Abstract
Pediatric developmental syndromes present with systemic, complex, and often overlapping clinical features that are not infrequently a consequence of Mendelian inheritance of mutations in genes involved in DNA methylation, establishment of histone modifications, and chromatin remodeling (the “epigenetic machinery”). The mechanistic cross-talk between histone modification and DNA methylation suggests that these syndromes might be expected to display specific DNA methylation signatures that are a reflection of those primary errors associated with chromatin dysregulation. Given the interrelated functions of these chromatin regulatory proteins, we sought to identify DNA methylation epi-signatures that could provide syndrome-specific biomarkers to complement standard clinical diagnostics. In the present study, we examined peripheral blood samples from a large cohort of individuals encompassing 14 Mendelian disorders displaying mutations in the genes encoding proteins of the epigenetic machinery. We demonstrated that specific but partially overlapping DNA methylation signatures are associated with many of these conditions. The degree of overlap among these epi-signatures is minimal, further suggesting that, consistent with the initial event, the downstream changes are unique to every syndrome. In addition, by combining these epi-signatures, we have demonstrated that a machine learning tool can be built to concurrently screen for multiple syndromes with high sensitivity and specificity, and we highlight the utility of this tool in solving ambiguous case subjects presenting with variants of unknown significance, along with its ability to generate accurate predictions for subjects presenting with the overlapping clinical and molecular features associated with the disruption of the epigenetic machinery.
Keywords: pediatric developmental disorders, epigenomic machinery, Kabuki syndrome, CHARGE syndrome, Floating Harbor syndrome, ATRX syndrome, Sotos syndrome, Claes-Jensen syndrome, machine learning, molecular diagnosis
Introduction
Genes encoding the epigenetic protein machinery that read, write, and erase post-translational signals on DNA and histones and remodel chromatin are implicated in a wide range of constitutional neurodevelopmental disorders.1, 2, 3 The pathogenesis of such disorders is likely caused by the downstream events orchestrated by the primary functional defect in these proteins of the so-called epigenetic machinery.1, 2, 3 Furthermore, specific mutations in these readers, writers, erasers, and chromatin remodelers are linked to the variability in clinical phenotype seen in their associated disorders. It is well established that histone modifications overlap and interact with genomic DNA methylation to affect chromatin remodeling,4 and thus, that mutations in the genes that are involved in histone modifications are expected to have an impact within the DNA methylome. Supporting this concept, we have previously reported DNA methylation epigenetic (epi-) signatures in the peripheral blood of subjects carrying mutations in genes involved in chromatin regulation, including the mutations in SRCAP (MIM: 611421) causing Floating-Harbor syndrome (MIM: 136140),5 DNMT1 (MIM: 126375) resulting in adult-onset autosomal-dominant cerebellar ataxia, deafness, and narcolepsy (ADCA-DN [MIM: 604121]),6 and ATRX (MIM: 300032), which is responsible for the alpha thalassemia/mental retardation X-linked (ATRX) syndrome (MIM: 300448).7 Epi-signatures in subjects with Sotos (MIM: 117550), CHARGE (MIM: 214800), and Kabuki (MIM: 147920) syndromes have also been reported.8, 9, 10
While the number of developmental and cancer-related conditions for which a DNA methylation epi-signature has been reported is increasing, the extent of overlap or distinction of these epi-signatures is not clear. Clinical overlap is a common finding in the diseases that result from such defects, and it is postulated that mechanistic overlap could be a basis for such phenotypic similarity. This is further acknowledged by noting that all of the proteins associated with these conditions regulate the epigenome through complex interactions with each other. Hence, the question has been raised as to how accurately one can use these epi-signatures as a tool in the molecular diagnosis of these conditions.9, 10 This is particularly important in the context of utilizing these epi-signatures as functional evidence to classify variants of uncertain clinical significance.
In the present study, we try to address these questions by concurrently examining 14 Mendelian conditions that result from direct or indirect disruptions of the proteins involved in the regulation of the epigenome. These conditions (see Table 1) include Rett syndrome (MIM: 312750) (methyl-CpG-binding protein 2; MeCP2 [MIM: 300005]), ADCA-DN (DNA methyltransferase 1; DNMT1), Kabuki syndrome (lysine-specific methyltransferase 2D; KMT2D [MIM: 147920]), ATRX syndrome (ATRX), Sotos syndrome (nuclear receptor binding SET domain protein 1; NSD1 [MIM: 117550]), Floating-Harbor syndrome (Snf2 related CREBBP activator protein; SRCAP), Weaver syndrome (MIM: 277590) (enhancer of zeste 2 polycomb repressive complex 2 subunit; EZH2 [MIM: 601573]), CHARGE syndrome (chromodomain helicase DNA binding protein 7; CHD7 [MIM: 608892]), Claes-Jensen syndrome (MIM: 300534) (lysine-specific demethylase 5C; KDM5C [MIM: 314690]), Genitopatellar syndrome (GTPTS [MIM: 606170]), and Say-Barber-Biesecker-Young-Simpson syndrome (SBBYSS [MIM: 603736]), both caused by mutations in lysine acetyltransferase 6B (KAT6B [MIM: 605880]), and Coffin-Siris syndrome (MIM: 135900) (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily B1 SMARCB1 [MIM: 601607], and AT-rich interaction domain 1B; AR1D1B [MIM: 614556]). These genes are directly involved in epigenomic regulation of the chromatin. We have also included two other conditions that result from mutations in genes that interact with the components of the epigenomic machinery. Saethre-Chotzen syndrome (MIM: 101400) is caused by mutations in the Twist family BHLH transcription factor 1 gene (TWIST [MIM: 601622]), encoding a transcription factor that binds to the p300 and p300/CREBBP-associated factor domains of histone acetyltransferases and regulates their activity,11 and Coffin-Lowry syndrome (MIM: 303600), which results from mutations in ribosomal protein S6 kinase (RSK2 [MIM: 300075]), a protein required for phosphorylation of histone H3 that regulates chromatin remodeling and gene expression.12
Table 1.
Structure and Demographics of the Study Cohort
| Syndrome | Total | Testing Cohort |
Discovery-Training Cohort |
Control Cohort |
Probes Passing QC | Probes Found | DMRs Found | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Individuals | Percentage Female | Mean Age ± SD | No. of Individuals | Percentage Female | Mean Age ± SD | ||||||
| Rett | 17 | 4 | 13 | 92% | 7.6 ± 11 | 52 | 92% | 7.4 ± 10 | 448,775 | N/A | no |
| Saethre-Chotzen | 25 | 6 | 19 | 63% | 10.6 ± 11 | 76 | 63% | 10.11 ± 10 | 442,079 | N/A | no |
| Weaver | 7 | 2 | 5 | 40% | age unknown | 20 | 40% | – | 455,427 | N/A | no |
| Coffin Siris | 9 | 3 | 6 | 50% | 5.9 ± 6 | 24 | 50% | 5.8 ± 5 | 453,321 | N/A | no |
| Coffin Lowry | 11 | 3 | 8 | 12% | 9.8 ± 6 | 32 | 12% | 9.7 ± 6 | 453,285 | N/A | no |
| ATRX | 19 | 4 | 15 | 0% | 11.4 ± 7 | 60 | 0% | 11.3 ± 7 | 450,748 | 1,112 | 41 |
| Floating-Harbor | 17 | 4 | 13 | 76% | 12.7 | 52 | 76% | 12.2 ± 11 | 453,189 | 1,078 | 54 |
| Sotos | 38 | 10 | 28 | 57% | 8.8 ± 4 | 112 | 57% | 8.9 ± 4 | 448,131 | 6,858 | 1,372 |
| ADCA-DN | 5 | 0 | 5 | 40% | age unknown | 20 | 40% | – | 330,788 | 3,562 | 52 |
| Claes-Jensen | 10 | 2 | 8 | 0% | 22.5 ± 14.2 | 32 | 0% | 21.2 ± 13 | 454,978 | 698 | 14 |
| Kabuki | 44 | 11 | 33 | 57% | 9.5 ± 6 | 132 | 57% | 9.7 ± 7 | 401,051 | 919 | 31 |
| CHARGE | 79 | 40 | 39 | 41% | 5.8 ± 6 | 156 | 48% | 5.9 ± 6 | 448,876 | 1,320 | 18 |
| GTPTS | 3 | 0 | 3 | 33% | 2.1 ± 5.8 | 20 | 33% | 2.2 ± 5.1 | 454,489 | 707 | 6 |
| SBBYSS | 1 | 0 | 1 | 100% | 6.3 ± 0 | 20 | 100% | 6.5 ± 0.8 | 456,134 | 864 | 6 |
In this study, we have identified DNA methylation profiles in peripheral blood samples from a large cohort of individuals who carry mutations in the associated candidate genes responsible for their respective developmental syndromes, including previously unreported signatures in cohorts for GTPTS and SBBYSS (KAT6B) and Claes-Jensen syndrome (KDM5C). We have also examined the degree of positional overlap between these epi-signatures and have evaluated whether a single classification tool, built on the combined epi-signatures of these conditions, can generate accurate predictions for subjects presenting with the overlapping clinical and molecular features associated with mutations in these genes that play an essential role in the epigenetic machinery.
Material and Methods
Source of Data
This study utilized data and specimens from multiple sources. Peripheral blood DNA samples from subjects with ADCA-DN, Coffin-Siris syndrome, SBBYSS, GTPTS, and Floating-Harbor syndrome were collected from the Care4Rare Canada Consortium. Samples from subjects with clinical characteristics of Kabuki syndrome, ATRX, Saethre-Chotzen syndrome, Coffin-Lowry syndrome, Rett syndrome, Claes-Jensen syndrome, and CHARGE syndrome were collected from the Greenwood Genetic Center (Greenwood, SC, USA). The Kabuki and CHARGE cohorts were supplemented by methylation array files publicly available from GEO (GSE97362).9 Epigenomic data from subjects with Sotos and Weaver syndromes were downloaded from GEO (GSE74432).8 All of these subjects had clinical features of the aforementioned syndromes and were screened for mutations in the related genes. The mutation report from every subject was reviewed according to the American College of Medical Genetics Guidelines for interpretation of genomic sequence variants,13 and only individuals confirmed to carry a pathogenic or likely pathogenic mutation were used to identify epi-signatures (subject-level data are summarized in Table S1 where available). Control subjects were selected from our lab reference cohort, which is composed of individuals with no known aberrant epigenomic change. This reference cohort was previously preselected from a larger cohort of approximately 1,000 individuals across a broad range of age, sex, and ethnicity distribution.
Methylation Array and Quality Assessment
Genomic DNA was extracted from peripheral blood using standard techniques. Following bisulfite conversion, DNA methylation analysis of the samples was performed using the Illumina Infinium bead chip array, according to the manufacturer’s protocol at the Genetic and Molecular Epidemiology Laboratory at McMaster University and the London Health Sciences Molecular Genetic Laboratory. Except for a cohort of subjects with CHARGE syndrome (n = 39), which was assayed using Illumina Infinium methylation EPIC array, all of the samples were assayed using the HumanMethylation450 bead chip. The two arrays harbor 96% overlap in CpG probes. Methylated and unmethylated intensity data were generated as idat files and imported into R 3.4.0 for analysis. Normalization was performed using Illumina normalization method with background correction using the minfi package. Probes with detection p value > 0.01 were excluded from the downstream analyses. For further quality improvement, probes located on chromosomes X and Y and probes known to contain SNPs at the CpG interrogation, or the single-nucleotide extension, were removed. As an additional quality-control step, the sex of the samples was predicted using the signal intensity of the X and Y chromosomes using the minfi package, and the files representing a discordance between the predicted and labeled sex were not used for identification of the DNA methylation profile. All of the samples were examined for genome-wide methylation density, and those deviating from a bimodal signal distribution were excluded. Files generated by the EPIC array were cast as 450k array, and the same analytical procedures used on the 450k array were applied.
Selection of Discovery/Training and Testing Cohorts and Controls
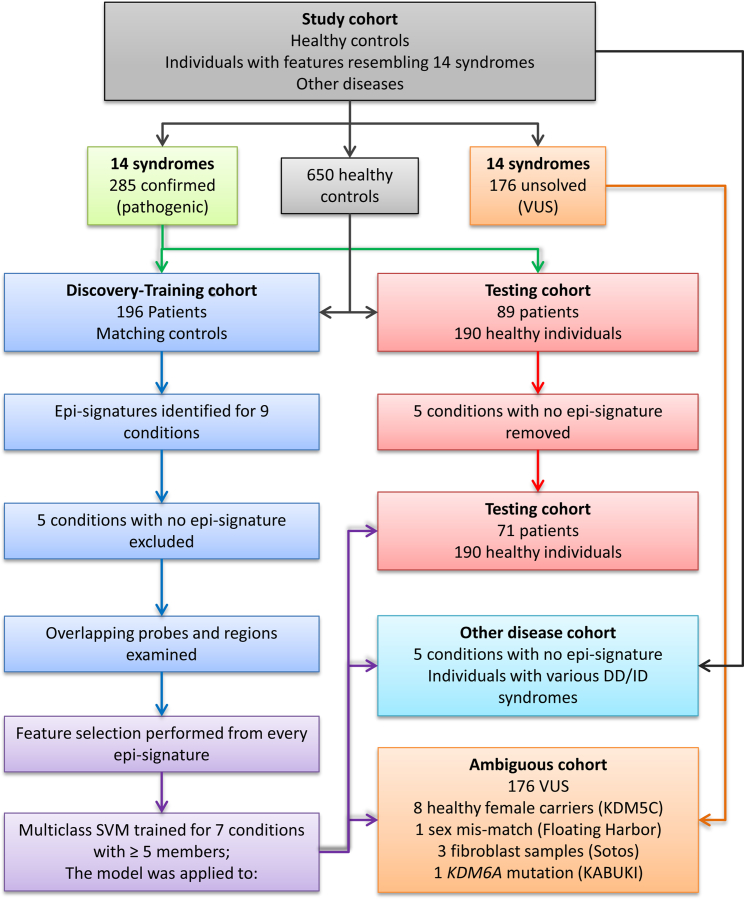
The identification of disease-specific epi-signatures was performed using a randomly selected 75% subset of the database (discovery/training set) using caTools package. The remaining samples were only used as a testing cohort to assess the performance of the classification model developed later in the study. This procedure was not performed when ≤5 samples were available for a disease group (ADCA-DN, GTPTS, and SBBYSS). Given probe differences and technology variations between the two array types (450k and EPIC), it was ensured that the entire discovery/training cohort is assayed using one array (450k). Therefore, all of the CHARGE-affected subjects who were assayed using the 450k array were selected as the discovery/training cohort and the rest were included in the testing cohort. For every disease group in the discovery cohort, a sex- and age-matched control group with a sample size at least four times larger (minimum n = 20) was selected from the reference control group using MatchIt package. The methylation profile of each disease group in the discovery cohort was compared with its matched control separately to identify the disease-specific epi-signature (Table 1). Figure 1 represents the flowchart of the study.
Figure 1.
Flowchart of the Study
Identification of Disease-Specific Methylation Epi-signatures
Analysis was performed using a modification of our previously published protocol.5, 6, 7, 10, 14 The methylation level for each probe was measured as a beta value, calculated from the ratio of the methylated signals versus the total sum of unmethylated and methylated signals, ranging between 0 (no methylation) and 1 (full methylation). This value was used for biological interpretation and visualization. For statistical analysis, wherever a normal distribution was required (linear regression modeling), beta values were logit transformed to M-values using the following equation: log2(beta/(1-beta)). A linear regression modeling using the limma package was used to identify the differentially methylated probes. The analysis was adjusted for blood cell type compositions predicted using minfi package. The generated p values were moderated using the eBayes function in the limma package and were corrected for multiple testing using Benjamini and Hochberg method. Probes with a corrected p value < 0.01 and a methylation difference greater than 10%–20% were considered significant. The effect size cutoffs (10%–20%) were determined separately for every condition following the examination of the volcano plots generated in every comparison as previously conducted by Butcher et al.9 The identified probes were examined using an unsupervised hierarchical clustering to ensure their ability in separating the subjects from controls. In the case of the Coffin-Siris syndrome, which was caused by two genes in our dataset, the cohort was first split based on the causing genes and then re-analyzed regardless of the gene.
Control for Batch Effect and Robustness of the Identified Epi-signatures
Given that the study cohort was composed of data generated by multiple centers, different dates, and via various instruments, several measures were utilized to minimize possible batch effects and other sources of variability. These steps involved: (1) whenever the sample size allowed, control subjects were selected from the same batch (as with ADCA-DN); (2) if case subjects and their control subjects in a comparison were assayed in multiple batches (as with Kabuki syndrome samples), the batch variable was included as a confounding factor in the regression model; (3) where a particular sample was assayed using both 450k and EPIC arrays (CHARGE syndrome), only files from 450k were used for identification of the signature; (4) for all other disease cohorts, a machine learning model (Support vector machine) was trained on the identified probes, and we examined how well it could distinguish the batch structure from the disease status. Failure to pass this step resulted in the findings being regarded as non-reliable.
Identification of Genomic Regions with Methylation Changes
To identify genomic regions harboring methylation changes (differentially methylated regions [DMRs]), a bump hunting approach was used by the bumphunter package.15 The analysis considered regions with greater than 10% change in the overall methylation between case and control subjects with gaps no more than 500 bp among neighboring CpGs. As suggested in the package, 1,000 bootstrapping procedure was performed to compute family-wise error rate (FWER). We selected regions containing a minimum of three consecutive probes and FWER < 0.01. The identified regions were mapped to CpG islands and coding genes. Gviz package was used for visualization of the DMRs.
Assessment of the Overlap between the Epi-signatures
Probes and regions differentially methylated in each disease group were examined to identify potential overlap. The number of probes shared in more than one disease group was visualized using a circos plot. Probes that were shared in >2 and 3 disease groups were used to measure the pairwise correlations in the disease cohort. Calculated correlation coefficients were visualized using a correlation plot. Genomic regions harboring differentially methylated probes were assessed for overlap using GenomicRanges package, and regions found in more than one disease type were reported. Functional annotation clustering and gene set enrichment analysis was performed using missMethyl and ReactomePA packages for the genes harboring the shared probes.
Construction and Validation of a Multi-class Prediction Model
The identified signatures were used to build a classification model with the ability to concurrently assess a given methylation profile belonging to any of the disease groups in the study. Caret package was used for feature selection from every signature. First, a receiver operating characteristic curve analysis was performed to identify the most differentiating probes. Those probes with an area under the curve above 0.8 were retained. Next, pairwise correlations among the remaining probes were measured to identify and exclude the redundant signals with R-squared > 0.8. A multi-class support vector machine (SVM) with linear kernel was trained on the remaining probes using e1071 package. To determine the best hyperparameters and to measure the accuracy of the model, a 10-fold cross-validation was performed. In this process, the training set was divided into ten folds. Nine folds were used for training the model and one fold for testing. After repeating this iteration for all of the ten folds, the mean accuracy was calculated and the hyperparameters with the optimal performance were selected. For every sample, the model was set to generate multiple classification scores between 0 and 1 as the probability of having a methylation profile related to every disease. To assess the sensitivity of the model, the testing cohort, which was not used for identification of the signature or construction of the SVM, was supplied to the model. To determine the specificity, we supplied all of the healthy subjects that were not used in the earlier stages of the study to the model. To understand whether this model is sensitive to other medical conditions representing developmental delay and intellectual disabilities, we tested, using the constructed model, a large number of subjects in our database with a confirmed clinical diagnosis of various diseases including autism spectrum disorders, imprinting defects, RASopathies, chromosomal aberrations, and Down syndrome. As well, we tested whether this classifier is sensitive to other diseases of epigenomic machinery for which no epi-signature was identified in this study. To further confirm that this classifier is not sensitive to the blood cell type compositions, we downloaded normalized methylation data from isolated cell populations of healthy individuals from GEO (GSE35069)16 and supplied them to our model for prediction.
Assessment of Ambiguous Case Subjects and Variants of Unknown Significance
The approved model was used to perform a prediction on the DNA methylation profiles of individuals with variants of unknown significance in the respective genes that were not previously included in the identification of the signature or in construction and validation of the classification model. In addition, a prediction was made on subjects with predicted sex discordance, samples obtained from tissues other than blood, healthy subjects carrying pathogenic mutations, and the single subject with Kabuki syndrome resulting from the less common KDM6A gene mutation.
Ethics Statement
This study has been approved by the Western University Research Ethics Boards (REB ID 106302) and the Hamilton Integrated Research Ethics Board (REB ID 13-653-T). All of the samples and records were de-identified before the study.
Results
Description of the Study Cohort
Figure 1 represents the flowchart of the study. Mutation analysis of the subjects with clinical features resembling each of the 14 syndromes (Table 1) identified a total of 285 subjects with pathogenic or likely pathogenic mutations, which were classified according to the American College of Medical Genetics (ACMG) guidelines. The remaining subjects (n = 176) carried benign variants or variants of unknown significance (VUS), leaving them unsolved. Our data also included healthy female carriers with pathogenic mutations in KDM5C (n = 8), fibroblast samples from individuals with Sotos syndrome (n = 3), and one individual affected with Kabuki syndrome with a pathogenic truncating mutation in KDM6A. Table S1 summarizes the mutation types and demographic characteristics where available. A sample of 196 randomly selected subjects from the 285 individuals with a pathogenic mutation was used as the discovery/training cohort for identification of epi-signatures as well as for training the classification model. The remaining 89 subjects were regarded as the testing cohort to be used for measuring the sensitivity of the classification model. For each disease group in the discovery/training cohort, a sex- and age-matched sample group four times larger (minimum n = 20) was selected from our reference healthy cohort (n = 650) for comparison. A total of 190 healthy samples never selected as a control for any of the diseases were later used to assess the specificity of the classification model. Table 1 shows the count, age, and sex distributions from every subject group in the discovery/training cohort along with the information from the matched control subjects.
Disease-Specific DNA Methylation Epi-signatures and Differentially Methylated Regions (DMRs)
The comparison of disease cohorts with their matched control subjects was performed using the subset of CpG probes that passed the quality assessment (Table 1). No reliable epi-signature was observed for five of the diseases tested here, including Saethre-Chotzen, Coffin-Siris, Coffin-Lowry, Rett, and Weaver syndromes, as the findings did not meet the criteria described in the methods. For other conditions (i.e., Floating-Harbor, ADCA-DN, Kabuki, ATRX, CHARGE, Sotos, GTPTS, SBBYSS, and Claes-Jensen syndromes), reliable epi-signatures were identified (Table 1). Of these, Sotos (n = 6,858) and ADCA-DN (n = 3,562) revealed the largest number of probes, mostly composed of hypomethylated CpGs. The identified probes from every cohort were confirmed to separate the subjects from the controls using hierarchical clustering (data not shown).
Consistent with the methylation profiles, the bump hunting approach did not identify any genomic segment to be differentially methylated in subjects with Saethre-Chotzen, Coffin-Siris, Coffin-Lowry, Rett, and Weaver syndromes. For the other nine syndromes, however, multiple genomic coordinates containing a minimum of 3 consecutive CpG probes, an average regional methylation difference > 0.10, and a family-wise error rate (FWER) < 0.01 were identified. Subjects with Sotos syndrome showed the largest number of identified regions (n = 1,372), mostly composed of hypomethylated segments (data not shown).
To further ensure that adjustment for blood cell type compositions has not masked a potential methylation profile in the five syndromes with negative results, we repeated the analysis without inclusion of the blood cell type estimates correction. Similar to the previous analyses, a significant methylation profile was not detected. At this stage, we concluded that either no epigenomic profile existed for these five conditions, or their methylation changes were too obscure to pass the thresholds and quality assessment criteria set in this study (see Material and Methods). Thus, our subsequent analyses described below focused on the syndromes for which a differential epigenomic profile was observed: i.e., Floating-Harbor, ADCA-DN, Kabuki, ATRX, GTPTS, SBBYSS, CHARGE, Sotos, and Claes-Jensen syndromes.
Overlap between the Epi-signatures and DMRs
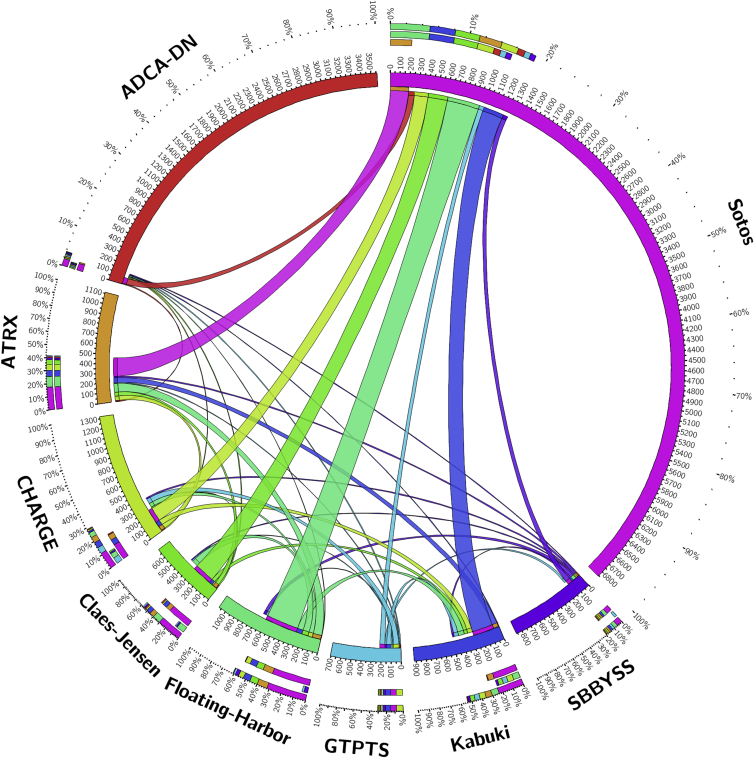
Of the total number of 15,408 probes that composed the epi-signatures of the 9 conditions, only 1,598 (∼10%) CpGs were shared across more than one disease group. Among these conditions, Floating-Harbor and Claes-Jensen syndromes were found to share the largest proportion of their epi-signatures with others (mainly with Sotos), whereas this number was minimal to negligible for other conditions, including ADCA-DN which, despite having a large number of CpGs in its epi-signature, shared less than 3% of its probes with other diseases (Figure 2). The number of probes that were shared by more than two conditions was limited to 217 probes. Table S2 shows the 217 probes and the related conditions. A gene ontology analysis of the 217 probes using missMethyl package found multiple ontology terms related to histone modifications to be enriched in the harboring genes, including S-methyltransferase activity and histone-lysine N-methyltransferase activity (Table S3). Next, genes overlapping these probes were evaluated using the ReactomePA package to identify pathways enriched in them (Table S4). Lysine histone transferase activity was found as the only enriched pathway with a multiple testing corrected p value < 0.05, which is composed of four genes (PRDM9, SETDB1, HIST1H3E, NSD1). These four genes also contained the most number of probes shared by different conditions (Table S2), which included PRDM9 with five probes found in all of the conditions except for ADCA-DN, GTPTS, and SBBYSS; HIST1H3E, with two probes shared by Floating-Harbor, ATRX, Claes-Jensen, GTPTS, and SBBYSS syndromes; and NSD1 and SETDB1 with three and two probes, respectively, shared by Sotos, Floating-Harbor, and Claes-Jensen syndromes. Table S2 also shows that 21 probes in HOXA5 (MIM: 142952) are shared by three conditions (Kabuki, CHARGE, and GTPTS).
Figure 2.
Quantity of Probes from the Epi-signatures of Every Nine Conditions that Are Shared with Each Other
Thickness of the bonds represents the number of the shared probes by every two diseases as shown by digits on the circumference of the plot. This plot does not visualize the 217 probes that are shared by more than two conditions.
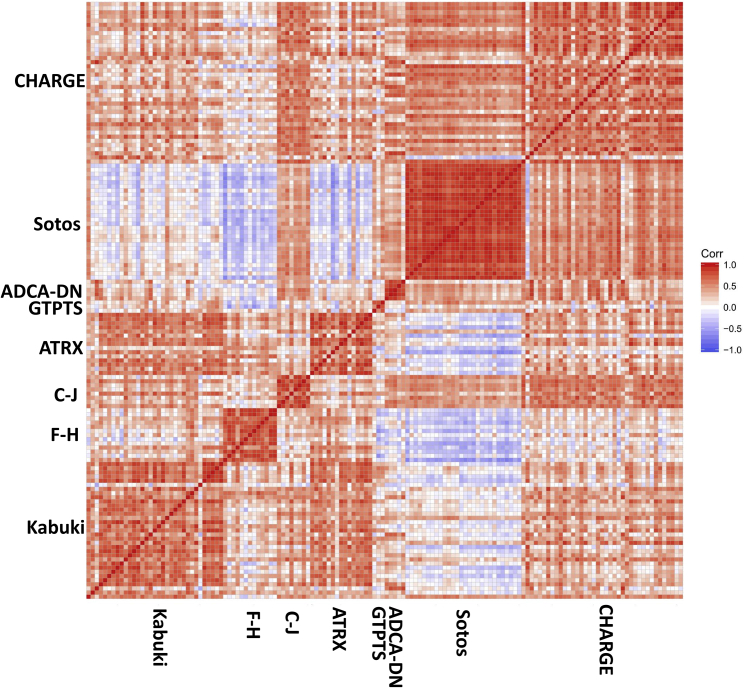
A pairwise correlation analysis on the methylation levels of all of these 217 probes for each affected subject in the disease group revealed that while samples from the same condition showed the strongest correlation with each other, only a weak to moderate positive correlation existed among the samples from across disease groups. The only exception was with subjects with Floating-Harbor and Sotos syndromes, which revealed a moderate negative correlation with each other, despite sharing the most number of probes of all. We further narrowed down the probes to 18 CpGs that were shared by more than three disease groups and re-evaluated the correlation of the subjects based on the methylation levels of these 18 probes. We observed a greater degree of negative correlations between different syndrome groups (Figure 3). The members of every condition, however, correlated well with each other in both of these analyses.
Figure 3.
Pairwise Correlation between Samples with Different Conditions using the Methylation Values of Ten Probes that Are Shared by More than Three Conditions
Red represents positive and blue represents negative correlation. Every visible square represents the correlations of one subject on x axis with its correspondence on y axis. Abbreviations: F-H, Floating-Harbor; C-J, Cales-Jensen
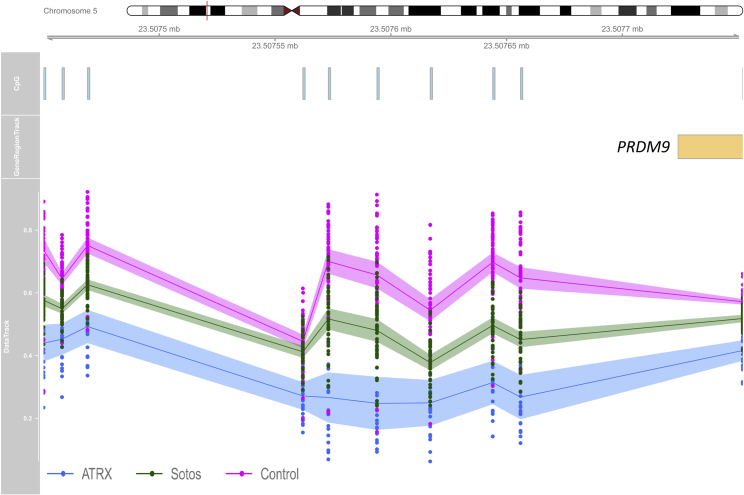
Table 2 shows the genomic coordinates that are shared between every two diseases. Similar to the results described at single probe analysis, Floating-Harbor and Sotos syndromes shared the most differentially methylated coordinates (n = 30), more than half of which showed an opposite direction of change in methylation levels by the two syndromes. As previously observed by the shared CpGs (Table S2), a segment in the promoter of PRDM9 was among the regions containing at least three consecutive hypomethylated probes in both ATRX and Sotos syndromes (Figure 4). The same segment contains disperse probes hypomethylated in Claes-Jensen syndrome but hypermethylated in Floating-Harbor syndrome (Table S2). The only region with multiple consecutive differentially methylated probes that was shared by more than two syndromes was a segment in the promoter of HOXA5. This segment is significantly hypomethylated in GTPTS but hypermethylated in both CHARGE and Kabuki syndromes (Table 2).
Table 2.
Overlapping Genomic Coordinates with Differential Methylation in Nine Conditions Compared to Control Subjects
| Chr | Disease 1 | Coordinates 1 | Methylation Difference 1 | Probes 1 | Disease 2 | Coordinates 2 | Methylation Difference 2 | Probes 2 | Direction of Change | Overlapping Gene(s) | Distance to CpG Island (bp) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 19 | Floating-Harbor | 8591364–8591776 | −0.4 | 4 | Kabuki | 8591364–8591776 | −0.28 | 4 | same | MYO1F | 0 |
| 11 | Floating-Harbor | 65360123–65360327 | −0.27 | 3 | Kabuki | 65360123–65360327 | −0.19 | 3 | same | KCNK7 | 0 |
| 1 | Sotos | 227746111–227747468 | −0.35 | 7 | Kabuki | 227746191–227746882 | 0.14 | 4 | opposite | – | 0 |
| 3 | Sotos | 49170496–49171051 | −0.22 | 7 | Kabuki | 49170599–49170794 | 0.16 | 4 | opposite | LAMB2 | 12,032 |
| 5 | Sotos | 1856325–1857828 | −0.2 | 7 | Kabuki | 1856713–1857477 | 0.16 | 4 | opposite | – | 0 |
| 12 | Sotos | 133179887–133180698 | −0.25 | 5 | Kabuki | 133179887–133180238 | 0.11 | 4 | opposite | LRCOL1 | 0 |
| 5 | Sotos | 101119084–101119766 | −0.24 | 5 | Kabuki | 101119084–101119566 | 0.11 | 4 | opposite | – | 512,283 |
| 7 | Sotos | 27170241–27170552 | 0.19 | 6 | Kabuki | 27170388–27170994 | −0.14 | 13 | opposite | HOXA4 | 0 |
| 19 | Sotos | 51330265–51330469 | −0.24 | 4 | Kabuki | 51330265–51330469 | 0.13 | 4 | opposite | KLK15 | 0 |
| 15 | Sotos | 29968032–29968195 | −0.3 | 3 | Kabuki | 29968032–29968195 | 0.18 | 3 | opposite | – | 410 |
| 7 | Sotos | 27170717–27171051 | 0.15 | 9 | Kabuki | 27170388–27170994 | −0.14 | 13 | opposite | – | 78 |
| 2 | Sotos | 109746691–109747003 | −0.21 | 5 | Kabuki | 109746691–109746754 | −0.14 | 4 | same | SH3RF3 | 0 |
| 7 | Sotos | 142494148 - 142494492 | −0.16 | 6 | Kabuki | 142494148–142494492 | −0.15 | 6 | same | – | 71 |
| 5 | Sotos | 176827082–176827697 | −0.16 | 5 | Kabuki | 176827392–176827697 | −0.16 | 3 | same | PFN3 | 0 |
| 10 | Sotos | 132099067–132100019 | −0.17 | 3 | Kabuki | 132099067–132100019 | 0.15 | 3 | opposite | – | 109,734 |
| 7 | CHARGE | 27182493–27183946 | 0.23 | 20 | Kabuki | 27182493–27183816 | 0.16 | 18 | same | HOXA5, HOXA-AS3 | 0 |
| 7 | CHARGE | 27184316–27184521 | 0.17 | 8 | Kabuki | 27184369–27184441 | 0.14 | 4 | same | HOXA-AS3 | 0 |
| 17 | Floating-Harbor | 7486551–7486874 | −0.24 | 7 | Claes–Jensen | 7486551–7486874 | −0.29 | 7 | same | – | 0 |
| 1 | Sotos | 247694041–247694531 | −0.25 | 6 | Claes–Jensen | 247694041–247694531 | −0.24 | 6 | same | GCSAML,GCSAML-AS1, OR2C3 | 0 |
| 5 | Sotos | 176559334–176559563 | −0.36 | 3 | Claes–Jensen | 176559334–176559563 | −0.32 | 3 | same | – | 0 |
| 13 | Sotos | 113242878–113243141 | −0.21 | 3 | Claes–Jensen | 113242878–113243141 | −0.32 | 3 | same | TUBGCP3 (396bp away) | 221 |
| 6 | Sotos | 32120625–32121433 | −0.31 | 27 | Floating-Harbor | 32120773–32120933 | −0.15 | 10 | same | – | 396 |
| 6 | Sotos | 31650735–31651249 | −0.17 | 17 | Floating-Harbor | 31650735–31650835 | −0.19 | 6 | same | – | 0 |
| 6 | Sotos | 31650735–31651249 | −0.17 | 17 | Floating-Harbor | 31650916–31651278 | −0.15 | 11 | same | – | 0 |
| 6 | Sotos | 32847702–32847845 | −0.29 | 10 | Floating-Harbor | 32847702–32847845 | 0.18 | 10 | opposite | – | 0 |
| 17 | Sotos | 36997420–36997740 | −0.38 | 7 | Floating-Harbor | 36997420–36997740 | −0.23 | 7 | same | C17orf98 | 0 |
| 8 | Sotos | 39171866–39172120 | −0.31 | 7 | Floating-Harbor | 39172020–39172120 | 0.2 | 6 | opposite | ADAM5 (61bp away) | 206,442 |
| 10 | Sotos | 50649666–50650248 | −0.39 | 5 | Floating-Harbor | 50649666–50650248 | 0.18 | 5 | opposite | – | 42,882 |
| 6 | Sotos | 33282867–33283055 | −0.16 | 12 | Floating-Harbor | 33282867–33283184 | −0.18 | 19 | same | – | 0 |
| 2 | Sotos | 129659018–129659946 | −0.24 | 7 | Floating-Harbor | 129659316–129659946 | 0.19 | 6 | opposite | – | 0 |
| 5 | Sotos | 83016779–83017553 | −0.25 | 6 | Floating-Harbor | 83017000–83017644 | 0.19 | 6 | opposite | HAPLN1 | 341 |
| 4 | Sotos | 11369349–11370872 | −0.18 | 8 | Floating-Harbor | 11370314–11370565 | 0.25 | 3 | opposite | MIR572 | 0 |
| 8 | Sotos | 102235927–102236831 | −0.23 | 6 | Floating-Harbor | 102236283–102236831 | 0.23 | 5 | opposite | – | 0 |
| 2 | Sotos | 165811816–165812159 | −0.26 | 5 | Floating-Harbor | 165811816–165812159 | 0.22 | 5 | opposite | SLC38A11 | 112,922 |
| 7 | Sotos | 92672812–92673176 | −0.25 | 5 | Floating-Harbor | 92672812–92673176 | 0.2 | 5 | opposite | – | 0 |
| 4 | Sotos | 155702409–155703138 | −0.18 | 6 | Floating-Harbor | 155702409–155703138 | 0.19 | 6 | opposite | RBM46 | 0 |
| 1 | Sotos | 1003126–1003529 | −0.25 | 4 | Floating-Harbor | 1003126–1003529 | −0.29 | 4 | same | – | 0 |
| 3 | Sotos | 159557552–159558031 | −0.24 | 4 | Floating-Harbor | 159557552–159558031 | 0.23 | 4 | opposite | SCHIP1, IQCJ-SCHIP1 | 74,528 |
| 2 | Sotos | 164204628–164204915 | −0.24 | 4 | Floating-Harbor | 164204752–164205343 | 0.31 | 6 | opposite | – | 0 |
| 22 | Sotos | 50737978–50738890 | −0.22 | 4 | Floating-Harbor | 50737978–50738890 | −0.29 | 4 | same | PLXNB2 | 0 |
| 12 | Sotos | 56617576–56617737 | −0.28 | 3 | Floating-Harbor | 56617576–56617737 | −0.23 | 3 | same | – | 258 |
| 9 | Sotos | 139258524–139259074 | −0.26 | 3 | Floating-Harbor | 139258524–139259074 | −0.23 | 3 | same | CARD9 | 0 |
| 19 | Sotos | 3480363–3480672 | −0.21 | 5 | Floating-Harbor | 3480363–3480672 | −0.2 | 5 | same | SMIM24 | 1,110 |
| 19 | Sotos | 49222892–49223278 | −0.2 | 5 | Floating-Harbor | 49222892–49223278 | −0.28 | 5 | same | MAMSTR | 0 |
| 4 | Sotos | 165898666–165898848 | −0.19 | 5 | Floating-Harbor | 165898666–165898848 | 0.19 | 5 | opposite | TRIM61 | 20,219 |
| 1 | Sotos | 45278971–45279349 | −0.21 | 4 | Floating-Harbor | 45278971–45279349 | −0.23 | 4 | same | BTBD19 | 0 |
| 19 | Sotos | 2428350–2428677 | −0.19 | 4 | Floating-Harbor | 2428350–2429209 | −0.2 | 6 | same | LMNB2 | 0 |
| 19 | Sotos | 1063624–1064218 | −0.19 | 3 | Floating-Harbor | 1063624–1064218 | −0.24 | 3 | same | ABCA7 | 0 |
| 4 | Sotos | 46126066–46126253 | −0.18 | 3 | Floating-Harbor | 46126066–46126448 | 0.25 | 7 | opposite | GABRG1 | 265,650 |
| 12 | Sotos | 47219737–47219958 | −0.14 | 9 | Floating-Harbor | 47219626–47219920 | 0.18 | 9 | opposite | SLC38A4 | 4,954 |
| 5 | Sotos | 110062384–110062837 | −0.16 | 6 | Floating-Harbor | 110062384–110062837 | 0.23 | 6 | opposite | TMEM232 | 11,768 |
| 11 | Sotos | 32449254–32449638 | −0.17 | 3 | ADCA-DN | 32449163–32450692 | 0.24 | 11 | opposite | WT1 | 0 |
| 7 | CHARGE | 27137922–27138712 | −0.15 | 4 | Sotos | 27137922–27138396 | −0.17 | 3 | same | HOTAIRM1 | 1,185 |
| 7 | CHARGE | 53254947–53255065 | 0.15 | 3 | Sotos | 53254947–53255065 | 0.18 | 3 | same | – | 0 |
| 5 | CHARGE | 161178574–161178796 | 0.14 | 3 | Sotos | 161178574–161178796 | 0.17 | 3 | same | – | 203,189 |
| 5 | ATRX | 23507450–23507752 | −0.26 | 10 | Sotos | 23507573–23507656 | −0.22 | 5 | same | PRDM9 | 1,682,751 |
| 15 | ATRX | 39871808–39872186 | −0.26 | 6 | Sotos | 39871876–39872186 | −0.16 | 5 | same | – | 341 |
| 3 | ATRX | 109056349–109056897 | 0.29 | 4 | Sotos | 109056349–109056897 | −0.26 | 4 | opposite | DPPA4 | 219,259 |
| 6 | ATRX | 34499314–34499504 | −0.28 | 4 | Sotos | 34499314–34499504 | −0.17 | 4 | same | PACSIN1 | 0 |
| 16 | ATRX | 58019866–58019984 | −0.37 | 3 | Sotos | 58019866–58019984 | −0.27 | 3 | same | TEPP | 0 |
| 10 | ATRX | 43698834–43699070 | −0.33 | 3 | Sotos | 43698834–43699070 | −0.32 | 3 | same | RASGEF1A | 656 |
| 18 | ATRX | 77905119–77905751 | −0.18 | 10 | Sotos | 77905663–77905751 | 0.16 | 4 | opposite | PARD6G-AS1 (56bp away) | 0 |
| 1 | ATRX | 228291486–228291705 | 0.23 | 5 | Sotos | 228291591–228291682 | 0.18 | 3 | same | – | 301 |
| 18 | ATRX | 74499372–74499584 | 0.2 | 5 | Sotos | 74499372–74499584 | −0.17 | 5 | opposite | – | 0 |
| 1 | ATRX | 3634867–3635100 | −0.22 | 4 | Sotos | 3634867–3635100 | −0.2 | 4 | same | TP73 | 0 |
| 7 | GTPTS | 27182493–27185282 | −0.36 | 51 | Kabuki | 27182493–27183816 | 0.15 | 18 | opposite | HOXA5 | 0 |
| 7 | GTPTS | 27182493–27185282 | −0.36 | 51 | CHARGE | 27182493–27183946 | 0.23 | 20 | opposite | HOXA5 | 0 |
| 14 | GTPTS | 54418728–54418851 | −0.38 | 3 | SBBYSS | 54418728–54418851 | −0.2 | 3 | same | BMP4 | 0 |
Figure 4.
Hypomethylation of the 5′ UTR of PRDM9 in Both Sotos-Affected and ATRX-Affected Subjects
The figure illustrates a 302-base pair region containing 10 CpG probes overlapping 5′ UTR of PRDM9. From top to bottom: chromosome ideogram, CpG probes, gene region, and methylation level data. Blue, ATRX; green, Sotos; pink, controls; line, average methylation; shadow, 95% confidence interval; dots, methylation values from every single sample (0-1).
Overall, our analyses showed limited overlap between the epi-signatures, which tend to occur only in a few genomic coordinates. Within these regions, the methylation levels do not correlate well across the diseases, and in many cases the direction of methylation change (hypo- versus hypermethylation) is opposite. These results suggested that it might be possible to combine all of the epi-signatures for building a single classification model for concurrent classification of all of the diseases.
Development and Validation of a Classification Model for Prediction of Disease Classes
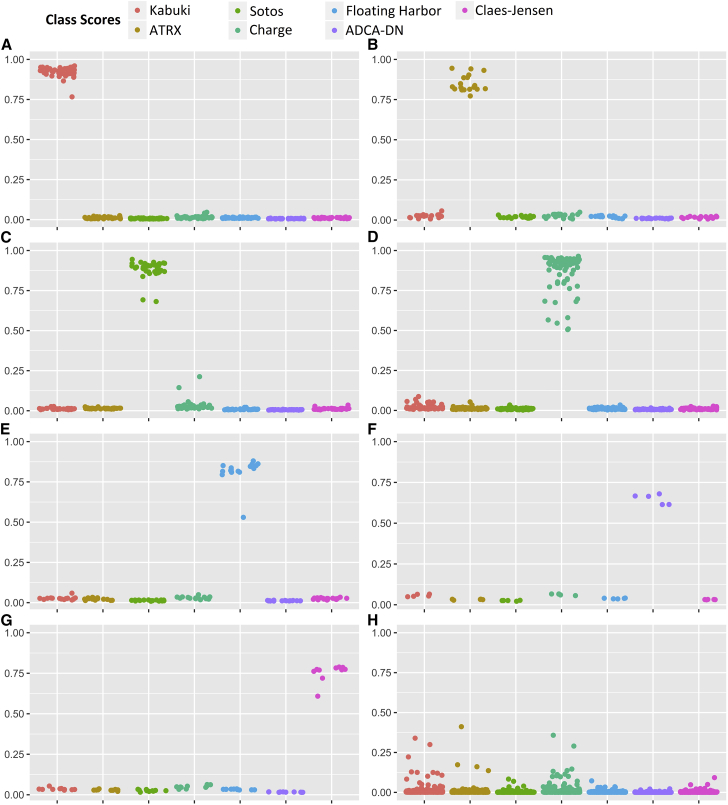
To develop a classification model, a multi-class support vector machine (SVM) with linear kernel was trained using a subset of 929 of the most differentiating and non-redundant probes selected from the epi-signature of every syndrome (Table S5). Due to small sample size for GTPTS (n = 3) and SBBYSS (n = 1), these conditions were not included in the model, and thus the training was performed only for the remaining seven syndromes. One model was trained for all of the affected subjects from these seven syndromes (n = 141) and the control subjects. Only the samples from the discovery/training cohort were used for training. The model was set to generate seven classification scores between 0 and 1 as the probability of having a methylation profile related to any of the seven syndromes. Ten-fold cross-validation of this model revealed an accuracy of 99.6%, and it correctly predicted the class of all of the 141 affected subject that were used for its training (Figures 5A–5G).
Figure 5.
Probability Scores Generated by the Classification Model
A 7-disease SVM classifier concurrently generates seven scores for every subject as the probability of having a DNA methylation profile similar to any of the seven diseases with a confirmed DNA methylation signature. y axis represents scores 0-1, with higher scores indicating a higher chance of carrying a methylation profile related to any of the seven conditions. x axis represents the seven classification scores generated for the same group of tested subjects. These include the probability of having a similar DNA methylation profile to Kabuki syndrome, ATRX, Sotos syndrome, CHARGE syndrome, Floating-Harbor syndrome, ADCA-DN, and Claes-Jensen, respectively. By default, the SVM classifier defines a cut-off of 0.5 for predicting the class; however, the vast majority of the tested individuals received a score close to 0 or 1. Therefore, for the purpose of better visualization, the points are jittered. Every point represents the probability score received for a single sample. This figure represents scores obtained by both the subjects in the training and testing cohorts. Shown are probability scores for belonging to any of the seven classes for: 44 subjects with Kabuki syndrome (A); 19 subjects with ATRX syndrome (B), 38 subjects with Sotos syndrome (C), 79 subjects with CHARGE syndrome (D), 17 subjects with Floating-Harbor syndrome (E), 5 subjects with ADCA-DN (F), and 10 subjects with intellectual disability due to KDM5C (G). The last panel (H) shows the probabilities of belonging to any of the seven disease groups for 436 subjects with other conditions presenting with DD/ID including diseases of epigenomic machinery for which no epi-signature was found, multiple chromosomal aberrations, Down syndrome, various forms of RASopathies, autism spectrum disorders, and imprinting defect conditions, together with 190 healthy control subjects which were not used in any previous step in the study.
To determine the sensitivity of our model, 71 subjects from the testing cohort were supplied to the classification model. All of these subjects received a score of close to 1 for the syndrome to which they belonged to but a low score for the other six conditions, showing that our model is 100% sensitive for detecting any of the seven conditions examined (Figures 5A–5G). To estimate the specificity of our method, we used 190 normal samples from our reference cohort that were not used for epigenomic profiling of any of the diseases in this study or training the model. All of these samples received scores close to zero by our model for all of the seven disease groups, showing a specificity of 100% (Figure 5H).
To determine whether this classification algorithm is sensitive to the composition of blood cell types, we downloaded a total of 60 methylation array data from 6 healthy individuals,16 each being assayed for whole blood, peripheral blood mononuclear cells (PBMCs), and granulocytes, as well as for seven isolated cell populations (CD4+ T cells, CD8+ T cells, CD56+ NK cells, CD19+ B cells, CD14+ monocytes, neutrophils, and eosinophils). The methylation data from these 60 samples were imported into our classification model. Our algorithm assigned a score of close to zero for having any of the seven conditions to all of the files, suggesting that the composition of blood cells will not influence the performance of our model. The scores obtained in this assessment are presented in Table S6.
The 7-Disease Classification Model Is Not Sensitive to Other Developmental Conditions
Next, we examined whether this classification model could distinguish between the epigenomic profile of the seven conditions and the five other previously tested syndromes (Saethre-Chotzen, Coffin-Siris, Coffin-Lowry, Rett, and Weaver syndromes) for which we did not find an epi-signature. Hence, the model was supplied with all of the 69 samples with pathogenic mutations in genes from these five syndromes, as well as to 55 subjects with clinical features similar to these conditions but with VUS variants in the related genes. All of these subjects received low scores for any of the seven disease classes in the algorithm. Next, to determine the performance of this classifier in subjects with developmental delay/intellectual disability (DD/ID) of other etiologies, the model was supplied with the methylation levels of additional cohorts composed of individuals with autism spectrum disorders (n = 146), various chromosomal abnormalities (n = 12), Down syndrome (n = 7), imprinting conditions (Angelman, Beckwith-Wiedemann, Prader-Willi syndrome, n = 50), and various forms of RASopathies (n = 97). All of these subjects had a confirmed clinical and molecular diagnosis of their respected conditions. We found that similar to our previous observations, each of these subjects received low scores for all of the seven diseases in our classification model, further demonstrating that the epi-signatures of these seven conditions are highly specific. These results are demonstrated in Figure 5H.
Classification of Variants of Unknown Significance, Healthy Carriers, Atypical Case Subjects, and Non-blood Methylation Profile
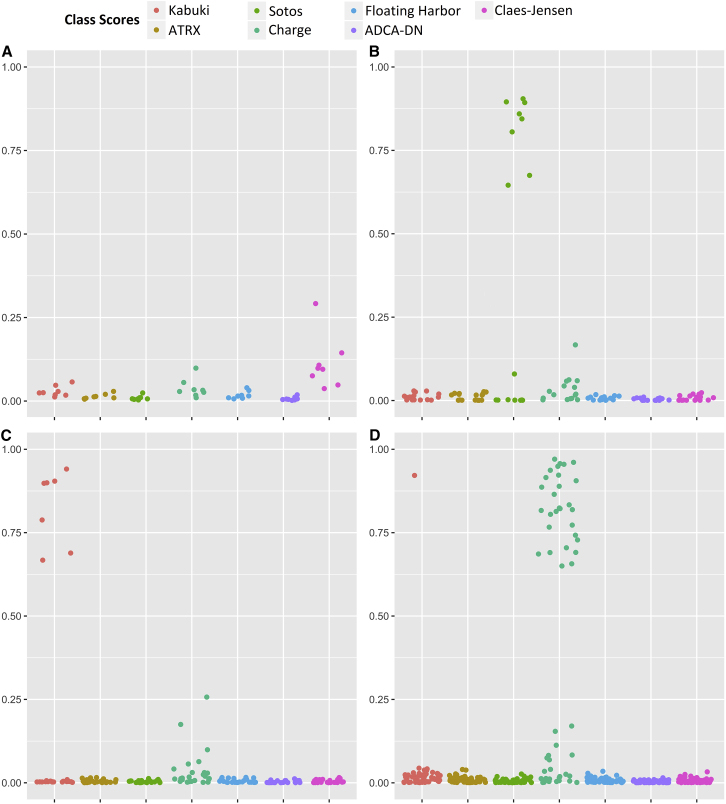
We next applied our classification model to a heterogeneous group of subjects (n = 176) from our cohort displaying some clinical features related to multiple of the aforementioned developmental conditions for which the variant pathogenicity was uncertain (Table S7; Figure 6), in addition to a few other samples from sources other than blood, healthy carriers, and a subject with Kabuki syndrome resulting from KDM6A mutations. Of the 36 subjects within this cohort with clinical features of Kabuki syndrome but with benign or VUS variants in KMT2D, seven were predicted to have a DNA methylation profile specific to Kabuki syndrome. The only subject with Kabuki syndrome with a KDM6A mutation also received a high classification score for Kabuki syndrome. In this cohort, there was also one individual with Floating-Harbor syndrome with a discordance between the predicted and the reported sex. This subject was predicted to belong to the Floating-Harbor class by our model. Later assessment confirmed that it was only a sample-labeling error. Our data contained healthy female carriers of pathogenic mutations in KDM5C (n = 8). All of these subjects received low scores for any of the seven conditions, including the Claes-Jensen syndrome. Two subjects with similar clinical presentations to ATRX but with no confirmed variants in the ATRX gene were predicted as not having any of the seven diseases including ATRX. Half of the 16 samples with VUS in NSD1 were predicted, based on the methylation score, as having ATRX. Our data contained four subjects suspected of having CHARGE syndrome for which sequence analysis results were not available at the time of the study, but who were all predicted to have CHARGE syndrome by our methylation prediction model. Subsequent sequence analysis reports of CHD7 for these four subjects showed that truncating exon-intron boundary mutations exist in two of them (GenBank: NG_007009.1, c.4533+1G>A [p.(=)]; GenBank: NG_007009.1, c.6937−1G>A [p.(=)]). The other two subjects shared one splice site mutation (GenBank: NG_007009.1, c.5405−17G>A [p.(=)]) which has previously been reported in CHARGE syndrome and confirmed to induce aberrant splicing using RNA-seq analysis in another subject with CHARGE syndrome. Of the remaining 51 subjects with similar phenotypic presentations to CHARGE syndrome, for whom CHD7 mutation screening had not provided a conclusive result, 23 were predicted as CHARGE, 27 were assigned low likelihood for all of the seven conditions, and 1 was predicted as having a methylation profile similar to Kabuki syndrome. Further follow-up and sequencing of this subject identified a pathogenic truncating variant (p.Arg5097∗ [c.15289C>T]; GenBank: NG_027827.1) in KMT2D, which confirmed the diagnosis of Kabuki syndrome. The three fibroblast samples from the Sotos-affected subjects were each predicted to have a similar methylation profile to the peripheral blood DNA epi-signature of Sotos syndrome. These findings recommended the high capacity of our method to assign new molecular diagnoses to subjects for whom the sequence analysis was not available or not interpretable, or the initial clinical suspicion was not correct. The complete classification outcome for these subjects is presented in Figure 6 and Table S7.
Figure 6.
Probability Scores Generated by the Classification Model for Case Subjects with Uncertain Diagnosis Carrying Benign or VUS Variants and Healthy Carriers
A 7-disease SVM classifier concurrently generates seven scores for every subject as the probability of having a DNA methylation profile similar to any of the seven diseases with a confirmed DNA methylation signature. y axis represents scores 0-1, with higher scores indicating a higher chance of carrying a methylation profile related to any of the seven conditions. x axis represents the seven classification scores generated for the same group of tested subjects. These include the probability of having a similar DNA methylation profile to Kabuki syndrome, ATRX, Sotos syndrome, CHARGE syndrome, Floating-Harbor syndrome, ADCA-DN, and Claes-Jensen, respectively. Every point represents the probability score obtained for a single sample. Shown are probability scores for belonging to any of the seven classes for: 8 healthy female carriers with pathogenic mutations in KDM5C (A), 16 subjects with VUS variants in NSD1 (B), 36 subjects with VUS and benign variants in KMT2D (C), and 55 subjects with similar features to CHARGE syndrome but no sequence data available or with VUS variants in CHD7 (D).
Discussion
Here we report syndrome-specific, minimally overlapping, DNA methylation epi-signatures in peripheral blood from a large cohort of individuals who possess pathogenic mutations in genes involved in the regulation of epigenomic machinery. Fourteen syndromes were represented in this cohort (Table 1), with reliable epi-signatures being observed for nine syndromes evaluated. We also describe here the development of a single classification algorithm for seven of these syndromes which has a highly sensitive and specific performance in predicting disease class, and which confidently rejects the probability of healthy individuals or other subjects with DD/ID to be affected by any of these seven conditions. Our results support the hypothesis that the disruption of normal functions by the genes that regulate epigenetic marks, particularly those associated with histone modifications, generate unique DNA methylation epi-signatures with minimal overlaps across the conditions.
Epigenetic marks are established and maintained by an intricate network of proteins that shape the epigenome. These proteins function as writers that establish the epigenetic marks, readers that interpret them, and erasers that remove the epigenetic marks.17 The fourth group of remodeller proteins comprises chromatin remodeling complexes that further regulate the epigenome to ensure the contextual (tissue-specific and temporal) accuracy of chromatin. Together, when properly integrated, these highly coordinated and interactive proteins act within multi-protein, multifunctional complexes on chromatin to ensure the tissue- and lineage-specific transcriptional activation and suppression that is essential for proper embryonic and fetal development.13 Mutations in the genes encoding these proteins can compromise the functional complexity and integration of the epigenetic machinery, lead to errors in the epigenetic signatures on chromatin, and result in a broad range of pediatric genetic disorders,2, 3 including those investigated in the present study. Thus, while the original genetic mutation in that particular epigenetic regulator may be primarily detrimental to normal neurodevelopment (and be a significant contributor to that clinical phenotype), all cells may carry an epigenetic echo (or legacy) of that original genetic defect in the form of a DNA methylation signature that may also contribute to the clinical phenotype. This association between a primary mutation in a gene encoding a regulator of chromatin accessibility and a secondary pattern of DNA methylation is likely due to the cross-talk between those interconnected pathways responsible for the post-translational histone modifications and DNA methylation.17
In the context of the cohort addressed in this paper, ADCA-DN and Rett syndrome resulted from mutations in proteins that write (DNMT1) and read (MeCP2) methylation marks on the DNA. Other syndromes resulted from errors in writers (KMT2D, NSD1, and KAT6B) and erasers (KDM5C) of the histone marks, and chromatin remodeling proteins (CHD7, SRCAP, ATRX). Our results show that syndromes caused by defects in chromatin remodeling proteins (CHARGE, Floating-Harbor, and ATRX), DNA methylation writer (ADCA-DN), histone mark erasers (Claes-Jensen syndrome), and histone mark writers (GTPTS, SBBYSS, Sotos, and Kabuki) generate specific DNA methylation signatures. On the other hand, in case subjects where the defect occurs in reading the methylation marks, such as in Rett syndrome, our study did not observe any consistent change in DNA methylation patterns, suggesting that the mutations in the multifunctional MeCP2 protein that lead to extensive temporal and spatial errors in levels of metabolites and gene expression across the entire genome, affect the function of multiple target pathways across cell lineages, rather than related to any lineage-specific epi-signature;18 however, a possibility of a methylation signature in an alternate tissue cannot be discounted.
Overlap in the DNA Methylation Signatures Is Limited to the Initiating Event
Phenotypic overlap is a common finding in various conditions resulting from the disruption of the epigenomic machinery. Clinical overlap between CHARGE and Kabuki syndrome and the difficulty in distinguishing between the two conditions by facial features alone has been reported.19 Sotos syndrome can be challenging to differentiate from Weaver or Beckwith-Wiedemann syndromes.20 Similarly, Floating-Harbor syndrome exhibits some clinical overlap with Rubinstein-Taybi syndrome (MIM: 180849) (another disease of epigenomic machinery) as a result of the molecular interaction between the genes causing the two syndromes.21 Bjornsson et al.2 suggest that the phenotypic overlap that is seen in some imprinting disorders and a group of multiple congenital anomalies and DD/ID syndromes also result from mutations in the genes encoding components of the epigenetic machinery. Thus, overlap in these shared molecular targets and biological pathways may be the basis for phenotypic overlap in many of such conditions. The shared presence of a small number of genes known to be crucial to embryonic development within these DNA methylation profiles of the conditions we evaluated has not been thoroughly investigated. Butcher et al.9 showed a gain of methylation in HOXA5 in both Kabuki and CHARGE syndromes, which was also observed in this study (Table 2). Additionally, we observed the same region to be significantly hypomethylated in subjects with GTPTS. Butcher et al.9 also report a segment in the MYO1F (MIM: 601480) gene body to be the most hypomethylated region in Kabuki syndrome-affected individuals. Our previous study on subjects with Floating-Harbor syndrome found the same region to be significantly hypomethylated.5 Our current data also shows that two CpG probes from the same MYO1F region are also differentially methylated in CHARGE syndrome (Table S2), although with an opposite direction of change (hypermethylation) to those seen in Kabuki and Floating-Harbor syndromes. Hence, the primary mutational events in epigenetic regulators associated with these syndromes may initiate aberrant DNA methylation and then hardwire these methylation changes into the epigenome as abnormal repressive (or activation) signals at loci encoding developmentally relevant transcription factors, many of which are observed in our study (Table 2) including WT1 (MIM: 607102) and MAMSTR (MIM: 610349) (Sotos), DPPA4 (MIM: 614125) (ATRX), and BMP4 (MIM: 112262) (GTPTS).
The concurrent screening of all of the single probes and regions with differential methylation levels in any of the diseases reported in this study have found that the existing overlap is limited to few specific regions and sites. Examination of DMRs, except for the segment mentioned above in HOXA5, did not reveal any region with a minimum of three consecutive probes to be shared by more than two conditions, and half of the shared regions (including HOXA5), showed an opposite direction of change in methylation levels. At the single CpG probe level, we found only 217 probes that were shared by more than two disease groups, the methylation levels of which were not strongly correlated in different conditions or were negatively correlated as observed between Sotos and Floating-Harbor, despite sharing the largest number of probes. Limiting the analysis to the probes shared by more than three conditions identified further discordance between the disorders (Figure 3). In addition, ADCA-DN shared the least number of probes and DMRs (only 1 DMR with opposite methylation change; Table 2) with other conditions, possibly due to the causative gene being a DNA methyltransferase versus other conditions resulting from histone modifications errors. Also, GTPTS and SBBYSS, which are caused by defects in the same gene, did not reveal any overlap in their epi-signatures and the total number of the probes they did share was less than a few dozen.
These limited shared probes were enriched and most frequently occurred in the genes related to laminin interaction (LAMB2/LAMC1), non-integrin membrane-ECM interactions (LAMB2/LAMC1/NRXN1), and synapse function (PTPRF/DLG4/NRXN1). Also, an intriguing finding was that multiple probes were shared across syndromes and were associated with genes functioning in pathways involving histone acetylation and methylation. Multiple ontology terms related to histone modifications were found to be enriched in the genes harboring these signatures, including S-methyltransferase activity and histone-lysine N-methyltransferase activities (Tables S3 and S4). These pathways involved three genes possessing histone methyltransferase activity (PRDM9, SETDB1, NSD1) and one coding for a subunit of histone H3 (HIST1H3E).
These results indicate that the shared pathways across these developmental syndromes include the initiating events leading to the disruption of the histone modification machinery. Once the initial mutation is established, each condition encompasses differential downstream paths leading to the generation of distinct epi-signatures. This is further supported by the single subject in our database with KDM6A mutation, whom we had predicted as having Kabuki syndrome, using the same model that is built on the epigenetic signature of KMT2D mutations. The immediate targets of these two genes are not overlapping; however, they mutually regulate a large number of genes downstream22 and their disruption leads to a single medical condition with indistinguishable clinical features. This further suggests that the epigenetic profile of these conditions is additionally composed of the downstream changes unique to the syndrome rather than the primary epigenomic event. In addition to the aforementioned components related to histone acetylation and methylation, the HOXA5 promoter was found to be differentially methylated in three syndromes, although with an opposite direction of methylation change. HOXA5 encodes a transcription factor that spatiotemporally regulates the body segmentation and morphogenesis during development. It is well recognized that its expression is tightly regulated by the methylation status of its promoter.23 Hypermethylation of HOXA5 promoter in CHARGE and Kabuki and its hypomethylation in GTPTS may partly explain the similarity in the phenotypic overlap between CHARGE and Kabuki syndromes, but not in GTPTS, supporting the concept that different epigenetic changes in a region might result in alternative phenotypic outcomes.
A Model Accounting for DNA Methylation Signatures as Legacies of Mutations in Genes of the Epigenetic Machinery
From our results, we propose a mechanistic model by which syndrome-specific DNA methylation signatures arise and are maintained. Central to our model is the concept that the initial dysfunction during early development, which is linked to a specific mutated epigenetic reader, writer, eraser, or remodeler, leaves a specific clinically identifiable and syndrome-related phenotype. In addition, a broader methylation legacy or “echo” remains throughout the genome. That epigenetic echo, reflected in (but not necessarily limited to) DNA methylation, may be a secondary contributor to the overall clinical phenotype through inappropriate activation (or repression) of gene expression in a cell-lineage-dependent manner. For example, Kabuki is among a number of syndromes (including CHARGE and Rubinstein-Taybi) that in addition to displaying clinical features of intellectual disability and distinct craniofacial features, can also present with immune dysfunction. This clinical feature can increase susceptibility to infections and an inability to generate or maintain immunological memory.24 Similarly, altered KMT2D function appears to impact cardiac development and cardiomyocyte function and may lead to defects in ion transport.24 Several of the target genes reported in these papers and associated with these physiological functions were detected in the epi-signatures reported here in our paper, supporting the efficacy and surrogacy of peripheral blood DNA as a source for identifying the Kabuki-specific epi-signature. Furthermore, our approach raises the potential to directly identify epigenetically silenced genes that could be the focus of targeted epigenetic or gene-editing therapies. Such therapies could be focused either on the primary mutation harbored by the epigenetic machinery gene25 or at the secondary targets that reside within the epi-signatures and display altered methylation within promoter or enhancer regions.
An additional application of the epigenetic signature detection extends beyond developmental syndromes to novel, targeted cancer therapy. In infrequent cases, young subjects with developmental syndromes including Kabuki, Coffin-Siris, and Sotos syndromes have been diagnosed with rare tumors at an early age.26, 27, 28 This is not surprising given that these very same epigenetic regulatory genes (KMT2D, NSD1, and SMARCB1) are frequently mutated in many somatic cancer types.17 Hence, such epigenetic information reflected by these epi-signatures has the potential to lead to the development of new targeted therapies for various adult cancers that are similarly associated with mutated genes encoding the epigenetic machinery.
DNA Methylation Signatures in Gene Defects across Different Tissues, Mutations, and Zygosity
A clinically applicable aspect of this study relates to whether DNA methylation signatures sourced from peripheral blood samples can act as a surrogate for aberrant DNA methylation patterns that have their primary functional consequences in the developing nervous system and brain. Previous studies have generated conflicting results on the correlation of DNA methylation patterns across tissues and cell types.29, 30, 31, 32 We were able to directly address this issue of tissue surrogacy, in that training our prediction algorithm on the DNA methylation signatures from peripheral blood of Sotos syndrome-affected individuals also assigned high scores for the three fibroblast DNA samples also obtained from Sotos syndrome-affected individuals. This suggests that easily accessible peripheral blood DNA can be functionally comparable to clinically relevant yet inaccessible target tissues such as the brain. Thus, from a developmental standpoint, some DNA methylation changes associated with primary defects in epigenetic machinery genes are well established before the differentiation of organs in embryonic development and are maintained in these particular cell lineages or have similar functions across tissues.
In any event, this has to be confirmed by studying multiple organs of the affected individuals in different disorders. The limited findings that we have presented in this regard comply with this observation. While ADCA-DN occurs due to one mutation present in all organs, the disease exclusively exhibits features of the nervous system involvement. Bjornsson et al.,2 based on this observation, postulates that various tissues show a specific and dosage-dependent response to disruption of epigenomic machinery. One observation in our study that may support this model is that the predictive algorithm that we trained using the epi-signature of the Claes-Jensen syndrome cannot detect any of the eight healthy female carriers of KDM5C pathogenic mutations. This indicates that the disruption of epigenetic machinery will be well represented in all tissues, but will lead to a functional disruption only when the dosage is beyond a critical point. This supports the clinical utility of peripheral blood as a surrogate tissue for screening for these conditions. Our study also suggests that the observed signature can be specific to the mutation type. GTPTS and SBBYSS are both caused by mutations in KAT6B. Depending on the location of the coding sequence and type of the mutation (i.e., missense versus frameshift), the gene product can be absent (loss of function) or abnormal (change of function), resulting in two distinct syndromes with overlapping features.33 Our data show that these two conditions, despite resulting from mutations in the same gene, have distinct epi-signatures. This finding suggests that epi-signatures can potentially be useful in differentiating distinct diseases with overlapping features that are caused by a shared gene defect, where the gene sequencing alone cannot always assign a diagnosis.
The Clinical Implications of the Multi-class DNA Methylation Classification Model
The minimal overlap in the DNA methylation signatures of each of the seven conditions has allowed for training a classifier with the capacity of concurrent assessment of any given subject for all of the seven disease groups with a complete accuracy, sensitivity, and specificity. Furthermore, using datasets generated across multiple institutions, we have demonstrated generalized applicability of this approach in clinical setting that is not sensitive to the confounders such as batch effect. The model not only distinguished related disorders of the epigenetic machinery from each other, but also assigned low probability scores to subjects with various forms of DD/ID ranging from autism spectrum disorders to chromosomal aberrations and imprinting defect conditions that are not caused by mutations in these genes. This method will have a practical value in the clinical diagnosis of such genetic conditions where the disease is rare, clinical features are overlapping, and sequence mutation screening does not always produce conclusive results. In Kabuki syndrome, for instance, the mutation screening of KDM6A and KMT2D, together, identifies pathogenic mutations in only 70% of subjects.34 A similar figure of 65%–70% is reported for atypical cases of CHARGE syndrome.35 The vast majority of rare missense and in-frame in/del variants are currently not interpretable, and as VUSs present a significant challenge in clinical interpretation in diagnostic laboratories and genetic clinics. Some subjects also may carry pathogenic mutations in noncoding regions which are often not screened (e.g., promoter, intronic regions).
The method presented here provides an innovative approach to assigning pathogenicity to these variants by using the epi-signatures of each condition studied. Although the findings would have to be complemented with further clinical assessment, our model has shown its capacity to resolve a large proportion of the unsolved cases in our dataset by assigning new classifications to more than half of the subjects for whom sequence variant assessment alone had not provided a definitive answer. Of interest was a 4-year-old male with cleft palate, dysmorphic ears, microcephaly, and developmental delay, who was suspected of having CHARGE syndrome and who was screened for mutations in CHD7 where only a heterozygous missense VUS variant (GenBank: NM_017780.3, c.2185A>G; GenBank: NM_017780.3, p.Lys729Glu) was found. The classification model we have described above predicted that this subject had an epigenomic profile similar, not to CHARGE syndrome (score = 0.02), but to Kabuki syndrome (score = 0.92). This subject was later confirmed to carry a pathogenic truncating variant in KMT2D and was subsequently diagnosed with Kabuki syndrome. It is worthwhile to note that in many of these conditions, such as Kabuki syndrome, the disease is often caused by de novo variants. Identification of a de novo variant increases the chance of the mutation being pathogenic, although this is not always sufficient to assign pathogenicity to a variant. Unlike the assessment of the “de novo” status of a genetic variant, DNA methylation analysis does not require parental sample DNA to assess the impact of the variant in the proband. However, where both analyses are available, combined evidence could be used to further strengthen the case for variant pathogenicity status.
With more samples becoming available from such conditions and more diseases being studied for the discovery of epi-signatures, the methodology that we have presented here can provide the basis of application in routine molecular diagnostics. Microarray technology has been shown to be an invaluable tool for the diagnosis of epigenetic conditions including imprinting disorders,36 and as we have shown in the present study, a combination of microarray technology with machine learning will have a great, and as yet relatively unexplored, potential in resolving a large number of subjects with unsolved etiology that are frequently seen in medical genetics practice.
Acknowledgments
We thank the families, the molecular genetics diagnostic laboratory at the Greenwood Genetic Center for identifying the mutations in the subjects, and the clinical geneticists at the Greenwood Genetic Center for making the clinical diagnosis. We also thank the Care4Rare Canada Consortium for the identification and molecular analyses of subjects with a subset of the neurodevelopmental conditions that were part of this study. E.A.-E. was supported by Children’s Health Research Institute Epigenetics Trainee Award, funded by the Children’s Health Foundation, London, Ontario, Canada. Dedicated to the memory of Ethan Francis Schwartz, 1996–1998.
Published: January 4, 2018
Footnotes
Supplemental Data include seven tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.12.008.
Web Resources
Supplemental Data
References
- 1.Fahrner J.A., Bjornsson H.T. Mendelian disorders of the epigenetic machinery: tipping the balance of chromatin states. Annu. Rev. Genomics Hum. Genet. 2014;15:269–293. doi: 10.1146/annurev-genom-090613-094245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjornsson H.T. The Mendelian disorders of the epigenetic machinery. Genome Res. 2015;25:1473–1481. doi: 10.1101/gr.190629.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berdasco M., Esteller M. Genetic syndromes caused by mutations in epigenetic genes. Hum. Genet. 2013;132:359–383. doi: 10.1007/s00439-013-1271-x. [DOI] [PubMed] [Google Scholar]
- 4.Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 5.Hood R.L., Schenkel L.C., Nikkel S.M., Ainsworth P.J., Pare G., Boycott K.M., Bulman D.E., Sadikovic B. The defining DNA methylation signature of Floating-Harbor syndrome. Sci. Rep. 2016;6:38803. doi: 10.1038/srep38803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kernohan K.D., Cigana Schenkel L., Huang L., Smith A., Pare G., Ainsworth P., Boycott K.M., Warman-Chardon J., Sadikovic B., Care4Rare Canada Consortium Identification of a methylation profile for DNMT1-associated autosomal dominant cerebellar ataxia, deafness, and narcolepsy. Clin. Epigenetics. 2016;8:91. doi: 10.1186/s13148-016-0254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schenkel L.C., Kernohan K.D., McBride A., Reina D., Hodge A., Ainsworth P.J., Rodenhiser D.I., Pare G., Bérubé N.G., Skinner C. Identification of epigenetic signature associated with alpha thalassemia/mental retardation X-linked syndrome. Epigenetics Chromatin. 2017;10:10. doi: 10.1186/s13072-017-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choufani S., Cytrynbaum C., Chung B.H.Y., Turinsky A.L., Grafodatskaya D., Chen Y.A., Cohen A.S.A., Dupuis L., Butcher D.T., Siu M.T. NSD1 mutations generate a genome-wide DNA methylation signature. Nat. Commun. 2015;6:10207. doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butcher D.T., Cytrynbaum C., Turinsky A.L., Siu M.T., Inbar-Feigenberg M., Mendoza-Londono R., Chitayat D., Walker S., Machado J., Caluseriu O. CHARGE and Kabuki syndromes: gene-specific DNA methylation signatures identify epigenetic mechanisms linking these clinically overlapping conditions. Am. J. Hum. Genet. 2017;100:773–788. doi: 10.1016/j.ajhg.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aref-Eshghi E., Schenkel L.C., Lin H., Skinner C., Ainsworth P., Paré G., Rodenhiser D., Schwartz C., Sadikovic B. The defining DNA methylation signature of Kabuki syndrome enables functional assessment of genetic variants of unknown clinical significance. Epigenetics. 2017;12:923–933. doi: 10.1080/15592294.2017.1381807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamamori Y., Sartorelli V., Ogryzko V., Puri P.L., Wu H.Y., Wang J.Y., Nakatani Y., Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 12.Sassone-Corsi P., Mizzen C.A., Cheung P., Crosio C., Monaco L., Jacquot S., Hanauer A., Allis C.D. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science. 1999;285:886–891. doi: 10.1126/science.285.5429.886. [DOI] [PubMed] [Google Scholar]
- 13.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schenkel L.C., Schwartz C., Skinner C., Rodenhiser D.I., Ainsworth P.J., Pare G., Sadikovic B. Clinical validation of fragile X syndrome screening by DNA methylation array. J. Mol. Diagn. 2016;18:834–841. doi: 10.1016/j.jmoldx.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Jaffe A.E., Murakami P., Lee H., Leek J.T., Fallin M.D., Feinberg A.P., Irizarry R.A. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int. J. Epidemiol. 2012;41:200–209. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinius L.E., Acevedo N., Joerink M., Pershagen G., Dahlén S.E., Greco D., Söderhäll C., Scheynius A., Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen H., Laird P.W. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehrhart F., Coort S.L., Cirillo E., Smeets E., Evelo C.T., Curfs L.M. Rett syndrome - biological pathways leading from MECP2 to disorder phenotypes. Orphanet J. Rare Dis. 2016;11:158. doi: 10.1186/s13023-016-0545-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel N., Alkuraya F.S. Overlap between CHARGE and Kabuki syndromes: more than an interesting clinical observation? Am. J. Med. Genet. A. 2015;167A:259–260. doi: 10.1002/ajmg.a.36804. [DOI] [PubMed] [Google Scholar]
- 20.Baujat G., Rio M., Rossignol S., Sanlaville D., Lyonnet S., Le Merrer M., Munnich A., Gicquel C., Colleaux L., Cormier-Daire V. Clinical and molecular overlap in overgrowth syndromes. Am. J. Med. Genet. C. Semin. Med. Genet. 2005;137C:4–11. doi: 10.1002/ajmg.c.30060. [DOI] [PubMed] [Google Scholar]
- 21.Hood R.L., Lines M.A., Nikkel S.M., Schwartzentruber J., Beaulieu C., Nowaczyk M.J., Allanson J., Kim C.A., Wieczorek D., Moilanen J.S., FORGE Canada Consortium Mutations in SRCAP, encoding SNF2-related CREBBP activator protein, cause Floating-Harbor syndrome. Am. J. Hum. Genet. 2012;90:308–313. doi: 10.1016/j.ajhg.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J.H., Sharma A., Dhar S.S., Lee S.H., Gu B., Chan C.H., Lin H.K., Lee M.G. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74:1705–1717. doi: 10.1158/0008-5472.CAN-13-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watson R.E., Curtin G.M., Hellmann G.M., Doolittle D.J., Goodman J.I. Increased DNA methylation in the HoxA5 promoter region correlates with decreased expression of the gene during tumor promotion. Mol. Carcinog. 2004;41:54–66. doi: 10.1002/mc.20043. [DOI] [PubMed] [Google Scholar]
- 24.Stagi S., Gulino A.V., Lapi E., Rigante D. Epigenetic control of the immune system: a lesson from Kabuki syndrome. Immunol. Res. 2016;64:345–359. doi: 10.1007/s12026-015-8707-4. [DOI] [PubMed] [Google Scholar]
- 25.Harmeyer K.M., Facompre N.D., Herlyn M., Basu D. JARID1 histone demethylases: emerging targets in cancer. Trends. Cancer. 2017;3:713–725. doi: 10.1016/j.trecan.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gossai N., Biegel J.A., Messiaen L., Berry S.A., Moertel C.L. Report of a patient with a constitutional missense mutation in SMARCB1, Coffin-Siris phenotype, and schwannomatosis. Am. J. Med. Genet. A. 2015;167A:3186–3191. doi: 10.1002/ajmg.a.37356. [DOI] [PubMed] [Google Scholar]
- 27.Kulkarni K., Stobart K., Noga M. A case of Sotos syndrome with neuroblastoma. J. Pediatr. Hematol. Oncol. 2013;35:238–239. doi: 10.1097/MPH.0b013e318279b232. [DOI] [PubMed] [Google Scholar]
- 28.Karagianni P., Lambropoulos V., Stergidou D., Fryssira H., Chatziioannidis I., Spyridakis I. Recurrent giant cell fibroblastoma: Malignancy predisposition in Kabuki syndrome revisited. Am. J. Med. Genet. A. 2016;170A:1333–1338. doi: 10.1002/ajmg.a.37584. [DOI] [PubMed] [Google Scholar]
- 29.Ma B., Wilker E.H., Willis-Owen S.A., Byun H.M., Wong K.C., Motta V., Baccarelli A.A., Schwartz J., Cookson W.O., Khabbaz K. Predicting DNA methylation level across human tissues. Nucleic Acids Res. 2014;42:3515–3528. doi: 10.1093/nar/gkt1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montaño C.M., Irizarry R.A., Kaufmann W.E., Talbot K., Gur R.E., Feinberg A.P., Taub M.A. Measuring cell-type specific differential methylation in human brain tissue. Genome Biol. 2013;14:R94. doi: 10.1186/gb-2013-14-8-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byun H.M., Siegmund K.D., Pan F., Weisenberger D.J., Kanel G., Laird P.W., Yang A.S. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum. Mol. Genet. 2009;18:4808–4817. doi: 10.1093/hmg/ddp445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barault L., Ellsworth R.E., Harris H.R., Valente A.L., Shriver C.D., Michels K.B. Leukocyte DNA as surrogate for the evaluation of imprinted loci methylation in mammary tissue DNA. PLoS ONE. 2013;8:e55896. doi: 10.1371/journal.pone.0055896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lonardo F., Lonardo M.S., Acquaviva F., Della Monica M., Scarano F., Scarano G. Say-Barber-Biesecker-Young-Simpson syndrome and Genitopatellar syndrome: lumping or splitting? Clin. Genet. 2017 doi: 10.1111/cge.13127. [DOI] [PubMed] [Google Scholar]
- 34.Bögershausen N., Gatinois V., Riehmer V., Kayserili H., Becker J., Thoenes M., Simsek-Kiper P.Ö., Barat-Houari M., Elcioglu N.H., Wieczorek D. Mutation update for Kabuki syndrome genes KMT2D and KDM6A and further delineation of X-linked Kabuki syndrome subtype 2. Hum. Mutat. 2016;37:847–864. doi: 10.1002/humu.23026. [DOI] [PubMed] [Google Scholar]
- 35.Zentner G.E., Layman W.S., Martin D.M., Scacheri P.C. Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am. J. Med. Genet. A. 2010;152A:674–686. doi: 10.1002/ajmg.a.33323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aref-Eshghi E., Schenkel L.C., Lin H., Skinner C., Ainsworth P., Paré G., Siu V., Rodenhiser D., Schwartz C., Sadikovic B. Clinical validation of a genome-wide DNA methylation assay for molecular diagnosis of imprinting disorders. J. Mol. Diagn. 2017;19:848–856. doi: 10.1016/j.jmoldx.2017.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.