Abstract
Crescent-shaped red blood cells, the hallmark of sickle cell disease, present a striking departure from the biconcave disc shape normally found in mammals. Characterized by increased mechanical fragility, sickled cells promote haemolytic anaemia and vaso-occlusions and contribute directly to disease in humans. Remarkably, a similar sickle-shaped morphology has been observed in erythrocytes from several deer species, without obvious pathological consequences. The genetic basis of erythrocyte sickling in deer, however, remains unknown. Here, we determine the sequences of human β-globin orthologs in 15 deer species and use protein structural modelling to identify a sickling mechanism distinct from the human disease, coordinated by a derived valine (E22V) that is unique to sickling deer. Evidence for long-term maintenance of a trans-species sickling/non-sickling polymorphism suggests that sickling in deer is adaptive. Our results have implications for understanding the ecological regimes and molecular architectures that have promoted convergent evolution of sickling erythrocytes across vertebrates.
Human sickling is caused by a single amino acid change (E6V) in the adult β-globin (HBB) protein1. Upon deoxygenation, steric changes in the haemoglobin tetramer enable an interaction between 6V and a hydrophobic acceptor pocket (known as the EF pocket) on the β-surface of a second tetramer2,3. This interaction promotes polymerization of mutant haemoglobin (HbS) molecules, which ultimately coerces red blood cells into the characteristic sickle shape. Heterozygote carriers of the HbS allele are typically asymptomatic4 whereas HbS homozygosity has severe pathological consequences and is linked to shortened lifespan5. Despite this, the HbS allele has been maintained in sub-Saharan Africa by balancing selection because it confers – by incompletely understood means – a degree of protection against the effects of Plasmodium infection and malaria6.
Sickling red blood cells were first described in 1840 – seventy years prior to their discovery in humans7 – when Gulliver8 reported unusual erythrocyte shapes in blood from white-tailed deer (Odocoileus virginianus). Subsequent research revealed that sickling is widespread amongst deer species worldwide8–11 (Fig. 1, Supplementary Table 1). It is not, however, universal: red blood cells from reindeer (Rangifer tarandus) and European elk (Alces alces, known as moose in North America) do not sickle; neither do erythrocytes from most North American wapiti (Cervus canadensis)11,12.
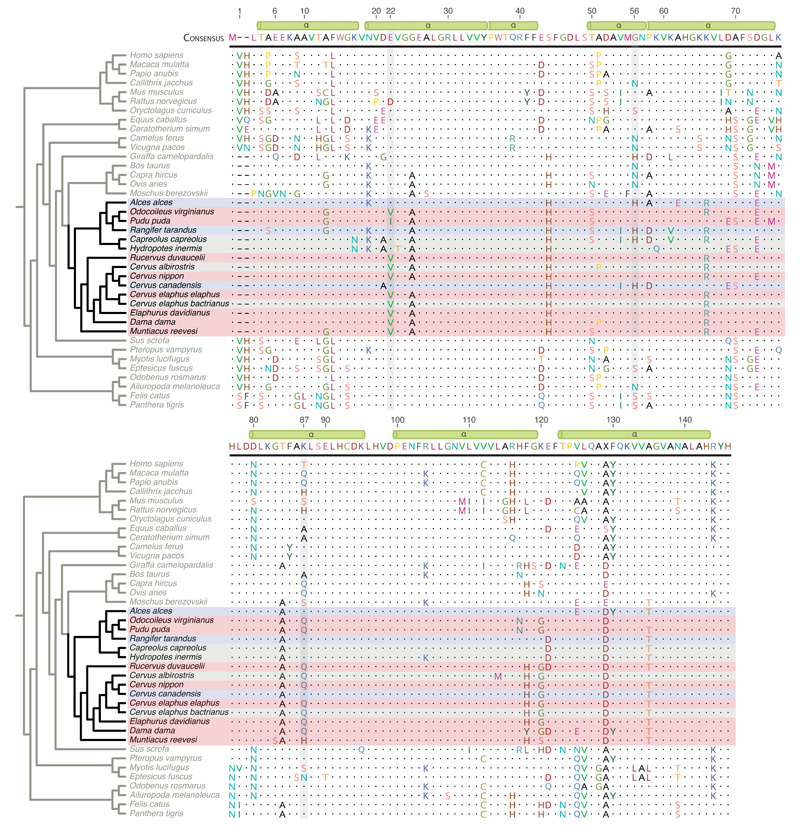
Fig. 1. Mammalian adult β-globin peptide sequences in phylogenetic context.
To facilitate comparisons with prior classic literature, residues here and in the main text are numbered according to human HBB, skipping the leading methionine. Dots represent residues identical to the consensus sequences, defined by the most common amino acid (X indicates a tie). Key residues discussed in the text are highlighted. HBBA sequences from deer are coloured according to documented sickling state: red = sickling, blue = non-sickling, grey = indeterminate (Supplementary Table 1). Green cylinders highlight the position of α-helices in the secondary structure of human HBB. See Methods and Supplementary Figure 11 for derivation of the accompanying cladogram.
Sickling deer erythrocytes are similar to human HbS cells with regard to their gross morphology and the tubular ultrastructure of haemoglobin polymers13–16. Moreover, as in humans, sickling is reversible through modulation of oxygen supply or pH9,17 and mediated by specific β-globin alleles18,19, with both sickling and non-sickling alleles segregating in wild populations of white-tailed deer20. As in humans, α-globin – two copies of which join two β-globin proteins to form the haemoglobin tetramer – is not directly implicated in sickling etiology18,21. Also as in humans, foetal haemoglobin molecules, which incorporate distinct β-globin paralogs in human and deer (see below), do not promote sickling under the same conditions19. But whereas HbS sickling occurs when oxygen tension is low, deer erythrocytes sickle under high pO2 and at alkaline pH17. Consequently, prime conditions for sickling in deer are likely found in lung capillaries (rather than in systemic capillaries were oxygen is unloaded), although in vivo sickling can also be observed in peripheral venous blood22, especially following exercise regimes that induce transient respiratory alkalosis23. Further, unlike in humans, sickled deer erythrocytes do not exhibit increased mechanical fragility in vitro17,18 and the sickling allele in white-tailed deer (previously labelled βIII) is the major allele, with ≥60% of individuals homozygous for βIII (REF. 20,24). Remarkably, βIII homozygotes do not display aberrant haematological values or obvious pathological traits25. Together, these observations are consistent with reduced physiological costs of sickling in deer. However, it is unknown whether sickling is simply innocuous, as previously suggested23, or plays an HbS-like adaptive role. In addition, partial peptide digests of sickling white-tailed deer β-globins did not recover the E6V mutation that causes sickling in humans24, leaving the genetic basis of sickling in deer unresolved.
Results
The molecular basis of sickling in deer is distinct from that in the human disease
To dissect the molecular basis of sickling in deer and elucidate its evolutionary history and potential adaptive significance, we used a combination of whole-genome sequencing, locus-specific assembly and targeted amplification to determine the sequence of the HBBA gene, which encodes the adult β-globin chain, in a phylogenetically broad sample of 15 deer species, including both sickling and non-sickling taxa (Fig. 1, Supplementary Table 1). Globin genes in mammals are located in paralog clusters, which – despite a broadly conserved architecture – constitute hotbeds of pseudogenization, gene duplication, conversion, and loss26,27. In ruminants, the entire β-globin cluster is triplicated in goat (Capra hircus)28 and duplicated in cattle (Bos taurus)29, where two copies of the ancestral β-globin gene sub-functionalized to become specifically expressed in adult (HBBA) and foetal (HBBF) blood. Based on a recent draft assembly of a white-tailed deer (O. virginianus texanus) genome, the architecture of the β-globin cluster mirrors that seen in cattle, consistent with the duplication event pre-dating the Bovidae-Cervidae split (Supplementary Figure 1). Primers designed before this assembly became available frequently co-amplified HBBA and HBBF (see Methods and Supplementary Figure 2). In the first instance, we therefore assigned foetal and adult status based on residues specifically shared with either HBBA or HBBF in cattle, which results in independent clustering of putative HBBA and HBBF genes on an HBBA/F gene tree (Supplementary Figure 3). To confirm these assignments, we sequenced mRNA from the red cell component of blood from an adult Père David’s deer (Elaphurus davidianus) and assembled the erythrocyte transcriptome de novo (see Methods). We identified a highly abundant β-globin transcript (>200,000 transcripts/million) corresponding precisely to the putative adult β-globin gene amplified from genomic DNA of the same individual (Supplementary Figure 4). Reads that uniquely matched the putative HBBA gene were >2000-fold more abundant than reads uniquely matching the putative HBBF gene, which is expressed at low levels. This is similar to the situation in humans, where transcripts of HBG, a distinct paralog that convergently evolved foetal expression, are found at low abundance in adult blood30. Finally, our assignments are consistent with partial peptide sequences for white-tailed deer24, fallow deer (Dama dama)31 and reindeer32 that were previously obtained from the blood of adult individuals.
We then considered deer HBBA orthologs in a wider mammalian context, restricting analysis to species with high-confidence HBB assignments (see Methods). Treating wapiti as non-sickling, and four species as indeterminate (no or insufficient phenotyping of sickling; see Supplementary Table 1), we find three residues (Fig. 1) that discriminate sickling from non-sickling species: 22 (non-sickling: E, sickling: V/I), 56 (n-s: H, s: G), and 87 (n-s: K, s: Q/H). The change at residue 22, from an ancestral glutamic acid to a derived valine (isoleucine in Pudu puda) is reminiscent of the human HbS mutation and occurs at a site that is otherwise highly conserved throughout mammalian evolution.
Structural modelling supports an interaction between 22V and the EF pocket
To understand how sickling-associated amino acids promote polymerization, we examined these residues in their protein structural context. Residue 22 lies on the surface of the haemoglobin tetramer, at the start of the second alpha helix (Fig. 2a). Close to residue 22 are residue 56 and two other residues that differ between non-sickling reindeer and moose (but not wapiti) and established sickling species: 19 (n-s: K, s: N) and 120 (n-s: K, s: G/S). Together these residues form part of a surface of increased hydrophobicity in sickling species (Fig. 2b). Distal to this surface, residue 87 is situated at the perimeter of the EF pocket, which in humans interacts with 6V to laterally link two β-globin molecules in different haemoglobin tetramers and stabilize the parallel strand architecture of the HbS fibre2,3,33,34. Mutation of residue 87 in humans can have marked effects on sickling dynamics35. For example, erythrocytes derived from HbS/Hb Quebec-Chori (T87I) compound heterozygotes sickle like HbS homozygotes36 while Hb D-Ibadan (T87K) inhibits sickling37.
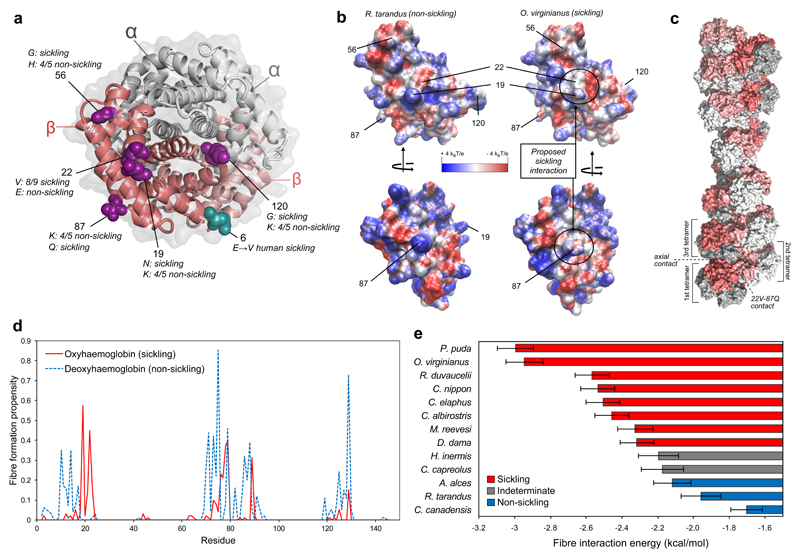
Fig. 2. Structural basis for sicking of deer haemoglobin.
a, Structure of oxyhaemoglobin (PDB ID: 1HHO), with the key residues associated with sickling highlighted in one of the β-globin chains. b, Comparison of the electrostatic surfaces of oxy HBBA from a non-sickling (R. tarandus) and sickling (O. virginianus) species. c, Example of a haemoglobin fibre formed via directed docking between residues 22V and 87Q of O. virginianus oxy HBBA. d, Fibre formation propensity derived from docking simulations centred at a given focal residue in O. virginianus oxy and deoxy HBBA. These values represent the fraction of docking models that result in HbS-like haemoglobin fibre structures. e, Fibre interaction energy for different deer species, determined by mutating the 270 22V-87Q docking models compatible with fibre formation and calculating the energy of the interaction. Error bars represent standard error of the mean.
Given the similarity between the human E6V mutation and E22V in sickling deer, we hypothesized that sickling occurs through an interaction in trans between residue 22 and the EF pocket. To test whether such an interaction is compatible with fibre formation, we carried out directed docking simulations centred on these two residues using a homology model of oxy β-globin from white-tailed deer (see Methods). We then used the homodimeric interactions from docking to build polymeric haemoglobin structures, analogous to how the 6V-EF interaction leads to extended fibres in HbS homozygotes. Strikingly, nearly half of our docking models resulted in HbS-like straight, parallel strand fibres (Fig. 2c). In contrast, when we performed similar docking simulations centred on residues other than 22V, nearly all were incompatible or much less compatible with fibre formation (Fig. 2d). Out of all 145 β-globin residues, only 19N, which forms a contiguous surface with 22V, has a higher propensity to form HbS-like fibres. By contrast, when docking is carried out using the deoxy β-globin structure, 22V is incompatible with fibre formation, consistent with the observation that sickling in deer occurs under oxygenated conditions. Importantly, when this methodology is applied to human HbS, we find that 6V has the highest fibre formation propensity out of all residues under deoxy conditions (Supplementary Figure 5), providing validation for the approach.
Next, we used a force field model to compare the energetics of fibre formation across deer species. We find that known non-sickling species (Fig. 1) and two species suspected to be non-sickling based on their β-globin primary sequence – Chinese water deer (Hydropotes inermis) and roe deer (Capreolus capreolus) – exhibit energy terms less favourable to fibre formation than sickling species (Fig. 2e). To elucidate the relative contribution of 22V and other residues to fibre formation, we introduced all single amino acid differences found amongst adult deer β-globins individually into a sickling (O. virginianus) and non-sickling (R. tarandus) background in silico and considered the change in fibre interaction energy. Changes at residue 22 have the strongest predicted effect on fibre formation, along with two residues – 19 and 21 – in its immediate vicinity (Supplementary Figure 6). Smaller effects of amino acid substitutions at residue 87, as well as residues 117 (N in P. puda and O. virginianus) and 118 (Y in D. dama) hint at species-specific modulation of sickling propensity. In silico residue swaps at a shorter evolutionary time-scale, between non-sickling C. canadensis and sickling sika deer (Cervus nippon), similarly implicate 22V as a key determinant of sickling (Supplementary Figure 6).
Taken together, the results support the formation of HbS-like fibres in sickling deer erythrocytes via surface interactions centred on residues 22V and 87Q in β-globin molecules of different haemoglobin tetramers. In contrast, previous attempts to model interactions in the deer haemoglobin fibre, based on preliminary crystallographic data for white-tailed deer haemoglobin24,38,39, either incorrectly assumed a hexagonal fibre architecture or proposed different relative orientations and contacts that fail to predict differences between sickling and non-sickling chains.
Evidence for incomplete lineage sorting during the evolution of HBBA
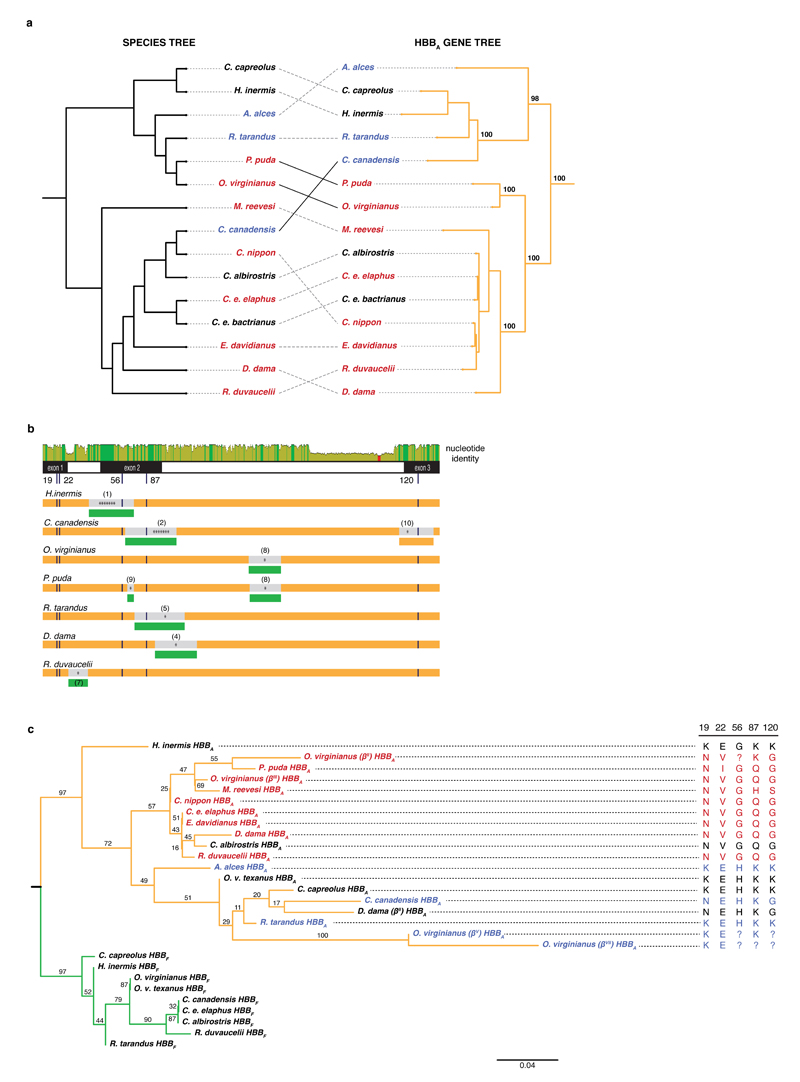
To shed light on the evolutionary history of sickling and elucidate its potential adaptive significance, we considered sickling and non-sickling genotypes in phylogenetic context. First, we note that the HBBA gene tree and the species tree (derived from 20 mitochondrial and nuclear genes) are significantly discordant (Approximately Unbiased test p<1e-61, see Methods). Notably, sickling and non-sickling genotypes are polyphyletic on the species tree but monophyletic on the HBBA tree where wapiti, an Old World deer, clusters with moose and reindeer, two New World deer (Fig. 3a). Gene tree-species tree discordance can result from a number of evolutionary processes, including incomplete lineage sorting, gene conversion, introgression, and classic convergent evolution, where point mutations arise and fix independently in different lineages. In our case, the convergent evolution scenario fits the data poorly. Discordant amino acid states are found throughout the HBBA sequence and are not limited to sickling-related residues. Furthermore, in many instances, amino acids shared between phylogenetically distant species are encoded by the same underlying codons. Conspicuously, this includes the case of residue 120 where all three codon positions differ between sickling species (GGT/AGT) and non-sickling relatives (AAG in reindeer, moose, and cattle; Supplementary Figure 7, Supplementary Data File 1). Even if convergence were driven by selection on a narrow adaptive path through genotype space, precise coincidence of mutational paths at multiple non-synonymous and synonymous sites must be considered unlikely. Rather, these patterns are prima facie consistent with incomplete lineage sorting, a process that might have prominently accompanied the rapid divergence of Old World from New World deer during the Miocene40.
Fig. 3. Evidence for incomplete lineage sorting, gene conversion, and a trans-species polymorphism in the evolutionary history of deer HBBA.
a, Discordances between the maximum likelihood HBBA gene tree and the species tree. Topological differences that violate the principal division into New World deer (NWD, Capreolinae) and Old World deer (OWD, Cervinae) are highlighted by solid black lines. Bootstrap values (% out of 1000 bootstrap replicates) are highlighted for salient nodes. b, Gene conversion and/or introgression. The top panel illustrates nucleotide identity between HBBA and HBBF orthologs (green: 100%, yellow: 30-100%, red: <30% identity). The low-identity segment towards the end of intron 2 marks repeat elements present in all adult but absent from all foetal sequences. Below, predicted recombination events affecting HBBA genes (orange), with either an adult ortholog (orange) or a foetal HBBF paralog (green) as the predicted source, suggestive of introgression or gene conversion, respectively. The number of asterisks indicates how many detection methods (out of a maximum of seven) predicted a given event (see Methods). Details for individual events (numbered in parentheses) are given in Supplementary Figure 8. c, Maximum likelihood protein tree of adult (orange) and foetal (green) β-globin. Alternate non-sickling D. dama (II) and O. virginianus (V, VII) alleles group with non-sickling species (coloured as in Fig. 1). Amino acid identity at key sites is shown on the right. ?: amino acid unresolved in primary source.
Gene conversion affects HBBA evolution but does not explain the phyletic pattern of sickling
To shore up this conclusion and rule out alternative evolutionary scenarios, we next asked whether identical genotypes, rather than originating from a common ancestor, might have been independently reconstituted from genetic diversity present in other species (via introgression) or in other parts of the genome (via gene conversion). To evaluate the likelihood of introgression and particularly gene conversion, which has been attributed a major role in the evolution of mammalian globin genes26, we first searched for evidence of recombination in an alignment of deer HBBA and HBBF genes. HBBF is the principal candidate to donate non-sickling residues to HBBA in a conversion event given that it is itself refractory to sickling19 and – as a recent duplicate of the ancestral HBBA gene – retains high levels of sequence similarity. Using a combination of phylogeny-based and probabilistic detection methods and applying permissive criteria that allow inference of shorter recombinant tracts (see Methods), we identify eight candidate HBBF-to-HBBA events, two of which, in Chinese water deer and wapiti, are strongly supported by different methods (Fig. 3b). Importantly, however, we find no evidence for gene conversion involving residue 22 (Fig. 3b, Supplementary Figure 8) even when considering poorly supported candidate events. Recombination between HBBF and/or HBBA genes therefore does not explain the distribution of glutamic acids and valines at residue 22 across Old World and New World deer. Consistent with this, removal of putative recombinant regions does not affect the HBBF/HBBA gene tree, with wapiti robustly clustered with other non-sickling species whereas white-tailed deer and pudu cluster with Old World sickling species (Supplementary Figure 8). We further screened raw genome sequencing data from white-tailed deer and wapiti for potential donor sequences beyond HBBF, such as HBE or pseudogenized HBD sequences, but did not find additional candidate donors. Thus, although gene conversion is a frequent phenomenon in the history of mammalian globins26 and contributes to HBB evolution in deer, it does not by itself explain the phylogenetic distribution of key sickling/non-sickling residues. Rather, gene conversion introduces additional complexity on a background of incomplete lineage sorting.
Balancing selection has maintained ancestral variation in HBBA
The presence of incomplete lineage sorting and gene conversion confounds straightforward application of rate-based (dN/dS-type) tests for selection, making it harder to establish whether the sickling genotype is simply tolerated or has been under selection. We therefore examined earlier protein-level data on HBBA allelic diversity. This allows us to include additional alleles previously identified from partial peptide digests, for which we have no nucleotide-level data. For white-tailed deer, this includes βII, which is associated with a different flavour of polymerization that results in matchstick-shaped erythrocytes41, and two rarer non-sickling alleles, βV and βVII. βII encodes 22V and expectedly clusters with other sickling HBBA sequences (Fig. 3c). More importantly, the non-sickling white-tailed deer alleles cluster with non-sickling HBBA orthologs rather than with the conspecific βII and βIII alleles (Fig. 3c), as does the HBBA sequence from O. v. texanus, for which we can also demonstrate clustering at the nucleotide level (Supplementary Figure 3). Similarly, an alternate adult β-globin chain previously observed in fallow deer31, a predominantly sickling Old World deer, clusters with non-sickling sequences (Fig. 3c). Finally, phenotypic heterogeneity in wapiti12 and sika deer42 sickling indicates that rare sickling and non-sickling variants, respectively, also segregate in these two species. Taken together, these findings point to the long-term maintenance of ancestral variation through successive speciation events dating back to the most common ancestor of Old World and New World deer, an estimated ~13.6 million years ago (mya) [CI: 9.84-17.33mya]43.
Might this polymorphism have been maintained simply by chance or must balancing selection be evoked to account for its survival? We currently lack information on broader patterns of genetic diversity at deer HBBA loci and surrounding regions that would allow us to search for footprints of balancing selection explicitly. However, we can estimate the probability P that a trans-species polymorphism has been maintained along two independent lineages by neutral processes alone as
where T is the number of generations since the two lineages split and Ne is the effective population size44,45. For simplicity, we assume Ne to be constant over T and the same for both lineages. In the absence of reliable species-wide estimates for Ne, we can nonetheless ask what Ne would be required to meet a given threshold probability. Conservatively assuming an average generation time of 1 year46,47 and a split time of 7.2mya (the lowest divergence time estimate in the literature43), Ne would have to be 2,403,419 to reach a threshold probability of 0.05. Although deer can have large census population sizes, an Ne >2,000,000 for both fallow and white-tailed deer is comfortably outside what we would expect for large-bodied mammals, >4-fold higher than estimates for wild mice48 and >2-fold higher even than estimates for African populations of Drosophila melanogaster49. Consequently, we argue that the HBBA trans-species polymorphism is inconsistent with neutral evolution and instead reflects the action of balancing selection.
A distinct genetic basis for sickling in sheep
While sickling in deer is particularly well-documented, the capacity for reversible haemoglobin polymerization has also been observed in a small coterie of other vertebrates10,50, including some species of fish50, mongoose51, and notably also goat and sheep (Ovis aries)11,52. For most of these species, we have no information on sickling-associated genotypes and allelic diversity. Sheep, where sickling has been found in a variety of domestic breeds52,53, are an exception in this regard. Two HBBA alleles, HbA and HbB, were previously identified54. HbA homozygotes and HbA/HbB heterozygotes sickle whereas HbB homozygotes do not52. We first compared the sheep reference sequence (Texel breed) included in Fig. 1 with partial peptide information for both alleles54 and found it to be fully consistent with the non-sickling HbB allele. We then surveyed amino acid variation at the β-globin gene across 75 breeds of sheep, selected to cover global sheep genetic diversity55. We observed all seven amino acids known to discriminate HbA from HbB but found no variation at residues 6 or 22 (Supplementary Figure 9), suggesting, first, that the genetic diversity panel captures HbA and, second, that HbA, lacking 6V and 22V, promotes polymerization by yet another mechanism. This conclusion is consistent with phenomenological differences in sickling dynamics between deer and sheep, including a) the finding that sickling in the latter only occurs when cells are suspended in hypertonic saline and incubated at 37ºC (REF. 11), making it less likely that sickling frequently takes place in vivo under physiological conditions, and b) the observation that the sickling allele is dominant in sheep but recessive in deer19. Importantly, these results also indicate that sickling evolved independently in deer (Cervidae) and their sister clade (Caprinae).
Discussion
Given the dramatic change in erythrocyte shape brought about by haemoglobin polymerization it is conspicuous that multiple vertebrate lineages have independently converged on this phenotype. In principle, recurrent emergence could be the result of non-adaptive forces. Recent findings suggest that symmetric protein complexes like haemoglobin exist at the edge of supramolecular self-assembly, often being a short mutational distance away from the propensity to form polymers56. However, in deer, the long-term maintenance of a trans-species polymorphism is inconsistent with selective neutrality and instead argues for fitness effects along multiple lineages. By direct implication, even though sickling is remarkably well tolerated in vivo, perhaps owing to unique properties of deer erythrocytes (Supplementary Discussion), it cannot be perfectly innocuous23. Rather, it must exert a physiological effect that is strong and frequent enough to be targeted by selection.
What are the ecological driving factors behind the maintenance of sickling (and non-sickling) alleles over evolutionary time? It has previously been suggested that haemoglobin polymerization might, by radically altering the intracellular environment of red blood cells, provide a generic defense mechanism against red blood cell parasites50. Deer certainly harbour a number of intra-erythrocytic parasites, including Babesia57 and Plasmodium58,59. The latter was recently found to be widespread in white-tailed deer, but, interestingly, associated with very low levels of parasitaemia59. Also worth noting in this regard is the marked geographic asymmetry in sickling status, where established non-sickling species are restricted to arctic and subarctic (elk, reindeer) or mountainous (wapiti) habitat. Might this indicate that the sickling allele loses its adaptive value in colder climates, perhaps linked to the lower prevalence of blood-born parasites? Although a general cross-species link between sickling and parasite burden is tantalizing, it is important to highlight that there is currently no concrete evidence for such a connection and alternative hypotheses should be considered. For example, with no evidence for heterozygote advantage, might it be that allelic diversity has been maintained by migration-selection balance? Or do the timescales involved render such a scenario improbable? Exploring geographic structure in the distribution of sickling and non-sickling alleles will be important in this regard and might point to the ecological factors involved in maintaining either allele. More generally, future epidemiological studies coupled to population genetic investigations will be required to unravel the evolutionary ecology of sickling in deer and establish whether parasites are indeed ecological drivers of between- and within-species differences in HBBA genotype. Ultimately, such analyses will determine whether deer constitute a useful comparative system to elucidate the link between sickling and protection from the effects of Plasmodium infection, which remains poorly understood in humans.
Methods
Sample collection and processing
Blood, muscle tissue, and DNA samples were acquired for 15 species of deer from a range of sources (Supplementary Table 1).
The white-tailed deer blood sample was heat-treated on import to the United Kingdom in accordance with import standards for ungulate samples from non-EU countries (IMP/GEN/2010/07). Fresh blood was collected into PAXgene Blood DNA tubes (PreAnalytix) and DNA extracted using the PAXgene Blood DNA kit (PreAnalytix). DNA from previously frozen blood samples was extracted using the QIAamp DNA Blood Mini kit (Qiagen). DNA from tissue samples was extracted with the QIAamp DNA Mini kit (Qiagen) using 25mg of tissue. Total RNA was isolated from an E. davidianus blood sample using the PAXgene Blood RNA kit (PreAnalytix) three days after collection into a PAXgene Blood RNA tube (PreAnalytix). All extractions were performed according to manufacturers’ protocols. For each sample, we validated species identity by amplifying and sequencing the cytochrome b (CytB) gene. With the exception of Cervus albirostris, we successfully amplified CytB from all samples using primers MTCB_F/R (Supplementary Figure 2) and conditions as described in REF. 60. Phusion High-Fidelity PCR Master Mix (ThermoFisher) was used for all amplifications. PCR products were purified using the MinElute PCR Purification Kit (Qiagen) and Sanger-sequenced with the amplification primers. The CytB sequences obtained were compared to all available deer CytB sequences in the 10kTrees Project61 using the ape package (function dist.dna with default arguments) in R62. In all cases, the presumed species identity of the sample was confirmed (Supplementary Table 2).
Whole genome sequencing
O. virginianus genomic DNA was prepared for sequencing using the NEB DNA library prep kit (New England Biolabs) and sequenced on the Illumina HiSeq platform. The resulting 229 million 100bp paired-end reads were filtered for adapters and quality using Trimmomatic63 with the following parameters: ILLUMINACLIP:adapters/TruSeq3-PE-2.fa:2:30:10 LEADING:30 TRAILING:30 SLIDINGWINDOW:4:30 MINLEN:50. Inspection of the remaining 163.5M read pairs with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) suggested that overrepresented sequences had been successfully removed.
Mapping and partial assembly of the O. virginianus β-globin locus
To seed a local assembly of the O. virginianus β-globin locus we first mapped O. virginianus trimmed paired-end reads to the duplicated β-globin locus in the hard-masked B. taurus genome (UMD 3.1.1; chr15: 48973631-49098735). The β-globin locus is defined here as the region including all B. taurus β-globin genes [HBE1, HBE4, HBBA (ENSBTAG00000038748), HBE2, HBBF (ENSBTAG00000037644)], the intervening sequences and 24kb either side of the two outer β-globins (HBE1, HBBF). The mapping was performed using bowtie2 (REF. 64) with default settings and the optional --no-mixed and --no-discordant parameters. 110 reads mapped without gaps and a maximum of one nucleotide mismatch. These reads, broadly dispersed across the B. taurus β-globin locus (Supplementary Figure 10), were used as seeds for local assembly using a customised aTRAM65 pipeline (see below). Prior to assembly, the remainder of the reads were filtered for repeat sequences by mapping against Cetartiodactyla repeats in Repbase66. The aTRAM.pl wrapper script was modified to accept two new arguments: max_target_seqs <int> limited the number of reads found by BLAST from each database shard; cov_cutoff <int> passed a minimum coverage cut-off to the underlying Velvet 1.2.10 assembler67. The former modification prevents stalling when the assembly encounters a repeat region, the latter discards low coverage contigs at the assembler level. aTRAM was run with the following arguments: -kmer 31 -max_target_seqs 2000 -ins_length 270 -exp_coverage 8 -cov_cutoff 2 -iterations 5. After local assembly on each of the 110 seed reads, the resulting contigs were combined using Minimo68 with a required minimum nucleotide identity of 99%. To focus specifically on assembling the adult β-globin gene, only contigs that mapped against the B. taurus adult β-globin gene ±500bp (chr15: 49022500-49025000) were retained and served as seeds for another round of assembly. This procedure was repeated twice. The final 59 contigs were compared to the UMD 3.1.1 genome using BLAT and mapped exclusively to either the adult or foetal B. taurus β-globin gene. From the BLAT alignment, we identified short sequences that were perfectly conserved between the assembled deer contigs and the B. taurus as well as the sheep assembly (Oar_v3.1). Initial forward and reverse primers (Ovirg_F1/Ovirg_R1, Supplementary Figure 2) for β-globin amplification were designed from these conserved regions located 270bp upstream (chr15:49022762-49022786) and 170bp downstream (chr15:49024637-49024661) of the B. taurus adult β-globin gene, respectively. Our local assembly is consistent with a recent draft genome assembly (https://www.ncbi.nlm.nih.gov/assembly/GCF_002102435.1/) from a white-tailed deer from Texas (O. virginianus texanus).
Globin gene amplification and sequencing
Amplification of β-globin from O. virginianus using primers Ovirg_F1 and Ovirg_R1 yielded two products of different molecular weights (~2000bp and ~1700bp; Supplementary Figure 2), which were isolated by gel extraction and Sanger-sequenced using the amplification primers. The high molecular weight product had higher nucleotide identity to the adult (93%) than to the foetal (90%) B. taurus β-globin coding sequence. Note that the discrepancy in size between the adult and foetal β-globin amplicons derives from the presence of two tandem Bov-tA2 SINEs in intron 2 of the adult β-globin gene in cattle, sheep, and O. virginianus and is therefore likely ancestral. We designed a second set of primers to anneal immediately up- and downstream, and in the middle of the adult β-globin gene (Ovirg_F2, Ovirg_R2, Ovirg_Fmid2, Supplementary Figure 2). Amplification from DNA extracts of other species with Ovirg_F1/Ovirg_R1 produced mixed results, with some species showing a two-band pattern similar to O. virginianus, others only a single band – corresponding to the putative adult β-globin (Supplementary Figure 2). Using these primers, no product could be amplified from R. tarandus, H. inermis, and C. capreolus. We identified a 3bp mismatch to the Ovirg_R1 primer in a partial assembly of C. capreolus (Genbank accession: GCA_000751575.1; scaffold: CCMK010226507.1) that is likely at fault. A re-designed reverse primer (Ccap_R1) successfully amplified the adult β-globin gene from the three deer species above as well as C. canadensis (Supplementary Figure 2). All amplifications were performed using Phusion High-Fidelity PCR Master Mix (ThermoFisher), with primers as listed in Supplementary Figure 2, and 50-100ng of genomic DNA. Annealing temperature and step timing were chosen according to manufacturer guidelines. Amplifications were run for 35 cycles. Gel extractions were performed on samples resolved on 1% agarose gels for 40 minutes at 90V using the MinElute Gel Extraction Kit (Qiagen) and following the manufacturer’s protocol. PCR purifications were performed using the MinElute PCR Purification Kit (Qiagen) following the manufacturer’s protocol. All samples were sequenced using the Sanger method with amplification primers and primer Ovirg_Fmid2.
Transcriptome sequencing and assembly
RNA was extracted from the red cell component of a blood sample of an adult Père David’s deer using the PAXgene Blood RNA kit (Qiagen). An mRNA library was prepared using a Truseq mRNA library prep kit and sequenced on the MiSeq platform, yielding 25,406,472 paired-end reads of length 150bp, which were trimmed for adapters and quality-filtered using Trim Galore! (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with a base quality threshold of 30. The trimmed reads were used as input for de novo transcriptome assembly with Trinity69 using default parameters. A blastn homology search against these transcripts, using the O. virginianus adult β-globin CDS as query, identified a highly homologous transcript (E-value = 0; no gaps; 97.5% sequence identity compared with 92.2% identity to the foetal β-globin). The CDS of this putative β-globin transcript was 100% identical to the sequence amplified from Père David’s deer genomic DNA (Supplementary Figure 4). We used emsar70 with default parameters to assess transcript abundances. The three most abundant reconstructed transcripts correspond to full or partial α- and β-globin transcripts, including one transcript, highlighted above, that encompasses the entire adult β-globin CDS. These transcripts are an order of magnitude more abundant than the fourth most abundant (Supplementary Figure 4), in line with the expected predominance of α- and β-globin transcripts in mature adult red blood cells. To investigate whether the foetal β-globin could be detected in the RNA-seq data and because amplification of the foetal β-globin from Père David’s deer genomic DNA was not successful, we mapped reads against the foetal β-globin gene of C. e. elaphus, the closest available relative. Given that the CDS of the adult β-globins in these species are 100% identical, we expected that the foetal orthologs would likewise be highly conserved. We therefore removed reads with more than one mismatch and assembled putative transcripts from the remaining 1.3M reads using the Geneious assembler v.10.0.5 (REF. 71) with default parameters (fastest option enabled). We recovered a single contig with high homology to the C. e. elaphus foetal β-globin CDS (only a single mismatch across the CDS). We then estimated the relative abundance of adult and the putative foetal transcripts by calculating the proportion of reads that uniquely mapped to either the adult or foetal CDS. 1820532 reads mapped uniquely to the adult sequence whereas 872 mapped uniquely to the foetal CDS, a ratio of 2088:1.
Structural analysis
Homology models were built for O. virginianus and R. tarandus β-globin sequences using the MODELLER-9v15 program for comparative protein structure modelling72 using both oxy (1HHO) and deoxy (2HHB) human haemoglobin structures as templates. The structures were used for electrostatic calculations using the Adaptive Poisson-Boltzmann Solver73 plug-in in the Visual Molecular Dynamics (VMD) program74. The surface potentials were visualised in VMD with the conventional red and blue colours, for negative and positive potential respectively, set at ±5 kT/e.
Modelling of haemoglobin fibres
We first used the program HADDOCK75 with the standard protein-protein docking protocol to generate ensembles of docking models of β-globin dimers. In each docking run, a different interacting surface centred around a specific residue was defined on each β-globin chain. All residues within 3Å of the central residue were defined as “active” and were thus constrained to be directly involved in the interface, while other residues within 8Å of the central residue were defined as “passive” and were allowed but not strictly constrained to form a part of the interface. We performed docking runs with the interaction centred between residue 87 and all other residues, generating at least 100 water-refined β-globin dimer models for each (although 600 O. virginianus oxy β-globin 22V-87Q models were built for use in the interaction energy calculations). The β-globin dimers were then evaluated for their ability to form HbS-like fibres out of full haemoglobin tetramers. Essentially, the contacts from the β-globin dimer models were used to build a chain of five haemoglobin molecules, in the same way that the contacts between 6V and the EF pocket lead to an extended fibre in HbS. HbS-like fibres were defined as those in which a direct contact was formed between the first and third haemoglobin tetramers in a chain (analogous to the axial contacts in HbS fibres, see Fig. 2c), and in which the chain is approximately linear. This linearity was measured as the distance between the first and third plus the distance between the third and the fifth haemoglobin tetramers, divided by the distance between the first and the fifth. A value of 1 would indicate a perfectly linear fibre, while we considered any chains with a value <1.05 to be approximately linear and HbS-like. Finally, chains containing significant steric clashes between haemoglobin tetramers (defined as >3% of Cα atoms being within 2.8Å of another Cα atom) were excluded. Fibre formation propensity was then defined as the fraction of all docking models that led to HbS-like fibres.
Interaction energy analysis
Using the 270 22V-87Q models of O. virginianus β-globin dimers that can form HbS-like fibres, we used FoldX76 and the ‘RepairPDB’ and ‘BuildModel’ functions to mutate each dimer to the sequences of all other adult deer species. Note that since C. e. elaphus, C. e. bactrianus and E. davidianus have identical amino acid sequence, only one of these was included here. The energy of the interaction was then calculated using the ‘AnalyseComplex’ function of FoldX, and then averaged over all docking models. The same protocol was then used for the analysis of the effects of individual mutations, using all possible single amino acid substitutions observed in the adult deer sequences, except that the interaction energy was presented as the change with respect to the wild-type sequence.
Deer species tree and wider mammalian phylogeny
The mammalian phylogeny depicted in Fig. 1 is principally based on the Timetree of Life43 with the order Carnivora regrafted to branch above the root of the Chiroptera and Artiodactyla to match findings in77. The internal topology of Cervidae was taken from the Cetartiodactyla consensus tree of the 10kTrees Project61. C. canadensis and Cervus elaphus bactrianus, not included in the 10kTrees phylogeny, were added as sister branches to C. nippon and C. e. elaphus, respectively, following REF. 78. Supplementary Figure 11 provides a graphical overview of these changes. To generate Fig. 1, we aligned adult deer β-globin coding sequences to a set of non-chimeric mammalian adult β-globin CDSs26.
Gene tree reconstruction
All trees were built using RAxML v8.2.10 based on alignments made with MUSCLE v3.8.1551. Unless stated otherwise, we used the RAxML joint maximum likelihood and bootstrap analysis (option –f a) with random seeds, a single partition, and 100 bootstrap replicates. The GTRGAMMA model was used for nucleotide alignments and the best fitting protein model was automatically chosen by RAxML using the PROTGAMMAAUTO option.
As some historical alleles (βII, βV, βVII) are only available at the peptide level, Fig. 3c was built at the protein level. HBBA (±HBBF) trees (Fig. 3a, Supplementary Figures 2&6), on the other hand, are nucleotide-level trees build from an alignment of coding exons and intervening introns. Note here, that intron 2, which is comparatively long and less constrained than coding sequence, contributes a comparatively large number of phylogenetically informative sites. In fact, the intron 2 tree re-capitulates the exon+intron tree almost perfectly, with a minor difference in the precise location of C. canadensis in the non-sickling cluster. Note further, that a large comparative contribution of intron 2 to the overall phylogenetic signal is fortuitous in this context. In order to understand patterns of lineage sorting and introgression, it is desirable to eliminate spurious phylogenetic signals introduced by gene conversion, which strongly affects exonic sequence (as evident in Fig. 3b) but is much less prevalent in intron 2. Since we explicitly demonstrate (in Supplementary Figure 6) that including sequence affected by gene conversion does not affect the overall tree topology, we present exon+intron (i.e. gene) trees throughout for simplicity.
Topology testing
To test for significant phylogenetic discordance between the HBBA gene tree and the species tree as depicted in Fig. 3a we compared both topologies using the Approximately Unbiased (AU) test79 implemented in CONSEL80. The unconstrained maximum likelihood (ML) HBBA gene tree was tested against an alternative ML tree (derived from 200 maximum likelihood starting trees) built under a single constraint: to recover the well-established monophyletic groups of Old World and New World deer. Branching patterns within these major clades were allowed to vary. With this approach, we conservatively test the significance of the incongruent placement of O. virginianus and P. pudu sickling alleles with the Old World deer (and C. canadensis with New World deer) without considering confounding signals from within-clade branching that might arise, for example, due to gene conversion. Both the constrained and unconstrained ML trees were calculated with RAxML as described above. Per site log-likelihoods were computed for the unconstrained and constrained ML trees with RAxML (option –f G).
Detection of recombination events
We considered two sources of donor sequence for recombination into adult β-globins: adult β-globin orthologs in other deer species and the foetal β-globin paralog within the same genome. H. inermis HBBF was omitted from this analysis since the sequence of intron 2 was only partially determined. We used the Recombination Detection Program (RDP v.4.83)81 to test for signals of recombination in an alignment of complete adult and foetal deer β-globin genes that were successfully amplified and sequenced, enabling all subtended detection methods (including primary scans for BootScan and SiScan) except LARD, treating the sequences as linear and listing all detectable events. In humans, conversion tracts of lengths as short as 110bp have been detected in the globin genes82 and tracts as short as 50bp in other gene conversion hotspots83,84. Given the presence of multiple regions of 100% nucleotide identity across the alignment of adult and foetal deer β-globins (Fig. 3b), we suspected that equally short conversion tracts might also be present. We therefore lowered window and step sizes for all applicable detection methods in RDP (Supplementary Figure 8) at the cost of a lower signal-to-noise ratio. As the objective is to test whether recombination events could have generated the phyletic distribution of sickling/non-sickling genotypes observed empirically, this is conservative.
Data availability
HBBA and HBBF full gene sequences (coding sequence plus intervening introns) have been submitted to GenBank with accession numbers KY800429-KY800452. An alignment of these sequences is also available as Supplementary Data. Père David’s deer RNA sequencing and white-tailed deer whole genome sequencing raw data has been submitted to the European Nucleotide Archive (ENA) with the accession numbers PRJEB20046 and PRJEB20034, respectively.
Supplementary Material
Acknowledgments
We thank the Zoological Society of London Whipsnade Zoo (F. Molenaar), Bristol Zoological Society (S. Dow, K. Wyatt), the Royal Zoological Society of Scotland Highland Wildlife Park (J. Morse), the Penn State Deer Research Center (D. Wagner), and the Northeast Wildlife DNA Laboratory (N. Chinnici) for samples, the MRC LMS Genomics Facility for DNA and RNA sequencing, B.N. Sacks, J. Mizzi, and T. Brown for access to Tule elk sequencing data, P.D. Butcher for discussions, and P. Sarkies, A. Brown, and B. Lehner for comments on the manuscript. This work was supported by an Imperial College Interdisciplinary Cross-Campus Studentship to A.E, an MRC Career Development Award (MR/M02122X/1) to J.A.M., a Leverhulme Trust Fellowship to V.S., and MRC core funding and an Imperial College Junior Research Fellowship to T.W.
Footnotes
Author contributions
A.E. performed laboratory experiments and evolutionary analyses and contributed to experimental design, data analysis and interpretation. L.T.B. and J.A.M. designed and performed structural modelling, and contributed to data analysis and interpretation. V.S. contributed tissue samples. T.W. conceived the study, contributed to experimental design, data analysis, and interpretation and wrote the manuscript with input from all authors.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Ingram VM. Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature. 1957;180:326–328. doi: 10.1038/180326a0. [DOI] [PubMed] [Google Scholar]
- 2.Harrington DJ, Adachi K, Royer WE., Jr The high resolution crystal structure of deoxyhemoglobin S. Journal of Molecular Biology. 1997;272:398–407. doi: 10.1006/jmbi.1997.1253. [DOI] [PubMed] [Google Scholar]
- 3.Wishner BC, Ward KB, Lattman EE, Love WE. Crystal structure of sickle-cell deoxyhemoglobin at 5 Å resolution. Journal of Molecular Biology. 1975;98:179–194. doi: 10.1016/s0022-2836(75)80108-2. [DOI] [PubMed] [Google Scholar]
- 4.Sears DA. The morbidity of sickle cell trait. The American Journal of Medicine. 1978;64:1021–1036. doi: 10.1016/0002-9343(78)90458-8. [DOI] [PubMed] [Google Scholar]
- 5.Platt OS, et al. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med. 1994;330:1639–1644. doi: 10.1056/NEJM199406093302303. [DOI] [PubMed] [Google Scholar]
- 6.Piel FB, et al. Global distribution of the sickle cell gene and geographical confirmation of the malaria hypothesis. Nature Communications. 2010;1:104. doi: 10.1038/ncomms1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrick JB. Peculiar elongated and sickle-shaped red blood corpuscles in a case of severe anemia. Arch Int Med. 1910;5:517. [PMC free article] [PubMed] [Google Scholar]
- 8.Gulliver G. Observations on certain peculiarities of form in the blood corpuscles of the mammiferous animals. Lond Edinb Dubl Phil Mag. 1840;17:325–327. [Google Scholar]
- 9.Undritz E, Betke K, Lehmann H. Sickling phenomenon in deer. Nature. 1960;187:333–334. doi: 10.1038/187333a0. [DOI] [PubMed] [Google Scholar]
- 10.Hawkey CM. Comparative Mammalian Haematology. Heinemann Educational Books; 1975. [Google Scholar]
- 11.Butcher PD, Hawkey CM. Haemoglobins and erythrocyte sickling in the artiodactyla: A survey. Comparative Biochemistry and Physiology Part A: Physiology. 1977;57:391–398. doi: 10.1016/0305-0491(77)90025-6. [DOI] [PubMed] [Google Scholar]
- 12.Weber YB, Giacometti L. Sickling Phenomenon in the Erythrocytes of Wapiti (Cervus Canadensis) Journal of Mammalogy. 1972;53:917–919. [PubMed] [Google Scholar]
- 13.Simpson CF, Taylor WJ. Ultrastructure of sickled deer erythrocytes. I. The typical crescent and holly leaf forms. Blood. 1974;43:899–906. [PubMed] [Google Scholar]
- 14.Schmidt WC, et al. The structure of sickling deer type III hemoglobin by molecular replacement. Acta Crystallogr Sect B Struct Crystallogr Cryst Chem. 1977;33:335–343. [Google Scholar]
- 15.Pritchard WR, Malewitz TD, Kitchen H. Studies on the mechanism of sickling of deer erythrocytes. Experimental and Molecular Pathology. 1963;2:173–182. doi: 10.1016/0014-4800(63)90050-9. [DOI] [PubMed] [Google Scholar]
- 16.Kitchen H, Easley CW, Putnam FW, Taylor WJ. Structural comparison of polymorphic hemoglobins of deer with those of sheep and other species. The Journal of Biological Chemistry. 1968;243:1204–1211. [PubMed] [Google Scholar]
- 17.Seiffge D. Haemorheological studies of the sickle cell phenomenon in european red deer (Cervus elaphus) Blut. 1983;47:85–92. doi: 10.1007/BF02482642. [DOI] [PubMed] [Google Scholar]
- 18.Kitchen H, Putnam FW, Taylor WJ. Hemoglobin Polymorphism: Its Relation to Sickling of Erythrocytes in White-Tailed Deer. Science. 1964;144:1237–1239. doi: 10.1126/science.144.3623.1237. [DOI] [PubMed] [Google Scholar]
- 19.Taylor WJ, Easley CW. Sickling phenomena of deer. Annals of the New York Academy of Sciences. 1974;241:594–604. doi: 10.1111/j.1749-6632.1974.tb21916.x. [DOI] [PubMed] [Google Scholar]
- 20.Harris MJ, Huisman THJ, Hayes FA. Geographic distribution of hemoglobin variants in the white-tailed deer. Journal of Mammalogy. 1973;54:270–274. [PubMed] [Google Scholar]
- 21.Harris MJ, Wilson JB, Huisman THJ. Structural studies of hemoglobin α chains from Virginia white-tailed deer. Archives of Biochemistry and Biophysics. 1972;151:540–548. doi: 10.1016/0003-9861(72)90531-0. [DOI] [PubMed] [Google Scholar]
- 22.Parshall CJ, Vainisi SJ, Goldberg MF, Wolf ED. In vivo erythrocyte sickling in the Japanese sika deer (Cervus nippon): methodology. Am J Vet Res. 1975;36:749–752. [PubMed] [Google Scholar]
- 23.Whitten CF. Innocuous Nature of the Sickling (Pseudosickling) Phenomenon in Deer. British Journal of Haematology. 1967;13:650–655. doi: 10.1111/j.1365-2141.1967.tb08830.x. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu K, et al. The primary sequence of the beta chain of Hb type III of the Virginia white-tailed deer (Odocoilus Virginianus), a comparison with putative sequences of the beta chains from four additional deer hemoglobins, types II, IV, V, and VIII, and relationships between intermolecular contacts, primary sequence and sickling of deer hemoglobins. Hemoglobin. 1983;7:15–45. doi: 10.3109/03630268309038399. [DOI] [PubMed] [Google Scholar]
- 25.Kitchen H, Taylor WJ. The sickling phenomenon of deer erythrocytes. Adv Exp Med Biol. 1972;28:325–336. doi: 10.1007/978-1-4684-3222-0_26. [DOI] [PubMed] [Google Scholar]
- 26.Gaudry MJ, Storz JF, Butts GT, Campbell KL, Hoffmann FG. Repeated evolution of chimeric fusion genes in the β-globin gene family of laurasiatherian mammals. Genome Biol Evol. 2014;6:1219–1234. doi: 10.1093/gbe/evu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardison RC. Evolution of Hemoglobin and Its Genes. Cold Spring Harb Perspect Med. 2012;2:a011627–a011627. doi: 10.1101/cshperspect.a011627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Townes TM, Fitzgerald MC, Lingrel JB. Triplication of a four-gene set during evolution of the goat beta-globin locus produced three genes now expressed differentially during development. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:6589–6593. doi: 10.1073/pnas.81.21.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schimenti JC, Duncan CH. Structure and organization of the bovine beta-globin genes. Mol Biol Evol. 1985;2:514–525. doi: 10.1093/oxfordjournals.molbev.a040369. [DOI] [PubMed] [Google Scholar]
- 30.Craig JE, Thein SL, Rochette J. Fetal hemoglobin levels in adults. Blood Reviews. 1994;8:213–224. doi: 10.1016/0268-960x(94)90109-0. [DOI] [PubMed] [Google Scholar]
- 31.Angeletti M, et al. Different functional modulation by heterotropic ligands (2,3-diphosphoglycerate and chlorides) of the two haemoglobins from fallow-deer (Dama dama) Eur J Biochem. 2001;268:603–611. doi: 10.1046/j.1432-1327.2001.01909.x. [DOI] [PubMed] [Google Scholar]
- 32.Petruzzelli R, et al. The primary structure of hemoglobin from reindeer (Rangifer tarandus tarandus) and its functional implications. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1991;1076:221–224. doi: 10.1016/0167-4838(91)90270-a. [DOI] [PubMed] [Google Scholar]
- 33.Adachi K, Reddy LR, Surrey S. Role of hydrophobicity of phenylalanine beta 85 and leucine beta 88 in the acceptor pocket for valine beta 6 during hemoglobin S polymerization. The Journal of Biological Chemistry. 1994;269:31563–31566. [PubMed] [Google Scholar]
- 34.Nagel RL, et al. Beta-chain contact sites in the haemoglobin S polymer. Nature. 1980;283:832–834. doi: 10.1038/283832a0. [DOI] [PubMed] [Google Scholar]
- 35.Adachi K, Konitzer P, Surrey S. Role of gamma 87 Gln in the inhibition of hemoglobin S polymerization by hemoglobin F. The Journal of Biological Chemistry. 1994;269:9562–9567. [PubMed] [Google Scholar]
- 36.Witkowska HE, et al. Sickle cell disease in a patient with sickle cell trait and compound heterozygosity for hemoglobin S and hemoglobin Quebec-Chori. N Engl J Med. 1991;325:1150–1154. doi: 10.1056/NEJM199110173251607. [DOI] [PubMed] [Google Scholar]
- 37.Watson-Williams EJ, Beale D, Irvine D, Lehmann H. A new haemoglobin, D Ibadan (beta-87 threonine -- lysine), producing no sickle-cell haemoglobin D disease with haemoglobin S. Nature. 1965;205:1273–1276. doi: 10.1038/2051273a0. [DOI] [PubMed] [Google Scholar]
- 38.Amma EL, Sproul GD, Wong S, Huisman THJ. Mechanism of sickling in deer erythrocytes. Annals of the New York Academy of Sciences. 1974;241:605–613. doi: 10.1111/j.1749-6632.1974.tb21917.x. [DOI] [PubMed] [Google Scholar]
- 39.Girling RL, Schmidt WC, Jr, Houston TE, Amma EL, Huisman THJ. Molecular packing and intermolecular contacts of sickling deer type III hemoglobin. Journal of Molecular Biology. 1979;131:417–433. doi: 10.1016/0022-2836(79)90001-9. [DOI] [PubMed] [Google Scholar]
- 40.Fernández MH, Vrba ES. A complete estimate of the phylogenetic relationships in Ruminantia: a dated species-level supertree of the extant ruminants. Biological Reviews. 2005;80:269–302. doi: 10.1017/s1464793104006670. [DOI] [PubMed] [Google Scholar]
- 41.Taylor WJ, Simpson CF. Ultrastructure of sickled deer erythrocytes. II. The matchstick cell. Blood. 1974;43:907–914. [PubMed] [Google Scholar]
- 42.Butcher PD, Hawkey CM. Red blood cell sickling in mammals. In: Montali RJ, Migaki G, editors. The Comparative Pathology of Zoo Animals. Smithsonian Institute; 1980. [Google Scholar]
- 43.Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. Tree of Life Reveals Clock-Like Speciation and Diversification. Mol Biol Evol. 2015;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiuf C, Zhao K, Innan H, Nordborg M. The Probability and Chromosomal Extent of trans-specific Polymorphism. Genetics. 2004;168:2363–2372. doi: 10.1534/genetics.104.029488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao Z, Przeworski M, Sella G. Footprints of ancient-balanced polymorphisms in genetic variation data from closely related species. Evolution. 2015;69:431–446. doi: 10.1111/evo.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baker KH, et al. Strong population structure in a species manipulated by humans since the Neolithic: the European fallow deer (Dama dama dama) Heredity. 2017;119:16–26. doi: 10.1038/hdy.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryman N, Baccus R, Reuterwall C, Smith MH. Effective Population Size, Generation Interval, and Potential Loss of Genetic Variability in Game Species under Different Hunting Regimes. Oikos. 1981;36:257. [Google Scholar]
- 48.Halligan DL, Oliver F, Eyre-Walker A, Harr B, Keightley PD. Evidence for Pervasive Adaptive Protein Evolution in Wild Mice. PLoS Genet. 2010;6:e1000825. doi: 10.1371/journal.pgen.1000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shapiro JA, et al. Adaptive genic evolution in the Drosophila genomes. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2271–2276. doi: 10.1073/pnas.0610385104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koldkjær P, McDonald MD, Prior I, Berenbrink M. Pronounced in vivo hemoglobin polymerization in red blood cells of Gulf toadfish: a general role for hemoglobin aggregation in vertebrate hemoparasite defense? American Journal of Physiology - Regulatory, Integrative and Comparative Physiology. 2013;305:R1190–R1199. doi: 10.1152/ajpregu.00246.2013. [DOI] [PubMed] [Google Scholar]
- 51.Hawkey CM, Jordan P. Sickle-cell erythrocytes in the mongoose Herpestes sanguineus. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1967;61:180–181. doi: 10.1016/0035-9203(67)90154-x. [DOI] [PubMed] [Google Scholar]
- 52.Butcher PD, Hawkey CM. The nature of erythrocyte sickling in sheep. Comparative Biochemistry and Physiology Part A: Physiology. 1979;64:411–418. [Google Scholar]
- 53.Evans ETR. Sickling Phenomenon in Sheep. Nature. 1968;217:74–75. doi: 10.1038/217074a0. [DOI] [PubMed] [Google Scholar]
- 54.Tucker EM. Genetic variation in the sheep red blood cell. Biol Rev Camb Philos Soc. 1971;46:341–386. doi: 10.1111/j.1469-185x.1971.tb01049.x. [DOI] [PubMed] [Google Scholar]
- 55.Kijas JW, et al. Genome-wide analysis of the world's sheep breeds reveals high levels of historic mixture and strong recent selection. Plos Biol. 2012;10:e1001258. doi: 10.1371/journal.pbio.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garcia-Seisdedos H, Empereur-Mot C, Elad N, Levy ED. Proteins evolve on the edge of supramolecular self-assembly. Nature. 2017;365:1596. doi: 10.1038/nature23320. [DOI] [PubMed] [Google Scholar]
- 57.Perry BD, Nichols DK, Cullom ES. Babesia odocoilei Emerson and Wright, 1970 in white-tailed deer, Odocoileus virginianus (Zimmermann), in Virginia. Journal of Wildlife Diseases. 1985;21:149–152. doi: 10.7589/0090-3558-21.2.149. [DOI] [PubMed] [Google Scholar]
- 58.Garnham PC, Kuttler KL. A malaria parasite of the white-tailed deer (Odocoileus virginianus) and its relation with known species of Plasmodium in other ungulates. Proc R Soc Lond, B Biol Sci. 1980;206:395–402. doi: 10.1098/rspb.1980.0003. [DOI] [PubMed] [Google Scholar]
- 59.Martinsen ES, et al. Hidden in plain sight: Cryptic and endemic malaria parasites in North American white-tailed deer (Odocoileus virginianus) Science Advances. 2016;2:e1501486. doi: 10.1126/sciadv.1501486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naidu A, Fitak RR, Munguia Vega A, Culver M. Novel primers for complete mitochondrial cytochrome b gene sequencing in mammals. Molecular Ecology Resources. 2012;12:191–196. doi: 10.1111/j.1755-0998.2011.03078.x. [DOI] [PubMed] [Google Scholar]
- 61.Arnold C, Matthews LJ, Nunn CL. The 10kTrees website: A new online resource for primate phylogeny. Evol Anthropol. 2010;19:114–118. [Google Scholar]
- 62.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 63.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langmead Ben, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Meth. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Allen JM, Huang DI, Cronk QC, Johnson KP. aTRAM - automated target restricted assembly method: a fast method for assembling loci across divergent taxa from next-generation sequencing data. BMC Bioinformatics. 2015;16:98. doi: 10.1186/s12859-015-0515-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA 2014 5:1. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Treangen TJ, Sommer DD, Angly FE, Koren S, Pop M. Next generation sequence assembly with AMOS. Curr Protoc Bioinformatics. 2011;Chapter 11 doi: 10.1002/0471250953.bi1108s33. Unit 11.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee S, et al. EMSAR: estimation of transcript abundance from RNA-seq data by mappability-based segmentation and reclustering. BMC Bioinformatics. 2015;16:278. doi: 10.1186/s12859-015-0704-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eswar N, et al. Comparative Protein Structure Modeling Using Modeller. Curr Protoc Bioinformatics. 2006 doi: 10.1002/0471250953.bi0506s15. 0 5 Unit– 5.6.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 75.Dominguez Cyril, Boelens Rolf A, Bonvin AMJJ. HADDOCK: A Protein-Protein Docking Approach Based on Biochemical or Biophysical Information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 76.Guerois R, Nielsen JE, Serrano L. Predicting Changes in the Stability of Proteins and Protein Complexes: A Study of More Than 1000 Mutations. Journal of Molecular Biology. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 77.Meredith RW, et al. Impacts of the Cretaceous Terrestrial Revolution and KPg Extinction on Mammal Diversification. Science. 2011;334:521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 78.Ludt CJ, Schroeder W, Rottmann O, Kuehn R. Mitochondrial DNA phylogeography of red deer (Cervus elaphus) Molecular Phylogenetics and Evolution. 2004;31:1064–1083. doi: 10.1016/j.ympev.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 79.Shimodaira H. An Approximately Unbiased Test of Phylogenetic Tree Selection. Systematic Biology. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 80.Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- 81.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Papadakis MN, Patrinos GP. Contribution of gene conversion in the evolution of the human beta-like globin gene family. Human Genetics. 1999;104:117–125. doi: 10.1007/s004390050923. [DOI] [PubMed] [Google Scholar]
- 83.Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet. 2004;36:151–156. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]
- 84.Bosch E, Hurles ME, Navarro A, Jobling MA. Dynamics of a human interparalog gene conversion hotspot. Genome Research. 2004;14:835–844. doi: 10.1101/gr.2177404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
HBBA and HBBF full gene sequences (coding sequence plus intervening introns) have been submitted to GenBank with accession numbers KY800429-KY800452. An alignment of these sequences is also available as Supplementary Data. Père David’s deer RNA sequencing and white-tailed deer whole genome sequencing raw data has been submitted to the European Nucleotide Archive (ENA) with the accession numbers PRJEB20046 and PRJEB20034, respectively.