineural hearing loss. Subsequent analysis of the clinical and biological records in all relatives affected by deafness showed variable thrombocytopenia, absence of inclusion bodies in neutrophils and mild neutropenia. These data did not support the suspicion of MYH9-RD, but prompted us to suspect an underlying molecular pathology in DIAPH1. In OMIM this gene is usually linked to autosomal dominant deafness with/without thrombocytopenia, and thus is considered to be a phenocopy of MYH9. Remarkably, our HTS test revealed, in both pedigrees, the variant p.Arg1213* in DIAPH1, with full penetrance and with deafness presenting as the main clinical abnormality. This is a gain-of-function variant affecting autoregulation of DIAPH1 activity and proplatelet formation, recently identified by the ThromboGenomics consortium in 2 unrelated pedigrees with a similar phenotype to that in our cases.34 Functional studies of our patients revealed a minor effect of the p.Arg1213* variant in platelet aggregation and secretion, while it associates with a mildly reduced platelet expression of the DIAPH1 protein and a slightly higher level of tubulin β1 (Online Supplementary Material). Our current data generally concord with the previously reported phenotype in p.Arg1213* carriers,34 and gives further support to the idea that DIAPH1-RD is a novel type of IPD.
Few other rare cases were identified with a novel molecular pathology affecting genes encoding cytoskeletal proteins involved in proplatelet formation, such as TUBB1 (Cases 51–55), ACTN1 (Case 64) and FLNA (Case 31).2 The later patient, a 13-year-old girl who displayed moderate to severe thrombocytopenia, had required sporadic blood transfusions and had been investigated for suspicion of WAS and CAMT, despite some clinical signs suggesting a filaminopathy (Table 2). Our identification of the novel missense mutation (c.3695C>T; p.Thr1232Ile) in exon 22 of FLNA supported the diagnosis of filaminopathy and warrants a more specific clinical investigation. This novel FLNA variant appears to be a de novo variant in the patient (Online Supplementary Figure S9), a phenomenon that occurs in about 20–30% of cases with sporadic bilateral periventricular nodular heterotopias.35 Additionally, 1 patient (Case 28) was characterized with STSL, a rare inherited sterol storage disorder.36 Functional in vitro studies are underway to explore the potential deleterious effect of these variants in platelet formation.
Three novel variants in RUNX1 were found in Cases 32, 73 and 74, who all presented with mild platelet dysfunction, whilst 2 of them also had thrombocytopenia. Only members of 1 of these families had a history of hematological malignancy. These variants are expected to affect the function of RUNX1, as 2 (p.Gln268* and p.Asn159Ser) lie within the RUNT homology domain which mediates DNA binding and heterodimerization with CBFβ and the remaining (p.Ser402Phe) in the C-terminal inhibitory domain of RUNX1.37 Comprehensive interpretation of variants in this transcription factor, like those in ANKRD26 (Case 72 in our series), is relevant as such variants may increase the risk of developing myeloid malignancies.37,38
In Case 40, the selective platelet dysfunction at the GPVI level, which likely had autosomal dominant inheritance, matched well with the finding of a novel in-frame deletion in GP6 detected by HTS. Among the subgroup of patients presenting with mild platelet dysfunction of uncertain etiology, 8 (44%) pedigrees exhibited likely pathogenic heterozygous variants in genes encoding other platelet receptors (ITGA2, TBXA2R, P2YR12) or enzymes involved in second messenger release and platelet signal transduction (GNAS, PLCB2, PTGS1, TBXAS1; Table 5). Further studies are required to determine the contribution, if any, of these genetic defects to the platelet dysfunction and bleeding tendency of these patients. Interestingly, other patients previously reported to have inherited defects in these platelet proteins were also heterozygous.39,40 Bleeding diathesis in these patients may be facilitated by co-inheritance with other genetic disorders of hemostasis, such as type 1 von Willebrand disease.41
A key protein for integrin signaling in platelets and neutrophils is the guanine nucleotide exchange factor CalDAG-GEFI. Recently a variant in RASGRP2, the gene encoding CalDAG-GEFI, was identified in 3 siblings with impaired platelet function and bleeding diathesis.42 Herein, our HTS test identified 3 novel variants in RASGRP2 (p.Ser381Phe, p. Gln236* and p. Cys296Tyr) from 3 unrelated children with lifelong severe bleeding complications and reduced platelet aggregation with most agonists. Further functional studies demonstrated that these novel variants severely affect CalDAG-GEFI expression and activity, leading to defective agonist-induced integrin activation in platelets and neutrophils.43,44 Of late other patients harboring pathogenic variants in RASGRP2 have been identified,45–47 indicating that this type of IPFD might occur more frequently than previously thought.
Conclusions
This study demonstrates that our HTS platform is an accurate, reproducible and reliable tool for the genetic characterization of IPDs. Using this approach, we can achieve a molecular diagnosis in most patients with a suspected etiology, and in about half the cases presenting with a disease of highly uncertain biological cause. Our findings reinforce the feasibility of introducing this technology into main-stream genetic testing for diagnosing IPDs. Patients with an IPD in which the HTS platform fails to identify the underlying molecular pathology are candidates for examination using less restrictive molecular approaches, such as WES or WGS. The use of human phenotype ontology codification, consensus guidelines for interpreting genetic variants, and in silico bioinformatics analysis tools to facilitate the identification of candidate causative variants will be important in aiding this process. However, definitive pathogenicity assignment of novel rare variants must be established on the basis of their identification in unrelated pedigrees with similar phenotype and/or demonstrative functional studies.
Supplementary Material
Acknowledgments
We acknowledge all the patients and their families for providing samples. We thank Dr Phil Mason for his help with technical aspects. We are grateful to the following clinicians: Members of the Castilla y León Society of Thrombosis and Haemostasis Group: RM Fisac (Hospital General, Segovia), MP Martínez-Badas (Complejo Asistencial de Ávila), LJ García-Frade and E Fontecha (Hospital Universitario Río Hortega, Valladolid), JM Martín-Antorán (Complejo Asistencial de Palencia), C Aguilera (Hospital de El Bierzo, Ponferrada), B Pérez (Complejo Asistencial de León) MJ Cebeira (Hospital Clínico de Valladolid), TJ González-López (Complejo Asistencial de Burgos), RM Henar-Cantalejo (Hospital General de Aranda de Duero), R Campos (Hospital de Jerez), E Pardal (Hospital Virgen del Puerto, Plasencia), R Ramos (Hospital Infanta Cristina, Badajoz), R Vidal and MP Llamas (Fundación Jiménez Díaz, Madrid), M Salces (Hospital Universitario La Paz, Madrid), P Olivera (Hospital Vall d’ Hebron, Barcelona), A Repáraz (Unidad de Citogenética y Genética Molecular, Hospital Álvaro Cunqueiro, Vigo), G Iruin (Hospital de Cruces, Bilbao), AR Cid (Hospital Universitario La Fe, Valencia), E Bardón (Hospital Universitario de Torrejón, Madrid), A Galera (Hospital Universitario Virgen de la Arrixaca, Murcia), JL Fuster and ME LLinares (Hospital Universitario Virgen de la Arrixaca, Murcia), S Riesco, MC Mendoza, A Benito and A Hortal (Hospital Universitario de Salamanca), MT Alonso (Hospital Universitario de Valladolid), J Huertas (Hospital Gregorio Marañón, Madrid), I Astigarraga (Hospital de Cruces, Bilbao), D Jaimes (Hospital de Donostia), H Cano (Hospital Los Arcos, Murcia), J Mateo (Hospital San Pablo, Barcelona), T Iturbe (Hospital Santa Lucia, Cartagena), R Berrueco (Hospital Sant Joan de Déu, Barcelona), M Lozano (Hospital Clinic, Barcelona), N Fernandez Mosteririn (Hospital Miguel Servet, Zaragoza), C Muñoz (Hospital Virgen de la Macarena, Sevilla), I Ancin (H. Cruces, Bilbao), T Jover (Hospital Universitario Virgen de la Arrixaca, Murcia), E Roselló (Hospital de Universitario de Bellvitge, Barcelona), EM Mingot, Hospital Universitario Carlos Haya de Málaga), RM Campos (Hospital de Jérez), JM Guinea (Hospital de Araba), M Trapero (Clínica Puerta de Hierro, Madrid), N Rollón (Hospital Virgen de la Salud, Toledo M) and Karkucak (Dpt. Medical Genetics, Sakarya University Training and Research Hospital, Turkey). We are also grateful to Irene Rodríguez, Sara González, Sandra Santos, Sandra Pujante, José Padilla, Ana Isabel Antón, Isabel Sánchez-Guiu, Eva Caparrós, Nerea Mota and Constantino Martínez for their help in isolating and processing DNA and for carrying out some of the platelet assays.
Footnotes
Funding
This study was supported by research grants from the Gerencia Regional de Salud (GRS 1370/A/16), ISCIII & Feder (PI14/01956), CIBERER CB15/00055, Fundación Séneca (19873/GERM/15) and Sociedad Española de Trombosis y Hemostasia (SETH). SPW holds a British Heart Foundation chair.
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/1/148
References
- 1.Nurden AT, Nurden P. Congenital platelet disorders and understanding of platelet function. Br J Haematol. 2014;165(2):165–178. [DOI] [PubMed] [Google Scholar]
- 2.Savoia A. Molecular basis of inherited thrombocytopenias: an update. Curr Opin Hematol. 2016;23(5):486–492. [DOI] [PubMed] [Google Scholar]
- 3.Gresele P. Diagnosis of inherited platelet function disorders: guidance from the SSC of the ISTH. J Thromb Haemost. 2015; 13(2):314–322. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez-Guiu I, Anton AI, Padilla J, et al. Functional and molecular characterization of inherited platelet disorders in the Iberian Peninsula: results from a collaborative study. Orphanet J Rare Dis. 2014;9:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watson SP, Lowe GC, Lordkipanidze M, Morgan NV. Genotyping and phenotyping of platelet function disorders. J Thromb Haemost. 2013;11 Suppl 1:351–363. [DOI] [PubMed] [Google Scholar]
- 6.Lentaigne C, Freson K, Laffan MA, Turro E, Ouwehand WH. Inherited platelet disorders: toward DNA-based diagnosis. Blood. 2016;127(23):2814–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westbury SK, Mumford AD. Genomics of platelet disorders. Haemophilia. 2016;22 Suppl 5:20–24. [DOI] [PubMed] [Google Scholar]
- 8.Sivapalaratnam S, Collins J, Gomez K. Diagnosis of inherited bleeding disorders in the genomic era. Br J Haematol. 2017; 179(3):363–376. [DOI] [PubMed] [Google Scholar]
- 9.Bastida JM, Del Rey M, Lozano ML, et al. Design and application of a 23-gene panel by next-generation sequencing for inherited coagulation bleeding disorders. Haemophilia. 2016;22(4):590–597. [DOI] [PubMed] [Google Scholar]
- 10.de Koning TJ, Jongbloed JD, Sikkema-Raddatz B, Sinke RJ. Targeted next-generation sequencing panels for monogenetic disorders in clinical diagnostics: the opportunities and challenges. Expert Rev Mol Diagn. 2015;15(1):61–70. [DOI] [PubMed] [Google Scholar]
- 11.Simeoni I, Stephens JC, Hu F, et al. A high-throughput sequencing test for diagnosing inherited bleeding, thrombotic, and platelet disorders. Blood. 2016;127(23):2791–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westbury SK, Turro E, Greene D, et al. Human phenotype ontology annotation and cluster analysis to unravel genetic defects in 707 cases with unexplained bleeding and platelet disorders. Genome Med. 2015;7(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freson K, Turro E. High-throughput sequencing approaches for diagnosing hereditary bleeding and platelet disorders. J Thromb Haemost. 2017;15(7):1262–1272. [DOI] [PubMed] [Google Scholar]
- 14.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stenson PD, Mort M, Ball EV, et al. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136(6):665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monteferrario D, Bolar NA, Marneth AE, et al. A dominant-negative GFI1B mutation in the gray platelet syndrome. N Engl J Med. 2014;370(3):245–253. [DOI] [PubMed] [Google Scholar]
- 19.Turro E, Greene D, Wijgaerts A, et al. A dominant gain-of-function mutation in universal tyrosine kinase SRC causes thrombocytopenia, myelofibrosis, bleeding, and bone pathologies. Sci Transl Med. 2016;8(328):328ra330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson B, Lowe GC, Futterer J, et al. Whole exome sequencing identifies genetic variants in inherited thrombocytopenia with secondary qualitative function defects. Haematologica. 2016;101(10):1170–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daber R, Sukhadia S, Morrissette JJ. Understanding the limitations of next generation sequencing informatics, an approach to clinical pipeline validation using artificial data sets. Cancer Genet. 2013;206(12):441–448. [DOI] [PubMed] [Google Scholar]
- 22.Albers CA, Paul DS, Schulze H, et al. Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat Genet. 2012; 44(4):435–439, S431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen RD, Wiedmeier SE, Yaish HM. A neonate with congenital amegakaryocytic thrombocytopenia associated with a chromosomal microdeletion at 21q22.11 including the gene RUNX1. J Perinatol. 2013;33(3):242–244. [DOI] [PubMed] [Google Scholar]
- 24.Kannan M, Saxena R. No genetic abnormalities identified in alpha2IIb and beta3: phenotype overcomes genotype in Glanzmann thrombasthenia. Int J Lab Hematol. 2017; 39(2):e41–e44. [DOI] [PubMed] [Google Scholar]
- 25.Amendola LM, Jarvik GP, Leo MC, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;98(6):1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savoia A, Kunishima S, De Rocco D, et al. Spectrum of the mutations in Bernard-Soulier syndrome. Hum Mutat. 2014; 35(9):1033–1045. [DOI] [PubMed] [Google Scholar]
- 27.Sivapalaratnam S, Westbury SK, Stephens JC, et al. Rare variants in GP1BB are responsible for autosomal dominant macrothrombocytopenia. Blood. 2017;129(4):520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nurden AT, Pillois X, Fiore M, et al. Expanding the mutation spectrum affecting alphaIIbbeta3 integrin in glanzmann thrombasthenia: screening of the ITGA2B and ITGB3 genes in a large international cohort. Hum Mutat. 2015;36(5):548–561. [DOI] [PubMed] [Google Scholar]
- 29.Buchbinder D, Nugent DJ, Fillipovich AH. Wiskott-Aldrich syndrome: diagnosis, current management, and emerging treatments. Appl Clin Genet. 2014;7:55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lozano ML, Rivera J, Sanchez-Guiu I, Vicente V. Towards the targeted management of Chediak-Higashi syndrome. Orphanet J Rare Dis. 2014;9:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez-Guiu I, Torregrosa JM, Velasco F, et al. Hermansky-Pudlak syndrome. Overview of clinical and molecular features and case report of a new HPS-1 variant. Hamostaseologie. 2014;34(4):301–309. [DOI] [PubMed] [Google Scholar]
- 32.Bastida JM, Del Rey M, Revilla N, et al. Wiskott-Aldrich syndrome in a child presenting with macrothrombocytopenia. Platelets. 2017;28(4):417–420. [DOI] [PubMed] [Google Scholar]
- 33.Pecci A, Klersy C, Gresele P, et al. MYH9-related disease: a novel prognostic model to predict the clinical evolution of the disease based on genotype-phenotype correlations. Hum Mutat. 2014;35(2):236–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stritt S, Nurden P, Turro E, et al. A gain-of-function variant in DIAPH1 causes dominant macrothrombocytopenia and hearing loss. Blood. 2016;127(23):2903–2914. [DOI] [PubMed] [Google Scholar]
- 35.Parrini E, Ramazzotti A, Dobyns WB, et al. Periventricular heterotopia: phenotypic heterogeneity and correlation with Filamin A mutations. Brain. 2006;129(Pt 7):1892–1906. [DOI] [PubMed] [Google Scholar]
- 36.Bastida JM, Benito R, Janusz K, et al. Two novel variants of the ABCG5 gene cause xanthelasmas and macrothrombocytopenia: a brief review of hematologic abnormalities of sitosterolemia. J Thromb Haemost. 2017;15(9):1859–1866. [DOI] [PubMed] [Google Scholar]
- 37.Daly ME. Transcription factor defects causing platelet disorders. Blood Rev. 2017;31(1):1–10. [DOI] [PubMed] [Google Scholar]
- 38.Noris P, Favier R, Alessi MC, et al. ANKRD26-related thrombocytopenia and myeloid malignancies. Blood. 2013;122(11): 1987–1989. [DOI] [PubMed] [Google Scholar]
- 39.Jones ML, Norman JE, Morgan NV, et al. Diversity and impact of rare variants in genes encoding the platelet G protein-coupled receptors. Thromb Haemost. 2015;113(4): 826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lecchi A, Razzari C, Paoletta S, et al. Identification of a new dysfunctional platelet P2Y12 receptor variant associated with bleeding diathesis. Blood. 2015;125(6):1006–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stockley J, Nisar SP, Leo VC, et al. Identification and characterization of novel variations in platelet G-protein coupled receptor (GPCR) genes in patients historically diagnosed with Type 1 von Willebrand Disease. PLoS One. 2015; 10(12):e0143913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Canault M, Ghalloussi D, Grosdidier C, et al. Human CalDAG-GEFI gene (RASGRP2) mutation affects platelet function and causes severe bleeding. J Exp Med. 2014;211(7): 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lozano ML, Cook A, Bastida JM, et al. Novel mutations in RASGRP2, which encodes CalDAG-GEFI, abrogate Rap1 activation, causing platelet dysfunction. Blood. 2016;128(9):1282–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sevivas T, Bastida JM, Paul DS, et al. Identification of two novel mutations in RASGRP2 affecting platelet CalDAG-GEFI expression and function in patients with bleeding diathesis. Platelets. 2017:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bermejo E, Alberto MF, Paul DS, et al. Marked bleeding diathesis in patients with platelet dysfunction due to a novel mutation in RASGRP2, encoding CalDAG-GEFI (p.Gly305Asp). Platelets. 2017:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kato H, Nakazawa Y, Kurokawa Y, et al. Human CalDAG-GEFI deficiency increases bleeding and delays alphaIIbbeta3 activation. Blood. 2016;128(23):2729–2733. [DOI] [PubMed] [Google Scholar]
- 47.Westbury SK, Canault M, Greene D, et al. Expanded repertoire of RASGRP2 variants responsible for platelet dysfunction and severe bleeding. Blood. 2017;130(8):1026–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bottega R, Nicchia E, Alfano C, et al. Gray platelet syndrome: Novel mutations of the NBEAL2 gene. Am J Hematol. 2017;92 (2):E20–E22. [DOI] [PubMed] [Google Scholar]
- 49.Yoon SH, Cho T, Kim HJ, et al. IVS6+5G>A found in Wiskott-Aldrich syndrome and X-linked thrombocytopenia in a Korean family. Pediatr Blood Cancer. 2012;58(2):297–299. [DOI] [PubMed] [Google Scholar]
- 50.Ambo H, Kamata T, Handa M, et al. Novel point mutations in the alphaIIb subunit (Phe289–>Ser, Glu324–>Lys and Gln747–>Pro) causing thrombasthenic phenotypes in four Japanese patients. Br J Haematol. 1998;102(3):829–840. [DOI] [PubMed] [Google Scholar]
- 51.Nurden AT, Breillat C, Jacquelin B, et al. Triple heterozygosity in the integrin alphaIIb subunit in a patient with Glanzmann’s thrombasthenia. J Thromb Haemost. 2004;2(5):813–819. [DOI] [PubMed] [Google Scholar]
- 52.Jallu V, Dusseaux M, Panzer S, et al. AlphaIIbbeta3 integrin: new allelic variants in Glanzmann thrombasthenia, effects on ITGA2B and ITGB3 mRNA splicing, expression, and structure-function. Hum Mutat. 2010;31(3):237–246. [DOI] [PubMed] [Google Scholar]
- 53.Bachli EB, Brack T, Eppler E, et al. Hermansky-Pudlak syndrome type 4 in a patient from Sri Lanka with pulmonary fibrosis. Am J Med Genet A. 2004; 127A(2):201–207. [DOI] [PubMed] [Google Scholar]
- 54.Gueguen P, Rouault K, Chen JM, et al. A missense mutation in the alpha-actinin 1 gene (ACTN1) is the cause of autosomal dominant macrothrombocytopenia in a large French family. PLoS One. 2013; 8(9):e74728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.