Abstract
DNA topoisomerases are essential enzymes involved in all the DNA processes and among them, type IA topoisomerases emerged as a key actor in the maintenance of genome stability. The hyperthermophilic archaeon, Sulfolobus solfataricus, contains three topoisomerases IA including one classical named TopA. SsoTopA is very efficient at unlinking DNA catenanes, grouping SsoTopA into the topoisomerase III family. SsoTopA is active over a wide range of temperatures and at temperatures of up to 85°C it produces highly unwound DNA. At higher temperatures, SsoTopA unlinks the two DNA strands. Thus depending on the temperature, SsoTopA is able to either prevent or favor DNA melting. While canonical topoisomerases III require a single-stranded DNA region or a nick in one of the circles to decatenate them, we show for the first time that a type I topoisomerase, SsoTopA, is able to efficiently unlink covalently closed catenanes, with no additional partners. By using single molecule experiments we demonstrate that SsoTopA requires the presence of a short single-stranded DNA region to be efficient. The unexpected decatenation property of SsoTopA probably comes from its high ability to capture this unwound region. This points out a possible role of TopA in S. solfataricus as a decatenase in Sulfolobus.
INTRODUCTION
DNA topoisomerases are the enzymes required to manage the topological constraints generated during all DNA transactions (1,2). Topoisomerases carry out this task by introducing transient breaks into DNA: type-II topoisomerases generate double-stranded breaks, whereas type-I topoisomerases generate single-stranded breaks. Type-I topoisomerases can be further subdivided into three subtypes, IA, IB and IC, the IA subtype being the only topoisomerase present in all three domains of life.
Structurally all type IA topoisomerases share a highly conserved toroidal architecture formed by two ‘topofold’ domains associated to a Toprim domain (3,4). Despite the structural similarities, it is possible to classify type IA enzymes into subfamilies based on their distinct catalytic activities (3,5,6). The subfamily corresponding to Escherichia coli protein ω (topoisomerase I, topo I) was the first described (7) and is principally responsible for relaxing negatively supercoiled DNA in mesophilic bacteria (8). The second subfamily of type IA topoisomerases, named topoisomerase III (topo III), was first described in E. coli and yeast (9–11). Unlike topo I, bacterial topo III is able to efficiently decatenates linked DNA (9,12–14), as long as one of the circles contains a break in one strand. The E. coli and yeast topoisomerase III enzymes are also able to act in coordination with partner proteins; the helicase RecQ for E. coli or Sgs1 and Rmi1 for yeast, to catalyze catenation and decatenation of dsDNA (14,15). Catenation occurs under conditions where the partner proteins are in excess, decatenation occurs at lower protein concentrations (14). Some topo III enzymes also unlink RNA catenanes (16–18). These diverse in vitro activities reflect the fact that topoisomerases III enzymes are implicated in the processing of a wide variety of linked nucleic acids substrates in vivo, including double Holliday junctions, hemicatenanes R-loops and D-Loops (14,19–23). Topo III enzymes, and together with helicases in the SF2 group such as RecQ, Sgs1 or BLM, also play key roles in maintaining genome stability both in Bacteria and in Eukarya (24–26).
Finally, the third subfamily of type IA topoisomerases are the reverse gyrases (TopR), a particular topoisomerase unique to and ubiquitous in hyperthermophiles sequenced to date (27,28). Structurally, this enzyme is a chimera between a classical type IA topoisomerase and an SF2 helicase (28,29). This chimera is responsible for adenosinetriphosphate (ATP)-dependent positive supercoiling of the DNA, which is thought to help prevent DNA melting at high temperature (28). However, the precise role of reverse gyrase is not yet understood.
While several organisms, including hyperthermophiles, have two type IA topoisomerases, most of the Crenarchaeota, such as Sulfolobus solfataricus P2 (30), have three type IA topoisomerases: a classical type IA topoisomerase (TopA), and two reverse gyrases (TopR1 and TopR2). The few studies that have been carried out on S. solfataricus TopA revealed that its relaxation activity on negatively supercoiled DNA is relatively inefficient, suggesting that S. solfataricus TopA (SsoTopA) may be more similar to topo III than to topo I enzymes (31,32).
In the present study, we characterized SsoTopA using ensemble analysis and single molecule experiments. Our data demonstrate that SsoTopA belongs to the topoisomerase III subfamily of type IA because it unlinks DNA. Surprisingly, we demonstrated that SsoTopA disentangles supercoiled catenanes, without requiring either a gap/nick in one of the DNA circles or additional partner proteins. SsoTopA needs only a short single-stranded DNA region to efficiently catalyze the topological conversions.
MATERIALS AND METHODS
Materials
Ethidium bromide was purchased from Boehringer Mannheim. Dithiothreitol (DTT), polyethylenimine, chloroquine and agarose were purchased from Sigma. Ammonium sulfate was from Fisher. Toluene was purchased from VWR Intl. All other chemicals were purchased from Merck. Thermophilic SSB, T5 exonuclease, Nb. BbvCI, Nt. BspQI and all the restriction enzymes were purchased from New England Biolabs (NEB). S1 nuclease was purchased from Fermentas.
Purification of SsoTopA
Expression and purification of SsoTopA were performed as described previously (33). A gel filtration step was included to isolate highly pure enzyme for the single molecule experiments. Briefly, we used a 24 ml Superdex 200 prepacked column (GE Healthcare Life Sciences) equilibrated in a buffer containing 40 mM Tris–Cl pH 8.0, 1 M NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.5 mM β-mercaptoethanol and 10% glycerol. The fractions corresponding to SsoTopA were pooled and stored at 4°C. The final preparation was pure, as judged by sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE), with only a single band visible by Coomassie blue staining (Supplementary Figure S1).
DNA relaxation and kinetoplast DNA decatenation
The standard reaction mixture (10 μl) contained 50 mM Tris–HCl, pH 8.0, 0.5 mM DTT, 0.5 mM EDTA, 100 mM NaCl (TR buffer), 15 mM MgCl2 and either 0.15 μg of negatively supercoiled plasmid pTZ18R DNA or 0.5 μg of kinetoplast DNA (from Topogen). After addition of the enzyme, the mixture was incubated at the indicated temperature for up to 30 min. The reaction was stopped by cooling the reaction mixture on ice and adding 0.1% SDS, 25 mg/ml bromophenol blue and 15% sucrose (final concentrations). The samples were loaded onto gels and analyzed by one-dimensional gel electrophoresis (1.2% agarose gel in TEP buffer: 36 mM Tris, 1 mM EDTA, 30 mM NaH2PO4) at room temperature and run at 3 V/cm for 6 h. The gel was washed in TEP buffer for 30 min and stained as described previously (34).
Nuclease assay
pTZ18R DNA was incubated with SsoTopA at 95°C for 8 min, then cooled at 4°C. DNA was extracted with chloroform/isoamyl alcohol (24:1), precipitated with ethanol, then resuspended in 10 mM Tris–HCl, pH 7, 0.1 mM EDTA and finally incubated with increasing amounts of S1 nuclease at 4°C overnight in the buffer provided by the manufacturer. The reaction products were analyzed by agarose gel electrophoresis as described above except that we used a 2% agarose gel.
T5 exonuclease assay
pTZ18R DNA was incubated with SsoTopA in the standard reaction mixture at 95°C for 5 min, then cooled at 4°C. After addition of 10 U of T5 exonuclease, the reactions were incubated for the indicated time at 4°C. The reaction products were analyzed as described above on a 2% agarose gel.
Unlinking intact supercoiled DNA circles
Catenated DNA molecules (catenated products, CP) were from Twister Biotech, Inc. (Houston, TX, USA). These DNA catenanes consist of a 3530 bp large circular DNA (large circle, LC) linked to a 339 bp minicircle (mC) and are generated by λ–integrase-mediated site-specific recombination on the parent plasmid (PP) pMC339 (35). As controls, three different DNA topological forms—supercoiled, linearized or nicked- of these multi-linked catenanes were obtained by incubating the DNA with the restriction enzymes BamHI or NdeI, or the nicking endonucleases Nt.BspQI or Nb.BbvCI, respectively. The superhelical density of the supercoiled multi-linked DNA was −0.07 (35,36). TopA was incubated in 10 μl TR buffer containing 10 mM MgCl2 and 0.27 μg of catenated DNA at various (as indicated) temperatures for 20 min. Reactions were stopped by cooling the reaction mixture on ice for at least 10 min. A total of 0.1% SDS (final concentration) with Purple Gel Loading Dye (no SDS) (from NEB) was added before loading onto gels. The reaction products were analyzed on 1.7 or 3.5% agarose gels in TEP buffer at room temperature and run at 3 V/cm for 2–4 h. The gels were stained by incubation in TEP buffer containing 2 μg/ml ethidium bromide for 30 min and stored in water before digitalization.
Preparation of streptavidin coated capillaries
To obtain single molecule tracking results at higher temperature, glass capillaries (1.00 mm ID * 0.20 mm Wall, VitroCom) were coated with PEG–Biotin–streptavidin. Briefly, after sonication in 1 M KOH for 15 min, capillaries were washed with water then dried with nitrogen gas and coated with MCC primer 80/20 at 110°C (MicroChem) for 10 min, rinsed with 0.5% azide-terminated polystyrene (Sigma) and dissolved in toluene. Capillaries were then dried with nitrogen gas again and incubated overnight with 2 μM DBCO–PEG–Biotin (5 KDa, Interchim) and 2 mM DBCO–PEG (5 KDa, Interchim) dissolved in phosphate-buffered saline (PBS) buffer pH 8.0 overnight at room temperature. After being washed with PBS, capillaries were incubated for 30 min at room temperature with 0.2 mg/ml streptavidin in 10 mM Tris pH 8.0 and 500 mM NaCl. Capillaries were rinsed and then filled with reaction buffer (10 mM Hepes pH 8.0, 200 mM NaCl, 10 mM MgCl2, 20 mM β-mercaptoethanol, 0.1% Tween 20 and 0.5 mg/ml bovine serum albumin (BSA)) and stored at 4°C before use.
DNA preparation for single molecule experiments
The DNA fragment used for the tethered-DNA assays (3 kb) corresponds to a part of the Thermus aquaticus rpoC gene (seq ID Y19223.3, from position 227 to 3190) cloned into the XbaI and SbfI sites of pUC18. The following oligonucleotide is inserted at the KpnI site of the rpoC gene: 5′-TGTCAGCCCGTGATATTCATTACTTCTTATCCTAA-3′. The recombinant plasmid was cut with XbaI and SbfI, gel purified and the corresponding insert DNA ligated with 1 kb biotin-labeled or digoxigenin-labeled DNA fragments. These 1 kb DNA were labeled as described previously (37).
For DNA containing a permanent 10 bp bubble, we used a 2.2 kb DNA fragment that corresponds to a part of the T. aquaticus rpoC gene (from position 2103 to 4125) cloned into XbaI and SbfI sites of pUC18. This DNA fragment was prepared as described above except that before the ligation with the 1 kb biotin-labeled or digoxigenin-labeled DNA fragments, the two following oligonucleotides were annealed and then inserted into the 2.2 kb DNA between the HindIII and SpeI sites: 5′-AGCTGGATACTTACAGCCATATCAGTTACGCCTACTCCATTCCATCCACTTCCTCATCAACTACCATATG-3′ and 5′-CTAGCATATGGTAGTTGATGAGGAAGTGGATGGAATGGAGTATCAGCCGTGTATATGGCTGTAAGTATCC-3′ (bases in bold correspond to the bubble region, see Supplementary Figure S6C). The labeled DNA fragments were attached first to the anti-digoxigenin-coated magnetic beads prepared as described (38), then to the streptavidin-coated capillaries, and finally placed on a temperature controlled home-built magnetic trap running the PicoTwist software suite (PicoTwist SARL, http://www.picotwist.com).
SsoTopA relaxation rate analysis during single molecule experiments
The assays were performed at 45°C in 10 mM Hepes pH 8.0, 200 mM NaCl, 10 mM MgCl2, 20 mM β-mercaptoethanol, 0.1% Tween 20 and 0.5 mg/ml BSA. The 3 kb DNA was torsionally constrained and extended in the magnetic field (with a 0.45 pN tensile force). After the addition of 600 pM of SsoTopA, DNA molecules were negatively supercoiled by magnetic tweezers (to a superhelical density (σ) ∼ −0.04). When most of the DNA molecules were partially or totally relaxed by the enzyme, a new round of negative supercoiling was performed again on the same DNA. The number of supercoils removed was obtained from the change in extension, compared to the calibration curve for each bead (39).
To measure the processivity of SsoTopA (number of supercoils removed during a single burst), the change of DNA extension in each single relaxation burst was measured and then divided by the average step-size achieved from the extension versus supercoiling curve in the linear part of the calibration curve (40), where the number of supercoils relaxed is proportional to the change of DNA extension during one burst.
To analyze the relaxation rate, the lifetimes of relaxation bursts were also measured. The total velocity of one burst was then calculated using the value of a relative processivity divided by the corresponding lifetime. The probability of relaxation (pr(t)) during a given time (t) fits with a single exponential:
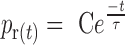 |
where, 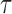 is the average time and C is a constant.
is the average time and C is a constant.
SsoTopA relaxation experiment with a DNA tether containing a single-stranded region
This experiment was conducted using a similar method as for the processivity and relaxation rate analysis but with the anti-digoxigenin-covered surface as described earlier (41) and anti-digoxigenin coated magnetic beads. A 2.2 kb DNA tether containing a 10 bp mismatch bubble with biotin- and digoxigenin-labeled ends was used, in the same reaction buffer mentioned above. The DNA molecules were extended with 0.42 pN magnetic force at 35°C. Lower SsoTopA concentration (6 pM) was used for this experiment. The step size for SsoTopA relaxation (number of turns removed by the enzyme in each relaxation step) was then measured by dividing the change of DNA extension in a single step to the relative step-size obtained from the extension versus supercoiling curve as mentioned above.
RESULTS
SsoTopA is a topo III
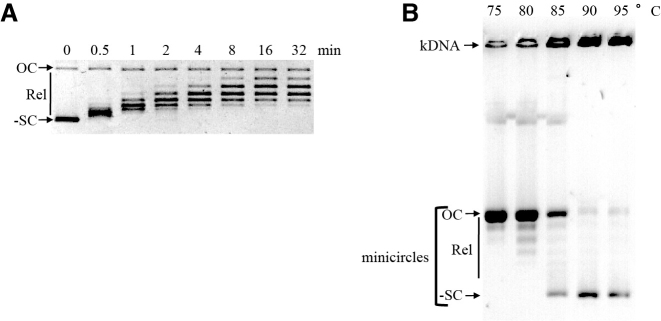
Recombinant SsoTopA was expressed and purified to homogeneity (Supplementary Figure S1). Temperature dependence (Supplementary Figure S2), MgCl2 dependence (Supplementary Figure S3) and NaCl dependence (not shown) of SsoTopA activity were identical to those obtained previously (31). By following the DNA relaxation reaction over time, we observed that at 75°C, the apparent optimal temperature (see Supplementary Figure S2), SsoTopA partially relaxed all of the highly negatively supercoiled DNA molecules after only 30 s of incubation (Figure 1A). However, further relaxation progressed more slowly and in a step-wise manner requiring at least 16 min for complete relaxation to be achieved (Figure 1A). This activity pattern suggests a higher efficiency of SsoTopA on highly negatively supercoiled DNA than on partially relaxed forms. It is consistent with the fact that this reaction is driven by the supercoiling energy stored in the DNA. The step-wise relaxation, with the gradual appearance of progressively more relaxed intermediates, is indicative of a progressive loss of this energy and a distributive mechanism (Figure 1A).
Figure 1.
Activity of SsoTopA in bulk experiments. (A) For relaxation, the protein/pTZ18R DNA molar ratio used was 1:1 and the incubation time was as indicated. The reaction products were analyzed by one dimensional agarose gel electrophoresis. (B) Decatenation of kDNA was carried out by adding SsoTopA just before the incubation at the indicated temperature for 30 min in the presence of kDNA. The reactions were stopped and kDNA products were separated on a 2% agarose gel. OC indicates open circular DNA, Rel, the relaxed topoisomers and -SC the negatively supercoiled DNA for the pTZ18R plasmid (A) or the mCs (B).
By using a kinetoplast DNA (kDNA) decatenation assay, we observed that SsoTopA is highly efficient in decatenating DNA, even at temperatures as high as 95°C (Figure 1B). The kDNA substrate naturally contains nicks or gaps in some of the catenated molecules, which, once decatenated, lead to the appearance of open circular form. This appearance was especially evident when the reaction was performed at 75 and 80°C (Figure 1B). More surprisingly, covalently closed DNA mCs (relaxed and/or supercoiled) were also observed depending on the temperature used for incubation (Figure 1B). Predominantly negatively supercoiled mC products were observed when the reaction was performed at 85–95°C (Figure 1B). The presence of supercoiled products means that, at higher temperatures, decatenation may not have required a nicked mC, suggesting that the presence of a nick or a gap on each catenated molecule is not a prerequisite for decatenation. A type I DNA-topoisomerase, such as SsoTopA, only cleaves one strand of the DNA molecule. Consequently, in the absence of a nicked or a gapped DNA region, the decatenation of kDNA catalyzed by SsoTopA at high temperature results from two sequential single strand passage events. The first reaction would lead to a transient hemicatenane stabilizing the single-stranded DNA region and consequently promoting the second strand passage reaction, thus providing either the decatenated product or the initial substrate.
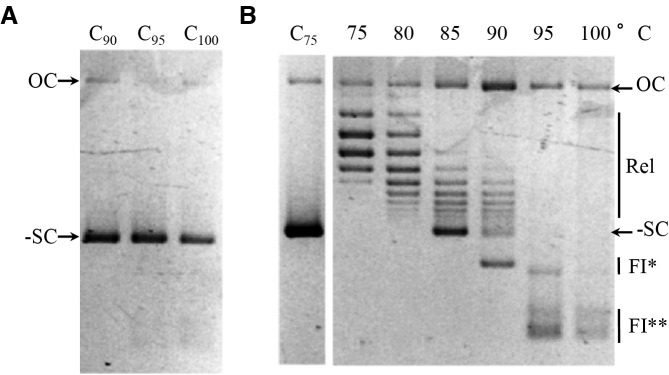
SsoTopA unlinks DNA at very high temperatures
To better understand the reactions carried out by SsoTopA above 75°C, we extended the range of temperature for analysis to as high as 100°C (Figure 2). At temperatures up to 85°C, SsoTopA relaxed negatively supercoiled plasmids (Figure 2B and Supplementary Figure S2). At 90°C and above, SsoTopA progressively converted the substrate into several fast migrating products, FI* and FI**, (Figure 2B) so named because of their apparent similarity with highly unwound species previously described (42). The appearance of these forms is not due to thermal denaturation or degradation of the DNA substrate alone, as they were not observed in the absence of enzyme (Figure 2A), nor were they observed in the presence of an inactive enzyme. Indeed, reactions lacking the magnesium cofactor essential for SsoTopA activity leave the DNA substrate unchanged (Supplementary Figure S3, lane 0). When the enzyme was inactivated with SDS before incubating with DNA, the FI* and FI** forms were no longer produced (Supplementary Figure S4, lane 1).
Figure 2.
Activity of SsoTopA at very high temperatures. (A) The DNA controls (lanes Ct°c) were incubated in the absence of SsoTopA for 8 min at the indicated temperatures. (B) The pTZ18R substrate was incubated at the temperature indicated in the presence of SsoTopA (at a molar ratio of 2:1). C75 corresponds to the DNA control incubated at 75°C for 8 min without SsoTopA. The reaction products were analyzed by one dimensional agarose gel electrophoresis: OC indicates open circular DNA, Rel, the relaxed topoisomers and -SC the negatively supercoiled DNA form. FI* and FI** correspond to unlinked DNA species and were apparently similar to highly unwound species first described by Parada and Marians (42).
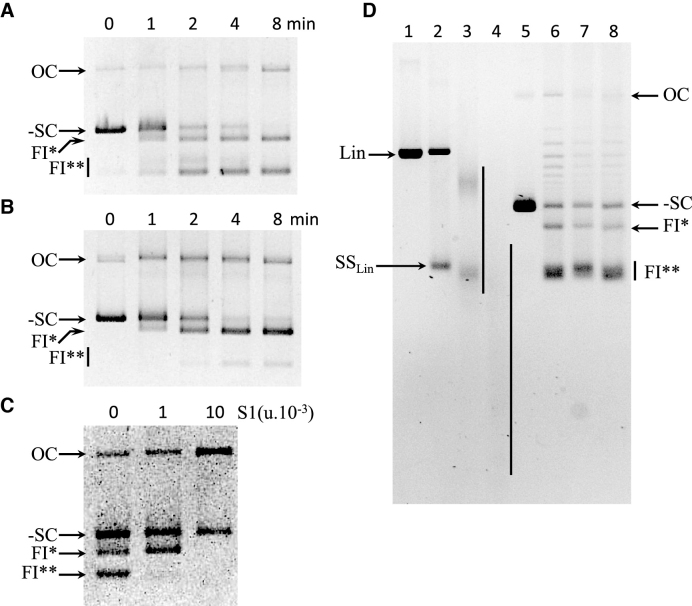
To investigate formation of the FI* and FI** forms, plasmid substrate was incubated with SsoTopA at 95°C over an 8 min time course, samples were removed at the time points indicated and incubated at 4°C prior to gel loading. We observed over time of incubation with SsoTopA the disappearance of the supercoiled DNA substrate and the appearance of FI* and FI** forms (Figure 3A). When the samples were further incubated 5 min at 25°C instead of 4°C prior to loading on the gel, the disappearance of the supercoiled DNA substrate was similar (Figure 3, compare -SC in panels A and B) while the accumulation kinetics were higher for FI* and lower for FI** (Figure 3, compare FI* and FI** in panels A and B). These observed differences reflect the conversion of FI** into FI* during the post-incubation at 25°C because the enzyme was inactivated just after the incubation at 95°C (Figure 3B). This conversion of FI** into FI* is reversible and is temperature dependent. Compared to 25°C, incubation at 65°C increased the conversion of FI** into FI* (Supplementary Figure S4, compare lanes 2 and 4). An additional heating of the samples at 95°C for 2 min before loading restored the presence of FI** with the concomitant disappearance of FI* (Supplementary Figure S4, lanes 3 and 5). In both experiments, we did not observe any linear double-stranded DNA or significant additional OC form (Figure 3, panel B and Supplementary Figure S4, compare lanes 2 and 4 with lanes 3 and 5). Therefore, no linear single-stranded form was produced during the unlinking reaction. These results indicate that the FI** corresponds to circular single-stranded DNA.
Figure 3.
Analysis of FI* and FI** DNA. (A) Generation of FI* and FI** over time during incubation with SsoTopA at 95°C. After incubation at 95°C for the indicated times, samples were cooled at 4°C for 5 min, centrifuged before adding 0.1% SDS, 25 mg/ml bromophenol blue and 15% sucrose (final concentrations). The samples were further incubated for an additional 5 min at 4°C. The samples were loaded onto an agarose gel at 4°C and run at 3 V/cm for 6 h at 4°C. (B) Spontaneous re-annealing of single-stranded forms FI** at 25°C. The samples were treated as in (A) except that they were further incubated for an additional 5 min at 25°C instead of 4°C prior to loading on the gel. (C) Sensitivity of the different DNA forms to S1 nuclease. Purified pTZ18R DNA obtained after incubation with TopA at 95°C for 4 min was further incubated at 4°C overnight without (0) or with 1 × 10−3 unit or 10 × 10−3 unit of S1 nuclease. (D) Sensitivity of the different DNA forms to T5 exonuclease. pTZ18R was linearized by BamHI (lanes 1–4) and then heated at 95°C for 5 min (lanes 2–4). Unlinked DNA forms (FI* and FI**) were obtained after incubation of pTZ18R with SsoTopA at 95°C for 5 min (lanes 6–8). After incubation at 95°C the DNA was further incubated at 4°C without (lanes 1, 2 and 6) or with 10 units of T5 exonuclease for 4 h (lanes 3 and 7) or 16 h (lanes 4 and 8). The SsoTopA treated samples (lanes 6–8) were additionally heated at 95°C for 2 min just before loading. Lane 5 is the control DNA substrate (pTZ18R). In the different experiments (A–D), the reaction products were analyzed by one dimensional gel electrophoresis: OC indicates open circular DNA, Lin the linear double stranded DNA, SSLin the linear single-stranded DNA, -SC the negatively supercoiled DNA form and the particular FI* and FI** forms.
The single-stranded nature of the FI** form was further confirmed by probing its sensitivity to S1 nuclease. We observed that form FI** was almost completely degraded when incubated at 4°C overnight with 1 × 10−3 units of S1 nuclease whereas form FI* remained intact (Figure 3C). Degradation of FI* required a 10-fold higher amount of S1 nuclease (Figure 3C). The negatively supercoiled form (-SC) contains limited single-stranded regions allowing SI nuclease at high concentration to introduce a nick, leading to an increase in the OC form (Figure 3C). The differential S1 nuclease sensitivity between FI* and FI** indicates that FI** contains a larger single-stranded region than that present in FI*. The circular form of FI* and FI** was further investigated by incubating the unlinked DNA with T5 exonuclease. No degradation of the different DNA forms, including FI* or FI** DNA, was seen (Figure 3D), indicating that the FI* and FI** DNA bands do not possess free ends. FI** were circular single-stranded DNA molecules, a substrate completely degraded by S1 nuclease (Figure 3C) but resistant to T5 exonuclease (Figure 3D). In contrast, FI* might correspond to two circular single-stranded molecules partially annealed, a substrate less susceptible to S1 nuclease (Figure 3C). Thus, we conclude that SsoTopA extensively unlinks the two complementary DNA strands at temperatures ≥85°C, leading to two distinct circular single-stranded DNA (FI**). FI** can convert to FI* depending on the temperature.
SsoTopA catalyzes both the melting and annealing of DNA molecules
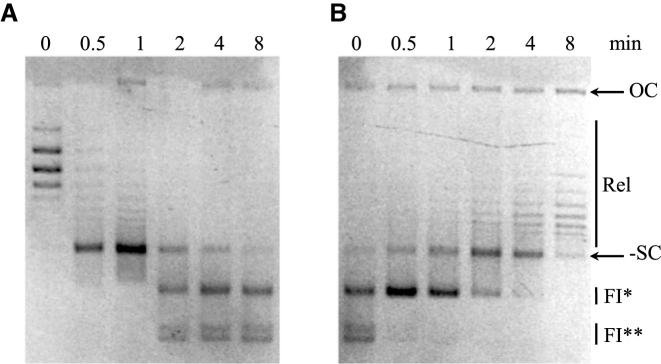
To further confirm the catalytic nature of the unlinking reaction performed by SsoTopA at high temperatures, we conducted a time course experiment at 95°C with a plasmid previously relaxed by SsoTopA at 75°C (Figure 4A, 0 min). We observed that the relaxed substrate was almost fully converted into the negatively supercoiled form within the first minute of incubation (Figure 4A, 1 min). The negatively supercoiled double-stranded DNA was subsequently converted to the two unlinked DNA forms FI* and FI** (Figure 4A, 2–8 min). After 8 min, the incubation time needed to convert all the double-stranded DNAs into forms FI* and FI**, the reaction mixture was transferred back to 75°C and the products monitored over time to assess the reversibility of the new species (Figure 4B). After just 30 s of incubation at 75°C, form FI** was almost completely converted into form FI* (Figure 4B, 0.5 min). With longer incubation times, form FI* was progressively converted into negatively supercoiled form before ultimately being relaxed (Figure 4B). To recover the covalently closed circular double-stranded DNA (cccDNA), the activity of SsoTopA was therefore required to restore the linking between two circular complementary DNA strands.
Figure 4.
Reversible activity of SsoTopA. (A) Kinetics of DNA melting catalyzed by SsoTopA. The pTZ18R substrate relaxed by SsoTopA (molar ratio of 2:1) at 75°C was further incubated at 95°C in the presence of TopA for the time indicated above each corresponding lane. (B) Kinetics of DNA re-annealing catalyzed by TopA. The reaction mix (SsoTopA and resulting products obtained after 8 min at 95°C, panel (A) was further incubated at 75°C. The incubation times are indicated above each corresponding lane. The reaction products were analyzed by one dimensional agarose gel electrophoresis: OC indicates open circular (nicked) DNA, Rel, the relaxed topoisomers and -SC the negatively supercoiled DNA. FI* and FI** are the unlinked DNA species.
We can conclude that (i) SsoTopA is quite thermostable as it remains active even after 8 min at 95°C, (ii) SsoTopA unlinks the strands of a circular covalently closed double-stranded DNA at higher temperatures, leading to single-stranded circular DNA at a temperature as high as 95°C, (iii) SsoTopA promotes strands annealing at lower temperature as previously described (32), (iv) SsoTopA is able to increase or decrease the linking number between the two DNA strands, demonstrating that SsoTopA is not able to discriminate between the two possible directions during strand passage. Most importantly, we conclude that the functional SsoTopA is able to catalyze the production of single-stranded DNA species as well as its reverse reaction (i.e. the re-annealing of two complementary circular single-stranded molecules back to double-stranded DNA species).
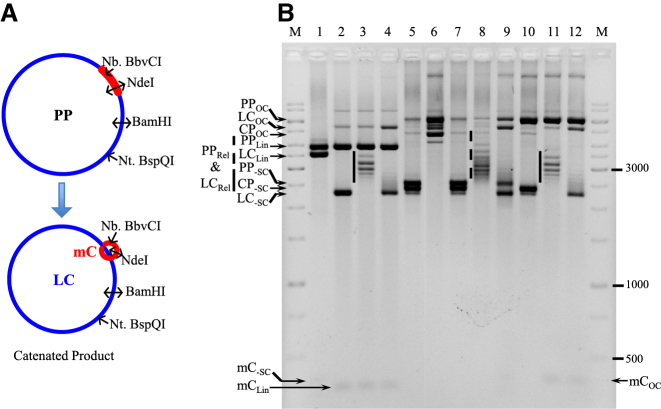
Sulfolobus solfataricus TopA decatenates two covalently closed circular double-stranded DNAs
To precisely analyze the decatenation reaction catalyzed by SsoTopA, we used multi-linked catenated double-stranded DNA substrate (35). Briefly, from a plasmid possessing λ-integrase recombination sites (PP, Figure 5A), it is possible to produce, after integrase-mediated recombination, an LC linked to a small DNA mC (Figure 5A). Following isolation of the catenane, three supercoiled DNA forms are present in the final product, corresponding to the parent plasmid (PP-SC), the catenane (CP-SC) and a trace amount of decatenated (unlinked) large circle (LC-SC) (Figure 5B, lanes 5 and 7). A faint band corresponding to the negatively supercoiled minicircle (mC-SC) was observed after linearization of the LC by BamH1 (LCLin), the parental plasmid being linearized as well (PPLin) (Figure 5B, lane 1).
Figure 5.
Decatenation of multi-linked DNA circles by SsoTopA. (A) The PP used to produce catenated molecules (CP) is schematically represented. The catenated DNA is composed of the LC (in blue color) and the mC (in red color) which are interlinked. The different enzymes used to characterize the different forms of these DNA are indicated. (B) The multi-linked DNA substrate (lanes 5 and 7) produced from the parent plasmid pMC339 (PP) includes two major species, the PP itself and the catenane (CP), and one minor species, the decatenated LC and mC. The multi-linked DNA substrate was incubated for 1 h at 37°C with BamHI (lane 1), NdeI (lanes 2–4), Nt.BspQI (lane 6) or Nb.BbvCI (lanes 10–12). Sso TopA was added in lanes 3, 4, 8, 9, 11 and 12 and incubated for 20 min at 80°C (lanes 3, 8 and 11) or 90°C (lanes 4, 9 and 12) producing relaxed DNA. The vertical plain bars correspond to the large plasmid relaxed at 80°C by TopA and the vertical dashed line corresponds to a mix of large plasmid and catenane relaxed at 80°C by SsoTopA. The DNA was loaded onto a 1.7% agarose gel and ran at about 2.5 V cm−1 for 3 h. The different DNA bands were attributed according to their size and topological forms (OC, open circular; Lin, linear; Rel, relaxed; -SC, negatively supercoiled) as deduced from the control reactions. M corresponds to the GeneRuler 1 kb ladders (ThermoFisher).
To facilitate the identification of the various bands in both the substrate and the reaction products, controls were set up with enzymes that cleave or nick at known positions on the PP, corresponding to the recognition sites on either the large plasmid or the mC component of the catenane (Figure 5A). The NdeI restriction enzyme cleaves both DNA strands within the mC sequence (Figure 5A), linearizing both the PP and the DNA mC (PPLin and mCLin, respectively) whereas the LC released from the catenated form remains supercoiled (LC-SC) (Figure 5B, lane 2; Supplementary Figure S5, lane 3). The nicking endonuclease Nt.BspQI cleaves at a specific position on only one DNA strand within the DNA sequence outside of the mC sequence on the PP (see Figure 5A). The incubation of the PP with Nt.BspQI resulted in a single nick generating an open circular form (PPOC) (Figure 5B, lane 6). With the catenated form (CP-SC) as substrate for Nt.BspQI, the LC was nicked but the mC remained supercoiled and linked, resulting in an open circular catenated form (CPOC) (Figure 5B, lane 6). Additional residual electrophoretic bands were also observed in lane 6, they correspond to different noded catenanes (i.e. catenanes with different number of crossovers), but it is difficult to definitively identify each band. The nicking endonuclease Nb.BbvCI cleaves at a specific position on only one DNA strand within the DNA sequence corresponding to the mC (see Figure 5A). The incubation with Nb.BbvCI led to nicking of the PP producing an open circular form (PPOC) (Figure 5B, lane 10 and Supplementary Figure S5, lane 2). With the catenated form substrate, the mC was nicked by Nb.BbvCI but remained linked to the LC. However because of the small size of the mC, there was no significant mobility change of the corresponding catenane compared with that of intact CPSC (Figure 5B, lane 10).
The decatenation of the negatively supercoiled catenated form by SsoTopA was performed at two different temperatures, 80°C (Figure 5B, lanes 3, 8 and 11) and 90°C (Figure 5B, lanes 4, 9 and 12), the upper temperature helping us to assign unambiguously the different DNA forms obtained. At 80°C, the catenane was unlinked and the decatenated LC was subsequently relaxed by SsoTopA and corresponding topoisomers were visualized after linearization of the PP and mC by NdeI (Figure 5B, lanes 3, LCrel). When incubated with SsoTopA at 90°C, these relaxed forms migrated exactly at the same position as the negatively supercoiled LC (LC-SC) in the control experiment (Figure 5B, lane 4 compared with lane 2) as we showed previously for both plasmid and kDNA substrates (Figure 2B, 85–90°C and Figure 1B, above 80°C, respectively). Similar profiles were obtained after incubation of the products with Nb.BbvC1: SsoTopA led to the decatenation of the linked rings into an LC and an mC (Figure 5B, lanes 11 and 12 compared with lanes 3 and 4, respectively). While the PP, catenane and decatenated LC were clearly visualized, the released mC, because of its small size was difficult to detect, except when a larger amount of DNA was used and loaded on a higher resolution gel (Supplementary Figure S5).
Importantly, by using supercoiled catenated DNA circle (CP-SC), we demonstrated that SsoTopA decatenated the linked circles and produced topoisomers corresponding to a mixture of the relaxed forms of both the decatenated LC (LCRel) and the PP (PPRel). These topoisomers were clearly visible as individual bands at 80°C (Figure 5B, lane 8) but as a single band at 90°C (Figure 5B, lane 9), as previously mentioned for the corresponding controls (Figure 5B, lanes 4 and 12). We observed the complete disappearance of catenanes (CP-SC), which were unlinked to release the LC that migrated as the supercoiled form (LP-SC) as illustrated by the higher intensity of the corresponding band (Figure 5B, compare lanes 9 and 7). This complete disappearance of the catenated circles indicates that SsoTopA is able to decatenate two linked covalently closed circular DNAs at 80 or 90°C (Figure 5B, lanes 8 and 9). The released DNA mCs are visualized as a single band at 90°C that migrates closely to open circular mC (Supplementary Figure S5, lane 1). By incubating these DNA products with the nicking endonuclease Nb.BbvCI or the restriction enzyme NdeI after the SsoTopA decatenation step, the topological pattern of the mCs was simplified: the relaxed mCs were converted to either open circular or linear forms (Supplementary Figure S5, lanes 2 and 3, respectively). Hence, for the first time, by using a multi-linked catenated DNA substrate for which the main DNA species are easily attributed, we demonstrate unambiguously that SsoTopA decatenates circular covalently closed double-stranded DNAs.
SsoTopA works on single-stranded DNA
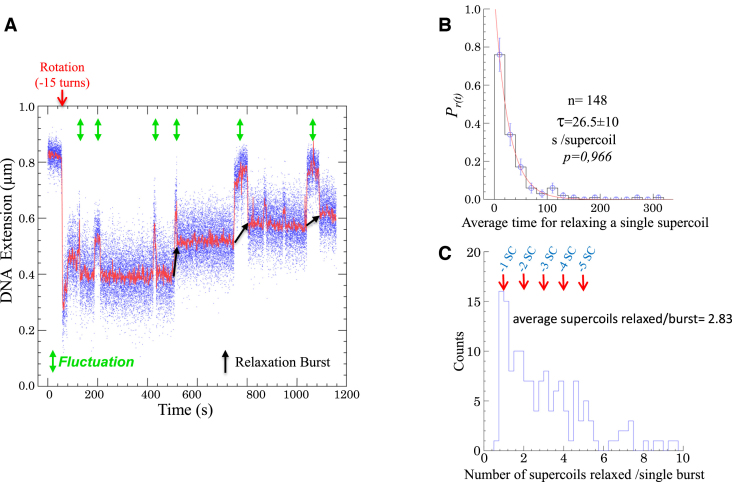
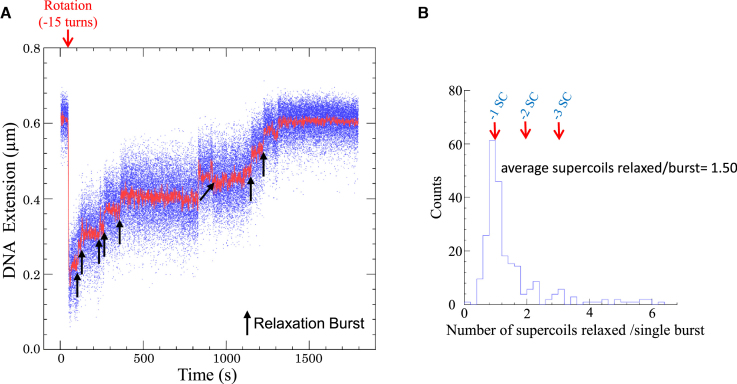
To characterize the mechanism catalyzed by SsoTopA, we analyzed the reaction using single-molecule manipulation. As shown for other thermophilic enzymes (43,44) including topoisomerases (45,46), SsoTopA is active at relatively low temperatures (31). This activity allowed us to perform single molecule assays at 45 or 35°C. In the absence of SsoTopA we observed a background of transient DNA extension fluctuations with a negatively supercoiled DNA substrate at 45°C with a magnetic tensile force of 0.45 pN (Supplementary Figure S6B). These fluctuations were not observed at a lower tensile force (e.g. 0.2 pN, not shown), suggesting that these transient extension changes might correspond to a reversible transition between a decrease of the plectoneme number and an introduction of a single-stranded DNA region (Supplementary Figure S6A) (47). In the absence of SsoTopA, these fluctuations were fairly short-lived and the DNA quickly reverted to its original extension (Supplementary Figure S6B). In the presence of SsoTopA, a transient extension change (corresponding approximately to one DNA helical repeat) was also observed and preceded each DNA relaxation event (Figure 6A). We hypothesized that SsoTopA binds and then operates on the transiently spontaneously formed single-stranded DNA region. This idea is supported by the faster relaxation velocity of SsoTopA on highly negatively supercoiled DNA than on partially relaxed forms in bulk experiment (Figure 1A). To test this hypothesis, we performed a similar experiment using DNA that contained a 10 bp mismatch bubble (Supplementary Figure S6C). The size of the bubble is similar to that of the transient melted region we observed (Figure 6A). At 45°C, the enzyme efficiency was dramatically increased with this DNA substrate, relative to the substrate lacking the single-stranded bubble (not shown). Therefore we lowered the enzyme concentration 100-fold and decreased the temperature down to 35°C to allow observation of individual DNA relaxation events (Figure 7A). In the presence of the permanent mismatch bubble, we did not observe the transient extension fluctuation prior to relaxation (Figure 7A and Supplementary Figure S6D). Moreover, SsoTopA relaxed the bubble-containing substrate at a higher rate than the fully base-paired substrate (compare Figures 7A and 6A). Indeed, with fully base-paired substrate the apparent SsoTopA rate is slow, 26.5 (±10) s to remove one negative supercoil (Figure 6B) compared to the maximum velocity of the T. maritima enzyme which is <1 s/supercoil (45). The apparent SsoTopA relaxation rate includes the time needed by the enzyme to bind a single-stranded DNA region. The measure of the enzyme velocity on double stranded DNA takes into account the single-stranded formation, the capture of this single-stranded DNA by SsoTopA, the strand passage reaction then the dissociation of the enzyme from DNA. In our experiments using DNA without bubble, this velocity is essentially dependent on the single-stranded formation frequency and in a lesser extent on the single strand capture ability. Moreover, we found that although most of the time SsoTopA relaxed only few supercoils per relaxation burst (Figure 6C) with an average of 2.83 supercoils/burst. The presence of a permanent bubble leads to a decrease of the SsoTopA processivity with an average of 1.50 supercoils relaxed/burst (Figure 7B). These results indicate that SsoTopA has very limited processivity contrary to the highly processive type I enzymes of T. maritima and Streptomyces coelicolor (45,48).
Figure 6.
Relaxation activity of SsoTopA on single molecule. (A) Time-trace of a DNA extension variation at 45°C of a 3 kb DNA extended by a force of 0.45 pN in the presence of 600 pM of SsoTopA. The time at which the negative supercoils are added to the DNA by rotating the magnet, is indicated by a red arrow. Extension length fluctuations are indicated by green arrows and the relaxation burst by black arrows. (B) Histograms of the average time in second per supercoil removed. The red line is the best fit (P-value = 0.966) using the single exponential equation 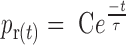 , where
, where 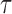 is the average time. A total of 148 relaxation events were used in this analysis. (C) Histograms of the burst processivity of SsoTopA. The number of supercoils removed was obtained from the extension variation using, for each bead, its calibration curve (39). The periodicity of one for the supercoil removal is indicated above the peaks. A total of 148 relaxation events were used in this analysis.
is the average time. A total of 148 relaxation events were used in this analysis. (C) Histograms of the burst processivity of SsoTopA. The number of supercoils removed was obtained from the extension variation using, for each bead, its calibration curve (39). The periodicity of one for the supercoil removal is indicated above the peaks. A total of 148 relaxation events were used in this analysis.
Figure 7.
Relaxation activity of SsoTopA on single molecule containing a permanent single-stranded DNA bubble. (A) Time-trace of the DNA extension variation at 35°C of the 2.2 kb DNA extended by a force of 0.42 pN and in the presence of 6 pM SsoTopA. (B) Histograms of the burst processivity of SsoTopA on a DNA containing a bubble. The number of supercoils removed was obtained from the extension variation using the calibration curve.
DISCUSSION
The type IA topoisomerases share a common structure and are able to perform multiple activities on a variety of DNA substrates. The efficiencies of these enzymes on their various substrates, however, are different. These differences allow to further subdivide the enzymes into two groups: the processive topo I enzymes efficiently remove negative supercoils at a high rate (45,48–50) whereas the topo III enzymes are distributive, relaxing negative supercoils at a much slower rate than the topo I enzymes (9,50). Unlike topoisomerase I, topoisomerase III is able to decatenate DNA, requiring at least one of the catenated circles to have a break in one strand (9,12,51). Moreover, decatenation reaction could occur with cccDNA in the presence of an excess of helicase (14,15). Finally, topoisomerase III is also much more efficient on DNA containing a single-stranded DNA region (51).
In the present report, we show that SsoTopA is efficient at relaxing and unlinking DNA, both activities requiring negatively supercoiled DNA. Indeed, like a topoisomerase III, SsoTopA decatenates linked DNA. Thus, we prove that SsoTopA is a topo III, as previously suggested by a phylogenetic analysis (31). kDNA is a network of catenated DNA circles, some of which naturally contain nicks or gaps. It was, thus, impossible to determine whether SsoTopA could decatenate cccDNA. Precisely topologically defined multi-linked catenanes were chosen to address this problem. With this substrate, we proved that SsoTopA decatenated two cccDNA. The decatenation reaction of cccDNA was an unexpected intrinsic property for a type I topoisomerase never reported before. This activity implies that two successive single-strand passage events should be performed on a single-stranded DNA region (cleavage, strand passage and resealing) to disentangle DNA.
Because SsoTopA is a type IA topoisomerase, its intrinsic activity must depend on single-stranded DNA regions for both relaxation and decatenation. Transient short-lived base-pair opening, even at temperatures below the DNA melting temperature, has been observed previously (47). Using a single molecule assay, we directly observed for the first time individual SsoTopA relaxation events. Indeed, SsoTopA uses the breathing of negatively supercoiled DNA to provide a transient single-stranded region to act upon to initiate relaxation. When a substrate engineered to contain a single-stranded DNA bubble region was used, the reaction rate increased drastically, verifying that single stranded DNA stimulates SsoTopA. We also observed that the stabilization of single-stranded regions by adding single strand binding proteins (SSBs) stimulates kDNA decatenation by SsoTopA (see Supplementary Figure S7), as observed previously for Saccharomyces cerevisiae topo III (14). A very recent report indicates that during DNA relaxation, E. coli topo I changes its conformation with a high frequency and successful strand passage events occur with a much lower frequency (52). Our single molecule experiments suggest that the rate-limiting step corresponds to the time required to trap single-stranded DNA. SsoTopA had previously been found to cleave and reseal single-stranded DNA oligonucleotides (32) or RNA molecules (18). Similarly, the Nanoarchaeum equitans hyperthermophilic enzyme, closely related to SsoTopA, is active on single-stranded DNA regions for dissolving hemicatenanes (22).
Unlinked DNA products were observed when the DNA substrate was incubated with SsoTopA at temperatures of ≥90°C. These DNA species have been reported during the initiation of pBR322 replication and referred to as FI* and FI** DNA (42). These two previously reported forms had similar increased electrophoretic mobility and S1 nuclease sensitivity as those we observed in the current study. For Desulfurococcus amyloliticus topo III such extensive unwinding was also observed but only after a long incubation time and only at very high temperatures (at least 90°C) (53). Unfortunately, the two highly unwound DNA forms observed in that study, also referred to as FI* and FI**, have not been fully characterized (53).
More recently, it has been shown that unwinding and unlinking of two complementary DNA strands can be performed by S. cerevisiae topo III in the presence of RPA or SSB, the eukaryotic and bacterial single-strand binding protein respectively, or Sgs1, the yeast SF2 helicase partner of topo III (14). Catenation of partially single-stranded DNA was also obtained by adding Rmi1, another partner of yeast topo III (14). It is likely that during the catenation/decatenation processes, topoisomerases III enzymes act on the single-stranded DNA region induced by these proteins. Indeed, all of these topo III partners are required for decatenation in numerous organisms and are essential to the maintenance of genome integrity (14,22,25). The unlinking feature is retained in the reaction catalyzed by SsoTopA except that no additional protein is required because single-stranded regions are, instead, favored by high temperature. Hence, depending on the temperature, SsoTopA is able either to promote or to prevent the unlinking of the two DNA strands. This observation implies that SsoTopA itself is not able to direct the strand passage and does not discriminate the direction of this strand passage as reported for E. coli ω protein (49,54), T. maritima topo I (45) or N. equitans topo III (22).
Recently, it was shown that yeast topo III partners with Sgs1 and Rmi1 or human topo IIIα partners with BLM and Rmi1 to form the dissolvasome which can resolve hemicatenanes (14,55). This complex decatenates kDNA, just as we observed for SsoTopA. Moreover, N. equitans, topo III is able to form or unlink hemicatenanes on its own (22) but it cannot be excluded that NeqTopo III possesses a true decatenation activity as its orthologous SsoTopA. Indeed, the decatenation activity that we observed for SsoTopA, could be a general property extended to all the topoisomerases III enzymes, either alone or in association with a SF2 helicase and proteins containing oligonucleotide-binding domain (OB fold).
SsoTopA is a topo III able to manipulate a wide variety of substrates, at least in vitro. It is obvious that single-stranded DNA regions required for the enzyme activity can be spontaneously formed in vivo due to the wide temperature range at which Sulfolobus cells divide (46,56). Considering that SsoTopA is fully competent to decatenate DNA, SsoTopA is licensed for, at least, the decatenation of hemicatenanes generated during replication, as reported for its homologue from N. equitans (22). Therefore, SsoTopA could be a relevant candidate to participate in chromosome segregation in Sulfolobus. This possibility is strengthened by the phenotype of the topA mutant obtained in S. islandicus (slow growth and defects in cell-cycle control), which strongly suggests that TopA plays a major role in resolving DNA entanglements during the cell cycle and in particular during the chromosome segregation step (57). Alternatively, TopoVI, the unique type II topoisomerase of Sulfolobus cells, may perform this action.
SsoTopA can perform a closely related decatenation reaction on knotted RNA suggesting another potential role of the enzyme in DNA transcription by acting on R-loops, as recently proposed (18). Finally, as demonstrated in this work, SsoTopA alone is unable to influence the direction of DNA strand transfer. Proteins known to associate with SsoTopA, such as Hel112 or SSB, could promote or limit the choice between the different possible reactions, as previously suggested (33). The action of other proteins could modulate SsoTopA activity, even without a direct interaction, but only by acting on DNA as proposed for topo III enzymes from other organisms (14,22). Thus, during the segregation process, the proteins involved in the chromosome tracking would allow SsoTopA to modulate the direction of strand transfer. To date, the importance of the different activities assumed by SsoTopA in DNA metabolism processes remains to be more fully characterized in vivo. Comparison of these activities catalyzed by the topoisomerase III enzymes throughout the three domains of life and organisms living in different conditions, especially those living at high temperature, will help us to understand their importance in the genomic stability and evolution.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Shuang Wang for providing us the DNA substrate containing a single-stranded DNA bubble used in the single molecule experiments and Jean-François Riou for helpful discussions.
Footnotes
Present address: Anna H. Bizard, University of Copenhagen, Institute of Cellular and Molecular Medicine (ICMM), Center for Chromosome Stability (CCS) and Center fo Healthy Ageing (CEHA), Blegdamsvej 3B, DK-2200 København N, Danmark.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique; National Institutes of Health [R56 AI054830, R01 GM115501 to L.Z., in part]. Funding for open access charge: Laboratory Grants.
Conflict of interest statement. None declared.
REFERENCES
- 1. Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001; 70:369–413. [DOI] [PubMed] [Google Scholar]
- 2. Wang J.C. Cellular roles of dna topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002; 3:430–440. [DOI] [PubMed] [Google Scholar]
- 3. Duguet M., Serre M.-C., Bouthier de La Tour C.. A universal type IA topoisomerase fold. J. Mol. Biol. 2006; 359:805–812. [DOI] [PubMed] [Google Scholar]
- 4. Viard T., Bouthier de La Tour C.. Type IA topoisomerases: a simple puzzle. Biochimie. 2007; 89:456–467. [DOI] [PubMed] [Google Scholar]
- 5. Tse-Dinh Y.C. Bacterial and archeal type I topoisomerases. Biochim. Biophys. Acta. 1998; 1400:19–27. [DOI] [PubMed] [Google Scholar]
- 6. Viard T., Lamour V., Duguet M., Bouthier de la Tour C.. Hyperthermophilic topoisomerase I from Thermotoga maritima. A very efficient enzyme that functions independently of zinc binding. J. Biol. Chem. 2001; 276:46495–46503. [DOI] [PubMed] [Google Scholar]
- 7. Wang J.C. Interaction between DNA and an Escherichia coli protein omega. J. Mol. Biol. 1971; 55:523–533. [DOI] [PubMed] [Google Scholar]
- 8. Zechiedrich E.L., Khodursky A.B., Bachellier S., Schneider R., Chen D., Lilley D.M., Cozzarelli N.R.. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2000; 275:8103–8113. [DOI] [PubMed] [Google Scholar]
- 9. DiGate R.J., Marians K.J.. Identification of a potent decatenating enzyme from Escherichia coli. J. Biol. Chem. 1988; 263:13366–13373. [PubMed] [Google Scholar]
- 10. DiGate R.J., Marians K.J.. Molecular cloning and DNA sequence analysis of Escherichia coli topB, the gene encoding topoisomerase III. J. Biol. Chem. 1989; 264:17924–17930. [PubMed] [Google Scholar]
- 11. Wallis J.W., Chrebet G., Brodsky G., Rolfe M., Rothstein R.. A hyper-recombination mutation in S-Cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989; 58:409–419. [DOI] [PubMed] [Google Scholar]
- 12. Hiasa H., DiGate R.J., Marians K.J.. Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J. Biol. Chem. 1994; 269:2093–2099. [PubMed] [Google Scholar]
- 13. Nurse P., Levine C., Hassing H., Marians K.J.. Topoisomerase III can serve as the cellular decatenase in Escherichia coli. J. Biol. Chem. 2003; 278:8653–8660. [DOI] [PubMed] [Google Scholar]
- 14. Cejka P., Plank J.L., Dombrowski C.C., Kowalczykowski S.C.. Decatenation of DNA by the S. cerevisiae Sgs1-Top3-Rmi1 and RPA complex: a mechanism for disentangling chromosomes. Mol. Cell. 2012; 47:886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harmon F.G., DiGate R.J., Kowalczykowski S.C.. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol. Cell. 1999; 3:611–620. [DOI] [PubMed] [Google Scholar]
- 16. Wang H., Di Gate R.J., Seeman N.C.. An RNA topoisomerase. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:9477–9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu D., Shen W., Guo R., Xue Y., Peng W., Sima J., Yang J., Sharov A., Srikantan S., Yang J. et al. Top3β is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 2013; 16:1238–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ahmad M., Xue Y., Lee S.K., Martindale J.L., Shen W., Li W., Zou S., Ciaramella M., Debat H., Nadal M. et al. RNA topoisomerase is prevalent in all domains of life and associates with polyribosomes in animals. Nucleic Acids Res. 2016; 44:6335–6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu L., Hickson I.D.. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003; 426:870–874. [DOI] [PubMed] [Google Scholar]
- 20. Plank J.L., Hsieh T.-S.. A novel, topologically constrained DNA molecule containing a double Holliday junction: design, synthesis, and initial biochemical characterization. J. Biol. Chem. 2006; 281:17510–17516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang J., Bachrati C.Z., Ou J., Hickson I.D., Brown G.W.. Human topoisomerase IIIalpha is a single-stranded DNA decatenase that is stimulated by BLM and RMI1. J. Biol. Chem. 2010; 285:21426–21436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee S.-H., Siaw G.E.-L., Willcox S., Griffith J.D., Hsieh T.-S.. Synthesis and dissolution of hemicatenanes by type IA DNA topoisomerases. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E3587–E3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fasching C.L., Cejka P., Kowalczykowski S.C., Heyer W.-D.. Top3-Rmi1 dissolve Rad51-mediated D Loops by a topoisomerase-based mechanism. Mol. Cell. 2015; 57:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Usongo V., Drolet M.. Roles of type 1A topoisomerases in genome maintenance in Escherichia coli. PLoS Genet. 2014; 10:e1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bizard A.H., Hickson I.D.. The dissolution of double Holliday junctions. Cold Spring Harb. Perspect. Biol. 2014; 6:a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bocquet N., Bizard A.H., Abdulrahman W., Larsen N.B., Faty M., Cavadini S., Bunker R.D., Kowalczykowski S.C., Cejka P., Hickson I.D. et al. Structural and mechanistic insight into Holliday-junction dissolution by Topoisomerase IIIα and RMI1. Nat. Struct. Mol. Biol. 2014; 21:261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Forterre P. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 2002; 18:236–237. [DOI] [PubMed] [Google Scholar]
- 28. Nadal M. Reverse gyrase: an insight into the role of DNA-topoisomerases. Biochimie. 2007; 89:447–455. [DOI] [PubMed] [Google Scholar]
- 29. Jaxel C., Bouthier de la Tour C., Duguet M., Nadal M.. Reverse gyrase gene from Sulfolobus shibatae B12: gene structure, transcription unit and comparative sequence analysis of the two domains. Nucleic Acids Res. 1996; 24:4668–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. She Q.Q., Singh R.K.R., Confalonieri F.F., Zivanovic Y.Y., Allard G.G., Awayez M.J.M., Chan-Weiher C.C.C., Clausen I.G.I., Curtis B.A.B., De Moors A.A. et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dai P., Wang Y., Ye R., Chen L., Huang L.. DNA topoisomerase III from the hyperthermophilic archaeon Sulfolobus solfataricus with specific DNA cleavage activity. J. Bacteriol. 2003; 185:5500–5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen L., Huang L.. Oligonucleotide cleavage and rejoining by topoisomerase III from the hyperthermophilic archaeon Sulfolobus solfataricus: temperature dependence and strand annealing-promoted DNA religation. Mol. Microbiol. 2006; 60:783–794. [DOI] [PubMed] [Google Scholar]
- 33. Valenti A., De Felice M., Perugino G., Bizard A., Nadal M., Rossi M., Ciaramella M.. Synergic and opposing activities of thermophilic RecQ-like helicase and topoisomerase 3 proteins in holliday junction processing and replication fork stabilization. J. Biol. Chem. 2012; 287:30282–30295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bizard A., Garnier F., Nadal M.. TopR2, the second reverse gyrase of sulfolobus solfataricus, exhibits unusual properties. J. Mol. Biol. 2011; 408:839–849. [DOI] [PubMed] [Google Scholar]
- 35. Fogg J.M., Kolmakova N., Rees I., Magonov S., Hansma H., Perona J.J., Zechiedrich E.L.. Exploring writhe in supercoiled minicircle DNA. J. Phys. Condens. Matter. 2006; 18:S145–S159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zechiedrich E.L., Khodursky A.B., Cozzarelli N.R.. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev. 1997; 11:2580–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Revyakin A., Liu C., Ebright R.H., Strick T.R.. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006; 314:1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Duboc C., Fan J., Graves E.T., Strick T.R.. Preparation of DNA substrates and functionalized glass surfaces for correlative nanomanipulation and colocalization (NanoCOSM) of single molecules. Methods Enzymol. 2017; 582:275–296. [DOI] [PubMed] [Google Scholar]
- 39. Strick T.R., Allemand J.-F., Bensimon D., Bensimon A., Croquette V.. The elasticity of a single supercoiled DNA molecule. Science. 1996; 271:1835–1837. [DOI] [PubMed] [Google Scholar]
- 40. Lionnet T., Allemand J.-F., Revyakin A., Strick T.R., Saleh O.A., Bensimon D., Croquette V.. Single-molecule studies using magnetic traps. Cold Spring Harb. Protoc. 2012; 2012:34–49. [DOI] [PubMed] [Google Scholar]
- 41. Fan J., Leroux-Coyau M., Savery N.J., Strick T.R.. Reconstruction of bacterial transcription-coupled repair at single-molecule resolution. Nature. 2016; 536:234–237. [DOI] [PubMed] [Google Scholar]
- 42. Parada C.A., Marians K.J.. Transcriptional activation of pBR322 DNA can lead to duplex DNA unwinding catalyzed by the Escherichia coli preprimosome. J. Biol. Chem. 1989; 264:15120–15129. [PubMed] [Google Scholar]
- 43. Lin H.-K.H., Chase S.F.S., Laue T.M.T., Jen-Jacobson L.L., Trakselis M.A.M.. Differential temperature-dependent multimeric assemblies of replication and repair polymerases on DNA increase processivity. Biochemistry. 2012; 51:7367–7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Choi J.-Y., Eoff R.L., Pence M.G., Wang J., Martin M.V., Kim E.-J., Folkmann L.M., Guengerich F.P.. Roles of the four DNA polymerases of the crenarchaeon Sulfolobus solfataricus and accessory proteins in DNA replication. J. Biol. Chem. 2011; 286:31180–31193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dekker N.H., Viard T., Bouthier de la Tour C., Duguet M., Bensimon D., Croquette V.. Thermophilic topoisomerase I on a single DNA molecule. J. Mol. Biol. 2003; 329:271–282. [DOI] [PubMed] [Google Scholar]
- 46. Couturier M., Bizard A.H., Garnier F., Nadal M.. Insight into the cellular involvement of the two reverse gyrases from the hyperthermophilic archaeon Sulfolobus solfataricus. BMC Mol. Biol. 2014; 15:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Phelps C., Lee W., Jose D., Hippel von P.H., Marcus A.H.. Single-molecule FRET and linear dichroism studies of DNA breathing and helicase binding at replication fork junctions. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:17320–17325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Szafran M.J., Strick T., Strzałka A., Zakrzewska-Czerwińska J., Jakimowicz D.. A highly processive topoisomerase I: studies at the single-molecule level. Nucleic Acids Res. 2014; 42:7935–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dekker N.H., Rybenkov V.V., Duguet M., Crisona N.J., Cozzarelli N.R., Bensimon D., Croquette V.. The mechanism of type IA topoisomerases. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:12126–12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Terekhova K., Gunn K.H., Marko J.F., Mondragón A.. Bacterial topoisomerase I and topoisomerase III relax supercoiled DNA via distinct pathways. Nucleic Acids Res. 2012; 40:10432–10440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Terekhova K., Marko J.F., Mondragón A.. Single-molecule analysis uncovers the difference between the kinetics of DNA decatenation by bacterial topoisomerases I and III. Nucleic Acids Res. 2014; 42:11657–11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gunn K.H., Marko J.F., Mondragón A.. An orthogonal single-molecule experiment reveals multiple-attempt dynamics of type IA topoisomerases. Nat. Struct. Mol. Biol. 2017; 24:484–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Slesarev A.I., Zaitzev D.A., Kopylov V.M., Stetter K.O., Kozyavkin S.A.. DNA topoisomerase III from extremely thermophilic archaebacteria. ATP-independent type I topoisomerase from Desulfurococcus amylolyticus drives extensive unwinding of closed circular DNA at high temperature. J. Biol. Chem. 1991; 266:12321–12328. [PubMed] [Google Scholar]
- 54. Kirkegaard K., Wang J.C.. Bacterial-DNA topoisomerase-i can relax positively supercoiled DNA containing a single-stranded loop. J. Mol. Biol. 1985; 185:625–637. [DOI] [PubMed] [Google Scholar]
- 55. Yang J., Bachrati C.Z., Hickson I.D., Brown G.W.. BLM and RMI1 alleviate RPA inhibition of TopoIIIα decatenase activity. PLoS One. 2012; 7:e41208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hjort K., Bernander R.. Changes in cell size and DNA content in sulfolobus cultures during dilution and temperature shift experiments. J. Bacteriol. 1999; 181:5669–5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li X., Guo L., Deng L., Feng D., Ren Y., Chu Y., She Q., Huang L.. Deletion of the topoisomerase III gene in the hyperthermophilic archaeon Sulfolobus islandicus results in slow growth and defects in cell cycle control. J. Genet. Genomics. 2011; 38:253–259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.