Abstract
CAGI (Critical Assessment of Genome Interpretation) conducts community experiments to determine the state of the art in relating genotype to phenotype. Here we report results obtained using newly-developed ensemble methods to address two CAGI4 challenges: enzyme activity for population missense variants found in NAGLU (Human N-acetyl-glucosaminidase) and random missense mutations in Human UBE2I (Human SUMO E2 ligase), assayed in a high throughput competitive yeast complementation procedure. The ensemble methods are effective, ranked 2nd for SUMO-ligase and 3rd for NAGLU, according to the CAGI independent assessors. However, in common with other methods used in CAGI, there are large discrepancies between predicted and experimental activities for a subset of variants. Analysis of the structural context provides some insight into these. Post-challenge analysis shows the ensemble methods are also effective at assigning pathogenicity for the NAGLU variants. In the clinic, providing an estimate of the reliability of pathogenic assignments is key. We have also used the NAGLU dataset to show that ensemble methods have considerable potential for this task, and are already reliable enough for use with a subset of mutations.
Keywords: Missense mutations, ensemble methods, monogenic disease, CAGI, NAGLU, SUMO-ligase
INTRODUCTION
The vast quantities of data generated by the high-throughput genotyping and next generation sequencing technologies (Soon et al. 2013; Reuter et al. 2015) have created a major demand for reliable methods of interpreting the phenotypic significance of genetic variation, particularly as it relates to human disease. Among various types of genetic variation, missense single nucleotide polymorphism (SNPs) and missense rare mutations in coding regions are of particular interest because of the major role these play in monogenic disease (Stenson et al. 2014), complex trait disease (Kryukov et al. 2007; Pal and Moult 2015), and cancer (Wood et al. 2007; Shi and Moult 2011).
Many computational methods have been developed to identify the relevance of missense variants to disease (Peterson et al. 2013). Most of these methods make use of sequence variation across species and within the human population to infer the likely fitness impact of an amino acid substitution, assumed to be related to disease relevance (Lichtarge et al. 1996; Ng and Henikoff 2003; Yue and Moult 2006; Thomas et al. 2006; Calabrese et al. 2009; Chun and Fay 2009; Schwarz et al. 2010; Choi et al. 2012; Katsonis and Lichtarge 2014; Kircher et al. 2014; Niroula et al. 2015). A few make use of three dimensional structure information, particularly to infer any thermodynamic destabilization of the structure (Yue et al. 2005; Redler et al. 2015), assuming that decreased protein activity implies a relationship to disease. Some methods combine both sequence and structure information (Li et al. 2009; Adzhubei et al. 2010; Carter et al. 2013; Hecht et al. 2015; Baugh et al. 2016; Folkman et al. 2016). Methods usually use supervised machine learning such as random forest (Li et al. 2009; Carter et al. 2013; Niroula et al. 2015), neural network (Hecht et al. 2015) and support vector machines (Yue and Moult 2006; Calabrese et al. 2009; Kircher et al. 2014), or models that do not need training (Lichtarge et al. 1996; Ng and Henikoff 2003; Thomas et al. 2006; Chun and Fay 2009; Choi et al. 2012).
Missense analysis methods have usually been evaluated by benchmarking against databases of known monogenic disease mutations and presumed benign species or population variants, and there have been very few independent tests. Critical Assessment of Genome Interpretation (CAGI), conducts community wide experiments to test these and other genome interpretation methods. CAGI participants are provided genetic variant information and asked to predict phenotypic consequences. Independent assessors then evaluate the results. The experiments are double blind in that participants do not know the phenotypes and the assessors do not know the identity of the participants. In the most recent CAGI round, CAGI4 (http://genomeinterpretation.org), there were two missense variant interpretation challenges: the NAGLU challenge (https://genomeinterpretation.org/content/4-NAGLU) and the SUMO-ligase challenge (https://genomeinterpretation.org/content/4-SUMO_ligase). Here we report our results for these.
NAGLU (MIM# 609701) encodes Human N-acetyl-glucosaminidase, an enzyme involved in the heparan sulfate degradation process, and is one of four (Valstar et al. 2008) lysosomal enzymes in which mutations may result in one of four corresponding types of Sanfilippo Syndrome (Sanfilippo et al. 1963). Mutations in NAGLU protein cause a rare neurological disease, Mucopolysaccharidosis IIIB or Sanfilippo B disease (O’Brien 1972; von Figura and Kresse 1972; Valstar et al. 2008). The NAGLU challenge utilized in vitro enzyme activity data for a set of 165 rare population missense mutations extracted from the ExAC exome database (60,706 individual genomes) (Lek et al. 2016), omitting 24 known disease mutations. CAGI challenge participants were asked to quantitatively predict the enzymatic activity of each mutant relative to that of the wild type enzyme. A unique feature of the NAGLU dataset is that it represents the distribution of protein function of rare variants present in a population. To our knowledge, this is the first test of this type for current missense analysis methods, and more relevant to variants encountered in the clinic than usual database benchmarking.
UBE2I (MIM# 601661) encodes the human small ubiquitin-like modifier proteins conjugating protein (SUMO E2 ligase) that catalyzes the covalent attachment of SUMO to a range of target proteins. The CAGI challenge data provider had generated a library of over 6,000 human SUMO-ligase UBE2I clones expressing nearly 2,000 unique missense mutations in various combinations. The competitive growth rate of each clone was deduced from deep sequencing of a yeast-based complementation system. CAGI participants were asked to predict the relative competitive growth rates of yeast cells carrying three different sets of random mutations. Unlike the NAGLU challenge, where enzyme activity is known to be directly related to pathogenicity (von Figura and Kresse 1972), the relationship between SUMO-ligase function and fitness is complicated by two factors – the multiple regulator and target proteins that interact with SUMO-ligase (Geiss-Friedlander and Melchior 2007), and the fact that the human SUMO-ligase was substituted for the native enzyme in yeast cells. These factors make this a complex system from the point of view of interpreting the CAGI results. Many similar high throughput mutational scans are now being undertaken, so it is of interest to use the CAGI experiment to begin to probe the strengths and limitations of this approach, both generally, and as a basis for CAGI challenges.
All submitted predictions in each challenge were evaluated by independent assessors, one for each challenge. Results reported here were ranked 2nd for the SUMO-ligase challenge and 3rd for NAGLU.
Most missense analysis methods assign each variant as either deleterious or benign. An unusual feature of both the NAGLU and SUMO-ligase challenges is that they require prediction of a continuous variable, in one case relative enzyme activity, and in the other, relative yeast growth rate. In other words, the challenges require a regression predictor rather than a classification predictor. To address this requirement, we made use of an ensemble approach, combining binary predictions or associated confidence scores from up to eleven different methods. In a number of fields, ensemble methods that combine results from multiple individual methods have proven effective (Dietterich 2000; Moult 2005; Abeel et al. 2010). A number of missense ensemble predictors, for example CONDEL (González-Pérez and López-Bigas 2011), PONP (Olatubosun et al. 2012), Meta-SNP (Capriotti et al. 2013) and most recently REVEL (Ioannidis et al. 2016) have also been developed for the more usual task of binary classification, but as far as we are aware, this is the first use for quantitative prediction of missense impact.
We also performed several post-challenge analyses on the NAGLU dataset, examining the usefulness of structure information for identification of deleterious mutations and comparing the performance of the new ensemble method with other missense methods for binary classification. In the clinic, a major concern is not just to have an accurate predictor of pathogenicity, but also to assign a reliable probability that an assignment of pathogenic or benign is correct. The NAGLU challenge data set provided an opportunity for testing methods of assigning such probabilities on a clinically relevant dataset.
METHODS
Challenge data and benchmark data
The challenge set of 165 NAGLU rare population missense mutations was provided by Jonathan H. LeBowitz (BioMarin). The SUMO-ligase CAGI challenge set was generated by the Fritz Roth lab using a competitive yeast complementation growth assay. Three sets of UBE2I (SUMO-ligase) mutations were provided – 1) a reliable (multiple measurements) set of 219 single missense mutations, 2) a less reliable set of 463 single missense mutations and 3) a set of 4427 double or more mutations per clone. The experimental NAGLU enzyme activity data and the SUMO-ligase yeast growth data were not released to CAGI participants until all predictions had been submitted. In addition, we also collected 90 NAGLU known disease-related variants from HGMD (Stenson et al. 2014), together with the 278 interspecies variants, as a benchmark set.
Data for training predictors of continuous activity
Methods training for both NAGLU enzyme activity and SUMO-ligase growth rates required data that are also on an appropriate continuous scale of biological activity (as opposed to the more usual pathogenic/benign classification). For this purpose, a set of enzyme activity data for 92 human Phenylalanine hydroxylase (PAH) variants from (http://www.biopku.org/pah/) was used, supplemented by a set of 139 PAH interspecies variants (identified by comparing the human sequence with those of seven PAH orthologs (HomoloGene, (NCBI Resource Coordinators 2015)) with sequence identities higher than 80%), assumed to have full activity. We also searched the literature for high throughput mutation datasets that might be appropriate for use as training data. Only one of these appeared suitable, a set of cell growth rate data for yeast ubiquitin (UBI4) mutations (Roscoe et al. 2013). In practice, methods trained on these data performed poorly, and so its use was discontinued.
Combining multiple missense analysis methods to predict relative protein activity
For the ensemble methods, up to eleven missense analysis methods were used: Polyphen-2 (Adzhubei et al. 2010), SIFT (Ng and Henikoff 2003), SNPs3D Profile (Yue and Moult 2006), CADD (Kircher et al. 2014), Panther (Thomas et al. 2006), PON-P2 (Niroula et al. 2015), SNAP2 (Hecht et al. 2015), PROVEAN (Choi et al. 2012), VEST3 (Carter et al. 2013), LRT (Chun and Fay 2009) and MutationTaster (Schwarz et al. 2010). The dbNSFP2.9 database (Liu et al. 2013) was used to obtain CADD, PROVEAN, LRT, VEST3 and MutationTaster results. SNPs3D Profile results were obtained using the standalone in-house software. Results of other methods were obtained from the corresponding web-servers.
Binary predictions and associated scores were collected when both were available. Polyphen-2 ‘Probably damaging’ and ‘Possibly damaging’ were merged as a deleterious assignment. The MutationTaster deleterious set was compiled by combining the ‘A’ and ‘D’ categories, and the benign set consisted of the ‘P’ and ‘N’ categories. Four methods (CADD, SNPs3D profile, Panther and VEST3) didn’t directly report binary assignments. The recommended threshold score of 15 was used for CADD and the standard score threshold of zero was used for SNPs3D profile. A ‘deleterious’ score of 0.5 and a score of 0.77 were chosen as the cutoffs for Panther and VEST3 respectively, the values at which the distribution curves of deleterious and benign training sets crossed each other.
For machine learning based prediction of protein activity, two sets of input features were tested: One set consists of the score values returned by each of the 11 missense methods listed above. The other set consists of the binary assignments of benign or deleterious, represented as 0 or 1. Both feature sets also included the fraction of agreement (FOA) for a deleterious assignment across predictors, calculated as following:
where the sum is over the number of missense methods included, and Ni is 1 if a binary assignment is available for the i-th method, and is 0 otherwise, Ci is 1 if the i-th method predicted deleterious and is 0 if the i-th method predicted benign or was not available.
Weka (Frank et al. 2016) with standard settings was used to test a number of machine learning models: logistic regression, linear regression, support vector machine (SVM) regression, multi-layer perceptron, M5 Rule, random tree and random forest. The overall best performance (as judged from Root mean square deviations (RMSD, see supplementary methods), Pearson, and Spearman) on the PAH training set with 10-fold cross validation was returned for an SVM regression with a RBF kernel with the default settings and using the 11 method scores and FOA as features. However, the spread of performance across the best combinations of the feature sets and the ML methods was small (Pearson’s r 0.84–0.87, RMSD 0.18–0.20, 10-fold cross validation) and so more extensive parameter optimization might have produced a different choice. In addition to the prediction of activity, CAGI4 rules also required estimated standard deviations for each activity value. We provided the RMSD on the PAH training set as the standard deviation for all predicted activities.
Scale calibration and manual adjustment for each challenge
The SVM regression model was used to predict the relative enzyme activity of each NAGLU mutation and the cell growth rate of each UBE2I (SUMO-ligase) mutation. Because the model was trained on a different gene (PAH) with enzyme activity measured using a different experimental assay, we expected some systematic bias in the predictions and assumed that results would require scaling for each challenge system. For NAGLU, a zero activity reference point was defined using 15 known disease mutations with reported zero enzyme activity (Weber et al. 1999; Tessitore et al. 2000; Lee-Chen et al. 2002; Beesley et al. 2004). A full activity reference point was defined by the 278 NAGLU interspecies variants compiled in the same way as the PAH interspecies variants described above. These reference points were used to linearly scale the NAGLU activity predictions. We also collected structural information on the NAGLU protein from SNPs3D stability (Yue et al. 2005) and FOLDX (Guerois et al. 2002) predictions, as well as information on the functional role of individual residues from UniProt (UniProt Consortium 2015). Two predictions affecting disulfide bonds were manually adjusted to 0.1 activity. Predictions for six residues were adjusted to lower predicted activity in an ad hoc manner, on the basis of predicted structure destabilization. The experimental data later showed that these manual adjustments did not improve overall prediction accuracy, and increased prediction error for three of the six residues. For SUMO-ligase, the distribution of experimental measurements was provided as part of the challenge. Two submissions were made using different calibration procedures. For the first, we used the closest experimental values to 0 and 1 as the zero and full growth rate reference points and applied a linear scaling procedure like that used for NAGLU. In the second submission, each predicted growth rate was uniquely matched to the corresponding ranked experimental value. We noted that the experimental distributions have a number of mutations with growth rates significantly higher than wild type. For each challenge set, for the submission not mapped to the distribution of experimental data, it was necessary to reassign some growth rates to values greater than wild type to match experiment. We increased the values for the top predicted growth rate subset, except for those that predicted destabilizing by SNPs3D Stability (Yue et al. 2005) and FOLDX (Guerois et al. 2002). We also took into account (Bernier-Villamor et al. 2002; UniProt Consortium 2015) several reports of mutations with enhanced growth rate. The experimental data showed that this procedure is less accurate than that without manual adjustments on most gain-of-function mutations (22 of 27 in set 1 and 47 of 52 in set 2). For Challenge set 3, where multiple mutations were present in each sample, we assumed that the highest impact prediction dominated, and assigned that predicted value. The results of each challenge presented throughout the rest of the manuscript are based on a final set of predictions that include the manual adjustments.
All final predictions were adjusted to be 0 if below 0, as required by the CAGI4 submission instructions.
Positive and negative controls
Positive and negative control models were used to further evaluate the continuous predictions of relative protein activity. The positive control model estimated the performance expected if the computational method were perfect so that the only discrepancies arose from experimental error. For this purpose, simulated experimental errors were randomly drawn from a Gaussian distribution using the reported experimental mean and standard deviation based on the experimental error for each mutation. The performance was averaged from 1000 repeats of this process. The negative control adopted the algorithm proposed by the CAGI SUMO-ligase assessor as follows:
Where Pw and Pm are the probability of the wild type and mutated residue type occurring at the mutated position in a multiple sequence alignment and Qw and Qm are the background frequencies of the wild type and mutated residue respectively in the entire sequence profile.
Analysis of the influence of training set type and size on performance
The continuous value prediction models used a small training set of mutations and that set was from an unrelated protein. Once the submissions were made and the experimental data were available, for each of the challenges, we tested the influence of these factors as follows. 15% of the data was set aside for testing and a series of subsets of different sizes were randomly selected from the remainder. The machine learning model was retrained on each of these subsets. The procedure was repeated 10 times, omitting a different 15% data each time. Performance was then evaluated as a function of training set size.
Training and testing data for the binary predictor
For training ensemble binary predictors of pathogenicity, all mutations in an earlier version of HGMD (Stenson et al. 2003) were used as true positives and a set of interspecies variants were used as true negatives (‘benign’ mutations), compiled by comparing homolog protein sequences across species with at least 90% sequence identity over at least 80% of the full length (Yue and Moult 2006). For testing pathogenicity models and assessing prediction reliability, we compiled two independent test data sets. The first set is composed of ClinVar (Landrum et al. 2016) variants with pathogenic or benign assignments, excluding all that are in HGMD (2014 version) (Stenson et al. 2014) and OMIM (http://omim.org/) in order to ensure independence from the commonly used training data. ClinVar ‘likely pathogenic’, ‘likely benign’ entries, and entries with conflicting ClinVar assignments were not included. The second is the challenge set of 165 NAGLU rare population missense mutations. A complication in this analysis is choosing an activity level below which all mutations are pathogenic (that is, penetrance is 100%). In other data referenced by the data provider, pathogenic mutations are found at activities up to 45% but most are below 15%. Because of this uncertainty, we evaluated methods performance using both 10% and 30% relative enzyme activity cutoffs for pathogenicity.
Pathogenicity prediction models
Three machine learning methods were tested for binary state (pathogenic/benign) prediction models: Logistic Regression (Weka), Random Forest (Weka) and SVM (RBF kernel, SVMlight (Joachims 1999)). Features sets were the same as those used for continuous value prediction except that Panther and SNAP2 predictions were removed due to the difficulty of collecting the large number of predictions required from the corresponding web-servers. Models were trained using the HGMD dataset with default parameters. REVEL (Ioannidis et al. 2016) predictions were downloaded from (https://sites.google.com/site/revelgenomics/). The dbNSFP2.9 database (Liu et al. 2013) was used to map REVEL results to individual protein mutations.
Measuring prediction reliability
In the clinic, variants are often accepted as pathogenic or benign if the confidence in that assignment is estimated as greater than some threshold, typically 90%. For each binary prediction method, we therefore evaluated the fraction of variants that were predicted with reliability (PPV, positive predictive value, see supplementary methods) at 95%, 90%, 85% and so on. To this end, for each method, the data were sorted by the associated prediction score, from highest confidence score to lowest. For prediction of pathogenicity, the fraction of highest confidence variants with a given PPV was then determined. The resulting fraction versus PPV curves were plotted using R ggplot2 (Wickham H 2009)(Wickham 2009). To reduce noise, the NAGLU dataset was expanded to 1000 variants by bootstrapping, and assessed by averaging over 1000 bootstrappings.
RESULTS
Comparison of predicted and experimental enzyme activities
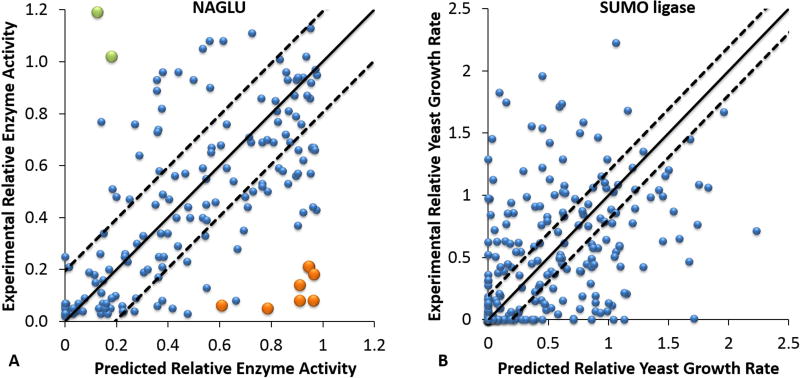
Figure 1A shows a scatter-plot for the NAGLU challenge mutations showing the relationship between all predicted and experimental enzyme activities. The overall RMSD between predicted and experimental values is 0.31, Pearson’s r is 0.55, and Spearman’s rho is 0.57. These values are worse than the cross validation results on the PAH training data, which are RMSD of 0.20, Pearson’s r of 0.82 and Spearman’s rho of 0.78. The NAGLU predicted values are also substantially worse than the positive control ‘perfect prediction’ RMSD of 0.12, 0.95 Pearson’s r and 0.94 Spearman’s rho (based on the reported experimental standard errors). There are a small number of serious outliers, and as the plot shows, most of these correspond to mutations identified by the assessor as ‘hard to predict’ on the basis of poor performance by all the top methods. A breakdown of performance by location in the structure (Supp. Figure S1) shows striking variations for the Pearson’s correlation coefficient of 0.83, 0.50 and 0.39 for buried, partially exposed and surface mutations respectively. (Variant location based on the STRIDE (Eisenhaber and Argos 1993; Eisenhaber et al. 1995; Frishman and Argos 1995) relative surface accessibility: buried core (≤ 0.05), partially exposed (> 0.05, ≤ 0.25) and surface (> 0.25)). The most serious outliers for both under and over-prediction of activity are in the partially or completely exposed subsets. Performance metrics are substantially improved omitting these ten, with RMSD of 0.24, Pearson’s r of 0.71 and Spearman’s rho of 0.71 (Table 1).
FIGURE 1. Prediction results for NAGLU and UBE2I (SUMO-ligase) mutations.
A. Scatter-plot comparing experimental NAGLU relative enzyme activities (Y axis) with the predicted values (X axis) for the CAGI challenge variant set. Dashed lines delineate the expected prediction RMSD from based on training results. 61% of the predicted values are within the range of the estimated RMSD, but a few mutations have very large deviations from the experimental measurements. The over-estimates shown in orange and the under-estimates shown in green are the ten mutations selected by the assessor as ‘hardest’ to predict. See text and Supp. Figure S1 and S2 for a discussion of these.
B. Scatter-plot comparing experimental relative yeast growth rates with the mapped predicted values for the SUMO-ligase CAGI challenge UBE2I mutation Set 1. Dashed lines delineate the expected prediction RMSD from the training on phenylalanine hydroxylase mutations. The correlation with experiment is substantially weaker than for the NAGLU challenge (Figure 1A). 39% of the predicted values are within the range of the estimated RMSD.
Table 1.
Metrics of prediction performance for NAGLU and SUMO-ligase
| Challengea | Prediction | Positive Controlb | Negative Controlc | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RMSD | Pearson's r | Spearman's rho |
RMSD | Pearson's r | Spearman's rho |
RMSD | Pearson’s r | Spearman’s rho |
||
| NAGLU | 0.31 | 0.55 | 0.57 | 0.12 | 0.95 | 0.94 | 0.42 | 0.45 | 0.48 | |
| NAGLU w/o outliers | 0.24 | 0.71 | 0.71 | 0.14 | 0.92 | 0.93 | 0.39 | 0.53 | 0.57 | |
| SUMO - Ligase Set 1 | No Map | 0.55 | 0.39 | 0.46 | 0.24 | 0.91 | 0.92 | 0.59 | 0.30 | 0.38 |
| Mapped | 0.55 | 0.39 | 0.46 | |||||||
| SUMO -Ligase Set 2 | No Map | 0.63 | 0.35 | 0.46 | 0.25 | 0.90 | 0.89 | 0.57 | 0.31 | 0.39 |
| Mapped | 0.56 | 0.33 | 0.46 | |||||||
| SUMO -Ligase Set 3 | No Map | 0.57 | 0.21 | 0.20 | 0.26 | 0.89 | 0.82 | 0.57 | 0.24 | 0.22 |
| Mapped | 0.59 | 0.18 | 0.20 | |||||||
In the SUMO-ligase challenge, there are two prediction sets, submission 1 with scaled prediction scores (No Map) and submission 2 (Mapped) with each predicted value mapped to the experimental value of closest rank.
In the positive control, the expected difference between experiment and prediction is estimated from the reported experimental errors. That is, a perfect prediction method could not be more accurate than this. See MATERIALS AND METHODS.
In the negative control, a prediction score was computed for each mutation based on amino acid frequency information only, using the equation described in MATERIALS AND METHODS. The resulting prediction scores were mapped to the experimental value of closest rank.
Are the ten outlier mutations cases where all the prediction methods systematically fail, or are these experimental artifacts of some sort? A definitive answer to this question is not possible without further experiments, but in some cases, likely explanations present themselves. For example, 10 out of 11 individual methods in the ensemble model and a structural method, SNPs3D Stability, predict mutation (NAGLU NP_000254.2:p.A627V) to be benign, but the reported experimental activity value is close to 0. Consistent with the prediction results, examination of a multiple sequence alignment shows A627 is at a variable position across species, where 15 different amino acid types are found. A627 is on the protein surface (Supp. Figure S2A) and the variant introduces a hydrophobic side chain (crystal structure from USPTO US08775146B2 (Meiyappan et al. 2014)). Under in vivo conditions, that may indeed have little impact, but in overexpression conditions of the experimental in vitro assay, aggregation may result. On the other hand, it is difficult to find any plausible explanation for some of the outliers. For example, one outlier (NAGLU NP_000254.2:p.P283L) is a partially exposed proline at an extremely conserved position (Supp. Figure S2B). All 11 individual prediction methods as well two structure based methods, FOLDX (Guerois et al. 2002; Schymkowitz et al. 2005) and SNPs3D Stability (Yue et al. 2005), predict this mutation deleterious. Inspection of the structure suggests no way in which the leucine side chain could be accommodated. The reported experimental activity is the highest of any of the variants, at 1.19.
Figure 1B is a scatter plot of the relationship between Submission 2 predicted and experimental growth rates for Set 1 UBE2I (SUMO-ligase) mutations. The performance is weaker (RMSD 0.55, Pearson’s r 0.39, Spearman’s rho 0.46) than the results for NAGLU, likely because of the complex relationship between aspects of SUMO-ligase function, its many substrates, and cell growth as well as effects from use of human protein in a yeast system. In contrast to NAGLU, the best performance is for surface residues (Pearson’s r 0.59), and it is less good for mutations of buried (Pearson’s r 0.35) and partially buried (0.29) residues. The results are worst for mutations in the substrate, SUMO, and SUMO-E3 ligase protein-protein interfaces ((Pearson’s r 0.24, Supp. Figure S3). For example, in the experimental structure with a human SUMOylation substrate, RANGAP1 (PDB code 3UIP), the wild type K74 forms a salt bridge with E526 of the substrate (Supp. Figure S4). Mutations (UBE2I NP_003336.1:p.K74S and UBE2I NP_003336.1:p.K74E) disrupt that interaction and in the case of K74E electrostatic repulsion is introduced. Both positions are conserved, and the mutations are overwhelmingly predicted deleterious, yet the experimental growth rates are higher than wild type. On the other hand, mutation (UBE2I NP_003336.1:p.K74R) appears to enhance the salt bridge with E526, and four out of ten sequence methods and the two structure methods predict it as benign. Yet the experimental value shows complete loss of growth. At the CAGI meeting the data provider, Fritz Roth, agreed that a possible complication here is that interfaces between human SUMO-ligase and its human partners may have significantly different properties from the equivalent yeast interfaces, and that in general the substantial number of gain of function mutations may be due to this cause. Some other SUMO-ligase substrates do not have exactly the same interface (Bernier-Villamor et al. 2002). Thus, in general, it is not clear how altering the interface with one substrate may affect interactions with other substrates, and therefore what the overall effect on growth may be.
Table 1 summarizes all the agreement statistics between prediction and experiment for the NAGLU mutations and the UBE2I (SUMO-ligase) set 1, set 2 and set 3 mutations, together with the values for the positive and negative controls. (Data are for the SVM regression models described in Materials and Methods). Supp. Table S1 shows the number of missense analysis methods reporting for each data set. The results show our models outperformed the (quite sophisticated) negative control in the NAGLU challenge (RMSD 0.31 versus 0.42, Pearson’s r 0.55 versus 0.45, and Spearman’s rho 0.57 versus 0.48). The model is also effective on the SUMO-ligase set 1 (the most reliable single mutations) when compared to the negative control (RMSD 0.55 versus 0.59, Pearson’s r 0.39 versus 0.30, and Spearman’s rho 0.46 versus 0.38). The large gap between the method’s performance and the positive control suggests that experimental error was likely not the limiting factor in the level of agreement with experiment.
NAGLU and SUMO-ligase challenge variant properties
The NAGLU challenge data are extracted from the ExAC database of population variants (Lek et al. 2016). In this respect it is a unique dataset – a set of variants found in a largely healthy population as opposed to the collections of known disease related mutations in databases such as HGMD (Stenson et al. 2003, 2014) and Clinvar (Landrum et al. 2016) and control sets of variants such as interspecies differences that are typically used for training and benchmarking methods. It is therefore of interest to ask how different the overall properties of these population variants are from the variants in the standard databases. Figure 2A shows that the predicted relative enzyme activity for the 90 NAGLU disease variants in HGMD and for 278 NAGLU interspecies variants have distinct distributions centered on 0 and 0.9~1 respectively, as expected. In contrast to this, the predictions for NAGLU CAGI challenge variants are approximately evenly distributed across the whole 0 to 1 range, in a manner similar to that of the experimental data.
FIGURE 2. Distributions of predicted and experimental enzyme activities.
A. Distribution of NAGLU relative enzyme activities 1) predicted for disease mutations in HGMD (HGMD, red); 2) predicted for inter-species variants (Interspecies, green); 3) predicted for mutations provided for the CAGI challenge (Prediction, blue), and 4) experimental activities for the challenge mutations (Experiment, purple). As expected, known disease mutations are predicted to have low activities and interspecies variant to have high activity. In contrast to these, the population variants have activities approximately equally distributed across the full range, for both prediction and experiment.
B. Relative yeast growth rate distributions for UBE2I (SUMO-ligase) mutation Set 1. The distribution of the unmapped predicted values (Submission 1, red) only approximately matches the experimental distribution (Experiment, black), available during the challenge. We submitted a second set of predictions in which each predicted value was mapped to the experimental value of closest rank (Submission 2, blue). This improves the overall match of the distributions (red and black) but not the prediction accuracy.
Figure 2B shows a comparison of the distribution of predicted yeast growth rates for SUMO-ligase challenge Set 1 mutations compared to the experimental distribution. An unusual feature of the experimental distribution is a substantial number (19%) of gain of function mutations, and this resulted in a poor overall fit from our prediction model. For submission 1, the distribution at low growth rates (below 0.2) is close to experiment, but between 0.2 and 1.0 there are too few predicted values and there are too many moderate gain of function values (in the 1.0 to 1.4 range). The second submission, which mapped each predicted value to the closest experimental value, corrects these distribution errors and produced a better overall distribution but doesn’t improve the prediction accuracy (Table 1). Set 2 showed similar results, whereas Set 3 shows many fewer gain-of-function mutations, presumably because of the presence of multiple mutations in each sample (Supp. Figure S5).
Role of structure destabilization
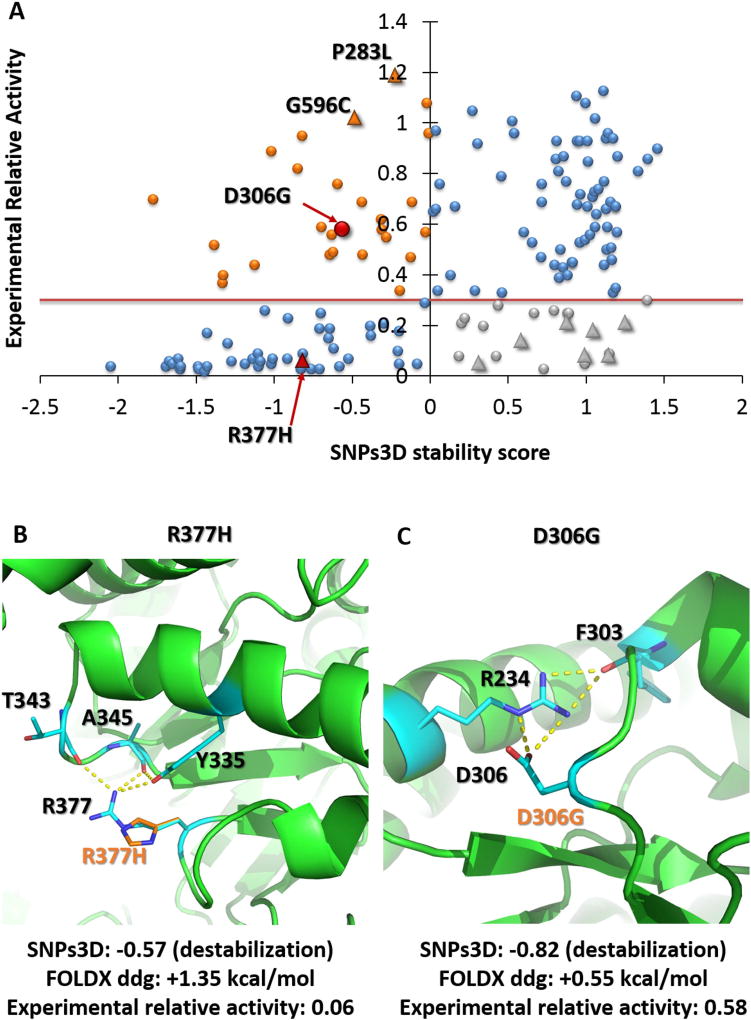
Thermodynamic destabilization of three-dimensional structure is established as playing a large role for monogenic disease causing mutations (Yue et al. 2005), so it was of interest to examine what part this factor plays for the challenge variants. (This analysis was undertaken after the results were known, and did not form part of our CAGI submissions). Figure 3A shows the distribution of destabilization scores from SNPs3D (Yue et al. 2005) for the NAGLU homo-trimer complex. At a NAGLU pathogenicity activity threshold of 0.3, a high fraction (68%) of the low activity variants are destabilizing, so, as in other monogenic disease, this factor plays a major role.
Figure 3. The role of thermodynamic destabilization in loss of function mutations.
A) Scatter plot comparing SNPs3D stability scores with experimental relative enzyme activity of NAGLU. Blue point variants in the lower left quadrant (68% of all those with low (> 0.3) activity) are predicted to destabilize the structure. Those at the upper right are predicted not destabilizing, consistent with their high activity. Those at the lower right (gray) are predicted to have low activity for reasons other than destabilization. The upper left quadrant variants (orange) are predicted destabilizing even though the experimental activity is high. Triangles show the location of the ten ‘hard to predict’ variants.
B) Structural context of NAGLU ‘hard’ outlier R377H (red). Predicted destabilization is consistent with the low experimental activity. A substantial fraction (5 out of 11) of sequence methods predict this variant to be benign.
C) Structural view of variant D306G (red), predicted to be destabilizing, inconsistent with the experimental activity. Although the variant disrupts some electrostatic interactions, these are likely compensated by greater solvation.
(Green: wild-type residues and interaction partners, orange: variants).
The structure analysis is independent of the sequence methods and so provides some evidence for whether or not the 10 ‘hard’ predictions are experimental artifacts or systematic failures of the sequence methods. Two of the ‘hard’ variants with high experimental activity (NAGLU NP_000254.2:p.P283L and NAGLU NP_000254.2:p.G596C) are predicted destabilizing, consistent with the sequence analysis results and inconsistent with experiment. One of the ‘hard’ very low activity (0.06) variants ((NAGLU NP_000254.2:p.R377H), Figure 3B) is found to be destabilizing though, consistent with experiment and in disagreement with some sequence methods (5 out of 11). Wild type R377 makes charge-dipole interactions with two main chain carbonyl groups (T343, A345) and a side chain hydroxyl group (Y335) so stabilizing a turn, and these interactions are absent for the variant (Figure 3B). The other seven ‘hard’ variants are all low activity and predicted to be not-destabilizing (lower right quadrant in Figure 3A). This could be because some other mechanism (for example involvement in catalysis) causes the low activity or because of experimental artifacts. Inspection of the structural environment does not reveal any such mechanisms, reinforcing the impression that these are experimental artifacts.
17% of the stability predictions disagree with the experimental data – predicted destabilizing but with higher than pathogenic activity. These partly reflect the shortcomings of present stability analysis methods as illustrated by the example of mutation (NAGLU NP_000254.2:p.D306G) (Figure 3C). Wild type D306 forms electrostatic interactions with R234 that is absent for the variant. In reality, loss of this interaction is likely largely compensated for by increased solvation energy, a factor poorly represented in the SNPs3D model. There is scope for improvement of these methods in this and a number of other ways.
Effect of training set size and choice of training data
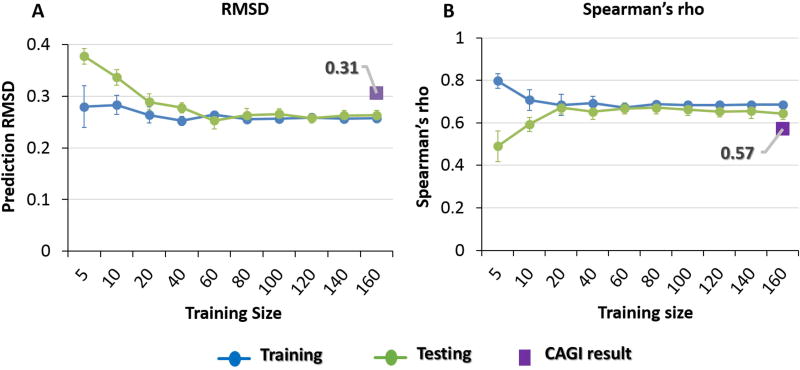
One obvious drawback to our approach is the limited number (activities for 231 phenylalanine hydroxylase mutations) of training data. Further, training on that single system may introduce systematic bias. In order to evaluate whether the performance of the model is restricted by these two factors, we retrained using the NAGLU enzyme activity data, after these were released to the CAGI community (see Materials and Methods). A range of training set sizes was used to determine the contribution of that factor to accuracy. For each size, we retrained and measured performance, and averaged over 10 repeats. For each training, 15% of the data were randomly chosen for evaluation, and omitted from training. Figure 4 shows that performance converged rapidly as the size of the training set increased beyond 100 mutations, showing that training set used in the CAGI challenges was large enough and not a factor limiting accuracy. Comparison between the converged performance and the performance in the blind CAGI challenges showed only a slight improvement of 0.05 RMSD and 0.07 Spearman’s rho for NAGLU and 0.08 RMSD for SUMO-ligase, so that the loss of performance from training on the phenylalanine hydroxylase system is small. Similar results were obtained for the SUMO-ligase challenge. Together, this analysis shows that the results were not substantially limited by either the training set size or training on a different system, and other factors must account for the worse than positive control performance.
Figure 4.
Blue: average training set performance, green: average test set performance (15% of data omitted from training). Averages over 10 runs. Purple rectangles show performance in the CAGI challenge with the model trained on PAH
4A) RMSD, 4B) Spearman rank correlation coefficient. Prediction performance converges rapidly as the training set size increases beyond 100 mutations. Training on the target protein rather than Phenylalanine hydroxylase (PAH) only slightly improves performance (0.05 RMSD and 0.07 Spearman’s rho). Thus, training set size and training on PAH are not limiting factors in performance.
Predicting pathogenicity using ensemble methods
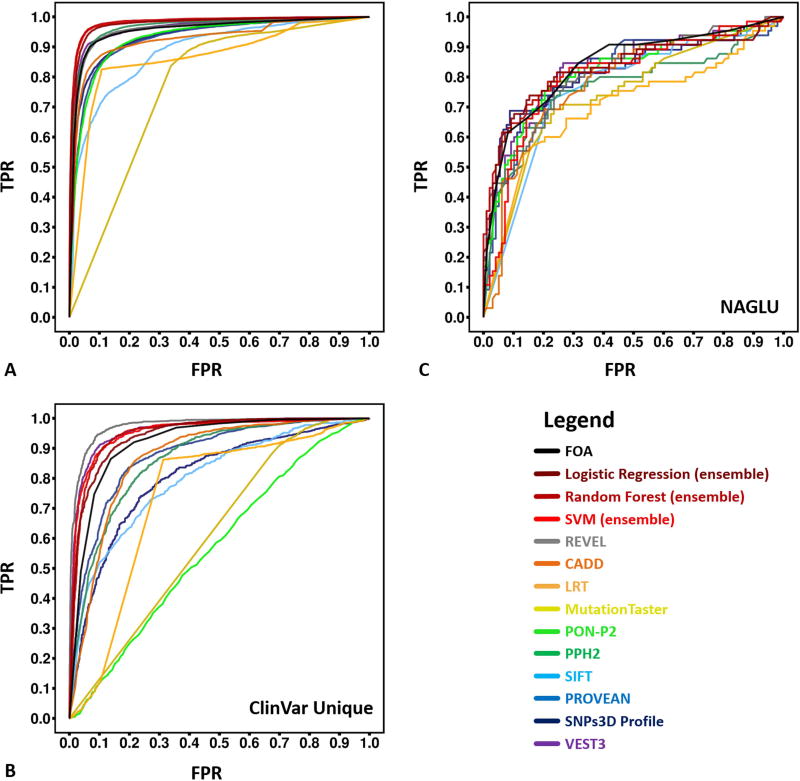
Post-challenge, we also investigated how well ensemble methods perform on assigning pathogenicity in the clinically relevant NAGLU data, compared with performance on standard benchmarking datasets. For these binary predictions (pathogenic/not pathogenic), we trained ensemble methods based on nine individual predictors (CADD (Kircher et al. 2014), LRT (Chun and Fay 2009), MutationTaster (Schwarz et al. 2010), PON-P2 (Niroula et al. 2015), PPH2 (Adzhubei et al. 2010), PROVEAN (Choi et al. 2012), SIFT (Ng and Henikoff 2003), SNPs3D Profile (Yue and Moult 2006) and VEST3 (Carter et al. 2013)) with three machine learning models (Logistic Regression, Random Forest, and SVM). Training was performed on a version of HGMD (Stenson et al. 2003) and a set of interspecies variants (see Materials and Methods). Results were evaluated using 10-fold cross-validation. When tested on HGMD, the ROC curves and AUCs of the ensemble machine learning predictors show better performance than any of the individual methods, with a highest AUC of 0.98 (Figure 5A and Table 2), although most perform extremely well. A number of individual predictors are partially or completely trained on HGMD, so to control for this factor, we also tested on a subset of ClinVar variants not in HGMD or OMIM (another common source of training data). (Figure 5B and Table 2). Though still better than most individual predictors, our ensemble predictors (best AUC 0.95) were slightly but significantly outperformed by VEST3 (Carter et al. 2013) (AUC 0.96) and the new ensemble method REVEL (Ioannidis et al. 2016) (AUC 0.97). As Figures 5C and Table 2 show, when the same methods were tested on the more relevant challenge NAGLU variant set, all showed substantially deteriorated performance (AUC up to 0.84 for the ensemble methods, slightly better than any other tested methods). Relative performance is insensitive to the exact activity threshold for pathogenic loss of activity (Table 2). We also converted the continuous NAGLU activity predictions to binary assignments and generated a ROC curve. That results in an AUC of 0.82, with both 0.1 and 0.3 activity cutoffs. Evidently, the distribution of activities found in the general population (all activities approximately equally likely to be encountered) are much more challenging for all methods than distinguishing between only pathogenic and interspecies variants.
Figure 5.
ROC (receiver operating characteristic) curves for predictions of pathogenicity by the new ensemble methods and other methods on HGMD, ClinVar unique and NAGLU challenge sets. For NAGLU, the pathogenicity threshold is an activity of 0.3 of wild type. The AUC (area under curve) of these ROC curves are listed in Table 2
A) For HGMD test data, the new ensemble models (Logistic Regression 0.98, Random Forest 0.98 and SVM 0.97) outperformed all constituent individual predictors on the HGMD test dataset. PPH2 and VEST3, which were also trained partially or completely on HGMD, have slight but significantly (P-value < 2.2e-6) worse AUCs.
B) For the unique ClinVar dataset (no overlap with HGMD or OMIM), another ensemble method, REVEL, outperformed all other methods. The next highest AUCs, for VEST3 and our ensemble models, are slightly but significantly (P-value < 0.05) smaller.
C) For the NAGLU rare population variants, all methods perform substantially worse than on HGMD and ClinVar. Our ensemble FOA (fraction of agreement) method has the best AUC of 0.84, followed by our Logistic Regression and Random Forest models, and VEST3. All four are not significantly different from each other (P-values > 0.05).
Table 2.
Metrics of binary prediction performance
| Methods | HGMD | ClinVar | NAGLUd | NAGLUc | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AUC | Fraction 1e | Fraction 2f | AUC | Fraction 1e | Fraction 2f | AUC | Fraction 1g | Fraction 2g | AUC | |
| Logistic Regressiona | 0.98 | 0.89 | 0.75 | 0.94 | 0.72 | 0.90 | 0.83 | 0.38 | 0.06 | 0.83 |
| Random Foresta | 0.98 | 0.90 | 0.75 | 0.95 | 0.78 | 0.93 | 0.83 | 0.43 | 0 | 0.83 |
| SVMa | 0.97 | 0.90 | 0.75 | 0.95 | 0.82 | 0.93 | 0.81 | 0.09 | 0.18 | 0.79 |
| FOAb | 0.96 | 0.85 | 0.73 | 0.92 | 0.27 | 0.88 | 0.84 | 0.28 | 0.56 | 0.83 |
| REVEL | 0.96 | 0.77 | 0.61 | 0.97 | 0.90 | 0.96 | 0.82 | 0.37 | 0.22 | 0.82 |
| CADD | 0.93 | 0.78 | 0.64 | 0.87 | 0.01 | 0.79 | 0.79 | 0.01 | 0 | 0.82 |
| LRT | 0.86 | 0.61 | 0.18 | 0.74 | 0 | 0 | 0.70 | 0 | 0.01 | 0.68 |
| MutationTaster | 0.77 | 0 | 0 | 0.61 | 0 | 0.22 | 0.74 | 0 | 0 | 0.79 |
| PON-P2 | 0.93 | 0.65 | 0.54 | 0.57 | 0 | 0.03 | 0.82 | 0.22 | 0 | 0.82 |
| PPH2 | 0.97 | 0.84 | 0.69 | 0.86 | 0.07 | 0.58 | 0.76 | 0 | 0 | 0.77 |
| PROVEAN | 0.93 | 0.76 | 0.63 | 0.88 | 0.26 | 0.68 | 0.82 | 0.08 | 0.50 | 0.82 |
| SIFT | 0.89 | 0.20 | 0.37 | 0.80 | 0 | 0.27 | 0.76 | 0 | 0 | 0.79 |
| SNPs3D Profile | 0.94 | 0.73 | 0.46 | 0.80 | 0.03 | 0 | 0.82 | 0.33 | 0.01 | 0.80 |
| VEST3 | 0.96 | 0.85 | 0.74 | 0.96 | 0.84 | 0.94 | 0.83 | 0.30 | 0.44 | 0.83 |
Ensemble model combining nine individual missense mutation analysis methods
Fraction of the nine methods making a deleterious assignment
Using NAGLU relative activity cutoff of 0.1
Using NAGLU cutoff of 0.3
The fraction of variants with a 0.9 PPV (i.e. less than a 10% error rate in predicting pathogenic)
The fraction of variants with a 0.9 NPV (i.e. less than a 10% error rate in predicting benign)
NAGLU dataset expanded by bootstrapping
Reliability of pathogenic assignments
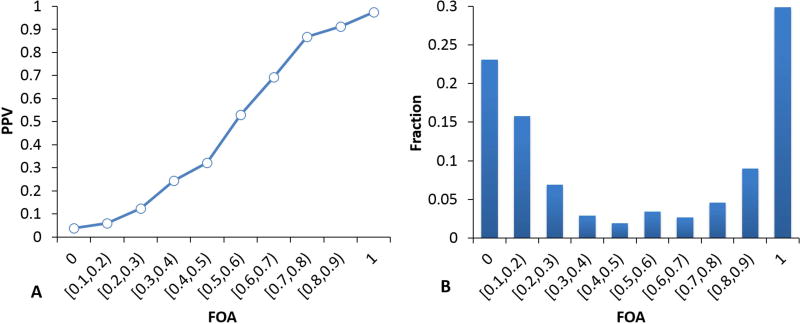
We investigated the effectiveness of ensemble methods for estimating the reliability of pathogenic assignments using the results from the binary pathogenicity analysis described above. To examine whether there is a useful ensemble signal to be exploited, we first examined the PPV as a function of the fraction of methods agreeing on a deleterious assignment (FOA) for the HGMD and interspecies dataset. Supp. Table S1 shows the number of methods included. There is strong dependence of PPV on FOA with the HGMD set (Figure 6A): For the set of variants where all nine methods predict deleterious, the PPV is 0.97 and the PPV is above 0.9 even when only 7 out of 9 methods predict deleterious. At the other end of the scale, the PPV is 0.04 when no method predicts deleterious and still below 0.1 even where two methods predict deleterious, so that in all 78% of mutations have better than 90% confidence assignments of either pathogenic or benign (Figure 6B). Thus even a very simple ensemble method shows promise for this purpose.
Figure 6.
A. Relationship between the fraction of methods that agree on a deleterious assignment (FOA) and the positive predictive value, PPV (fraction of predicted pathogenic variants that are pathogenic), for HGMD and interspecies variants. 6B, fraction of variants in each bin. Approximately 39% of variants can be predicted pathogenic with 90% or greater confidence (PPV) and 39% can be predicted benign with 90% or greater confidence (NPV). This simple analysis demonstrated a potential usefulness of ensemble methods in assigning prediction reliability.
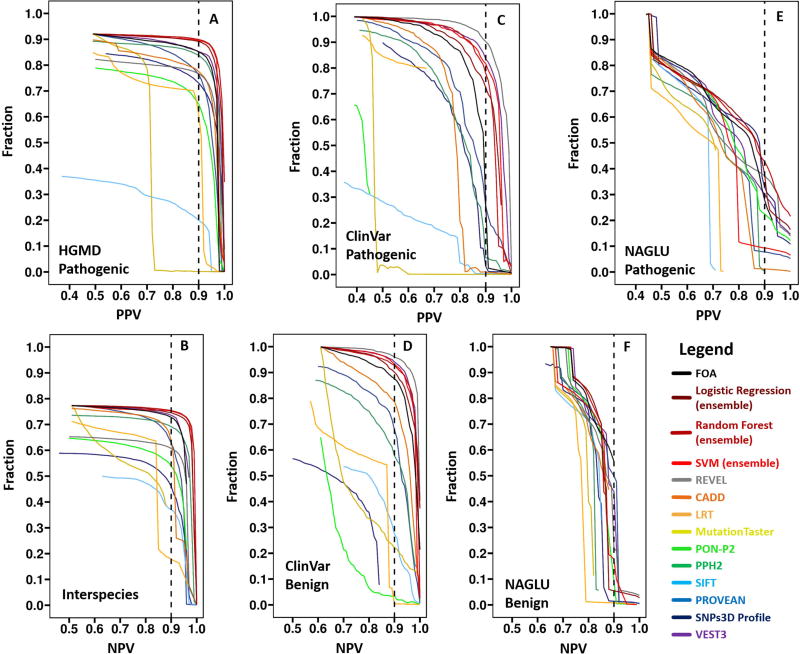
A fuller analysis is shown in Figure 7. Here the fraction of variants meeting a given reliability threshold is plotted as a function of the threshold, for both confidence in pathogenicity (left panels) and non-pathogenicity (right panels). As with the pathogenicity assignment results above, our ensemble methods and REVEL perform best on the HGMD and ClinVar sets respectively. Also as with the pathogenicity assignment, performance is substantially better on the HGMD and ClinVar test sets than on the NAGLU data. For HGMD, the best methods assign pathogenicity with 90% or greater confidence for 90% of the data, and benign assignments with equal confidence are made for about 75% of data. Pathogenicity confidence on the ClinVar set is similar, with a higher fraction meeting 90% confidence criterion (96%) for benign assignments. For the more realistic NAGLU dataset using an activity of 0.3 as the pathogenicity threshold, 43% of the pathogenic variants are predicted with 90% or better accuracy, and 56% benign assignments are 90% or better correct. However, the dependence of accuracy on threshold is steep for both these numbers, and precise values are likely to be dataset specific. Overall, the results do show that ensemble methods are advantageous for assigning reliability to pathogenicity assignments, and that the fraction of variants for which 90% confidence can be reached in the clinic is likely quite high. More realistic datasets such as the NAGLU one are needed to further investigate these properties.
Figure 7.
Fraction of data for which pathogenicity or benign status is predicted at a specified level of confidence, as a function of the confidence level, for HGMD (7A, 7B), ClinVar (7C, 7D) and the NAGLU challenge dataset (pathogenicity cutoff of 0.3, 7E, 7F). Vertical dashed lines show the 0.9 reliability threshold. For each dataset, the left panel shows the fraction of pathogenic variants meeting a reliability (PPV) threshold as a function of threshold and the right panel shows the equivalent data for reliability of benign assignment (NPV). Our ensemble methods and REVEL perform best on the HGMD and ClinVar sets respectively. Overall, even in the demanding NAGLU dataset, a substantial fractions of variants can be assigned as pathogenic or benign with high confidence.
DISCUSSION
Ensemble methods for the NAGLU and SUMO-ligase challenges
The NAGLU and SUMO-ligase challenges are unusual in that CAGI participants were asked to predict a continuous variable – in the case of NAGLU, relative enzyme activity, and in the case of SUMO-ligase, relative growth rate in a yeast complementation assay. Most missense analysis methods are designed to make a binary assignment of pathogenic or non-pathogenic, and so are not immediately applicable to the challenges. To address this, we explored the use of an ensemble strategy, incorporating up to 11 of the binary assignment methods. Ensemble methods have already been shown to be effective for the binary pathogenicity assignment task (González-Pérez and López-Bigas 2011; Olatubosun et al. 2012; Capriotti et al. 2013; Ioannidis et al. 2016). Here we assume that the more single methods make a pathogenic assignment for a given variant, the lower the corresponding protein activity will be. As the simple FOA (fraction of agreement between methods) approach demonstrates, this is the case. Use of confidence scores for each contributing method rather than binary values makes the procedure more nuanced, and machine learning provides a means of combining the methods in a balanced way. A potential limitation was the lack of suitable enzyme activity training data, but post-challenge analysis showed that as few as 100 phenylalanine hydroxylase variant activities were sufficient, and also that there was no significant bias from training on that system. The ensemble approach was successful in that it performed well, although it was slightly behind the best performers. In the NAGLU challenge, the ensemble approach was marginally outperformed by MutPred2 (unpublished, −0.005 in RMSD, +0.05 in Pearson’s r, +0.04 in Spearman’s rho and +0.00 in AUC) and by Evolution Action (Katsonis and Lichtarge 2014, −0.028 in RMSD, +0.001 in Pearson’s r, −0.019 in Spearman’s rho and +0.03 in AUC). In the SUMO ligase challenge, our two submissions of the ensemble approach performed best on set 1 and set 2 respectively, but were outperformed by most other methods on set 3 (multiple mutation set), probably due to our assumption that growth would be determined by the most deleterious mutation for each sample, rather than affected additively. However, neither our ensemble approach nor other best performers provided revolutionary accuracy. As discussed below, limitations in all contemporary approaches probably ensure that is not possible.
Accuracy
Although the methods used here and others in CAGI produce very strong statistical significance in terms of the relationship between predicted and experimental activity values, the agreement appears substantially less than expected, given the reported experimental accuracy. What limits the accuracy? – Some part of the disagreement may be due to experimental artifacts. For example as noted earlier, for one of the 10 NAGLU ‘hard’ variants the conditions of expression in the cell line may contribute to aggregation not encountered in vivo. For SUMO-ligase, as discussed in Results, differences between yeast and human proteins contribute to discrepancies.
Overall though, most of the discrepancy likely comes from the inherent deficiencies of the methods. Nearly all primarily attempt to relate sequence conservation patterns to pathogenicity (some also incorporate partial structure information (Adzhubei et al. 2010; Carter et al. 2013; Hecht et al. 2015)). Although there clearly is a qualitative relationship of this type, there is no theoretical framework providing a quantitative relationship. Such a framework would need to relate phylogenic profiles to fitness, something which the molecular evolution community has not succeeded in doing after many years of effort (Orr 2009). Further, the relationship between fitness and disease relevance is also not straightforward. As a consequence, all current pathogenicity prediction methods are ad hoc, using calibration or machine learning to achieve some level of quantitation. Given that, they are surprisingly effective. There are number of ways in which accuracy may improve in the future. In our results there is markedly different accuracy for surface and interior residues, so that treating these classes of residues differently may be useful. Other structural and functional information may also help. Specific training only on variants where individual methods do not correlate well might be helpful, if there are sufficient data and an appropriate algorithm for training. More generally, at present, most methods are completely non-specific, and are applied to different proteins without incorporating information pertinent to each case. In future, we envision that protein specific models will be built. There is also major requirement for more realistic training and testing datasets, such as NAGLU.
Assigning pathogenicity
As noted earlier, the NAGLU challenge data set is so far unique in that it consists of protein activity data drawn from a background population representative of that expected in the clinic. The commonly used HGMD and ClinVar databases, although useful compilations of clinically relevant data, are usually paired with highly benign controls for training and testing purposes, and so not very representative of clinical encounters. Therefore, we also tested an ensemble approach for assigning pathogenicity in the NAGLU dataset, compared to standard benchmarks. The new ensemble method and many others tested here perform extremely well on two standard benchmark sets, HGMD (Stenson et al. 2014) and a unique subset of ClinVar (Landrum et al. 2016), many with AUCs of over 95%. Both our ensemble method and another recent ensemble approach, REVEL (Ioannidis et al. 2016) have relatively good performance on the NAGLU data, but overall, all methods are strikingly less effective (best AUCs up to 0.84). The results suggest that we need many more clinically relevant datasets like NAGLU in order to realistically evaluate the pathogenicity assignment methods.
Utilization of protein structure information
As demonstrated here and in other work (Yue et al. 2005; Adzhubei et al. 2010; Carter et al. 2013; Hecht et al. 2015; Baugh et al. 2016; Folkman et al. 2016; Redler et al. 2016), analysis based on protein structure provides an orthogonal approach that, in spite of its own accuracy limitations, can sometimes provide valuable insight into the atomic level mechanisms in play. In particular, as with other monogenic disease related mutations (Yue et al. 2005), for NAGLU, structure analysis shows a large fraction operate by destabilizing protein three-dimensional structure. There is considerable scope for further improvement of these approaches, using more biophysical approaches (Seeliger and de Groot 2010).
Reliability for pathogenicity assignments
In the clinic a major concern is not just to have an accurate predictor of pathogenicity, but also to be able to have a reliable probability that an assignment of pathogenic or benign is correct: a method may be highly accurate some of the time and fail on a subset of variants, and it is important to know when the prediction can be trusted and with what confidence. Because of a lack of well tested reliability estimates, present clinical guidelines allow computational methods of predicting pathogenicity only secondary status as evidence for establishing a genetic cause for disease symptoms (Richards et al. 2015). The challenge NAGLU data set provided an opportunity for testing methods of assigning such probabilities on a clinically relevant dataset. The ensemble methods reported here, as well as other ensemble approaches such as REVEL (Ioannidis et al. 2016), are among the best for this purpose. Encouragingly, even on the realistic NAGLU population variants, a substantial fraction (up to 40%) of pathogenicity assignments can be made with greater than 90% confidence. More testing on diverse mutation sets is needed to establish clinical applicability.
Supplementary Material
Acknowledgments
This work was supported in part by NIH R01GM104436 and R01GM120364 to JM. The CAGI experiment coordination is supported by NIH U41 HG007446 and the CAGI conference by NIH R13 HG006650. We are grateful to the NAGLU (Jonathan H. LeBowitz, Wyatt T. Clark and G. Karen Yu) and SUMO ligase (Fritz Roth) dataset providers for making these challenges possible.
References
- Abeel T, Helleputte T, Van de Peer Y, Dupont P, Saeys Y. Robust biomarker identification for cancer diagnosis with ensemble feature selection methods. Bioinformatics. 2010;26:392–8. doi: 10.1093/bioinformatics/btp630. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh EH, Simmons-Edler R, Müller CL, Alford RF, Volfovsky N, Lash AE, Bonneau R. Robust classification of protein variation using structural modelling and large-scale data integration. Nucleic Acids Res. 2016;44:2501–13. doi: 10.1093/nar/gkw120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beesley C, Moraitou M, Winchester B, Schulpis K, Dimitriou E, Michelakakis H. Sanfilippo B syndrome: molecular defects in Greek patients. Clin Genet. 2004;65:143–149. doi: 10.1111/j.0009-9163.2004.00210.x. [DOI] [PubMed] [Google Scholar]
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–56. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat. 2009;30:1237–1244. doi: 10.1002/humu.21047. [DOI] [PubMed] [Google Scholar]
- Capriotti E, Altman RB, Bromberg Y. Collective judgment predicts disease-associated single nucleotide variants. BMC Genomics. 2013;14(Suppl 3):S2. doi: 10.1186/1471-2164-14-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter H, Douville C, Stenson PD, Cooper DN, Karchin R. Identifying Mendelian disease genes with the Variant Effect Scoring Tool. BMC Genomics. 2013;14(Suppl 3):S3. doi: 10.1186/1471-2164-14-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–61. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietterich TG. Multiple Classifier Systems. Springer; Berlin Heidelberg: 2000. Ensemble Methods in Machine Learning; pp. 1–15. [Google Scholar]
- Eisenhaber F, Argos P. Improved strategy in analytic surface calculation for molecular systems: Handling of singularities and computational efficiency. J Comput Chem. 1993;14:1272–1280. [Google Scholar]
- Eisenhaber F, Lijnzaad P, Argos P, Sander C, Scharf M. The double cubic lattice method: Efficient approaches to numerical integration of surface area and volume and to dot surface contouring of molecular assemblies. J Comput Chem. 1995;16:273–284. [Google Scholar]
- Folkman L, Stantic B, Sattar A, Zhou Y. EASE-MM: Sequence-Based Prediction of Mutation-Induced Stability Changes with Feature-Based Multiple Models. J Mol Biol. 2016;428:1394–1405. doi: 10.1016/j.jmb.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Frank E, Hall MA, Witten IH. Online Appendix for “Data Mining: Practical Machine Learning Tools and Techniques”. Morgan Kaufmann; 2016. The WEKA Workbench. [Google Scholar]
- Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins Struct Funct Genet. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- González-Pérez A, López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88:440–9. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerois R, Nielsen JE, Serrano L. Predicting Changes in the Stability of Proteins and Protein Complexes: A Study of More Than 1000 Mutations. J Mol Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- Hecht M, Bromberg Y, Rost B. Better prediction of functional effects for sequence variants. BMC Genomics. 2015;16(Suppl 8):S1. doi: 10.1186/1471-2164-16-S8-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, Musolf A, Li Q, Holzinger E, Karyadi D, Cannon-Albright LA, Teerlink CC, et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am J Hum Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsonis P, Lichtarge O. A formal perturbation equation between genotype and phenotype determines the Evolutionary Action of protein-coding variations on fitness. Genome Res. 2014;24:2050–8. doi: 10.1101/gr.176214.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Pennacchio LA, Sunyaev SR. Most Rare Missense Alleles Are Deleterious in Humans: Implications for Complex Disease and Association Studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, Jang W, Katz K, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Chen GJ, Lin SP, Lin SZ, Chuang CK, Hsiao KT, Huang CF, Lien WC. Identification and characterisation of mutations underlying Sanfilippo syndrome type B (mucopolysaccharidosis type IIIB) J Med Genet. 2002;39:E3. doi: 10.1136/jmg.39.2.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, Mooney SD, Radivojac P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25:2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtarge O, Bourne HR, Cohen FE. An Evolutionary Trace Method Defines Binding Surfaces Common to Protein Families. J Mol Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiyappan M, Concino MF, Norton AW. USPTO; USA: US 8,775,146 B2 Crystal Structure of Human Alpha-N-Acetylglucosaminidase. 2014
- Moult J. A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction. Curr Opin Struct Biol. 2005;15:285–289. doi: 10.1016/j.sbi.2005.05.011. [DOI] [PubMed] [Google Scholar]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015;44(D1):D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niroula A, Urolagin S, Vihinen M. PON-P2: Prediction Method for Fast and Reliable Identification of Harmful Variants. PLoS One. 2015;10(2):e0117380. doi: 10.1371/journal.pone.0117380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien JS. Sanfilippo syndrome: profound deficiency of alpha-acetylglucosaminidase activity in organs and skin fibroblasts from type-B patients. Proc Natl Acad Sci USA. 1972;69:1720–2. doi: 10.1073/pnas.69.7.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olatubosun A, Väliaho J, Härkönen J, Thusberg J, Vihinen M. PON-P: Integrated predictor for pathogenicity of missense variants. Hum Mutat. 2012;33:1166–1174. doi: 10.1002/humu.22102. [DOI] [PubMed] [Google Scholar]
- Orr HA. Fitness and its role in evolutionary genetics. Nat Rev Genet. 2009;10:531–539. doi: 10.1038/nrg2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal LR, Moult J. Genetic Basis of Common Human Disease: Insight into the Role of Missense SNPs from Genome-Wide Association Studies. J Mol Biol. 2015;427:2271–89. doi: 10.1016/j.jmb.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TA, Doughty E, Kann MG. Towards precision medicine: advances in computational approaches for the analysis of human variants. J Mol Biol. 2013;425:4047–63. doi: 10.1016/j.jmb.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redler RL, Das J, Diaz JR, Dokholyan NV. Protein Destabilization as a Common Factor in Diverse Inherited Disorders. J Mol Evol. 2016;82:11–16. doi: 10.1007/s00239-015-9717-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JA, Spacek DV, Snyder MP. High-Throughput Sequencing Technologies. Mol Cell. 2015;58:586–597. doi: 10.1016/j.molcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscoe BP, Thayer KM, Zeldovich KB, Fushman D, Bolon DN. Analyses of the Effects of All Ubiquitin Point Mutants on Yeast Growth Rate. J Mol Biol. 2013;425:1363–1377. doi: 10.1016/j.jmb.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfilippo SJ, Podosin R, Langer LO, Jr, Good RA. Mental retardation associated with acid mucopolysacchariduria (heparitin sulfate type) J Pediatr. 1963;63:837–838. [Google Scholar]
- Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33(Suppl 2):W382–388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger D, de Groot BL. Protein thermostability calculations using alchemical free energy simulations. Biophys J. 2010;98:2309–16. doi: 10.1016/j.bpj.2010.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Moult J. Structural and Functional Impact of Cancer-Related Missense Somatic Mutations. J Mol Biol. 2011;413:495–512. doi: 10.1016/j.jmb.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon WW, Hariharan M, Snyder MP. High-throughput sequencing for biology and medicine. Mol Syst Biol. 2013;9:640. doi: 10.1038/msb.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessitore A, Villani GR, Di Domenico C, Filocamo M, Gatti R, Di Natale P. Molecular defects in the α-N-acetylglucosaminidase gene in Italian Sanfilippo type B patients. Hum Genet. 2000;107:568–576. doi: 10.1007/s004390000429. [DOI] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–W650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium TU. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valstar MJ, Ruijter GJ, van Diggelen OP, Poorthuis BJ, Wijburg FA. Sanfilippo syndrome: A mini-review. J Inherit Metab Dis. 2008b;31:240–252. doi: 10.1007/s10545-008-0838-5. [DOI] [PubMed] [Google Scholar]
- von Figura K, Kresse H. The Sanfilippo B corrective factor: A N-acetyl-α-D-glucosaminidase. Biochem Biophys Res Commun. 1972;48:262–269. doi: 10.1016/s0006-291x(72)80044-5. [DOI] [PubMed] [Google Scholar]
- Weber B, Guo XH, Kleijer WJ, Kamp JJ van de, Poorthuis BJ, Hopwood JJ. Sanfilippo type B syndrome (mucopolysaccharidosis III B): allelic heterogeneity corresponds to the wide spectrum of clinical phenotypes. Eur J Hum Genet. 1999;7:34–44. doi: 10.1038/sj.ejhg.5200242. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer; 2009. [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, et al. The Genomic Landscapes of Human Breast and Colorectal Cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Yue P, Li Z, Moult J. Loss of Protein Structure Stability as a Major Causative Factor in Monogenic Disease. J Mol Biol. 2005;353:459–473. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Yue P, Moult J. Identification and Analysis of Deleterious Human SNPs. J Mol Biol. 2006;356:1263–1274. doi: 10.1016/j.jmb.2005.12.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.