ABSTRACT
Expression of virulence factors in Staphylococcus aureus is regulated by a wide range of transcriptional regulators, including proteins and small RNAs (sRNAs), at the level of transcription and/or translation. The sarA locus consists of three overlapping transcripts generated from three distinct promoters, all containing the sarA open reading frame (ORF). The 5′ untranslated regions (UTRs) of these transcripts contain three separate regions ∼711, 409, and 146 nucleotides (nt) upstream of the sarA translation start, the functions of which remain unknown. Recent transcriptome-sequencing (RNA-Seq) analysis and subsequent characterization indicated that two sRNAs, teg49 and teg48, are processed and likely produced from the sarA P3 and sarA P1 transcripts of the sarA locus, respectively. In this report, we utilized a variety of sarA promoter mutants and cshA and rnc mutants to ascertain the contributions of these factors to the generation of teg49. We also defined the transcriptional regulon of teg49, including virulence genes not regulated by SarA. Phenotypically, teg49 did not impact biofilm formation or affect overall SarA expression significantly. Comparative analyses of RNA-Seq data between the wild-type, teg49 mutant, and sarA mutant strains indicated that ∼133 genes are significantly upregulated while 97 are downregulated in a teg49 deletion mutant in a sarA-independent manner. An abscess model of skin infection indicated that the teg49 mutant exhibited a reduced bacterial load compared to the wild-type S. aureus. Overall, these results suggest that teg49 sRNA has a regulatory role in target gene regulation independent of SarA. The exact mechanism of this regulation is yet to be dissected.
KEYWORDS: RNA-Seq, S. aureus, SarA, biofilms, gene expression, gene regulation, mouse model, small RNAs
INTRODUCTION
Staphylococcus aureus is both a commensal and an opportunistic pathogen that causes a broad range of human and animal infections. The diseases can range from acute food poisoning, pneumonia, meningitis, superficial skin infections (e.g., acne, boils, or cellulitis), arthritis, osteomyelitis, or endocarditis to toxic shock syndrome (1, 2). Besides demonstrating a capacity to rapidly develop resistance to many antibiotics, including vancomycin, S. aureus has an ability to thwart the innate host defense systems, included those provided by neutrophils and macrophages (3, 4). In addition, staphylococcal infections are common after viral infections. Not surprisingly, S. aureus infections are frequently associated with high mortality, morbidity, and health-related costs.
The pathogenesis of S. aureus is a complex process that involves multiple virulence factors, specifically, surface proteins, invasive enzymes and proteases, immunological disguises, membrane-damaging toxins, exotoxins, and inherent and acquired resistance to antimicrobial agents (1, 5). The expression of many virulence factors is primarily controlled by global regulatory systems, including two-component regulatory systems (TCS) (e.g., agr, lytSR, and saeRS) and transcriptional regulators (e.g., at least 10 SarA family proteins, tcaR, and sigB) (6–9). Among these, the best characterized are the agr locus, which represents a prototypic two-component quorum-sensing system (10, 11), and the sarA locus (12), which can regulate directly and indirectly a large number of genes involved in virulence, autolysis, biofilm formation, stress responses, antibiotic resistance, and metabolic processes (6, 8, 13–15).
The sarA locus is a complex regulatory system comprising three promoters (P2, P3, and P1) that yield three distinct but overlapping transcripts (sarA P2, sarA P3, and sarA P1), each containing the sarA open reading frame (ORF) (16). The expression of these transcripts is known to be growth phase dependent, with the P2 and P1 promoters being σA dependent and the σB-dependent P3 promoter expressed during the postexponential phase of growth (16, 17). Although the expression of the three transcripts varies with the growth phase, the overall level of SarA production remains relatively constant until the cell culture reaches the stationary phase of growth, when SarA expression appears to be higher, presumably as the summation of SarA translation from the combined sarA transcripts (18, 19). The complexity of sarA regulation is due in part to the presence of an unusually long (∼850-bp) region upstream of the sarA coding sequence that is required for optimal expression of SarA and its target genes throughout the growth cycle (20).
Recently, it was shown that a number of small RNAs (sRNAs) are involved in the regulation of virulence genes in various organisms, including S. aureus (21–24). Small RNAs are classified based on their target regulation: cis-acting sRNAs, such as in binding to the complementary mRNA (antisense), or trans-acting, such as those that bind target RNA or proteins at distal sites (21). There are at least 303 potential sRNAs in various S. aureus genomes, only 92 of which have been positively confirmed by independent experimental methods (24, 25). The most studied sRNA in S. aureus is agr RNAIII, which is the effector of the agr system through competitive binding to mRNA of the target genes (26), including mgrA, hla, and spa mRNAs (22, 27). There are several well-characterized sRNAs, including rsaE, which coordinates the downregulation of oppB and opp-3A (amino acid and peptide transporter) and sucC (succinyl-coenzyme A synthase) mRNAs to inhibit translation (28); artR, which is involved in activating alpha toxin expression by targeting the 5′ untranslated region (UTR) of sarT mRNA (29); and sprD, which represses the translation of sbi mRNA (an immunoglobulin binding protein) (30). Another unique sRNA is the dual-function PSM-mec gene, which confers methicillin resistance through the cytolytic toxin PSMα and inhibits the translation of agrA mRNA through 5′-end base pairing (31). Transcriptome-sequencing (RNA-Seq) analysis of the sarA locus disclosed two sRNAs, teg49 and teg48, with teg49 residing within the sarA P3 and P1 promoter region and involved in virulence gene expression (25, 32).
In this study, we constructed various sarA promoter and cshA and rnc (RNase III) mutant strains to understand the mechanism of teg49 generation in S. aureus. We also evaluated the transcriptome of the teg49 mutant, showing both SarA-dependent and SarA-independent gene regulation. These results indicated that the overall expression of SarA and biofilm formation were not significantly altered due to deletion of the teg49 region. RNA-Seq analyses of the teg49 mutant compared to wild-type (wt) and sarA mutant strains indicated that about 133 upregulated genes and 97 downregulated genes are specifically regulated by teg49. RNA-Seq data also revealed that 10 putative regulatory (e.g., saeRS and rbsU) and 34 putative virulence (e.g., sbi, map, lukSF, coa, setnM, and smc) genes are regulated by teg49, in addition to a large number of metabolic genes and genes with unknown functions. Validation of potential teg49-regulated genes by quantitative real-time PCR (qRT-PCR) confirmed the validity of the alteration of gene expression. The in vivo significance of the teg49 mutant, along with the hairpin loop 1 (HP1) mutant, of teg49 was assessed using the murine model of skin abscess infection; the results indicated that the teg49 mutant showed a reduced bacterial load in infected skin tissues versus the parental strain. Overall, these results suggest that teg49 sRNA has a regulatory role in target virulence gene regulation in S. aureus, although the exact mechanism of its action has yet to be dissected in detail.
RESULTS
Analysis of factors involved in the generation of teg49 sRNA.
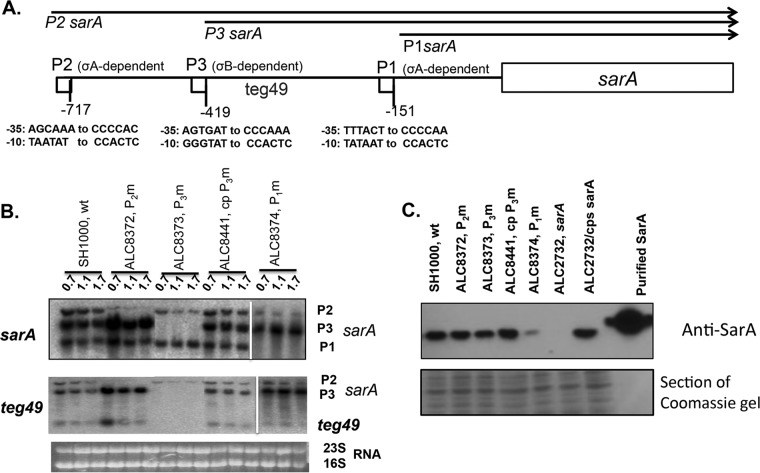
In prior studies, Beaume et al. (25) identified, by RNA-Seq of strain N315, two sRNAs, teg49 and teg48, within the sarA locus. In subsequent studies, we demonstrated the existence of teg49 and teg48 sRNAs within the sarA locus by Northern blotting and primer extension studies in strain Newman (32). The teg49 sRNA, located within the sarA P3-P1 promoter region (Fig. 1A), is 196 nucleotides (nt) in length. To determine precisely the origin of the teg49 sRNA, we first constructed various sarA promoter mutants in the SH1000 background. Accordingly, the −35 and −10 promoter sequences of all three sarA transcripts, P2, P3, and P1, were altered by site-directed mutagenesis, as shown in Fig. 1A. Promoter-specific knockout mutants were then constructed, utilizing the temperature-sensitive origin-of-replication shuttle vector pMAD, and verified by PCR, DNA sequencing, and Northern analysis, as described in Materials and Methods. To verify the mutation, we ascertained that the corresponding P2, P3, or P1 transcript was absent or markedly reduced in the respective promoter mutants (ALC8372 [P2m], ALC8373 [P3m], and ALC8374 [P1m] mutants) when probed with a fragment containing the sarA ORF (Fig. 1B, top). These mutants were then analyzed for expression of the teg49 sRNA. Importantly, a band corresponding to the teg49 sRNA was absent in ALC8373 (the P3 promoter mutant) but was present in ALC8372 (the P2 promoter mutant) and ALC8374 (the P1 promoter mutant), while the restored P3 promoter strain (ALC8441) was able to reestablish both the P3 sarA mRNA and the teg49 sRNA (Fig. 1B, bottom). These data indicate that the teg49 transcript likely originates from the P3 sarA mRNA, presumably via specific cleavage.
FIG 1.
Expression of the sarA transcripts and teg49 sRNA from sarA promoter mutants at various phases of growth in S. aureus. (A) Schematic representation of the sarA locus showing various transcripts (sarA P2, sarA P3, and sarA P1) that originate from the P2, P3, and P1 promoters (arrows), respectively. The sarA open reading frame is indicated by a box, and the promoters are indicated by small boxes. The numbers of the promoter locations are based on the start codon (ATG) of the sarA gene. (B) Northern blot analysis of total RNA isolated from the wild-type SH1000 and the various isogenic promoter mutants at various phases of growth recorded as OD600 values (OD600 values of 0.7, 1.1, and 1.5 represent mid-log, late log, and early stationary phases, respectively). In all the gels, 10 μg of total cellular RNA was loaded onto each lane, and the blots were probed with a 375-bp sarA DNA probe containing the sarA ORF (top) or a 180-bp DNA fragment containing teg49 (middle). The 16S and 23S rRNAs of the ethidium bromide-stained gel used for blotting is also shown as a loading control at the bottom. The spliced images were taken from different regions of the same image. (C) Western blot analysis for SarA, with anti-SarA antibody, of the wild type, its isogenic promoter mutants, the sarA mutant, and a complemented mutant. Equivalent amounts of extracts (10 μg) from the late exponential phase of growth (OD600 ≈ 1.1) were used to detect SarA protein. (Bottom) A Coomassie blue-stained duplicate-run gel used for blotting is shown as a loading control.
The lack of teg49 in the sarA P3 promoter mutant suggests that the sarA P3 transcript may be processed to yield teg49, a stable sRNA species that can be detected by Northern, primer extension, and real-time PCR (RT-PCR) analyses (32). One possibility is that this processing step may involve a specific endoribonuclease(s). To verify this possibility, we examined the expression of the teg49 sRNA in putative RNase mutants in the S. aureus JE2 library from the University of Nebraska (33). As shown in Fig. 2A, only the RNase III (rnc) mutant of JE2 failed to express teg49 among these putative RNase mutants, while an elevated level of expression was observed in ΔnrdD (anaerobic ribonucleotide reductase large subunit; SauUSA300_2551), Δrny (RNaseY; SAOUHSC_01179), and Δendo-RNase L-PSP (putative endoribonuclease L-PSP; SauUSA300_0474) mutants. There were no significant reductions in the levels of teg49 in nine other mutants, i.e., ΔSauUSA300_1184 (a conserved hypothetical protein similar to thiamine binding protein), ΔpnpA (polyribonucleotidyltransferase; SauUSA300_1167), ΔnrdG (anaerobic ribonucleotide reductase small subunit; SauUSA300_2550), ΔSauUSA300_2328 (conserved hypothetical protein), ΔSauUSA300_2021 (S1 RNA binding domain protein Tex), ΔxseA (exodeoxyribonuclease VII large subunit; SauUSA300_1472), ΔSauUSA300_1692 (conserved hypothetical protein), Δrnr (RNase R, SauUSA300_0764) mutnts and a control ΔSauUSA300_0308 (putative ABC transporter; permease) mutant. For confirmation, we constructed an rnc deletion mutant, as well as a trans-complemented mutant, in the SH1000 background. As can be seen in Fig. 2B (top), the expression of teg49 was markedly diminished in the rnc mutants and was restored to nearly parental level in the complemented mutant (ALC8445).
FIG 2.

Northern blots of diverse mutants to determine the expression of teg49 and teg23 sRNAs. (A) Northern blot analysis for the teg49 sRNA in the wild-type JE2 and its isogenic RNase and other putative mutants as indicated. (B) Northern blot analyses to determine the expression of sRNAs (teg49, teg23, and teg35) in the wild-type, cshA, hfq, rnc, and complemented rnc strains hybridized with radiolabeled 180-bp teg49, 190-bp teg23, and 225-bp teg35 probes. A total of 10 μg of cellular RNA from the various phases of growth was loaded onto each lane. An ethidium bromide-stained gel used for blotting, showing 16S and 23S rRNA bands, is shown as the loading control. The spliced images are taken from different regions of the same image. The numbers above the gels are OD600 values.
In an earlier study, we showed that CshA, a DEAD box RNA helicase, protects sarA mRNAs from the endoribonuclease cleavage of the mazF toxin in strain Newman (34). Contrary to the usual role of CshA as part of the degradosome to degrade mRNA, CshA has been linked to the expression and stability of teg49 and possibly 21 other sRNAs (out of 85 sRNAs examined), as detected by NanoString analysis in strain Newman (34). To confirm this finding in SH1000, we verified by Northern analysis that expression of teg49, and also teg23, another sRNA protected by CshA, was diminished in the cshA mutant versus the parent while expression of teg35, the control sRNA not affected by CshA (34), was found to be elevated in the cshA mutant versus the parent, as one would predict if teg35 is processed as part of the degradosome (Fig. 2B, bottom). Hfq, a chaperone that binds and stabilizes many sRNAs in Gram-negative species, did not have any major impact on the expression of teg49, teg23, and the control sRNA, teg35 (Fig. 2B). Notably, the expression of teg23 was also diminished in the Δrnc mutant of SH1000 and was restored to the parental level in the complemented mutant, while rnc had no effect on the expression of teg35.
Effect of the sarA P3 promoter on SarA expression.
The generation of teg49 from the sarA P3 transcript implied that a P3 promoter mutant would have a dual effect on reducing expression of sarA P3 mRNA and teg49. In our previous studies (32), our data seemed to suggest that teg49 may have regulatory activity on its own independent of sarA. To assess the dual effect of sarA P3 mRNA and teg49 in the P3 promoter mutant on SarA protein expression, immunoblots of whole-cell lysates of various promoter mutants, along with the wild-type and P3m revertant strains, were probed with monoclonal anti-SarA antibody. As shown in Fig. 1C, the P3m promoter mutant (ALC8373) exhibited a moderate reduction in SarA expression of 30% ± 15% at an optical density at 600 nm (OD600) of ∼1.1 compared to the wild type (set at 100% by ImageJ analysis), while the SarA protein level was restored in the revertant mutant (ALC8441). As a comparison, there was no significant decrease in the SarA protein level for the P2m promoter mutant (ALC8372) compared to the wild-type strain. However, a significant decrease in the SarA protein level was observed in the P1m promoter mutant (ALC8374), with a reduction of 75% ± 15% at an OD600 of ∼1.1 with respect to the wild-type strain (100%). As a control, the sarA deletion mutant did not exhibit any SarA protein expression, but this defect was restored to the parental level upon complementation (ALC2732). Together, these data revealed a moderate decrease in the SarA protein level in a P3 promoter mutant in which the sarA P3 mRNA and teg49 are absent. Given that the sarA P1 promoter mutant can express teg49 at a normal level (Fig. 1B), accompanied by significantly reduced SarA expression (Fig. 1C), it is unlikely that SarA is involved in the production of teg49.
Effects of teg49 on the sarA P3 transcript and SarA expression.
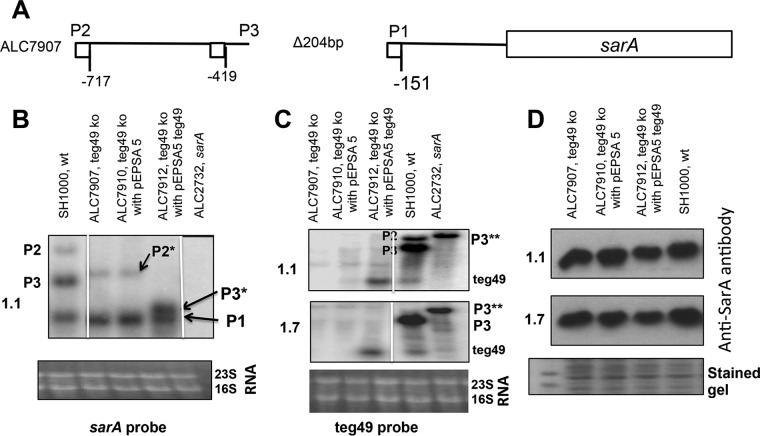
Given our finding on the SarA protein level in the sarA P3 promoter mutant, we wanted to dissect the role of teg49 in SarA expression. Accordingly, we constructed a 204-bp deletion mutant of teg49 (ALC7907) (Fig. 3A) and a trans-complemented mutant (ALC7912) by introducing a xylose-inducible promoter vector, pEPSA5, driving a 196-bp teg49 fragment (32). While the P1 sarA mRNA can be detected in assorted strains of the teg49 mutant, the P3 mRNA is noticeably absent in the teg49 mutants (ALC7907 and ALC7910) (Fig. 3B). To rule out the possibility that deletion of teg49 may disrupt the sarA P3 transcript, we trans-complemented the Δteg49 strain with the pEPSA5 plasmid driving the expression of teg49 (ALC7912). As shown in Fig. 3B, this strain expressed both the sarA P1 transcript (591 nt long) and a truncated P3 sarA transcript (∼650 nt long) due to the chromosomal teg49 deletion. This result indicated that deletion of teg49 affects the expression of the sarA P3 transcript and that overexpression of teg49 may help stabilize the P3 sarA transcript in a teg49 mutant strain; alternatively, teg49 may somehow affect sarA P3 promoter activity. As expected, the teg49 transcript was detected in the trans-complemented teg49 strain (ALC7912), but not in the Δteg49 mutant strain (Fig. 3C). To ascertain if deletion of teg49 would alter SarA protein expression independently of the sarA P3 promoter activity, we performed Western blot analysis of the whole-cell lysate from ALC7907 (Δteg49) and its derivative strains containing pEPSA5 (vector; ALC7910) or the complemented plasmid, pEPSA5::teg49 (ALC7912), along with the wild type, using monoclonal anti-SarA antibody 1F1. As shown in Fig. 3D, the changes in the SarA protein level for these strains in two growth phases were not significant (15% ± 10%), as measured by densitometric scanning with ImageJ. Overall, these results support the notion that the sarA P3 transcript contributed moderately to the expression of SarA inside the cell and that teg49 contained within the sarA P3 transcript is not a major contributor to SarA expression.
FIG 3.
Expression of sarA transcripts and teg49 sRNA in the teg49 mutant and its derivative strains at various phases of growth in S. aureus. (A) Schematic representation of the teg49 mutant showing the region deleted to construct the mutant (ALC7907). (B and C) Northern analysis of RNA isolated from the wild-type SH1000, teg49 mutant (7907), teg49 mutant with pEPSA5 (7910), and teg49 mutant with pEPSA5::teg49 and the sarA deletion mutant (ALC2732) at different growth phases. The blots were probed with a 375-bp DNA probe containing the open reading frame of the sarA gene (B) or a 180-bp DNA fragment containing teg49 (C). The 16S and 23S rRNA bands in an ethidium bromide-stained gel served as loading controls. Various sarA transcripts (P2, P3, and P1) are marked, while P2* and P3* (B) indicate the corresponding truncated transcripts due to deletion of the teg49 region. P3** (C) denotes the size of the P3 transcript in the sarA mutant of SH1000, where the sarA gene has been replaced by ermC. The spliced images were taken from different regions of the same image. (D) Western blot analyses for SarA with anti-SarA antibody of the wild type, the teg49 strain, and its isogenic derivative strains from panels B and C. Equivalent amounts of extracts (10 μg) from the different phases of growth (OD600 ≈ 1.1, late exponential phase; OD600 ≈ 1.7, postexponential phase) were used to detect SarA protein. (Bottom) A section of Coomassie blue-stained duplicate-run gel used for blotting is shown as a loading control for all the gels. All the strains containing pEPSA5 and its derivatives were grown in the presence of 2% xylose to induce teg49 expression. The numbers at the left of the gels are the OD600 values.
Biofilm formation and expression of some virulence genes in the teg49 mutant.
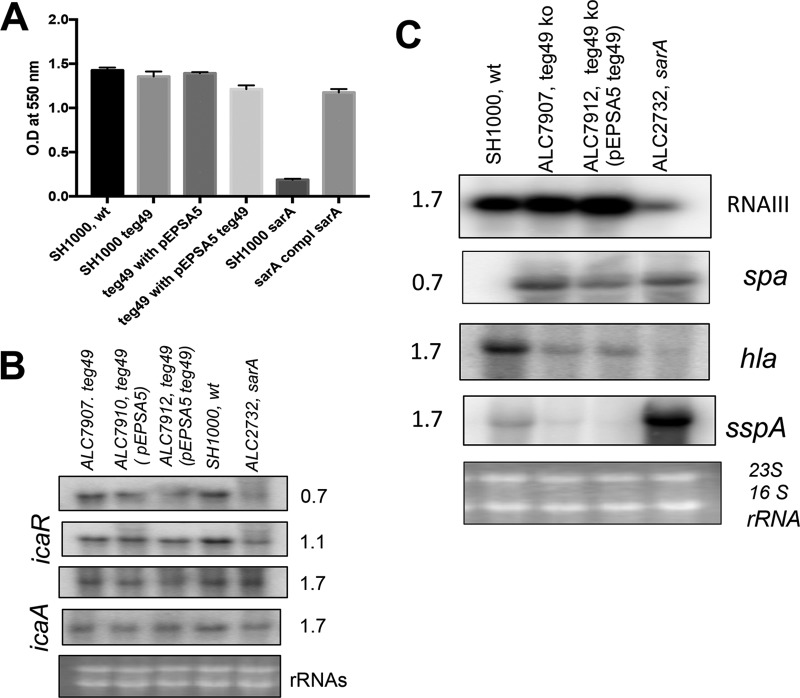
Several studies have disclosed the critical role of SarA in regulation of biofilm formation in Staphylococcus species (13, 35). To determine the role of the teg49 mutant in biofilm formation, biofilm assays were performed (Fig. 4A). Concordant with a lack of effect on the SarA protein level, the teg49 mutant exhibited biofilm formation similar to that of the parental strain, SH1000, and the complemented mutant. Expression of biofilm-related genes, such as the icaRA genes, was also found not to be affected by inactivation or overexpression of teg49 (Fig. 4B). Collectively, these data indicate that teg49 is not a major contributor to biofilm formation in strain SH1000.
FIG 4.
Phenotypic and genetic analyses for biofilm formation and selective virulence gene expression for the teg49 mutant and its derivative strains. (A) Biofilm formation by various strains in microtiter wells containing TSB and 0.25% glucose. The cells were grown for 24 h, and the biofilms were stained with crystal violet and solubilized with acetic acid to read OD550. The sarA mutant and complemented mutant were used as negative and positive controls, respectively. The results are expressed as means and standard errors of the mean. (B) Northern blots for the wild-type, teg49 mutant, teg49 containing pEPSA5, teg49 mutant containing pEPSA5::teg49, and isogenic sarA mutant strains at various phases of growth. The probes used for hybridization were icaR and icaA for biofilm phenotype-related genes. (C) Northern blots of the regulatory locus (agr RNAIII), protein A (spa), α-hemolysin (hla), and V8 protease (sspA) genes for the wild-type, teg49 mutant (ALC7907), teg49 mutant with pEPSA5::teg49 (ALC7912), and isogenic sarA mutant strains from various growth phases as indicated. All the strains containing pEPSA5 and its derivatives were grown in the presence of 2% xylose for the induction of teg49. A total of 10 μg of cellular RNA from various phases of growth was loaded onto each lane. The 23S and 16S rRNAs of an ethidium bromide-stained gel used for blotting are shown as the loading control. The numbers beside the gels are OD600 values.
The effect of teg49 on virulence genes, such as agr, spa, hla, and sspA, in the SH1000 background was also examined. As shown in Fig. 4C, RNAIII of the agr locus was moderately increased in the complemented mutant (27% ± 5%) versus the parent (set at 100%) and the teg49 mutant (9% ± 5%), while RNAIII expression was much lower in the sarA deletion mutant (56% ± 5%) than in the parental strain, SH1000. However, the effect of teg49 on spa, hla, and sspA was more subtle, with lower hla expression and higher spa expression in the teg49 mutant versus the parent; these phenotypes are inconsistent with higher RNAIII expression. A slight decrease in sspA mRNA was detected in the mutant versus the parent, concordant with a lack of change in the SarA protein level in the mutant. As expected, the control sarA deletion mutant expressed lower levels of agr and hla and higher spa and sspA levels versus the parent (Fig. 4C). Plate-based phenotypic assays were performed to determine the zone of clearance on tryptic soy agar (TSA) containing sheep blood to determine the overall hemolysin activity. As shown in Table 1, there were very slight changes in the zone of clearances for hemolysis: 15 ± 2 mm for ALC7907 (teg49 mutant) compared to 18 ± 2 mm for the wild type. Further characterization indicated that there was no growth difference between the wild-type and the teg49 mutant strains. In addition, DNA sequence analysis of the sarA loci of both strains suggested that there were no additional base changes in the wild-type and teg49 mutant strains. Therefore, the observed results from these experiments were likely attributable to the deletion of the teg49 region within the sarA locus.
TABLE 1.
Plate assays for hemolysins and proteases
| Straina | Zone of clearance (mm) |
|
|---|---|---|
| Sheep blood agar | 5% skim milk | |
| SH1000 (wt) | 18 ± 2 | 5 ± 1 |
| ALC7907 (teg49 KO) | 15 ± 2 | 4 ± 1 |
| ALC7910 (ALC7907 containing pEPSA5) | 16 ± 2 | 4 ± 1 |
| ALC7912 (ALC7907 containing pEPSA5 plus teg49) | 15 ± 2 | 5 ± 1 |
| ALC2732 (sarA KO) | 12 ± 2 | 15 ± 1.5 |
KO, knockout.
RNA-Seq analysis of the teg49 mutant.
As deletion of teg49 did not significantly alter SarA protein expression and overexpression of teg49 seemed to augment agr expression independently of SarA, we wanted to gain insight into the overall spectrum of genes controlled by teg49 via transcriptional profiling using Illumina RNA-Seq. To refine the expression profile of the teg49 mutant, a minimum read value of 50 reads per kilobase per million (RPKM) and a 4-fold change were considered to reduce the large number of genes attributable to teg49 deletion. In addition, phage-borne genes were also eliminated from our analysis. Using 4-fold change as the cutoff, transcriptional profiling by RNA-Seq revealed that 155 genes were upregulated in the teg49 mutant versus the wild-type strain, while 140 genes were downregulated. As the sarA P3 mRNA was altered in the teg49 mutant (Fig. 3B), the RNA-Seq profile of teg49 was further refined by subtracting sarA-mediated genes of SH1000 deduced from the RNA-Seq data of the sarA mutant. In all, 133 genes were upregulated for expression and 97 genes were downregulated in the teg49 mutant versus the wild-type strain (see Tables S2 and S3 in the supplemental material). There were 47 increases and 70 decreases in metabolism- and enzyme-related transcripts, 33 increases and 13 decreases in cell wall- and transport-related gene products, 40 increases and 3 decreases in mRNAs involved in cellular processes and regulatory and virulence factors, and 13 increases and 11 decreases in expression of genes with unknown functions in the teg49 mutant compared to the wild-type strain. Table 2 shows a few selected categories of genes, such as those involved in cellular function, regulatory processes, and virulence factors, that were altered in the teg49 mutant versus the parental strain. Interestingly, two known regulatory genes (saeRS and lytS), as well as four unknown genes, were upregulated in the teg49 mutant versus the parent, while three putative regulatory genes were downregulated. Notably, the saeRS regulatory system, which is not a part of the sarA regulon, was upregulated (∼104-fold for saeR and ∼52-fold for saeS transcripts) in the teg49 mutant versus the parent, indicating strong saeRS repression by teg49. The exact role of this regulatory relationship is not clear and is under investigation. Among 33 upregulated virulence genes in the teg49 mutant, 13 genes are related to toxins (e.g., hlgA, hlgB, lukS, lukF, SAOUHSC_00354, SAOUHSC_00383, SAOUHSC_00392 to SAOUHSC_00395, SAOUHSC_01124, SAOUHSC_01125, and SAOUHSC_01127), 7 genes are related to autolysis and proteases (e.g., lrgA, SAOUHSC_02166, SAOUHSC_02170, SAOUHSC_02023, SAOUHSC_02019, and SAOUHSC_01576), and 13 genes are related to cell wall factors (e.g., map, sbi, SAOUHSC_02167, SAOUHSC_02160, SAOUHSC_01114, SAOUHSC_01115, SAOUHSC_01110, and SAOUHSC_01112), while the expression of one virulence gene (smc) is reduced among downregulated genes in the teg49 mutant (Table 2). Consistent with our finding that the SarA protein level did not change significantly between the teg49 mutant and the wild type, the RNA-Seq data revealed no significant differences in the expression of several well-known sarA-regulated genes between the isogenic mutant strains. For instance, some of the regulatory genes known to be repressed by SarA, e.g., sarT (1 versus 2), sarU (1 versus 1), and sarV (117 versus 93) did not differ in expression read values (RPKM) between the teg49 mutant and the wild type, implying that there may be two distinct pathways for the regulation of target genes by SarA and teg49 sRNAs in S. aureus.
TABLE 2.
Upregulated and downregulated expression of selected genes in the teg49 mutant versus the wild type
| Gene designation | Gene name or description | Expression value |
Fold change | |
|---|---|---|---|---|
| Wild-type SH1000 | ALC7907 teg49 mutant | |||
| Upregulated genes | ||||
| Cellular process and regulatory systems | ||||
| SAOUHSC_01575 | Helix-turn-helix domain-containing protein | 0 | 166 | 166 |
| SAOUHSC_02053 | Transcriptional activator RinB-like protein | 0 | 143 | 143 |
| SAOUHSC_00715 | Response regulator; SaeR | 298 | 31,113 | 104.40 |
| SAOUHSC_02077 | Mga helix-turn-helix domain protein | 0 | 72 | 72 |
| SAOUHSC_02235 | Repressor | 0 | 55 | 55 |
| SAOUHSC_00714 | Sensor histidine kinase; SaeS | 425 | 22,405 | 52.72 |
| SAOUHSC_00230 | Two-component sensor histidine kinase | 92 | 384 | 4.17 |
| Virulence factors | ||||
| SAOUHSC_02167 | Staphylococcal complement inhibitor | 10 | 32,147 | 3214.7 |
| SAOUHSC_02160 | Map domain protein | 27 | 20,766 | 769.11 |
| SAOUHSC_02160 | MHCa class II analog protein (Map) | 30 | 20,205 | 673.5 |
| SAOUHSC_00401 | Staphylococcal complement inhibitor; SCIN | 77 | 36,003 | 467.57 |
| SAOUHSC_01115 | Staphylococcal complement inhibitor | 26 | 7,600 | 292.30 |
| SAOUHSC_01114 | Fibrinogen-binding protein | 26 | 7,399 | 284.57 |
| SAOUHSC_02166 | Putative holing-like protein | 0 | 254 | 254 |
| SAOUHSC_02706 | Immunoglobulin G-binding; Sbi | 19 | 3,767 | 198.26 |
| SAOUHSC_02170 | Peptidoglycan hydrolase | 0 | 193 | 193 |
| SAOUHSC_02708 | Gamma-hemolysin H-gamma-II subunit; HlgA | 12 | 1,902 | 158.5 |
| SAOUHSC_01110 | Fibrinogen-binding protein-like protein | 21 | 3,142 | 149.61 |
| SAOUHSC_01112 | Formyl peptide receptor-like 1-inhibitory protein | 34 | 4,726 | 139 |
| SAOUHSC_00354 | Staphylococcal/streptococcal toxin, beta-grasp domain | 11 | 1,119 | 101.72 |
| SAOUHSC_02710 | Leukocidin F subunit; HlgB | 37 | 3,416 | 92.32 |
| SAOUHSC_02243 | Leukotoxin; LukS | 84 | 5,299 | 63.08 |
| SAOUHSC_02171 | Staphylokinase | 0 | 55 | 55 |
| SAOUHSC_02023 | Bifunctional autolysin | 0 | 42 | 42 |
| SAOUHSC_02019 | Autolysin | 0 | 34 | 34 |
| SAOUHSC_01576 | Exonuclease family protein | 0 | 27 | 27 |
| SAOUHSC_02241 | Leukotoxin; LukF | 130 | 3,494 | 26.87 |
| SAOUHSC_00393 | Superantigen-like protein 8 | 4 | 91 | 22.75 |
| SAOUHSC_00816 | Extracellular matrix and plasma binding protein; Ssp | 20 | 407 | 20.35 |
| SAOUHSC-00394 | Superantigen-like protein 9 | 4 | 74 | 18.5 |
| SAOUHSC_00383 | Superantigen-like protein 1 | 27 | 464 | 17.18 |
| SAOUHSC_00191 | Staphylococcal complement inhibitor | 5 | 68 | 13.6 |
| SAOUHSC_00395 | Superantigen-like protein 10 | 11 | 132 | 12 |
| SAOUHSC_01127 | Superantigen-like protein | 10 | 100 | 10 |
| SAOUHSC_01125 | Superantigen-like protein | 8 | 79 | 9.87 |
| SAOUHSC_00392 | Superantigen-like protein 7 | 8 | 72 | 9 |
| SAOUHSC_00192 | Staphylocoagulase; Coa | 7 | 46 | 6.57 |
| SAOUHSC_01124 | Superantigen-like protein | 11 | 68 | 6.18 |
| SAOUHSC_00241 | Cold shock protein B | 7,372 | 30,223 | 4.09 |
| SAOUHSC_00232 | Murein hydrolase regulator; LrgA | 141 | 602 | 4.26 |
| Downregulated genes | ||||
| Cellular process, transcriptional regulators, and virulence factors | ||||
| SAOUHSC_01196 | Fatty acid biosynthesis transcriptional regulator | 1,075 | 115 | 9.34 |
| SAOUHSC_01204 | SMC domain-containing protein | 209 | 35 | 5.97 |
| SAOUHSC_01206 | DNA-binding protein | 259 | 47 | 5.51 |
MHC, major histocompatibility complex.
Validating the expression of selective target genes of teg49 sRNA.
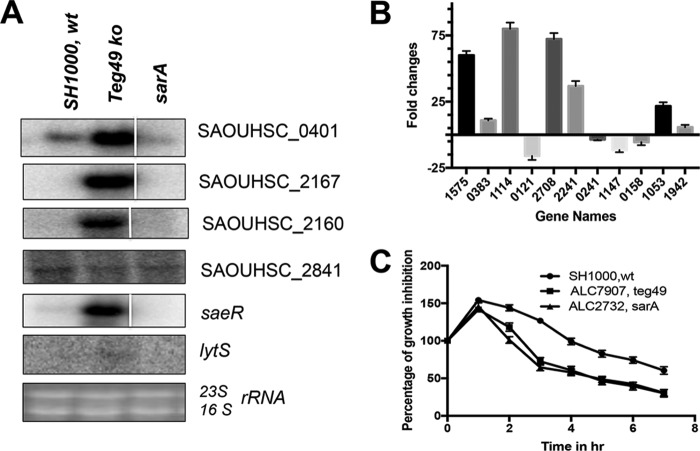
To validate our RNA-Seq data, we analyzed by Northern analysis six known and putative genes that were altered in the teg49 mutant but not in the sarA mutant (SAOUHSC_00401, SAOUHSC_02160, SAOUHSC_02167, SAOUHSC_02841, saeR, and lytS). As shown in Fig. 5A, the expression of the SAOUHSC_401, SAOUHSC_02167, and SAOUHSC_2160 mRNAs was significantly elevated in the teg49 mutant versus the wild type, consistent with the RNA-Seq data (read values of 36,003 versus 77, 32,147 versus 10, and 20,766 versus 27, respectively, for SAOUHSC_401, SAOUHSC_02167, and SAOUHSC_2160) (Table 2). In contrast, the expression of the SAOUHSC_02841 transcript was reduced in the teg49 mutant, concordant with a 32-fold reduction in read values (RPKM) in the teg49 mutant versus the parental strain (see Table S2 in the supplemental material). Similarly, the expression of two regulatory genes, saeR and lytS, was found to be increased in the teg49 mutant versus the parent by Northern analysis (Fig. 5A), in agreement with the results of RNA-Seq (Table 2). To further validate the RNA-Seq results, quantitative RT-PCR was performed with 13 putative genes, using the gyrB gene as the control (Fig. 5B). Among these analyzed genes, nine were upregulated, as determined by RNA-Seq (see Table S2 in the supplemental material) (SAOUHSC_02235, SAOUHSC_01575, SAOUHSC_00383/set1 nm, SAOUHSC_01925, SAOUHSC_01114, SAOUHSC_02708/hlgA, SAOUHSC_02241/lukF, SAOUHSC_01053/mntH, and SAOUHSC_01942/splA) while four were downregulated (SAOUHSC_00121/capH, SAOUHSC_00241/rbsU, SAOUHSC_01147/murD, and SAOUHSC_00158), covering a wide range of genetic pathways ranging from metabolism to proteins with unknown functions (see Table S3 in the supplemental material). qRT-PCR analysis confirmed changes in gene expression in the teg49 mutant (Fig. 5B), with the exception of two genes (SAOUHSC_02235 and SAOUHSC_01925) that failed to produce any product either by qRT-PCR or by normal PCR using the first-strand synthesized DNA (cDNA) as the template, indicating that detection of these two genes by RNA-Seq may be due to artifacts. Thus, Northern blots and qRT-PCR results validated the RNA-Seq data and indicated that teg49 sRNA has an important role in regulating the stability of these transcripts. Whether teg49 is involved directly or indirectly has yet to be determined. The increase in transcript levels for genes encoding putative autolysin (SAOUHSC_02019 and SAOUHSC_02023), putative peptidoglycan hydrolase (SAOUHSC_02170), a putative holin-like protein (SAOUHSC_02166), and regulator-like LrgA in the teg49 mutant (but not in the sarA mutant) suggested that teg49 may have some role in autolysis of S. aureus. Accordingly, we tested the teg49 mutant for susceptibility to penicillin-induced lysis (Fig. 5C). Penicillin G (0.04 μg/ml; the MIC for strain SH1000 is ≤0.004 μg/ml) was added to a growing culture at the early exponential phase of growth (OD650, ∼0.25), and the OD650 was monitored over a 7-h period. As shown in Fig. 5C, the teg49 mutant exhibited sensitivity to penicillin-induced lysis compared to the wild-type SH1000. As a positive control, the sarA mutant displayed high sensitivity to penicillin-induced lysis, consistent with previously published data (36). This sensitivity to penicillin-induced lysis in the teg49 mutant is notable because the SarA protein level in the teg49 mutant was only slightly reduced compared with the wild type (Fig. 3D). Overall, this phenotypic observation validates the changes in the levels of transcription of autolysis-related genes in the teg49 mutant compared with the parent.
FIG 5.
Validation of RNA-Seq data by Northern blot and phenotypic assays. (A) Northern blots of RNA-Seq data on deduced teg49 target genes, including SAOUHSC_00401 (467-fold upregulated), SAOUHSC_02167 (3,214-fold upregulated), SAOUHSC-02160 (769-fold upregulated), SAOUHSC_02841 (32-fold downregulated), saeR (104-fold upregulated), and lytS (4-fold upregulated) for the wild-type, teg49 mutant (ALC7907), and isogenic sarA mutant strains at the exponential phase of growth. A total of 10 μg of cellular RNA was loaded onto each lane. The 23S and 16S bands of the ethidium bromide-stained gel used for blotting are shown as the loading control. The spliced images were taken from different regions of the same image. (B) Thirteen randomly selected RNA-Seq-analyzed genes, along with the gyrB control, were selected for qRT-PCR using total RNA isolated from the respective strains at the exponential phase of growth. The data were normalized against gyrB as the reference transcript for qRT-PCR. All the numbers on the x axis are the gene names from the S. aureus strain NCTC8325 genome (SAOUHSC). (C) Effect of teg49 inactivation on penicillin-induced lysis or growth inhibition. Penicillin G-induced lysis of wild-type SH1000, teg49 mutant (ALC7907), and isogenic sarA mutant (ALC2732 as the control) strains was measured as a decrease in the optical density at 650 nm over time. Penicillin G (0.04 μg/ml, or ∼10-fold subinhibitory concentration) was added to early-exponential-phase growing cultures (OD650 ≈ 0.25), and changes in the OD650 were monitored over a 7-h period. The percentage of growth inhibition or lysis by penicillin G over time was plotted and was calculated as the OD over time divided by the OD at the time penicillin G was added multiplied by 100. The experiments were repeated at least three times. The results are expressed as means and standard errors of the mean.
Murine model of skin abscess with the teg49 mutant.
To seek in vivo correlation, the virulence of the teg49 mutant, along with those of the wild type and the HP1 mutant of teg49 (32), was evaluated in a mouse skin abscess model of infection (Fig. 6). Using the number of CFU per gram of skin abscess tissue as a readout, the results indicated that there was a significant difference (P ≈ 0.0272) between the bacterial cell density of teg49 mutant and those of the wild-type strains at 5 days after initial infection. Similarly, mutation in HP1 of teg49, a crucial structure of teg49 (32), also showed a reduced bacterial load (P ≈ 0.0128) compared to the wild-type strain. It should be mentioned that, despite the finding that teg49 did not affect the overall SarA expression in a significant way, RNA-Seq studies suggested that teg49 might affect virulence genes independently of SarA. This in vivo study using the murine model of cutaneous abscess seems to support that hypothesis.
FIG 6.
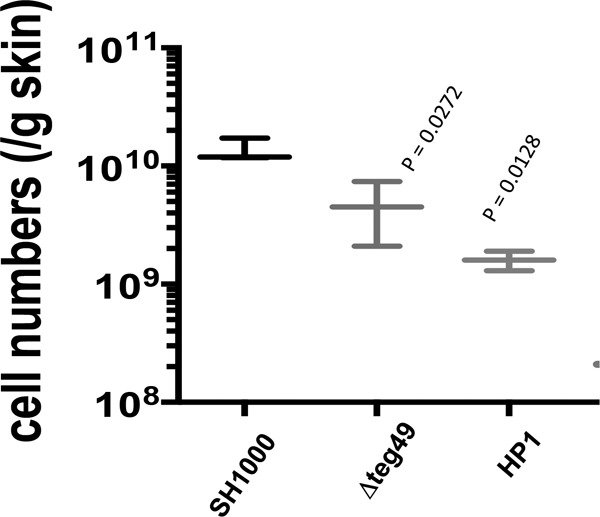
The teg49 and HP1 mutants exhibited reduced virulence in a mouse model of skin abscess infection. Mice (6 in each group) were challenged with ∼1 × 108 CFU of S. aureus SH1000 or its isogenic teg49 or HP1 mutant and observed daily for the presence of skin abscesses. Shown are the mean numbers of colonies counted per gram of infected skin tissue with standard errors of the mean. The mean counts per gram of tissue for the teg49 and HP1 mutants were considered to be statistically significantly different from that of the parent.
DISCUSSION
The discovery of sRNAs in bacteria in the last 2 decades has provided new insights into gene regulation. sRNAs have been found to be ubiquitous and represent the most abundant class of posttranscriptional regulators in prokaryotes (21–23). Most of the sRNAs act by base pairing with target mRNAs. However, the majority of sRNA studies have been limited to Escherichia coli and other Gram-negative bacteria (23). With the exception of RNAIII, most of the sRNA studies in Gram-positive bacteria, including S. aureus, are not well delineated mechanistically (22). The RNAIII molecule, a 516-nt-long RNA, is an effector molecule of the agr system and activates translation of the hla gene product (26) and stabilizes mgrA transcripts (27), as well as inhibiting translation of many transcripts, such as rot, sa1000 (fibrinogen binding protein), sa2261 (ABC transporter), spa (protein A), lytM (peptidoglycan hydrolase), map (major histocompatibility complex class II-analogous protein), and coa (coagulase) (21, 22). Thus, RNAIII of the agr system has established the role of sRNA in stability and translation of mRNA in S. aureus.
Besides agr, sarA is another major regulatory locus involved in the expression of toxins, enzymes, and cell wall adhesins. The sarA locus is peculiar in that the 372-bp sarA coding region is preceded by an ∼850-bp-long 5′ UTR that consists of three distinct promoters, yielding three overlapping transcripts (16, 17). However, complementation studies suggest that the 5′ UTR may play an important role in optimal expression of the SarA protein (20). Recent identification of two sRNAs in this region by RNA-Seq analysis (25) provided the impetus to ascertain the role of sRNA teg49 in gene regulation in SarA-dependent and SarA-independent manners (32).
In this report, we have shown via mutation analysis of the −35 and −10 promoters of the sarA locus that teg49 is most likely produced and processed from the P3 sarA transcript (Fig. 1). Most notably, in the absence of teg49 in SH1000, the transcription of P3 sarA mRNA ceased (Fig. 3B) but was restored upon trans-complementation from a plasmid expressing teg49 in the teg49 mutant (ALC7912), thus indicating that teg49 has an important role in the stability of the P3 sarA transcript. Surprisingly, the overall SarA level was, at best, slightly altered (Fig. 3D) despite the relative absence of the sarA P3 and, to a certain extent, sarA P2 transcripts in the teg49 mutant, signifying that overall SarA expression is mostly due to the contribution of the P1 sarA transcript in S. aureus. Previous studies have shown in strain Newman that the production of teg49 was cshA dependent (32). This result was also confirmed in the SH1000 background. Traditionally, CshA has been known to be involved in ribosome biogenesis, ribosome assembly, and mRNA decay in a degradosome involving RNase J, RNase Y, and polynucleotide phosphorylase (PNPase) (37). Our unpublished in vitro assay with purified CshA and in vitro-transcribed P3 sarA mRNA demonstrated that CshA can bind the sarA P3 transcript and protect it from endonuclease (e.g., RNase III). In this regard, we surmise that CshA may be involved in the initial stability and eventual processing of the sarA P3 transcript by an RNase to yield teg49 sRNA. Surprisingly, inactivation of Hfq, an sRNA chaperone well described in Gram-negative bacteria, did not yield any alterations in the levels of transcripts of three analyzed sRNAs (i.e., teg49, teg23, and teg35). This finding indicated that Hfq does not have a major role in stabilizing sRNA in S. aureus, consistent with other reports of Gram-positive bacteria (23).
Analysis of the putative RNase mutants in the JE2 mariner transposon library indicated that RNase III, an endoribonuclease that cleaves double-stranded RNA (38, 39), may be required for processing of the sarA P3 transcript to produce teg49. The specificity of RNase III in this cleavage was demonstrated with teg23, another sRNA known to be stabilized by CshA, while the control, teg35, an sRNA not stabilized by CshA (34), was not altered in the rnc mutant. Previous studies have shown that RNase III is normally involved in regulating the turnover of mRNAs and noncoding RNAs (40, 41). RNase III also likely targets the 5′ overlapping regions of divergent mRNAs to generate species with shorter or even leaderless 5′ UTRs (42).
Among several other mutants tested for putative endoribonuclease activity to cleave sarA P3 mRNA (Fig. 2A), we found that inactivation of the conserved hypothetical-protein genes (e.g., SauUSA300_1184, SauUSA300_2328, and SauUSA300_1692), ABC permease gene (SauUSA_308), and polyribonucleotide transferase (pnp) gene did not have any effect on expression of teg49. Only inactivation of three genes (nrdD, rny, and endo-RNase L-PSP) showed increased expression; the reason for this increase is not clear at this point. Notably, RNase E, an important part of the degradosome that processes mRNA in many Gram-negative bacteria, is absent in S. aureus, while RNase Y (encoded by rny), which is involved in processing of monophosphorylated RNA (43), appears to have a role in destabilizing teg49 (Fig. 2A). Collectively, these results indicated that endoribonucleases are likely involved in the generation and stabilization of teg49 sRNA. Consistent with a lack of major alteration in the overall level of the SarA protein, a major player in regulating biofilm formation, in the teg49 mutant, there was no major change in biofilm formation in the teg49 mutant. It should be noted that regulation of biofilm formation is a complex process, and several known regulatory systems, such as Rbf, SarX, and SarR, also participate in regulation at the transcriptional level in Staphylococcus species (44). In contrast to the biofilm-related genes (e.g., icaA and icaR), the spa transcript is upregulated while the hla and sspA transcripts are downregulated in the teg49 mutant, as supported by RNA-Seq and Northern analyses, accompanied by slight phenotypic alterations in zones of clearance for hemolysis and proteases (Table 1). This pattern of regulation is inconsistent with moderate upregulation of RNAIII and mild alteration of the SarA protein level in the teg49 mutant. Coincidentally, we determined by RNA-Seq that saeRS, a genetic locus not directly linked to agr and sarA, is significantly upregulated in the teg49 mutant. Elevated saeRS would be expected to result in elevated spa and hla and curtailed sspA expression, which does not completely explain the teg49 virulence genotype/phenotype that we have observed. It is conceivable that inactivation of teg49 leads to a combination of regulatory events (e.g., elevated agr and sae and other factors) that may be able explain what we observed here in the teg49 mutant.
By setting various criteria, such as minimum read values, discarding phage-borne genes, eliminating sarA-regulated genes, and designating 4-fold changes for significance, our RNA-Seq analysis also showed that 133 and 97 genes are up- and downregulated, respectively, in the teg49 mutant compared to the wild type. Several virulence genes, such as those encoding complement inhibitor proteins (SAOUHSC_02167, SAOUHSC_00401, SAOUHSC_01115, SAOUHSC_02160, map, and sbi), leukotoxins and superantigen-like proteins, and fibrinogen-binding protein-like proteins, were altered due to teg49 deletion in the S. aureus genome (Table 2).
It has been shown that antisense base pairing of sbi mRNA (encoding an immunoglobulin binding protein) with SprD, an sRNA expressed from an S. aureus pathogenicity island (PIΦ), led to an impaired host immune response (30). It would be interesting to investigate the existence of similar mechanisms between teg49-regulated virulence genes and teg49 in the future. Besides direct base-pairing with target mRNA, several other mechanisms, including dual-function sRNA that acts as an antisense molecule and codes for a small peptide (e.g., Hld in RNAIII), have been proposed to act on the same or other pathway genes, and also riboswitches that exhibit a structured receptor domain specifically recognized by a small molecule or metabolite (22, 45). Together, our data suggest that teg49 sRNA has a regulatory role in S. aureus, but the exact mode of regulation is yet to be determined.
Overall, our results presented here indicated that teg49 sRNA is processed and produced from the sarA P3 transcript of the sarA locus and that its stability depends on the cshA and rnc gene products. Inactivation of teg49 has no major effect on the overall level of SarA expression, and teg49 appears to be required for the stability of the P3 sarA transcript. Transcriptome profiling indicated about 230 genes with at least a 4-fold change were affected in the teg49 mutant in a sarA-independent manner. The sRNA teg49 appears to be involved in penicillin-mediated lysis but not in biofilm formation. A mouse skin abscess model of infection revealed a modest but significant reduction in the bacterial load in the infected skin tissue, thus demonstrating that teg49 plays a noticeable role in virulence gene regulation in S. aureus. Whether teg49 acts as a riboswitch in S. aureus is not clear at this point. Whether teg49 can be considered a target for the development of novel antibacterial compounds to overcome the current mechanisms of resistance remains to be determined.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
The bacterial strains and plasmids used in this study are listed in Table 3. S. aureus strain RN4220, a restriction-deficient derivative of strain 8325-4, was used as the initial recipient for the transformation of plasmid constructs. The S. aureus strains were grown in tryptic soy broth (TSB) or on TSA supplemented with antibiotics when necessary (5 μg/ml of erythromycin and 10 μg/ml of chloramphenicol). The cells were grown with continuous aeration in a shaker at either 37°C or the required permissive temperatures. Luria-Bertani (LB) broth or agar, supplemented with suitable antibiotics when necessary (100 μg/ml of ampicillin and 40 μg/ml of kanamycin), was used for growing E. coli. Growth was monitored by measuring changes in turbidity at 600 nm in a spectrophotometer (Spectronic 20D).
TABLE 3.
Bacterial strains and plasmids used in this study
| S. aureus strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| RN4220 | Restriction-deficient transformation recipient | 52 |
| SH1000 | Wild-type laboratory strain with a functional rsbU+ derivative of the 8325-4 rsbU-defective strain | 53 |
| ALC7907 | teg49 mutant of SH1000; chromosomal deletion of 204-bp teg49 from bp 182 to 386 from the ATG start codon of the sarA gene | This study |
| ALC7910 | ALC7907 containing an empty xylose-inducible vector, pEPSA5 | This study |
| ALC7912 | ALC7907 containing pEPSA5 carrying 183-bp teg49 fragment DNA under a xylose-inducible promoter | 32 |
| ALC8070 | teg48 mutant of SH1000; chromosomal deletion of 68 bp from the teg48 region of the sarA locus | This study |
| ALC8372 | SH1000 chromosomal P2 promoter mutant at the −35 and −10 sequences of the sarA locus | This study |
| ALC8373 | SH1000 chromosomal P3 promoter mutant at the −35 and −10 sequences of the sarA locus | This study |
| ALC8374 | SH1000 chromosomal P1 promoter mutant at the −35 and −10 sequences of the sarA locus | This study |
| ALC 8441 | ALC8373 mutated P3 promoter chromosomally repaired to wild-type P3 promoter at the −35 and −10 sequences of the sarA locus | This study |
| ALC 2732 | sarA mutant of SH1000; sarA::erm | 14 |
| AM 1262 | Complemented with sarA into the geh locus on chromosome of the sarA mutant of SH1000; sarA::erm | 14 |
| ALC8243 | rnc or RNase III mutant of SH1000; rnc::erm | This study |
| ALC8445 | Complemented ALC8243 with pSK236 carrying a 1.5-kb fragment containing the rnc gene region | This study |
| ALC8235 | cshA mutant of SH1000, chsA::kan | This study |
| ALC8245 | hfq mutant of SH1000; hfq::erm | This study |
| NE1494 | rnc RNase III of JE2 (SAUUSA300_01126) | 33 |
| NE1707 | SAUSA300_1184::erm of JE2 | 33 |
| NE0581 | nrdD mutant of JE2; nrdD::erm (SAUUSA_2551) | 33 |
| NE0259 | pnpA mutant of JE2; pnpA::erm (SAUSA300_1167) | 33 |
| NE1205 | nrdG mutant of JE2; nrdG::erm (SAUSA_2550) | 33 |
| ALC8428 | rnaseY mutant of JE2 constructed by transduction from Newman rnaseY::erm (SAUSA300_01179) | This study |
| NE0291 | SAUSA300_2328::erm of JE2 | 33 |
| NE1079 | SAUSA300_2021::erm of JE2 | 33 |
| NE0458 | xseA mutant JE2; xseA::erm (SAUSA300_1472) | 33 |
| NE1340 | SAUSA300_1692::erm of JE2 | 33 |
| NE0501 | rnr mutant of JE2; rnr::erm (SAUSA300_0764) | 33 |
| NE0404 | Endo-RNase L-PSP mutant of JE2; endo-RNase L-PSP::erm (SAUSA300_0474) | 33 |
| NE0931 | SAUSA300_0308::erm of JE2 | 33 |
| Plasmids | ||
| pMAD | E. coli-S. aureus shuttle vector containing temperature-sensitive origin of replication, bgaB; Ermr Apr | 47 |
| pEPSA5 | E. coli-S. aureus xylose-inducible shuttle vector containing xylose-inducible promoter for conditional expression of the desired gene; Cmr Apr | 32 |
| pEPSA5::teg49 | pEPSA5 containing 196-bp teg49 sRNA region | 32 |
| pSK236 | Shuttle vector containing pUC19 cloned into the HindIII site of pC194 | 14 |
Erm, erythromycin; Ap, chloramphenicol; Cm, chloramphenicol.
Genetic manipulation.
Standard procedures for DNA manipulations were performed using routine cloning procedures (46). Mutagenesis of various sarA promoters (P2, P3, and P1) was performed using pCR2.1 containing a P2 sarA fragment flanking BamHI sites at both ends as a template and various mutagenized primer pairs as listed in Table S1 in the supplemental material (P2 sarA, −35 AGCAAA to CCCCAC and −10 TAATAT to CCACTC; P3 sarA, −35 AGTGAT to CCCAAA and −10 GGGTAT to CCACTC; and P1 sarA, −35 TTTACT to CCCCAA and −10 TATAAT to CCACTC). PCR-mediated plasmid mutagenesis was performed using Pfu and Taq polymerases, and clones were authenticated by DNA sequencing. The 1.5-kb BamHI fragments containing various individual promoter mutations were cloned into the BamHI site of the temperature-sensitive shuttle vector pMAD (47). The recombinant pMAD plasmids were first electroporated into strain RN4220 and then into S. aureus strain SH1000 for construction of various mutants. Construction, selection, and authentication of the putative mutants were performed as described previously (46, 47). These mutant strains were labeled ALC8372 (P2m), ALC8373 (P3m), and ALC8374 (P1m) for sarA P2, P3, and P1 promoter knockout mutants, respectively. To restore the promoter in the P3m promoter knockout strain, a 1.5-kb fragment containing the native P3 sarA fragment was cloned into pMAD and transformed into strain ALC8373 to construct a revertant P3m/P3wt strain, ALC8441, following the same procedure as previously described for mutant strain construction. Construction of rnc::erm (ALC8243), cshA::kan (ALC8235), hfq::erm (ALC8245) of SH1000 and rny::erm (ALC8428) of JE2 was performed by phage transduction from the respective Newman background mutant strains. To complement the rnc mutation, a 1.5-kb DNA fragment encompassing the rnc gene region was cloned in shuttle plasmid pSK236 and transformed into ALC8243 (rnc::erm) to construct a trans-complemented strain (ALC8445).
Construction of the teg49 mutant was performed using the same PCR-mediated method of mutagenesis with the primers used in Table S1 in the supplemental material to delete 204 bp from bp 385 to 182 upstream from the ATG start codon of the sarA gene using pCR2.1 carrying a 2.3-kb fragment encompassing the sarA locus as the template. A 2.2-kb DNA fragment containing the deletion was cloned into pMAD to construct the teg49 deletion mutant (ALC7907), as described previously for the construction of various promoter mutants. Final authentication of various mutant strains was performed by DNA sequencing of the PCR products and Northern hybridization. Complementation of the teg49 mutant was performed in trans using pEPSA5::teg49 in the presence of 2% xylose in broth (32).
Isolation of total cellular RNA and Northern blot hybridization.
Isolation of total cellular RNA from various growth phases and subsequent analysis by Northern blot hybridization were performed as described previously (46). DNA fragments containing the open reading frames of sarA, icaA, icaR, spa, hla, and sspA or sRNA teg49, teg23, and teg35 and other RNA-Seq-validated genes were amplified by PCR or excised from the plasmids containing the desired genes with restriction endonucleases and then gel purified. For detection of specific transcripts, gel-purified DNA probes were radiolabeled with [α-32P]dCTP by using the random-primed DNA-labeling kit (Roche Diagnostics GmbH) and hybridized under aqueous-phase conditions at 65°C. The blots were subsequently washed, exposed to a phosphorimager screen, and scanned. Each of the experiments was repeated at least three times independently.
Western blotting and immunodetection.
Whole-cell extracts from S. aureus wild-type SH1000 or various mutant strains were prepared from cells grown to different growth phases, as described previously (46). The concentration of total proteins from clear lysates was determined by using a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, USA) using bovine serum albumin as the standard. Western blotting and detection with primary antibody (anti-SarA) and donkey anti-mouse secondary antibody conjugated with horseradish peroxidase (HRP) and enhanced chemiluminescence (ECL) Western blotting substrate (Thermo Scientific) were performed as described previously (46). The experiments were repeated at least three times independently.
Biofilm assay.
Quantification of biofilm formation on abiotic surfaces was performed as described previously (35, 48). Briefly, S. aureus strains grown overnight in TSB were diluted (1:50) in TSB supplemented with 0.25% glucose. This cell suspension was used to inoculate sterile 96-well U-bottom polystyrene microtiter plates (Costar, Corning Inc., USA) in triplicate. After 24 h of incubation at 37°C, the wells were gently washed three times with water, air dried in an inverted position, and stained with 0.1% crystal violet dye in water for 15 min at room temperature. The wells were rinsed 3 or 4 times by submerging them in water, dried for several hours to overnight in an inverted position, and solubilized in 30% acetic acid in water for 15 min. The solubilized dye was transferred to flat-bottom microtiter plates, and the absorbance at 550 nm was determined (Infinite M1000 Pro; Tecan, Austria). Each assay was repeated at least three times in separate experiments.
Protease and hemolysin assays.
A protease assay for all the tested strains was performed using TSA supplemented with 5% skim milk. Similarly, a hemolysis assay was done using 5% sheep blood TSA plates (Remel, Kansas, USA). Equal volumes (3 μl) of equivalent optical density (OD600) of various strains were spotted onto plates in triplicate and incubated for 20 to 36 h at 37°C. The zones of clearance from three independent experiments were measured and compared.
RNA-Seq experiments.
Samples for RNA-Seq were prepared from the following strains: the wild-type SH1000, its isogenic derivative teg49 mutant (ALC7907), and the isogenic sarA mutant (ALC2732). Briefly, overnight cultures were diluted 1:100 in fresh TSB (initial OD600, about 0.04 to 0.05) and grown at 37°C for 3 h. Two replicate cultures for each sample were grown, and total cellular RNAs from two biological replicates were extracted as described previously (32). RNA was quantified by using Qubit (Life Technologies), and RNA integrity was assessed with a 2100 Bioanalyzer (Agilent Technologies). A 1-μg amount of total RNA was ribodepleted with a bacterial Ribo-Zero kit from Illumina. A TruSeq RNA stranded kit from Illumina was used for library preparation. The library quantity was measured with Qubit, and quality was assessed on a Tapestation on a DNA high-sensitivity chip (Agilent Technologies). The libraries were pooled at equimolarity and loaded at 2 nM for clustering. Oriented 50-base single-read sequencing was performed on an Illumina HiSeq 4000 sequencer, yielding a minimum of 9 million mapped reads per sample. Final RNA-Seq analysis and data analysis were carried out using previously described procedures (32). Statistical analyses were done in R v3.2.3 using the edgeR package (49) following a bioinformatics protocol described previously (50). Briefly, genes whose expression was not at least one read per million in two replicates of the samples were filtered out. Counts of the retained genes were normalized according to the weighted trimmed mean of M values (TMM) method. Read counts for each transcript in each sample were modeled according to a negative binomial (NB) distribution. Finally, pairwise comparisons between read libraries from various strains were performed with the exact test (49, 50) to detect differentially expressed transcripts. P values were corrected according to the false-discovery rate (FDR).
qRT-PCR analysis.
qRT-PCR was performed as previously described (34). Briefly, cDNA was obtained using GoScript cDNA synthesis mix (Promega, USA) and DNase I-treated purified total RNA as a template. Maxima SYBR green/ROX qRT-PCR master mix (Thermo Scientific) was used to generate standard curves for the cDNA concentration/crossing point (Cp) for the target genes and the reference gene, gyrB. The mean target transcriptional expression level for the three transcript measurements was calculated. The threshold cycle (2−ΔΔCT) method was used to calculate relative changes in gene expression, using triplicate samples. Applied Biosystems StepOne Plus software was used to analyze the qRT-PCR results. Control reaction mixtures containing master mix and primers but no cDNA were also analyzed.
Penicillin G-mediated lytic assay.
The penicillin G-mediated lytic assay was performed as described previously (36). In brief, to assess the sensitivity of the teg49 mutant and its isogenic derivative strains, along with the sarA mutant, to penicillin, inoculations were done in TSB from overnight cultures to yield a starting OD650 of 0.05. Penicillin G was added to a concentration of 0.04 μg/ml (the MIC determined for strain SH1000 is ≤0.0039 μg/ml) at the early exponential phase of growth (OD650 ≈ 0.25). Cultures were incubated at 37°C with shaking, and the OD650 was measured every hour for 7 h.
Murine cutaneous model of infection.
A murine skin infection model was used to determine the effect of the teg49 mutation on virulence in the SH1000 background. Animal experiments were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee at the Geisel School of Medicine at Dartmouth, Hanover, NH. Female BALB/c mice were held, shaved on the back, and inoculated by subcutaneous injection in the right flank with 1 × 108 S. aureus bacteria in 50 μl of PBS, using a 23-gauge needle. The mice were weighed before inoculation, and abscess formation was monitored at 24-h intervals for 5 days. The sizes of the abscesses were calculated using a standard formula for area [A = (π/2) × length × width], as described previously (51). Dermonecrosis was scored as present or absent. Six mice per group were used for each bacterial strain or control group. After 5 days of infection, at which time abscesses were evident in most mice, the mice were sacrificed and abscess tissues were collected, weighed, homogenized in saline containing 0.1% Triton X-100, and plated in triplicate in mannitol salt agar for incubation at 37°C for 48 h to derive the number of CFU per gram of abscess tissue for various strains.
Statistical analyses.
Statistics for the abscess or lesion area were performed using 1-way or 2-way analysis of variance (ANOVA), as indicated. The results are expressed as means ± standard errors of the mean unless otherwise indicated. All data were analyzed using Graph Pad Prism 7. The data represent the standard errors of the means from at least three independent experiments with multiple replicates unless otherwise indicated. For normally distributed data, comparisons were tested with Student's t test.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants AI106937 from the National Institutes of Health (to A.C.) and 31003A_153474/1 from the Swiss National Science Foundation (to P.F.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00635-17.
REFERENCES
- 1.Crossley KB, Archer GL. 1997. The staphylococci in human disease. Churchill Livingston, New York, NY. [Google Scholar]
- 2.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Foster TJ. 2005. Immune evasion by staphylococci. Nat Rev Microbiol 3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy AD, DeLeo FR. 2009. Neutrophil apoptosis and the resolution of infection. Immunol Res 43:25–61. doi: 10.1007/s12026-008-8049-6. [DOI] [PubMed] [Google Scholar]
- 5.Diep BA, Otto M. 2008. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol 16:361–369. doi: 10.1016/j.tim.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung AL, Nishina KA, Trotonda M-P, Tamber S. 2008. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol 40:355–361. doi: 10.1016/j.biocel.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novick RP. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 8.Ballal A, Ray B, Manna AC. 2009. sarZ, a sarA family gene, is transcriptionally activated by MgrA and is involved in the regulation of genes encoding exoproteins in Staphylococcus aureus. J Bacteriol 191:1656–1665. doi: 10.1128/JB.01555-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arya R, Princy SA. 2016. Exploration of modulated genetic circuits governing virulence determinants in Staphylococcus aureus. Indian J Microbiol 56:19–27. doi: 10.1007/s12088-015-0555-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornblum J, Kreiswirth B, Projan SJ, Ross H, Novick RP. 1990. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus, p 403–420. In Novick RP. (ed), Molecular biology of the staphylococci. VCH Publishers, New York, NY. [Google Scholar]
- 11.Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu Rev Genet 42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 12.Cheung AL, Koomey JM, Butler CA, Projan SJ, Fischetti VA. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci U S A 89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kavanaugh JS, Horswill AR. 2016. Impact of environmental cues on staphylococcal quorum sensing and biofilm development. J Biol Chem 291:12556–12564. doi: 10.1074/jbc.R116.722710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ballal A, Manna AC. 2010. Control of thioredoxin reductase (trxB) transcription by SarA in Staphylococcus aureus. J Bacteriol 192:336–345. doi: 10.1128/JB.01202-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. 2001. Transcriptional profiling based identification of S. aureus genes regulated by the agr and/or sarA loci. J Bacteriol 183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bayer MG, Heinrichs JH, Cheung AL. 1996. The molecular architecture of the sar locus in Staphylococcus aureus. J Bacteriol 178:4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manna AC, Bayer MG, Cheung AL. 1998. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J Bacteriol 180:3828–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ballal A, Manna AC. 2009. Expression of sarA-family genes in S. aureus strains. Microbiology 155:2342–2352. doi: 10.1099/mic.0.027417-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blevins JS, Beenken KE, Elasri MO, Hurburt BK, Smeltzer MS. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect Immun 70:470–480. doi: 10.1128/IAI.70.2.470-480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung AL, Bayer MG, Heinrichs JH. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J Bacteriol 179:3963–3971. doi: 10.1128/jb.179.12.3963-3971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guillet J, Hallier M, Felden B. 2013. Emerging functions for the Staphylococcus aureus RNome. PLoS Pathog 9:e1003767. doi: 10.1371/journal.ppat.1003767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomasini A, Francois P, Howden BP, Fechter P, Romby P, Caldelari I. 2014. The importance of regulatory RNAs in Staphylococcus aureus. Infect Genet Evol 21:616–626. doi: 10.1016/j.meegid.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 23.Pitman S, Cho KH. 2015. The mechanisms of virulence regulation by small noncoding RNAs in low GC Gram-positive pathogens. Int J Mol Sci 16:29797–29814. doi: 10.3390/ijms161226194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carroll RK, Weiss A, Broach WH, Wiemels RE, Mogen AB, Rice KC, Shaw LN. 2016. Genome-wide annotation, identification, and global transcriptomic analysis of regulatory or small RNA gene expression in Staphylococcus aureus. mBio 7:e01990-15. doi: 10.1128/mBio.01990-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaume M, Hernandez D, Farinelli L, Deluen C, Linder P, Gaspin C, Romby P, Schrenzel J, Francois P. 2010. Cartography of methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions. PLoS One 5:e10725. doi: 10.1371/journal.pone.0010725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morfeldt E, Taylor D, von Gabain A, Arvidson S. 1995. Activation of alpha-toxin in S. aureus by trans-encoded antisense RNA, RNAIII. EMBO J 14:4569–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta RK, Luong TT, Lee CY. 2015. RNAIII of the Staphylococcus aureus agr system activates global regulator MgrA by stabilizing mRNA. Proc Natl Acad Sci U S A 112:14036–14041. doi: 10.1073/pnas.1509251112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorski P, Borezee-Durant E, Barbet R, Jacquet E, Jacq A, Gautheret D, Felden B, Vogel J, Bouloc P. 2010. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res 38:6620–6636. doi: 10.1093/nar/gkq462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue T, Zhang X, Sun H, Sun B. 2014. ArtR, a novel sRNA of Staphylococcus aureus, regulates α-toxin expression by targeting the 5′-UTR of sarT mRNA. Med Microbiol Immunol 203:1–12. doi: 10.1007/s00430-013-0307-0. [DOI] [PubMed] [Google Scholar]
- 30.Chabelskaya S, Gaillot O, Felden B. 2010. A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule. PLoS Pathog 6:e1000927. doi: 10.1371/journal.ppat.1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaito C, Saito Y, Ikuo M, Omae Y, Mao H, Nagano G, Fujiyuki T, Numata S, Han X, Obata K, Hasegawa S, Yamaguchi H, Inokuchi K, Ito T, Hiramatsu K, Sekimizu K. 2013. Mobile genetic element SCCmec-encoded psm-mec RNA suppresses translation of agrA and attenuates MRSA virulence. PLoS Pathog 9:e1003269. doi: 10.1371/journal.ppat.1003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S, Reyes D, Beaume M, Francois P, Cheung A. 2014. Contribution of teg49 small RNA in the upstream transcriptional region of sarA to virulence in Staphylococcus aureus. Infect Immun 82:4369–4379. doi: 10.1128/IAI.02002-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fey FD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S, Corvaglia A-R, Leo S, Cheung A, Francois P. 2016. Characterization of RNA helicase CshA and its role in protecting mRNAs and small RNAs of Staphylococcus aureus strain Newman. Infect Immun 84:833–844. doi: 10.1128/IAI.01042-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trotonda MP, Manna AC, Cheung AL, Lasa I, Penades JR. 2005. SarA control Bap-dependent biofilm formation in Staphylococcus aureus. J Bacteriol 187:5790–5798. doi: 10.1128/JB.187.16.5790-5798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manna AC, Ingavale SS, Maloney M, van Wamel W, Cheung AL. 2004. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J Bacteriol 186:5267–5280. doi: 10.1128/JB.186.16.5267-5280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oun S, Redder P, Didier JP, Francois P, Corvaglia AR, Buttazzoni E, Giraud C, Girard M, Schrenzel J, Linder P. 2013. The CshA DEAD-box RNA helicase is important for quorum sensing control in Staphylococcus aureus. RNA Biol 10:157–165. doi: 10.4161/rna.22899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gottesman S, Storz S. 2011. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 3:a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stead MB, Marshburn S, Mohanty BK, Mitra J, Pena Castillo L, Ray D, van Bakel H, Hughes TR, Kushner SR. 2011. Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res 39:3188–3203. doi: 10.1093/nar/gkq1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romilly C, Chevalier C, Marzi S, Masquida B, Geissmann T, Vandenesch F, Westhof E, Romby P. 2012. Loop-loop interactions involved in antisense regulation are processed by the endoribonuclease III in Staphylococcus aureus. RNA Biol 9:1461–1472. doi: 10.4161/rna.22710. [DOI] [PubMed] [Google Scholar]
- 41.Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. 2006. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol 61:1038–1048. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 42.Lioliou E, Sharma CM, Caldelari I, Hefer A-C, Fechter P, Vandenesch F, Romby P. 2012. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet 8:e1002782. doi: 10.1371/journal.pgen.1002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richards J, Liu Q, Pellegrini O, Celesnik H, Yao S, Bechhofer DH, Conon C, Belasco JG. 2011. An RNA pyrophosphohydrolase triggers 5′-exonucleolytic degradation of mRNA in Bacillus subtilis. Mol Cell 43:940–949. doi: 10.1016/j.molcel.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowe SE, Campbell C, Lowry C, O'Donnell ST, Olson ME, Lindgren JK, Waters EM, Fey PD, O'Gara JP. 2016. AraC-type regulator Rbf controls the Staphylococcus epidermidis biofilm phenotype by negatively regulating the icaADBC repressor SarR. J Bacteriol 198:2914–2924. doi: 10.1128/JB.00374-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gimpel M, Brantl S. 2017. Dual-function small regulatory RNAs in bacteria. Mol Microbiol 103:387–397. doi: 10.1111/mmi.13558. [DOI] [PubMed] [Google Scholar]
- 46.Manna AC, Cheung AL. 2001. Characterization of sarR, a modulator of sarA expression in Staphylococcus aureus. Infect Immun 69:885–896. doi: 10.1128/IAI.69.2.885-896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnaud M, Chastanet A, Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC content Gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Toole GA. 2011. Microtiter dish biofilm formation assay. J Vis Exp 41:2437. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson MD, McCarthy DJ, Smyth GK. 2010. Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. 2013. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 51.Bunce C, Wheeler L, Reed G, Musser J, Barg N. 1992. Murine model of cutaneous infection with Gram-positive cocci. Infect Immun 60:2636–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 12:3967–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. 2002. σB modulates virulence determinants expression and stress resistance: characterization of a functional rsbU strain of Staphylococcus aureus 8325-4. J Bacteriol 184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.