Abstract
The human CST (Ctc1, Stn1 and Ten1) complex binds the telomeric overhang and regulates telomere length by promoting C-strand replication and inhibiting telomerase-dependent G-strand synthesis. Structural and biochemical studies on the human Stn1 and Ten1 complex revealed its mechanism of assembly and nucleic acid binding. However, little is known about the structural organization of the multi-domain Ctc1 protein and how each of these domains contribute to telomere length regulation. Here, we report the structure of a central domain of human Ctc1. The structure reveals a canonical OB-fold with the two identified disease mutations (R840W and V871M) contributing to the fold of the protein. In vitro assays suggest that although this domain is not contributing directly to Ctc1’s substrate binding properties, it affects full-length Ctc1 localization to telomeres and Stn1-Ten1 binding. Moreover, functional assays show that deletion of the entire OB-fold domain leads to significant increase in telomere length, frequency of internal single G-strands and fragile telomeres. Our findings demonstrate that a previously unknown OB-fold domain contributes to efficient Ctc1 telomere localization and chromosome end maintenance.
INTRODUCTION
Telomeres are the nucleoprotein structures that localize to the ends of eukaryotic chromosomes allowing for the faithful replication and stability of our genome (1–3). There are two well-known complexes that bind specifically to the telomeric DNA, namely the shelterin and the CST (Ctc1, Stn1 and Ten1). Shelterin binds single and double stranded telomeric DNA providing chromosome end protection by repressing Ataxia-telangiectasia mutated (ATM)- and Rad3-related (ATR)-dependent DNA damage response that could lead to a host of undesirable effects at the ends of chromosomes (4).
The CST complex was originally thought to belong exclusively to yeast, but conserved components have been subsequently identified in a wide range of species including plants, vertebrates and humans (5–8). Like in yeast, the human CST complex (hCST), composed of hCtc1, hStn1 and hTen1, is essential for the proper capping and maintenance of telomeres (7,9). Current evidence shows that the vertebrate and yeast CST (yCST composed of Cdc13, yStn1 and yTen1) localize to telomeres through association with the telomeric overhang (5,10). They regulate access of telomerase and DNA polymerase alpha (Polα) to the end of chromosomes for G- and C-strand synthesis, respectively (10–16) and the human complex has been shown to facilitate telomere replication restart after fork stalling (17–19).
Saccharomyces cerevisiae Cdc13, considered a Ctc1 homolog, contains four Oligonucleotide/Oligosaccharide-Binding (OB) folds, which are involved in a wide array of functions, including Cdc13 homo-dimerization, as well as telomerase, Polα-primase and single-stranded DNA binding (20–28). Despite the similarities between the yeast and human CST complexes, lack of sequence identity between Cdc13 and Ctc1 has made it difficult to predict the fold, domain organization and full function of hCtc1 although preliminary studies predict that Ctc1, like Cdc13, most likely consists of multiple OB-folds (5,20,29).
hCtc1, like Cdc13, forms a stable complex with hStn1-Ten1 and binds single-stranded, telomeric DNA with high affinity and specificity for G-rich sequences (30). However, unlike Cdc13, there is currently no evidence to suggest that hCtc1 is involved in telomerase recruitment to telomeres (8,31–33). In contrast, hCtc1 has been shown to downregulate telomerase activity via sequestering the telomeric overhang and possibly via its interaction with TPP1 and is currently known as telomerase activity terminator (34–36).
Naturally occurring mutations of hCtc1 are associated with the rare genetic disorders aplastic anemia, pulmonary fibrosis, dyskeratosis congenita and Coats plus syndrome (37–41). Current evidence suggests that the disease associated mutations are the result of a dysfunctional hCtc1, unable to carry out its DNA, Polα or hStn1–Ten1 binding properties (34,36). However, the mechanism of action of these mutations in human disease remains unknown.
To better understand hCtc1 domain organization and function, we sought to identify and express soluble domains of hCtc1 for structural and biochemical studies. Our effort generated a hCtc1 construct consisting of residues 716-880 (hCtc1(OB)). The crystal structure revealed a classical OB-fold with extended surface loops. Biochemical studies showed that this domain is monomeric in solution and does not interact with nucleic acid, Stn1-Ten1 or Polα in vitro. Mutation or deletion of the hCtc1(OB) resulted in telomere replication defects including elongated and fragile telomeres revealing that this domain is critical to hCtc1 function.
MATERIALS AND METHODS
Protein expression and purification
We identified a stable construct of hCtc1, consisting of residues 716-880, using limited proteolysis and mass spectrometry. Limited proteolysis was carried out with the endoproteinase Glu-C (V8 protease - Sigma 10791156001). A hCtc1 fragment consisting of residues 716–938, identified by sequence alignment (Clustal Omega (42)) and secondary structure prediction (PHYRE (43)) was subjected to limited proteolysis with the V8 protease. A total of 15 μl of 2 mg/ml protein was treated with 1/100 ratio of V8/hCtc1 at room temperature for 5, 30 and 60 min. The resulting stable band was analyzed by the Wistar mass spectrometry facility and consisted of residues 716-880 (Supplementary Figure S1). The wild-type (WT) and mutant (R840W and V871M) hCtc1(OB) were cloned into a pET28b vector containing an N-terminal hexahistidine-pMocr fusion tag, with a flexible linker cleavable by tobacco etch virus (TEV) protease. The hCtc1(OB) construct was overexpressed in Escherichia coli ScarabXpress T7 lac competent cells (Scarab Genomics) at 30°C for 4 h using 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside; Gold Biotechnology). The cells were harvested by centrifugation and lysed in a buffer containing 25 mM Tris, pH 7.5, 1.0 M KCl, 1.0 M Urea, 5% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 mM benzamidine (Ni Buffer A) via sonication. The protein was then purified over a nickel-nitrilotriacetic acid (Ni-NTA) column and buffer exchanged while on the Ni-NTA column with 25 mM Tris, pH 7.5, 0.2 M KCl and 5% glycerol (Ni Buffer C). The complex was eluted with 300 mM imidazole onto a tandem HS(poros)-HQ(poros) columns (Applied Biosystems) equilibrated with Ni Buffer C. The HS-HQ columns were then detached and the hCtc1(OB) protein was eluted from the HQ column with a salt gradient of 0.2 M KCl to 1.0 M KCl in a buffer also containing 25 mM Tris, pH 7.5, 1 mM dithiothreitol (DTT) and 5% glycerol. The fusion tag was cleaved by TEV protease overnight at 4°C and both TEV and the fusion tag were removed from the sample by an additional step of Poros-HS and HQ columns equilibrated with 0.15 M KCl, 25 mM Tris, pH 7.5, 1 mM DTT and 5% glyercol. The sample was then concentrated and separated over a Superdex S200 (GE Healthcare), to remove any aggregates, with a buffer containing 0.1 M KCl, 10 mM Tris, pH 7.5, 1mM tris(2-carboxyethyl)phosphine (TCEP) and 5% glycerol.
Protein crystallization and structure determination
Crystallization screening for hCtc1(OB) produced a crystal hit under sitting-drop vapor diffusion and room temperature in a crystallization buffer containing 0.1 M MES, pH 6.0, and 20% w/v poly(ethylene glycol) 2000 mono-methyl ether. The crystals diffracted to 1.86 Å in a crystal form that belongs to the P212121 space group. Data were collected at the National Synchrotron Light Source (NSLS) X25 beamline and processed using HKL2000 (44) (Table 1). The crystals contained two hCtc1(OB) molecules in the asymmetric unit. We used the method of single-wavelength anomalous diffractions (SAD) and a mercury derivative crystal to obtain the initial phases (Table 1). Mercury derivatives were obtained by soaking hCtc1(OB) crystals with mercury chloride (Sigma) for several minutes. The structure was built in Phenix (45) using AutoSolve and refined with Refmac (46). Building was done in Coot (47) and figures were made in Pymol (http://www.pymol.org). Amino acid conservation was performed by Consurf (http://consurf.tau.ac.il/).
Table 1. Data collection, phasing and refinement statistics.
| Native | Phasing | |
|---|---|---|
| Hg derivative | ||
| Data collection | ||
| Wavelength (Å) | 1.1 | 1.1 |
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 47.9 49.8 147.9 | 47.5 50.8 144.5 |
| Resolution (Å) | 20–1.96 (2.02–1.96) | 40–1.86 (1.90–1.86) |
| I/sI | 21.1 (2) | 24.08 (2.3) |
| Completeness (%) | 99.1 (98.0) | 100 (100) |
| Redundancy | 3.3 (2.7) | 6.7 (6.7) |
| Phasing Analysis | ||
| Resolution (Å) | 40–1.86 | |
| Number of sites | 6 | |
| BAYES-CC*: (Phenix) | 48.2 ± 18.7 | |
| Mean figure of merit (FOM) | 0.32 | |
| Refinement | ||
| Resolution (Å) | 20–1.95 | |
| No. reflections | 25217 (1874) | |
| R work/ R free | 21.4/25.1 (21.8/25.5) | |
| No. atoms | ||
| Protein | 2293 | |
| Water | 90 | |
| B-factors | ||
| Protein | 40.4 | |
| Water | 52.7 | |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.01 | |
| Bond angles (°) | 1.25 | |
| Ramachandran plot (%) | ||
| Most favored | 98.2 | |
| Allowed | 1.8 | |
*Estimate of the quality of the experimental electron density map.
Crosslinking
WT and mutant hCtc1(OB) proteins were diluted to 0.1 mg/ml in a buffer containing 25 mM Hepes, pH 7.5, 0.1 M KCl, 1 mM TCEP and 5% glycerol. Crosslinking was performed by mixing 4 μl of 1.5% formaldehyde or 5 μl of 2.5% glutaraldehyde in 20 μl of protein sample and incubating at room temperature for 90 mins. A total of 5 μl of 1.5 M Tris.HCl (pH 8.0) was added to the reaction mix to stop the crosslinking and the samples were then run on a 16% sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gel.
Fluorescence polarization (FP) assay
Fluorescence polarization (FP) assays were carried out in 20 μl binding reactions in a buffer containing 20 mM Hepes, pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM ethylenediaminetetraacetic acid (EDTA), 2 mM DTT, 1 mg/ml bovine serum albumin (BSA), 5% v/v glycerol and 75 nM polyT50 competitor (IDT). The 12mer DNA probe (TTAGGGTTAGGG) and 18mer DNA probe (TTAGGGTTAGGGTTAGGG) was purchased with a 5′ 6-FAM label from Integrated DNA Technologies. The final probe concentration used was 2.5 nM, while the hCtc1(OB) protein concentration ranged from 0 to 10 μM. Each reaction was performed in triplicate and incubated at room temperature for 30 min, pipetted into a black 384 well optiplate (PerkinElmer) and scanned with an Envision Xcite Multilabel Plate Reader (Perkin Elmer). Each well was excited with 480 nm light and the emissions were measured at 535 nm light. The Envision operating software (PerkinElmer) were used to calculate the milipolarization (mP) values.
Isothermal titration calorimetry (ITC)
Isothermal titration calorimetry (ITC) experiments were performed on a MicroCal iTC200 (Malvern) and proteins were purified and buffer exchanged into a buffer containing 25 mM Hepes, pH 7.5, 0.1 M KCl, 5% glycerol and 1 mM TCEP. The concentration of each protein sample was measured using a Bradford Assay (48). Full-length hStn1 and hStn1C (C-terminal domain of hStn1) was purified as described previously (49). Full-length hStn1 was used at a concentration of 125 μM in the syringe and was injected into a cell containing 25 μM of hCtc1(OB). hStn1C was used at a concentration of 100 μM (75 μl) in the syringe and was injected into a cell containing 10 μM (200 μl) of hCtc1(OB). For the binding experiments, the calorimeter was kept at 25°C (298.15 K) and there was a total of 17 injections with the volume of 2.47 μl used for each injection.
Size-exclusion chromatography and multi-angle light scattering (SEC-MALS)
Size-exclusion chromatography and multi-angle light scattering (SEC-MALS) experiments were performed in the laboratory of Dr Gideon Dreyfuss and as described previously (50). A DAWN HELEOS II 18-angle light scattering instrument (Wyatt Technologies) was used in tandem with a Superdex 200 10/300 GL column (GE Healthcare) to calculate the absolute molecular mass at 20°C in a buffer consisting of 0.1 M KCl, 25 mM Hepes, pH 7.5, 1 mM TCEP, and 5% glycerol. The molecular mass of hCtc1(OB) was monitored by UV at 280 nm (refractive index) as it eluted of the SEC column and molecular mass was measured by MALS. ASTRA software (version 5.2, Wyatt Technologies) was used within the defined chromatographic peaks in order to calculate the average molecular weight.
SYPRO orange assay
WT and mutant hCtc1(OB) proteins were diluted in a buffer consisting of 20 mM HEPES at pH 7.5, 0.15 M KCl, 5% glycerol and 1 mM TCEP-HCl to two final concentrations of 5 μM, and 16 μM. The protein samples were transferred into a MicroAmp Optical 384 well plate (Applied Biosystems). To each of the protein samples we added 4 µl of Sypro Orange (5000 × stock, ThermoFisher Scientific - diluted 1:300). The plate was spun down and heated from 20 to 95°C using a quantitative polymerase chain reaction (qPCR) (ABI 7900 RealTime PCR) with a 1% ramp rate and fluorescent readings were recorded every 2 min. Data were analyzed and plotted using Microsoft Excel version 15.33.
Differential scanning calorimetry
We further tested the thermostability of the WT and mutant hCtc1(OB) proteins using a MicroCal VP-Capillary DSC (Malvern Instruments Ltd). The hCtc1(OB) proteins were diluted in a buffer consisting of 20 mM HEPES at pH 7.5, 0.15 M KCl, 5% glycerol and 1 mM TCEP to a final concentration of 1 mg/ml, and heated from 10 to 90°C at a scan rate of 60°C per hour. The thermogram for each protein sample was normalized to the buffer and data analysis was performed using the MicroCal Origin 7.0 software.
Plasmids
We cloned the full length, WT, double mutants (F801A/R840A and L732A/V871A) and deletion Δ726-876, human Ctc1 (NM_025099.5) into the lentiviral vector pLU-EF1A-iBlast (pLU) with an N-terminal 1× Flag tag using standard cloning procedures while the mutations and deletion where introduced using QuickChange II site-directed mutagenesis kit (Agilent). The sequences of all plasmids were confirmed by DNA sequencing.
Lentivirus production and cell culture
We produced WT and mutant human Ctc1 lentivirus by co-transfecting 293T cells with 12 μg of Ctc1 plasmids and the viral packaging vectors PMDG (4 μg), pRSV (4 μg) and pRRE (4 μg). The medium containing lentivirus were harvested at 96 h post-transfection. Human 293T cells were grown in Dulbecco’s modified Eagle’s medium media supplemented with 10% (vol/vol) fetal bovine serum (Atlanta Biologics Ltd) and 1 mM penicillin-streptomycin. All cells were cultured in a 5% CO2 incubator at 37°C. 293T cell lines stably expressing 1× Flag-hCtc1 WT and mutants, were established using lentiviral infection with 2 μg/ml polybrene (Sigma). A pLKO.1 vector, with puromycin resistance, carrying the shRNA (TRCN0000330094) targeting the 3′ UTR of endogenous hCtc1 (shCtc1) was obtained from the Sigma Mission shRNA library and delivered to the 293T cells with lentiviral infection. The hCtc1 WT and mutant cell lines were maintained with media containing 2 μg/ml Puromycin and 5 μg/ml blasticidin.
Reverse transcription and quantitative PCR analysis
To test the effectiveness of the shRNA targeting the 3′UTR of endogenous hCtc1, we isolated and quantified the mRNA from cells treated with shRNA. The mRNA was isolated from pelleted cells using TRIzol (Life Technologies) and the RNA was purified using the Direct-zol RNA mini-prep kit (Zymo Research). Reverse transcription PCR was performed using random hexamer primers followed by quantitative PCR using primers targeting the 3′ UTR of the hCtc1 gene. The following primer pairs were used to measure hCtc1 and GAPDH mRNA levels after knockdown: hCtc1 (5′ GTGATTGAACCAAGGACTCCAGAT 3′ and 5′CAGGCTGGCACCAGAACAC3′); GAPDH (5′ATGGAAATCCCATCACCATCTT3′ and 5′CGCCCCACTTGATTTTGG3′).
Immunoprecipitation and western blotting
For the pulldown assays by immunoprecipitation, we used lysed ∼1 × 107 cells expressing Flag-tagged, WT and mutant hCtc1 proteins. Lysing was done in 1ml ice-cold non-denaturation lysis buffer (NDLB) [300 mM NaCl, 20 mM Tris–HCl (pH 7.4), 2 mM EDTA, 1% NP-40, 10% glycerol, 0.5% Sodium deoxycholate, 1 mM PMSF and 1× protease inhibitor cocktail (Sigma)] for 30 min at 4°C with gentle agitation. The lysates were then centrifuged at 10 000 × g for 10 min at 4°C and the supernatants were collected. The lysates were precleared with protein G-Sepharose beads for 2 h at 4°C and inputs were collected prior to immunoprecipitation. The precleared lysates were immunoprecipitated with anti-Flag M2 affinity gel (Sigma) overnight at 4°C. The next day we centrifuged the lysates at 2000 rpm for 2 min and washed the beads six times with ice-cold NDLB buffer. Immunoprecipitates were eluted with 200 μg/ml of 3× flag tag peptide (Sigma) and the input and eluents were electrophoresed on an 8-16% (wt/vol) Tris-Glycine gel (Life Technologies). The gel was transferred to a nitrocellulose membrane (BioRad) and blocked in 5% nonfat dry milk in Tris-buffered saline with 0.05% Tween 20 for 2 h. The membrane was then probed with the primary HRP-conjugated antibodies against Flag (Sigma), Ctc1 (HPA044349, Sigma), Stn1 (HPA037924, Sigma), Ten1 (HPA043486, Sigma) and GAPDH (2118S, Cell Signaling technologies) and visualized by Luminata HRP detection reagent (Millipore). The levels of Stn1 and Ten1 interacting with hCtc1 were calculated (using imageJ) by taking the sum intensity of Stn1+Ten1 and divided by the intensity of hCtc1. The calculated values were then normalized to 1 by dividing with the mutant calculated levels by the WT ones.
Chromatin immunoprecipitation and DotBlot analysis
Crosslink chromatin immunoprecipitation was performed in 293T cell lines stably expressing endogenous or shCtc1 treated, ectopic WT and mutant hCtc1 proteins. The 5 × 107 cells were treated with 1% formaldehyde for 20 min for protein-DNA crosslinking and the reaction was stopped with 0.125 M glycine. The cells were lysed with a buffer consisting of 50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton-X, 1 mM PMSF and 1× protease inhibitor cocktail (Sigma) for 10 min at 4°C with gentle agitation. The lysates were centrifuged at 1350 × g for 5 min at 4°C and the supernatant was discarded. The pellet further lysed in buffer containing 10 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1 mM PMSF and 1× protease inhibitor cocktail (Sigma). The lysates were centrifuged at 1350 × g for 5 min at 4°C and the supernatant was discarded. The resulting pellet was resuspended in lysis buffer containing 10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Sodium deoxycholate, 0.5% Sodium laurysarcosine 1 mM PMSF and 1× protease inhibitor cocktail (Sigma) and sonicated to yield DNA fragments of between 200 and 500 bp in length. A total of 1/10 volume of 10% triton-X was added to the sonicated lysate and centrifuged at 20000 × g for 10 min at 4°C to pellet the debris. The supernatants were precleared with 25 μl of Protein A and 25 μl of protein G beads along with 2.5 μg of bacterial DNA for 2 h at 4°C. Flag-M2 (Sigma) and mouse IgG (Sigma) antibodies were added to the precleared lysates and incubated overnight at 4°C with rotation. We then added 25 μl of Protein A and 25 μl of protein G beads along with 2.5 μg of bacterial DNA and incubated at 4°C with rotation for 2 h. We pelleted the beads by spinning at 2000 rpm for 2 min at 4°C and the supernatant was carefully removed. We washed the beads five times with RIPA buffer containing 50 mM HEPES-KOH, pH 7.5, 500 mM LiCl, 1 mM EDTA, 1% NP-40, 0.7% Sodium deoxycholate, 1 mM PMSF and 1× protease inhibitor cocktail. We then resuspended the beads in 150 μl of elution buffer containing Tris-HCl, pH 8.0, 1% SDS and 10 mM EDTA, vortexed and incubated at 65°C for 15 min. The eluents were collected and decrosslinked overnight at 65°C with final concentration of 200 mM NaCl. The samples were then treated with RNAse A and proteinase K and the DNA was purified using the QIAquick PCR purification kit (Qiagen). We subsequently performed dot blot experiments and probed the membrane with a 32P-labeled probe (CCCATT)4 in Church buffer (0.5 N Na-phosphate, pH 7.2, 7% SDS, 1 mM EDTA, 1% BSA) overnight at 42°C. The blot was washed twice, at room temperature for 5 min, with 0.2 M buffer containing 0.2 N Na-phosphate, 2% SDS and 1 mM EDTA, and once with 0.1 M buffer of 0.1 N Na-phosphate, 2% SDS and 1 mM EDTA at 50°C. Radioactive signals were detected by phosphor-imager (GE Healthcare), and visualized with a Typhoon 9410 Imager (GE Healthcare). The images were quantified with ImagQuant 5.2 software (Molecular Dynamics).
Telomere length assay and Southern blotting
For the Southern blot assays, we first isolated the genomic DNA from 293T cells stably expressing the WT and mutant 1× Flag-Ctc1 at four different passages (0, 3, 6 and 9) using the QIAamp DNA mini kit (Qiagen). All cell lines carried the shRNA targeting the 3′ UTR of the endogenous hCtc1. To isolate the telomeric DNA, we treated 10 μg of genomic DNA with the restriction endonucleases AluI and MboI overnight at 37°C. A total of 2 μg of the digested DNA were run on a 0.7% agarose gel. After electrophoresis the DNA was denatured by incubating the gel for 20 min in 0.25 N HCl followed by 2 × 20 min in 0.8 M NaOH, 150 mM NaCl and by incubation in neutralization buffer 0.5 M Tris.HCl, pH 7.5, 3 M NaCl. The gel was then transferred to a GeneScreen Plus blotting membrane (Perkin-Elmer) in the presence of 20 × saline-sodium citrate (SSC) buffer (3 M NaCl and 0.3 M Sodium Citrate pH 7) and was left to incubate overnight for the DNA transfer to occur. The DNA was UV cross-linked onto the membrane at 1250 eV using the UV Stratalinker 2400 (Stratagene). The blot was hybridized with a 32P-labeled probe containing the telomeric oligonucleotide (TTAGGG)4 in Church buffer (0.5 N Na-phosphate, pH 7.2, 7% SDS, 1 mM EDTA, 1% BSA) overnight at 42°C. The blot was washed twice at room temperature for 5 min with 0.2 M buffer containing 0.2 N Na-phosphate, 2% SDS, and 1 mM EDTA and once with 0.1M buffer of 0.1 N Na-phosphate, 2% SDS and 1 mM EDTA at 50°C. Radioactive signals were detected by phosphor-imager (GE Healthcare), and visualized with a Typhoon 9410 Imager (GE Healthcare). The data were analyzed using TeloTool version 1.3 (Matlab) (51).
Metaphase telomere FISH
We cultured 293T cells expressing WT and mutant Ctc1 with fresh medium for 24 h prior to metaphase arrest. The 0.1 μg/ml colcemid (Gibco) was added for 4 h to arrest cells in mitotic phase. Cells were trypsinized, resuspended in 75 mM KCl hypotonic solution and incubated for 30 min at 37°C. Cells were immediately fixed by adding 10 ml fresh 3:1 methanol/acetic acid dropwise three times. Fixed cells were dropped onto cold wet glass microscope slides, and dried overnight in a humid environment. Metaphase chromosome spreads were fixed in 4% formaldehyde in 1 × phosphate-buffered saline (PBS) for 3 min, treated with 1 mg/ml pepsin for 10 min at 37°C, dehydrated in 70, 95, 100% (vol/vol) ethanol successively and air-dried. Slides were denatured for 5 min at 80°C in hybridization mix (10 mM Tris–HCl (pH 7.2), 70% formamide and 0.5% blocking solution (Roche)) containing the telomeric PNA-Tamra-(TTACCC)3 probe. The slide was then put in a dark, humid chamber at room temperature for 2 h to allow the probe to hybridize to the telomeric DNA. After hybridization, slides were washed twice for 15 min each with buffer containing 10 mM Tris-HCl (pH 7.2), 70% formamide and 0.1% BSA and three times for 5 min each with buffer consisting of 0.15 M NaCl, 0.1 M Tris-HCl (pH 7.2) and 0.08% Tween-20. Chromosomal DNA was counterstained with 0.1 μg/ml DAPI in 1 × PBS for 5 min and slides were mounted with Fluoromount-G (Southern Biotech). Images were taken with a 100 × lens on a Nikon E600 Upright Microscope (Nikon Instruments) using ImagePro Plus software (Media Cybernetics) for image processing. Statistical analysis was performed using two-tailed unpaired t-test.
Telomeric overhang analysis
Telomeric overhang analysis was performed by non-denaturing in-gel hybridization as described (Feng X et al. (13)) with minor modifications. Briefly, genomic DNA was purified by genomic DNA purification kit (Promega) and 20 μg of purified genomic DNA was treated with 100U Exo I (New England Biolabs) in Exonuclease I reaction buffer containing 67 mM Glycine-KOH, 6.7 mM MgCl2, 10 mM β-mercaptoethanol for 48 h at 37°C. A total of 15 μg of Exo I treated and untreated genomic DNA was then digested with 15U of AluI and MboI restriction endonucleases (New England Biolabs) overnight at 37°C, and purified by phenol-chloroform extraction. A total of 15 μg of Exo I-treated and untreated samples were run on a 0.7% agarose gel. The gel was dried at 28°C by a vacuum gel dryer (BioRad). The dried gel was hybridized with a 32P-labeled (5′-CCCTAA-3′)4 probe in Church buffer overnight and at 42°C. The gel was then washed twice with 0.2 N Na-phosphate, 2% SDS and 1 mM EDTA buffer at room temperature for 5 min each and once for 5 min with 0.1 N Na-phosphate, 2% SDS and 1 mM EDTA buffer at 42°C. For the detection of the telomeric overhangs, radioactive signals were detected by phosphor-imager (GE Healthcare) and visualized with a Typhoon 9410 Imager (GE Healthcare). To re-probe the blot for total telomeric DNA signals, the gel was denatured in alkaline solution (0.5 M NaOH, 0.15 M NaCl) twice for 30 min each at 42°C, neutralized in neutralization solution (0.5M Tris-HCl (pH 7.5), 3M NaCl) twice for 20 min each at 42°C and hybridized with a 32P-labeled (5′-CCCTAA-3′)4 probe as described above. The images were quantified with ImagQuant 5.2 software (Molecular Dynamics).
RESULTS
Structure of the human hCtc1(OB)
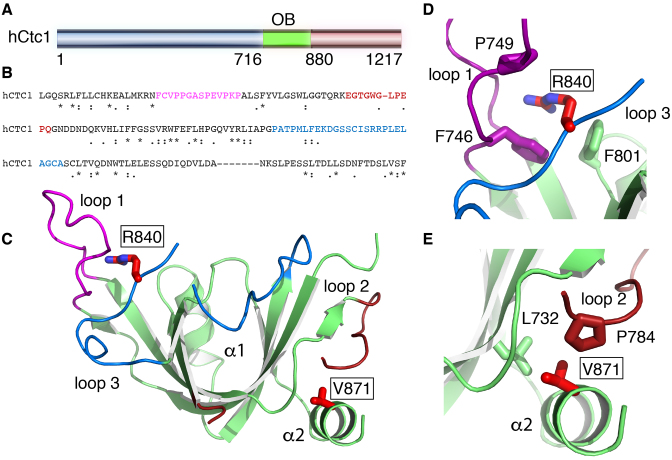
We used limited proteolysis and mass spectrometry to identify a soluble domain of hCtc1 spanning residues 716–880 (hCtc1(OB)) (Figure 1A and Supplementary Figure S1). We solved the hCtc1(OB) crystal structure to 1.86 Å resolution using a mercury chloride derivative and the method of SAD (Table 1). The hCtc1(OB) adopts a canonical OB-fold that is structurally similar to the DNA-binding domain A of Ustilago maydis RPA70 (52) (PDB ID: 4GOP - RMSD 2.8 Å). However, unlike the RPA70 OB fold, the hCtc1(OB) does not contain a defined canonical DNA binding pocket and the residues located in this region of the OB fold are not conserved (Supplementary Figure S2). An interesting feature of hCtc1(OB) is three unusually long loops located on the surface of the protein consisting of amino acids 745-759 (loop 1), 777-795 (loop 2) and 824-849 (loop 3) (Figure 1C). Sequence alignment indicates that these three loops comprise non-conserved regions of the protein and their precise function in hCtc1 is currently unclear (Figure 1B).
Figure 1.
X-ray crystal structure of a central OB-fold of human Ctc1. (A) Primary sequence of hCtc1. (B) Sequence conservation of hCtc1(OB); the following genes were used for the alignment; Human, zebrafish, mallard, Vitis vinifera, mouse, rat, Spermophilus tridecemlineatus, rabit, killifish, bovine, African bush elephant, orangutan. Color font represents the surface loops of hCtc1(OB). (C) Crystal structure of hCtc1(OB) with surface loops highlighted in color. The disease-associated mutations R840W and V871M are shown in red stick. (D) Zoom-in representation of R840 and the residues interacting with it. (E) Zoom in representation of V871 and the residues interacting with it.
Interestingly, two naturally occurring mutations (R840W and V871M) implicated in the rare genetic disorders dyskeratosis congenita and Coats plus are located within this hCtc1(OB). R840 comprises part of loop 3 and is solvent exposed. Although the guanidinium portion of this side chain is not involved in direct contacts with the rest of the protein the aliphatic portion is sandwiched by the hydrophobic side chains of F746 and P749 that form part of loop 1 and F801 that comprises part of the core of the protein. V871 on the other hand is located at the very C-terminus of the OB fold and comprises part of helix α2. The side chain of V871 is buried within the protein fold and is involved in extensive interactions with the side chains of L732 and P784.
The hCtc1(OB) is not involved in substrate binding in vitro
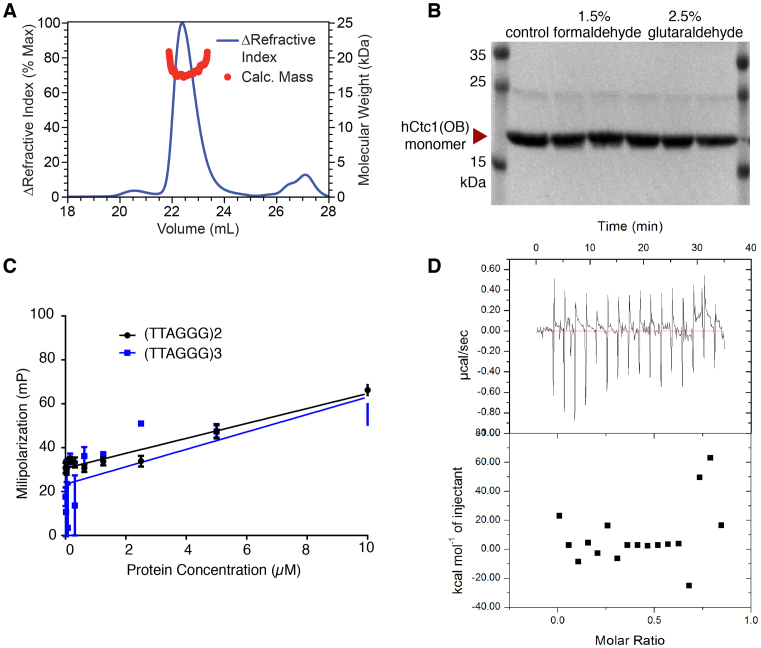
Interestingly, the OB2 of Cdc13, the closest homolog of hCtc1, also contains extensive loops on its surface which are involved in Cdc13 dimerization (26). To determine if the hCtc1(OB) is involved in hCtc1 dimerization we first performed multi-angle light scattering (SEC-MALS) experiments with the purified hCtc1(OB). The results indicate that this domain of hCtc1 is monomeric in solution (Figure 2A and B).
Figure 2.
Oligomerization and substrate binding assays of hCtc1. (A) The oligomeric state of hCtc1(OB) was analyzed by SEC-MALS. The blue line corresponds to the Refractive Index (RI) of the hCtc1(OB) eluting from the SEC column. The red circles correspond to the molecular mass of hCtc1(OB) measured by multi-angle, light scattering (MALS: red). The data suggest that hCtc1(OB) is monomeric in solution. (B) Cross linking experiments of WT hCtc1(OB) using formaldehyde or glutaraldehyde also shows that this domain hCtc1 is monomeric in solution. (C) FP assays of hCtc1(OB) with 5′ 6-FAM (Fluorescein) labeled, single-stranded telomeric DNA (two or three repeats) shows that this domain of hCtc1 is not involved in DNA binding. (D) ITC assay of hCtc1(OB) with the full length Stn1–Ten1 complex show no measurable interaction.
To further understand the function of hCtc1(OB) we asked if this portion of hCtc1 is involved in telomeric DNA binding, a known substrate of hCtc1. To address this question, we performed fluorescent polarization (FP) assays using the purified hCtc1(OB) with a fluorescent labeled DNA probe consisting of two or three telomeric repeats. The results indicate that this domain of hCtc1 does not bind telomeric DNA by itself, even at 10 μM concentration (Figure 2C).
We then asked if the hCtc1(OB) binds the Stn1–Ten1 complex, the two interacting partners of hCtc1 that together form the CST complex. To address this question, we performed ITC experiments of the purified hCtc1(OB) with the hStn1–Ten1 complex. The data indicates that the hCtc1(OB) does not bind the hStn1–Ten1 complex (Figure 2D).
hCtc1(OB) disease mutations contribute to the stability of the OB fold
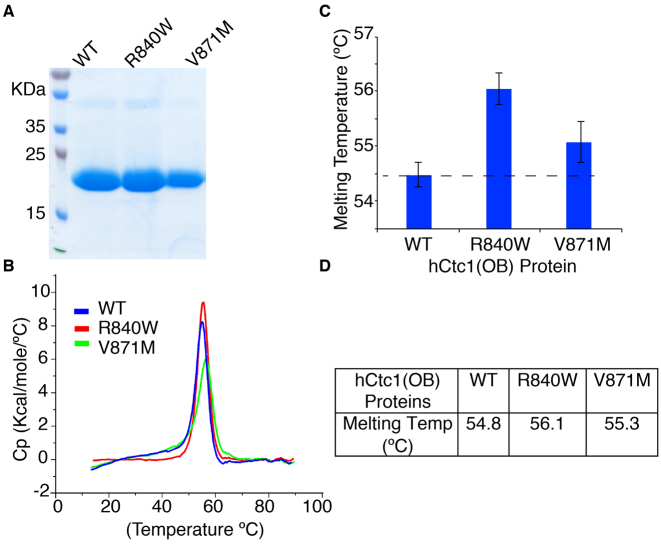
To assess the effect of the hCtc1 naturally occurring mutations R840W and V871M in the stability of the proteins we performed thermal denaturation experiments. The WT and mutant proteins were expressed and purified to homogeneity (Figure 3A) and the stability of the protein samples was tested using the SYPRO® Orange assay and differential scanning calorimetry (DSC) (Molecular Probes by Life Technologies). The results clearly show that the melting temperature (Tm) of both mutants is higher than that of the WT protein (Figure 3B and Supplementary Figure S3). The Tms for the WT and mutant hCtc1(OB) R840W and V871M are 54.8, 56.1 and 55.3°C respectively (Figure 3C and Supplementary Figure S3).
Figure 3.
The hCtc1(OB) disease mutant proteins are more stable than the WT protein. (A) SDS-PAGE gel showing the WT and the mutant R840W and V871M hCtc1(OB) purified proteins used in the experiments carried out for this study. (B) Normalized thermal denaturation curves derived from three independent DSC experiments for each of the WT and mutant R840W and V871M purified proteins shown in panel A. Detailed analysis of the data and Tm calculation is provided in Supplementary Figure S3. (C) Quantification of the Tm of the WT and the mutant R840W and V871M proteins using DSC. (D) Table of the melting temperatures of the WT and mutant hCtc1(OB).
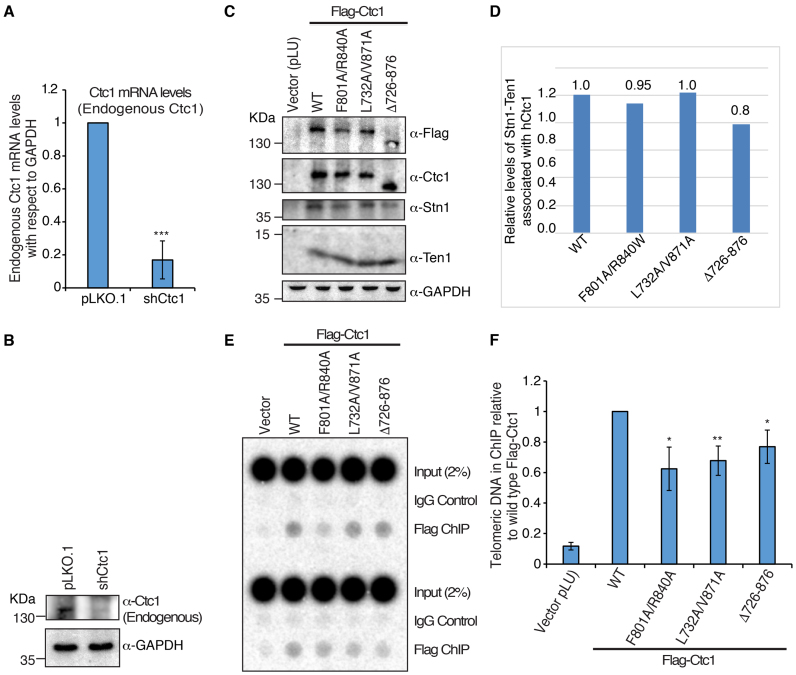
Immunoprecipitation analysis of hCtc1 in mammalian cells
To investigate the contribution of the hCtc1(OB) in living cells, we generated mammalian lentivirus expression vectors (pLU; EF-1 alpha - Takara) with Flag-tagged hCtc1 containing either full-length WT hCtc1 or hCtc1 with the hCtc1(OB) (Δ726–876) deletion. We also generated double alanine mutations at F801A/R840A and L732A/V871A to characterize naturally occurring mutations, but designed to accentuate their otherwise subtle phenotypes (34). The non-disease mutations F801A and L732A were selected using the crystal structure based on their proximity to the disease mutations R840A and V871A (Figure 1D and E). WT and mutant Flag-hCtc1 genes and an shRNA targeting the 3′UTR region of hCtc1 were introduced into 293T cells by lentivirus transduction. The levels of endogenous hCtc1 were reduced by 80% by the shRNA targeting the 3′UTR region of the gene (Figure 4A and B). qRT-PCR and western blot assays show a significant decrease (∼80%) in endogenous hCtc1 mRNA (Figure 4A) and protein (Figure 4B) following knockdown. Flag-IP assays using the ectopic WT and double mutant, Flag-hCtc1 successfully pull downs its interacting partners Stn1 and Ten1, while the deletion mutant shows a 20% decrease in Stn1–Ten1 binding (Figure 4C and D).
Figure 4.
Mutations in the hCtc1(OB) domain of hCtc1 affect its localization to telomeres. (A and B) qRT-PCR and western blot analysis of endogenous hCtc1 mRNA and protein levels, respectively, in 293T after shRNA knockdown. For qRT-PCR, results are the average of three independent experiments. Standard deviations are shown in the form of error bars; ***P < 0.0001 (calculated by two-tailed paired Student’s t-test). (C) Immunoprecipitation of hCtc1 stable cell lines established from 293T. Flag-tagged hCtc1 were isolated by flag beads and assayed by western blotting. Antibody for hCtc1 was used to detect the full-length hCtc1, hCtc1-(F801A/R840A), hCtc1-(L732A/V871A) and hCtc1-(Del726–876). GAPDH was used as the input control. (D) Bar graph showing the relative protein levels of Stn1–Ten1 complex pulled down by the full length, WT and mutant (double and deletion) hCtc1. Results are the average of three independent experiments. *P < 0.05 (calculated by two-tailed paired Student’s t-test). (E) Telomere localization of full length, WT and mutant (double and deletion) hCtc1 using crosslinking ChIP experiments in 293T cells expressing these proteins. ChIP DNA was assayed by dot blotting and hybridization with 32P-labeled probed specific for TTAGGG. (F) Bar graph showing the relative levels of full length, WT and mutant (double and deletion) hCtc1 proteins at telomeres. Results are the average of three independent experiments. *P < 0.05, **P < 0.001 (calculated by two-tailed paired Student’s t-test).
The hCtc1(OB) contributes to full length hCtc1 localization to telomeres
We also examined the ability of full-length hCtc1 to bind telomeric DNA. To address this question, we used crosslink chromatin immunoprecipitation assays in 293T cell lines stably expressing endogenous or shCtc1 treated, ectopic WT and mutant hCtc1 proteins. Protein–DNA crosslinking was achieved by 1% formaldehyde treatment of the cells. The crosslinked cells were gently lysed twice and the resulting pellet was sonicated to yield DNA fragments of between 200 and 500 bp in length. The substrates were isolated with Flag-M2 (Sigma) and mouse IgG (Sigma) antibodies and Protein A and G beads incubation. The de-crosslinked samples were treated with RNAse A and proteinase K and the DNA was purified. Dot blot experiments performed in triplicate clearly show a decrease in the localization of full length hCtc1 double mutants and the hCtc1(OB) deletion compared to the WT protein (Figure 4E and F).
The hCtc1(OB) contributes to Ctc1-dependent telomere length regulation
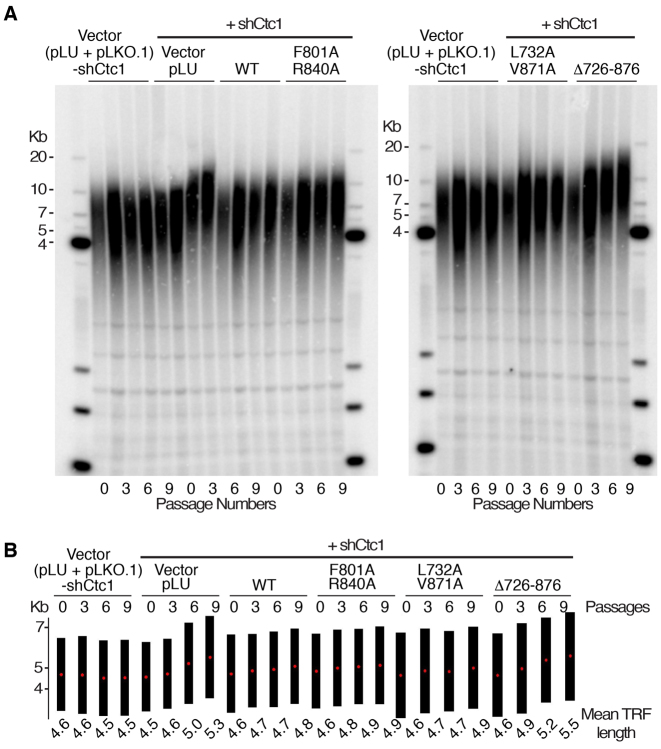
We next assessed telomere length in 293T cells with endogenous hCtc1 knockdown that are stably expressing hCtc1 WT, F801A/R840A, L732A/V871A and Δ726–876 (Figure 4A and B). We used Southern blot analysis of telomeric restriction fragment probed with G-rich telomere repeat sequence (TTAGGG)4 and found no consistent alterations in cells that express the vector, the WT and double mutants (F801A/R840A and L732A/R871A) after nine population doublings. However, we found that the hCtc1 knockdown and the Δ726-876 mutant produced a consistent increase in telomere repeat length after nine population doublings (Figure 5).
Figure 5.
hCtc1(OB) deletion leads to telomere length elongation. (A) Southern blot analysis of 293T cells expressing shCtc1 and WT or mutant (double or deletion) hCtc1 genes. DNA from 0, 3, 6 and 9 passages is shown. DNA length standards are indicated along the left and right of the blots. (B) Quantitative representation of the mean telomere length (kb) calculated from A using TeloTool. Black bars indicate the range of telomere restriction fragments (TRFs) and the red dots represent the mean TRF length.
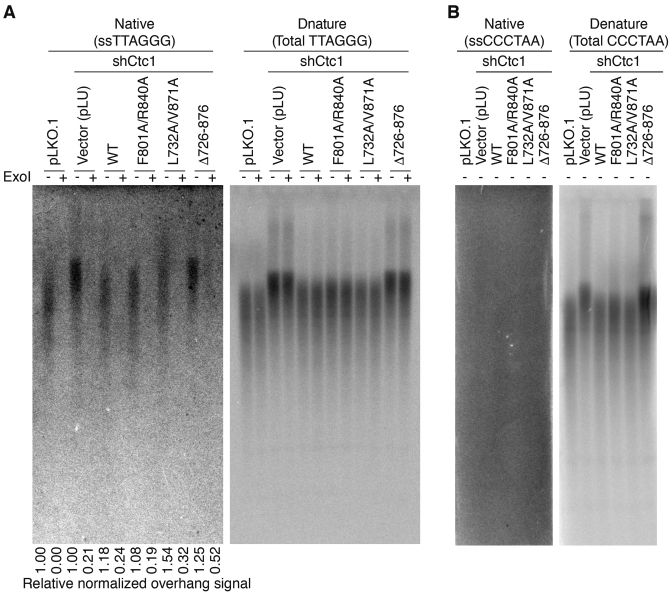
hCtc1(OB) contributes to hCtc1 dependent C-strand synthesis
hCtc1 has been implicated in both telomeric DNA replication and single-strand DNA protection. To understand the role of the hCtc1(OB) in either of these two functions of hCtc1, we examined the sensitivity of telomeric DNA to exonuclease I (ExoI) in gels hybridized either under denaturing or native conditions (Figure 6A and B). Comparison of the telomere signal intensity between these two conditions was used to distinguish between the relative abundance of single-stranded regions at the termini (ExoI sensitive) or internal single stranded DNA stretches (ExoI resistance). The native gel shows an increase in telomeric G-overhang length in shCtc1 knockdown and hCtc1 deletion (Δ726–876) mutation. The gel also shows that the cells with the deletion (Δ726–876) are enriched (∼2.0-fold) for ExoI resistant single-stranded telomeric DNA relative to WT control (Figure 6A). These findings suggest that hCtc1(OB) deletion accumulates internal single-stranded telomeric DNA consistent with defective DNA replication at telomeres.
Figure 6.
hCtc1(OB) deletion increases ExoI-resistant single-stranded telomere DNA. (A) Analysis of single-stranded telomeric DNA from 293T cell lines expressing full length, WT and mutant (double and deletion) hCtc1 with ExoI prior to AluI/MboI digestion. ExoI treated (+) and untreated (−) (ss)TTAGGG signal was detected with 32P-labeled (CCCTAA)4 probe under native condition (left) and denaturing conditions (right). The ssTTAGGG signal was normalized to the total TTAGGG signal in the same lane as indicated at the bottom. (B) Probing for the presence of single stranded ssCCCTAA signal in the same DNA samples as of panel (A). DNA samples were probed with 32P-labeled (TTAGGG)4 under native and denaturing conditions; the results show no CCCTAA signal under native conditions (left) which indicates the absence of single-stranded internal CCCTAA DNA.
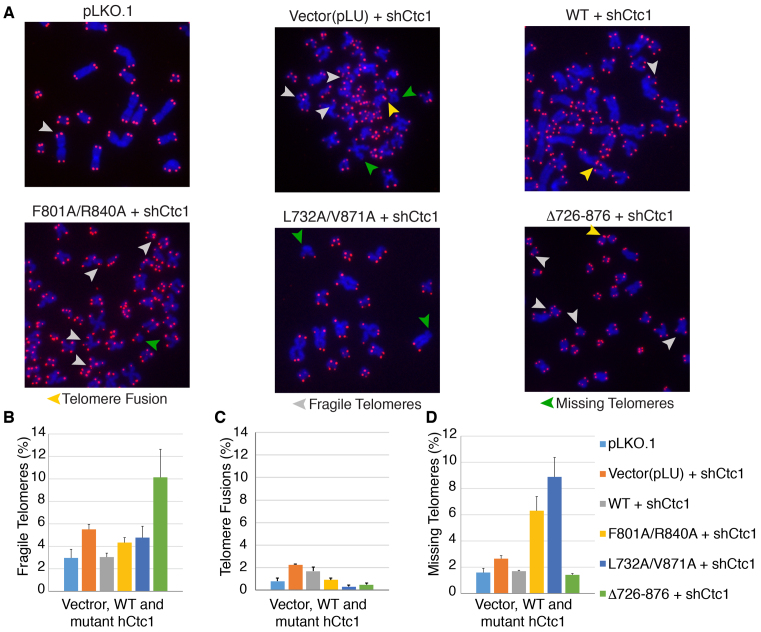
hCtc1(OB) is required for telomere integrity
The effect of hCtc1(OB) mutations in telomere integrity was next assessed by microscopy of metaphase chromosome spreads combined with telomere DNA FISH (Figure 7A and B). Using the same cell lines as those used for the Southern blot analysis, we found that the vector (pLU) and WT hCtc1 did not show any evidence of gross telomere defects, such as fusions, telomere signal loss or endo-duplicated fragile sites in metaphase FISH. However, we observed significant increase in missing telomere signal for the double mutants F801A/R840A (∼3-fold) and L7823A/R871A (∼5-fold) and significant (∼2-fold) increase in fragile sites in Δ726-876. The shRNA knockdown shows a significant increase in chromosome fusions (5-fold increase compared to WT hCtc1).
Figure 7.
Metaphase telomere FISH reveals that mutation or deletion of hCtc1(OB) leads to telomeres defects. (A) Telomeric DNA FISH analysis on metaphase spreads in 293T cells stably expressing full length, WT and mutant (double and deletion) hCtc1 proteins. Missing telomere signal, fragile telomeres and telomere fusions are indicated by green, gray and yellow arrowheads respectively. (B) The bar graphs in panels B-D show the various levels of dysfunctional telomeres for the hCtc1 double and deletion mutants. A total of 1000 chromosomes were used for each hCtc1 mutant. Standard deviation for 1000 chromosome measurements for WT and mutant hCtc1 are shown as error bars.
DISCUSSION
Here, we present the structure of a central domain of hCtc1 (Figure 1) identified by limited proteolysis (Supplementary Figure S1). The structure reveals a canonical OB-fold with unusual long loops on its surface. Interestingly, hCtc1(OB) does not contain a defined canonical substrate binding pocket and the residues that form this pocket are variable (Supplementary Figure S2). It is therefore not surprising that extensive biochemical studies show that this domain of hCtc1 is not directly involved in known substrate binding (DNA or Stn1–Ten1) in vitro, consistent with what has been previously reported (34) (Figure 2C and D). We also find no biochemical evidence that it is involved in hCtc1 dimerization (Figure 2A and B), a property of its yeast homolog Cdc13 (25–28,33).
Two naturally occurring hCtc1 mutations, R840W and V871M, are implicated in the genetic disorder Coats plus and are found within the hCtc1(OB) (36,37). Interestingly, both of these mutations are buried within the OB-fold and most likely affect the stability of the hCtc1(OB) protein. WT and mutant proteins subjected to DSC show that the mutant proteins are more stable (R840W - 56.1°C and V871M - 55.3°C) than the WT (54.8°C) protein (Figure 3 and Supplementary Figure S3). Although the temperature changes, and in particular for mutant V871A, are subtle, in the context of a disease the onset of which may occur over a long period of time, the observed subtle changes in Tm for these mutants may be significant.
The DSC results can be explained in the context of the structure. R840 is located on the surface of the protein and the aliphatic portion of this arginine is involved in contacts with F746 and P749 that form part of loop1 (Figure 1C and D). Introduction of the larger hydrophobic tryptophan side chain (R840W) to this position of the OB-fold would increase the contacts of this residue with the hydrophobic side chains of F746 and P749, thus increasing the stability of the protein. Similarly, V871 comprises part of the C-terminal helix of hCtc1(OB). V871 makes extensive hydrophobic interactions with the side chains of L732 and P784 and is important for the structural organization of this region of the protein. Replacement of the small, branched valine side chain with the larger hydrophobic methionine (V871M), would increase contacts with the surrounding residues and promote stability. Moreover, the introduction of the larger methionine side chain in V871M would most likely perturb the position of the C-terminal helix of the OB fold. Perturbation of the C-terminal helix may have significant consequences in the context of the full-length protein and therefore the onset of bone marrow failure syndromes associated with this mutation.
To investigate the effect of the disease mutations in cells we generated cell lines stably expressing full length, ectopic, WT, double and deletion (F801A/R840A and L7823A/R871A, Δ726-876) hCtc1 and in an endogenous hCtc1 knockdown background, which eliminates ∼80% of the endogenous hCtc1 (Figure 4A and B). Pull-down assays clearly show that the double mutants interact efficiently with their interacting partners Stn1 and Ten1 (Figure 4C and D) suggesting that these mutations do not alter the ability of mutant hCtc1 to form the trimeric CST complex. However, the deletion mutant shows only an 80% efficiency in Stn1-Ten1 binding (Figure 4C and D), clearly suggesting a role of this domain is efficient CST assembly.
Moreover, ChIP assays show a reduction in the levels of full-length, hCtc1 at telomeres for the double and deletion mutants, suggesting an indirect role of hCtc1(OB) in full-length hCtc1 telomere localization (Figure 4E and F). In addition, Southern blot and ExoI analysis clearly indicate an increase in telomere length for the hCtc1 knockdown and deletion mutant (Figures 5 and 6). These data suggest a role of this domain of hCtc1 in regulation of telomerase access to the telomeric overhang consistent with reports of CST as a terminator of telomerase activity. Moreover, ExoI digest shows a 2-fold increase for the deletion hCtc1 mutant in internal, single stranded, telomeric DNA compared to the WT protein. The data suggest the presence of replication stress for the deletion mutant (Figure 6A and B) and therefore a role for the hCtc1(OB) in Polα recruitment to telomeres and C-strand synthesis consistent with what has been previously reported (34).
Although we observe loss of telomere localization for the mutant and deletion hCtc1 proteins (Figure 4E and F), we only see a defect in telomere length regulation for the deletion mutant (Figure 5A and B). This could be explained by the fact that only the deletion mutant is partially defective of CST assembly (efficient hCtc1 binding to Stn1–Ten1 - Figure 4C and D) and telomere length regulation. A defective CST complex, unable to efficiently inhibit telomerase dependent telomere replication and efficient recruitment of Polα to telomeres leads to telomere defects associated with missing telomere signal, fragile telomeres and chromosome fusions (Figure 7).
Taken together our structural and biochemical data suggest that the hCtc1(OB) is not involved in direct substrate binding, but plays a critical role in the structural and domain organization of the protein. While we cannot rule out the possibility that this domain may contribute to DNA binding in the context of the full-length protein in vivo, lack of conserved substrate binding amino acids argues that this domain does not contact DNA. We therefore suggest that the naturally occurring, disease mutations, located within the hCtc1(OB) alter the structural properties of the domain and most likely alter the structural and functional properties of the full-length protein. Although the effects of these mutations are subtle over the period of nine populations doublings, in the context of bone marrow syndromes, the onset of which may occur over a long period of time, these subtle changes may be highly significant leading to disease. These studies corroborate the role of hCtc1 in telomere length regulation and DNA replication, and further reveal the diverse roles of OB-fold domains in genome maintenance.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structure have been deposited to the Protein Data Bank (RCSB: http://www.rcsb.org/pdb/home/home.do) under accession number 5W2L.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Eric Babiash and Jingqi Duan for assistance with the SEC-MALS experiment and Michael Grasso for assistance with the protein thermostability assays.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NCI [1 RO1 CA201312–01]; NIGMS [5 R01 GM088332–03]; Wistar Cancer Center Support Grant [P30 CA10815]; NIH [RO1CA140652 to P.M.L.]. Funding for open access charge: NCI [RO1 CA201312]; NIH [RO1CA140652]; Wistar Cancer Center Support Grant [P30 CA10815].
Conflict of interest statement. None declared.
REFERENCES
- 1. de Lange T. How telomeres solve the end-protection problem. Science. 2009; 326:948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Viscardi V., Clerici M., Cartagena-Lirola H., Longhese M.P.. Telomeres and DNA damage checkpoints. Biochimie. 2005; 87:613–624. [DOI] [PubMed] [Google Scholar]
- 3. O'Sullivan R.J., Karlseder J.. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010; 11:171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005; 19:2100–2110. [DOI] [PubMed] [Google Scholar]
- 5. Miyake Y., Nakamura M., Nabetani A., Shimamura S., Tamura M., Yonehara S., Saito M., Ishikawa F.. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell. 2009; 36:193–206. [DOI] [PubMed] [Google Scholar]
- 6. Martin V., Du L.L., Rozenzhak S., Russell P.. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:14038–14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Surovtseva Y.V., Churikov D., Boltz K.A., Song X., Lamb J.C., Warrington R., Leehy K., Heacock M., Price C.M., Shippen D.E.. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell. 2009; 36:207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Linger B.R., Price C.M.. Conservation of telomere protein complexes: shuffling through evolution. Crit. Rev. Biochem. Mol. Biol. 2009; 44:434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Song X., Leehy K., Warrington R.T., Lamb J.C., Surovtseva Y.V., Shippen D.E.. STN1 protects chromosome ends in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:19815–19820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakaoka H., Nishiyama A., Saito M., Ishikawa F.. Xenopus laevis Ctc1-Stn1-Ten1 (xCST) protein complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J. Biol. Chem. 2012; 287:619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang C., Dai X., Chai W.. Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res. 2012; 22:1681–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakaoka H., Nishiyama A., Saito M., Ishikawa F.. Xenopus laevis Ctc1-Stn1-Ten1 (xCST) protein complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J. Biol. Chem. 2012; 287:619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng X., Hsu S.J., Kasbek C., Chaiken M., Price C.M.. CTC1-mediated C-strand fill-in is an essential step in telomere length maintenance. Nucleic Acids Res. 2017; 45:4281–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Derboven E., Ekker H., Kusenda B., Bulankova P., Riha K.. Role of STN1 and DNA polymerase alpha in telomere stability and genome-wide replication in Arabidopsis. PLoS Genet. 2014; 10:e1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang C., Dai X., Chai W.. Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res. 2012; 22:1681–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lue N.F., Chan J., Wright W.E., Hurwitz J.. The CDC13-STN1-TEN1 complex stimulates Pol alpha activity by promoting RNA priming and primase-to-polymerase switch. Nat. Commun. 2014; 5:5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stewart J.A., Wang F., Chaiken M.F., Kasbek C., Chastain P.D. 2nd, Wright W.E., Price C.M.. Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 2012; 31:3537–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gu P., Min J.N., Wang Y., Huang C., Peng T., Chai W., Chang S.. CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 2012; 31:2309–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bhattacharjee A., Stewart J., Chaiken M., Price C.M.. STN1 OB Fold Mutation Alters DNA Binding and Affects Selective Aspects of CST Function. PLoS Genet. 2016; 12:e1006342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Flynn R.L., Zou L.. Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Crit. Rev. Biochem. Mol. Biol. 2010; 45:266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Theobald D.L., Wuttke D.S.. Prediction of multiple tandem OB-fold domains in telomere end-binding proteins Pot1 and Cdc13. Structure. 2004; 12:1877–1879. [DOI] [PubMed] [Google Scholar]
- 22. Garvik B., Carson M., Hartwell L.. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. 1995; 15:6128–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin J.J., Zakian V.A.. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:13760–13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin Y.C., Shih J.W., Hsu C.L., Lin J.J.. Binding and partial denaturing of G-quartet DNA by Cdc13p of Saccharomyces cerevisiae. J. Biol. Chem. 2001; 276:47671–47674. [DOI] [PubMed] [Google Scholar]
- 25. Mitchell M.T., Smith J.S., Mason M., Harper S., Speicher D.W., Johnson F.B., Skordalakes E.. Cdc13 N-terminal dimerization, DNA binding, and telomere length regulation. Mol. Cell. Biol. 2010; 30:5325–5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mason M., Wanat J.J., Harper S., Schultz D.C., Speicher D.W., Johnson F.B., Skordalakes E.. Cdc13 OB2 dimerization required for productive Stn1 binding and efficient telomere maintenance. Structure. 2013; 21:109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu E.Y., Sun J., Lei M., Lue N.F.. Analyses of Candida Cdc13 orthologues revealed a novel OB fold dimer arrangement, dimerization-assisted DNA binding, and substantial structural differences between Cdc13 and RPA70. Mol. Cell. Biol. 2012; 32:186–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun J., Yang Y., Wan K., Mao N., Yu T.Y., Lin Y.C., DeZwaan D.C., Freeman B.C., Lin J.J., Lue N.F. et al. . Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha. Cell Res. 2011; 21:258–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rice C., Skordalakes E.. Structure and function of the telomeric CST complex. Comput Struct Biotechnol J. 2016; 14:161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hom R.A., Wuttke D.S.. Human CST Prefers G-Rich but Not Necessarily Telomeric Sequences. Biochemistry. 2017; 56:4210–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chandra A., Hughes T.R., Nugent C.I., Lundblad V.. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001; 15:404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Evans S.K., Lundblad V.. Est1 and Cdc13 as comediators of telomerase access. Science. 1999; 286:117–120. [DOI] [PubMed] [Google Scholar]
- 33. Mason M., Skordalakes E.. Insights into Cdc13 dependent telomere length regulation. Aging. 2010; 2:731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen L.Y., Majerska J., Lingner J.. Molecular basis of telomere syndrome caused by CTC1 mutations. Genes Dev. 2013; 27:2099–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen L.Y., Redon S., Lingner J.. The human CST complex is a terminator of telomerase activity. Nature. 2012; 488:540–544. [DOI] [PubMed] [Google Scholar]
- 36. Gu P., Chang S.. Functional characterization of human CTC1 mutations reveals novel mechanisms responsible for the pathogenesis of the telomere disease Coats plus. Aging Cell. 2013; 12:1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anderson B.H., Kasher P.R., Mayer J., Szynkiewicz M., Jenkinson E.M., Bhaskar S.S., Urquhart J.E., Daly S.B., Dickerson J.E., O'Sullivan J. et al. . Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat. Genet. 2012; 44:338–342. [DOI] [PubMed] [Google Scholar]
- 38. Keller R.B., Gagne K.E., Usmani G.N., Asdourian G.K., Williams D.A., Hofmann I., Agarwal S.. CTC1 Mutations in a patient with dyskeratosis congenita. Pediatr. Blood Cancer. 2012; 59:311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Polvi A., Linnankivi T., Kivela T., Herva R., Keating J.P., Makitie O., Pareyson D., Vainionpaa L., Lahtinen J., Hovatta I. et al. . Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. Am. J. Hum. Genet. 2012; 90:540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Romaniello R., Arrigoni F., Citterio A., Tonelli A., Sforzini C., Rizzari C., Pessina M., Triulzi F., Bassi M.T., Borgatti R.. Cerebroretinal microangiopathy with calcifications and cysts associated with CTC1 and NDP mutations. J. Child Neurol. 2012; 28:1702–1708. [DOI] [PubMed] [Google Scholar]
- 41. Walne A.J., Bhagat T., Kirwan M., Gitiaux C., Desguerre I., Leonard N., Nogales E., Vulliamy T., Dokal I.S.. Mutations in the telomere capping complex in bone marrow failure and related syndromes. Haematologica. 2013; 98:334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J. et al. . Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011; 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kelley L.A., Sternberg M.J.. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009; 4:363–371. [DOI] [PubMed] [Google Scholar]
- 44. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 45. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 47. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 48. Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976; 72:248–254. [DOI] [PubMed] [Google Scholar]
- 49. Bryan C., Rice C., Harkisheimer M., Schultz D.C., Skordalakes E.. Structure of the human telomeric Stn1-Ten1 capping complex. PLoS One. 2013; 8:e66756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gupta K., Martin R., Sharp R., Sarachan K.L., Ninan N.S., Van Duyne G.D.. Oligomeric properties of survival motor neuron.gemin2 complexes. J. Biol. Chem. 2015; 290:20185–20199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gohring J., Fulcher N., Jacak J., Riha K.. TeloTool: a new tool for telomere length measurement from terminal restriction fragment analysis with improved probe intensity correction. Nucleic Acids Res. 2014; 42:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fan J., Pavletich N.P.. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012; 26:2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structure have been deposited to the Protein Data Bank (RCSB: http://www.rcsb.org/pdb/home/home.do) under accession number 5W2L.