Abstract
Controlling DNA nanostructure interaction with protein is essential in developing nanodevices with programmable function, reactivity, and stability for biological and medical applications. Here, we show that the sequence-specific action of restriction endonucleases towards sharp triangular or rectangular DNA origami exhibits a novel, binary ‘on/off’ behaviour, as canonical recognition sites are either essentially fully reactive, or strongly resistant to enzymatic cutting. Moreover, introduction of structural defects in the sharp triangle can activate an otherwise unreactive site, with a site-to-defect distance of ∼50 nm. We argue that site reactivity is dependent upon programmable, mechanical coupling in the two-dimensional DNA origami, with specific structural elements, including DNA nicks and branches proximal to the sites that can function as negative(anti) determinants of reactivity. Empirically modelling the constraints to DNA degrees of freedom associated with each recognition site in the sharp triangle can rationalize the pattern of suppressed reactivity towards nine restriction endonucleases, in substantial agreement with the experimental results. These results provide a basis for a predictive understanding of structure-reactivity correlates of specific DNA nanostructures, which will allow a better understanding of the behaviour of nucleic acids under nanoscale confinement, as well as in the rational design of functional nanodevices based on self-assembling nucleic acids.
INTRODUCTION
Structural DNA nanotechnology has achieved an unprecedented level of precision in the geometric positioning of diverse biomolecules on physical supports by using DNA hybridization as a programmable scaffolding process (1–8). The nanoscale spatial proximity of complementary, reactive biomolecules can overcome limits imposed by diffusion and chemical equilibrium (9–12), and in turn can support reactions that are otherwise unachievable in solution (13). Such an approach has spurred innovative directions in biophysics (14–16), optics (17,18), synthetic biology (11,19,20) and nanomedicine (21–26).
While DNA nanostructures clearly provide a central function as scaffolds, their potential to interact with other biomolecular components, using a defined physicochemical lexicon, has yet to be fully realised (27–29). As one aspect, it has been shown that soluble 2D and 3D DNA origami nanostructures exhibit a generally higher resistance to nucleolytic degradation than the corresponding linear sequences (25,30). This is a fundamental requirement for many if not most of the intended applications of DNA nanotechnology (31). DNA nanostructures can function as nanosensors for RNA detection in cell lysates (25), while drug delivery (23) and in vivo imaging (32) studies in the mouse showed that the half-life of a DNA nanostructure in the bloodstream can be up to 4-fold greater than that of the linear dsDNA counterpart. The enhanced bio-stability may reflect the high molecular packing density in the DNA nanostructures that significantly diminishes local accessibility of DNA (30), due to reduced electrostatic screening (25).
In this study, we assess the site-specific nucleolytic reactivity of specific DNA nanostructures, which reveals unanticipated structure-reactivity correlations. We provide evidence that variation of localized structural features of a 2D DNA origami nanostructure, but without alteration of basic molecular density, influences restriction enzyme (REase) action in an unprecedented ‘on/off’ fashion. An empirical model, incorporating structural elements that function as negative(anti) determinants (33) of restriction site reactivity, is used to rationalize the regulated interaction of an REase with DNA nanostructures. These results demonstrate nucleolytic reaction control, encompassing unreactive and fully reactive states of specific sites, through programmable changes in physical parameters that reflect the confinement and local features unique to DNA nanostructures.
MATERIALS AND METHODS
Preparation of DNA origami objects
DNA origami structures were designed using Cadnano V0.2 (34), with M13mp18 ssDNA (New England BioLabs, Ipswich, USA) used as scaffold. Oligodeoxynucleotide staples were purchased in cartridge-purified form from Biomers.net GmbH (Ulm, Germany). DNA origami objects were produced in one-pot reaction mixtures, containing staples (50 nM each) and scaffold DNA (10 nM) in a final volume of 50 μl in 40 mM Tris–HCl, 12.5 mM MgCl2 (pH 8.5). Self-assembly was initiated by heating the solution at 95°C for 3 min, then decreasing the temperature from 95°C to 20°C at a rate of 1°C/min in a thermal cycler (Bio-rad, Hercules, CA, USA). The DNA origami objects were purified from excess staples using a PEG-based method (35). Briefly, the solution with DNA origami objects at a concentration of 10 nM was mixed 1:1 (v/v) with a precipitation solution containing 15% PEG8000 (w/v), 5 mM Tris, 1 mM EDTA and 500 mM NaCl at pH 8. The solution was mixed by inversion then centrifuged at 16 000 × g for 25 min at 4°C in tabletop microcentrifuge (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The supernatant was discarded and the DNA origami pellet resuspended in a 40 mM Tris (pH 8.5) solution to a volume equivalent to the initial volume of DNA origami solution, then incubated for ∼15 h at room temperature. The DNA origami objects were stored for maximum 1 day at 4°C before being used in experiments. If not otherwise stated, all reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Restriction endonuclease reactions
Reactions were prepared that contained either purified DNA origami, or M13mp18 dsDNA plasmid at a concentration of 5 nM, 10 units of the restriction enzyme in Tango Buffer for AluI, BsuRI, DraI, Hin1II, HhaI, MspI and RsaI (ThermoFisher Scientific, Waltham, Massachusetts, USA) or in CutSmart Buffer for BamHI, HaeIII, NlaIII and HpyCH4IV enzymes (New England Biolabs). The reaction was incubated at 37°C for 1 h, or for the indicated times in time-course experiments in a water bath (Julabo GmbH, Seelbach, Germany) and was stopped using a 20 min incubation at –80°C.
Gel electrophoresis
TriTrack DNA Loading Dye (Thermo Fisher Scientific, Waltham, Massachusetts, USA) was added to the DNA origami samples, then subjected to a thermal shock by heating the solution for 3 min at 95°C, followed by cooling on ice. The GeneRuler 1 kb plus DNA ladder (Thermo Fisher Scientific, Waltham, Massachusetts, USA) was added to one gel lane. Approximately 200 ng of the DNA was electrophoresed in a 1% agarose gel in TAE buffer (pH 8) at 60 V for ∼3 h. Gels were stained with 0.5 μg/mL ethidium bromide solution for 15 min and rinsed in ddH2O for 15 min. Gel images were generated using the Molecular Imager Chemi Doc (Bio-Rad, Hercules, CA, USA). Images were analysed using ImageJ V2.0. The intensity value of each peak was determined by measuring the distance from the peak apex to the peak base.
RESULTS
Restriction endonuclease action on DNA origami sharp triangle and rectangle
REases remain a highly relevant as well as convenient class of nucleases in understanding mechanisms of recognition and catalysis in protein-DNA interactions (36), as their action leaves a highly specific physical mark on DNA molecules as a consequence of cleavage within or near a short DNA recognition sequence (down to four bases, ∼1.4 nm) (37). While little is known of protein–nanostructure interactions, previous studies have shown the dependence of REase action upon crowding conditions (38–40). In addition, due to their exquisite DNA sequence specificity, REases are routinely used in DNA manipulation and analysis, with applications in biotechnology (41) and synthetic biology (42). For this study, we selected a panel of eleven REases (BamHI, DraI, Hin1II, NlaIII, HaeIII, BsuRI, MspI, RsaI, Hpy4CH4, HhaI and AluI) with restriction sites represented in the bacteriophage M13mp18 DNA (M13 DNA) sequence (Supplementary Figure S1). REase action was examined using two representative and well-characterised DNA origami structures: the sharp triangle (1), and the ‘twist-corrected’ rectangle (43). The REase recognition site contents are identical for the two structures since they have identical scaffold sequences.
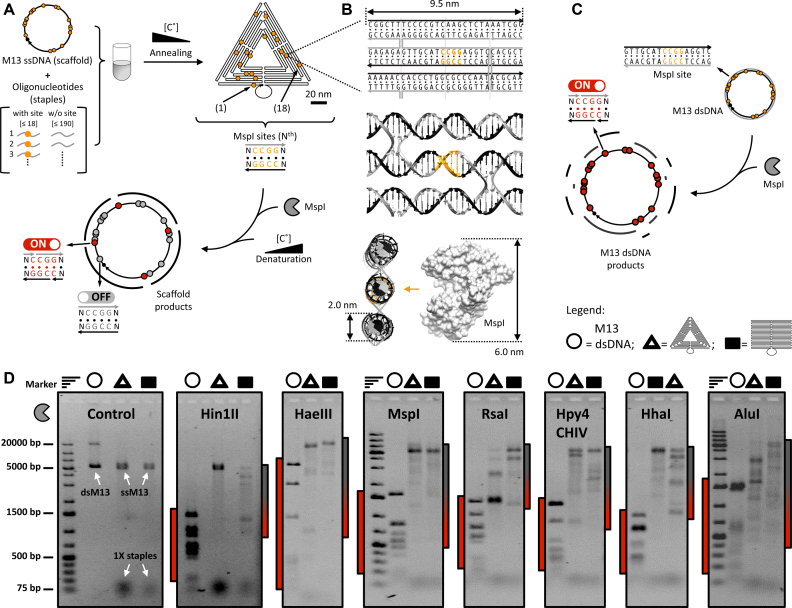
The diagram in Figure 1A (top) shows the location of restriction sites within the sharp triangle, which was assembled as described (1) using single-stranded(ss) M13 DNA as scaffold. Figure 1B shows a representative structural motif in the triangle that carries an REase recognition site. The nanostructures were purified from excess staples as described (35) (see Methods), to avoid interference from unincorporated staples. The purified structures were incubated with a selected REase, then disassembled by thermal treatment (see Figure 1A bottom), allowing gel electrophoretic separation of scaffold fragments generated by enzymatic cleavage(s). As a reference, double-stranded(ds) M13 DNA was reacted with the REases and the products fractionated by gel electrophoresis (Figure 1C). To ensure equivalent reaction conditions, the number of enzyme units per mole of restriction site was the same for all experiments. Figure 1D shows gel electropherograms of the fragments produced by seven different REases with 4-bp recognition specificity: Hin1II, HaeIII, MspI, RsaI, Hpy4CHIV, HhaI and AluI, on the circular M13 dsDNA, sharp triangle, and rectangle. Supplementary Figure S2 provides the results obtained with the 6 bp recognition site enzymes BamHI and DraI, as well as NlaIII and BsuRI that are isoschizomers of Hin1II and HaeIII.
Figure 1.
REase reactivity of the DNA sharp triangle and rectangle. (A) Schematic illustration of DNA origami sharp triangle formation and REase cleavage. (Top) the M13 ssDNA (‘scaffold’, solid-black line in the triangle), naturally provided with restriction sites (yellow dots indicate MspI sites), is hybridised to a pool of oligodeoxynucleotides (‘staples’) (Supplementary Figures S5 and S6). The spatial positions of the REase sites are staple-dependent (Supplementary Figure S7). (Bottom) after treatment with a REase (1 h at 37°C), the reaction products are separated by thermally denaturing the nanostructure, which releases ssDNA scaffold fragments delimited by the cleaved restriction sites, along with ssDNA staples (not shown). (B) (Top) diagram illustrating a restriction site (yellow) within a specific nanostructure element comprising several staple strands (grey arrow pointing to the 3′), and a scaffold strand (black arrow pointing to the 3′). (Centre) a double-helical model of the same nanostructure element (Supplementary Figure S18). (Bottom) the same double-helical diagram, rotated by 90° to illustrate the dimensional constraints encountered by MspI REase (53) acting on the nanoconfined site. (C) Schematic illustration of REase cleavage of the double-stranded form of M13 DNA. After REase treatment (1 hour at 37°C) the products are specific dsDNA fragments ending with cleaved restriction sites. (D) The products of (i) M13 dsDNA, (ii) sharp triangle, and (iii) rectangle reaction with several REases, as analysed by 1% agarose gel electrophoresis followed by ethidium fluorescence visualization. The control experiment lacks enzyme. For each REase, the bars flanking the gel indicate the molecular weight range of DNA fragments, and the presence of cleaved dsDNA in red (left) and partially cleaved nanostructures in grey/red (right). The gel bands with lowest molecular weight in each lane correspond to the DNA staples carried by the reacted DNA origami (absent for dsDNA). Experimental replicates are supplied in Supplementary Figure S2. Marker, 1 kb DNA molecular weight marker; dsM13, M13 dsDNA; ssM13, M13 ssDNA.
For both the sharp triangle and rectangle each REase produced a limited set of fragments that have a higher molecular weight than the products of cleavage of circular M13 dsDNA, the latter substrate exhibiting complete cleavage of all the sites (Supplementary Figure S3). The results indicate that for each REase, only a subset of restriction sites can be recognised and cleaved with high efficiency, albeit with some variation (e.g. in Figure 1D the gel bands produced by RsaI acting on the sharp triangle have different intensities). This is to be contrasted with the scenario in which the REases cleave each site with similar low efficiency, leaving the DNA nanostructure partially digested. In such a case, a REase would produce a large number of fragments, derived from all possible combinations of cleavage of the available sites. For instance, the stochastic action of Hin1II on its 15 sites would lead to 211 fragments of differing molecular weight, generated by ∼30 000 combinations of cutting (see Supplementary Figure S4). For each REase this behaviour would lead to a continuum of bands in the gel electropherograms, which is not observed.
Positional selectivity of HhaI REase action on DNA sharp triangle and DNA rectangle
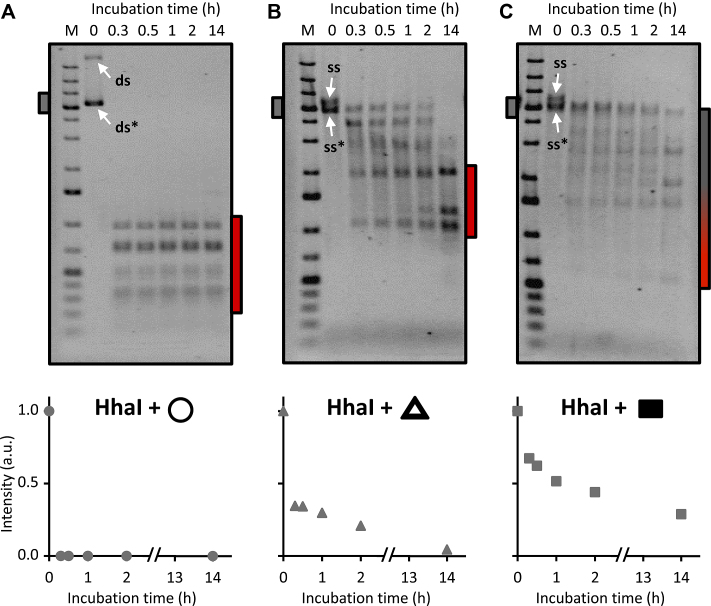
To characterise the structure-reactivity relationships underlying the results shown in Figure 1, and to gain a measure of relative reactivity we determined the extent of HhaI cleavage of the folded forms of M13 DNA at specific reaction times. Circular M13 dsDNA is completely cleaved within 0.3 h (Figure 2A), indicating that the conditions used, including the amount of HhaI were optimal for full reaction. For both the sharp triangle and rectangle there was a relatively rapid appearance of defined fragments that remained stable at extended reaction times (Figure 2B and C). After a 2 h incubation of the sharp triangle with enzyme, ∼20% of the DNA scaffold remained intact, while complete reaction was achieved by fourteen hours (Figure 2B). In contrast, the HhaI reaction with the rectangle (Figure 2C) does not apparently reach completion by fourteen hours, with ∼30% of the starting DNA scaffold amount unreacted (see Figure 2C plot). The number of large scaffold fragments produced in the gels shown in Figure 2B and C coincides with the number of cleaved HhaI sites (bottom-left diagram, Figure 1A). With the sharp triangle, three major fragments were generated as stable end products (Figure 2B), which shows that this structure possesses only three significantly reactive HhaI sites. For the DNA rectangle, despite the incomplete reaction, four large fragments are generated that correspond to a different set of significantly reactive HhaI sites (Figure 2C).
Figure 2.
Time-dependent action of HhaI REase towards the sharp triangle and rectangle. (top) Ethidium-bromide-stained 1% agarose gel analysis of HhaI enzymatic reactions on (from left to right) M13 dsDNA (A), sharp triangle (M13 ssDNA) (B) and rectangle (M13 ssDNA) (C), carried out between 0 and 14 h. The incubation time ‘0’ was an hour-long incubation lacking HhaI. For each DNA form, the graph below plots the relative intensity of the gel band, which corresponds to the residual, unmodified M13 DNA sequence (dsDNA, ds* in A, and ssDNA, ss* in B and C) with respect to reaction time. M, 1 kb DNA molecular weight marker; ds, relaxed form of M13 circular dsDNA; ds*, supercoiled form of M13 circular dsDNA; ss and ss*, respectively, circular and linear forms of M13 ssDNA. Likely, the ss* form (also visible in the control in Figure 1B) is the product of non-specific brekage induced by the thermal treatments depicted in Figure 1A.
The selective reactivity of the sharp triangle and rectangle is maintained at long incubation times. This indicates the existence of only a limited set of restriction sites that can be recognised and cleaved with high (albeit variable) efficiency as compared to the reactivity of the circular dsDNA. In summary, REase sites embedded in a DNA nanostructure can exhibit a qualitative binary (‘on/off’) reactivity. In addition, for the same REase the pattern of product fragments can differ between two different DNA structures having the same scaffold, indicating that specific local structures, reflective of structurally intertwined DNA, rather than DNA density per se, is a key determinant of REase recognition sequence reactivity. We show below that long-range effects also influence reactivity.
On/Off behaviour of HhaI recognition sites in the sharp triangle
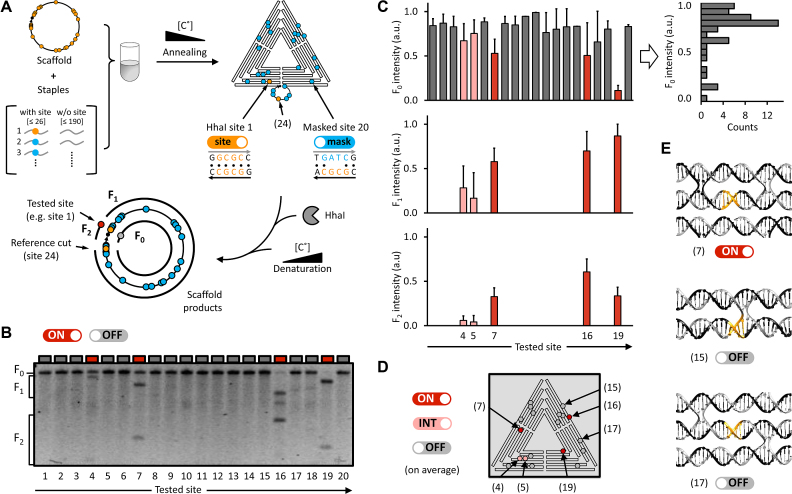
To provide a method for determining the reactivity of individual REase sites in a DNA nanostructure, we focused on the action of HhaI towards the sharp triangle. The approach, depicted in Figure 3A, involved the generation of a set of sharp triangle variants through separate self-assembly reactions (Figure 3A, right top), each of which contains only two reactive sites (hereafter termed ‘2 sites state’, or ‘2SS’), with one of them serving as a control site. The control site is in a short helical sequence protruding from the triangle (portrayed by the loop in the diagram in Figure 3A, bottom right). The remaining HhaI sites were masked by using staples that introduced base mismatches. Using this approach, the reactivity of each site in a DNA nanostructure can be separately assessed through comparison with the control site. Accordingly, a successful HhaI reaction with a site in a 2SS triangle variant should provide two distinct scaffold fragments as output (F1 and F2 in Figure 3A, bottom left). In the case of a site that is resistant to HhaI cutting, the reaction would provide as output a linearized form of the circular scaffold (F0 in Figure 3A, bottom left).
Figure 3.
HhaI REase reactivity of the sharp triangle at the single-site level. (A) (Left) strategy for assessing reactivity of individual restriction sites, as based on the experiments described in Figure 1 for the sharp triangle: twenty sharp triangle variants were generated by folding the M13 scaffold using variant-specific staple sets, and which contained only two HhaI sites of correct sequence (‘2 sites state’, or ‘2SS’). (Right) The HhaI recognition sequence (5′-GCGC-3′, in orange) in all staples except two was changed to 5′-GATC-3′ (in blue), thus masking the corresponding HhaI sites. Site 24 is one of the two unperturbed HhaI sites in each 2SS sharp triangle variant, and serves as the reference (control) site since it is external to the nanostructure, thereby ensuring constant reactivity across all variants. (Bottom) HhaI reaction of reactive 2SS triangle variants leads to two, variant-specific M13 fragments, termed F1 (longer), and F2 (shorter). In contrast, the HhaI-resistant 2SS variants produce linearized forms (F0) of the circular scaffold (as only the reference site is cleaved). (B) Agarose gel analysis of the HhaI reaction end products for all the 2SS triangle variants, illustrating the appearance of visible F1 and F2 fragments only for sites 4, 7, 16, and 19 (colour code: red is reactive, ‘on’, and grey is unreactive, ‘off’). (C) Data analysis with triplicate experiments (data in Supplementary Figure S12). The graphs show the average intensities of bands F0 (top), F1 (centre), and F2 (bottom), respectively, for each sharp triangle variant (1–20). (Right) The F0 intensity distribution is too dispersed to allow accurate detection of cleaved variants, while the distributions of F1 and F2 fragments unequivocally show an on/off behaviour in HhaI activity. Data are means ± S.D. (D) Shown is a sharp triangle diagram of the reactive HhaI sites according to the analysis in C. Site reactivity legend: ‘on’ (red), reactive sites (i.e. for which F1 and F2 occur in all experimental repeats); ‘off’ (grey), unreactive sites (i.e. for which only the F0 fragment occur in all experimental repeats); ‘int’ (pink), intermediate reactivity (for which the F1 and F2 pair occurs intermittently across the different experimental repeats). (E) Double-helical models for three representative HhaI sites. Sites 7 and 17 have similar motifs but exhibit opposite HhaI reactivity.
Each HhaI restriction site in the sharp triangle was identified and numbered (Figure 3A right top; and Supplementary Figures S7 and S8), with site 24 as the control site. Staples were identified that participate in each HhaI restriction site, and were mutated when the site was masked. The staple mutation changed the two central nucleotides in the HhaI recognition sequence (5′-GCGC-3′) to a 5′-GATC-3′ sequence, creating a mismatched duplex with the scaffold sequence, that is not recognised by HhaI (44,45) (Supplementary Figures S9 and S10). Atomic force microscopy (AFM) characterization of the modified 2SS triangles shows that the structures are properly formed (Supplementary Figure S11). The sharp triangle variant with all HhaI sites masked was essentially completely resistant to HhaI cleavage (see Supplementary Figure S12a).
Figure 3B displays the HhaI reactivity of the 2SS sharp triangle variants. Strikingly, among the variants, only the reactions with triangles carrying site 7, 16 or 19 provide highly visible product pairs (F1 and F2 fragments), thus indicating high HhaI reactivity of the sites. 2SS variant carrying site 4, instead, exhibits reduced reactivity. The molecular masses of the DNAs in each product pair are consistent with the predicted lengths of the F1 and F2 fragments (Supplementary Table S1), and are labelled with red rectangles in the respective gel lanes in Figure 3B. In contrast, for all the other 2SS sharp triangle variants, the reaction product corresponds to fragment F0 (see grey rectangles on top of the respective gel lanes in Figure 3B). The diagram in Figure 3D summarizes the results of three experimental repeats (Figure 3B, Supplementary Figure S12). We define three site reactivities according to the reproducibility of F1 and F2 fragments among the repeats. For reactive sites (‘on’), F1 and F2 occur in all experiments; for unreactive sites (‘off’), only the F0 fragment occur in all experiments; the remaining sites are defined to have ‘intermediate reactivity’ (‘int’), meaning that, the F1 and F2 pair occurs intermittently across the different experiments. The diagram in Figure 3D shows that reactive and unreactive HhaI sites are broadly distributed throughout the structure.
Of note, the DNA fragments associated with the three, fully reactive sites evidenced in Figure 3B match the DNA fragments produced by HhaI cleavage of the unmodified sharp triangle (Figure 1D, Supplementary Figure S12a). These results therefore indicate the absence of cross-talk between sites within the unmodified structure, meaning that the reactivity of each site is independent upon that of the others. A closer examination of the precise placement of the recognition sites (Supplementary Figure S7) reveals that sites 10, 11 and 15 directly overlap crossover junctions (see Figure 3E, middle). The major structural perturbation imposed by a crossover structure can rationalize why these sites are unreactive, most likely reflecting an inability of the enzyme to bind the sequence (46). In contrast, sites 7 (Figure 3E, top) and 17 (Figure 3E bottom) are contained within similar, regular double-helical segments, but their reactivities are completely opposite. Thus, the ‘on/off’ behaviour of HhaI restriction sites can reflect subtle structural differences. In addition, for the unmodified sharp triangle, a significant gel band corresponding to F0 is found (Figure 1D, Supplementary Figure S12), meaning incomplete REase digestion. Conversely, F0 is absent for the 2SS sharp triangle variants carrying sites 16 and 19 (Figure 3B, Supplementary Figure S12), meaning full REase digestion. These results therefore show that HhaI REase cleaves the unmodified sharp triangle less efficiently than the sharp triangle variants, thus suggesting absence of cooperativity in the enzymatic action over the reactive sites.
Effect of sharp triangle structural defects on HhaI REase action
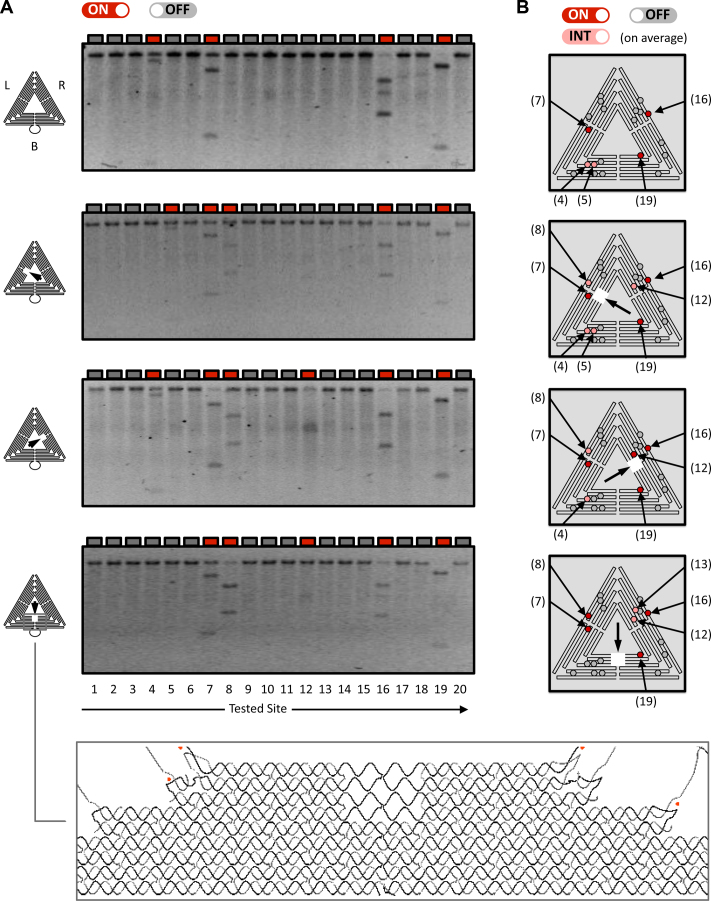
To further define the role of structure on REase site reactivity, three sets of sharp triangle variants were prepared, with each triangle exhibiting a large structural defect in one of the three trapezoidal substructures (Figure 4A left in column). The defect was created by omitting from the self-assembly reaction four staples that contribute to stabilizing the structure of the central area of a trapezoid, in effect creating a square ‘hole’ (Figure 4A inset, and Supplementary Figures S13–S15). The omitted staples do not directly participate in the formation of HhaI restriction sites, and it was anticipated that the large defect would introduce significant changes to the mechano-structural behaviour of the sharp triangle.
Figure 4.
Effect of sharp triangle structural defects on REase reactivity. (A) The single-site resolution analysis of HhaI reactivity (see also Figure 3A) was repeated using sharp triangle variants with a defect in one of the three trapezoidal components (see the square hole in each diagram on the left). Variants were generated by omitting four specific staples (see detailed diagram at the bottom). Agarose gel electrophoresis was used to analyse HhaI reaction products of the three triangle variants (L, R and B). Results for the unmodified triangle (Figure 2B) are shown on top for comparison. (B) Diagrams showing the reactive sites in the defective sharp triangles, according to the analysis shown in A, and based on duplicate experiments (Supplementary Figure S16). ‘on’ (red), ‘int’ (pink), and ‘off’ (grey) labels are defined by the legend in Figure 3D.
The 2SS sharp triangle variants were prepared and treated with HhaI, with Figure 4A showing the results for the unmodified triangle (top) and, in descending order, the results for the triangles with the defect localised in trapezoid L (left), R (right), or B (bottom). AFM characterization of the R variant shows that the structure is properly formed (Supplementary Figure S11). Figure 4b summarises the reactivity status (‘on/int/off') of the HhaI sites in the defective sharp triangles, as determined in two separate experiments (see also Supplementary Figure S16). The high reactivity of sites 7, 16 and 19 is unaffected by the defect (Figure 4A, top), while sites 8, 12 and 13 (see Supplementary Figure S16, top) exhibit significant reactivity only in the presence of the defect. Intriguingly, sites 8 and 12 become reactive irrespective of the placement of the defect, while sites 4, 5 and 13 change their reactivity only when the defect is in a separate trapezoid substructure, with sites 4 and 5 already displaying a moderate reactivity in the unmodified triangle (‘int’). In all cases, the activated site-to-defect distance ranges from 15 to 50 nm. In summary, these results suggest that a structural defect can increase the number of reactive REase sites, while maintaining the overall binary behaviour of HhaI action (Figure 3).
Parameterization of REase recognition site reactivity in a DNA nanostructure
The HhaI reactivities of the sharp triangle structural variants suggest a capacity of DNA nanostructures for allosteric control of nuclease site reactivity. Allostery would necessary reflect the mechano-structural properties, and the specific form of allostery would reflect the basic structural form, such as a sharp triangle, and would not necessarily function in the same manner, across other nanostructures.
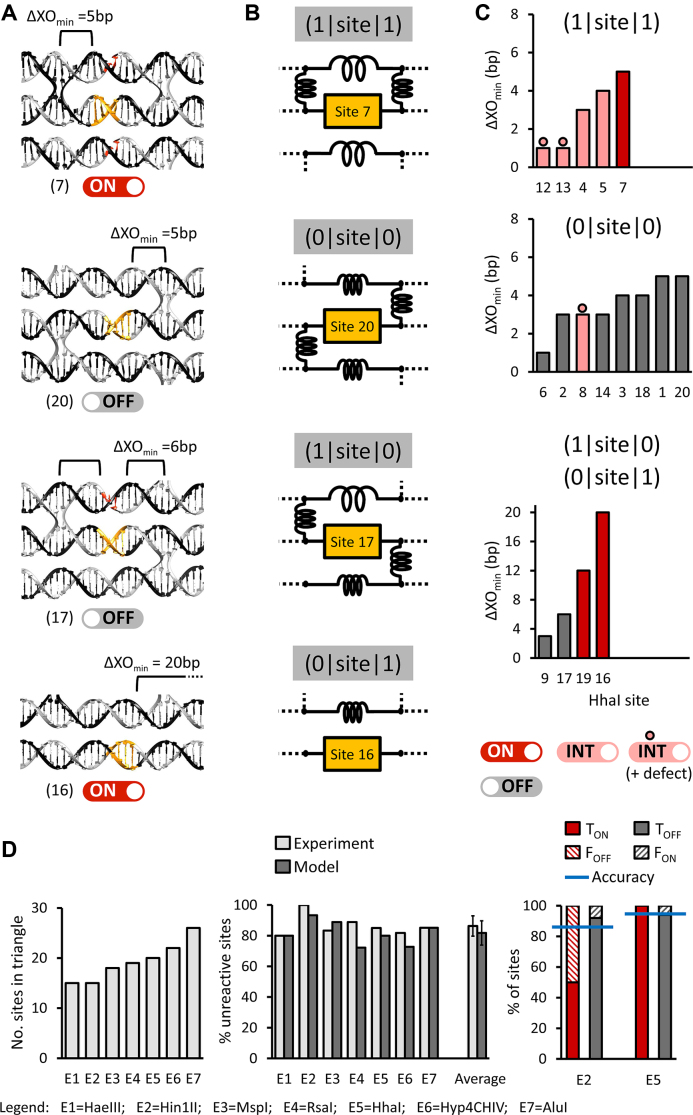
Predicting the mechanical properties of DNA nanostructures is challenging (47–51). For the sharp triangle the helices are predicted to exhibit an average helical rise of 10.67 bp per turn (1), whereas for the twist-corrected rectangle the average helical rise is of 10.44 bp per turn (43), with position-dependent values varying between 10.00 and 10.67 bp per turn (see Supplementary Figure S17). In both cases therefore the unnatural helical rise likely introduces conformational stress that is not uniformly distributed throughout the structure (30,52). It is therefore possible that helix conformational perturbations would contribute to restriction site reactivity. Structural features proximal to the restriction site also are expected to contribute to reactivity. Figure 5A displays sites 7, 20, 17 and 16 as representative sharp triangle sites. Here, each 16-bp double-helical segment flanking the restriction site is modelled as a spring (see corresponding diagrams in Figure 5B) that either is ‘relaxed’ (with a ‘mechanical index’ = 1) or ‘tight’ (with a mechanical index = 0), depending on whether the segment is nicked (see nucleotides labelled in red in Figure 5A diagrams) or is fully intact, respectively. In addition, a crossover connecting adjacent dsDNA segments is represented as a rigid spring orthogonal to the helical segments (Figure 5B). Using this mechanical analogy, the HhaI sites can be classified in three different structural states: (1|site|1), (1|site|0) or (0|site|1), and (0|site|0), with the digits as the mechanical indexes assigned to the flanking segments. If a site is located on a nanostructure edge (for instance, site 16; see Figure 5A, bottom), a mechanical index of ‘1’ is assigned to the (absent) flanking segment (see also the corresponding diagram in Figure 5B). Finally, the model incorporates the distance between the edge of a REase recognition site and the nearest crossover junction (ΔXOmin) as shown in Figure 5A. The graphs associate each HhaI site with an ΔXOmin value, and the inherent reactivity (Figure 5C). Pooling the results for the sharp triangle with and without large defects, the (1|site|1) and (1|site|0)/(0|site|1) configurations include nearly all HhaI sites that are reactive (either ‘on’or ‘int’), whether or not the structure contains a large defect. In contrast, the configuration (0|site|0) includes only the unreactive sites in the unmodified triangle, which (with the exception of site 8) also are unresponsive to the introduced defects. The graphs also show that, in the absence of a sharp triangle defect, HhaI highly efficiently cleaves sites with an ΔXOmin value of at least 5 bp (i.e. site 7 in Figure 5C, top and sites 16 and 19 in Figure 5C, bottom). When a defect is introduced in the sharp triangle, the ΔXOmin of the essentially reactive (1|site|1) sites is reduced to 1 bp (Figure 5C, top).
Figure 5.
Empirical modelling of sharp triangle REase reactivity. (A) Double-helical models for four HhaI sites (reactive sites 7 and 16, and unreactive sites 20 and 17) in the sharp triangle. For each site, ΔXOmin is defined as the minimum distance (in bp) between the REase recognition sequence and nearest crossover junction. (B) Site-specific diagrams relative to the adjacent double-helical models, with structural states defined as (a|site|b), and with ‘a’ and ‘b’ as ‘mechanical’ indexes. A ‘tight’ spring has a mechanical index = 0, while a ‘relaxed’ (or absent) spring has a mechanical index = 1. (C) For each mechanical state, ΔXOmin is plotted in ascending order as a function of HhaI site index (see Supplementary Figure S8). ‘on’ (red), ‘ int’ (pink), and ‘off’ (grey) labels are defined by the legend in Figure 3D. Dotted columns are for those sites whose REase activity is significantly influenced by the presence of a structural defect in the sharp triangle (Figure 4). The configuration (0|site|0) is found to correspond to unreactive sites only, except for site 8, while the configurations (1|site|1) and (1|site|0)/(0|site|1) include nearly all HhaI reactive sites. (D) (Left) number of recognition sites in the sharp triangle for seven REases (the E1 and E2 isoschizomers are not shown) with 4-bp specificities. (Centre) percentage of unreactive sites in the unmodified sharp triangle estimated experimentally (pale grey) and predicted (grey) by assuming that, for any REase, the reactive sites are either (1|site|1) or (1|site|0)/(0|site|1) configuration, and have ΔXO ≥ 5 bp. Experimental estimates are obtained by counting the number of M13 fragments produced by each REase (Figure 1B, Supplementary Table S3). On the left in the histogram, average percentages are calculated from the values on the right as means ± S.D. (Right) Site-specific analysis of model accuracy for Hin1II and HhaI. TON, ‘true ON rate’, fraction of reactive sites predicted ‘on’ (red); FOFF, ‘False OFF rate’, fraction of reactive sites prediction ‘off’ (pale red); TOFF, ‘True OFF rate’, fraction of unreactive sites predicted ‘off’ (grey); FON, ‘False ON rate’, fraction of unreactive sites predicted ‘on’ (pale grey). Model accuracy is calculated as (TON + TOFF) / (TON + TOFF + FON + FOFF) (blue line) (see Supplementary Figure S23).
Additional local structural features shown in Figure 5A were examined as possible determinants of site reactivity. These include (i) the relative angular orientation of a site with respect to neighbouring structures (data not shown), (ii) the presence of a nick (corresponding to juxtaposed staple termini), either near to or within the recognition sequence (Supplementary Figures S18 and S19), and (iii) the distance between flanking crossover junctions, having the same orientation (see also Supplementary Figures S18 and S20). While the contributions of (i) and (ii) to site reactivity would appear to be minimal, with respect to (iii), the majority of unreactive sites are in a 32 bp segment bounded by crossover junctions, which is the standard helical segment length in the sharp triangle (Supplementary Figures S18 and S20).
The similar behaviour of other REases with 4-bp recognition sequences (Hin1II, HaeIII, MspI, RsaI, Hpy4CHIV, HhaI and AluI; Figure 1D), prompted the question whether the mechanical model based on HhaI site reactivity could be used to predict the reactivities for other REase sites. The ΔXOmin value and the site configuration were determined for every site for the listed REases (Supplementary Table S2), providing a predicted percentage of reactive and unreactive sites. It was assumed that a site would be reactive if it belongs to either of two configurations: (1|site|1) or (1|site|0)/(0|site|1), and having a ΔXOmin ≥ 5 bp. This model, does not take into account the effect of structural defects of the type presented above. The model also does not take into account that different REases would most likely have different ΔXOmin and steric requirements that could influence site reactivity. Analysis of available REase crystal structures indicates that REases often interact with base pairs that flank the restriction site, and that the sizes and shapes of REases vary, creating different steric volumes (53,54).
The percentage of unreactive sites for each REase (142 sites in total, see Figure 5D, left) was qualitatively determined by gel electrophoretic analysis (Supplementary Table S3). Biases in the estimates include the possibility that the number of the unreactive sites may be an underestimate, due to the presence of uncharacterised, ‘background’ reaction products. On the other hand, the number of the unreactive sites may be an overestimate if there are two, very close and reactive sites on the scaffold, producing fragments that are not distinguishable with the resolution provided by the gel electrophoresis. The results of the analysis (see middle histogram of Figure 5D) reveal that an average of 86 ± 7% of the sites are unreactive, while the model predicts a slightly lower value of 82 ± 8%. We also evaluated the accuracy of the model in predicting REase site reactivity, on a site-by-site basis. To this purpose, in addition to HhaI, we examined the action of Hin1II towards 2SS triangle variants, each of which contains only one of the inherent 15 sites (CATG). The results (Supplementary Figures S21 and S22) show that there is only a single reactive Hin1II site, that has the configuration (1|site|1), and has a ΔXOmin of 4 bp, which is near but within the proposed threshold. All other sites with the configurations (1|site|1), (1|site|0) or (0|site|1), and (0|site|0) were unreactive. The histogram on the right in Figure 5D presents the fraction of sites that are experimentally determined to be reactive, with their prediction as either ‘on’ (TON) or ‘off’ (FOFF) (in red), or are unreactive, while predicted to be either ‘off’ (TOFF) or ‘on’ (FON) (in grey). 92% and 94% of the unreactive sites of Hin1II and HhaI, respectively, are correctly accounted for by the empirical model, while the accuracy is 86% for Hin1II and 95% for HhaI (see Supplementary Figure S23).
DISCUSSION
This study demonstrates the structure-dependent, programmable nucleolytic reactivity of model DNA nanostructures. We found that the sharp triangle and DNA rectangle exhibit a high degree of selective reactivity towards REases, compared to the corresponding linear M13 dsDNA, in substantial agreement with the results of others (25,30). Second, the REase sites in the sharp triangle are either essentially highly reactive (‘on’), thus demonstrating full accessibility, or are unreactive (‘off’), indicative of hindered recognition. The data also show that protein recognition of multiple sites in a DNA nanostructure can occur in an independent, non-interfering manner. Third, the presence of large defects in the trapezoids that form the sharp triangle can selectively activate otherwise unreactive sites, with a defect-to-site distance of up to 50 nm, demonstrating an involvement of long-range allostery in control of reactivity.
Recent studies examined the effect of DNA density on REase reactivity, using 15 nm thick ‘brushes’ of surface-immobilised dsDNA (39,40). It was shown that dsDNA surface density directly impacts REase access to, as well as diffusion within, the DNA brushes, indicating that REase reactivity is controlled primarily by steric accessibility at the DNA brush-solution interface (40). Since three-dimensional DNA nanostructures can exhibit complex solid-liquid interfaces, for which an understanding of the functional impact remains a significant challenge, the study of two-dimensional nanostructures provides a significant simplification. With a thickness equal to the diameter of a double helix (∼2 nm), a 2D nanostructure maximises the liquid interface contact area (48). This in turn implies that a two-dimensionally confined and twisted DNA structure should exhibit comparable accessibility to protein in solution as the corresponding unstructured (linear) form. However, the differential reactivity of restriction sites shown here, and the previously described behaviour of the surface-arrayed DNA brushes, indicates that a simple relationship between DNA density and REase action cannot fully explain the reactivity of soluble DNA nanostructures.
Keum and Bermudez investigated the site-specific action of DdeI REase on a DNA tetrahedron with 20 bp edges (55). The tetrahedron contained one DdeI site and several nicks, and was prepared in two variant forms, with the site located in two different positions along the same dsDNA edge. Applying an enzymatic ligation reaction prior to DdeI action caused an increased resistance to REase cleavage, and it was suggested that the overall rigidity of the tetrahedron increases following ligation, thus impairing DdeI binding, presumably reflecting specific features of the intact 3D structure.
Here, we argue that the predominant ‘on/off’ reactivity of the cleavage sites in the sharp triangle reflects constrained DNA degrees of freedom near the REase recognition sequences, in the absence of any constraints imposed by a three-dimensional structure. Such local, structural/mechanical constraints downregulate REase recognition and cleavage in an essentially digital fashion, and can therefore be regarded as antideterminants (33) of nucleolytic reactivity. The constraints include the distance between an REase recognition sequence border and the nearest DNA branch (crossover junction), and the presence of one or more nicks in the double-helical segments flanking a recognition sequence (Figure 5A). Intriguingly, the mechanical constraints can be relaxed as a consequence of distant structural defects in the nanostructure. Understanding the emergence of allostery, reflecting the type of defect and its distance from a recognition site will require further analysis. For instance, sites 4 and 5, which are partially reactive (‘int’) when the other sites are inactivated by bp mismatches, are localised in the region of the sharp triangle with the highest density of sites (Figure 3D). It is therefore possible that the presence of base mismatches in the nearby ‘masked’ sites can alter the structure and impair reactivity of a site.
While the empirical model was developed to specifically rationalize the action of a specific REase towards a specific DNA nanostructure, it is anticipated to be applicable to a wider range of REases and DNA nanostructures. The latter are formed and stabilized by similar hybridisation of multiple DNAs, forming branched motifs (e.g. crossover junctions) and nicks. For instance, the model explains the reactivity of a DNA tetrahedron towards DdeI REase (55). Here, the observed reduction in DdeI reactivity following nick ligation would change the REase site type from (1|site|1) (predicted as reactive) into a (0|site|0) (predicted as unreactive). A more complex model would be required, however, to determine the reactivity of multilayer, 3D DNA origami structures. Since the REase sites on the nanostructure surface will likely be more accessible to REases, additional antideterminants may include the position of the REase site with respect to the surface, and the nanostructure's global mechanical rigidity, which can be much higher than that of single-layer 2D DNA origami (30).
In addition, our empirical model largely reproduces the behaviour of a total of nine REases investigated in this study, and with an ascertained accuracy of ≥90% for two REases in particular (Figure 5D). Further refinement of the model should improve the accuracy.
This study opens the door to the opportunity of designing soluble DNA nanostructures that exhibit controlled enzymatic reactivity through the deliberate programming of physical parameters associated with nanoscale density and local structural features. The study also shows that a defined set of criteria could be applied to design functional DNA nanostructures. The unprecedented, even counterintuitive results presented here, however, also underscore the need for high-level computational modelling of DNA nanostructures in order to understand nucleic acid nanostructure structure-function correlates, as well as optimally design nanodevices with biotechnological and medical applications (49,51). Studies are underway to model and experimentally evaluate the enzymatic and non-enzymatic interactions of proteins with model nucleic acid nanostructures.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Prof. Allen W. Nicholson, Prof. Vincenzo Carnevale and Prof. Giacinto Scoles (Temple University, Philadelphia, PA), Prof. Paul W. K. Rothemund (Caltech, CA), Prof. Darko Stefanovic (University of New Mexico, NM), Dr Adrian Keller (Paderborn University, DE) and Dr Enrico Ferrari (University of Lincoln, UK) for their insight and contribution in data interpretation and manuscript revision. The experimental work was performed in the MONALISA Laboratories in the Department of Medical and Biological Sciences at the University of Udine, Udine, Italy, and the School of Food Science and Nutrition at the University of Leeds, Leeds, UK.
Footnotes
Present addresses:
Alex Stopar, Department of Biology, Temple University, Philadelphia, PA 19122-6078, USA.
Lucia Coral, Regional Referral Centre for Rare Diseases, Azienda Sanitaria Universitaria Integrata di Udine, 33100 Udine, Italy.
Stefano Di Giacomo, Institute of Life Sciences (ISV), Biochemistry, Biophysics and Genetics of Microorganisms (BBGM), Université catholique de Louvain, Louvain-La-Neuve 1348, Belgium.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council [ERC Ideas 2010 n.269051, Quidproquo]; Italian Minister of Health [Ricerca Finalizzata e Giovani Ricercatori n. WFR GR-2013-02356714]. Funding for open access charge: University of Leeds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Rothemund P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature. 2006; 440:297–302. [DOI] [PubMed] [Google Scholar]
- 2. Douglas S.M., Dietz H., Liedl T., Högberg B., Graf F., Shih W.M.. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009; 459:414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dietz H., Douglas S.M., Shih W.M.. Folding DNA into twisted and curved nanoscale shapes. Science. 2009; 325:725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Han D., Pal S., Yang Y., Jiang S., Nangreave J., Liu Y., Yan H.. DNA gridiron nanostructures based on four-arm Junctions. Science. 2013; 339:1412–1415. [DOI] [PubMed] [Google Scholar]
- 5. Benson E., Mohammed A., Gardell J., Masich S., Czeizler E., Orponen P., Högberg B.. DNA rendering of polyhedral meshes at the nanoscale. Nature. 2015; 523:441–444. [DOI] [PubMed] [Google Scholar]
- 6. Wei B., Dai M., Yin P.. Complex shapes self-assembled from single-stranded DNA tiles. Nature. 2012; 485:623–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ke Y., Ong L.L., Shih W.M., Yin P.. Three-dimensional structures self-assembled from DNA bricks. Science. 2012; 338:1177–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Veneziano R., Ratanalert S., Zhang K., Zhang F., Yan H., Chiu W., Bathe M.. Designer nanoscale DNA assemblies programmed from the top down. Science. 2016; 352:1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fu J., Liu M., Liu Y., Woodbury N.W., Yan H.. Interenzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures. J. Am. Chem. Soc. 2012; 134:5516–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yin P., Yan H., Daniell X.G., Turberfield A.J., Reif J.H.. A unidirectional DNA walker that moves autonomously along a track. Angew. Chem. Int. Ed. 2004; 43:4906–4911. [DOI] [PubMed] [Google Scholar]
- 11. Shaw A., Lundin V., Petrova E., Fördős F., Benson E., Al-Amin A., Herland A., Blokzijl A., Högberg B., Teixeira A.I.. Spatial control of membrane receptor function using ligand nanocalipers. Nat. Methods. 2014; 11:841–846. [DOI] [PubMed] [Google Scholar]
- 12. Zhao Z., Fu J., Dhakal S., Johnson-Buck A., Liu M., Zhang T., Woodbury N.W., Liu Y., Walter N.G., Yan H.. Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat. Commun. 2016; 7:10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gopinath A., Rothemund P.W.K.. Optimized assembly and covalent coupling of single-molecule DNA origami nanoarrays. ACS Nano. 2014; 8:12030–12040. [DOI] [PubMed] [Google Scholar]
- 14. Shrestha P., Jonchhe S., Emura T., Hidaka K., Endo M., Sugiyama H., Mao H.. Confined space facilitates G-quadruplex formation. Nat. Nanotechnol. 2017; 12:582–588. [DOI] [PubMed] [Google Scholar]
- 15. Stein I.H., Steinhauer C., Tinnefeld P.. Single-molecule four-color FRET visualizes energy-transfer paths on DNA origami. J. Am. Chem. Soc. 2011; 133:4193–4195. [DOI] [PubMed] [Google Scholar]
- 16. Funke J.J., Ketterer P., Lieleg C., Korber P., Dietz H.. Exploring nucleosome unwrapping using DNA origami. Nano Lett. 2016; 16:7891–7898. [DOI] [PubMed] [Google Scholar]
- 17. Kuzyk A., Schreiber R., Fan Z., Pardatscher G., Roller E.-M., Högele A., Simmel F.C., Govorov A.O., Liedl T.. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature. 2012; 483:311–314. [DOI] [PubMed] [Google Scholar]
- 18. Acuna G.P., Bucher M., Stein I.H., Steinhauer C., Kuzyk A., Holzmeister P., Schreiber R., Moroz A., Stefani F.D., Liedl T. et al. Distance dependence of single-fluorophore quenching by gold nanoparticles studied on DNA origami. ACS Nano. 2012; 6:3189–3195. [DOI] [PubMed] [Google Scholar]
- 19. Angelin A., Kassel O., Rastegar S., Strähle U., Niemeyer C.M.. Protein-functionalized DNA nanostructures as tools to control transcription in zebrafish embryos. ChemistryOpen. 2017; 6:33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Langecker M., Arnaut V., Martin T.G., List J., Renner S., Mayer M., Dietz H., Simmel F.C.. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science. 2012; 338:932–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiang Q., Song C., Nangreave J., Liu X., Lin L., Qiu D., Wang Z.-G., Zou G., Liang X., Yan H. et al. DNA origami as a carrier for circumvention of drug resistance. J. Am. Chem. Soc. 2012; 134:13396–13403. [DOI] [PubMed] [Google Scholar]
- 22. Liu X., Xu Y., Yu T., Clifford C., Liu Y., Yan H., Chang Y.. A DNA nanostructure platform for directed assembly of synthetic vaccines. Nano Lett. 2012; 12:4254–4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee H., Lytton-Jean A.K.R., Chen Y., Love K.T., Park A.I., Karagiannis E.D., Sehgal A., Querbes W., Zurenko C.S., Jayaraman M. et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012; 7:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Douglas S.M., Bachelet I., Church G.M.. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012; 335:831–834. [DOI] [PubMed] [Google Scholar]
- 25. Mei Q., Wei X., Su F., Liu Y., Youngbull C., Johnson R., Lindsay S., Yan H., Meldrum D.. Stability of DNA origami nanoarrays in cell lysate. Nano Lett. 2011; 11:1477–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang H., Chao J., Pan D., Liu H., Qiang Y., Liu K., Cui C., Chen J., Huang Q., Hu J. et al. DNA origami-based shape IDs for single-molecule nanomechanical genotyping. Nat. Commun. 2017; 8:14738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hernandez-Garcia A., Estrich N.A., Werten M.W.T., Van Der Maarel J.R.C., LaBean T.H., de Wolf F.A., Cohen Stuart M.A., de Vries R.. Precise coating of a wide range of DNA templates by a protein polymer with a DNA binding domain. ACS Nano. 2017; 11:144–152. [DOI] [PubMed] [Google Scholar]
- 28. Estrich N.A., Hernandez-Garcia A., de Vries R., LaBean T.H.. Engineered diblock polypeptides improve DNA and gold solubility during molecular assembly. ACS Nano. 2017; 11:831–842. [DOI] [PubMed] [Google Scholar]
- 29. Castronovo M., Stopar A., Coral L., Redhu S.K., Vidonis M., Kumar V., Del Ben F., Grassi M., Nicholson A.W.. Effects of nanoscale confinement on the functionality of nucleic acids: implications for nanomedicine. Curr. Med. Chem. 2013; 20:3539–3557. [DOI] [PubMed] [Google Scholar]
- 30. Castro C.E., Kilchherr F., Kim D.-N., Shiao E.L., Wauer T., Wortmann P., Bathe M., Dietz H.. A primer to scaffolded DNA origami. Nat. Methods. 2011; 8:221–229. [DOI] [PubMed] [Google Scholar]
- 31. Chen Y.-J., Groves B., Muscat R.A., Seelig G.. DNA nanotechnology from the test tube to the cell. Nat. Nanotechnol. 2015; 10:748–760. [DOI] [PubMed] [Google Scholar]
- 32. Jiang D., Sun Y., Li J., Li Q., Lv M., Zhu B., Tian T., Cheng D., Xia J., Zhang L. et al. Multiple-armed tetrahedral DNA nanostructures for tumor-targeting, dual-modality in vivo imaging. ACS Appl. Mater. Interfaces. 2016; 8:4378–4384. [DOI] [PubMed] [Google Scholar]
- 33. Zhang K., Nicholson A.W.. Regulation of ribonuclease III processing by double-helical sequence antideterminants. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:13437–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Douglas S.M., Marblestone A.H., Teerapittayanon S., Vazquez A., Church G.M., Shih W.M.. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009; 37:5001–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stahl E., Martin T.G., Praetorius F., Dietz H.. Facile and scalable preparation of pure and dense DNA origami solutions. Angew. Chem. 2014; 126:12949–12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dehé P.-M., Gaillard P.-H.L.. Control of structure-specific endonucleases to maintain genome stability. Nat. Rev. Mol. Cell Biol. 2017; 18:315–330. [DOI] [PubMed] [Google Scholar]
- 37. Loenen W.A.M., Dryden D.T.F., Raleigh E.A., Wilson G.G., Murray N.E.. Highlights of the DNA cutters: a short history of the restriction enzymes. Nucleic Acids Res. 2014; 42:3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rivas G., Minton A.P.. Macromolecular crowding in vitro, in vivo, and in between. Trends Biochem. Sci. 2016; 41:970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Castronovo M., Radovic S., Grunwald C., Casalis L., Morgante M., Scoles G.. Control of steric hindrance on restriction enzyme reactions with surface-bound DNA nanostructures. Nano Lett. 2008; 8:4140–4145. [DOI] [PubMed] [Google Scholar]
- 40. Castronovo M., Lucesoli A., Parisse P., Kurnikova A., Malhotra A., Grassi M., Grassi G., Scaggiante B., Casalis L., Scoles G.. Two-dimensional enzyme diffusion in laterally confined DNA monolayers. Nat. Commun. 2011; 2:210–297. [DOI] [PubMed] [Google Scholar]
- 41. Barrangou R., Doudna J.A.. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 2016; 34:933–941. [DOI] [PubMed] [Google Scholar]
- 42. Hutchison C.A., Chuang R.-Y., Noskov V.N., Assad-Garcia N., Deerinck T.J., Ellisman M.H., Gill J., Kannan K., Karas B.J., Ma L. et al. Design and synthesis of a minimal bacterial genome. Science. 2016; 351:aad6253. [DOI] [PubMed] [Google Scholar]
- 43. Woo S., Rothemund P.W.K.. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat. Chem. 2011; 3:620–627. [DOI] [PubMed] [Google Scholar]
- 44. Roberts R.J., Myers P.A., Morrison A., Murray K.. A Specific Endonuclease from Haemophilus haemolyticus. J. Mol. Biol. 1976; 103:199–208. [DOI] [PubMed] [Google Scholar]
- 45. Reckmann B., Krauss G.. The cleavage of single-stranded DNA by the isoschizomeric restriction endonuclease HhaI and CfoI. Biochim. Biophys. Acta - Gene Struct. Expr. 1987; 908:90–96. [DOI] [PubMed] [Google Scholar]
- 46. Lu M., Guo Q., Seeman N.C., Kallenbach N.R.. DNase I cleavage of branched DNA molecules. J. Biol. Chem. 1989; 264:20851–20854. [PubMed] [Google Scholar]
- 47. Kim D.N., Kilchherr F., Dietz H., Bathe M.. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 2012; 40:2862–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arbona J.M., Aim J.. Modeling the mechanical properties of DNA nanostructures. Phys. Rev. 2012; 51912:1–11. [DOI] [PubMed] [Google Scholar]
- 49. Yoo J., Aksimentiev A.. In situ structure and dynamics of DNA origami determined through molecular dynamics simulations. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:20099–20104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Maffeo C., Yoo J., Aksimentiev A.. De novo reconstruction of DNA origami structures through atomistic molecular dynamics simulation. Nucleic Acids Res. 2016; 44:3013–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nicholson A.W., Redhu S.K., Stopar A., Coral L., Carnevale V., Castronovo M.. Erdmann V, Jurga S, Barciszewski J. Emergent properties and functions of nanoconfined nucleic acid architectures. RNA and DNA Diagnostics. 2015; Cham: RNA Technologies. Springer; 183–204. [Google Scholar]
- 52. Li Z., Wang L., Yan H., Liu Y.. Effect of DNA hairpin loops on the twist of planar DNA origami tiles. Langmuir. 2012; 28:1959–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu Q.S., Roberts R.J., Guo H.-C.. Two crystal forms of the restriction enzyme MspI-DNA complex show the same novel structure. Protein Sci. 2005; 14:2590–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Newman M., Strzelecka T., Dorner L.F., Schildkraut I., Aggarwal a.K.. Structure of Bam HI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science. 1995; 269:656–663. [DOI] [PubMed] [Google Scholar]
- 55. Keum J.-W., Bermudez H.. Enhanced resistance of DNA nanostructures to enzymatic digestion. Chem. Commun. (Camb.). 2009; 45:7036–7038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.