Abstract
Factors that contribute to the onset of atherosclerosis may be elucidated by bioinformatic techniques applied to multiple sources of genomic and proteomic data. The results of genome wide association studies, such as the CardioGramPlusC4D study, expression data, such as that available from expression quantitative trait loci (eQTL) databases, along with protein interaction and pathway data available in Ingenuity Pathway Analysis (IPA), constitute a substantial set of data amenable to bioinformatics analysis. This study used bioinformatic analyses of recent genome wide association data to identify a seed set of genes likely associated with atherosclerosis. The set was expanded to include protein interaction candidates to create a network of proteins possibly influencing the onset and progression of atherosclerosis. Local average connectivity (LAC), eigenvector centrality, and betweenness metrics were calculated for the interaction network to identify top gene and protein candidates for a better understanding of the atherosclerotic disease process. The top ranking genes included some known to be involved with cardiovascular disease (APOA1, APOA5, APOB, APOC1, APOC2, APOE, CDKN1A, CXCL12, SCARB1, SMARCA4 and TERT), and others that are less obvious and require further investigation (TP53, MYC, PPARG, YWHAQ, RB1, AR, ESR1, EGFR, UBC and YWHAZ). Collectively these data help define a more focused set of genes that likely play a pivotal role in the pathogenesis of atherosclerosis and are therefore natural targets for novel therapeutic interventions.
Keywords: Bioinformatics, Coronary disease, Atherosclerosis, SNPs, Genomics
Introduction
Atherosclerosis is a multifactorial disease with a strong genetic component. Genome wide association studies for coronary artery disease (CAD) related phenotypes have identified at least 56 susceptibility loci at genome wide significance 1;2, and a study into the role of low-frequency (frequency 1% – 5%) and rare (frequency < 1%) DNA sequence variants in early onset myocardial infarction (MI) identified additional candidate genes 3. Investigation of proteins encoded by genes in close proximity to the susceptibility loci or implicated in the analysis of rare variants may lead to an enhanced understanding of the molecular mechanisms of atherosclerosis, and thereby facilitate the identification of novel candidates for targeted therapeutic interventions.
As part of the Genomic and Proteomic Architecture of Atherosclerosis (GPAA) project, we plan to utilize sensitive and highly accurate targeted mass spectrometry to quantify and thereby validate proteins identified as putative pathogenic candidates driving coronary artery disease. Multiple reaction monitoring (MRM) experiments will be performed on arterial tissue samples from individuals with and without extensive premature atherosclerosis collected as part of the Pathobiological Determinants of Atherosclerosis in Youth (PDAY) study 4. The PDAY study measured the extent and prevalence of atherosclerosis in 2,876 subjects between the ages of 15 and 34 who died of non-cardiac related causes. In order to utilize this precious resource to its full potential, we must first identify candidate proteins for assay development, and we seek to identify these candidates by combining discovery proteomics with bioinformatic data mining of network and pathway analysis of SNPS and genes associated with coronary disease from previous GWAS and rare variant association studies. Our goal is to expand the list of candidate proteins beyond the handful of well-known atherosclerosis proteins to include additional and novel proteins that represent the full spectrum of pathogenic molecular events underlying atherosclerosis development. Within the context of the GPAA project, the purpose of the current analysis is to identify relevant proteins, encoded by genes near susceptibility loci, to define an expanded set of candidate proteins hypothesized to contribute to the onset or development of atherosclerosis.
Graph theory and pathway analysis of protein interactions has proven useful for identifying essential proteins in complex protein networks 5;6 and elucidating physiologic mechanisms for complex traits, such as familial combined hyperlipidemia 7. Likewise, epigenetic feature analysis, based on publically available Encyclopedia of DNA Elements (ENCODE) data 8, has the potential to identify regulatory regions of the genome controlling expression of members of such networks, and the likelihood that SNPs in these regions are involved in this regulation. In this work, we used the results of genome wide association studies 2 and gene regulation data to identify a seed set of CAD associated genes. We then constructed the gene interaction network using Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood City, CA) to include other genes that interact with the seed set. We performed the network analysis to identify key gene nodes in the interaction network. To complement similar analyses that have been performed previously 2;9;10, we focused on two network properties in particular: centrality and betweenness 6. Betweenness is a measure of the number of shortest paths in a network that pass through the node; this is an indication of the importance that node has in connecting subnetworks within the network. Centrality can be measured in several ways; we used eigenvector centrality, which measures importance of a node as a function of that node’s links to other important nodes 11;12. We hypothesized that gene nodes with high betweenness scores may be links between functional modules, whereas gene nodes with high centrality scores may participate in multiple functional modules. Changes in the functioning of these high scoring gene nodes may disrupt functional modules and ultimately effect variability in phenotypes. In addition, we used the local average connectivity based method, LAC, for identifying essential proteins from the network level 13. LAC determines a protein’s essentiality by evaluating the relationship between a protein and its neighbors. LAC has been applied to predict the essentiality of proteins in yeast protein interaction networks and has been shown to outperform Eigenvector Centrality, Betweenness Centrality, Closeness Centrality, Bottle Neck, Information Centrality, Neighborhood Component, and Subgraph Centrality for identifying yeast essential proteins based on the different validations of sensitivity, specificity, and accuracy 13. However, the LAC method has not yet been applied to cardiovascular disease gene network analysis. In this study, we applied LAC in combination with the two commonly used network analysis methods, eigenvalue centrality and betweenness, to identify top gene candidates that are potentially playing key roles in the atherosclerosis disease network.
Materials and Methods
Selection and Curation of CAD Associated Genes. We included the genes assigned to the SNPs in the original CARDIoGRAM publication (“positional candidates”), as well as any genes linked to these SNPs in previously published expression quantitative trait loci (eQTL) analyses. The initial set of target genes was based on 162 unique SNPs identified by the CARDIoGRAM GWAS meta-analysis 2. These included the “known CAD susceptibility loci” (Table 1 in Deloukas et al, 2013 2), “Additional loci showing genome-wide significant association with CAD” (Table 2 in Deloukas et al, 2013 2), and “SNPs at an FDR≤5% and LD threshold of r2 < 0.2 used in estimating heritability” (Supplementary Table 9 in Deloukas et al, 2013 2). To identify potential eQTLs, we first expanded the list of 162 candidate SNPs using linkage disequilibrium (LD) to identify proxy SNPs. LD was determined with the Broad Institute’s SNP Annotation and Proxy (SNAP) search tool (http://archive.broadinstitute.org/mpg/snap) using an r2 > 0.8 in either the 1000 Genomes or HapMap data sets, based on the CEU population, within 500kb. All SNPs within the LD regions, including the original SNPs, were searched for eQTLs using the University of Chicago eQTL browser (eqtl.uchicago.edu), which contains data from 17 published studies. For each candidate SNP, the eQTLs with the highest score (−log10 p-value) are shown along with the proxy SNP (Supplemental Table S1).
Construction of Gene Interaction Networks. The selected CAD associated genes from above were used as the initial set of genes to construct gene interaction networks using IPA. IPA constructs networks based on extensive molecular interaction records maintained in the Ingenuity Pathways Knowledge Base (IPKB)14;15. IPKB is the largest curated database of biological networks, created from millions of relationships between genes and gene products. Given a list of genes/proteins, IPA can identify a set of relevant networks that these genes/proteins are involved in. IPA can merge the smaller networks into larger ones by using linker genes/proteins (common genes/proteins shared by the smaller networks). In this study, the larger merged network was used for the centrality and betweenness analysis to identify the key players in the network.
The experimentally observed relationships, such as protein-protein interactions, protein-DNA interactions, protein-RNA interactions, co-expression, translocation, activation, inhibition, molecular cleavage, membership, and phosphorylation were used to bring in other interacting molecules from the Ingenuity Knowledge Base to the network, and the additional molecules were used to specifically connect two or more smaller networks by merging them into a larger one. The resulting multiple networks were then merged into one network. The following parameters were used in the network construction: 1) All genes and chemicals in the Ingenuity Knowledge Base were used as the reference set and the species was set to human; 2) Only the direct relationships were considered; 3) The confidence level was set to be “Experimentally Observed” to retrieve the relationships that have been experimentally observed; 4) The number of molecules per network and the number of networks were set to the maximum allowed, 140 and 25, respectively.
Gene Interaction Network Analysis. Network analysis was performed using Cytoscape (www.cytoscape.org, version 3.1.1) and the CytoNCA plugin 16. Local average connectivity (LAC), eigenvector centrality and betweenness scores were calculated for each gene in the gene interaction network using CytoNCA. The direction of the edges is not considered in the network analysis. Parallel edges between two gene nodes represent different types of relationships that were observed between those two nodes. To reduce redundancy, these parallel edges and self-loops were removed in the network analysis.
Pathway Analysis Methods. Candidate genes selected from the network analysis were again analyzed with IPA for biological functions, cellular locations, signaling and metabolic canonical pathways, and associated diseases. The p-values for the identified canonical pathways, disease associations and functions were calculated using Fisher’s exact test. The Benjamini-Hochberg method was used to estimate the false discovery rate (FDR), and an FDR-corrected p-value of 0.05 was used to select significantly enriched pathways.
Availability of data and materials
Additional data used in this study is available in Supplemental Tables 1 through 5.
Ethics and Consent to participate
The original data used in this manuscript was obtained from published material, and no additional human subjects were included.
Results
CAD Associated Gene Prioritization. The 162 CARDIoGRAMplusC4D SNPs were associated with 160 unique genes, based on proximity alone. eQTLs were prioritized by selecting cis SNPs with a minimum eQTL score of 6 (p=10−6 in their respective, original study). eQTL analysis with the 162 SNPs and their LD proxies identified an additional 34 unique genes that were not included in the previous publication. Seventeen of the original positional candidates were also eQTLs (Supplemental Table S1). Twelve SNPs were associated with expression of at least two nearby genes, with a maximum of four genes for rs602633 (CELSR2, SORT1, PSRC1, and PSMA5). The strongest overall eQTL was with rs1412444, a proxy for the original SNP rs2246833 (r2=1.0) and LIPA expression in monocytes (eQTL score = 163.21). The original 160 positional genes and the 34 unique eQTL genes were combined for all downstream analyses, for a total of 194 unique genes.
Construction of the Gene Interaction Network. Of the 194 unique, CAD-associated genes curated from the CardioGramPlusC4D study and the eQTL analysis combined, 185 of these were found and mapped in the IPA database. These genes were used as seeds for the network construction. IPA network construction identified four major networks (Supplemental Table S2). These four networks were then merged into one large network, which included 422 connected nodes (molecules) with 1890 edges (relationships) (Supplemental Table S3).
Gene Interaction Network Analysis. Supplemental Table S4 shows the LAC, eigenvector centrality, and betweenness results from the CytoNCA network analysis. The top thirty network nodes ranked by each of the analysis methods, LAC, eigenvector centrality, and betweenness, are listed in Table 1. These nodes include genes, gene groups and chemicals. Among the top genes ranked by LAC, 10 were from the original seed set (highlighted in red; CDKN1A, APOE, SMARCA4, APOA1, APOC2, TERT, APOB, APOC1, APOA5 and SCARB1). Among the top genes ranked by eigenvector centrality, five were from the original seed set (highlighted in red; CDKN1A, SMARCA4, APOA1, APOE and TERT). Among the top genes ranked by betweenness, four were from the original seed gene set (highlighted in red; APOA1, CDKN1A, SMARCA4 and CXCL12). Three seed genes CDKN1A, SMARCA4 and APOA1 (red text and underlined) were the common, top-ranked genes identified by all three methods (LAC, eigenvector centrality, and betweenness), indicating the importance of these genes in the network. In addition to these three common seed genes, ten genes not in the original seed set were also identified by all three methods. These 10 new genes are TP53, MYC, PPARG, YWHAQ, RB1, AR, ESR1, EGFR, UBC and YWHAZ.
Table 1.
Top network nodes ranked by LAC, eigenvector centrality and betweenness scores. (The genes from the original seed set are highlighted in red. The common seed genes identified by all three methods are in red text and underlined
| Gene/Chemical | LAC | Gene/Chemical | Eigenvector | Gene/Chemical | Betweenness |
|---|---|---|---|---|---|
| CDKN1A | 9 | TP53 | 0.293992 | APP | 40725.25 |
| TP53 | 8.441559 | APP | 0.242665 | HNF4A | 24356.68 |
| APOE | 8 | ESR1 | 0.235588 | ELAVL1 | 23250.77 |
| MYC | 7.107143 | MYC | 0.225831 | Gpcr | 21763.95 |
| PPARG | 7.032258 | UBC | 0.194388 | TP53 | 20339.18 |
| YWHAQ | 6.571429 | CDKN1A | 0.188231 | ESR1 | 15732.38 |
| SMARCA4 | 6.5625 | HNF4A | 0.179053 | UBC | 13405.76 |
| RNA polymerase II | 6.5 | AR | 0.167393 | MYC | 10966.7 |
| RB1 | 6.307693 | ELAVL1 | 0.163778 | CREB1 | 9632.628 |
| AR | 6.243902 | EGFR | 0.159149 | NXF1 | 7906.177 |
| HSPA8 | 6.24 | PPARG | 0.15383 | EGFR | 7797.91 |
| APOA1 | 6.214286 | SMARCA4 | 0.150114 | VHL | 7234.26 |
| Hsp70 | 6 | YWHAZ | 0.149565 | YWHAZ | 7099.747 |
| APOC2 | 6 | YWHAQ | 0.144655 | AR | 7044.75 |
| ESR1 | 5.768116 | RB1 | 0.134934 | DLG4 | 6159.478 |
| TERT | 5.666667 | HSPA8 | 0.134347 | PPARG | 5938.707 |
| Histone h3 | 5.6 | CREB1 | 0.132037 | GRB2 | 5115.18 |
| EGFR | 5.55 | APOA1 | 0.130066 | VCP | 5050.226 |
| APOB | 5.5 | RNA polymerase II | 0.121485 | REL | 4567.824 |
| NFkB (complex) | 5.375 | APOE | 0.118555 | APOA1 | 4350.606 |
| APOC1 | 5.333334 | GRB2 | 0.116974 | GPR12 | 3997.739 |
| APOA5 | 5.333334 | VHL | 0.115261 | collagen | 3765.427 |
| SCARB1 | 5.142857 | Hsp70 | 0.114036 | CDKN1A | 3632.875 |
| UBC | 5.107143 | VCP | 0.110983 | SMARCA4 | 3361.238 |
| Histone h4 | 4.888889 | Histone h3 | 0.110957 | F2R | 3271.263 |
| estrogen receptor | 4.769231 | TERT | 0.106133 | YWHAQ | 3228.63 |
| HDL | 4.666667 | ZFP36 | 0.09736 | CXCL12 | 3088.951 |
| Akt | 4.571429 | PPARA | 0.096878 | ZFP36 | 2985.264 |
| YWHAZ | 4.444445 | NFkB (complex) | 0.090869 | LATS2 | 2786.828 |
| N-cor | 4.4 | REL | 0.085653 | RB1 | 2672.204 |
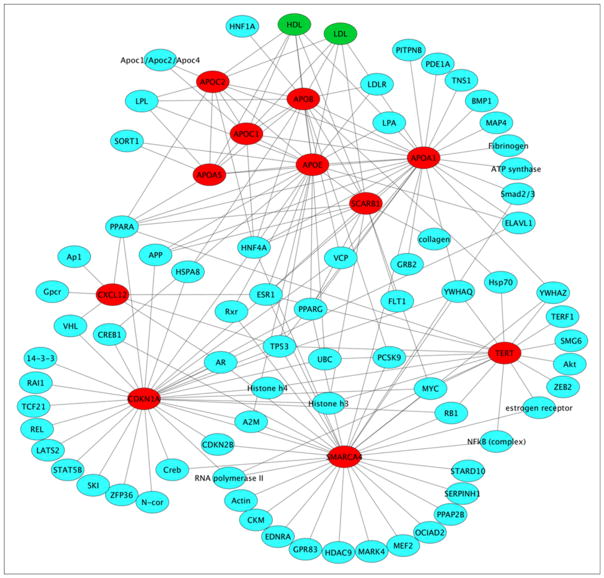
Combining the LAC, eigenvector centrality, and betweenness lists in Table 1, a total of 10 genes (CDKN1A, APOE, SMARCA4, APOA1, APOC2, TERT, APOB, APOC1, APOA5 and SCARB1) are from the original seed set, which suggests that these CAD associated genes are important in the gene interaction network. Figure 1 shows the interactions between these 10 genes (in red) and their interacting genes (in blue) and chemicals (in green) in the gene interaction network. Most of these top genes are highly connected in the sub-network.
Figure 1.
The interactions between 10 top ranking genes (red nodes) and their interacting genes (blue nodes) and chemicals (green nodes) in the sub-network. The graph was generated with Cytoscape 35.
Pathway Analysis. The top-ranked proteins from Table 1 were selected to perform metabolic and signaling canonical pathways analysis using IPA. The result is shown in Supplemental Table S5. The top ten pathway hits were FXR/RXR Activation, Clathrin-mediated Endocytosis Signaling, Telomerase Signaling, IL-12 Signaling and Production in Macrophages, Prostate Cancer Signaling, ERK/MAPK Signaling, Myc Mediated Apoptosis Signaling, LXR/RXR Activation, Atherosclerosis Signaling and Estrogen-mediated S-phase Entry (Table 2).
Table 2.
Top pathway hits of the selected network genes
| Ingenuity Canonical Pathways | B-H p-value | Genes |
|---|---|---|
| FXR/RXR Activation | 4.68E-10 | PPARG,PPARA,APOE,APOB,APOA1,SCARB1,APOC1,APOC2,HNF4A |
| Clathrin-mediated Endocytosis Signaling | 7.76E-09 | HSPA8,APOE,APOB,APOA1,F2R,GRB2,APOC1,APOC2,UBC |
| Telomerase Signaling | 5.01E-08 | TP53,MYC,RB1,GRB2,TERT,CDKN1A,EGFR |
| IL-12 Signaling and Production in Macrophages | 3.39E-07 | PPARG,APOE,APOB,APOA1,APOC1,APOC2,REL |
| Prostate Cancer Signaling | 4.68E-07 | TP53,RB1,AR,GRB2,CREB1,CDKN1A |
| ERK/MAPK Signaling | 2.51E-06 | PPARG,YWHAQ,MYC,GRB2,CREB1,YWHAZ,ESR1 |
| Myc Mediated Apoptosis Signaling | 2.88E-06 | YWHAQ,TP53,MYC,GRB2,YWHAZ |
| LXR/RXR Activation | 2.88E-06 | APOE,APOB,APOA1,APOC1,APOA5,APOC2 |
| Atherosclerosis Signaling | 2.88E-06 | APOE,APOB,APOA1,CXCL12,APOC1,APOC2 |
| Estrogen-mediated S-phase Entry | 2.88E-06 | MYC,RB1,CDKN1A,ESR1 |
Discussion
In this study, protein-protein interaction networks were analyzed to identify proteins with potentially essential roles (high centrality) and those with minimal functional redundancy (high betweenness). Starting with known susceptibility loci, we identified proteins encoded by genes near susceptibility loci and identified those proteins most likely to act as hubs and bottlenecks. Ranking proteins by local average connectivity, betweenness, and centrality scores provides a method for prioritizing targets for future MRM mass spectrometry experiments, designed to identify proteins contributing to the onset or development of atherosclerosis. Proteins with high ranks in LAC, eigenvector centrality, and betweenness scores are considered top candidates for further investigation with experimental proteomics techniques.
Our network analysis using LAC, eigenvector centrality, and betweenness methods identified a set of 49 high ranking molecules based on their importance and connectivity within the interaction network we constructed. Among these 49 molecules, several already have a very well established and known association with cardiovascular disease risk, including APOA1, APOA5, APOB, APOC1, APOC2, APOE, CDKN1A, CXCL12, SCARB1, SMARCA4 and TERT (e.g., 17–21). While these well-established proteins serve as an important validation for our approach, of potentially more biological interest are the additional and more novel candidates identified with our expanded network approach. These included TP53, MYC, PPARG, YWHAQ, RB1, AR, ESR1, EGFR, UBC and YWHAZ, which were identified by all three analysis methods, but do not have the same level of prior literature evidence supporting a known association with cardiovascular disease. These proteins also rank highly by betweenness scores, indicating they may be involved in multiple pathways, and fewer proteins may perform their function within pathways. In our study, each of these novel proteins interacted with at least three of our seed proteins (Figure 1), supporting the plausible importance of their role in the biology of coronary artery disease and atherosclerosis progression.
Four of these 10 highly-connected novel genes (TP53, MYC, YWHAQ, and YWHAZ) were also identified recently in an independent publication as “Predicted CVD genes” using a different pathway-based approach 22. Both TP53 and MYC are well-known for their role in cancer and may also be involved in the regulation of smooth muscle cell proliferation during neointima formation in coronary artery disease 23;24. Much less is known about YWHAQ and YWHAZ, which are highly conserved scaffolding proteins of the 14-3-3 family, involved in multiple signal transduction pathways including those linked to p53 apoptosis signaling 25 and Epidermal Growth Factor Receptor (EGFR) signaling 26. The EGFR protein was another of the 10 novel top proteins identified in this analysis, and is a well-known activator of ERK/MAPK signaling which was among the top canonical pathways from the IPA analysis of these data. While EGFR is known to be expressed in atherosclerotic plaques 27;28, its mechanistic role in coronary artery disease pathogenesis is as yet unclear. Interestingly, another cell-signaling scaffold protein, Growth Factor Receptor Binding Protein 2 (GRB2), was also detected among our top 49 candidate proteins, and together with YWHAZ, has been shown to be involved in the clathrin-endocytosis mediated internalization of EGFR 29. Furthermore, GRB2 has been identified as a critical protein for neointima and atherosclerotic lesion formation in ApoE −/− mouse models of coronary artery disease 30;31. These connections become rather interesting in light of our observation of “clathrin-mediated endocytosis” as a top pathway in the IPA analysis (Table 2) connecting several of our candidate proteins. Taken together, these data indicate that the multifunctional signaling scaffold proteins YWHAZ, YWHAQ, and GRB2, may represent critical hubs for the EGFR, and other growth factor, signaling networks and may represent important nodes in the molecular cascades that become dysregulated in coronary artery disease.
Interesting potential links to atherosclerosis can also be found among the remaining 10 novel proteins identified in the LAC, Eigenvector, and betweenness rankings. The Retinoblastoma-associated protein (RB1) is a component of a transcriptional-repressor complex that interacts with the well-known cardiovascular disease protein SMARCA4, which was also top ranked in our analysis. Another transcriptional regulator, peroxisome proliferator-activated receptor gamma (PPARG), which regulates genes involved in fatty acid metabolism and inflammation, is expressed in atherosclerotic lesions and is thought to negatively regulate pro-atherosclerotic processes, suggesting the potential use of PPAR-activators for atherosclerosis treatment 32. The combined observation of androgen receptor (AR) and estrogen receptor (ESR1) suggest that the reproductive steroid hormones testosterone and estradiol may play intriguing roles in coronary artery disease progression and thus may also represent important sex-dependent mechanisms in atherosclerosis pathogenesis 33. Finally, in addition to poly-ubiquitin (UBC) identified in our top 10 novel proteins, two other components of ubiquitin-proteasomal degradation, valosin-containing protein (VCP) and von-hippel lindau tumor suppressor (VHL) were also found among the top 49 molecules in our expanded network. Together these three proteins are consistent with an emerging hypothesis regarding the importance of the ubiquitin-proteasomal degradation pathway in the pathogenesis of atherosclerosis 34;35.
To summarize, there are numerous biological connections between the top ranked proteins identified in this expanded network analysis of coronary artery disease genes, and these connections support the inclusion of these molecules as candidates for follow-up analysis in the GPAA project. Furthermore, these discoveries support the utility of this expanded approach to the analysis of genomic scale datasets for the identification of candidate disease proteins. The validity of our approach can be illustrated by the APOA1 node in our predicted network. Mutations that alter the functioning of APOA1 could adversely impact the functioning of several interacting proteins, as indicated by the high hub score of the APOA1 node. In addition, APOA1 interacts strongly with other apolipoproteins (e.g., APOB, APOE) that also have high node scores. LDLR interacts with all three of these proteins (Figure 1), and exome sequencing recently identified a marked increased risk of myocardial infarctions in individuals with rare mutations in LDLR 3, further highlighting the utility of evaluating proteins targeted within the biological hub.
As further validation of biological relevance, our pathway analysis of the top ranked proteins in the network analysis identified a list of pathways that are known to influence atherosclerosis (Table 2). In addition to the four pathways, Atherosclerosis signaling, LXR/RXR activation, FXR/RXR activation and Acute phase response signaling, which were previous identified by Deloukas et al 2, we identified additional disease related pathways such as Clathrin-mediated Endocytosis Signaling, Telomerase Signaling, IL-12 Signaling and Production in Macrophages, Prostate Cancer Signaling, ERK/MAPK Signaling, Myc Mediated Apoptosis Signaling, and Estrogen-mediated S-phase Entry.
Our analysis had some similarities with previous analyses 2;9;10;22;36, in that we focused on the top SNP associations, and then expanded that list with eQTL findings. While some of these studies also used pathway and gene ontology analyses, our analyses went considerably beyond previous work by focusing on the interactions of the seed proteins with others, based primarily on the centrality and betweenness of the molecules. This was done independent of the role of the additional proteins, allowing us to identify several proteins that have not received serious attention as candidates to monitor in studying the pathophysiology of CVD-related processes.
Our study, like other protein-protein interaction analyses, was limited by the current state of knowledge of protein interactions. The lack of evidence for interactions between proteins should not be interpreted as evidence for lack of such an interaction. Proteins with high betweenness scores may be actual bottlenecks in metabolic or regulatory pathways, or they may be understudied macromolecules that warrant further investigation. A risk of using literature-based interaction analysis is that well-published proteins or genes may appear more commonly. This may account for the identification of a portion of our newly identified proteins (e.g., TP53, MYC), but not for others, where little published work is available (e.g., YWHAQ, YWHAZ). The set of protein interactions analyzed in this study were not filtered based on location of expression, and some interactions may only occur in tissues unrelated to atherosclerosis. Including such interactions may lead to overestimates in the centrality scores. However, filtering based on known expression locations may also eliminate relevant interactions if the proteins are not included in tissue expression databases; this could lead to over estimates in the betweenness scores. Finally, our approach used the genes nearest to the associated SNPs when eQTLs were not identified. More distal genes may be regulated by these SNPs, but without additional functional data these loci were difficult to identify and we used the most likely genes to be involved in each region.
Conclusion
Using a protein-protein interaction network approach, we have identified the most likely genes involved in CAD-related phenotypes using the CARDIoGRAM GWAS meta-analysis as a starting point 2. In addition to the well-known candidates, we identified a subset of genes that interact with these likely contributors, but have not otherwise been associated with CAD. These new candidates represent novel targets for assay development and MRM-based monitoring to determine their expression profile and its correlation to atherosclerotic disease in the PDAY sample set. Ultimately, the goal of this project is to prioritize these proteins in terms of their likely effectiveness as targets for therapeutic intervention, and perhaps offer the opportunity to develop novel as well as repurpose existing drugs for cardiovascular and atherosclerosis related conditions.
Supplementary Material
Acknowledgments
This work was funded by NIH grant R01HL111362.
Conflict of Interest: The authors declare that they have no conflicts of interest.
References
- 1.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Do R, Stitziel NO, Won HH, Jorgensen AB, Duga S, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strong JP, Malcom GT, McMahan CA, Tracy RE, Newman WP, III, et al. Prevalence and extent of atherosclerosis in adolescents and young adults: implications for prevention from the Pathobiological Determinants of Atherosclerosis in Youth Study. JAMA. 1999;281:727–735. doi: 10.1001/jama.281.8.727. [DOI] [PubMed] [Google Scholar]
- 5.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 6.Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol. 2007;3:e59. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang YL, Xue F, Liu LZ, He ZH. Pathway analysis detect potential mechanism for familial combined hyperlipidemia. Eur Rev Med Pharmacol Sci. 2013;17:1909–1915. [PubMed] [Google Scholar]
- 8.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barth AS, Tomaselli GF. Gene scanning and heart attack risk. Trends Cardiovasc Med. 2016;26:260–265. doi: 10.1016/j.tcm.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braenne I, Civelek M, Vilne B, Di Narzo A, Johnson AD, et al. Prediction of Causal Candidate Genes in Coronary Artery Disease Loci. Arterioscler Thromb Vasc Biol. 2015;35:2207–2217. doi: 10.1161/ATVBAHA.115.306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonacich P. Power and Centrality - A Family of Measures. American Journal of Sociology. 1987;92:1170–1182. [Google Scholar]
- 12.Borgatti SP. Centrality and network flow. Social Networks. 2005;27:55–71. [Google Scholar]
- 13.Li M, Wang J, Chen X, Wang H, Pan Y. A local average connectivity-based method for identifying essential proteins from the network level. Comput Biol Chem. 2011;35:143–150. doi: 10.1016/j.compbiolchem.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, et al. Inflamm and Host Response to Injury Large Scale Collab. Res. Program. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. Erratum in: Nature. 2005; 438:696. [DOI] [PubMed] [Google Scholar]
- 15.Ficenec D, Osborne M, Pradines J, Richards D, Felciano R, et al. Computational knowledge integration in biopharmaceutical research. Brief Bioinform. 2003;4:260–278. doi: 10.1093/bib/4.3.260. [DOI] [PubMed] [Google Scholar]
- 16.Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Guardiola M, Cofan M, Castro-Oros I, Cenarro A, Plana N, et al. APOA5 variants predispose hyperlipidemic patients to atherogenic dyslipidemia and subclinical atherosclerosis. Atherosclerosis. 2015;240:98–104. doi: 10.1016/j.atherosclerosis.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Lange LA, Hu Y, Zhang H, Xue C, Schmidt EM, et al. Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am J Hum Genet. 2014;94:233–245. doi: 10.1016/j.ajhg.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartmann P, Schober A, Weber C. Chemokines and microRNAs in atherosclerosis. Cell Mol Life Sci. 2015;72:3253–3266. doi: 10.1007/s00018-015-1925-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanislovaitiene D, Lesauskaite V, Zaliuniene D, Smalinskiene A, Gustiene O, et al. SCARB1 single nucleotide polymorphism (rs5888) is associated with serum lipid profile and myocardial infarction in an age- and gender-dependent manner. Lipids Health Dis. 2013;12:24. doi: 10.1186/1476-511X-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bressler J, Franceschini N, Demerath EW, Mosley TH, Folsom AR, et al. Sequence variation in telomerase reverse transcriptase (TERT) as a determinant of risk of cardiovascular disease: the Atherosclerosis Risk in Communities (ARIC) study. BMC Med Genet. 2015;16:52. doi: 10.1186/s12881-015-0194-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarajlic A, Janjic V, Stojkovic N, Radak D, Przulj N. Network topology reveals key cardiovascular disease genes. PLoS ONE. 2013;8:e71537. doi: 10.1371/journal.pone.0071537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speir E, Modali R, Huang ES, Leon MB, Shawl F, et al. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 24.Napoli C, Lerman LO, de Nigris F, Sica V. c-Myc oncoprotein: a dual pathogenic role in neoplasia and cardiovascular diseases? Neoplasia. 2002;4:185–190. doi: 10.1038/sj.neo.7900232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang HY, Wen YY, Chen CH, Lozano G, Lee MH. 14-3-3 sigma positively regulates p53 and suppresses tumor growth. Mol Cell Biol. 2003;23:7096–7107. doi: 10.1128/MCB.23.20.7096-7107.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oksvold MP, Huitfeldt HS, Langdon WY. Identification of 14-3-3zeta as an EGF receptor interacting protein. FEBS Lett. 2004;569:207–210. doi: 10.1016/j.febslet.2004.05.068. [DOI] [PubMed] [Google Scholar]
- 27.Miyagawa J, Higashiyama S, Kawata S, Inui Y, Tamura S, et al. Localization of heparin-binding EGF-like growth factor in the smooth muscle cells and macrophages of human atherosclerotic plaques. J Clin Invest. 1995;95:404–411. doi: 10.1172/JCI117669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamb DJ, Modjtahedi H, Plant NJ, Ferns GA. EGF mediates monocyte chemotaxis and macrophage proliferation and EGF receptor is expressed in atherosclerotic plaques. Atherosclerosis. 2004;176:21–26. doi: 10.1016/j.atherosclerosis.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 29.Tomassi L, Costantini A, Corallino S, Santonico E, Carducci M, et al. The central proline rich region of POB1/REPS2 plays a regulatory role in epidermal growth factor receptor endocytosis by binding to 14-3-3 and SH3 domain-containing proteins. BMC Biochem. 2008;9:21. doi: 10.1186/1471-2091-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S, Ren J, Khan MF, Cheng AM, Abendschein D, et al. Grb2 is required for the development of neointima in response to vascular injury. Arterioscler Thromb Vasc Biol. 2003;23:1788–1793. doi: 10.1161/01.ATV.0000085015.49110.85. [DOI] [PubMed] [Google Scholar]
- 31.Proctor BM, Ren J, Chen Z, Schneider JG, Coleman T, et al. Grb2 is required for atherosclerotic lesion formation. Arterioscler Thromb Vasc Biol. 2007;27:1361–1367. doi: 10.1161/ATVBAHA.106.134007. [DOI] [PubMed] [Google Scholar]
- 32.Neve BP, Fruchart JC, Staels B. Role of the peroxisome proliferator-activated receptors (PPAR) in atherosclerosis. Biochem Pharmacol. 2000;60:1245–1250. doi: 10.1016/s0006-2952(00)00430-5. [DOI] [PubMed] [Google Scholar]
- 33.den Ruijter HM, Haitjema S, Asselbergs FW, Pasterkamp G. Sex matters to the heart: A special issue dedicated to the impact of sex related differences of cardiovascular diseases. Atherosclerosis. 2015;241:205–207. doi: 10.1016/j.atherosclerosis.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Herrmann J, Ciechanover A, Lerman LO, Lerman A. The ubiquitin-proteasome system in cardiovascular diseases-a hypothesis extended. Cardiovasc Res. 2004;61:11–21. doi: 10.1016/j.cardiores.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 35.Wang F, Lerman A, Herrmann J. Dysfunction of the ubiquitin-proteasome system in atherosclerotic cardiovascular disease. Am J Cardiovasc Dis. 2015;5:83–100. [PMC free article] [PubMed] [Google Scholar]
- 36.Makinen VP, Civelek M, Meng Q, Zhang B, Zhu J, et al. Integrative genomics reveals novel molecular pathways and gene networks for coronary artery disease. PLoS Genet. 2014;10:e1004502. doi: 10.1371/journal.pgen.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional data used in this study is available in Supplemental Tables 1 through 5.