Abstract
Tyrosyl–DNA Phosphodiesterases 1 (TDP1) and 2 (TDP2) are eukaryotic enzymes that clean-up after aberrant topoisomerase activity. While TDP1 hydrolyzes phosphotyrosyl peptides emanating from trapped topoisomerase I (Top I) from the 3′ DNA ends, topoisomerase 2 (Top II)-induced 5′-phosphotyrosyl residues are processed by TDP2. Even though the canonical functions of TDP1 and TDP2 are complementary, they exhibit little structural or sequence similarity. Homozygous mutations in genes encoding these enzymes lead to the development of severe neurodegenerative conditions due to the accumulation of transcription-dependent topoisomerase cleavage complexes underscoring the biological significance of these enzymes in the repair of topoisomerase–DNA lesions in the nervous system. TDP1 can promiscuously process several blocked 3′ ends generated by DNA damaging agents and nucleoside analogs in addition to hydrolyzing 3′-phosphotyrosyl residues. In addition, deficiency of these enzymes causes hypersensitivity to anti-tumor topoisomerase poisons. Thus, TDP1 and TDP2 are promising therapeutic targets and their inhibitors are expected to significantly synergize the effects of current anti-tumor therapies including topoisomerase poisons and other DNA damaging agents. This review covers the structural aspects, biology and regulation of these enzymes, along with ongoing developments in the process of discovering safe and effective TDP inhibitors.
INTRODUCTION
The double-helical structure of DNA is paramount for the storage of genetic information and its transmission through DNA metabolic processes such as replication, transcription, recombination and chromatin remodeling. Local unwinding of DNA induced by these DNA metabolic processes causes supercoiling of DNA and topological entanglements that need to be resolved in order to maintain cellular function and genomic stability. Fortunately, cells have evolved special, highly conserved biological tools called topoisomerases to resolve these genomic disruptions.
Topoisomerases I and II regulate DNA topology using a cleavage-religation mechanism in which they induce transient single-strand breaks (SSBs) and double-strand breaks (DSBs) in the DNA respectively (1). The transient cleavage concomitantly links Top I to the 3′ end of DNA (or Top II to the 5′ end) via the active site tyrosine residue forming DNA-enzyme covalent intermediates commonly referred as Top I (or Top II) cleavage-complexes (TopIcc or TopIIcc) (2). These intermediates are briskly religated in the final step of the transesterification reaction thereby causing relaxation of the supercoiled DNA. However, this relaxation can be rather perilous as modifications like abasic sites, nicks or gaps, mismatches, modified bases, nucleotide analogs and almost all kinds of DNA lesions as well as topoisomerase poisons like camptothecin interfere with the ligation reaction and result in trapped covalent DNA-enzyme intermediates with SSBs or DSBs (3). Consequently, these trapped covalent complexes (TopIcc or TopIIcc) pose a risk to the integrity of the genome.
Persistence of these covalent complexes with SSBs or DSBs leads to the activation of the DNA damage response (DDR) cascade allowing recruitment of specialized enzymes called tyrosyl–DNA phosphodiesterases. These enzymes precisely release the tyrosyl-linked covalent topoisomerase peptides from the DNA and channel the accompanying SSBs or DSBs to the respective repair pathways in the cell. These tyrosyl–DNA phosphodiesterases thus help in rescuing the genome from the perils of atypical relaxation brought about due to aberrant topoisomerase activity. In this review, we give an account of the discovery of tyrosyl–DNA phosphodiesterases and present insights into the structural aspects, functional diversity, regulation and the current state of development of inhibitors of these precision biological tools.
TYROSYL–DNA PHOSPHODIESTERASE 1 (TDP1)
Discovery
In 1996, it was observed that a synthetic analogue of a reaction intermediate in certain recombination reactions, namely, an oligonucleotide bearing a phosphotyrosine residue in an ester linkage with the 3′ end of the DNA, was processed in an unpredicted manner upon its incubation with extracts of several eukaryotic cells. Treatment of the substrate resulted in the formation of a product with mobility similar to that expected from the hydrolytic loss of terminal tyrosine (4). Fortunately, these seminal experiments serendipitously provided the first evidence of an enzymatic activity that could hydrolyze the phosphodiester bond that joins the tyrosyl residue of Top I to the 3′ end of the DNA. The specificity of this tyrosyl–DNA phosphodiesterase activity, its conservation across a range of eukaryotic species and the fact that 3′-phosphotyrosyl substrates mimic trapped Top I cleavage complexes suggested that this enzyme might be a part of the pathway for the repair of TopIcc (4). Subsequently, the Saccharomyces cerevisiae gene encoding TDP1 was isolated by random mutagenesis and screening of clones for loss of TDP1 activity. TDP1-defective mutants were found to be hypersensitive to camptothecin (CPT), an anticancer chemotherapeutic drug that specifically traps Top I, only when the TDP1 mutation was combined with mutation in other proteins such as Rad9 or Rad1–Rad10, further drawing attention to its role in the repair of TopIcc in the absence of other backup pathways (5,6). The human gene for TDP1 was soon cloned and it was found by mutational and sequence analysis that TDP1 was a member of the phospholipase D (PLD) superfamily. Subsequent work established the crystal structure of TDP1 and the mechanism of its action (3,7). Shortly after, it was determined by linkage analysis, physical mapping and a positional candidate gene approach in a Saudi Arabian family that mutation in TDP1, and thereby a deficiency in repairing the stalled Top I complexes, caused an extremely rare genetic disease Spinocerebellar Ataxia with Axonal Neuropathy (SCAN1) (8). With the TDP1 gene cloned and its crystal structure solved, new avenues opened allowing researchers to investigate the biochemistry and the molecular biology of a previously challenging niche of DNA repair.
Structural insights
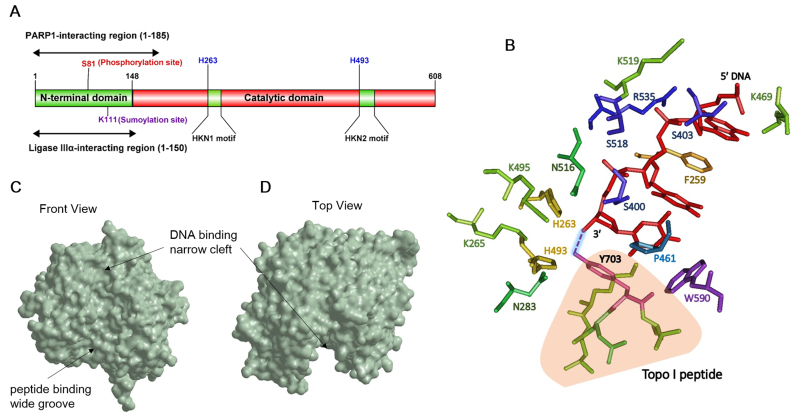
Human tyrosyl–DNA phosphodiesterase 1 is a predominantly nuclear protein comprising 608 amino acids with a molecular weight of 68.5 kDa. The protein can be subdivided into two domains – an N terminal regulatory domain extending up to amino acid 148 and a C-terminal catalytic domain extending from 149 to 608 amino acids (Figure 1A) (3). The N-terminal domain is dispensable for the catalytic function but is important for the recruitment of TDP1 to the sites of damaged chromatin. Indeed, an N-terminal deletion mutant (Δ1–148) of human TDP1 (hTDP1) in vitro retained wild type levels of processing of the Top I peptide from the 3′ end of an oligonucleotide substrate (7). TDP1 has two nuclear localization sequences (NLS) for its transport into the nucleus, NLS1–H56 to P74 and NLS2–P216 to P223 (9,10). Although TDP1 has been shown to localize to the mitochondria and play a role in mitochondrial base excision repair, the protein lacks the putative mitochondrial signal peptide sequence needed for mitochondrial transport (9). How TDP1 translocates to the mitochondria remains a mystery to date (11).
Figure 1.
Structure of TDP1. (A) Domain structure of human TDP1. Sites shown in blue are key residues in the active site of TDP1. (B) Critical TDP1-substrate interactions in the TDP1 active site. Crystal structure of TDP1 (1NOP) obtained from Protein Data Bank (PDB) website was used to generate a model of the TDP1 active site in complex with the DNA substrate and the Top I peptide using Cn3D. Amino acids in the active site are represented as sticks and color-coded. DNA substrate is shown in red. 3′-phosphotyrosyl bond is indicated as a dashed line between 3′ end of the substrate and Y703 and highlighted. Top I peptide is shown in light orange. (C and D) Surface representation of TDP1 substrate binding channel was generated using Chem3D on crystal structure of TDP1 (1JY1) obtained from PDB (C) Front view and (D) Top view of the substrate binding channel with the DNA and peptide binding regions highlighted.
Sequence alignments of TDP1 orthologs from different species have demonstrated that TDP1 represents a unique subclass within the PLD superfamily of proteins (7). The uniqueness of TDP1 and its orthologs is attributed to the two ‘HKN’ catalytic motifs instead of the characteristic HKD motifs found in the other members of the PLD superfamily (Figure 1A). Each of the sequence motifs contain a highly conserved histidine, lysine and asparagine (H263, K265 and N283 in the N-terminal motif and H493, K495 and N516 in the C-terminal motif). Site-directed mutagenesis established that both H263 and H493 are the key catalytic residues as H263A, H493A and H493N mutants were 10,000×, 3,000× and 15,000× less active than the wild-type protein (7). The other conserved residues in the ‘HKN’ motifs K265, N283, K495 and N516 are key for substrate binding and the stabilization of the transition state (12). These two HKN motifs together make up the active site at the bottom and near the center of the protein allowing it to function as a monomer (3). This active site is embedded in a substrate-binding channel whose molecular architecture is highly asymmetrical with respect to the shape and charge distribution as expected for an enzyme whose canonical substrate is a protein–DNA hybrid covalent complex. Whereas the channel is ∼8 Å narrow on one side with a positively charged electrostatic surface potential, it enlarges into a 20 Å wide groove with a more even charge distribution as a result of a blend of both acidic and basic amino acid residues. This asymmetry allows the protein moiety of the substrate to fit in the wider passage of the channel while the negatively charged DNA substrate is received by the narrow positively charged region (Figure 1C and D). Despite the positive charge of the narrow DNA binding groove, quite surprisingly, the critical interactions of DNA are with the polar and hydrophobic residues in the groove while the basic amino acids play only a minor role. S400, S403 and S518 DNA form hydrogen bonds with the phosphate groups on the DNA and thus, are the key amino acid residues that interact with the DNA substrate (12). Additionally, F259, P461 and W590 make hydrophobic contacts with the DNA (13) with the phenylalanine intercalating between the DNA bases -3 and -2 (between bases 3 and 2 upstream of the phosphotyrosine bond). K410, K469, K519 and R535 stabilize the DNA via ionic interactions as it exits the DNA binding channel (Figure 1B) (14). The interaction of the enzyme with the phosphodiester backbone of the DNA and not with the bases allows TDP1 to function at global genomic locations without any sequence specificity, an important feature for a DNA repair protein.
The structure of yeast TDP1 (yTDP1) has been solved recently and is remarkably similar to its human counterpart despite a modest 24% sequence identity between the two proteins (Human TDP1—NCBI Accession # Q9NUW8.2 compared with Yeast TDP1—NCBI Accession # KZV13308.1). A small difference between the otherwise indistinguishable active sites of the two proteins is seen in the size of the active site-clefts; yTDP1 cleft is larger, more open and less restricted than that of hTDP1 (15). This larger active site cleft possibly explains why yeast but not human TDP1 can process 5′-phosphotyrosyl adducts; the narrow, tighter active site of hTDP1 likely prevents the non-specific interaction with the substrate whereas the larger cleft reduces the specificity of the yeast enzyme.
Catalytic mechanism
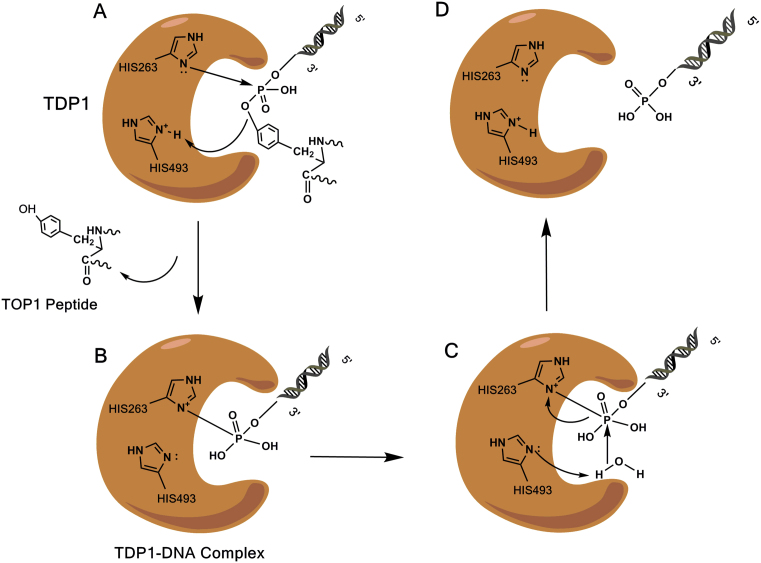
TDP1 belongs to the phospholipase D superfamily whose other few members include phospholipase D, a Salmonella nuclease (Nuc), cardiolipin synthase and phosphatidyl serine synthases (3). Although TDP1 differs from the other members of the superfamily in that it has ‘HKN’ instead of ‘HKD’ consensus motifs, it is similar to all the members in its mechanism of catalyzing a two-step ‘ping pong’-type phosphoryl transfer reaction via a covalent phosphoenzyme intermediate (16). The first step involves a nucleophilic attack on the tyrosyl–DNA 3′-phosphate by the imidazole N2 atom of H263 of the N-terminal HKN motif. H493 of the C-terminal HKN motif acts as a general acid catalyst to protonate the tyrosyl moiety of the departing Top I peptide. This results in the formation of the covalent intermediate in which the cleaved substrate is temporarily linked to TDP1 (Figure 2A and B). In the second step, H493 acts as a general base catalyst and activates a water molecule which subsequently hydrolyzes the phosphoenzyme intermediate (Figure 2C) (13,17). This leads to the release of 3′-phosphate-ended DNA (Figure 2D) which is converted by polynucleotide kinase/ phosphatase (PNKP) into 3′-OH DNA.
Figure 2.
Catalytic cycle of TDP1. (A) Nucleophilic attack on the tyrosyl–DNA 3′-phosphate by the imidazole N2 atom of H263. H493 donates a proton to the outgoing Top I peptide. (B) Formation of the TDP1-DNA covalent intermediate. (C) H493 activates a water molecule which attacks the 3′-P breaking the N-P bond and hydrolyzing the phosphoenzyme intermediate. (D) Release of DNA 3′-phosphate from TDP1.
A missense mutation in the TDP1 gene (A1478G) causing a substitution of histidine 493 with an arginine (TDP1H493R) residue is the genetic basis behind the pathology of SCAN1, an extremely rare autosomal recessive neurodegenerative disorder, characterized by a series of symptoms with progressing cerebellar ataxia leading to wheelchair dependency, cerebellar atrophy and peripheral neuropathy (8). The TDP1 H493R mutant manifests around 25 times decreased rate of hydrolysis of the tyrosyl containing peptide from the DNA and ironically itself becomes covalently trapped with a relatively long half-life of around 13 min (17,18). Whereas in normal cells the TDP1-dependent single-strand break repair (SSBR) pathway briskly repairs Top I-DNA lesions formed in response to Top I poisons (Figure 7A), in SCAN1 cells the TDP1H493R mutation leads to an accumulation of residual unrepaired Top I-DNA lesions. This accumulation is attributed to the loss-of-function of TDP1. Thus, the failure of mutant TDP1 in repairing SSBs formed as a result of oxidative stress and stalled Top I reactions is the molecular basis behind the manifestation of this disease in post-mitotic neurons (19). Mutations in PNKP have also been linked to neurological conditions including microcephaly with seizures (MCSZ) and ataxia with oculomotor apraxia (AOA4) (20).
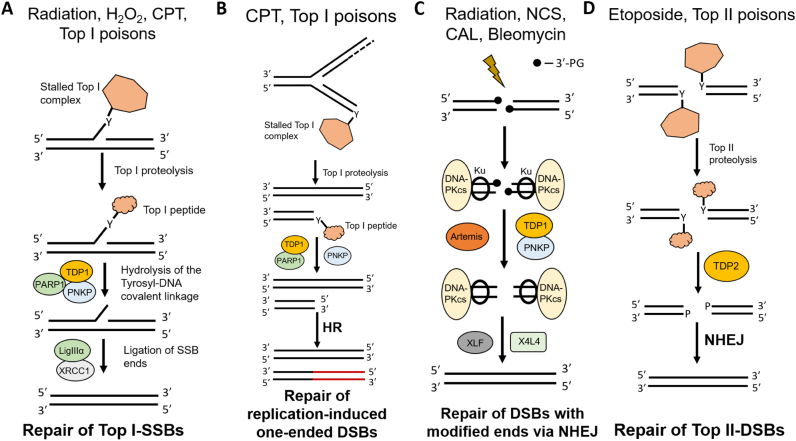
Figure 7.
Biological pathways that engage TDP1 and TDP2. (A) Canonical role in the repair of Top I cleavage complexes. DNA damage-induced stalling of Top I leads to the formation of a single-strand break with a covalent linkage between the active site tyrosine of Top I and the 3′-end of the DNA. Upon Top I proteolysis, TDP1, in conjunction with PARP1, resolves the tyrosyl–DNA phosphodiester bond leaving a 3′-phosphate which is further processed by PNKP to a 3′-OH. The SSB ends are subsequently ligated by LigIIIα in the presence of XRCC1. (B) Stalled Top I complexes can also form during the DNA synthesis phase. Collision of these complexes with the approaching replication fork can produce one-ended DSBs with a 3′-Top I peptide fragment. These breaks are processed by TDP1 in conjunction with PNKP and PARP1 and are then shunted to the HR pathway. (C) TDP1 in DSB repair. Radiation and radiomimetic agents produce double-strand breaks with modified ends, a significant fraction of which bear 3′-PG termini. These DSBs are repaired by the NHEJ machinery. TDP1 can resolve these 3′-PG termini which are further processed by PNKP to a 3′-OH. In addition, TDP1 may also suppress insertions by mutagenic polymerases during NHEJ. The ends are subsequently ligated by XRCC4-LigIV complex in the presence of XLF. The 3′-PG resolution can be alternatively carried out by other nucleases such as Artemis. (D) Stalled Top II complexes formed due to aberrant Top II activity are repaired by TDP2. Upon Top II proteolysis, TDP2 removes the 5′-phosphotyrosyl Top II peptide leaving a 5′-phosphate-ended DSB which is a substrate for NHEJ-mediated DSB repair.
Functional diversity
Canonical role in the repair of Top I cleavage complexes
TDP1 was initially described as a clean phosphodiesterase activity that explicitly removed a tyrosyl-containing peptide from a DNA end (Figure 3A) leaving a 3′-phosphate and was shown to be critical for the repair of TopIcc (4,5). Since redundant pathways exist for the repair of replicative Top I DNA damage, TDP1-deficient yeast cells show hypersensitivity to CPT only in the presence of mutations in additional proteins such as DNA damage checkpoint protein, Rad9, or the nucleotide excision repair (NER) endonuclease Rad1–Rad10 (5,6). TDP1-deficient human cells also show hypersensitivity to CPT (17,19,21) and its clinical derivative irinotecan (22). Furthermore, as observed by single-cell gel electrophoresis experiments, human embryonic kidney 293 (HEK293) cells overexpressing WT but not mutant TDP1H263A exhibit significantly reduced DNA damage induced by CPT (23). The critical role of TDP1 in repairing spontaneous Top I-mediated DNA damage is further highlighted by the fact that in fission yeast S. pombe, TDP1 is essential for viability in the absence of an alternative SUMO-targeted Ubiquitin Ligase (STUbL)-, Rad60- and Nse2-dependent pathway for the repair of Top I-mediated DNA damage (24).
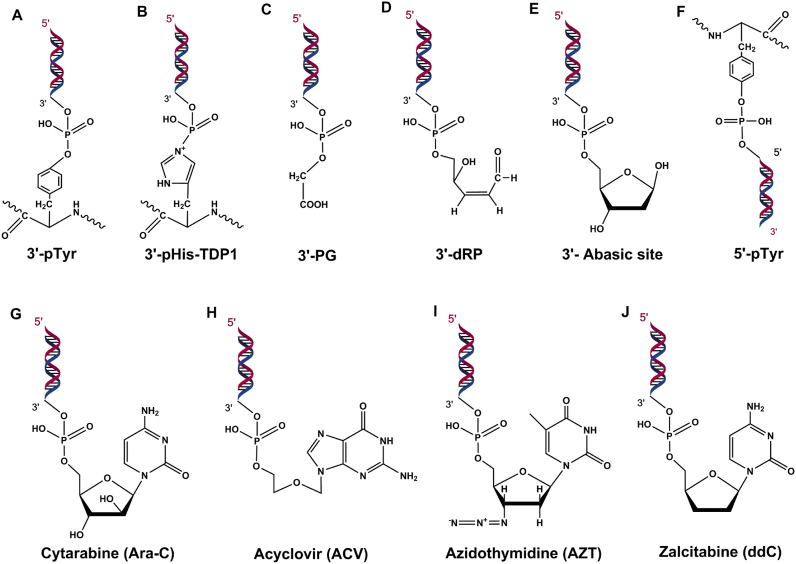
Figure 3.
TDP1 substrates. (A) 3′-phosphotyrosyl peptide - canonical substrate of TDP1. (B) 3′-phosphoamide adducts formed by H493R-mutant TDP1 causing covalent linkage of H263-TDP1 to 3′-DNA end. (C) 3′-phosphoglycolates. (D) 3′-deoxyribose phosphate and (E) 3′-abasic sites produced because of oxidative DNA damage. (F) 5′-phosphotyrosyl peptide. (G–J) Chain terminating nucleoside analogs.
To date, TDP1 remains one of the very few enzymes that specifically remove a 3′-block from the DNA end without actually resecting the DNA by even a single base. Although it shows a weak activity in removing a normal nucleoside from a 3′ DNA end (25), the inability of TDP1 to act on the resulting 3′-phosphate terminus prevents TDP1 from functioning as a general 3′-exonuclease. For this reason, activity of TDP1 in the removal of 3′-phosphotyrosyl residues in human cells is coupled with a specific DNA 3′-phosphatase, PNKP, to generate 3′-OH, which can then be readily acted upon by DNA polymerases and ligases (Figure 7A) (26).
A number of studies have defined, in some detail, the substrate requirements of TDP1. Yeast and human TDP1 process 3′-phosphotyrosyl blocks more efficiently on single-stranded DNA and on 3′-overhanging- or blunt-ended duplex DNA compared to tailed or nicked duplex DNA substrates (27,28). Studies on the protein component have shown that TDP1 cannot remove a full-length Top I protein bound to DNA and that Top I must be proteolyzed prior to cleavage by TDP1. Gel-shift assays have shown that whereas Top I -derived peptides in size up to 108 amino acids are efficiently cleaved by TDP1, the efficiency of cleavage varies inversely with the size of the peptide (29,30). On the other hand, the cleavage efficiency varies directly with the size of the oligonucleotide although TDP1 retains activity for DNA substrates with as few as 4 nucleotides (13,29).
The sensitivity of TDP1-deficient cells to Top I poisons was initially hypothesized to be specific to cells in the DNA synthesis phase where collision of approaching replication forks with TopIcc would lead to the formation of DSBs (Figure 7B). However, TDP1’s involvement in the pathology of SCAN1, a disease of terminally differentiated post-mitotic neurons, and the fact that SCAN1 cells show hypersensitivity to Top I poisons led to the emergence of a new question: Why does TDP1 mutation and Top I poisoning kill post-mitotic neurons, cells that do not enter S-phase? An explanation to this discrepancy was provided when it was shown that sensitivity of SCAN1 cells to CPT was abrogated by DRB (5,6-dichloro-1-β-d-ribofuranosylbenzimidazole), a transcription inhibitor, but not by aphidicolin, a replication inhibitor suggesting that SCAN1 cells are defective for the repair of transcription-induced TopIcc (21). In addition, these transcription-induced cleavage complexes cause the formation of transcription-dependent DSBs after Top I proteolysis prior to TDP1’s action leading to the activation of the DNA damage response via Ataxia-telangiectasia mutated (ATM) and DNA-dependent protein kinase (DNA-PK), and these co-transcriptional DSBs kill quiescent cells (31,32). Thus, the highly elevated transcription rates and increased oxygen demands in neuronal cells lacking TDP1 produce increased levels of unrepaired TopIcc and oxidative damage due to enhanced topoisomerase activity, providing molecular insights in the pathogenesis of SCAN1.
TDP1−/− mice, independently generated via either targeted active site deletion or insertional mutagenesis to model various SCAN1 features, do not exhibit embryonic or neonatal lethality (33–35). These mice as well as neurospheres and embryonic fibroblasts derived from them display hypersensitivity to CPT and bleomycin, but not to etoposide, an anticancer chemotherapeutic Top II poison. TDP1−/− mouse embryonic fibroblasts (MEFs) were completely deficient in the processing of overhanging 3′-PG termini on model DSB substrates but only partially deficient in removing blunt 3′-PG DSB termini (35). This result suggested that the hypersensitivity shown by the TDP1−/− animals to bleomycin was likely due to persistent unrepaired 3′-PG DSBs. In one of the studies, an age-dependent reduction in the cerebellar size as well as the non-neurological symptom of hypoalbuminemia was observed in the TDP1−/− mice akin to symptoms seen in human SCAN1 pathogenesis (34). Independently, TDP1−/− female mice were also shown to be significantly less active than TDP1+/+ mice suggesting symptoms of ataxia. However, this effect was transient and not observed at later times (35). In contrast, another report demonstrated that there were no electrophysiological changes or neuropathological SCAN1 symptoms like ataxia or cerebellar degeneration even upon treatment with CPT or Topotecan (33).
Role in SSB repair
One of the first studies highlighting the role of TDP1 in SSB repair was reported by Caldecott and colleagues when it was shown that TDP1 repairs chromosomal SSBs arising from aberrant Top I activity or oxidative stress (19). In addition, TDP1 was shown to interact directly with DNA Ligase IIIα, forming SSB repair complexes which are rendered catalytically inactive in SCAN1 cells due to the loss-of-function mutation in TDP1 (Figure 7A) (19). Subsequent studies have brought to light the versatility of TDP1 in the repair of various 3′-blocking groups. TDP1 eliminates 3′-phosphoglycolate (PG) ends both in vitro and in cells formed as a result of oxidative DNA damage although the efficiency of this processing is one hundred times less than that of its canonical 3′-phosphotyrosyl substrate (Figure 3C) (35–37). SCAN1 cells are defective in repairing ionizing radiation-induced SSBs (38). The alkylating agent methyl methane sulfonate (MMS) produces N7-methyl guanine adducts which are processed by DNA N-glycosylases/AP lyases to form abasic (AP) sites and 3′-deoxyribose phosphate (3′-dRP) ends (Figure 3D) (39,40). TDP1−/− DT40 chicken cells and human TDP1 knockdown (KD) cells show hypersensitivity to alkylating agent MMS and this sensitivity can be almost fully averted by complementing with human TDP1 (39). Additional depletion of apurinic/ apyrimidinic endonuclease 1 (APE1) in TDP1 KD cells enhances the hypersensitivity of these cells to MMS (41). Thus, these results suggest involvement of TDP1 in the base excision repair (BER) pathway in removing AP sites (Figure 3E) independently of APE1. Interestingly, these lesions also trap TopIcc potentially explaining the involvement of TDP1 in their repair.
In addition to nuclear BER, insights from confocal microscopy and biochemical analysis are indicative of TDP1’s involvement in mitochondrial BER for the efficient repair of oxidative damage in mitochondrial DNA (9). A recent study further strengthened the evidence of TDP1’s role in the repair of mitochondrial protein DNA breaks (mtPDBs). TDP1 promotes the repair of mitochondrial TopIcc and is upregulated in the presence of Top I mtT554A/N558H, a mutant mitochondrial Top I that does not exhibit religation activity, produces increased mtPDBs and decreases mitochondrial transcription. This TDP1 upregulation leads to a compensatory increase in the release of mtPDBs balancing mitochondrial gene transcription. TDP1 also promotes oxidative phosphorylation as seen from bioenergetics profiling and protects mammalian cells from H2O2-induced oxidative damage as TDP1−/− mouse embryonic fibroblasts (MEFs) showed hypersensitivity to H2O2 (42).
Chain-terminating nucleoside analogs (CTNAs) lack a 3′-OH group and thus block DNA synthesis after being incorporated into DNA. These CTNAs are extensively used as anti-viral and anti-cancer agents especially in treating HIV and adult T-cell leukemia (ATL) respectively (43). TDP1 repairs nuclear and mitochondrial DNA damage induced by CTNAs including acyclovir (ACV), cytarabine (Ara-C), zidovudine (AZT) and zalcitabine (ddC) (Figure 3G–J) (44). TDP1−/− DT40 and MEF cells are hypersensitive to ACV and Ara-C and show enhanced depletion of mitochondrial DNA in response to AZT and ddC. ATL cells are deficient in TDP1-dependent repair and are thus selectively killed by anti-HIV drug Abacavir (ABC) (45). Very recently, in the first published evidence of successful inactivation of the human TDP1 by genetic manipulation, it has been shown that TDP1−/− HCT116 and TSCER2 cells display enhanced sensitivity to 2′-C-cyano-2′-deoxy-1-β-d-arabino-pentofuranosylcytosine (CNDAC), an analog of Ara-C further underscoring the importance of TDP1 in the repair of CTNA-induced DNA damage (46). Interestingly, knockout of ATM, Fanconi anemia complementation group D2 protein (FANCD2) or Breast cancer type 1/2 susceptibility protein (BRCA1/2) but not of Poly (ADP-Ribose) Polymerase 1 (PARP1) in DT40 cells showed hypersensitivity to CNDAC. It is an interesting prospect to examine, by creating double mutants, whether TDP1 and these repair proteins function in the same pathway for the repair of these nucleoside analogs. In addition, TDP1 cleaves several synthetic substrates attached to the 3′-P end of DNA which have been valuable in the screening of inhibitors for TDP1 (13).
Role in DSB repair
The biochemical competency of TDP1 in the resolution of glycolate ends first suggested the possible involvement of TDP1 in the repair of DSBs bearing terminally-occluded 3′-overhangs. Whole cell extracts (WCEs) from lymphoblastoid cells derived from SCAN1 patients are deficient in catalyzing the conversion of 3′-PG termini on 3′-overhanging model DSB substrates in vitro with no measurable processing for several hours. In comparison, normal cells from unaffected relatives show substantial processing within a few minutes (37). As mentioned above, TDP1−/− MEFs are completely inept in removing protruding 3′-PG termini from similar substrates suggesting that the processing of DSBs with protruding 3′-PG termini is entirely dependent on TDP1 (Figure 7C) (35).
Ionizing radiation-induced DNA damage is extremely heterogeneous as DSBs are often accompanied by additional single-stranded lesions and base modifications. On the other hand, the radiomimetic antitumor antibiotics, namely bleomycin and the enediynes, calicheamicin (CAL) and neocarzinostatin (NCS), induce highly specific DSBs with modified ends (47). CAL yields bistranded lesions, a substantial portion of which contain a 3′-PG on a 2-base overhang; NCS-induced DSBs are similar except they bear the 3′-PG on a 1-base overhang while DSBs produced by bleomycin predominantly have the 3′-PG on blunt-ends (48). Experiments with these radiomimetic agents have implicated TDP1 in the repair of modified DSBs in cells. TDP1−/− mice and TDP1−/− DT40 chicken cells both show hypersensitivity to bleomycin (33,39). Clonogenic survival assays performed in HeLa cells depleted for TDP1 demonstrate slight sensitivity to calicheamicin (49). Treatment with calicheamicin leads to increased chromosomal aberrations in SCAN1 cells, particularly dicentric chromosomes suggesting that absence of TDP1 leads to mis-joining of DSB ends (49). By performing a ligation-mediated PCR assay to track the processing of damaged DSB ends in cells, it was shown that 3′-PG termini formed as a result of NCS treatment were more persistent in SCAN1 cells and were processed 2–3-fold slower as compared to normal cells suggesting a significant role of TDP1 in the repair of NCS-induced 3′-PG ended DSBs (50). With the success in generation of human TDP1 knockout cell lines, this assay or any variation thereof can prove invaluable in tracking the processing of 3′-end blocking groups and examining the kinetics of their removal.
In yeast, TDP1 has been shown to be an accessory component of the Non-Homologous End Joining (NHEJ) pathway (51). Clean DSB ends generated by linearizing plasmid substrates with restriction enzymes are mis-repaired leading to inaccurate repair joints with insertions in the absence of TDP1. Additional deletion of yeast NHEJ proteins such as Ku and Ligase IV does not increase the frequency of mis-repair suggesting that yTDP1 promotes repair fidelity in the context of NHEJ (51). Furthermore, human TDP1 has been suggested to play a role in the early stages of mammalian NHEJ by promoting the assembly of NHEJ protein complexes on DNA (52,53). TDP1 associates with the NHEJ protein complexes by directly interacting with X-ray repair cross complementing protein 4 (XRCC4)-like factor (XLF) (Figure 1A) and Ku70/80. This contrasts with other end-processing factors like PNKP, Aprataxin and Aprataxin and PNKP-like factor (APLF) that play a role in NHEJ by interacting with XRCC4. XLF stimulates activity of TDP1 on double-stranded DNA (dsDNA). Additionally, TDP1 promotes DNA binding by Ku70/80 and modestly stimulates the kinase activity of DNA-PK (52). Furthermore, it was recently reported that TDP1 was required for efficient NHEJ in human cells as HEK293 cells deficient in TDP1 showed an increase in insertions at I-SceI-induced DSB repair joints (54). Finally, TDP1 has also been shown to play a role in the repair of etoposide-generated DSBs (55). Human TDP1 KD cells are hypersensitive to etoposide and show increased number of chromosomal breaks and mis-joining events which are further enhanced by DNA-PK depletion. However, equal number of Rad51 foci and sister-chromatid exchanges in WT and TDP1 KD cells suggest that depletion of TDP1 disrupts classical as well as the alternative end joining pathways but not Homologous Recombination (HR) for the repair of TopIIcc (53,55). Thus, taken together, these results present persuasive evidence for the involvement of TDP1 in DSB repair.
Regulation and interplay with other DNA repair factors
Like most other DNA repair proteins, the biology of TDP1 in the repair of TopIcc lesions as well as oxidative DNA damage is elegantly regulated by its interactions with other DNA repair factors and post-translational modifications (PTMs) including poly ADP-ribosylation (PARylation), phosphorylation and SUMOylation. The N-terminal region of TDP1, which is dispensible for catalytic function, is the target for these modifications. Thus, these PTMs do not play a role in enhancing the catalytic function of the protein but merely increase stabilization and recruitment to the sites of DNA damage.
PARP1 is a highly conserved multifunctional enzyme that catalyzes the polymerization of ADP-ribose moieties derived from nicotinamide adenine dinucleotide (NAD+) onto itself or other target proteins (56). It was established almost 30 years ago that PARP-deficient Chinese hamster V79 cell line shows hypersensitivity to CPT suggesting an involvement of PARP in the repair of TopIcc which was confirmed by several subsequent studies (57). CPT sensitivity in WT MEFs was enhanced by Veliparib (ABT-888)-induced PARP inhibition but not by knockout of PARP1 suggesting that presence of PARP1 is critical for this sensitization (58). It is now known that PARP1 and TDP1 are epistatic for the repair of TopIcc as TDP1−/−. PARP1−/− double mutant avian DT40 cells show similar sensitivity to CPT as their single mutant counterparts (59). The C-terminal region of PARP1 binds the N-terminal region of TDP1 (Figure 1A) and poly ADP-ribosylates TDP1 without inhibiting its catalytic activity and promotes its recruitment to TopIcc-induced DNA damage sites. Micro-irradiation with live-cell microscopy and biochemical analysis show that PARylation of TDP1 promotes the recruitment of both itself and X-ray repair cross complementing protein 1 (XRCC1) to the sites of Top I-induced DNA damage and leads to the stabilization of TDP1 in response to TopIcc-induced DNA damage (Figure 7A) (59).
In human cells, TDP1 is phosphorylated at serine 81 by ATM and DNA-PK following ionizing radiation and CPT treatment (Figure 1A) (60). As this site is located in the N-terminal region which is dispensable for enzyme activity, phosphomutants show no difference in enzymatic activity in yeast in vitro (61). However, pS81-TDP1 forms nuclear foci that co-localize with γH2AX foci which presumably are sites where Top I-induced SSBs are converted to DSBs following replication fork collision (60). In addition, phosphorylation of TDP1 is important for its stabilization and promotes binding to XRCC1 and Ligase IIIα as seen from co-immunoprecipitation (Co-IP) and immunofluorescence microscopy (60,61). XRCC1 is a scaffolding protein that interacts with several BER factors and is also known to play a role in the repair of TopIcc presumably by recruiting TDP1 and PNKP (62). TDP1 also directly interacts with Ligase IIIα forming a multi-protein SSB repair complex for the repair of SSBs arising out of aberrant Top I activity or oxidative stress (19). The interaction of TDP1 with Ligase IIIα also likely contributes to mitochondrial BER (19,63).
SUMOylation is another form of PTM that regulates TDP1’s function in the repair of TopIcc. SUMOylation is similar to ubiquitination in the overall scheme and the types of enzymes but instead of ubiquitin (Ub) conjugation, it involves conjugation with SUMOs (Small Ubiquitin-like MOdifiers). TDP1 is SUMOylated at lysine 111 (Figure 1A) which promotes its accumulation at sites of SSBs as seen from live-cell microscopy (64). A SUMOylation K111R mutant does not show diminished enzymatic function or inhibit TDP1’s interaction with Ligase IIIα (64). However, the mutant displays reduced rate of repair of transcription-associated SSBs suggesting the importance of this modification in preserving post-mitotic neurons from oxidative stress.
TDP1 Inhibitors
A very common early event in evolution of cancer cells is the genetic dysregulation of one or more DNA damage repair pathways, such as HR, making the cells heavily reliant on compensatory repair mechanisms for survival (65). As the genetic inactivation is typically restricted to the tumor cells and not present in normal cells, pharmacological inhibition of the compensatory mechanism provides an exciting opportunity to specifically sensitize cancer cells. This approach has been referred to as ‘synthetic lethality’ and has been a cardinal principle in the development of DDR therapeutics. This selective cytotoxicity principle has led to the development of the several DDR inhibitors and is best exemplified by the effectiveness of PARP inhibitors in HR-deficient breast and ovarian cancers (66–68).
The effectiveness of TDP1 against a broad spectrum of substrates ordains inhibition of TDP1 as an attractive target for tumor cell sensitization in combination with Top I inhibitors, radiation or radiomimetic drugs, nucleoside analogs or alkylating agents. Given the prominence of TDP1 in the repair of TopIcc, inhibition of TDP1 can provide a convenient approach in exacerbating the sensitivity of cancer cells to Top I poison like CPT. The observation that non-small cell lung cancer (NSCLC) cells develop resistance to CPT by virtue of overexpressing TDP1 buttresses the argument for the development of TDP1 inhibitors which can lead to the reversal of resistance in these cancers (69,70). In addition to TDP1-dependent repair, alternative xeroderma pigmentosum group F complementing protein – excision repair cross complementing protein 1 (XPF-ERCC1) (Rad1–Rad10)-dependent or Mre11-dependent pathways for the repair of TopIcc exist in cells. PARP inhibition in XPF-depleted U2OS cells enhances their sensitivity to CPT suggesting that PARP1 and XPF-ERCC1 function in parallel pathways for the repair of TopIcc (71). Thus, inhibiting TDP1 in cancers expressing low levels of XPF-ERCC1 (e.g. testicular cancers) (72,73) could provide a synthetic lethal opportunity to target these cancers. Similarly, MRE11-deficient tumors (including mismatch repair (MMR)-deficient colon cancers that often acquire mutations at short repetitive sequences in the Mre11 gene) might be effectively sensitized to Top I poisons by TDP1 inhibitors (74). Glioblastoma cancer cells become resistant to Temozolomide (TMZ) by overexpressing methyl guanine methyl transferase (MGMT). Depletion of TDP1 in these cells sensitizes them to TMZ thus opening new avenues for the use of TDP1 inhibitors to be used in combination with alkylating agents in cancer chemotherapy (41). Inhibition of TDP1 in HR-deficient tumors can provide a synthetic lethal approach in their treatment on the grounds that persistent one-ended DSBs generated in the absence of TDP1 post replication fork collapse will remain mis-repaired thereby enhancing genomic instability and limiting cancer survival (Figure 7B). Additionally, as PARP enzymes affect several cellular processes and given the epistatic interplay between PARP and TDP1, inhibitors of TDP1 can provide a more direct alternative for use either in HR-deficient tumors or in synergy with Top I anticancer drugs.
Research on TDP1 inhibitors has come a long way since the early reports on highly non-specific, wide-range action compounds including transition metals like vanadate and tungstate, and aminoglycoside antibiotics like neomycin (75). Several studies have reported the development of specific TDP1 inhibitors since then. Many chemically synthesized compounds including but not limited to diamidines, phosphotyrosine mimetics, benzopentathiepines and indenoisoquinolines as well as naturally-occurring plant and fungal metabolites have been tested as candidates for TDP1 inhibition (13,76–94). Of note, modification of indenoisoquinolines, originally discovered as exclusive non-CPT Top I inhibitors, has led to the emergence of a new class of triple Top I-TDP1–TDP2 inhibitors. These triple inhibitors have shown potent cytotoxicity in a panel of 60 human cancer cell lines and elicited significantly enhanced DNA damage in human lymphoblastic leukemia CCRF-CEM cancer cells as evident from the intense γ-H2AX staining after 2 h of treatment compared to normal lymphocytes (PBMCs) (94). However, due to a lack of pre-clinical animal model studies and less than compelling evidence obtained from cellular models and in-vitro assays, none of the inhibitors developed so far are close to clinical trials.
TYROSYL–DNA PHOSPHODIESTERASE 2 (TDP2)
Discovery
TRAF and TNF receptor associated protein (TTRAP) was originally identified as a novel intracellular protein shown to inhibit the activation of Nuclear factor- kappa beta (NF-κB) by associating with various members of the tumor necrosis factor receptor (TNFR) family (95). Soon after this discovery, it was observed by peptide sequence and secondary structure analysis that TTRAP belonged to the Mg+2/Mn+2-dependent family of phosphodiesterases having a significant amino acid sequence similarity with the human DNA repair enzyme APE1 (96). Although this suggested the possibility that TTRAP could be involved in certain aspects of DNA metabolism, it wasn’t until several years later that its role in DNA repair was unearthed.
The failure of TDP1 in the processing of the 5′-phosphotyrosyl residues (4,97) and the increasing importance of Top II poisons in cancer chemotherapy (98) had led researchers to believe the existence of a separate enzyme which was biochemically competent for the resolution of 5′-phosphotyrosyl residues generated by aberrant Top II-mediated DNA metabolism. It was known that, in yeast, two distinct pathways were involved in the removal of Top I from the 3′ end, one involving TDP1 and the other involving the structure-specific NER factor Rad1–Rad10 endonuclease. In order to identify alternative 3′ human tyrosyl DNA phosphodiesterases, Caldecott and colleagues employed a novel experimental strategy. CPT-sensitive tdp1Δ rad1Δ double mutant yeast cells were transformed with a human cDNA library followed by isolation of transformants displaying resistance to CPT. Some transformants isolated contained cDNA clones encoding human TTRAP and indeed TTRAP processed 3′-phosphotyrosyl residues albeit more weakly than TDP1. However, in contrast to its weak ability to hydrolyze 3′-phosphotyrosyl bonds, TTRAP possessed a strong 5′-tyrosyl–DNA phosphodiesterase activity which was dependent on Mg2+. Additionally, human cell extracts depleted for TTRAP showed a marked reduction in the processing of 5′-phosphotyrosyl ends and increased DSB accumulation (97). Based on the complementary functions of TDP1 and TTRAP in removing the 3′- and 5′-phosphotyrosyl bonds respectively, they designated TTRAP as tyrosyl–DNA phosphodiesterase 2 (TDP2) and hypothesized that this enzyme might play a role in the resolution of Top II cleavage complexes. Subsequently, it was shown that TDP2 was the primary 5′-tyrosyl DNA phosphodiesterase in vertebrate cells (99). Moreover, genetic inactivation of the TDP2 locus resulted in cells being hypersensitive to etoposide, but not to CPT or MMS which identified TDP2 as a crucial factor in the repair of Top2 lesions and a useful target for inhibition during cancer chemotherapy (99).
Structural insights
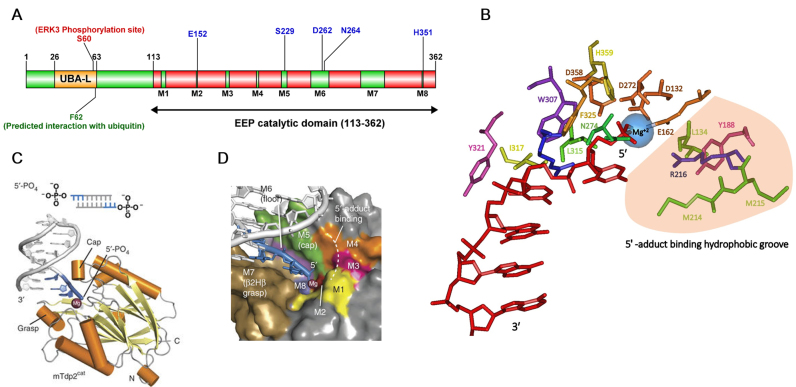
Human TDP2 is composed of 362 amino acids with a molecular weight of 41 kDa. Similar to TDP1, it is a two-domain repair protein; an N-terminal ubiquitin-associated like (UBA-L) domain from amino acids 26–63 and a C-terminal exonuclease/ endonuclease/ phosphodiesterase (EEP) catalytic domain extending from amino acids 113–362 (Figure 4A) (100). The UBA-L domain in TDP2 differs from the canonical UBA domain seen in other Ubiquitin (Ub) receptor family proteins. Whereas the UBA domain in most proteins is characterized by the presence of a three α-helix bundle with a highly conserved ‘MGF’ sequence motif, the atypical UBA-L domain seen in TDP2 has four short α-helices instead of three and lacks the conserved ‘MGF’ sequence (101,102). Despite this difference, nuclear magnetic resonance (NMR) studies show that the TDP2 UBA-L domain binds Ub in a canonical fashion (101).
Figure 4.
Structure of TDP2. (A) Domain structure of TDP2. Sites shown in blue are key residues in the active site of TDP2. (B) Critical TDP2-substrate-Mg+2 interactions in the TDP2 active site. Crystal structure of TDP2 (4GZ1) obtained from PDB was used to generate a model of the TDP2 active site in complex with the DNA substrate and the Mg+2 ion using Cn3D. Amino acids in the active site are represented as sticks and color-coded. DNA substrate is shown in red with the indicated 5′ and 3′ ends. Mg+2 is shown as a blue sphere. 5′-tyrosyl binding region lined by hydrophobic residues is shown in light orange. (C) Structure of mTDP2cat in complex with Mg2+ and 5′-phosphate DNA [Image reproduced with permission from (103)]. (D) Interaction of 5′-phosphate DNA end shown in blue with the color-coded active site motifs is shown. [Image reproduced with permission from (103)].
Peptide sequence analysis shows that TDP2 (previously known as TTRAP) belongs to the Mg2+/Mn2+ dependent phosphodiesterase superfamily (96). Crystal structures of zebrafish, C. elegans, mouse and human TDP2 catalytic domain (TDP2cat) have been obtained in independent studies (100,102,103). Small-angle X-ray scattering (SAXS) analyses in these studies show that the catalytic domain of TDP2 from human and C. elegans is greatly ordered and adopts a globular structure in solution whereas full length TDP2 is present in an elongated conformation in solution at physiological conditions (102,103). Thus, the N-terminal region of the protein appears to be flexibly connected to the ordered catalytic domain. In addition, modelling of full-length cTDP2, Guinier analysis of the SAXS data and size-exclusion chromatography suggests that the N-terminal region is important for dimerization of TDP2 in solution (102).
The structure of mTDP2cat in complex with the 5′-phosphate dsDNA and a magnesium ion, which resembles the catalytic product formed after TDP2-mediated removal of the 5′-tyrosyl adduct, outlined several key features of the TDP2-dsDNA-Mg+2 interaction (Figure 4C). Structure-based sequence alignment of TDP2 homologs demonstrates the presence of eight highly conserved motifs (M1-M8) in the catalytic domain (Figure 4D). Motifs M5, M6 and M7 are critical in binding the DNA substrate and together form the ‘grasp’, ‘cap’ and ‘floor’ of a highly conserved DNA binding cleft. M7 assumes a novel β-2helix-β (β2Hβ) DNA binding fold comprising a series of highly conserved hydrophobic residues. This fold protrudes outwards from the TDP2 active site mediating sequence-independent interactions that ‘grasp’ the terminal three nucleotides of the uncovered 5′ DNA end. W307, F325 and L315 form a cleft that interacts with the terminal 5′ nucleoside (N1) via Van der Waals forces (all amino acid residue numbering is for the mouse protein unless otherwise mentioned). I317 and Y321 (C311 in humans) interact specifically with the penultimate base (N2) and the sugar moiety of N3 respectively to stabilize the 5′ DNA end structure (103). Motif M5 ‘caps’ the DNA from the opposite side of the grasp and orients the 5′ end toward the active site using the conserved H236, S239 and R241 residues. Finally, D272, N274 and R276 of motif M6 form the ‘floor’ between the cap and the grasp.
The crystal structure of TDP2 complexed with 5′ adduct-dsDNA revealed that the 5′ adduct occupies a hydrophobic groove in the TDP2 active site created by the conserved L134, Y188, M214, M215 and R216 residues in motifs M1, M3 and M4. Modelling of the 5′-tyrosine in this hydrophobic pocket revealed key Van der Waals interactions between the hydrophobic pocket and the tyrosine ring and alanine substitutions of these key hydrophobic residues led to defects in 5′-tyrosyl processing. Lastly, D132, E162 and D358 in motifs M1, M2 and M8 respectively interact with the magnesium in the active site (Figure 4B) (103).
Catalytic mechanism
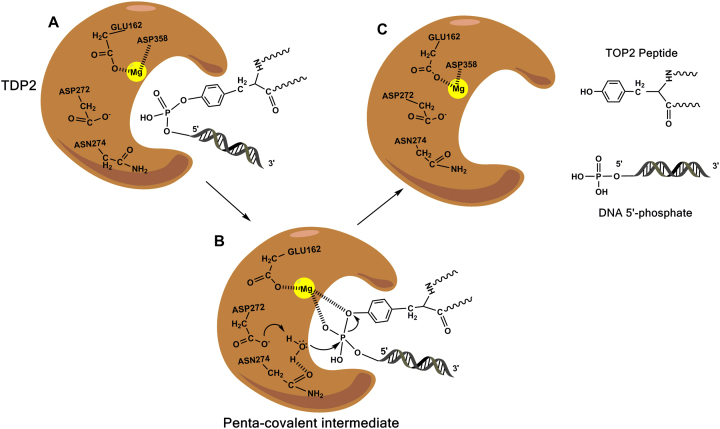
Although TDP2 shares little structural and sequence identity with APE1 (around 15%), its closest relative in the EEP family of nucleases, both these proteins have evolved a similar enzymatic reaction mechanism (103). X-ray structural studies and biochemical analyses have been valuable in deciphering the catalytic mechanism of TDP2. Metal-titration analysis performed on TDP2, prior to obtaining its crystal structure, had suggested that the TDP2-mediated reaction involved a two-metal ion catalytic mechanism (104). However, there is strong evidence of the catalytic reaction of TDP2 proceeding via a one-metal ion mechanism, as crystal structures of TDP2 obtained to date have detected the presence of only one metal in the active site (103). High-resolution structural analysis followed by quantum mechanics/ molecular mechanics (QM/MM) modelling of the TDP2 reaction coordinate has been performed in mice and has further strengthened this one-metal reaction hypothesis (105).
The reaction catalyzed by TDP2 is an SN2 displacement reaction. A nucleophilic water molecule is stabilized by strong hydrogen-bonding with D272 and N274 (All amino acid residue numbering is for the mouse protein unless otherwise mentioned). When the 5′-phosphotyrosyl of the substrate is close enough to the water molecule (around 2.18 Å between the water ‘O’ and the ‘P’ of the substrate), D272 activates the water which in turn makes a nucleophilic attack on the incoming 5′-phosphate of the tyrosyl–DNA adduct. Conserved residues H236, S239, H359 and the divalent Mg+2 ion, stabilized by E162, are optimally positioned to interact with the 5′-phosphate forming a penta-covalent transition state intermediate (Figure 5B) (103,105). In addition, D358 is also predicted to bind Mg+2 and stabilize the DNA-bound conformation by hydrogen bonding with W307. As the reaction proceeds, the O-P bond between the water and the substrate lengthens and ultimately breaks forming the product – DNA with a 5′-phosphate end (Figure 5C). This product formation concomitantly involves the movement of a proton from the nucleophilic water to D272 (105). Unlike TDP1 which produces 3′-phosphate that needs to be further processed, 5′-phosphate formed after a TDP2-mediated reaction is a direct substrate for DNA Ligases. Consistent with this mechanism, extensive mutational analyses have been performed on the active site residues in the human TDP2 protein (D262H/L/M/N/A, E152A, S229A, H351A/Q) and all substitutions fully abolish enzyme activity (104,105). An SNP in the active site of human TDP2 was identified very recently through the NCBI SNP database. This SNP causes a missense mutation hD350N (mD358) markedly abrogating the enzyme activity due to its inability to bind Mg+2 ion (105).
Figure 5.
Catalytic cycle of TDP2. (A) Active site of TDP2 accommodates the 5′-phosphotyrosyl–DNA adduct. (B) Nucleophilic water is activated by D272. ‘O–P’ bond formed between the water ‘O’ and DNA ‘P’ forming the penta-covalent intermediate. Mg2+ ion represented as a yellow sphere stabilizes the 5′-phosphate of the DNA. (C) Breakage of the phosphotyrosyl bond causes the release of the Top II peptide and DNA 5′-phosphate.
Functional diversity
Canonical function in the repair of Top II cleavage complexes
TDP2 is primarily involved in catalyzing the hydrolysis of 5′-phosphotyrosyl bonds thereby releasing trapped Top II or fragments thereof from the 5′ end of aberrant Top II-induced DSBs (Figures 6A and 7D). Consistent with this, TDP2-depleted human A549 cells show hypersensitivity to etoposide and a higher number of etoposide-induced γ-H2AX foci whereas overexpression of wild-type TDP2 but not catalytically inactive mutants (TDP2D262A or TDP2E152A) in yeast ISE2 cells leads to a significant increase in resistance to etoposide (97,106). Similarly, in DT40 chicken cells, knocking out all three alleles of TDP2 results in a profound hypersensitivity to etoposide as well as other structurally diverse Top II poisons, doxorubicin and amsacrine (m-AMSA) which can be complemented by expressing recombinant WT hTDP2. TDP2 knockout mice have been generated and are viable. These mice show lymphoid and intestinal toxicities in response to low concentrations of etoposide, and MEFs derived from them are also hypersensitive to etoposide (107). In addition, TDP2 also protects transcription from Top II-induced DSBs formed by abortive Top II activity (108).
Figure 6.
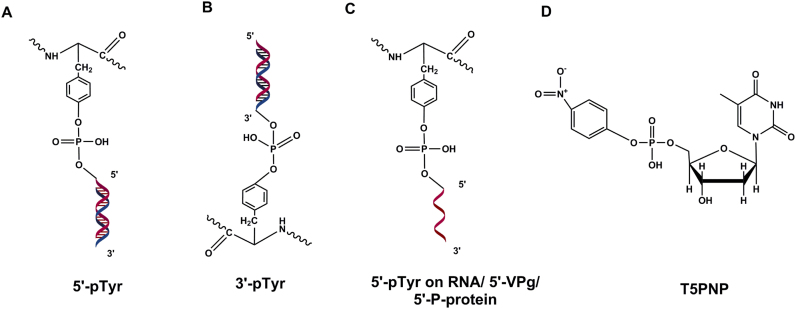
TDP2 substrates. (A) 5′-phosphotyrosyl–DNA—canonical substrate of TDP2. (B) 3′-phosphotyrosyl–DNA. (C) 5′-phosphotyrosyl-RNA, 5′-VPg from picornaviridae family of viruses, 5′-P-protein from Hepatitis B virus. (D) p-Nitrophenyl-thymidine-5′-phosphate (T5PNP) is processed by TDP2 producing thymidine-5′-monophosphate and p-nitrophenol, which can be used in a simple colorimetric TDP2 activity assay.
Three brothers from a consanguineous Irish family were identified to suffer from intellectual disability, epilepsy and progressive ataxia as result of a splice donor mutation in the TDP2 gene resulting in the insertion of a premature stop codon deleting the catalytic domain of TDP2 (109). Separately, an Egyptian individual with intellectual disability, fits and ataxia was also identified to have a homozygous truncating mutation in TDP2 (109). Protein extracts isolated from the blood of both the Irish and the Egyptian patients were deficient in 5′-TDP activity. Lymphoblastoid cells from these patients were hypersensitive to etoposide and show delayed DSB repair kinetics (109). These cases underscore the importance of the role of TDP2 in the maintenance of neural tissues and any imbalance in this maintenance is associated with neurological conditions.
Being a relatively recently discovered protein, substrate specificities of TDP2 have not been as extensively studied as TDP1. TDP2 is more efficient in the removal of the 5′-phosphotyrosine adduct from a single-stranded or 5′ overhanging dsDNA substrate than from nicked or blunt-ended duplex DNA (104,110). This result is expected given that its canonical substrate in cells is a 4-base 5′-phosphotyrosyl overhang. Additionally, it retains activity on DNA substrates ranging in size from as short as five nucleotides to as big as 37 nucleotides long. Similar to TDP1, the efficiency of 5′-phosphotyrosyl processing by TDP2 varies directly with size of the DNA substrate (104). Crystal structures of TDP2 from several species have demonstrated that TDP2 has a relatively tight catalytic site. Accordingly, TDP2 cannot remove full length native Top II from DSBs and therefore, Top II needs to be proteolyzed, presumably via the 26S proteasome, prior to its removal by TDP2 (110–112).
Other functions
Despite the lack of sequence or structural similarity to TDP1, TDP2 can repair DNA damage induced by abortive Top I activity (Figure 6B) (13,99). In fact, it was originally discovered based on its ability to rescue the sensitivity of rad1Δtdp1Δ yeast cells to CPT (97). TDP1−/− MEFs and DT40 cells are hypersensitive to CPT, but additional deletion of TDP2 in these cells enhances this sensitivity even further. Similarly, TDP1−/−. TDP2−/− DT40 cells overexpressing hTDP2 show a level of resistance to CPT higher than that shown by TDP1−/− single mutant cells, suggesting that overexpression of TDP2 can partially complement the defect inflicted by the loss of TDP1 and that TDP2 contributes to TopIcc repair in the absence of TDP1 (99).
In contrast to the promiscuous substrate interaction of TDP1, activity of TDP2 is reserved specifically for substrates containing 5′-phosphotyrosyl bonds. For example, TDP2 possesses no activity toward synthetic 5′-fluorescein, 5′-biotin or 5′-digoxigenin substrates unless the digoxigenin is linked via a phosphotyrosyl linkage to the DNA substrate (104). In addition, TDP2 lacks any detectable activity on abasic sites or 5′-AMP substrates (97). TDP2 processes p-nitrophenyl-thymidine-5′-phosphate (T5PNP) (Figure 6D) producing thymidine-5′-monophosphate and p-nitrophenol which can be used in a simple colorimetric assay to measure TDP2 activity precluding the use of the cumbersome gel-based assay (113). It has been predicted that more than a million ribonucleotides are incorporated in the nuclear genome per replication cycle which are usually excised by the RNase H2-dependent pathway (114). The incorporated ribonucleotides can lead to the formation of TopIIcc and promote genomic instability. It has been recently shown by co-crystalizing TDP2 bound to a tyrosyl-RNA substrate that TDP2 hydrolyzes the 5′-tyrosine covalently linked to a single- or poly-ribonucleotide substrate albeit less efficiently compared to that on a deoxyribonucleotide substrate (Figure 6C) (110). This also suggests that TDP2 might play a role in the processing of DNA damage induced by Top3β (RNA Topoisomerase) (13).
Recent studies have suggested an involvement of TDP2 in the life cycle of several viruses. The RNA genome of viruses belonging to the picornaviridae family, e.g. the poliovirus, rhinovirus and coxsackievirus lacks a 7-methylguanine cap at their 5′ end but instead has a small 20–22 amino acid protein called VPg covalently linked to the 5′ terminus by a O4-(5′-uridylyl) tyrosine-phosphodiester bond (115). Upon infection and polysome association, VPg must be unlinked from the viral RNA to allow efficient translation of viral proteins. Interestingly, TDP2 was shown to function as this host cellular VPg unlinkase cleaving the 5′-tyrosyl–DNA phosphodiester bond (Figure 6C) (116,117). Additionally, it was also shown recently that TDP2 potentiates viral replication in enterovirus infection. TDP2 deletion in MEFs significantly decreases viral titers of coxsackievirus B3 (CVB3) and to a lesser extent that of poliovirus and human rhinovirus. Hepatitis B virus (HBV) contains a relaxed circular (RC) dsDNA genome in which one strand is covalently linked to the viral polymerase protein (P protein) via a 5′-tyrosyl–DNA phosphodiester bond. Upon infection of hepatocytes, the RC DNA is transported to the nucleus and converted to episomal covalently closed circular (ccc) DNA. This conversion requires the removal of P-protein from the 5′ end. cccDNA serves as a persistence reservoir for the replication of Hepatitis B virus and is refractory to current anti-HBV treatments. It was shown that human and avian TDP2 specifically cleave the tyrosyl–DNA phosphodiester bond and release P-protein of duck hepatitis B virus (DHBV) in vitro and that small hairpin RNA (shRNA)-mediated depletion of TDP2 in HepG2 cells significantly delays the conversion of RC-DNA to ccc DNA whereas ectopic expression of TDP2 restored the conversion kinetics (118,119). These viral pathogens present a major dilemma for public health worldwide and as such it is tempting to speculate that inhibitors targeting TDP2 could potentially be effective as anti-viral agents against these infections.
TDP2 was also independently identified as EAPII (ETS1-Associated Protein II) and TTRAP and in those capacities, acts as a transcriptional regulator through interaction with several proteins and is involved in the NF-κB, mitogen-activated protein kinase (MAPK) and transforming growth factor beta (TGF-β) signal transduction pathways (120–122). TDP2 (TTRAP) is an intracellular TNF receptor binding protein that inhibits the activation of NF-KB mediated by TRAFs but not IKKα or p65/RelA (95). In zebrafish, TTRAP interacts with Smad3 and controls gastrulation movements and nodal signaling (120). TDP2, as an ETS1-Associated protein, is also thought to contribute to lung cancer development via the activation of the MAPK-ERK signaling pathway as its overexpression increased proliferation whereas its knockdown promoted apoptosis of NSCLC cell lines (123). How are the nuclear functions of TDP2 in DNA repair and regulation of transcription correlated? It is tempting to speculate that in theory, TDP2 may increase transcription efficiency by protecting actively transcribed regions from Top II-induced breaks and to that effect, the phosphodiesterase and transcription regulatory roles of TDP2 (as EAPII) may be correlated in the nucleus.
Regulation and interplay with other DNA repair factors
Post-translational modifications play an essential role in the activation and recruitment of a plethora of DNA repair proteins. Very recently, it was shown that extracellular signal-related kinase 3 (ERK3), an atypical MAP kinase, phosphorylates TDP2 at S60 residue and critically regulates its phosphodiesterase activity (Figure 4A). TDP2S60A mutant completely fails in processing a 5′-phosphotyrosyl substrate whereas TDP2S60D phosphomimetic shows a slight increase in processing compared to WT protein. Additionally, TDP2S60A A549 cells are hypersensitive to etoposide (124). Given that this site is in the N-terminal UBA-L domain and mutation of this site completely abolishes catalytic function, it will be interesting to see if this site is important for the intramolecular interaction between the N-terminal regulatory domain and the C-terminal catalytic domain of TDP2 (103). However, it is important to note that mutation of S60 to alanine did not abolish the phosphorylation of TDP2 by ERK3 suggesting that there is/are other additional ERK3 target site/s in TDP2 (124). It is an interesting proposition to identify which additional sites on TDP2 are ERK3 substrates and establish their functional relevance in the overall scheme of DNA repair.
Another level of regulation of TDP2 that has been recently reported involves its interaction with Ub. The N-terminal domain of TDP2 contains the non-canonical UBA-like domain capable of binding to Ub moieties (101). The hypersensitivity of TDP2−/-/− DT40 cells to etoposide was rescued completely by WT TDP2 but only partially by TDP2(Δ1–100) mutant with an N-terminal deletion or by mutants which are predicted to lose binding capability with Ub namely TDP2F62R or TDP2F62R/V35R/R56D (101). These results suggest that the N-terminal domain is important for TDP2’s function, possibly via interaction with ubiquitinated histones, Top II or other ubiquitinated proteins during DSB repair. Top II is SUMOylated in response to Top II poisons. Very recently, it was reported that Zinc finger protein 451 (ZNF451), a SUMO2 E3/E4 ligase, binds to Top II and TDP2 and preferentially SUMOylates TopIIcc over Top II. In addition, ZNF451 directly stimulates the catalytic activity of TDP2 in hydrolyzing SUMOylated TopIIcc (125). Surprisingly, the SUMO2 binding region in TDP2 is mapped to the catalytic domain of TDP2 which lacks a canonical SUMO-interaction motif. The interaction of TDP2 with SUMO2, Top II and ZNF451 possibly promotes its recruitment at the sites of TopIIcc.
DSBs generated by aberrant Top II activities bear 4-base 5′-phosphotyrosyl overhangs which can be processed by TDP2 to give rise to 5′-phosphate ends that then serve as a direct substrate for ligation by the core NHEJ machinery (Figure 7D). On these lines, TDP2 and Ku are epistatic and thus, function in the same pathway for the repair of etoposide-induced DSBs (Figure 7D) (107). However, LigIV−/− and Ku−/− DT40 are more sensitive to Top II poisons, VP-16 and ICRF-193 than TDP2−/− cells arguing that Ku-dependent NHEJ employs other additional functions in the repair of Top II-induced DSBs (126). In addition, deletion of TDP2 leads to a compensatory increase in etoposide-induced HR as seen by an increase in Rad51 foci and sister chromatid exchanges in TDP2-deleted MEFs (107). Importantly, the ‘clean’ phosphodiesterase function of TDP2 precludes the need for other error-prone nucleases in NHEJ from acting at DSB sites and thus, accurately preserves the DNA sequence. Consequently, TDP2 has been shown to suppress chromosomal translocations and promote genomic stability, as TDP2-deleted MEFs and lymphoblastoid cells from patients with mutant TDP2 show increased formation of micronuclei, nucleoplasmic bridges and chromosomal aberrations upon etoposide treatment (105,107,108). This result suggests that blocking the TDP2-mediated repair pathway channels DSBs toward alternative pathway(s) that are more prone to mis-joining of exchanged DSB ends.
TDP2 inhibitors
As a relatively newly discovered DNA repair protein, only a few inhibitors of TDP2 have been reported. Deazaflavin and its derivatives have been identified recently to act as anti-cancer agents by activating tumor suppressor p53 and preventing its ubiquitination-induced degradation (127). These compounds inhibit TDP2 in vitro in the nanomolar range and also synergize the effects of etoposide in DU145 human prostate cancer cells, TK6 human lymphoblastoid cells and avian DT40 cells in a TDP2-dependent manner (128,129). Very recently, crystal structures of a deazaflavin derivative, 163, were obtained in complex with a ‘humanized’ form of murine TDP2 (mutating four residues in mTDP2 to their human counterparts). This study demonstrated that compound 163 competitively inhibits TDP2 by occupying a position equivalent to the second incoming nucleotide and thereby interfering with the DNA substrate (100). Isoquinoline-1,3-dione was recently identified as a novel TDP2 inhibitor with an IC50 of 2.2 μM against TDP2 in hTDP2-complemented DT40 TDP2−/− WCEs (130). The discovery of NSC111041 as a TDP2 inhibitor that inhibits TDP2-DNA binding without intercalating in the DNA is promising. NSC111041 also enhanced the etoposide-induced cytotoxicity in a TDP2-dependent manner and led to increased etoposide-induced γ-H2AX foci by affecting DSB repair, suggesting that this small molecule inhibitor can enhance the efficacy of Top II poisons (106). However, similar to inhibitors of TDP1, efforts at development of TDP2 inhibitors are in the preliminary stage. Development of effective and safe candidate compounds followed by rigorous pre-clinical evidence from in vitro as well as in vivo model systems is required before considering clinical trials.
Concluding remarks
Despite substantial advancements made in the understanding of TDP1 and TDP2 since their discovery, several questions remain. The role of TDP1 in repair of double-strand breaks needs to be more clearly defined. Given the interaction of TDP1 with key NHEJ proteins and its biochemical competency in the repair of 3′-PG-modified ends, it will be an interesting proposition to examine whether TDP1 plays a catalytic role during the resolution step of NHEJ. The underlying pathology of SCAN1 involves the nuclear transcription-dependent TopIcc that remain unrepaired in the absence of a normal TDP1 protein. Considering the extremely high energy demands on the mitochondria of neuronal cells, it will be interesting to dissect how important a role does accumulation of mitochondrial TopIcc play in SCAN1 pathology. Topoisomerases have been observed to be mutated in a subset of people with autism spectrum disorder (ASD). Topotecan treatment as well as knockdown of Top I and Top2β in neuronal cells reduced the transcription of several long genes linked with ASD due to increased torsional stress and enhanced supercoiling (131). Thus, it will be interesting to see what roles TDP1 and TDP2 play, if any, in the pathogenesis of ASD.
It is also tempting to speculate that a potential resistance mechanism to the anti-cancer (in ATL treatment) and anti-HIV chain terminating analogs involves overexpression of TDP1 causing the removal of these modified nucleosides from the genome. As such, use of TDP1 inhibitors in combination with these drugs might provide an efficient therapeutic response in treatment for HIV or ATL. Interestingly, as PARP1-deficient DT40 cells show no sensitivity to nucleoside analogs (46), it can be surmised that TDP1-mediated repair of nucleoside analogs does not require PARylation of TDP1. On the other hand, TDP1 is epistatic with PARP1 and PARylated for the repair of CPT-induced TopIcc (59). This differential post-translational modification of TDP1 in response to different blocked 3′ ends needs further investigation.
Although a great deal of research has been performed on TDP2 in its capacity as a phosphodiesterase, several outstanding questions are yet to be answered. How is TDP2 recruited to the sites of DSBs? Like TDP1, is TDP2 PARylated for the repair of TopIIcc? Do Phosphatidylinositol-3 kinase-related kinases (PI3KK) including ATM and DNA-PK regulate the function of TDP2 in DSB repair by phosphorylation? In addition to answering these critical questions about the regulation of TDP2, it is also important to decipher key synthetic lethal relationships of TDP2 with other proteins for the effective therapeutic use of TDP2 inhibitors. TDP2 suppresses chromosomal translocations and genomic instability in cells and thus use of TDP2 inhibitors in tumors already deficient in DNA repair could potentially provide effective therapeutic targeting of these tumors.
Elucidation of regulation of TDP1 and TDP2 and their functional interaction with other DNA repair proteins will add to our understanding of the complex web of networks to which DNA repair pathways belong, with the ultimate aim of making it easier to develop DNA repair inhibitors, functioning either clinically or in research to unravel myriad DNA repair mechanisms.
FUNDING
National Institutes of Health [CA166264] (in part). The open access publication charge for this paper has been waived by Oxford University Press - NAR.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ashour M., Atteya R., El-Khamisy S.. Topoisomerase-mediated chromosomal break repair: an emerging player in many games. Nat. Rev. Cancer. 2015; 15:137–151. [DOI] [PubMed] [Google Scholar]
- 2. Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem. Biol. 2013; 8:82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davies D., Interthal H., Champoux J., Hol W.. The crystal structure of human tyrosyl–DNA phosphodiesterase, Tdp1. Structure. 2002; 10:237–248. [DOI] [PubMed] [Google Scholar]
- 4. Yang S.-W., Burgin A., Huizenga B., Robertson C., Yao K., Nash H.. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerase. PNAS. 1996; 93:11534–11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pouliot J., Yao K., Robertson C., Nash H.. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999; 286:552–555. [DOI] [PubMed] [Google Scholar]
- 6. Wilson T., Vance J.. Yeast Tdp1 and Rad1–Rad10 function as redundant pathways for repairing Top1 replicative damage. PNAS. 2002; 99:13669–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Interthal H., Pouliot J., Champoux J.. The tyrosyl–DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. PNAS. 2001; 98:12009–12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takashima H., Boerkoel C., John J., Saifi G.M., Salih M., Armstrong D., Mao Y., Quiocho F., Roa B., Nakagawa M. et al. Mutation of TDP1, encoding a topoisomerase I–dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 2002; 32:267–272. [DOI] [PubMed] [Google Scholar]
- 9. Das B., Dexheimer T., Maddali K., Pommier Y.. Role of tyrosyl–DNA phosphodiesterase (TDP1) in mitochondria. PNAS. 2010; 107:19790–19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dexheimer T., Antony S., Marchand C., Pommier Y.. tyrosyl–DNA phosphodiesterase as a target for anticancer therapy. Adv. Anticancer Agents Med. Chem. 2008; 8:381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holt I. Mitochondrial DNA replication and repair: all a flap. Trends Biochem. Sci. 2009; 34:358–365. [DOI] [PubMed] [Google Scholar]
- 12. Davies D., Interthal H., Champoux J., Hol W.. Crystal structure of a transition state mimic for Tdp1 assembled from vanadate, DNA, and a topoisomerase I-derived peptide. Cell Chem. Biol. 2003; 10:139–147. [DOI] [PubMed] [Google Scholar]
- 13. Pommier Y., Huang S.-y., Gao R., Das B., Murai J., Marchand C.. tyrosyl–DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair. 2014; 19:114–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He X., van Waardenburg R., Babaoglu K., Price A., Nitiss John, Nitiss K., Bjornsti M.-A., White S.. Mutation of a conserved active site residue converts tyrosyl–DNA phosphodiesterase I into a DNA topoisomerase I-dependent poison. J. Mol. Biol. 2007; 372:1070–1081. [DOI] [PubMed] [Google Scholar]
- 15. Gajewski S., Comeaux E., Jafari N., Bharatham N., Bashford D., White S., Waardenburg R.. Analysis of the active site mechanism of tyrosyl–DNA phosphodiesterase I: a member of the phospholipase D superfamily. J. Mol. Biol. 2012; 415:741–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gottlin E., Rudolph A., Zhao Y., Matthews H., Dixon J.. Catalytic mechanism of the phospholipase D superfamily proceeds via a covalent phosphohistidine intermediate. PNAS. 1998; 95:9202–9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Interthal H., Chen H.J., Kehl-Fie T., Zotzmann J., Leppard J., Champoux J.. SCAN1 mutant Tdp1 accumulates the enzyme–DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 2005; 24:2224–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Povirk L.F. Processing of damaged DNA ends for double-strand break repair in mammalian cells. ISRN Mol. Biol. 2012; 2012:345805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. El-Khamisy S., Saifi G., Weinfeld M., Johansson F., Helleday T., Lupski J., Caldecott K.. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005; 434:108–113. [DOI] [PubMed] [Google Scholar]
- 20. Jiang B., Glover J., Weinfeld M.. Neurological disorders associated with DNA strand-break processing enzymes. Mech. Ageing Dev. 2017; 161:130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miao Z.-H., Agama K., Sordet O., Povirk L., Kohn K., Pommier Y.. Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes. DNA Repair. 2006; 5:1489–1494. [DOI] [PubMed] [Google Scholar]
- 22. Meisenberg C., Gilbert D., Chalmers A., Haley V., Gollins S., Ward S., El-Khamisy S.. Clinical and cellular roles for TDP1 and TOP1 in modulating colorectal cancer response to irinotecan. Mol. Cancer Ther. 2015; 14:575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barthelmes H., Habermeyer M., Christensen M., Mielke C., Interthal H., Pouliot J., Boege F., Marko D.. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J. Biol. Chem. 2004; 279:55618–55625. [DOI] [PubMed] [Google Scholar]
- 24. Heideker J.P., Prudden John, Perry J. Jefferson, Tainer John, Boddy Michael. SUMO-targeted ubiquitin ligase, Rad60, and Nse2 SUMO ligase suppress spontaneous Top1–mediated DNA damage and genome instability. PLoS Genet. 2011; 7:e1001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Interthal H., Chen H.J., Champoux J.. Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J. Biol. Chem. 2005; 280:36518–36528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weinfeld M., Mani R., Abdou I., Acetuno R.D., Glover J.. Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem. Sci. 2011; 36:262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Raymond A., Staker B., Burgin A.J.. Substrate specificity of tyrosyl–DNA phosphodiesterase I (Tdp1). J. Biol. Chem. 2005; 280:22029–22035. [DOI] [PubMed] [Google Scholar]
- 28. Pouliot J., Robertson C., Nash H.. Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells. 2001; 6:677–687. [DOI] [PubMed] [Google Scholar]
- 29. Debethune L., Kohlhagen G., Grandas A., Pommier Y.. Processing of nucleopeptides mimicking the topoisomerase I–DNA covalent complex by tyrosyl–DNA phosphodiesterase. Nucleic Acids Res. 2002; 30:1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Interthal H., Champoux J.. Effects of DNA and protein size on substrate cleavage by human tyrosyl–DNA phosphodiesterase 1 (TDP1). Biochem. J. 2011; 436:559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sordet O., Redon C., Guirouilh-Barbat J., Smith S., Solier S., Douarre C., Conti C., Nakamura A., Das B.B., Nicolas E. et al. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep. 2009; 10:887–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Christini A., Park J.-H., Capranico G., Legube G., Favre G., Sordet O.. DNA-PK triggers histone ubiquitination and signaling in response to DNA double-strand breaks produced during the repair of transcription-blocking topoisomerase I lesions. Nucleic Acids Res. 2016; 44:1161–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hirano R., Interthal H., Huang C., Nakamura T., Deguchi K., Choi K., Bhattacharjee M., Arimura K., Umehara F., Izumo S. et al. Spinocerebellar ataxia with axonal neuropathy: consequence of a Tdp1 recessive neomorphic mutation. EMBO J. 2007; 26:4732–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Katyal S., el-Khamisy S., Russell H., Li Y., Ju L., Caldecott K., McKinnon P.. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007; 26:4720–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hawkins A., Subler M., Akopiants K., Wiley J., Taylor S., Rice A., Windle J., Valerie K., Povirk L.F.. In vitro complementation of Tdp1 deficiency indicates a stabilized enzyme-DNA adduct from tyrosyl but not glycolate lesions as a consequence of the SCAN1 mutation. DNA Repair. 2009; 8:654–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Inamdar K., Pouliot J., Zhou T., Lees-Miller S., Rasouli-Nia A., Povirk L.. Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double strand breaks by the human tyrosyl–DNA phosphodiesterase hTdp1. J. Biol. Chem. 2002; 277:27162–27168. [DOI] [PubMed] [Google Scholar]
- 37. Zhou T., Lee J.W., Tatavarthi H., Lupski J., Valerie K., Povirk L.F.. Deficiency in 3′-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl–DNA phosphodiesterase (TDP1). Nucleic Acids Res. 2005; 33:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. El-Khamisy S., Hartsuiker E., Caldecott K.. TDP1 facilitates repair of ionizing radiation-induced DNA single-strand breaks. DNA Repair. 2007; 6:1485–1495. [DOI] [PubMed] [Google Scholar]
- 39. Murai J., Huang S.-y., Das B.B., Dexheimer T., Takeda S., Pommier Y.. tyrosyl–DNA phosphodiesterase 1 (TDP1) repairs DNA damage induced by topoisomerases I and II and base alkylation in vertebrate cells. J. Biol. Chem. 2012; 287:12848–12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lebedeva N., Rechkunova N., Lavrik O.. AP-site cleavage activity of tyrosyl–DNA phosphodiesterase 1. FEBS Lett. 2011; 585:683–686. [DOI] [PubMed] [Google Scholar]
- 41. Alagoz M., Wells O., El-Khamisy S.. TDP1 deficiency sensitizes human cells to base damage via distinct topoisomerase I and PARP mechanisms with potential applications for cancer therapy. Nucleic Acids Res. 2014; 42:3089–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chiang S.-C., Meagher M., Kassouf N., Hafezparast M., McKinnon P., Haywood R., El-Khamisy S.. Mitochondrial protein-linked DNA breaks perturb mitochondrial gene transcription and trigger free radical-induced DNA damage. Sci. Adv. 2017; 3:e1602506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamamoto J., Takahata C., Kuraoko I., Hirota K., Iwai S.. Chemical incorporation of chain-terminating nucleoside analogs as 3′-blocking DNA damage and their removal by human ERCC1-XPF endonuclease. Molecules. 2016; 21:e766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang S.-y., Murai J., Rosa I., Dexheimer T., Naumova A., Gmeiner W., Pommier Y.. TDP1 repairs nuclear and mitochondrial DNA damage induced by chain-terminating anticancer and antiviral nucleoside analogs. Nucleic Acids Res. 2013; 41:7793–7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tada K., Kobayashi M., Takiuchi Y., Iwai F., Sakamoto T., Nagata K., Shinohara M., Io K., Shirakwa K., Hishizawa M. et al. Abacavir, an anti–HIV-1 drug, targets TDP1-deficient adult T cell leukemia. Sci. Adv. 2015; 1:e1400203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Al Abo M., Sasanuma H., Liu X., Rajapakse V., Huang S.-y., Kiselev E., Takeda S., Plunkett W., Pommier Y.. TDP1 is critical for the repair of DNA breaks induced by sapacitabine, a nucleoside also targeting ATM- and BRCA-deficient tumors. Mol. Cancer Ther. 2017; 16:2543–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Povirk L. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat. Res., Fundam. Mol. Mech. Mutagen. 1996; 355:71–89. [DOI] [PubMed] [Google Scholar]
- 48. Dedon P., Goldberg I.. Free-radical mechanisms involved in the formation of sequence-dependent bistranded DNA lesions by the antitumor antibiotics bleomycin, neocarzinostatin, and calicheamicin. Chem. Res. Toxicol. 1992; 5:311–332. [DOI] [PubMed] [Google Scholar]
- 49. Zhou T., Akopiants K., Mohapatra S., Lin P.-S., Valerie K., Ramsden D., Lees-Miller S., Povirk L.F.. tyrosyl–DNA phosphodiesterase and the repair of 3′-phosphoglycolate-terminated DNA double-strand breaks. DNA Repair. 2009; 8:901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Akopiants K., Mohapatra S., Menon V., Zhou T., Valerie K., Povirk L.. Tracking the processing of damaged DNA double-strand break ends by ligation-mediated PCR: increased persistence of 3′-phosphoglycolate termini in SCAN1 cells. Nucleic Acids Res. 2014; 42:3125–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bahmed K., Nitiss K., Nitiss J.. Yeast Tdp1 regulates the fidelity of nonhomologous end joining. PNAS. 2010; 107:4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heo J., Li J., Summerlin M., Hays A., Katyal S., McKinnon P., Nitiss K., Nitiss J., Hanakahi L.. TDP1 promotes assembly of non-homologous end joining protein complexes on DNA. DNA Repair. 2015; 30:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Menon V., Povirk L.. End-processing nucleases and phosphodiesterases: an elite supporting cast for the non-homologous end joining pathway of DNA double-strand break repair. DNA Repair. 2016; 43:57–68. [DOI] [PubMed] [Google Scholar]
- 54. Li J., Summerlin M., Nitiss K., Nitiss J., Hanakahi L.. TDP1 is required for efficient non-homologous end joining in human cells. DNA Repair. 2017; 60:40–49. [DOI] [PubMed] [Google Scholar]
- 55. Borda M., Palmitelli M., Veron G., Gonazalez-Cid M., de Campos Nebel M.. tyrosyl–DNA-phosphodiesterase I (TDP1) participates in the removal and repair of stabilized-Top2α cleavage complexes in human cells. Mutat. Res. 2015; 781:37–48. [DOI] [PubMed] [Google Scholar]
- 56. Ray Chaudhuri A., Nussenzweig A.. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017; 18:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chatterjee S., Cheng M., Trivedi D., Petzold S., Berger N.. Camptothecin hypersensitivity in poly(adenosine diphosphate-ribose) polymerase-deficient cell lines. Cancer Commun. 1989; 1:39–394. [DOI] [PubMed] [Google Scholar]
- 58. Patel A., Flatten K., Schenider P., Dai N., McDonald J., Poirier G., Kaufmann S.. Enhanced killing of cancer cells by poly(ADP-ribose) polymerase inhibitors and topoisomerase I inhibitors reflects poisoning of both enzymes. J. Biol. Chem. 2012; 287:4198–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Das B.B., Huang S.-y., Murai J., Rehman I., Ame J.-C., Sengupta S., Das S., Majumdar P., Zhang H., Biard D. et al. PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage. Nucleic Acids Res. 2014; 42:4435–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Das B.B., Antony S., Gupta S., Dexheimer T., Redon C., Garfield S., Shiloh Y., Pommier Y.. Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. EMBO J. 2009; 28:3667–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chiang S.-C., Caroll J., El-Khamisy S.. TDP1 serine 81 promotes interaction with DNA ligase IIIα and facilitates cell survival following DNA damage. Cell Cycle. 2010; 9:588–595. [DOI] [PubMed] [Google Scholar]
- 62. Plo I., Liao Z.-Y., Barcelo J., Kohlhagen G., Caldecott K., Weinfeld M., Pommier Y.. Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair. 2003; 2:1087–1100. [DOI] [PubMed] [Google Scholar]
- 63. Tomkinson A., Sallmyr A.. Structure and function of the DNA ligases encoded by the mammalian LIG3 gene. Gene. 2013; 531:150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hudson J., Shih-Chieh C., Wells O., Rookyard C., El-Khamisy S.. SUMO modification of the neuroprotective protein TDP1 facilitates chromosomal single-strand break repair. Nat. Commun. 2012; 3:733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. O’Connor M. Targeting the DNA damage response in cancer. Mol. Cell. 2015; 60:547–560. [DOI] [PubMed] [Google Scholar]
- 66. Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J. Clin. Oncol. 2008; 26:3785–3790. [DOI] [PubMed] [Google Scholar]
- 67. Curtin N. DNA repair dysregulation from cancer driver to therapeutic target. Nat. Rev. Cancer. 2012; 12:801–817. [DOI] [PubMed] [Google Scholar]
- 68. Pearl L., Schierz A., Ward S., Al-Lazikani B., Pearl F.. Therapeutic opportunities within the DNA damage response. Nat. Rev. Cancer. 2015; 15:166–180. [DOI] [PubMed] [Google Scholar]
- 69. Liu C., Zhou S., Begum S., Sidransky D., Westra W., Brock M., Califano J.. Increased expression and activity of repair genes TDP1 and XPF in non-small cell lung cancer. Lung Cancer. 2007; 55:303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Abugable A., Awwad D., Fleifel D., Ali M., El-Khamisy S., Elserafy M.. Advances in Experimental Medicine and Biology. 2017; Cham: Springer; 157–178. [DOI] [PubMed] [Google Scholar]
- 71. Zhang Y.-W., Regairaz M., Seiler J., Agama K., Doroshow J., Pommier Y.. Poly(ADP-ribose) polymerase and XPF–ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 2011; 39:3607–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Usanova S., Piee-Staffa A., Sied U., Thomale J., Schneider A., Kaina B., Koberle B.. Cisplatin sensitivity of testis tumour cells is due to deficiency in interstrand-crosslink repair and low ERCC1-XPF expression. Mol. Cancer. 2010; 9:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. McNeail E.A.M., Melton D.W.. DNA repair endonuclease ERCC1-XPF as a novel therapeutic target to overcome chemoresistance in cancer therapy. Nucleic Acids Res. 2012; 40:9990–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wen Q., Scorah J., Phear G., Rodgers G., Rodgers S., Meuth M.. A mutant allele of MRE11 found in mismatch repair-deficient tumor cells suppresses the cellular response to DNA replication fork stress in a dominant negative manner. Mol. Biol. Cell. 2008; 19:1693–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Laev S., Salakhutdinov N., Lavrik O.. tyrosyl–DNA phosphodiesterase inhibitors: progress and potential. Bioorg. Med. Chem. 2016; 24:5017–5027. [DOI] [PubMed] [Google Scholar]
- 76. Antony S., Marchand C., Stephen A., Thibaut L., Agama K., Fisher R., Pommier Y.. Novel high-throughput electrochemiluminescent assay for identification of human tyrosyl–DNA phosphodiesterase (Tdp1) inhibitors and characterization of furamidine (NSC 305831) as an inhibitor of Tdp1. Nucleic Acids Res. 2007; 35:4474–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ngueyn T., Morrell A., Conda-Sheridan M., Marchand C., Agama K., Bermingam A., Stephen A., Chergui A., Naumova A., Fisher R. et al. Synthesis and biological evaluation of the first dual tyrosyl–DNA phosphodiesterase I (Tdp1)–topoisomerase I (Top1) inhibitors. J. Med. Chem. 2012; 55:4457–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Marchand C., Lea W., Jadhav A., Dexheimer T., Austin C., Inglese J., Pommier Y., Simeonov A.. Identification of phosphotyrosine mimetic inhibitors of human tyrosyl–DNA phosphodiesterase I by a novel AlphaScreen high-throughput assay. Mol. Cancer Ther. 2009; 8:240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dexheimer T., Gediya L., Stephen A., Weidlich I., Antony S., Marchand C., Interthal H., Niklaus M., Fisher R., Njar V. et al. 4-Pregnen-21-ol-3,20-dione-21-(4-bromobenzenesufonate) (NSC 88915) and related novel steroid derivatives as tyrosyl–DNA phosphodiesterase (Tdp1) inhibitors. J. Med. Chem. 2009; 52:7122–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Comeaux E., van Waardenburg R.C.. tyrosyl–DNA phosphodiesterase I resolves both naturally and chemically induced DNA adducts and its potential as a therapeutic target. Drug Metab. Rev. 2014; 46:494–507. [DOI] [PubMed] [Google Scholar]
- 81. Sirivolu V., Vernekar S.K., Marchand C., Naumova A., Chergui A., Renaud A., Stephen A., Chen F., Sham Y., Pommier Y. et al. 5-Arylidenethioxothiazolidinones as inhibitors of tyrosyl–DNA phosphodiesterase I. J. Med. Chem. 2012; 55:8671–8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zakharenko A., Khomenko T., Zhukova S., Koval O., Zakharova O., Anarbaev R., Lebedeva N., Korchagina D., Komarova N., Vasiliev V.. Synthesis and biological evaluation of novel tyrosyl–DNA phosphodiesterase 1 inhibitors with a benzopentathiepine moiety. Bioorg. Med. Chem. 2015; 9:2044–2052. [DOI] [PubMed] [Google Scholar]
- 83. Khomenko T., Zakharenko A., Odarchenko T., Arabshahi H., Sannikova V., Zakharova O., Korchagina D., Reynisson J., Volcho K., Salakhutdinov N. et al. New inhibitors of tyrosyl–DNA phosphodiesterase I (Tdp 1) combining 7-hydroxycoumarin and monoterpenoid moieties. Bioorg. Med. Chem. 2016; 24:5573–5581. [DOI] [PubMed] [Google Scholar]
- 84. Dean R., Fam H.K., An J., Choi K., Shimizu Y., Jones S., Boerkoel C., Interthal H., Pfeifer T.. Identification of a putative Tdp1 inhibitor (CD00509) by in vitro and cell-based assays. SLAS Discov. 2014; 19:1372–1382. [DOI] [PubMed] [Google Scholar]
- 85. Nguyen T.X., Abdelmalak M., Marchand C., Agama K., Pommier Y., Cushman M.. Synthesis and biological evaluation of nitrated 7-, 8-, 9-, and 10-hydroxyindenoisoquinolines as potential dual topoisomerase I (Top1)–tyrosyl–DNA phosphodiesterase I (TDP1) inhibitors. J. Med. Chem. 2015; 58:3188–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Conda-Sheridan M., Reddy P.V.N., Morrell A., Cobb B., Marchand C., Agama K., Chergui A., Renaud A., Stephen A., Bindu L. et al. Synthesis and biological evaluation of indenoisoquinolines that inhibit both tyrosyl–DNA phosphodiesterase I (Tdp1) and topoisomerase I (Top1). J. Med. Chem. 2015; 56:182–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lv P.-C., Agama K., Marchand C., Pommier Y., Cushman M.. Design, synthesis, and biological evaluation of O-2-modified indenoisoquinolines as dual topoisomerase I–tyrosyl–DNA phosphodiesterase I inhibitors. J. Med. Chem. 2014; 57:4324–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Conda-Sheridan M., Reddy P., Morrell A., Cobb B., Marchand C., Agama K., Chergui A., Renaud A., Stephen A., Bindu L. et al. Synthesis and biological evaluation of indenoisoquinolines that inhibit both tyrosyl–DNA phosphodiesterase I (Tdp1) and topoisomerase I (Top1). J. Med. Chem. 2013; 56:182–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Takagi M., Ueda J.-y., Hwang J.-H., Hashimoto J., Izumikawa M., Murakami H., Sekido Y., Shin-ya K.. tyrosyl–DNA phosphodiesterase 1 inhibitor from an anamorphic fungus. J. Nat. Prod. 2012; 75:764–767. [DOI] [PubMed] [Google Scholar]
- 90. Tian L.-W., Feng Y., Tran T., Shimizu Y., Pfeifer T., Forster P., Quinn R.. tyrosyl–DNA Phosphodiesterase I Inhibitors from the Australian Plant Macropteranthes leichhardtii. J. Nat. Prod. 2015; 78:1756–1760. [DOI] [PubMed] [Google Scholar]
- 91. Zakharenko A., Luzina O., Koval O., Dmitry N., Nilov D., Gushchina I., Dyrkheeva N., Švedas V., Salakhutdinov N., Lavrik O.. Tyrosyl–DNA phosphodiesterase 1 inhibitors: usnic acid enamines enhance the cytotoxic effect of camptothecin. J. Nat. Prod. 2016; 79:2961–2967. [DOI] [PubMed] [Google Scholar]
- 92. Bermingham A., Price E., Marchand C., Chergui A., Naumova A., Whitson E., Krumpe L., Goncharova E., Evans J., McKee T. et al. Identification of natural products that inhibit the catalytic function of human tyrosyl–DNA phosphodiesterase (TDP1). SLAS Discov. 2017; 22:1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tian L.-W., Feng Y., Tran T., Shimizu Y., Pfeifer T., Vu H., Quinn R.. Achyrodimer F, a tyrosyl–DNA phosphodiesterase I inhibitor from an Australian fungus of the family Cortinariaceae. Bioorg. Med. Chem. Lett. 2017; 27:4007–4010. [DOI] [PubMed] [Google Scholar]
- 94. Wang P., Elsayed M., Plescia C., Ravji A., Redon C., Kiselev E., Marchand C., Zeleznik O., Agama K., Pommier Y. et al. Synthesis and biological evaluation of the first triple inhibitors of human topoisomerase 1, tyrosyl–DNA phosphodiesterase 1 (Tdp1), and tyrosyl–DNA phosphodiesterase 2 (Tdp2). J. Med. Chem. 2017; 60:3275–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Pype S., Declercq W., Ibrahimi A., Michiels C., Van Rietschoten J., Dewulf N., de Boer M., Vandenabeele P., Huylebroeck D., Remacle J.. TTRAP, a novel protein that associates with CD40, tumor necrosis factor (TNF) receptor-75 and TNF receptor-associated factors (TRAFs), and that inhibits nuclear factor-kappa B activation. J. Biol. Chem. 2000; 275:18586–18593. [DOI] [PubMed] [Google Scholar]
- 96. Rodrigues-Lima F., Josephs M., Katan M., Cassinat B.. Sequence analysis identifies TTRAP, a protein that associates with CD40 and TNF receptor-associated factors, as a member of a superfamily of divalent cation-dependent phosphodiesterases. Biochem. Biophys. Res. Commun. 2001; 285:1274–1279. [DOI] [PubMed] [Google Scholar]
- 97. Cortes Ledesma F., El-Khamisy S., Zuma M., Osborn K., Caldecott K.. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009; 461:674–678. [DOI] [PubMed] [Google Scholar]
- 98. Nitiss J.L. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer. 2009; 9:338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zeng Z., Sharma A., Ju L., Murai J., Umans L., Vermeire L., Pommier Y., Takeda S., Huylebroeck D., Caldecott K. et al. TDP2 promotes repair of topoisomerase I-mediated DNA damage in the absence of TDP1. Nucleic Acids Res. 2012; 40:8371–8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hornyak P., Askwith T., Walker S., Komulainen E., Paradowski M., Pennicott L., Bartlett E., Brissett N., Raoof A., Watson M. et al. Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2. Biochem. J. 2016; 473:1869–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rao T., Gao R., Takada S., Abo M., Chen X., Walters K., Pommier Y., Aihara H.. Novel TDP2-ubiquitin interactions and their importance for the repair of topoisomerase II-mediated DNA damage. Nucleic Acids Res. 2016; 44:10201–10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Shi K., Kurahashi K., Gao R., Tsutakawa S., Tainer J., Pommier Y., Aihara H.. Structural basis for recognition of 5′-phosphotyrosine adducts by TDP2. Nat. Struct. Mol. Biol. 2012; 19:1372–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Schellenberg M., Appel C., Adhikari S., Robertson P., Ramsden D., Williams R.. Mechanism of repair of 5′-topoisomerase II-DNA adducts by mammalian tyrosyl–DNA phosphodiesterase 2. Nat. Struct. Mol. Biol. 2012; 19:1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Gao R., Huang S.-y., Marchand C., Pommier Y.. Biochemical characterization of human tyrosyl–DNA phosphodiesterase 2 (TDP2/TTRAP): a Mg(2+)/Mn(2+)-dependent phosphodiesterase specific for the repair of topoisomerase cleavage complexes. J. Biol. Chem. 2012; 287:30842–30852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Schellenberg M., Perera L., Strom C., Waters C., Monian B., Appel C.D., Vilas C., Williams J., Ramsden D., Williams R.S.. Reversal of DNA damage induced Topoisomerase 2 DNA-protein crosslinks by Tdp2. Nucleic Acids Res. 2016; 44:3289–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kont Y., Dutta A., Mallisetty A., Mathew J., Minas T., Kraus C., Dhopeshwarkar P., Kallakury B., Mitra S., Uren A. et al. Depletion of tyrosyl DNA phosphodiesterase 2 activity enhances etoposide-mediated double-strand break formation and cell killing. DNA Repair. 2016; 43:38–47. [DOI] [PubMed] [Google Scholar]
- 107. Gómez-Herreros F., Romero-Granados R., Zeng Z., Álvarez-Quilón A., Quintero C., Ju L., Umans L., Vermeire L., Huylebroeck D., Caldecott K. et al. TDP2–dependent non-homologous end-joining protects against topoisomerase II–induced DNA breaks and genome instability in cells and in vivo. PLoS Genet. 2013; 9:e1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Gómez-Herreros F., Zagnoli-Vieira G., Ntai I., Martínez-Macías M., Anderson R., Herrero-Ruíz A., Caldecott K.. TDP2 suppresses chromosomal translocations induced by DNA topoisomerase II during gene transcription. Nat. Commun. 2017; 8:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Gomez-Herreros F., Schuurs-Hoeijmakers J., McCormack M., Greally M., Rulten S., Romero-Granados R., Counihan T., Chaila E., Conroy J., Ennis S. et al. TDP2 protects transcription from abortive topoisomerase activity and is required for normal neural function. Nat. Genet. 2014; 46:516–521. [DOI] [PubMed] [Google Scholar]
- 110. Gao R., Schellenberg M., Huang S.-y., Abdelmalak M., Marchand C., Nitiss K., Nitiss J., Williams R.S., Pommier Y.. Proteolytic degradation of topoisomerase II (Top2) enables the processing of Top2·DNA and Top2·RNA covalent complexes by tyrosyl–DNA-phosphodiesterase 2 (TDP2). J. Biol. Chem. 2014; 289:17960–17969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Mao Y., Desai S., Ting C.-Y., Hwang J., Liu L.. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J. Biol. Chem. 2001; 276:40652–40658. [DOI] [PubMed] [Google Scholar]
- 112. Zhang A., Lyu Y.L., Lin C.-P., Zhou N., Azarova A., Wood L., Liu L.. A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J. Biol. Chem. 2006; 281:35997–60003. [DOI] [PubMed] [Google Scholar]
- 113. Adhikari S., Karmahapatra S., Elias H., Dhopeshwarkar P., Williams R.S., Byers S., Uren A., Roy R.. Development of a novel assay for human tyrosyl DNA phosphodiesterase 2. Anal. Biochem. 2011; 416:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Clausen A., Zhang S., Burgers P., Lee M., Kunkel T.. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase δ. DNA Repair. 2013; 12:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Langereis M., Feng Q., Nelissen F., Virgen-Slane R., van der Heden van Noort G., Maciejewski S., Filippov D., Semler B., van Delft F., van Kuppeveld F.. Modification of picornavirus genomic RNA using ‘click’ chemistry shows that unlinking of the VPg peptide is dispensable for translation and replication of the incoming viral RNA. Nucleic Acids Res. 2014; 42:2473–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Maciejewski S., Nguyen J., Gómez-Herreros F., Cortés-Ledesma F., Caldecott K., Semler B.. Divergent requirement for a DNA repair enzyme during enterovirus infections. mBio. 2016; 7:e01931–01915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Virgen-Slane R., Rozovics J., Fitzgerald K., Ngo T., Wayne C., van der Heden van Noort G., Filippov D., Gershon P., Semler B.. An RNA virus hijacks an incognito function of a DNA repair enzyme. PNAS. 2012; 109:14634–14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Königer C., Wingert I., Marsmann M., Rösler C., Beck J., Nassal M.. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. PNAS. 2014; 111:E4244–E4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Cui X., McAllister R., Boregowda R., Sohn J., Cortes Ledesma F., Caldecott K., Seeger C., Hu J.. Does tyrosyl DNA phosphodiesterase-2 play a role in hepatitis B virus genome repair. PLoS One. 2015; 10:e0128401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Esguerra C., Nelles L., Vermeire L., Ibrahimi A., Crawford A., Derua R., Janssens E., Waelkens E., Carmeliet P., Collen D. et al. Ttrap is an essential modulator of Smad3-dependent Nodal signaling during zebrafish gastrulation and left-right axis determination. Development. 2007; 134:4381–4393. [DOI] [PubMed] [Google Scholar]
- 121. Li C., Sun S.-Y., Khuri F., Li R.. Pleiotropic functions of EAPII/TTRAP/TDP2: cancer development, chemoresistance and beyond. Cell Cycle. 2011; 10:3274–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Nitiss J.N., Nitiss K.C.. Tdp2: a means to fixing the ends. PLoS Genet. 2013; 9:e1003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Li C., Fan S., Owonikoko T., Khuri F., Sun S., Li R.. Oncogenic role of EAPII in lung cancer development and its activation of the MAPK-ERK pathway. Oncogene. 2011; 30:3802–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Bian K., Muppani N.R., Elkhadragy L., Wang W., Zhang C., Chen T., Jung S., Seternes O.M., Long W.. ERK3 regulates TDP2-mediated DNA damage response and chemoresistance in lung cancer cells. Oncotarget. 2015; 7:6665–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Schellenberg M., Lieberman J., Herrero-Ruiz A., Butler L., Jason W., Muñoz-Cabello A., Mueller G., London R., Cortés-Ledesma F., Williams R.S.. ZATT (ZNF451)–mediated resolution of topoisomerase 2 DNA-protein cross-links. Science. 2017; 357:1412–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Adachi N., Suzuki H., Iiizumi S., Koyama H.. Hypersensitivity of nonhomologous DNA end-joining mutants to VP-16 and ICRF-193: implications for the repair of topoisomerase II-mediated DNA damage. J. Biol. Chem. 2003; 278:35897–35902. [DOI] [PubMed] [Google Scholar]
- 127. Dickens M., Roxburgh P., Hock A., Mezna M., Kellam B., Vousden K., Fischer P.. 5-Deazaflavin derivatives as inhibitors of p53 ubiquitination by HDM2. Bioorg. Med. Chem. 2013; 21:6868–6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Marchand C., Abdelmalak M., Kankanala J., Huang Shar-yin, Kiselev E., Fesen K., Kurahashi K., Sasanuma H., Takeda S., Aihara H., Wang Z. et al. Deazaflavin inhibitors of tyrosyl–DNA phosphodiesterase 2 (TDP2) specific for the human enzyme and active against cellular TDP2. ACS Chem. Biol. 2016; 11:1925–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Raoof A., Depledge P., Hamilton N., Hamilton N., Hitchin J., Hopkins G., Jordan A., Maguire L., McGonagle A., Mould D. et al. Toxoflavins and deazaflavins as the first reported selective small molecule inhibitors of tyrosyl–DNA phosphodiesterase II. J. Med. Chem. 2013; 56:6352–6370. [DOI] [PubMed] [Google Scholar]
- 130. Kankanala J., Marchand C., Abdelmalak M., Aihara H., Pommier Y., Wang Z.. Isoquinoline-1,3-diones as selective inhibitors of tyrosyl DNA phosphodiesterase II (TDP2). J. Med. Chem. 2016; 59:2734–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. King I., Yandava C., Mabb A., Hsiao J., Huang H.-S., Pearson B., Calabrese J.M., Starmer J., Parker J., Magnuson T. et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013; 501:58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]