Abstract
Modified Bloom–Richardson (mBR) grading is known to have prognostic value in breast cancer (BCa), yet its use in clinical practice has been limited by intra- and interobserver variability. The development of a computerized system to distinguish mBR grade from entire estrogen receptor-positive (ER+) BCa histopathology slides will help clinicians identify grading discrepancies and improve overall confidence in the diagnostic result. In this paper, we isolate salient image features characterizing tumor morphology and texture to differentiate entire hematoxylin and eosin (H and E) stained histopathology slides based on mBR grade. The features are used in conjunction with a novel multifield-of-view (multi-FOV) classifier—a whole-slide classifier that extracts features from a multitude of FOVs of varying sizes—to identify important image features at different FOV sizes. Image features utilized include those related to the spatial arrangement of cancer nuclei (i.e., nuclear architecture) and the textural patterns within nuclei (i.e., nuclear texture). Using slides from 126 ER+ patients (46 low, 60 intermediate, and 20 high mBR grade), our grading system was able to distinguish low versus high, low versus intermediate, and intermediate versus high grade patients with area under curve values of 0.93, 0.72, and 0.74, respectively. Our results suggest that the multi-FOV classifier is able to 1) successfully discriminate low, medium, and high mBR grade and 2) identify specific image features at different FOV sizes that are important for distinguishing mBR grade in H and E stained ER+ BCa histology slides.
Index Terms: Breast cancer (BCa), digital pathology, image analysis, modified Bloom–Richardson (mBR) grade, multi-field-of-view (multi-FOV), nuclear architecture, nuclear texture
I. Introduction
WHILE breast cancer (BCa) is an increasingly common cancer diagnosis in women [1], advancements in screening, diagnostic, and therapeutic techniques have improved survival rates in recent years [2]. A number of prognostic criteria have been developed to characterize the level differentiation of BCa tumor cells via visual analysis of hematoxylin and eosin (H and E) stained histopathology, including the Bloom–Richardson and Nottingham grading schemes [3]. In particular, the modified Bloom–Richardson (mBR) system has gained popularity due to the integration of different morphological signatures that are related to BCa aggressiveness [4]. Recently, the close relationship between BCa grade and prognosis (i.e., patient outcome) has been explored [5], [6]; yet clinical usage of mBR grade is often limited by concerns about intra-and interrater variability [7]–[9]. Meyer et al. [9] showed that agreement between seven pathologists is only moderately reproducible (κ = 0.50–0.59), while Dalton et al. [8] further illustrated the suboptimal treatment that can result from incorrect mBR grading. Boiesen et al. [7] demonstrated similar levels of reproducibility (κ = 0.50–0.54) across a number of pathology departments. A possible reason for this discrepancy is that pathologists currently lack the automated image analysis tools to accurately, efficiently, and reproducibly quantify mBR grade in histopathology.
The primary goal of this paper is to identify a quantitative image signature that allows for discrimination of low versus high, low versus intermediate, and intermediate versus high mBR grade on whole-slide estrogen receptor-positive (ER+) BCa histopathology images. The mBR grading system encompasses three visual signatures (degree of tubular formation, nuclear pleomorphism, and mitotic activity), each of which is scored on a scale of 1–3 to produce a combined mBR scale of 3–9 [4]. We quantify various aspects of mBR grade by focusing on the architectural and textural descriptors in BCa tissue. Variations in nuclear architecture (i.e., the 2-D spatial arrangement of cancer nuclei in histopathology) are important in clinical practice because they allow pathologists to distinguish between normal and cancerous tissues as well as between levels of differentiation and tubule formation in BCa tumor cells [4]. Textural information from nuclear regions (i.e., nuclear texture) represents the variation in chromatin arrangement [10], which is generally more heterogeneous in rapidly dividing, higher grade BCa cells. Computerized modeling of the phenotypic appearance of BCa histopathology has traditionally focused on quantifying nuclear morphology [11]–[14] as well as various textural representations of image patches [10], [11], [15]–[17]. In this paper, we address some of the shortcomings in previous works, including 1) comprehensive analysis of whole-slide histology rather than individual nuclei [10], [11] and 2) consideration of the intermediate mBR grade rather than a limited low- versus high-grade evaluation [13]. Recently, researchers have used also fractals to describe the variations architectural complexity of epithelial tissue with respect to the level of differentiation of cells in BCa tumors [18]–[21]. While these studies are extremely promising, their results are still preliminary because evaluation has generally been limited to isolated fields-of-view (FOVs) (e.g. individual cells in [19] and tissue microarrays (TMAs) in [20]), relatively small cohorts [19], and specialized stains [20].
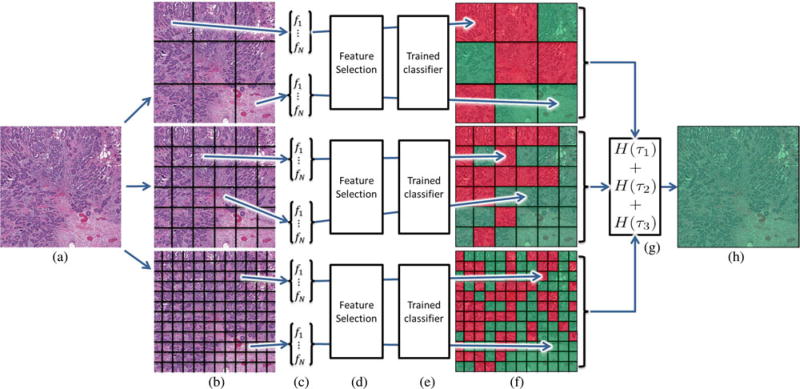
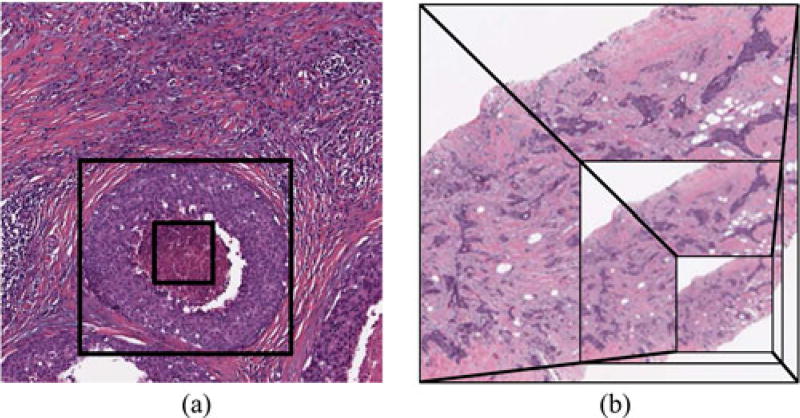
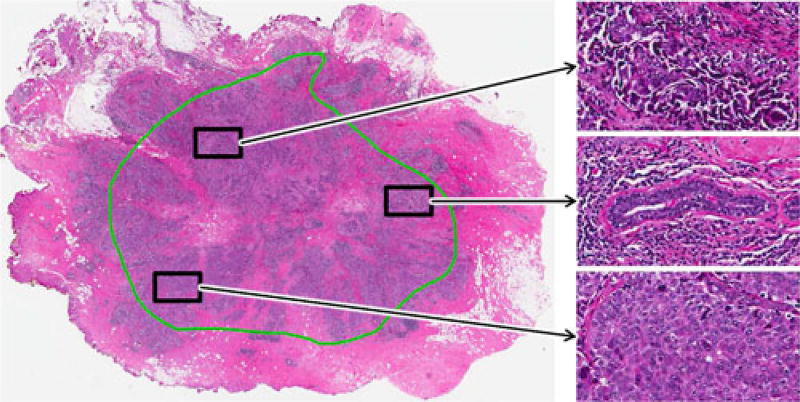
In order to differentiate entire ER+BCa histopathology slides based on their mBR grades, we utilize a multi-FOV classifier that automatically integrates image features from multiple FOVs at various sizes [22], [23] (see Fig. 3). While clinicians perform this task implicitly, the a priori selection of an optimal FOV (i.e., image patch) size for computerized analysis of entire histopathology slides is not straightforward. For example, in Fig. 1(a), while the smallest FOV simply looks like necrotic tissue, the medium-sized FOV would be accurately classified as ductal carcinoma in situ (DCIS). At the other end of the spectrum, the largest FOV (i.e., entire image) containing both DCIS and invasive cancer would be classified ambiguously since it is too heterogeneous. It is important to note that the multi-FOV framework differs from traditional multiscale (i.e., multiresolution) classifiers that operate on a fixed FOV at multiple spatial resolutions [24]–[26] [see Fig. 1(b)]. While this approach is often useful for evaluating large images in a hierarchical manner [26], it may not be able to capture the local heterogeneity found in large BCa histopathology slides [27], [28] (see Fig. 2).
Fig. 3.
Flowchart outlining the methodological steps of the automated BCa grading system, whereby (a) an ER+ BCa histopathology slide is first divided into (b) FOVs of various sizes. (c) Image features that quantify mBR grade phenotype are extracted from each FOV and (d) a feature selection scheme is used to identify salient features at each FOV size. (e) Pretrained classifiers are used to predict (f) mBR grade for each FOV (illustrated by red and green squares). (g) Predictions for individual FOVs are aggregated to achieve a class prediction H(τ) for an entire FOV size τ. (h) Class predictions from FOV sizes are combined to achieve a final classification result for the entire ER+ BCa histopathology slide.
Fig. 1.
(a) Multi-FOV framework presented in this paper operates by maintaining a fixed scale while analyzing several FOV sizes. (b) Conversely, a multiscale framework would operate by analyzing a fixed FOV at different spatial resolutions.
Fig. 2.
FOVs taken from a single histopathology slide illustrate the high level of intratumoral heterogeneity in ER+ BCa. The green annotation represents invasive cancer as determined by an expert pathologist. Note the disorganized tissue structure of FOVs with higher malignancy (top, bottom) compared to the FOV containing benign tissue (middle).
The main novel contributions of this study are the following.
A multi-FOV classifier able to apply a single operator across a multitude of FOV at different sizes in order to extract relevant histomorphometric information.
The incorporation of a robust feature selection scheme into the multi-FOV framework to independently identify salient image features at each FOV size.
The first image-based classifier specifically correlating with BCa grade that comprehensively analyzes digitized whole-slide histopathology images rather than arbitrarily selected FOVs.
The rest of the paper is organized as follows. Previous work and novel contributions are explained in Section II. Section III details the methods used for feature extraction, feature selection, and patient classification. The experimental design is presented in Section IV, followed by quantitative results in Section V, and concluding remarks in Section VI.
II. Previous Related Work
In this paper, we employ multiple classes of quantitative image features characterizing both the architecture and texture of BCa cancer nuclei, both of which reflect various aspects of the mBR grading system. Here, this concept is modeled by using individual nuclei as vertices for the construction of graphs (Voronoi Diagram (VD), Delaunay Triangulation (DT), and Minimum Spanning Tree (MST) and, subsequently, extracting relevant statistics related to the size, shape, and length of the graphs. Such graph-based features have previously been used to accurately distinguish variations in lymphocytic infiltration [14], cancer type [29], tumor grade [13], [30], and prognosis [30] in digitized BCa histopathology, as well as hierarchical tissue structure in glioma [31] and tumor grade in prostate [32]. In addition, researchers have recently demonstrated the ability to identify high-grade regions within individual BCa histopathology slides via sparse analysis of VDs [33]. Hence, the applicability of graph-based features for a wide range of diseases and classification tasks suggests that they are able to quantify the general large-scale patterns that reflect varying levels of tissue organization across different disease states.
The diagnostic importance of nuclear texture has been widely studied [15], [34]–[36]; yet recent work in differentiating BCa grade via analysis of nuclear texture has been limited. For example, Weyn et al. performed a limited study that explored the ability of wavelet, Haralick, and densitometric features to distinguish nuclei from low, intermediate, and high BCa tissues [10]. More recently, Petushi et al. [15]. found that the extent of cell nuclei with dispersed chromatin is related to BCa tumor grade. Note that this differs from studies that have relied on the extraction of textural statistics from entire FOVs (i.e., global texture) [13], [26]. Doyle et al. utilized gray-level, Gabor, and Haralick texture features extracted from entire FOVs to discriminate low- and high-grade tumors in both prostate [26] and BCa [13] histopathology. In this paper, Haralick texture features [37] (i.e., second-order statistics calculated from a graylevel co-occurrence matrix) are calculated within segmented nuclear regions. Haralick features have previously been used in both nuclear and global textural analysis for classification of tumor grade in numerous cancers, including the breast [10], [15], prostate [26], and thyroid [36].
The identification of relevant image features is undoubtedly important; yet, the selection of appropriate FOVs must also be considered in the analysis of large histopathology slides. Prior work in histological image analysis has traditionally involved empirical selection of individual FOVs at a fixed size based on experimental results [13]–[15], [30], [38], [39], leading to potential bias and cost (in terms of both time and money) associated with user intervention. Note that manual FOV selection is also intrinsic to the TMA analysis, in which small tissue “spots” are sampled from larger regions of interest by an expert pathologist [40]. Some researchers have utilized randomized FOV selection in an effort to address the bias associated with manual FOV selection [10], [19], [41]. However, random sampling introduces variability into the classification results that can be difficult to overcome, especially in heterogeneous cancers such as BCa where random FOVs may not be representative of the overall histopathology slide. More recently, Huang et al. have approached FOV selection from a holistic perspective through the use of dynamic sampling, which incorporates domain information into the identification of salient regions of interest [33]. By contrast, the multi-FOV approach does not require empirical selection of FOVs or an optimal FOV size for classification; rather this approach combines class predictions from image features across all FOV sizes.
III. Methods
For all methods, an image scene 𝒞 = (C, g) is defined as a 2-D set of pixels c ∈ C with associated vectorial function g assigning the red, green, and blue (RGB) color space and class label 𝒴(𝒞) ∈ {0, 1}.
For each 𝒞 and FOV size τ ∈ T, a grid containing FOVs is constructed, where , m ∈ {1, 2, …, M (τ)} is a square FOV with edge length of τ pixels and M(τ) is the total number of FOVs for a given τ. We define as the function that extracts features from each . Grid construction and feature extraction are repeated likewise for each τ ∈ T.
A. Nuclear Detection and Segmentation
The extraction of features describing nuclear architecture and nuclear texture first require: 1) identification of the centroids of individual cancer nuclei; and 2) the segmentation of nuclear regions, respectively. For both tasks, we take advantage of the fact that hematoxylin primarily stains nuclear structures.
1) Color Deconvolution
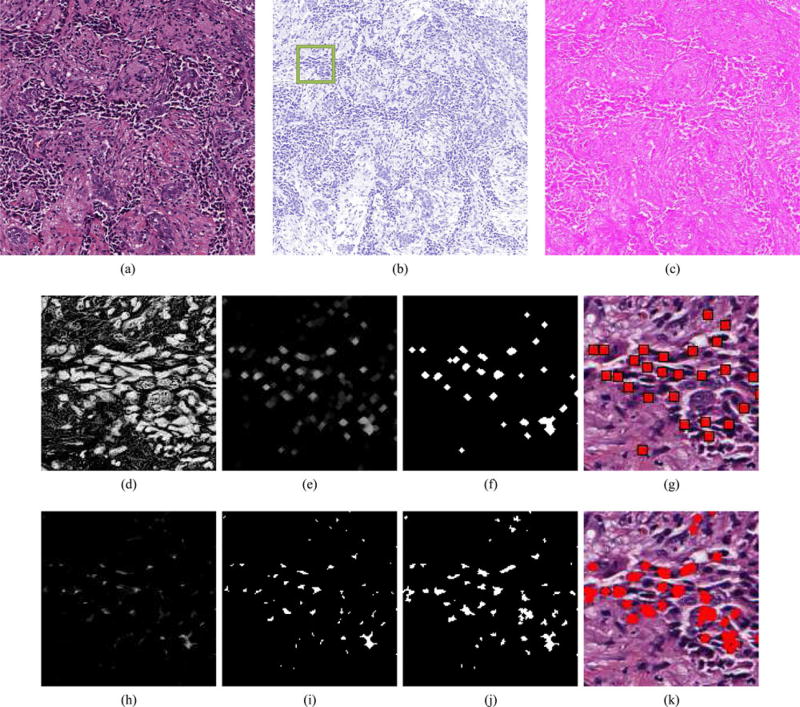
First, color deconvolution [42], [43] is used to convert each FOV from the RGB color space g to a new color space ḡ defined by hematoxylin H, eosin E, and background K (i.e., white) channels [see Fig. 4(b) and (c)]. The relationship between color spaces g and ḡ is defined as g = Aḡ, where the transformation matrix is given by
| (1) |
where ĤR, ĤG, and ĤB denote the predefined, normalized RGB values, respectively, for the H channel. The second and third rows of A are defined analogously for the E and K channels, respectively. In this paper, the predefined values in A are selected based on published values by Ruifrok and Johnston [42]. The intensity of a pixel c in the new color space is defined as ḡ(c) = A−1 (c)g(c), where g(c) and ḡ(c) are 3 × 1 column vectors. The extent of hematoxylin staining H(C) = {H(c): ∀c ∈ C} is subsequently isolated [see Fig. 4(d)]. Centroids of individual nuclei are identified by applying morphological opening to the hematoxylin channel and thresholding the result [see Fig. 4(e)–(g)]. Note that this method does not detect each and every nucleus; instead, it identifies a sufficient number of nuclei to reflect variations in their spatial arrangement in the entire FOV.
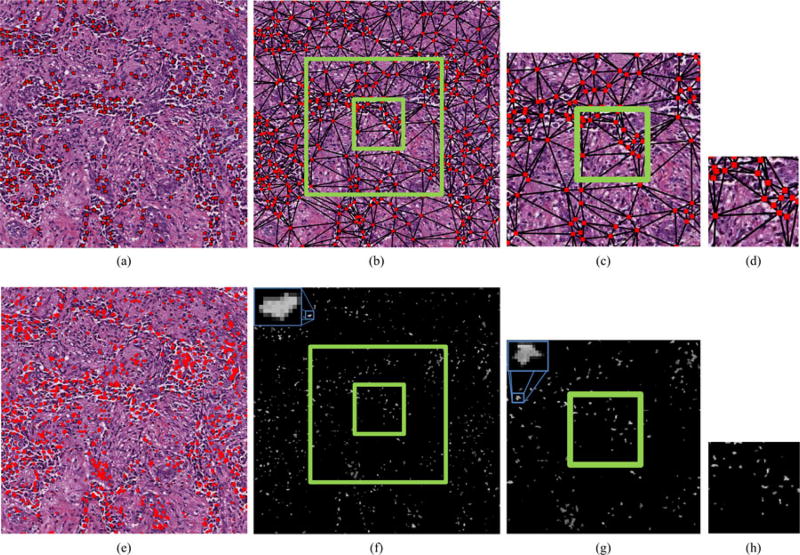
Fig. 4.
(a) High-grade histopathology image with its (b) hematoxylin and (c) eosin channels separated by color deconvolution. The green box in (b) denotes an inset providing more detailed visualization of the nuclear detection and segmentation process in (d)–(k). For nuclear detection, (d) the intensity of the hematoxlyin channel undergoes, (e) morphological opening, and (f) thresholding. (g) Centroids of the individual nuclei are later used for graph construction. The nuclear segmentation process also uses (d) the intensity of the hematoxylin channel, applying (h) morphological erosion, (i) thresholding, and (j) the color gradient based active contour model (CGAC), to achieve (k) a final segmentation result that is used for extraction of nuclear texture.
2) Color Gradient Based Geodesic Active Contour
To segment nuclear regions, the hematoxylin channel [see Fig. 4(d)] is used to initialize a color gradient based geodesic active contour (CGAC) model [44]. Assuming the image plane Ω ∈ ℝ2 is partitioned into two nonoverlapping regions, the foreground Ωf and background Ωb, by a zero level set function φ. The optimal partition can be obtained through minimizing the energy functional as follows:
| (2) |
where the first and second terms are the energy functional of a traditional GAC model [45] and the balloon force [46], respectively. An additional third term is added to the energy functional to remove the reinitialization phase which is required as a numerical remedy for maintaining stable curve evolution in traditional level set methods [47]. The edge-detector function in the traditional GAC model and the balloon force are based on the calculation of the gray scale gradient of the image [45]. In this paper, the edge-detector function is based on the color gradient which is defined as . Here, s(g(c)) is the local structure tensor based color gradient which is defined as [48], where λ+ and λ− are the maximum and minimum eigenvalues of the local structure tensor of each pixel in the image. By locally summing the gradient contributions from each image channel, this term is able to represent the extreme rates of change in the direction of the corresponding eigenvectors. The final boundaries of the CGAC model are used to define a mask denoting nuclear regions [see Fig. 4(j)]. Note that we aim to segment only nuclei belonging to cancerous epithelial cells while avoiding the darker nuclei representing lymphocytes and fibroblasts.
B. Feature Extraction
For each FOV , we extract two sets of quantitative image features [i.e., nuclear architecture and nuclear texture ] that reflect the phenotypic variations seen across BCa grades.
1) Quantification of Nuclear Architecture via Graph-Based Features
Utilizing individual nuclei for the construction of graphs allows for the quantification of tissue architecture. A graph is defined as a set of vertices (i.e., BCa nuclei) with corresponding edges connecting all nuclei. In this paper, we consider three graphs: VD, DT, and MST). For a particular FOV, the VD constructs a polygon around each nucleus such that each pixel in the image falls into the polygon associated with the nearest nucleus. The DT is simply the dual graph of the VD and is constructed such that two nuclei are connected by an edge if their associated polygons share an edge in the VD [see Fig. 5(b)–(d)]. The MST connects all nuclei in the image while minimizing the total length of all edges. A total of 25 features describing variations in these graphs are extracted (see Table I). An additional 25 features are calculated directly from individual nuclei to quantify nearest neighbor (NN) and global density statistics (see Table I), resulting in a total of 50 features describing nuclear architecture for each FOV .
Fig. 5.
(a) Centroids of nuclei are isolated from the hematoxylin channel and used as vertices for the construction of graphs such as the (b)–(d) DT at various FOV sizes, from which features describing nuclear architecture are extracted. (e) The segmentation of nuclei allows for extraction of Haralick texture features such as (f)–(h) Contrast Variance in FOVs of various sizes. For all images, note that green boxes represent FOVs of different sizes and blue boxes represent insets to enhance visualization.
TABLE I.
Fifty Nuclear Architecture Features Used in This Paper, Derived From VD, DT, and MST Graphs, as Well as NN Statistics
| Type | Name |
|---|---|
| VD (13) | Total area of polygons |
| Polygon area: mean, std dev, min/max ratio, disorder | |
| Polygon perimeter: mean, std dev, min/max ratio, disorder | |
| Polygon chord length: mean, std dev, min/max ratio, disorder | |
| DT (8) | Triangle side length: mean, std dev, min/max ratio, disorder |
| Triangle area: mean, std dev, min/max ratio, disorder | |
| MST (4) | Edge length: mean, std dev, min/max ratio, disorder |
| NN (25) | Nuclear density |
| Distance to 3 nearest nuclei: mean, std dev, disorder | |
| Distance to 5 nearest nuclei: mean, std dev, disorder | |
| Distance to 7 nearest nuclei: mean, std dev., disorder | |
| # nuclei in 10 µm radius: mean, std dev, disorder | |
| # nuclei in 20 µm radius: mean, std dev, disorder | |
| # nuclei in 30 µm radius: mean, std dev, disorder | |
| # nuclei in 40 µm radius: mean, std dev, disorder | |
| # nuclei in 50 µm radius: mean, std dev, disorder |
2) Quantification of Nuclear Texture via Haralick Features
Using the nuclear mask to restrict analysis to the desired region, Haralick co-occurrence features [26], [37] are extracted from each FOV. First, the FOV is transformed from the RGB color space to the HSV color space since the latter is more similar to the manner in which humans perceive color [49]. At each relevant pixel, a co-occurrence matrix is constructed to quantify the frequency of pixel intensities in a fixed neighborhood. A set of 13 Haralick features [37] are extracted from the co-occurrence matrices (Contrast Energy, Contrast Inverse Moment, Contrast Average, Contrast Variance, Contrast Entropy, Intensity Average, Intensity Variance, Intensity Entropy, Entropy, Energy, Correlation, and two Information Measures of Correlation), from which the mean, standard deviation, and disorder statistics are calculated for each FOV [see Fig. 5(f)–(h)]. This task is repeated for each of the three channels in the HSV color space, resulting in a total of 117 nuclear texture features for each FOV .
C. Feature Selection via Minimum Redundancy Maximum Relevance
Conceptually, a large number of descriptive features are highly desirable in terms of distinguishing patients based on mBR grade. In reality, however, large feature sets present problems in data classification such as 1) the curse of dimensionality [50], which calls for an exponential growth in the data cohort for each additional feature used; and 2) the presence of redundant features that do not provide additional class discriminatory information.
We address both issues by using minimum redundancy maximum relevance (mRMR) feature selection [51], which has previously been used in various biomedical applications ranging from the isolation of salient genes in microarray data [52] to insight into drug interactions of various protein groups [53]. Given a feature set f, the mRMR scheme identifies a subset f̄ ⊂ f that maximizes “relevance” and minimizes “redundancy” between individual features. In practice, feature fj is incrementally included in f̄ based on the criteria
| (3) |
where I is mutual information, 𝒴 is the class label associated with a given sample, and |f̄| represents the cardinality of selected feature set. In this paper, relevant features are isolated from both nuclear architecture f̄NA ⊂ fNA and nuclear texture f̄NT ⊂ fNT feature sets based on their ability to distinguish BCa histopathology slides with low, intermediate, and high mBR grades.
D. Multi-FOV Approach for Whole-Slide Classification
The multi-FOV framework [22] (see Fig. 3) is employed in terms of its ability to classify large, heterogeneous images in an automated and unbiased fashion as described in the algorithm below. For a single slide 𝒞, a pretrained Random Forest [54] classifier is first used to assign an initial class prediction for each individual FOV with associated features f. Predictions are aggregated (i.e., mean prediction) for all FOVs Dτ at a single size τ ∈ T to achieve a combined prediction H(Dτ ; τ, f). Subsequently, the multi-FOV classification H(D; f), where D = {Dτ : ∀τ ∈ T} is the collective data over all FOV sizes, is achieved via a consensus prediction across all FOV sizes. In this paper, consensus is achieved via averaging of H(Dτ ; τ, f), ∀ τ ∈ T.
Input: Image 𝒞. FOV sizes T = {t1, t2, …, tN}. Classifier for each τ ∈ T.
Output: Multi-FOV classification H(D; f) for image 𝒞.
for all τ ∈ T do
From 𝒞, define M(τ) FOVs .
Extract features f from , .
Initial classification of each .
For all FOVs Dτ at size τ, make class prediction .
end for
Across all FOV sizes τ ∈ T, make multi-FOV prediction .
IV. Experimental Design
A. Data Cohort
BCa histopathology slides were obtained from 126 patients (46 low mBR, 60 intermediate mBR, and 20 high mBR) at the Hospital of the University of Pennsylvania and The Cancer Institute of New Jersey. All slides were digitized via a whole-slide scanner at 10× magnification (1 µm/pixel resolution). Each slide is accompanied by mBR grade as determined by an expert pathologist. Note that commonly accepted clinical cutoffs are used to define the low (mBR 3–5), intermediate (mBR 6–7), and high (mBR 8–9) grade classes used as ground truth in this paper. For each experiment, our BCa grading system is evaluated via a series of two-class classification tasks to distinguish slides with low versus high mBR grade, low versus intermediate mBR grade, and intermediate versus high mBR grade. In addition, a wide range of FOVsizes T = 4000, 2000, 1000, 500, 250}µm was selected empirically based on classification of individual FOV sizes in previous work [22], [23].
B. Experiment 1: Whole-Slide Classification Using Nuclear Architecture
We first evaluate the ability of our BCa grading system to discriminate entire BCa histopathology slides based on mBR grade via architectural features f̄NA. Since the multi-FOV classifier utilizes a trained classifier, it is susceptible to the arbitrary selection of training and testing data. A threefold cross-validation scheme is used to mitigate this bias by splitting the data cohort into three subsets in a randomized fashion, from which two subsets are used for training and the remaining subset is used for evaluation. The subsets are subsequently rotated until a multi-FOV prediction H(D;f̄NA) is made for each slide. The multi-FOV predictions for all slides are thresholded to create receiver operating characteristic (ROC) curves using the respective mBR grades as ground truth. The entire cross-validation procedure is repeated 20 times, with the mean and standard deviation of the area under the ROC curve (AUC) reported.
C. Experiment 2: Whole-Slide Classification Using Nuclear Texture
Similar to the procedure outlined in Experiment 1, the BCa grading system is evaluated using the Haralick co-occurrence texture features f̄NT. A multi-FOV prediction H(D;f̄NT) is made for each slide and results over 20 trials of threefold cross-validation are reported, as described in Section IV-B.
D. Experiment 3: Comparison to Classification Across Multiple Image Resolutions
Although this paper focuses on the combination of FOVs of different sizes, the ability to integrate image information at various spatial resolutions is also important for the characterization of digitized histopathology slides [22]. For comparison to the multi-FOV approach, a multiresolution classifier is constructed by reextracting each FOV of size τ = 1000 µm at spatial resolutions of κ ∈ {0.25, 0.5, 1, 2, 4} µm/pixel. A consensus multiresolution prediction is achieved for each histopathology slide in a manner analogous to the multi-FOV approach (see Section III-D), whereby data are aggregated over all spatial resolutions rather than FOV sizes.
V. Results and Discussion
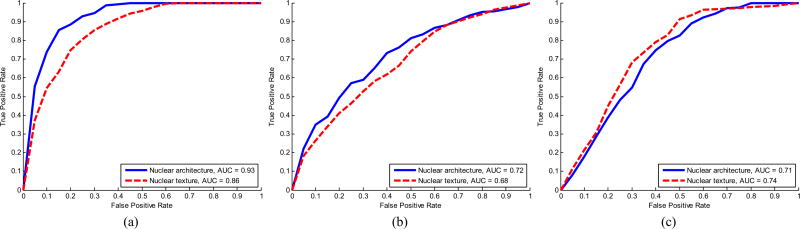
Quantitative results (see Table VIII) suggest that predictions made by nuclear architecture H(D;f̄NA) and nuclear texture H(D;f̄NT) both perform well in characterizing mBR grade in entire ER+ BCa histopathology slides (see Fig. 6). Specifically, nuclear architecture appears to yield higher AUC values than nuclear texture (AUC of 0.93 and 0.86) in terms of discriminating low- versus high mBR grade. By contrast, both nuclear architecture and nuclear texture yield similar results for distinguishing low versus intermediate (AUC of 0.72 and 0.68) and intermediate versus high mBR grade (AUC of 0.71 and 0.74) slides, respectively.
TABLE VIII.
AUC Values for the Comparison of Low-, Intermediate-, and High-Grade Cancers Using Both Multi-FOV and Multiresolution Classifiers
| Experiment | Feature Set | Multi-FOV | Multi-Res. |
|---|---|---|---|
| Low vs. high | f̄NA | 0.93 ± 0.012 | 0.86 ± 0.035 |
| f̄NT | 0.86 ± 0.036 | 0.84 ± 0.036 | |
| Low vs. intermed. | f̄NA | 0.72 ± 0.037 | 0.67 ± 0.049 |
| f̄NT | 0.68 ± 0.028 | 0.67 ± 0.074 | |
| Interned, vs. high | f̄NA | 0.71 ± 0.051 | 0.65 ± 0.054 |
| f̄NT | 0.74 ± 0.036 | 0.66 ± 0.075 |
Fig. 6.
Mean ROC curves over 20 trials of threefold cross-validation for (a) low versus high grade, (b) low versus intermediate grade, and (c) intermediate versus high grade classification tasks. For each task, ROC curves are shown for both nuclear architecture and nuclear texture feature sets along with associated AUC values.
A. Experiment 1: Feature Selection in Nuclear Architecture
To mitigate the challenges associated with large feature sets (as discussed in Section III-C), the ROC curves in Fig. 6 were constructed using feature subsets selected by the mRMR algorithm. For each experiment, Tables II–VI show the features selected at each FOV size along with the cumulative classification accuracy of the multi-FOV approach with the inclusion of each additional feature. Note that some experiments, e.g., nuclear architecture for low versus high grading (see Table II) and nuclear texture for intermediate versus high grading (see Table VII), demonstrate considerable improvement in classification accuracy with the addition of relevant features, while other experiments, e.g., nuclear texture for low versus intermediate grading (see Table VI) reach a plateau with the selection of only one or two features.
TABLE II.
Selected Nuclear Architecture Features for Low versus High mBR Grade Classification
| Rank | τ | Feature Description | Cum. Acc. |
|---|---|---|---|
| 1 | 4000 | VD chord length: min/max ratio | 0.85 ± 0.033 |
| 2000 | DT side length: min/max ratio | ||
| 1000 | # nuclei in 10 µm radius: mean | ||
| 500 | # nuclei in 10 µm radius: mean | ||
| 250 | # nuclei in 10 µm radius: mean | ||
| 2 | 4000 | DT area: min/max ratio | 0.86 ± 0.056 |
| 2000 | DT area: disorder | ||
| 1000 | # nuclei in 10 µm radius: disorder | ||
| 500 | DT area: min/max ratio | ||
| 250 | Dist. to 5 nearest nuclei: disorder | ||
| 3 | 4000 | VD area: min/max ratio | 0.88 ± 0.038 |
| 2000 | # nuclei in 10 µm radius: mean | ||
| 1000 | DT area: min/max ratio | ||
| 500 | # nuclei in 10 µm radius: disorder | ||
| 250 | DT area: min/max ratio | ||
| 4 | 4000 | # nuclei in 10 µm radius: mean | 0.91 ± 0.023 |
| 2000 | MST edge length: min/max ratio | ||
| 1000 | MST edge length: min/max ratio | ||
| 500 | DT side length: min/max ratio | ||
| 250 | Dist. to 7 nearest nuclei: disorder | ||
| 5 | 4000 | VD perimeter: min/max ratio | 0.91 ± 0.015 |
| 2000 | # nuclei in 10 µm radius: disorder | ||
| 1000 | DT side length: min/max ratio | ||
| 500 | Dist to 7 nearest nuclei: disorder | ||
| 250 | DT side length: min/max ratio |
TABLE VI.
Selected Nuclear Texture Features for Low versus Intermediate mBR Grade Classification
| Rank | τ | Feature Description | Cum. Acc. |
|---|---|---|---|
| 1 | 4000 | Hue: Contrast variance - disorder | 0.69 ± 0.024 |
| 2000 | Sat: Contrast variance - mean | ||
| 1000 | Val: Contrast average - std. dev. | ||
| 500 | Sat: Contrast variance - std. dev. | ||
| 250 | Sat: Info, measure 1 - std. dev. | ||
| 2 | 4000 | Val: Contrast variance - std. dev. | 0.69 ± 0.027 |
| 2000 | Val: Contrast variance - std. dev. | ||
| 1000 | Sat: Contrast inv. moment - std. dev. | ||
| 500 | Sat: Info, measure 1 - std. dev. | ||
| 250 | Sat: Contrast variance - std. dev. | ||
| 3 | 4000 | Val: Contrast entropy - disorder | 0.70 ± 0.024 |
| 2000 | Sat: Contrast average - std. dev. | ||
| 1000 | Hue: Intensity average - disorder | ||
| 500 | Val: Info, measure 1 - std. dev. | ||
| 250 | Sat: Contrast inv. moment - std. dev. |
TABLE VII.
Selected Nuclear Texture Features for Intermediate versus High mBR Grade Classification
| Rank | τ | Feature Description | Cum. Acc. |
|---|---|---|---|
| 1 | 4000 | Hue: Contrast variance - std. dev. | 0.68 ± 0.082 |
| 2000 | Hue: Contrast variance - mean | ||
| 1000 | Sat: Contrast variance - std. dev. | ||
| 500 | Val: Contrast variance - std. dev. | ||
| 250 | Val: Contrast variance - std. dev. | ||
| 2 | 4000 | Hue: Contrast variance - mean | 0.75 ± 0.044 |
| 2000 | Val: Contrast entropy - disorder | ||
| 1000 | Hue: Contrast variance - mean | ||
| 500 | Sat: Contrast variance - std. dev. | ||
| 250 | Sat: Contrast variance - std. dev. | ||
| 3 | 4000 | Sat: Contrast variance - mean | 0.74 ± 0.040 |
| 2000 | Hue: Contrast variance - std. dev. | ||
| 1000 | Hue: Contrast variance - std. dev. | ||
| 500 | Val: Contrast entropy - disorder | ||
| 250 | Hue: Info, measure 1 - std. dev. | ||
| 4 | 4000 | Sat: Contrast variance - std. dev. | 0.74 ± 0.030 |
| 2000 | Sat: Contrast variance - mean | ||
| 1000 | Val: Contrast variance - std. dev. | ||
| 500 | Sat: Contrast inv. moment - std. dev. | ||
| 250 | Sat: Contrast inv. moment - std. dev. | ||
| 5 | 4000 | Hue: Contrast variance - disorder | 0.75 ± 0.035 |
| 2000 | Sat: Contrast variance - std. dev. | ||
| 1000 | Sat: Contrast variance - mean | ||
| 500 | Val: Entropy - std. dev. | ||
| 250 | Val: Contrast entropy - disorder |
In addition to improved classification accuracy, the feature selection process also reveals the specific features that best distinguish low- and high-grade cancers. For example, Table II suggests that the average number of neighboring nuclei in a 10-µm radius around each nucleus is the most discriminating feature in smaller FOVs (1000, 500, and 250 µm), but has lesser importance in larger FOV sizes of 2000 and 4000 µm, where it is ranked third and fourth, respectively. Conversely, graph-based features derived from the VD and DT appear to play a greater role in larger FOVs, where variations in VD chord length, DT side length, and DT area are more important than NN statistics. This pattern is further reinforced in the features selected for distinguishing low versus intermediate grades (see Table III) and intermediate versus high grades (see Table IV).
TABLE III.
Selected Nuclear Architecture Features for Low Versus Intermediate mBR Grade Classification
| Rank | τ | Feature Description | Cum. Acc. |
|---|---|---|---|
| 1 | 4000 | VD perimeter: min/max ratio | 0.71 ± 0.0042 |
| 2000 | DT area: disorder | ||
| 1000 | DT area: disorder | ||
| 500 | # nuclei in 10 µm radius: disorder | ||
| 250 | DT side length: min/max ratio | ||
| 2 | 4000 | VD chord length: min/max ratio | 0.71 ± 0.011 |
| 2000 | DT side length: min/max ratio | ||
| 1000 | VD chord length: min/max ratio | ||
| 500 | Dist. to 7 nearest nuclei: disorder | ||
| 250 | # nuclei in 10 µm radius: mean | ||
| 3 | 4000 | DT area: disorder | 0.73 ± 0.028 |
| 2000 | # nuclei in 10 µm radius: mean | ||
| 1000 | # nuclei in 10 µm radius: mean | ||
| 500 | Dist. to 5 nearest nuclei: disorder | ||
| 250 | Dist. to 3 nearest nuclei: disorder | ||
| 4 | 4000 | MST edge length: min/max ratio | 0.74 ± 0.037 |
| 2000 | VD perimeter: min/max ratio | ||
| 1000 | VD perimeter: min/max ratio | ||
| 500 | DT area: min/max ratio | ||
| 250 | # nuclei in 10 µm radius: disorder |
TABLE IV.
Selected Nuclear Architecture Features for Intermediate Versus High mBR Grade Classification
| Rank | τ | Feature Description | Cum. Acc. |
|---|---|---|---|
| 1 | 4000 | VD area: min/max ratio | 0.70 ± 0.035 |
| 2000 | DT area: disorder | ||
| 1000 | DT area: disorder | ||
| 500 | VD area: std. dev. | ||
| 250 | DT area: min/max ratio | ||
| 2 | 4000 | DT area: disorder | 0.71 ± 0.054 |
| 2000 | VD perimeter: min/max ratio | ||
| 1000 | VD chord length: min/max ratio | ||
| 500 | # nuclei in 10 µm radius: mean | ||
| 250 | Dist. to 7 nearest nuclei: disorder | ||
| 3 | 4000 | DT side length: min/max ratio | 0.72 ± 0.048 |
| 2000 | # nuclei in 10 µm radius: mean | ||
| 1000 | MST edge length: min/max ratio | ||
| 500 | DT area: disorder | ||
| 250 | # nuclei in 40 pm radius: mean | ||
| 4 | 4000 | VD chord: min/max ratio | 0.73 ± 0.056 |
| 2000 | DT side length: min/max ratio | ||
| 1000 | DT side length: min/max ratio | ||
| 500 | DT area: min/max ratio | ||
| 250 | # nuclei in 10 µm radius: mean |
B. Experiment 2: Feature Selection in Nuclear Texture
By examining the features selected for nuclear texture (see Tables V–VII), the dominant role of contrast statistics (especially variance and entropy) is immediately apparent. In addition, the information measure of correlation is shown to have importance for discriminating smaller FOVs (τ ∈ {250, 500}) and data across all three channels (hue, saturation, and intensity) appear to be equally relevant in terms of meaningful feature extraction.
TABLE V.
Selected Nuclear Texture Features for Low versus High mBR Grade Classification
| Rank | τ | Feature Description | Cum. Acc. |
|---|---|---|---|
| 1 | 4000 | Val: Contrast variance - std. dev. | 0.80 ± 0.047 |
| 2000 | Hue: Contrast variance - mean | ||
| 1000 | Sat: Contrast variance - std. dev. | ||
| 500 | Val: Contrast variance - std. dev. | ||
| 250 | Val: Contrast entropy - disorder | ||
| 2 | 4000 | Sat: Contrast variance - std. dev. | 0.81 ± 0.044 |
| 2000 | Sat: Contrast variance - mean | ||
| 1000 | Hue: Contrast variance - mean | ||
| 500 | Hue: Info, measure 1 - std. dev. | ||
| 250 | Hue: Info, measure 1 - std. dev. | ||
| 3 | 4000 | Hue: Contrast variance - std. dev. | 0.84 ± 0.040 |
| 2000 | Hue: Contrast variance - std. dev. | ||
| 1000 | Val: Contrast variance - std. dev. | ||
| 500 | Val: Contrast entropy - disorder | ||
| 250 | Val: Contrast average - std. dev. |
C. Experiment 3: Comparison to Multiresolution Classifier
Using all selected features from each classification task (see Tables II–VII), the multi-FOV approach is further evaluated via comparison to a multiresolution scheme. A comparison of AUC values between the two methods (see Table VIII) suggests that the aggregation of image features at multiple FOVs (i.e., multi-FOV classifier) is able to outperform the aggregation of image features at multiple spatial resolutions (i.e., multiresolution classifier) for the grading of BCa histopathology slides. For nuclear architecture fNA, the superiority of the multi-FOV approach in terms of differentiating low versus high grades (AUC = 0.93 ± 0.012), low versus intermediate grades (AUC = 0.72 ± 0.037), and intermediate versus high grades (AUC = 0.71 ± 0.051) is expected since the spatial arrangement of nuclei is invariant to changes in image resolution. In addition, the ability of a nuclear textural features fNT to perform comparably to nuclear architecture in distinguishing low versus high grades (AUC = 0.84 ± 0.036) and low versus intermediate grades (AUC = 0.67 ± 0.074) is also unsurprising since textural representations of nuclei will reveal different types of class discriminatory information at various image resolutions. These results suggest that an intelligent combination of the multi-FOV and multiresolution approaches may yield improved classification of tumor grade in whole-slide BCa histology.
VI. Concluding Remarks
The development of a quantitative, reproducible grading system for whole-slide histopathology will be an indispensable diagnostic and prognostic tool for clinicians and their patients. In this paper, we demonstrate a computerized grading scheme for ER+ BCa that uses only image features from entire H and E stained histopathology slides. Specifically, we exhibit 1) a multi-FOV classifier with robust feature selection for classifying entire ER+BCa histopathology slides into low, intermediate, and high mBR grades; 2) a BCa grading system that utilizes all image information on a digitized histopathology slide rather than arbitrarily selected FOVs; and 3) image features describing both nuclear architecture (i.e., spatial arrangement of nuclei) and nuclear texture (i.e., textural patterns within nuclei) that are able to quantify mBR grade. It is important to note that, while the initial multi-FOV scheme was formulated in [22], the implementation presented in this paper involves a much larger cohort as well as experiments involving the classification of intermediate mBR grades. This paper also introduces the addition of a feature selection scheme, yielding both improvements in classifier performance and specific image features that play an important role in computerized BCa grading.
Unsurprisingly, the grading system performs best when differentiating patients with low- and high-grade tumors. We were also able to achieve reasonable performance for distinguishing patients with low- and intermediate-grade tumors and patients with intermediate- and high-grade tumors. Future work will involve the incorporation of additional feature sets (e.g., tubule formation patterns, fractal-based, and textural) as well as novel methods for producing an intelligent combination of various feature sets. In addition, we will explore the possibility of integrating the multi-FOV and multiresolution approaches, which may yield improved results with respect to features that vary at different image resolutions (e.g., textural representations).
An issue worth investigating in future work will be as to how class balance affects the performance of the classifier and the corresponding features. Class balance refers to the differences in the number of low-, intermediate-, and high-grade studies, an issue that has been known to bias classifier performance [55]. The lack of class balance in our cohort is representative of the ER+ BCa population as a whole, in which women are more likely to be diagnosed in the earlier stages of the disease. While we have previously discussed its importance in the classification of digital pathology [56], a more rigorous analysis of the effect of class balance on the multi-FOV classifier will need to be addressed in future work.
Acknowledgments
This work was supported by the National Institute of Health under Grant R01CA136535, Grant R01CA140772, Grant R43EB015199, and Grant R21CA167811), the National Science Foundation under Grant IIP-1248316, and the QED Award from the University City Science Center and Rutgers University.
Footnotes
Color versions of one or more of the figures in this paper are available online at http://ieeexplore.ieee.org.
Contributor Information
Ajay Basavanhally, Department of Biomedical Engineering, Rutgers University, Piscataway, NJ 08854 USA.
Shridar Ganesan, Cancer Institute of New Jersey, New Brunswick, NJ 08903-2681 USA.
Michael Feldman, Department of Surgical Pathology, Hospital of the University of Pennsylvania, Philadelphia, PA 19104 USA.
Natalie Shih, Department of Surgical Pathology, Hospital of the University of Pennsylvania, Philadelphia, PA 19104 USA.
Carolyn Mies, Department of Surgical Pathology, Hospital of the University of Pennsylvania, Philadelphia, PA 19104 USA.
John Tomaszewski, Department of Pathology and Anatomical Sciences, The State University of New York, Buffalo, NY 08854 USA.
Anant Madabhushi, Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH 44106 USA.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. Cancer J. Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Ward E, Thun M. Declining death rates reflect progress against cancer. PLoSOne. 2010;5(3):e9584. doi: 10.1371/journal.pone.0009584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genestie C, Zafrani B, Asselain B, Fourquet A, Rozan S, Validire P, Vincent-Salomon A, Sastre-Garau X. Comparison of the prognostic value of Scarff-Bloom-Richardson and Nottingham histological grades in a series of 825 cases of breast cancer: Major importance of the mitotic count as a component of both grading systems. Anticancer Res. 1998;18(1B):571–576. [PubMed] [Google Scholar]
- 4.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology. 1991 Nov;19(5):403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 5.Flanagan MB, Dabbs DJ, Brufsky AM, Beriwal S, Bhargava R. Histopathologic variables predict oncotype dx recurrence score. Mod. Pathol. 2008 Oct;21(10):1255–1261. doi: 10.1038/modpathol.2008.54. [DOI] [PubMed] [Google Scholar]
- 6.Weigelt B, Reis-Filho JS. Molecular profiling currently offers no more than tumour morphology and basic immunohistochemistry. Breast Cancer Res. 2010;12(4):S5. doi: 10.1186/bcr2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boiesen P, Bendahl PO, Anagnostaki L, Domanski H, Holm E, Idvall I, Johansson S, Ljungberg O, Ringberg A, Ostberg G, Fernö M, et al. Histologic grading in breast cancer–reproducibility between seven pathologic departments. South Sweden Breast Cancer Group. Acta Oncol. 2000;39(1):41–45. doi: 10.1080/028418600430950. [DOI] [PubMed] [Google Scholar]
- 8.Dalton LW, Pinder SE, Elston CE, Ellis IO, Page DL, Dupont WD, Blamey RW. Histologic grading of breast cancer: Linkage of patient outcome with level of pathologist agreement. Mod. Pathol. 2000 Jul;13(7):730–735. doi: 10.1038/modpathol.3880126. [DOI] [PubMed] [Google Scholar]
- 9.Meyer JS, Alvarez C, Milikowski C, Olson N, Russo I, Russo J, Glass A, Zehnbauer BA, Lister K, Parwaresch R, C. B. C. T. Resource Breast carcinoma malignancy grading by bloom-richardson system vs proliferation index: Reproducibility of grade and advantages of proliferation index. Mod. Pathol. 2005 Aug;18(8):1067–1078. doi: 10.1038/modpathol.3800388. [DOI] [PubMed] [Google Scholar]
- 10.Weyn B, van de Wouwer G, van Daele A, Scheunders P, van Dyck D, vanMarck E, Jacob W. Automated breast tumor diagnosis and grading based on wavelet chromatin texture description. Cytometry. 1998 Sep;33(1):32–40. doi: 10.1002/(sici)1097-0320(19980901)33:1<32::aid-cyto4>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 11.Wolberg WH, Street WN, Heisey DM, Mangasarian OL. Computer-derived nuclear features distinguish malignant from benign breast cytology. Human Pathol. 1995 Jul;26(7):792–796. doi: 10.1016/0046-8177(95)90229-5. [DOI] [PubMed] [Google Scholar]
- 12.Rajesh L, Dey P, Joshi K. Automated image morphometry of lobular breast carcinoma. Anal. Quant. Cytol. Histol. 2002 Apr;24(2):81–84. [PubMed] [Google Scholar]
- 13.Doyle S, Agner S, Madabhushi A, Feldman M, Tomaszewski J. Automated grading of breast cancer histopathology using spectral clustering with textural and architectural image features. Proc. IEEE 5th Int. Symp. Biomed. Imag. Nano Macro. 2008 May;:496–499. [Google Scholar]
- 14.Basavanhally AN, Ganesan S, Agner S, Monaco JP, Feldman MD, Tomaszewski JE, Bhanot G, Madabhushi A. Computerized image-based detection and grading of lymphocytic infiltration in her2+ breast cancer histopathology. IEEE Trans. Biomed. Eng. 2010 Mar;57(3):642–653. doi: 10.1109/TBME.2009.2035305. [DOI] [PubMed] [Google Scholar]
- 15.Petushi S, Garcia FU, Haber MM, Katsinis C, Tozeren A. Large-scale computations on histology images reveal grade-differentiating parameters for breast cancer. BMC Med. Imag. 2006;6(14) doi: 10.1186/1471-2342-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karaçali B, Tözeren A. Automated detection of regions of interest for tissue microarray experiments: An image texture analysis. BMC Med. Imag. 2007;7(2) doi: 10.1186/1471-2342-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall BH, Ianosi-Irimie M, Javidian P, Chen W, Ganesan S, Foran DJ. Computer-assisted assessment of the human epidermal growth factor receptor 2 immunohistochemical assay in imaged histologic sections using a membrane isolation algorithm and quantitative analysis of positive controls. BMC Med. Imag. 2008;8(11) doi: 10.1186/1471-2342-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cross SS. Fractals in pathology. J. Pathol. 1997 May;182(1):1–8. doi: 10.1002/(SICI)1096-9896(199705)182:1<1::AID-PATH808>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 19.Dey P, Mohanty SK. Fractal dimensions of breast lesions on cytology smears. Diagn. Cytopathol. 2003 Aug;29(2):85–86. doi: 10.1002/dc.10324. [DOI] [PubMed] [Google Scholar]
- 20.Tambasco M, Eliasziw M, Magliocco AM. Morphologic complexity of epithelial architecture for predicting invasive breast cancer survival. J. Trans. Med. 2010;8:140. doi: 10.1186/1479-5876-8-140. [Online] Available: http://dx.doi.org/10.1186/1479-5876-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tay CH, Mukundan R, Racoceanu D. Multifractal analysis of histopathological tissue images; presented at the Proc. Imag. Vis. Comput; Auckland, New Zealand. 2011. [Google Scholar]
- 22.Basavanhally A, Ganesan S, Shih N, Mies C, Feldman M, Tomaszewski J, Madabhushi A. A boosted classifier for integrating multiple fields of view: Breast cancer grading in histopathology. Proc. IEEE Int. Symp. Biomed. Imag. Nano Macro. 2011 Mar;:125–128. [Google Scholar]
- 23.Basavanhally A, Feldman M, Shih N, Mies C, Tomaszewski J, Ganesan S, Madabhushi A. Multi-field-of-view strategy for image-based outcome prediction of multi-parametric estrogen receptor-positive breast cancer histopathology: Comparison to oncotype dx. J. Pathol. Inf. 2011;2 doi: 10.4103/2153-3539.92027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong J, Sertel O, Shimada H, Boyer K, Saltz J, Gurcan M. Computer-aided grading of neuroblastic differentiation: Multi-resolution and multi-classifier approach. Proc. IEEE Int. Conf. Imag. Process. 2007 Sep-Oct;5:525–528. [Google Scholar]
- 25.Boccignone G, Napoletano P, Caggiano V, Ferraro M. A multiresolution diffused expectation-maximization algorithm for medical image segmentation. Comput. Biol. Med. 2007;37(1):83–96. doi: 10.1016/j.compbiomed.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Doyle S, Feldman M, Tomaszewski J, Madabhushi A. A boosted Bayesian multi-resolution classifier for prostate cancer detection from digitized needle biopsies. IEEE Trans. Biomed. Eng. 2012 May;59(5):1205–1218. doi: 10.1109/TBME.2010.2053540. [DOI] [PubMed] [Google Scholar]
- 27.Connor AJM, Pinder SE, Elston CW, Bell JA, Wencyk P, Robertson JFR, Blarney RW, Nicholson RI, Ellis IO. Intratumoural heterogeneity of proliferation in invasive breast carcinoma evaluated with mibi antibody. Breast. 1997;6(4):171–176. [Google Scholar]
- 28.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. Engl. J. Med. 2012 Mar;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loménie N, Racoceanu D. Point set morphological filtering and semantic spatial configuration modeling: Application to microscopic image and bio-structure analysis. Pattern Recognit. 2012 Aug;45(8):2894–2911. [Online] Available: http://dx.doi.org/10.1016/j.patcog.2012.01.021. [Google Scholar]
- 30.Basavanhally A, Xu J, Madabhushi A, Ganesan S. Computer-aided prognosis of er+ breast cancer histopathology and correlating survival outcome with oncotype dx assay. Proc. IEEE Int. Symp. Biomed. Imag. Nano Macro. 2009 Jun-Jul;:851–854. [Google Scholar]
- 31.Demir C, Gultekin SH, Yener B. Augmented cell-graphs for automated cancer diagnosis. Bioinformatics. 2005 Sep;21(2):ii7–i12. doi: 10.1093/bioinformatics/bti1100. [DOI] [PubMed] [Google Scholar]
- 32.Doyle S, Hwang M, Shah K, Madabhushi A, Tomaszewski J, Feldman M. Proc. IEEE Int. Symp. Biomed. Imag. Washington, DC, USA: Apr, 2007. Automated grading of prostate cancer using architectural and textural image features; pp. 1284–1287. [Google Scholar]
- 33.Huang C-H, Veillard A, Roux L, Loménie N, Racoceanu D. Time-efficient sparse analysis of histopathological whole slide images. Comput. Med. Imag. Graph. 2011;35(7–8):579–591. doi: 10.1016/j.compmedimag.2010.11.009. [Online] Available: http://dx.doi.org/10.1016/j.compmedimag.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 34.Dawson AE, Austin R, Jr, Weinberg DS. Nuclear grading of breast carcinoma by image analysis. classification by multivariate and neural network analysis. Amer. J. Clin. Pathol. 1991 Apr;95(4, Suppl. 1):S29–S37. [PubMed] [Google Scholar]
- 35.Palcic B. Nuclear texture: Can it be used as a surrogate endpoint biomarker? J. Cell. Biochem. Suppl. 1994;19:40–46. [PubMed] [Google Scholar]
- 36.Wang W, Ozolek JA, Rohde GK. Detection and classification of thyroid follicular lesions based on nuclear structure from histopathology images. Cytometry A. 2010 May;77(5):485–494. doi: 10.1002/cyto.a.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973 Nov;SMC-3(6):610–621. [Google Scholar]
- 38.Gurcan MN, Boucheron L, Can A, Madabhushi A, Rajpoot N, Yener B. Histopathological image analysis: A review. IEEE Rev. Biomed. Eng. 2009;(2):147–171. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sertel O, Kong J, Shimada H, Catalyurek UV, Saltz JH, Gurcan MN. Computer-aided prognosis of neuroblastoma on wholeslide images: Classification of stromal development. Pattern Recognit. 2009 Jun;42(6):1093–1103. doi: 10.1016/j.patcog.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, West RB, van de Rijn M, Koller D. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci. Trans. Med. 2011 Nov;3(108):108–113. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 41.Tambasco M, Magliocco AM. Relationship between tumor grade and computed architectural complexity in breast cancer specimens. Human Pathol. 2008 May;39(5):740–746. doi: 10.1016/j.humpath.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 2001 Aug;23(4):291–299. [PubMed] [Google Scholar]
- 43.Basavanhally A, Yu E, Xu J, Ganesan S, Feldman M, Tomaszewski J, Madabhushi A. Incorporating domain knowledge for tubule detection in breast histopathology using o’callaghan neighborhoods. Proc. SPIE Med. Imag. 2011;7963(1):1–15. [Google Scholar]
- 44.Xu J, Janowczyk A, Chandran S, Madabhushi A. A high-throughput active contour scheme for segmentation of histopathological imagery. Med. Imag. Anal. 2011;15:851–862. doi: 10.1016/j.media.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caselles V, Kimmel R, Sapiro G. Geodesic active contours. Int. J. Comput. Vis. 1997;22(1):61–79. [Google Scholar]
- 46.Cohen LD. On active contour models and balloons. CVGIP: Imag. Underst. 1991;53(2):211–218. [Google Scholar]
- 47.Li C, Xu C, Gui C, Fox MD. Distance regularized level set evolution and its application to image segmentation. IEEE Trans. Imag. Process. 2010 Dec;19(12):3243–3254. doi: 10.1109/TIP.2010.2069690. [DOI] [PubMed] [Google Scholar]
- 48.Xu J, Janowczyk A, Chandran S, Madabhushi A. A weighted mean shift, normalized cuts initialized color gradient based geodesic active contour model: Applications to histopathology image segmentation. Proc. SPIE. 2010;7623(1):76230Y. [Google Scholar]
- 49.Jain AK. Fundamentals of Digital Image Processing. Englewood Cliffs, NJ, USA: Prentice-Hall; 1989. [Google Scholar]
- 50.Bellman R, Corporation R. Dynamic Programmin. Princeton, NJ, USA: Princeton Univ. Press; 1957. (ser. Rand Corporation research study) [Google Scholar]
- 51.Peng H, Long F, Ding C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005 Aug;27(8):1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 52.Ding C, Peng H. Minimum redundancy feature selection from microarray gene expression data. J Bioinf. Comput. Biol. 2005 Apr;3(2):185–205. doi: 10.1142/s0219720005001004. [DOI] [PubMed] [Google Scholar]
- 53.He Z, Zhang J, Shi X-H, Hu L-L, Kong X, Cai Y-D, Chou K-C. Predicting drug-target interaction networks based on functional groups and biological features. PLoS ONE. 2010 Mar;5(3):e9603. doi: 10.1371/journal.pone.0009603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- 55.Japkowicz N. The class imbalance problem: Significance and strategies. Proc. Int. Conf. Artif. Intell. 2000:111–117. [Google Scholar]
- 56.Doyle S, Monaco J, Feldman M, Tomaszewski J, Madabhushi A. An active learning based classification strategy for the minority class problem: Application to histopathology annotation. BMC Bioinformat. 12:424. doi: 10.1186/1471-2105-12-424. [Online] Available: http://dx.doi.org/10.1186/1471-2105-12-424. [DOI] [PMC free article] [PubMed] [Google Scholar]