Abstract
13C labeling experiments in aerobic glucose limited cultures of Saccharomyces cerevisiae at four different growth rates (0.054; 0.101, 0.207, 0.307 h−1) are used for calculating fluxes that include intracellular cycles (e.g., storage carbohydrate cycles, exchange fluxes with amino acids), which are rearranged depending on the growth rate. At low growth rates the impact of the storage carbohydrate recycle is relatively more significant than at high growth rates due to a higher concentration of these materials in the cell (up to 560-fold) and higher fluxes relative to the glucose uptake rate (up to 16%). Experimental observations suggest that glucose can be exported to the extracellular space, and that its source is related to storage carbohydrates, most likely via the export and subsequent extracellular breakdown of trehalose. This hypothesis is strongly supported by 13C-labeling experimental data, measured extracellular trehalose, and the corresponding flux estimations.
Abbreviations: 2PG, 2-phosphoglycerate; 3PG, 3-phosphoglycerate; 6PG, 6-phospho gluconate; α-KG, oxoglutarate; ALA, alanine; ASP, aspartate; CoA, coenzyme-A; DHAP, dihydroxy acetone phosphate; E4P, erythrose-4-phosphate; F6P, fructose-6-phosphate; FBP, fructose-1,6-bis-phosphate; FUM, fumarate; G1P, glucose-1-phosphate; G6P, glucose-6-phosphate; GAP, glyceraldehyde-3-phosphate; GLN, glutamine; GLU, glutamate; GLY, glycine; Iso-Cit, isocitrate; LEU, leucine; LYS, lysine; MAL, malate; METH, methionine; OAA, oxaloacetate; PEP, phospho-enol-pyruvate; PRO, proline; PYR, pyruvate; Rib5P, ribose-5-phosphate; Ribu5P, ribulose-5-phosphate; SER, serine; S7P, sedoheptulose-7-phosphate; SUC, succinate; T6P, trehalose-6-phosphate; UDPG, UDP-glucose; UDP, uridine-5-diphosphate; UTP, uridine-5-triphosphate; X5P, xylulose-5-phosphate; ACO, aconitate hydratase; AK, adenylate kinase; ENO, phosphopyruvate hydratase; FBA, fructose-bisphosphate aldolase; FMH, fumarate hydratase; G6PDH, glucose-6-phosphate dehydrogenase; GAPDH&PGK, glyceraldehyde-3-phosphate dehydrogenase+phosphoglycerate kinase; GPM, phosphoglycerate mutase; PFK, 6-phosphofructokinase; PGI, glucose-6-phosphate isomerase; PGM, phosphoglucomutase; PMI, mannose-6-phosphate isomerase; PYK, pyruvate kinase; RPE, ribulose-phosphate 3-epimerase; RPI, ribose-5-phosphate isomerase; TPP, trehalose- phosphatase; TPS, alpha,alpha-trehalose-phosphate synthase; DO, dissolved oxygen; IDMS, Isotope dilution mass spectrometry; OUR, Oxygen uptake rate; PPP, pentose phosphate pathway; TCA, tricarboxylic acid cycle.
Keywords: Non-stationary 13C labeling, Flux estimation, Trehalose, Glycogen, Amino acids
Highlights
-
•
Cyclic fluxes between large metabolite pools and central C-metabolism are studied.
-
•
Cyclic fluxes are rearranged depending on growth rate.
-
•
Impact of storage carbohydrate recycle is more significant at low growth rates.
-
•
Impact of exchange fluxes with amino acids is more significant at high growth rates.
-
•
Trehalose export and degradation seem to play a major role in glucose recycle.
1. Introduction
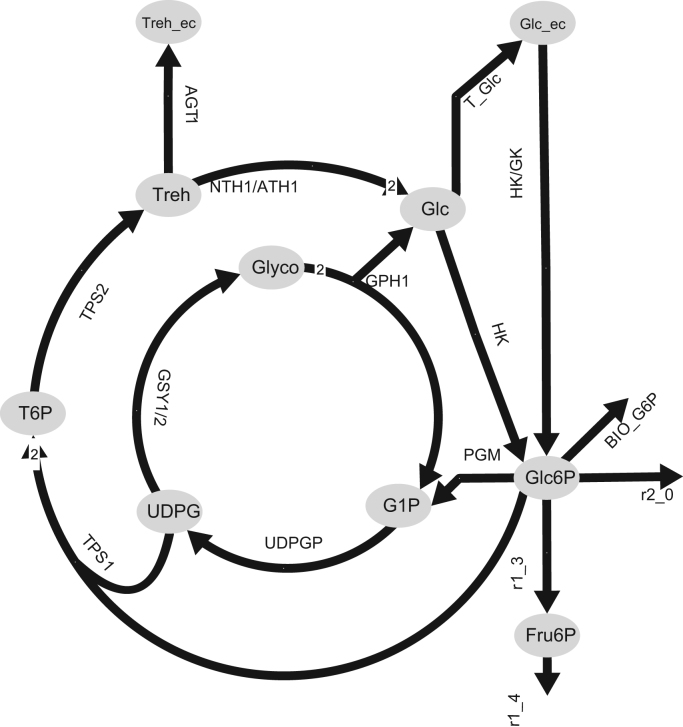
The yeast Saccharomyces cerevisiae is one of the major workhorses used in biotechnology for producing pharmaceuticals, biofuels (Hashem and Darwish, 2010), and bulk chemicals (Willke and Vorlop, 2004). S. cerevisiae is also an important model organism for studying the physiology (Pereira et al., 2001), genetics and metabolic mechanisms of eukaryotes (Castrillo et al., 2007). Estimating fluxes of metabolic network reactions accurately is crucial for metabolic applications. Recently, the metabolic flux of the different storage nodes has received more attention as its interaction with the central carbon metabolism seems to have a significant impact when estimating flux distributions and studying intracellular dynamics (Aboka et al., 2009, van Heerden et al., 2014). Trehalose and glycogen are reported to be the largest carbohydrate pools in S. cerevisiae (François and Parrou, 2001), and can represent up to 30% of the dry cell weight (Parrou et al., 2005). They mainly function as energy storage and stress protectant (Parrou et al., 2005, Stambuk et al., 1996). The pathways for synthesis and degradation of both, trehalose and glycogen, are well documented (Daran et al., 1995, Peng et al., 1990), indicating that these two processes may occur simultaneously (Fig. 1), generating a futile cycle that consumes ATP (Mashego et al., 2004). Glycogen and trehalose concentrations are found to be negatively correlated with growth rate, and linked with cell cycle progression (Paalman et al., 2003, Shi et al., 2010). Aboka et al. (2009) observed that in chemostat cultivations a shift up in the glucose uptake (i.e., increased growth rate from 0.05 to 0.075 h−1) counter-intuitively triggered a temporary mobilization of storage carbohydrates into the glycolysis that reached about one third of the glycolytic flux. In addition, Shi et al. (2010) suggested that trehalose was a key metabolite for the entrance of Saccharomyces cerevisiae into the quiescent metabolic state (G0 phase) when growing under limiting substrate conditions. Although, only a fraction of the cells entered into this state. These observations suggest that non-homogeneous populations (i.e., cells at different stages of the cell cycle) could be relevant when studying the metabolism of trehalose and glycogen in yeast.
Fig. 1.
Metabolic reactions of the storage carbohydrates pathway in S. cerevisiae. Letters in italics represent the enzyme/reaction. All metabolites are intracellular except those identified with –ec, which stands for extracellular. Double arrows indicate reversible reactions.
Despite significant molecular knowledge on storage carbohydrate metabolism, quantitative measurements of the involved rates at different growth rates are not yet available. Especially, it remains unclear how this glucose recycle influences fluxes of the central carbon metabolism. Metabolic Flux Analysis (MFA) is commonly used to characterize the in vivo steady-state (flux) of a metabolic network (Vallino and Stephanopoulos, 1993, Van Gulik and Heijnen, 1995, Varma and Palsson, 1994). While frequently applied, there are intrinsic limitations that have to be compensated by additional assumptions, like optimizing the biomass yield, minimizing the redox potential or maximizing the ATP yield. In practice, it has been observed that these assumptions do not predict the phenotypes of organisms correctly, probably because the organism's objective is more complex (Fischer and Sauer, 2005, Schmidt et al., 1998, Schuetz et al., 2007).
To minimize these assumptions while also quantifying intracellular cycles, tracer experiments are performed (Wiechert and De Graaf, 1997). 13C MFA does not rely on cofactor balances, but on the 13C enrichment of metabolites (Dauner et al., 2000, van Winden et al., 2002, Wiechert, 2001). State of the art 13C MFA relies on transient labeling enrichment measurements of intracellular metabolites combined with isotopomer modeling (Crown and Antoniewicz, 2013, Murphy et al., 2013, Nöh et al., 2007, Noh et al., 2006, Wahl et al., 2008, Young et al., 2011). Nevertheless, depending on the network complexity and available measurements, a priori assumptions can be required for anaplerotic reactions, transamination reactions, storage metabolism, protein turnover and mRNA degradation (Wiechert and Nöh, 2013).
Focus of this this work are the storage carbohydrate recycle fluxes and its relation with the central metabolism in dependence of growth rate (here we used D=0.307, 0.207, 0.101 and 0.054 h−1). Additionally, other cyclic fluxes that interact with the central metabolism are quantified. Especially, exchange fluxes with large pools of amino acids, RNA and lipids. To do so, 13C labeling experiments with uniformly labeled glucose were performed on cultures of Saccharomyces cerevisiae at metabolic steady state.
2. Material and methods
2.1. Strain and culture conditions
A cryopreserved stock culture (glycerol, −80 °C) of the haploid yeast Saccharomyces cerevisiae CEN PK 113-7D obtained from the Centraalbureau van Schimmelcultures (Fungal Biodiversity Centre, Utrecht, The Netherlands) was used. All cultivations were performed using a low-salt Verduyn minimal medium (Canelas et al., 2009) with a glucose concentration of 7.5 g L−1. Ethanol was not added to the medium since no oscillations were observed under these conditions. Microorganisms from one cryo-vial were used for the seed culture and grown for 10 h in 1L-Erlenmeyer flasks containing 100 mL medium at a temperature of 30 °C and a shaker speed of 200 rpm. The pre-culture was used to inoculate a 2L-bioreactor (Applikon, Delft-The Netherlands) operated with a constant working volume of 0.952 L (D=0.307, 0.207 and 0.101 h−1) and 0.917 L in case of D=0.054 h−1. The aeration was performed using pressurized air at 0.249 L min−1 (approx. 0.25 vvm) and a stirrer speed of 600 rpm, which were sufficient to keep the DO between 20% at D=0.307 h−1 and 80% at D=0.054 h−1. The broth was maintained at pH 5.0 by adding either 4 M KOH or 2 M H2SO4. The temperature was controlled at 30 °C and the pressure was kept at 0.3 bar (overpressure.)
2.2. Chemostat cultivation
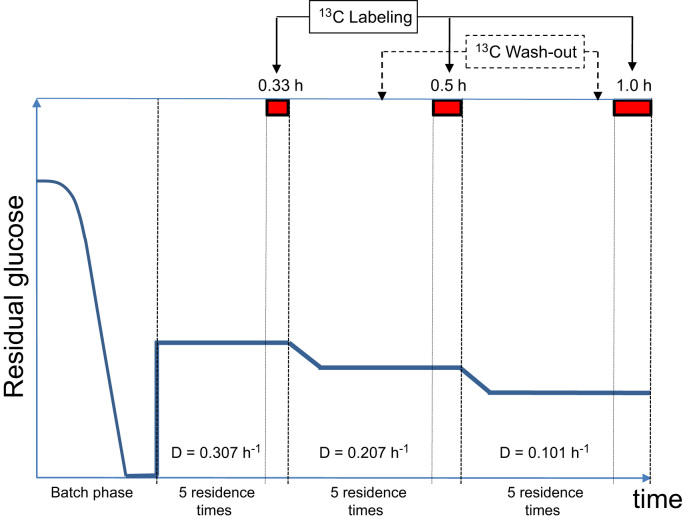
Once the batch phase was complete (indicated by a strong and fast CO2 decrease and dissolved oxygen (DO) increase to almost saturation), the chemostat phase was started at a dilution rate of 0.307 h−1. Sampling for concentrations of extra and intracellular metabolites at steady-state was performed after 5 residence times, followed by the 13C-labeling experiment by switching to a medium that contained U-13C-glucose with the same molar concentration. The duration of the 13C-labeling experiment depended on the dilution rate (Fig. 2). After the end of each labeling experiment, the medium was switched back to 12C medium and the flow rate was shifted down (i.e., D=0.207; 0.101 h−1). In between dilution rate shifts, the culture was run for at least five residence times in order to remove at least 99% of the 13C material.
Fig. 2.
Experimental design using dilution rate shifts. Solid lines represent the residual glucose concentration. Bars plus numbers at the top indicate the length of the labeling period. Vertical dotted lines indicate the start of the 13C labeling experiment. After the end of the 13C wash-in (indicated by the vertical dashed lines) the medium was switched to the non-labeled substrate and the dilution rate was shifted down.
Mashego et al. (2005) reported that significant changes in the metabolome of Saccharomyces cerevisiae were observed after about 20 residence times, most likely due to evolution. In this work, about 15 generations were reached after running the culture at three different dilution rates (D=0.307, 0.207, 0.101 h−1). Therefore, a new experiment was started to perform the 13C flux analysis at D=0.054 h−1 following a similar scheme (batch, 5 residence times to reach steady state and 13C labeling for 3 hours).
2.3. Data acquisition, sample processing and analysis
The O2 and CO2 volume fractions in the dried off-gas were measured on-line by a combined paramagnetic/infrared NGA2000 off-gas analyzer (Rosemount Analytics, CA, USA). Dissolved oxygen (DO) was measured by a polarographic ADI dO2 sensor (AppliSens, Applikon Dependable Instruments, Delft-The Netherlands) and monitored on-line. Biomass concentration was determined by a gravimetrical method (dry weight): 15 mL of broth were filtered through a pre-dried and pre-weighed membrane (Supor-450, 0.45 μm, 47 mm, Pall Corporation). Membranes containing the biomass cake were dried at 70 °C for 72 h and cooled to room temperature in a desiccator before weighing.
2.3.1. Extracellular metabolites
Broth samples for extra- and intracellular metabolite concentration measurements were rapidly withdrawn using two separate rapid-sampling ports. For quantification of extracellular metabolite concentrations, 1.5 mL of broth were withdrawn into a syringe containing pre-cooled (−20 °C) stainless steel beads (approx. 26 g). The cooled broth (~1 °C) was immediately filtered as described by Mashego et al. (2006). Extracellular (filtrate) glucose, ethanol, acetate, glycerol and trehalose concentrations were determined by one/all of the following three methods: GC–MS, HPLC, or enzymatic assays (Canelas et al., 2011). We found that glucose determination by HPLC was not sufficiently precise for concentrations below 0.5 mM. Based on a series of glucose standards we found that the enzymatic and GC–MS methods performed well for concentrations below 0.5 mM with the latter being the most accurate method when the concentration was below 0.2 mM.
For GC–MS analysis, 100 μL of filtrate and 20 μL of 13C cell extract were transferred into a GC-glass vial. The mixture was subsequently frozen and kept at −80 °C until further processing. The IDMS method described in Cipollina et al. (2009) and (Wahl et al., 2014) was used for metabolite quantification. For HPLC analysis, 10 μL of the sample were automatically injected into the HPLC (Bio-Rad AminexHPX-87H column, at 60 °C, 0.6 mL min−1 1.7 mM phosphoric acid as eluent, coupled to a Waters 2414 RI detector and a Waters 2489 UV detector at 210 nm). For the enzymatic analysis an assay kit (d-Glucose, Boehringer Mannheim) was used according to the manufacturer's instructions but scaled down to microtiter plates.
2.3.2. Intracellular metabolites
For quantification of intracellular metabolite concentrations, 1.0 mL-broth samples were rapidly withdrawn and quenched in pre-weighed tubes containing 5 mL cold (−40 °C) pure methanol (Canelas et al., 2009) followed immediately by vigorous vortexing. The quenched samples were rapidly weighed and poured into a filtration device containing a cellulose membrane (Supor-200, 0.2 μm, 47 mm, Pall Corporation) and previously layered with 15 mL of cold methanol (−40 °C). Subsequently, a vacuum was applied followed by an immediate additional washing step with 15 mL cold methanol (−40 °C). The filter containing the cold washed biomass was then transferred into a 50 mL-falcon tube (BD Biosciences) containing 30 mL of preheated (75 °C) aqueous ethanol solution (75% v/v). 100 μL of 13C cell extract was added to the tube as an internal standard (Wu et al., 2005). The tube containing the sample was then tightly closed, shaken vigorously, and placed into a water bath at 95 °C during 3 min for metabolite extraction. The tubes were then cooled using an ice bath and the filter was removed. This extract was then concentrated by complete evaporation of the ethanol–water mixture under vacuum (Mashego et al., 2004), and resuspended in 500 μL milliQ water. After a first centrifugation at 15000 g for 5 min at 1 °C, the supernatant was transferred into a centrifugal filter unit (Ultrafree MC-ML, Millipore, MA, USA) and centrifuged again at the same conditions. The filtrate was placed into a screw-capped polypropylene vial and stored at −80 °C until further analysis. Samples were analyzed by GC–MS (Cipollina et al., 2009, de Jonge et al., 2011, Mashego et al., 2004, Wu et al., 2005) and/or LC–MS (Dam et al., 2011, Seifar et al., 2009).
2.3.3. Mass isotopomer measurements
Samples for quantification of mass isotopomer enrichments were taken following the same procedure described above for concentrations of extra- and intracellular metabolites with the exception that no 13C cell extract was added. The mass isotopomer samples were then analyzed by LC–MS (Van Winden et al., 2005) and GC–MS (Canelas et al., 2009, Cipollina et al., 2009, Dam et al., 2011, de Jonge et al., 2011). The measured data was processed to correct for the natural isotopologues of non-carbon atoms (Wahl et al., 2004).
2.3.4. Glycogen determination
For glycogen determination, 1 mL broth was withdrawn from the reactor and quenched in 5 mL cold methanol (−40 °C). The suspension was centrifuged (−19 °C, 2000 rpm, 5 min) and the supernatant was discarded. The pellet was processed according to the method of Parrou et al. (1997) with adjustments by Conte (2011) using amyloglucosidase (Sigma 10115). The enzymatic digestion lasted for 15.5 h at 57 °C, and the glycogen concentration was determined by enzymatic assay based measurement of glucose (as described above).
2.3.5. Rate reconciliation
The biomass specific uptake and secretion rates were reconciled using the approach of Verheijen (2010). A constant elemental biomass composition and molar weight of 26.4 gDW C mol−1 (Lange and Heijnen, 2001) was used for all conditions.
2.4. Metabolic network and model structure
The metabolic network (Supplementary material 1) used for the flux estimation was constructed based on a published stoichiometric model (Daran-Lapujade et al., 2004). This model was extended with reactions of the storage metabolism (trehalose and glycogen), reactions of the pentose phosphate pathway (PPP), and exchange fluxes with the large amino acid pools (alanine, valine, aspartate and glutamate). For parameter estimation and modeling, the following assumptions were made:
-
i.
The extracellular fluxes were fixed to the reconciled specific uptake and secretion rates;
-
ii.
To account for a compartmentalization of pyruvate, this pool was split into two parts: mitochondrial and cytosolic. A variable ‘yPyr_frac’ was introduced representing the cytosolic pyruvate fraction that was included in the parameter estimation;
-
iii.
To reduce the amount of cumomers, the molecule trehalose-6-phosphate (T6P) was considered as two separate molecules: T6Pa produced from G6P and T6Pb produced from UDPG. Both were produced via reaction Tre1: G6P+UDPG>T6Pa+T6Pb. Then the total T6P pool consisted of (T6Pa+T6Pb), where T6Pa=T6Pb;
-
iv.
Production of trehalose was represented by two reactions: Tre2: T6Pa <> Trehalose, and Tre2c: T6Pb <> Trehalose. Trehalose was then considered as a 6-C molecule produced via Tre2 and Tre2c. Trehalose degradation to intracellular glucose is described by the Tre3 reaction (Treh>Glc_ic). Extracellular trehalose is produced via Treh_trans and degraded via Tre4;
-
v.
In the model, conversion of T6P to trehalose (Tre2 and Tre2c) was considered to operate bidirectional in order to account for a yet unknown mechanism producing unlabeled T6P (see later a discussion on this reaction);
-
vi.
Two reactions named deg_prod_in (for forward flux, prod>ACCOA ) and deg_prod_out (for backward flux, ACCOA>prod) were used to account for unknown sources and sinks of acetyl-CoA. The combined two reactions had a net flux equal to zero;
-
vii.
Fluxes (TCA5, TCA6), which produce symmetrical metabolites were split equally (i.e. 1:1);
-
viii.
To obtain a better reproduction of the Rib5P and G3P mass isotopomers, two exchange reactions were introduced: 1- mRNA_exchange (Rib5P <> mRNA) and 2- FA_metabolism (G3P <> Fatty_acid), which accounted for the net result of degradation and synthesis of mRNA and lipids respectively.
The final network consisted of 49 balanced metabolites and 109 fluxes including 30 backward fluxes. The mass isotopomer measurements of 31 metabolites were used for the parameter estimation of 34 free fluxes. A cytosolic fraction of pyruvate (yPyr_frac) and citrate (yCit_frac) were used to account for compartmentalization of these pools. Of the 49 intracellular metabolite concentrations, 39 have been measured, 8 concentrations (intracellular pools of ethanol, acetate, CO2, mRNA, lipids, OAA, Glyoxylate and acetaldehyde) were assumed based on a previous work (Jordà et al., 2013). Effluxes for biomass formation were calculated based on the biomass composition reported in Lange and Heijnen (2001). These authors measured the content of protein, carbohydrate, lipids and RNA/DNA under substrate limited conditions for D=0.052, 0.107 and 0.211 h−1. It was assumed that the biomass composition at D=0.307 h−1 was comparable to D=0.211 h−1. Detailed values can be found in the Supplementary material 1.
The cumomer representation was used (Wiechert, 2001) for simulation and parameter estimation, and gPROMS Model Builder 4.0 (PSE Limited, London) was used as the computing platform. The gPROMS parameter estimation routine is based on maximum likelihood (NLPSQP iterative algorithm). The accuracy of the mass isotopomer distribution measurements was set to a constant variance of 0.005 corresponding to a constant standard deviation of 7.1%. Note that this standard deviation is an estimate including variance originating from minimal deviations in sampling time, processing, and analytical reproducibility, which was found in the order of 0.5–2% for most metabolites (Niedenführ et al., 2015).
The parameter optimization was initialized from different starting values. The result was considered acceptable when the obtained solutions converged for key fluxes, i.e., the values for storage degradation and the flux into pentose phosphate pathway. The residual sum of squares was higher than the chi-square value with a 95% confidence for the corresponding degrees of freedom (total number of experimental points minus number of parameters), which suggests that some experimental data are not sufficiently reproduced by the model. It has been noted that most of the contribution to the deviation is originated in some metabolites of the TCA cycle, which may be related to compartmentalization. Although this issue needs to be resolved in future projects, here, we accepted those deviations since they have no impact on the fluxes that are relevant for this study, i.e., trehalose and glycogen recycling as well as the oxidative pentose phosphate pathway.
3. Results and discussion
3.1. Metabolite concentrations and extracellular rates
The carbon and degree of reduction balances closed with a recovery of 100±2% for all the dilution rates (Table 1). From D=0.054 h−1 up to 0.307 h−1 the glucose uptake rate changed about 7.5-fold (see Table 2 and Supplementary material 2 containing a list of metabolite concentrations and data quality check). The biomass yield was significantly lower at D=0.307 h−1 due to the secretion of ethanol, which is in agreement with previous reports (Van Hoek et al., 1998). A small amount of acetate was produced at all dilution rates, which did not follow a specific trend, especially the concentration measured at D=0.101 h−1 deviated from the other dilution rates. In all cases neither extracellular glycerol nor succinate were observed. All other reconciled specific rates were in agreement with previous reports (Aboka et al., 2012, Daran-Lapujade et al., 2004, Gombert et al., 2001).
Table 1.
Macroscopic measurements for the different dilution rates.
| Dilution rate (h−1) | 0.054 | 0.101 | 0.207 | 0.307 |
|---|---|---|---|---|
| Biomass concentration () | 3.51±0.01 | 3.64±0.01 | 3.60±0.01 | 2.60±0.01 |
| CO2_out (%) | 0.86±0.01 | 1.51±0.01 | 3.11±0.02 | 4.13±0.02 |
| O2_out (%) | 20.6±0.1 | 19.6±0.1 | 18.2±0.1 | 18.1±0.1 |
| Ethanol concentration (mM) | b.d. | b.d. | b.d. | 23.46±0.66 |
| Residual Glucose concentration (mM) | 0.071±0.001 | 0.183±0.002 | 0.185±0.006 | 0.420±0.011 |
| Acetate concentration (mM) | 1.64±0.02 | 3.58±0.07 | 1.14±0.05 | 1.68±0.02 |
| Extracellular trehalose concentration (μM) | 4.31±0.01 | 5.96±0.14 | 0.41±0.16 | 0.35±0.14 |
b.d: below detection limit. Values are the average of three different samples taken at different times after 5 residence times had elapsed for each chemostat or dilution rate.
Table 2.
Reconciled biomass specific conversion rates during stepwise experiment (all rates in μmol h−1).
| Dilution rate (h−1) | 0.054 | 0.101 | 0.207 | 0.307 |
|---|---|---|---|---|
| Biomass | 2050±8 | 3822±9 | 7838±12 | 11618±13 |
| Glucose (−qS) | 632±15 | 1141±25 | 2321±45 | 4755±68 |
| Ethanol (qEth) | 0±0 | 0±0 | 0±0 | 2781±110 |
| Acetate (qAcet) | 25±1 | 99±5 | 65±4 | 195±4 |
| Oxygen (−qO) | 1596±87 | 2660±140 | 5600±270 | 7650±140 |
| CO2 (qC) | 1688±87 | 2830±140 | 5960±270 | 10960±210 |
3.2. The storage carbohydrates and free amino acids are the major C-storage pools at different growth rates
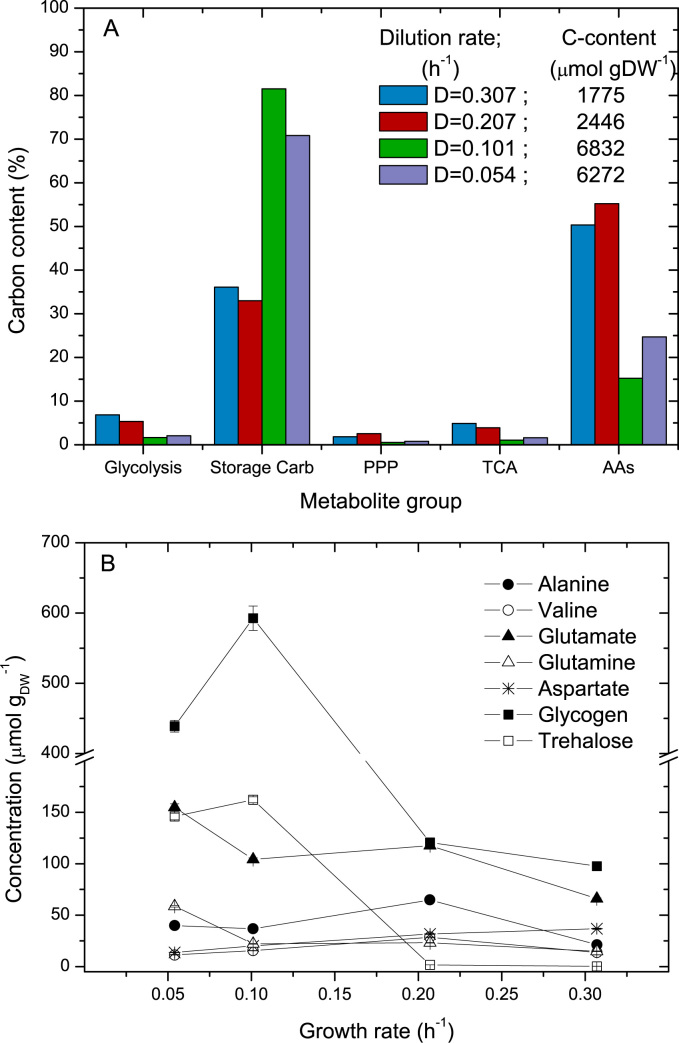
Storage carbohydrates as well as free amino acids pools are closely connected with central carbon metabolites and exhibit a significantly higher concentrations. We consider that these large, connected pools can act as buffers that maintain concentrations of the central carbon metabolites balanced. Hence, rapid changes in metabolic flux are counterbalanced by consuming or producing carbon from storage pools (Fig. 3A). At low growth rates storage carbohydrates are present at high concentrations and their level decreases with growth rate, while amino acids become the largest pools at high growth rates. In case of a sudden lack of carbon/energy supply in the system, these metabolites can help microorganisms to keep their function over a period of time.
Fig. 3.
Buffering capacity of the different metabolite groups. A: Relative carbon content grouped by metabolite class with respect to the total carbon contained in all the measured free intracellular metabolites at the different growth rates. B: Intracellular concentration of storage carbohydrates (trehalose and glycogen) and selected free amino acids. Glycogen concentration is given as glucose equivalents.
Trehalose and glycogen varied in concentration up to 560-fold (Fig. 3B). For instance, trehalose ranged between 0.29±0.02 μmol at D=0.307 h−1 up to 162±3 μmol at 0.101 h−1. The size of the storage pool was expected to be higher at lower dilution rates (Guillou et al., 2004), surprisingly, in our experiment we found that at D=0.054 h−1 both glycogen and trehalose were slightly lower than at D=0.101 h−1. Although, no hypothesis is yet available for this behavior, the values obtained in this experiment for D=0.054 h−1 are in agreement with previous observations (Guillou et al., 2004, Mashego et al., 2005, Mashego et al., 2007).
Though the amino acid pools were not as large as storage carbohydrates, these pools deserve attention, especially with respect to the buffering role they may have when interacting with metabolites of the central carbon metabolism (e.g., alanine – pyruvate, glutamate – α-ketoglutarate, and aspartate – oxaloacetate). In particular, we observed that the largest amino acid pools (alanine, valine, aspartate, glutamine and glutamate) showed a tendency to increase with decreasing growth rates. Contrary to the other amino acids, aspartate increased with the growth rate indicating that this amino acid was consistently growth rate related. Given the pool size of these amino acids and their known interactions with organic acids via transamination reactions, they should be considered when estimating carbon fluxes from dynamic 13C experiments, especially at high growth rates where the pools of free amino acids contain about 5 times more carbon than the sum of all central carbon metabolites and nearly 2 times larger than the storage carbohydrates pools.
3.3. Dynamics of the 13C enrichment
The transient mass isotopomer measurements are given in Supplementary material 3. Inspection of the labeling pattern revealed the presence of cyclic fluxes of unlabeled carbon from large intracellular pools (e.g., storage carbohydrates and amino acids). These results include the time profiles of the experimentally measured mass isotopomers, as well as the time profiles resulting from the simulation using the best flux estimate.
3.3.1. Extracellular glucose
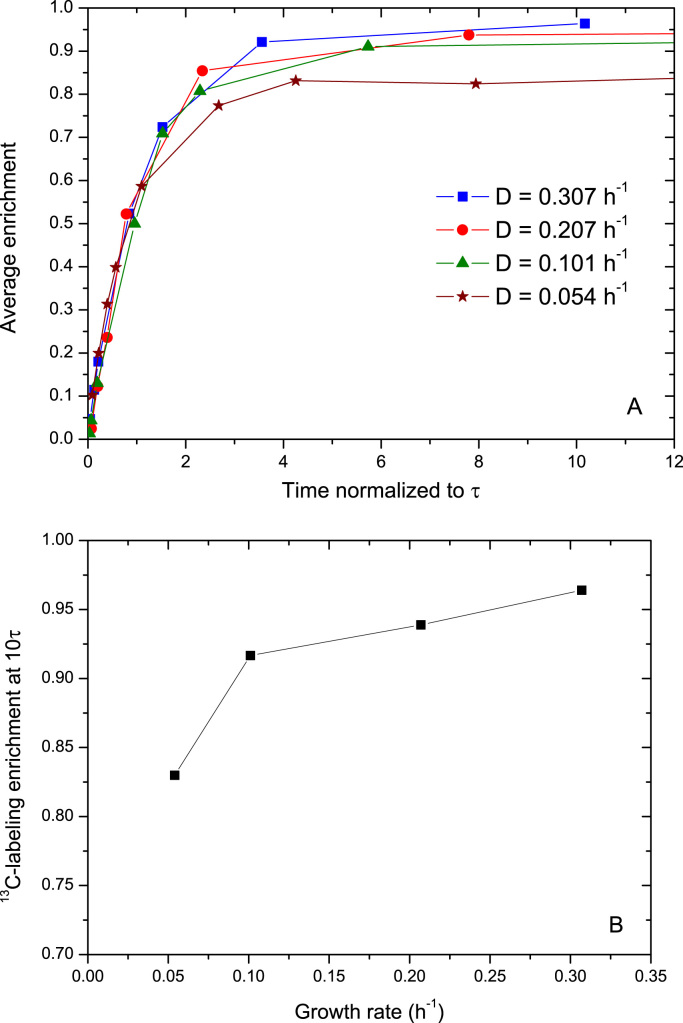
The carbon enrichment of extracellular glucose in time is influenced by: the 13C feed flow rate, the specific glucose uptake rate, and the residual 12C-glucose concentration. Considering that the measured enrichment of the glucose added in the fresh medium was 98.75%, it is expected that once the 12C-glucose is fully replaced (after about 5 turnover times), this enrichment level will be reached. Turnover times (τ) were 118, 77, 157 and 114 seconds for D=0.307, 0.207, 0.101 and 0.054 h−1, respectively. Accordingly, the fastest extracellular glucose enrichment was observed at D=0.207 h−1, while at D=0.307 and D=0.054 h−1 were similar and the slowest was at D=0.101 h−1 (Supplementary material 3).
While the observed transients until about 1.5τ seem to be consistent with the expectations (Fig. 4A), the 13C-labeling enrichment reached is lower and seems to correlate with the growth rate (i.e., the higher the growth rate, the higher the maximum level of enrichment, see Fig. 4B). For instance, after 10τ, the maximum enrichment reached was 96.4% at D=0.307 h−1, while it was only 83.0% at D=0.054 h−1. A number of tests (Supplementary material 4) were performed to discard measurement artifacts like contamination of samples with unlabeled glucose, cold-shock effect when quenching the samples, etc. All tests indicated that the observations must be the result of an unknown mechanism that provides unlabeled glucose, since no other extra source was present.
Fig. 4.
Carbon enrichment of extracellular glucose. A: Dynamics of the 13C enrichment at different dilution rates as a function of time normalized by the turnover time of the extracellular glucose pool (experimental time/τ); B: 13C-labeling enrichment at 10τ as a function of the growth rate.
Moreover, given the large discrepancy, a biological source of unlabeled glucose must be present. We hypothesize that this source is originating from the export and subsequent hydrolysis of trehalose. Our observations that the intracellular trehalose concentration was higher at low dilution rates (Fig. 3B) with a lower level of enrichment (Supplementary material 3) support this hypothesis, since it is consistent with a higher trehalose efflux leading to a lower level of glucose enrichment (Fig. 4B). In addition, we found trehalose in the extracellular space (Table 1), although this metabolite was not detected in the fresh medium. The ratio of intracellular to extracellular trehalose concentration (expressed as M/M) varied from 487 at D=0.307 h−1 to almost 2×104 at D=0.054 h−1 indicating that trehalose export would be favored at low growth rates. To test this mechanism, we included an export of unlabeled glucose from hydrolysis of trehalose in our model.
3.3.2. Metabolites of the glycolysis
Though slightly slower at low dilution rates, the 13C-labeling of metabolites in the upper glycolysis showed a pattern that was consistent with the extracellular glucose labeling, e.g., with similar dynamics and similar maximum level of enrichment. The enrichment dynamics of G6P, F6P and M6P were nearly the same indicating that glucose-6-phosphate isomerase (PGI) and mannose-6-phosphate isomerase (PMI) reactions operate bidirectional. This was also supported by the corresponding mass action ratios (see Supplementary material 2).
At D=0.101 and 0.054 h−1, the enrichment of FBP was slightly lower than those metabolites in both, the upper and the lower glycolysis. Nevertheless, this behavior may be the consequence of very low FBP concentrations at low growth rates, which might bias the MS measurements. The nearly identical enrichment transients of the lower glycolytic metabolites (3PG, 2PG and PEP) suggest that they are linked through pseudo equilibrium reactions (e.g., GPM and ENO), as confirmed by the corresponding mass action ratios (see Supplementary material 2). Pyruvate showed a slightly lower 13C-enrichment than metabolites upstream, especially at higher growth rates (D=0.207 and 0.307 h−1). In the model we therefore included an exchange flux with alanine (cytosolic) and separated the pool into a cytosolic and mitochondrial compartments (see Supplementary material 1).
3.3.3. Metabolites of the storage carbohydrate branches (glycogen and trehalose)
Glycogen is produced from G1P and UDPG (Voit, 2003) with the latter being a common metabolite for the synthesis of trehalose. Unfortunately, in this study we did not measure the enrichment of glycogen. Nevertheless, there is some indication that glycogen recycling takes place and could be estimated. We observed that the transient G1P enrichment was slower than its precursor G6P, which could originate from glycogen degradation. Moreover, the difference between G1P and G6P was more pronounced at low growth rates where storage carbohydrate pools were higher.
The 13C-labeling of UDPG showed a behavior similar to G1P, but trehalose-6-phosphate remarkably deviated from this transient. T6P is the lowest concentrated pool, varying from about 0.34±0.01 μmol at D=0.054 h−1 to 0.016±0.001 μmol at D=0.307 h−1. T6P is synthesized from UDPG and G6P by trehalose-6-phosphate synthase, and consumed by T6P phosphatase. With only these reactions in place, the observed enrichment pattern of T6P could not be explained, i.e., the T6P enrichment would reach a level comparable to UDPG and G6P. A lower level of enrichment (e.g., about 45% at D=0.054 h−1) can only be obtained if there is a source of unlabeled T6P. Moreover, we have observed a similar behavior in other labeling experiments (data not shown) indicating that the low T6P enrichment is a recurrent phenomenon. Although there is no reported evidence, we included a putative reaction that produces T6P from trehalose.
3.3.4. Metabolites of the pentose phosphate pathway, TCA cycle and amino acids
With the exception of Rib5P, metabolites of the pentose phosphate pathway followed the dynamics of upper glycolysis. Rib5P showed a slower and lower enrichment, which we assumed to originate from RNA degradation (either messenger or ribosomal). Metabolites of the TCA cycle enriched very slow showing some peculiar behavior. Especially, a higher labeling enrichment was observed for iso-citrate compared to its precursor citrate. These observations suggest differences in compartmentalization and interactions between carboxylic acids (e.g., pyruvate, α-KG) with corresponding amino acids via transamination reactions. Pool compartmentalization alone cannot fully explain the observed behavior. Next to it there has to be compartmentalization of the metabolic activity. For instance, some metabolites of the TCA cycle located in a compartment different from mitochondria can remain inactive while the mitochondrial pool is metabolically active. Unfortunately, from whole cell measurements it is not yet possible to discriminate the actual amount of a compartmentalized metabolite in each particular compartment, neither is possible to determine its actual activity. In regard to interaction with amino acids, we could observe rates of enrichment of amino acids that were faster than their respective turnover times, which is an indication of the presence of active exchange fluxes with metabolites of the central metabolism.
3.4. Non-stationary 13C metabolic flux analysis
Flux maps were derived from parameter estimation for the four different dilution rates and the flux values with their corresponding standard deviations are provided in Supplementary material 5. In general, we observed that most of the carbon was channeled through glycolysis and the TCA cycle with the exception of D=0.3 h−1 where ethanol was produced. Based on the flux estimation, a detailed discussion follows with focus on the cyclic flux interaction of central carbon metabolism with storage carbohydrates and amino acid pools.
3.5. Glucose recycle
To investigate the source of the unexpected reduced labeling enrichment in extracellular glucose, we implemented putative reactions for the degradation of trehalose as the main source of unlabeled glucose. This route involves the transport of trehalose and its subsequent hydrolysis (reactions treh_trans and tre4_fwd), which is based on reports suggesting that S. cerevisiae is capable of transporting trehalose to the periplasm by an Agt1p/H+- symporter and degradation by an acid trehalase Ath1p at the cell surface (Jules et al., 2008). We found that although this mechanism partly explained the incomplete labeling of extracellular glucose (Supplementary material 3), a deviation in the enrichment pattern is still present, especially at low growth rates. In order to fully reproduce the time course of the EC-glucose enrichment, it may be necessary to include an additional source of secreted unlabeled glucose. We calculated if cell lysis could explain the amount of unlabeled glucose found in the extracellular space and concluded that given the experimental results this option is unlikely (see Supplementary material 4 for a more detailed argumentation). Thus, we included the intracellular degradation of glycogen or trehalose into glucose, which is subsequently exported through a yet unknown mechanism as an alternative route.
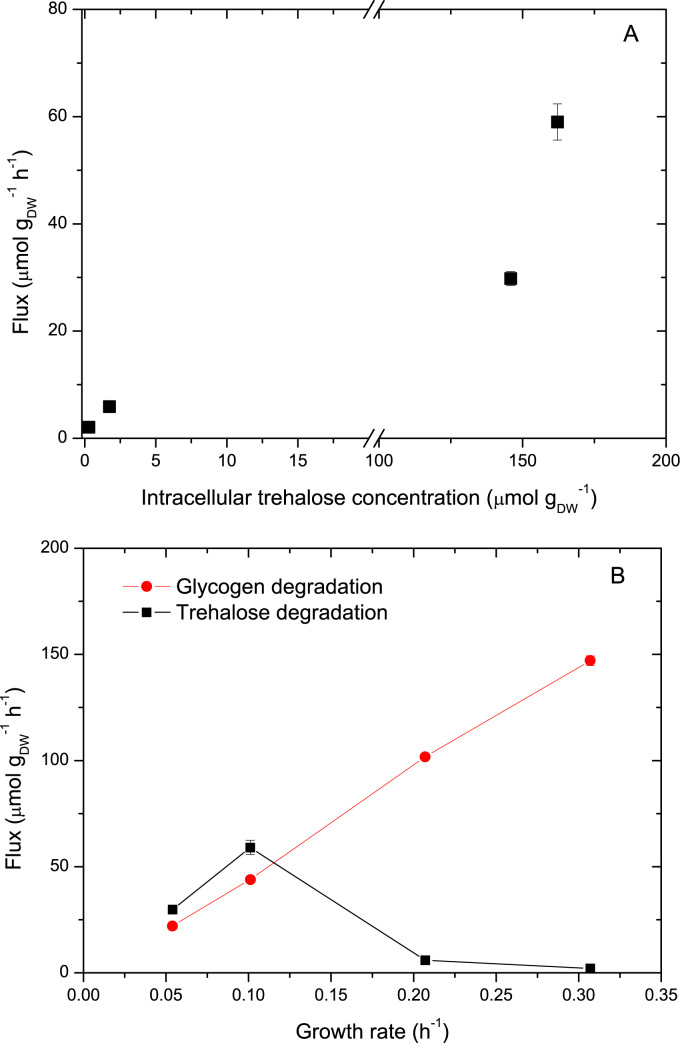
We found that trehalose degradation increased with the intracellular trehalose concentration (Fig. 5A), whereas it negatively correlates with growth rate (Fig. 5B). On the contrary, glycogen degradation increased with growth rate. These observations support the hypothesis that trehalose degradation can largely contribute to the observed unlabeled glucose in the extracellular space at low growth rates. Although the absolute flux of glycogen degradation exhibit an almost linear correlation with growth rate (Fig. 5B), the relative flux with respect to glucose uptake was almost constant about 3–4%. Thus, the apparently larger fluxes of glycogen degradation seem to be absorbed by the larger glycogen concentrations. Turnover times for trehalose and glycogen were estimated to be: 6.39 and 6.47 h at D=0.054 h−1; 3.52 and 4.01 h at D=0.101 h−1; 0.52 and 0.53 h at D=0.207 h−1; 0.25 and 0.30 h at D=0.307 h−1, respectively.
Fig. 5.
Glucose recycle. A: Trehalose degradation (μmol h−1) as a function of intracellular trehalose concentration; B: flux of trehalose and glycogen degradation as a function of growth rate (μmol h−1).
3.6. Impact of trehalose and glycogen metabolism on cellular ATP conservation
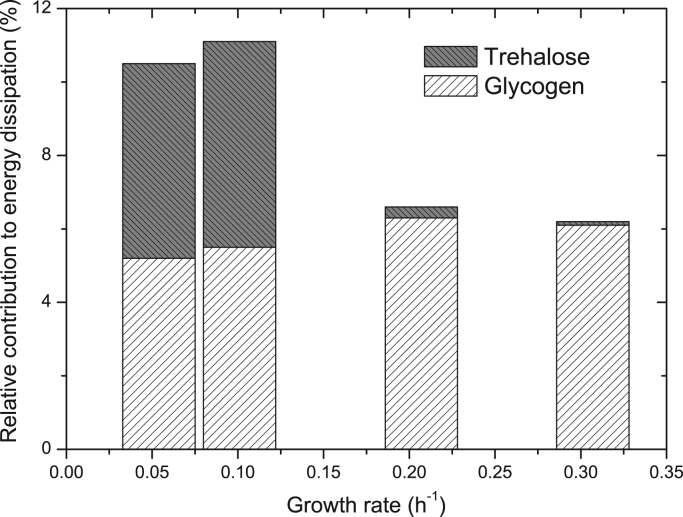
Recycling of glucose via trehalose or glycogen as discussed above implies a net consumption of ATP. In particular, 3 mol of ATP per 2 mol of glucose are required for the synthesis of trehalose, which is not recovered by the degradation reactions back to glucose. The contribution of glucose recycle to the dissipation of energy tends to be higher at low growth rates (Fig. 6) mainly due to the recycle through the trehalose node. Whereas the contribution of glucose recycle via glycogen seems to be almost constant at about 5–6% for all the dilution rates, the contribution via trehalose vary from about 5% at D=0.054 h−1 to almost zero at D=0.307 h−1. In case the export of trehalose proceeds via a proton symporter there would be reduced ATP costs since less protons would be exported via other mechanisms. Though glycogen synthesis also involves the consumption of ATP (2 mol ATP/mol glucose), its degradation does not necessarily involve a complete loss of ATP since the degradation can yield G1P via glycogen phosphorylase that can be converted back to G6P via phosphoglucomutase (Wilson et al., 2010). Nevertheless, glycogen hydratases are also present in yeast (Wilson et al., 2010), leading to increased energetic losses (2 ATP/mol glucose).
Fig. 6.
Relative contribution of glucose recycle to energy dissipation. The values in the figure correspond to the percentage of ATP that would be dissipated by recycle of glucose through trehalose and glycogen, where 100% corresponds to a maintenance energy factor of 0.415 mol ATP C mol−1 (Vanrolleghem et al., 1996).
3.7. Analysis of fluxes at the T6P node
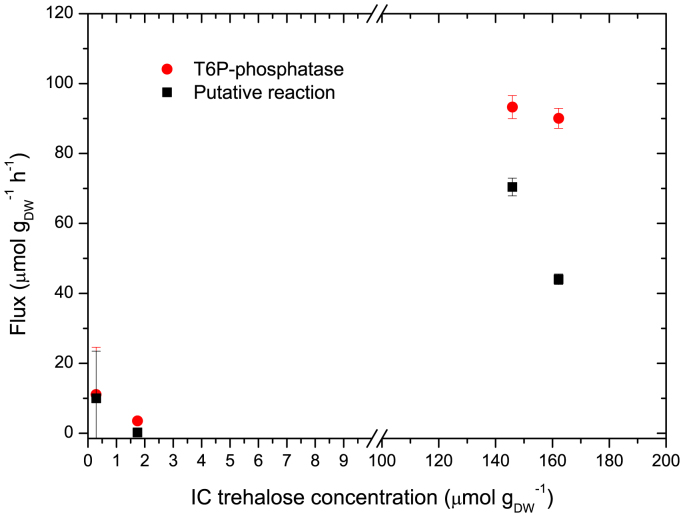
To explain the low 13C enrichment of T6P from the estimated fluxes, we calculated the flux ratio between the putative Tre→T6P reaction and the T6P-synthase and T6P-phosphatase reactions. We found that this putative mechanism is more active at low growth rates when the concentration of trehalose is higher (Fig. 7). The observed inverse correlation between trehalose concentration and T6P enrichment supports the hypothesis of the putative reaction. Note that at D=0.307 h−1 T6P concentration was the lowest but its enrichment was the highest, about 90%.
Fig. 7.
Analysis of fluxes at the T6P node. Comparison between degradation of T6P by T6P-phosphatase and production of T6P via the putative reaction that converts trehalose into T6P. Fluxes are presented as a function of the intracellular trehalose concentration.
The T6P phosphatase reaction (T6P+H2O→Trehalose+Pi) is basically an irreversible reaction. Under the assumption of a cytosolic Pi concentration of 25.5 μmol , the in vivo Gibbs free energy for this reaction is −8.6, −14.2, and −16.4 kJ mol−1 at D=0.101 and 0.054 h−1, and 0.207, 0.307 h−1, respectively (see Supplementary material 6). To reverse the direction of this reaction an unrealistic cytosolic Pi concentration of at least 850 μmol would be required. S. cerevisiae may have other enzymatic mechanisms that produce T6P from trehalose, for example unspecific kinase that uses ATP for the phosphorylation of trehalose into T6P, or a trans-phosphorylation mechanism. Although, to our knowledge, there are no reports on the presence of these mechanisms in S. cerevisiae, there are other organisms that can degrade trehalose via trehalose phosphorylase (Pichia fermentans (Schick et al., 1995); Caldanaerobacter subterraneous (Van der Borght et al., 2011)) or a reversible trehalose synthase from Pyrococcus horikoshii (Ryu et al., 2011).
3.8. Carbon flux distribution at the G6P branch
Most of the carbon influx at the G6P node was directed to glycolysis (Table 3), with a maximum of 75% at D=0.307 h−1. At this growth rate the carbon funneled towards trehalose was almost negligible as compared with the glucose uptake, while the flux to glycogen represented about 3.5% of the incoming flux. The remaining (14.7%) was diverted to the oxidative pentose phosphate pathway. A similar pattern was observed at D=0.207 h−1. An interesting deviation from the trend was observed at D=0.101 h−1 where less flux was directed towards PPP, most likely due to the increasing carbon flux diverted to storage pools. At this growth rate, about 3.8% of the carbon influx to G6P was directed to PPP, while about 16% was invested as storage. This behavior was foreseen by Van Winden et al. (2005) when evaluating the confidence interval of the PPP split ratio. They found the split ratio to vary between 0.05 and 0.52 with a maximum probability at 0.24. However, they also suggested that the storage carbohydrate turnover rate had a close, negative correlation with the PPP split ratio. Therefore, it would be necessary to consider the turnover of storage carbohydrate for a better estimation of the PPP split ratio, as performed here with high coverage of the intracellular metabolites.
Table 3.
Carbon flux distribution at the G6P node (all rates in μmol h−1).
| Reaction/Pathway | D=0.054 h−1 | D=0.101 h−1 | D=0.207 h−1 | D=0.307 h−1 |
|---|---|---|---|---|
| Influx | ||||
| Glucose uptake | 670±1 | 1149±2 | 2391±1 | 4857±1 |
| Intracellular Glucose recycle | 22±1 | 103±2 | 102±1 | 149±2 |
| Efflux | ||||
| Glycolysis | 426±15 | 921±196 | 1741±44 | 3758±132 |
| PPP | 118±2 | 47±1 | 366±4 | 698±22 |
| Glycogen | 69±1 | 150±8 | 130±5 | 178±8 |
| Trehalose | 23±1 | 46±1 | 4±1 | 1±0.1 |
| Biomass | 56±0 | 88±0 | 252±0 | 371±0 |
In a different work, Zhao et al. (2008) found the PPP split ratio to be 41% in Penicillium chrysogenum by using 13C labeling and local metabolic analysis around 6PG. Earlier, Bruinenberg et al. (1983), performed a theoretical analysis and estimated the PPP split ratio to be about 3% of the total glucose metabolized for yeast. In the present study, by using non-stationary 13C flux analysis, we found values between 3.8% and 17.1% at different growth rates.
3.9. Exchange fluxes between central metabolism and free amino acids, mRNA, and lipids
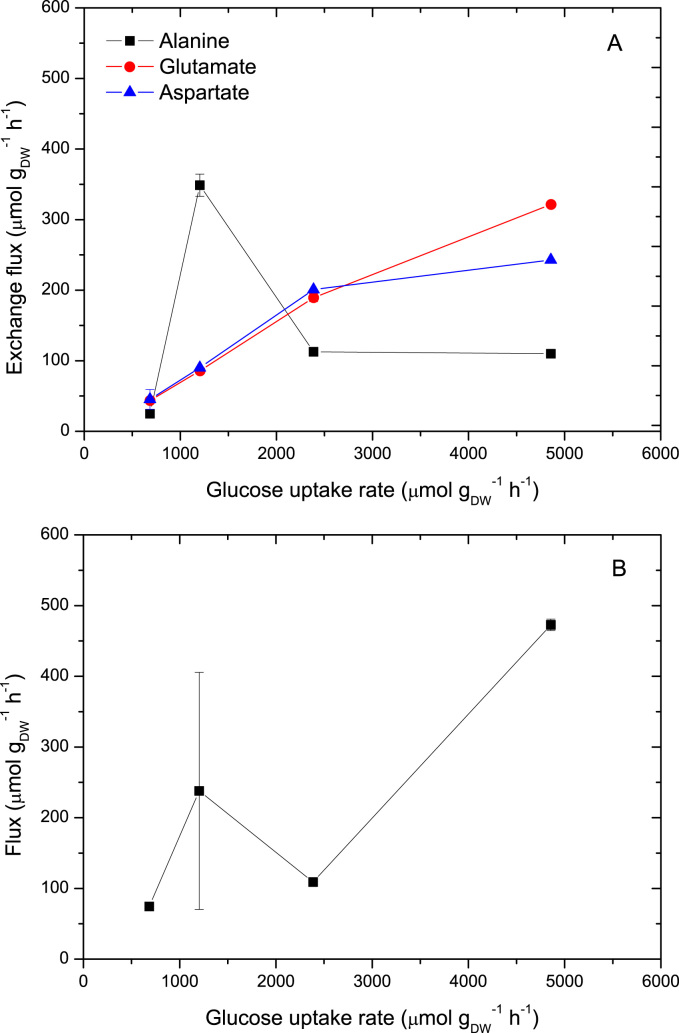
We found that the exchange fluxes of alanine, glutamate and aspartate with metabolites of the central carbon metabolism were in the same order of magnitude as the exchange fluxes of storage carbohydrates. In general, exchange fluxes with glutamate and aspartate were about 6–8% relative to the glucose uptake rate (Fig. 8A). Remarkably, the exchange flux with alanine at D=0.101 h−1 was about one third of the glucose uptake while it was about 2–5% for the other rates. The turnover times for alanine were 85–250 times faster than the turnover time of biomass. The Gibbs free energy of alanine transaminase was calculated as −0.2 kJ mol−1 at the lowest dilution rate and −0.6, −2.2 and 0 kJ mol−1 at D=0.101, 0.207 and 0.307 h−1, respectively. These values for the Gibbs free energy indicate that this reaction operates near equilibrium and exchange flux is likely to occur. Note that this estimation is based on whole cell concentrations, nevertheless, the obtained values are consistent with the estimated exchange flux from 13C analysis. Similarly, turnover times of glutamate and aspartate ranged from 200 up to 2700 times faster than biomass turnover times, which indicate that exchange fluxes of free amino acids with the central carbon metabolism are significant.
Fig. 8.
Comparison of exchange fluxes of amino acids and RNA-pools with central carbon metabolism and the corresponding glucose uptake rate. A: Exchange fluxes with amino acids; B: Exchange flux with RNA-pools. Error bars indicate the standard deviation of the fluxes based on linearized error.
The exchange flux between RNA-pools and Rib5P showed a magnitude in the same order of magnitude as exchange fluxes with amino acids (Fig. 8B). Assuming an RNA content of 7% (w/w) the turnover time would be 1.8, 0.8, 1.8, 0.4 h at D=0.054, 0.101, 0.207 and 0.307 h−1, respectively. This means about 10 times faster than the biomass turnover time. If we compare turnover times of free amino acids and RNA-pools with the fast turnover time of G6P, we find that turnover of amino acids is about 1 order of magnitude slower than the one of G6P, while those of RNA-pools are about 2 orders of magnitude slower.
3.10. Reproduction of cellular co-factor balances
The model used in this study and the results derived from the flux estimations provide a flux representation of the microorganism's physiology. Especially, the calculated extracellular rates (qO2, qCO2, qS, qEtOH, qAcet) as well as the NADH and NADPH balances can be reconstructed based on the independently determined 13C flux values.
3.10.1. NADPH balance
The sum of NADPH producing reactions of the network fulfill NADPH requirements (Table 4) when we considered reaction TCA4_fwd, which represents the NADP-dependent isocitrate dehydrogenase reaction. It has been reported that Saccharomyces cerevisiae has three enzymes that catalyze the conversion of isocitrate to α-ketoglutarate (Haselbeck and McAlister-Henn, 1993, Minard and McAlister-Henn, 2005). Two of them are mitochondrial enzymes (IDH and IDP1), while the other is reported to be cytosolic (IDP2). IDH is a NAD-dependent isozyme, while IDP1 and IDP2 are NADP-dependent. When isocitrate dehydrogenase is considered to be NAD-dependent only, NADPH supply is far below requirements from biomass reaction. Hence, it is imperative to consider isocitrate dehydrogenase as a NADP-dependent reaction. The NADPH contribution from IDP1 and IDP2 cannot be discriminated from this study, however, it is generally accepted that NADPH required for biosynthesis should be present in the cytosol since yeast has not yet been reported to possess any transhydrogenase enzyme.
Table 4.
Redox supply and demand (NADH and NADPH) based on estimated fluxes as well as flux based reconstruction of qO2 and qCO2 (all rates in μmol h−1).
| Dilution rate (h−1) | 0.054 | 0.101 | 0.207 | 0.307 |
|---|---|---|---|---|
| NADPH requirements (biomass synthesis) | 570 | 1020 | 2259 | 3347 |
| NADPH supply from oxidative PPP | 237 | 95 | 733 | 1396 |
| NADPH supply from acetaldehyde dehydrogenase | 25 | 99 | 65 | 197 |
| Putative NADPH supply from IDP (NADP-dependent isocitrate dehydrogenase) | 507 | 872 | 1865 | 1995 |
| Required IDP flux, and flux ratio IPD/(IDP+IDH) to fulfill NADPH requirements | 308 | 826 | 1461 | 1754 |
| 0.61 | 0.95 | 0.78 | 0.88 | |
| Estimated qO2 | 1606 | 2646 | 5769 | 7386 |
| Experimental and reconciled oxygen uptake rate (qO2) | 1742±87 | 2898±140 | 5696±285 | 8175±409 |
| 1596±87 | 2660±140 | 5600±270 | 7650±140 | |
| Estimated qCO2 | 1716 | 2805 | 6120 | 10526 |
| Experimental and reconciled carbon dioxide rate (qCO2) | 1691±87 | 2820±141 | 5972±270 | 11027±210 |
| 1688±87 | 2830±140 | 5960±270 | 10960±210 |
Measurements to quantify proteins were performed in a separate experiment with a similar setup (i.e., step down in dilution rate from D=0.3 to 0.2 and 0.1 h−1, see Supplementary material 7). Though the proteomics results were semi-quantitative, they suggest that at high growth rates the amount of IDP2 was lower, while the mitochondrial enzymes (IDH and IDP1) steadily increased with the growth rate. Up to a certain extent, those results were in agreement with the findings of Loftus et al. (1994), who suggested that IDP2 is repressed by glucose. Further support for a significant contribution of isocitrate dehydrogenase to NADPH supply is that a disruption of the Saccharomyces cerevisiae ZWF1 gene encoding glucose-6-phosphate dehydrogenase only resulted in a mild growth phenotype. In addition, the co-disruption of ZWF1 and IDP2 produced a rapid loss in cell viability following shifts from medium containing glucose to medium containing either oleate or acetate as the carbon source (Minard and McAlister-Henn, 2005).
Given the calculated flux distribution at the G6P node (Table 3), NADPH supply by PPP alone would not be sufficient to meet all the NADPH requirements. Our results are in line with the suggestions of Bruinenberg et al. (1983), who found that the PPP should be an extra source of NADPH, since isocitrate dehydrogenase alone would not be able to produce all the necessary reducing equivalents for biosynthesis. Provided that yeasts showed no fructose-1,6-biphosphatase activity under the conditions tested in this experiment, it would be expected that about 3% of the total glucose metabolized would be oxidized via PPP (in case IDP oxidizes all the produced isocitrate following Bruinenberg's theoretical analysis). In conclusion, our results suggest that IDP, and most likely IDP2, is the main reaction for the oxidation of isocitrate providing NADPH for biosynthesis.
In addition, although the acetaldehyde dehydrogenase tend to increase with growth rate, the contribution to NADPH-supply via pyruvate decarboxylase remains minor (4.4%, 9.7%, 2.9% and 5.9% at D=0.054, 0.101, 0.207 and 0.307 h−1, respectively) compared to the main sources, PPP and IDP.
3.10.2. NADH balance, O2 consumption rate and CO2 production rate
Comparable to the reconstruction of NADPH, all NADH producing and consuming reactions were balanced. The O2 consumption rate by oxidative phosphorylation can be better matched to the reconciled rates for all the dilution rates tested if a NADP-dependent isocitrate dehydrogenase reaction is assumed. Following the study of Bruinenberg et al. (1983) it is possible to estimate the contribution of IDP to isocitrate dehydrogenase by meeting all the NADPH requirements (Table 4). We found that more than 60% of the isocitrate dehydrogenase activity can be accounted by IDP. Taking this into consideration it is possible to correct the estimated oxygen consumption rate, which results in better estimates while fulfilling redox balances. Thus, the corrected qO2 deviated by less than 1% from the reconciled rates.
Similarly, based on the sum of all the CO2-producing/consuming reactions we found that the model estimates the carbon dioxide production rate reasonably well for all growth rates (with a deviation of less than 2% with the exception of D=0.307 h−1 where 4% deviation was observed).
4. Conclusions
Using 13C labeling experiments in combination with aerobic glucose limited cultures of Saccharomyces cerevisiae at four different growth rates, we have estimated fluxes that include intracellular cycles (e.g., storage carbohydrate cycles), some of which appeared to be rearranged depending on the growth rate. First, we found that at low growth rates the impact of the storage carbohydrate recycle was more significant due to a higher concentration of these materials in the cell (up to 560-fold) and higher fluxes relative to the glucose uptake rate (up to 16%). Second, to reproduce the experimental observations, a biological source of unlabeled glucose was required. Especially, storage carbohydrate mobilization is a putative source i.e., via the export and subsequent break down of trehalose into glucose through the system formed by the Agt1p/H+ symporter and the acid trehalase (Ath1p). This hypothesis was strongly supported by the 13C-labeling experimental data of extracellular glucose obtained at four different growth rates (0.054; 0.101, 0.207, 0.307 h−1), the observed trehalose in the extracellular space and the corresponding flux estimations, indicating that at D=0.101 h−1 up to 9% of the glucose uptake can be recycled via this route. Because the microbial cultures were not synchronized in this work (i.e., some of the cells could be at different phase of the cell cycle), the results obtained here should be regarded as an average culture response. Nevertheless, our findings are supported by the general agreement on the key role that growth rate plays in the accumulation of these reserve materials. Thus, we believed that the results presented here contribute to the understanding of the interaction of storage carbohydrates with the central carbon metabolism.
The evidence of the existence of a glucose recycle through storage carbohydrates, implies an ATP-loss. ATP invested in the early phosphorylation step of glycolysis would be lost since G6P is a precursor for both, trehalose and glycogen. At low growth rates (0.054 and 0.101 h−1), about 11% of the ATP dissipation (maintenance) can be explained by the estimated glucose recycle through the storage carbohydrates, while at high growth rates (0.207 and 0.307 h−1) this loss accounts for about 6–7%. The storage recycle has a significant impact when estimating fluxes in the central metabolism. For instance, the flux distribution at the G6P node can be distorted due to a higher or lower turnover of glucose from the storage pools. At D=0.101 h−1, the flux towards PPP was about 3.8% of the glucose uptake, most likely due the higher turnover of both trehalose and glycogen. The interaction between the PPP split ratio and the storage recycle was suggested by Van Winden et al. (2005) and confirmed in this study. Based on the NADPH balance, we believe that this research presents an in vivo evidence for the activity of a NADP-dependent isocitrate dehydrogenase as was earlier suggested by Bruinenberg et al. (1983).
Next to carbon recycling, there are other exchange fluxes between free amino acids and the central carbon metabolism reaching a flux of 15–45% relative to the glucose uptake rate. Similarly, we considered interactions between Rib5P and RNA-pools with exchange fluxes ranging from 5% to 20% relative to the glucose uptake rate depending on the growth rate, indicating that the turnover time of RNA is about 10 times higher than biomass turnover. For flux analysis and interpretation of metabolic activity it is recommended to consider the implications of carbon recycling, as it has consequences on the energy and flux balance of the microorganism.
Acknowledgments
The authors express their gratitude to Cor Ras, Dr. Reza Seifar and Patricia van Dam for the outstanding analytical work. Dr. Peter Verheijen, Dr. Ana Luisa da Cruz, Dr. Amit Deshmukh and Hugo Cueto-Rojas for valuable discussions and support. This work was carried out within the research programme of the Kluyver Centre for Genomics of Industrial Fermentation which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.meteno.2016.01.001.
Contributor Information
C.A. Suarez-Mendez, Email: casuarezmendez@unal.edu.co.
S.A. Wahl, Email: s.a.wahl@tudelft.nl.
Appendix A. Supplementary material
Supplementary material SM1. Metabolic network and system definitions
Supplementary material SM2. Metabolite data and quality check
Supplementary material SM3. Dynamics of mass isotopomers
Supplementary material SM4. Glucose tests
Supplementary material SM5. Calculated fluxes and flux maps
Supplementary material SM6. Thermodynamic analysis of the T6P-phosphatase reaction
Supplementary material SM7. Quantification of proteins-dehydrogenases
References
- Aboka F.O., Heijnen J.J., Van Winden W.A. Dynamic 13C-tracer study of storage carbohydrate pools in aerobic glucose-limited Saccharomyces cerevisiae confirms a rapid steady-state turnover and fast mobilization during a modest stepup in the glucose uptake rate. FEMS Yeast Res. 2009;9:191–201. doi: 10.1111/j.1567-1364.2008.00465.x. [DOI] [PubMed] [Google Scholar]
- Aboka F.O., van Winden W.A., Reginald M.M., van Gulik W.M., van de Berg M., Oudshoorn A., Heijnen J.J. Identification of informative metabolic responses using a minibioreactor: a small step change in the glucose supply rate creates a large metabolic response in Saccharomyces cerevisiae. Yeast. 2012;29:95–110. doi: 10.1002/yea.2892. [DOI] [PubMed] [Google Scholar]
- Bruinenberg P.M., Van Dijken J.P., Scheffers W.A. A theoretical analysis of NADPH production and consumption in yeasts. J. Gen. Microbiol. 1983;129:953–964. doi: 10.1099/00221287-129-4-965. [DOI] [PubMed] [Google Scholar]
- Canelas A.B., Ras C., ten Pierick A., van Gulik W.M., Heijnen J.J. An in vivo data-driven framework for classification and quantification of enzyme kinetics and determination of apparent thermodynamic data. Metab. Eng. 2011;13:294–306. doi: 10.1016/j.ymben.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Canelas A.B., ten Pierick A., Ras C., Seifar R.M., van Dam J.C., van Gulik W.M., Heijnen J.J. Quantitative evaluation of intracellular metabolite extraction techniques for yeast metabolomics. Anal. Chem. 2009;81:7379–7389. doi: 10.1021/ac900999t. [DOI] [PubMed] [Google Scholar]
- Castrillo J., Zeef L., Hoyle D., Zhang N., Hayes A., Gardner D., Cornell M., Petty J., Hakes L., Wardleworth L. Growth control of the eukaryote cell: a systems biology study in yeast. J. Biol. 2007;6:4. doi: 10.1186/jbiol54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipollina C., ten Pierick A., Canelas A.B., Seifar R.M., van Maris A.J.A., van Dam J.C., Heijnen J.J. A comprehensive method for the quantification of the non-oxidative pentose phosphate pathway intermediates in Saccharomyces cerevisiae by GC-IDMS. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2009;877:3231–3236. doi: 10.1016/j.jchromb.2009.07.019. [DOI] [PubMed] [Google Scholar]
- Conte, R. L., 2011. Optimization of a Glycogen and Trehalose Determination Method for Whole Yeast Cells. TU Delft
- Crown S.B., Antoniewicz M.R. Publishing 13C metabolic flux analysis studies: a review and future perspectives. Metab. Eng. 2013;20:42–48. doi: 10.1016/j.ymben.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daran-Lapujade P., Jansen M.L.A., Daran J.-M., van Gulik W., de Winde J.H., Pronk J.T. Role of Transcriptional Regulation in Controlling Fluxes in Central Carbon Metabolism of Saccharomyces cerevisiae: a chemostat culture study. J. Biol. Chem. 2004;279:9125–9138. doi: 10.1074/jbc.M309578200. [DOI] [PubMed] [Google Scholar]
- Daran J.M., Dallies N., Thines-Sempoux D., Paquet V., Francois J. Genetic and biochemical characterization of the UGP1 gene encoding the UDP-glucose pyrophosphorylase from saccharomyces cerevisiae. Eur. J. Biochem. 1995;233:520–530. doi: 10.1111/j.1432-1033.1995.520_2.x. [DOI] [PubMed] [Google Scholar]
- Dauner M., Bailey I., Wiechert W., Witholt B., Sauer U. ETH Zurich; Zurich: 2000. Intracellular Carbon Flux Analysis by 13C-Tracer Experiments. [Google Scholar]
- de Jonge L.P., Buijs N.A.A., ten Pierick A., Deshmukh A., Zhao Z., Kiel J.A.K.W., Heijnen J.J., van Gulik W.M. Scale-down of penicillin production in Penicillium chrysogenum. Biotechnol. J. 2011;6:944–958. doi: 10.1002/biot.201000409. [DOI] [PubMed] [Google Scholar]
- Fischer E., Sauer U. Large-scale in vivo flux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism. Nat. Genet. 2005;37:636–640. doi: 10.1038/ng1555. [DOI] [PubMed] [Google Scholar]
- François J., Parrou J.L. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2001;25:125–145. doi: 10.1111/j.1574-6976.2001.tb00574.x. [DOI] [PubMed] [Google Scholar]
- Gombert A.K., Dos Santos M.M., Christensen B., Nielsen J. Network identification and flux quantification in the central metabolism of Saccharomyces cerevisiae under different conditions of glucose repression. J. Bacteriol. 2001;183:1441–1451. doi: 10.1128/JB.183.4.1441-1451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou V., Plourde-Owobi L., Parrou J.L., Goma G., François J. Role of reserve carbohydrates in the growth dynamics of Saccharomyces cerevisiae. FEMS Yeast Res. 2004;4:773–787. doi: 10.1016/j.femsyr.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Haselbeck R.J., McAlister-Henn L. Function and expression of yeast mitochondrial NAD- and NADP-specific isocitrate dehydrogenases. J. Biol. Chem. 1993;268:12116–12122. [PubMed] [Google Scholar]
- Hashem M., Darwish S.M.I. Production of bioethanol and associated by-products from potato starch residue stream by Saccharomyces cerevisiae. Biomass Bioenergy. 2010;34:953–959. [Google Scholar]
- Jordà J., Suarez C., Carnicer M., ten Pierick A., Heijnen J.J., van Gulik W., Ferrer P., Albiol J., Wahl A. Glucose-methanol co-utilization in Pichia pastoris studied by metabolomics and instationary 13C flux analysis. BMC Syst. Biol. 2013:7. doi: 10.1186/1752-0509-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jules M., Beltran G., François J., Parrou J.L. New insights into trehalose metabolism by Saccharomyces cerevisiae: NTH2 encodes a functional cytosolic trehalase, and deletion of TPS1 reveals Ath1p-dependent trehalose mobilization. Appl. Environ. Microbiol. 2008;74:605–614. doi: 10.1128/AEM.00557-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H., Heijnen J. Statistical reconciliation of the elemental and molecular biomass composition of Saccharomyces cerevisiae. Biotechnol. Bioeng. 2001;75:334–344. doi: 10.1002/bit.10054. [DOI] [PubMed] [Google Scholar]
- Loftus T.M., Hall L.V., Anderson S.L., McAlister-Henn L. Isolation, characterization, and disruption of the yeast gene encoding cytosolic NADP-specific isocitrate dehydrogenase. Biochemistry. 1994;33:9661–9667. doi: 10.1021/bi00198a035. [DOI] [PubMed] [Google Scholar]
- Mashego M.R., Jansen M.L.A., Vinke J.L., van Gulik W.M., Heijnen J.J. Changes in the metabolome of Saccharomyces cerevisiae associated with evolution in aerobic glucose-limited chemostats. FEMS Yeast Res. 2005;5:419–430. doi: 10.1016/j.femsyr.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Mashego M.R., Van Gulik W.M., Heijnen J.J. Metabolome dynamic responses of Saccharomyces cerevisiae to simultaneous rapid perturbations in external electron acceptor and electron donor. FEMS Yeast Res. 2007;7:48–66. doi: 10.1111/j.1567-1364.2006.00144.x. [DOI] [PubMed] [Google Scholar]
- Mashego M.R., van Gulik W.M., Vinke J.L., Visser D., Heijnen J.J. In vivo kinetics with rapid perturbation experiments in Saccharomyces cerevisiae using a second-generation BioScope. Metab. Eng. 2006;8:370–383. doi: 10.1016/j.ymben.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Mashego M.R., Wu L., van Dam J.C., Ras C., Vinke J.L., Van Winden W.A., van Gulik W.M., Heijnen J.J. MIRACLE: mass isotopomer ratio analysis of U-13C-Labeled Extracts. A new method for accurate quantification of changes in concentrations of intracellular metabolites. Biotechnol. Bioeng. 2004;85:620–628. doi: 10.1002/bit.10907. [DOI] [PubMed] [Google Scholar]
- Minard K.I., McAlister-Henn L. Sources of NADPH in yeast vary with carbon source. J. Biol. Chem. 2005;280:39890–39896. doi: 10.1074/jbc.M509461200. [DOI] [PubMed] [Google Scholar]
- Murphy T.A., Dang C.V., Young J.D. Isotopically nonstationary 13C flux analysis of Myc-induced metabolic reprogramming in B-cells. Metab. Eng. 2013;15:206–217. doi: 10.1016/j.ymben.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenführ S., ten Pierick A., van Dam P.T.N., Suarez-Mendez C.A., Nöh K., Wahl S.A. Natural isotope correction of MS/MS measurements for metabolomics and 13C fluxomics. Biotechnol. Bioeng. 2015 doi: 10.1002/bit.25859. [DOI] [PubMed] [Google Scholar]
- Nöh K., Grönke K., Luo B., Takors R., Oldiges M., Wiechert W. Metabolic flux analysis at ultra short time scale: isotopically non-stationary 13C labeling experiments. J. Biotechnol. 2007;129:249–267. doi: 10.1016/j.jbiotec.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Noh K., Wahl A., Wiechert W. Computational tools for isotopically instationary 13C labeling experiments under metabolic steady state conditions. Metabolic Eng. 2006;8:554–577. doi: 10.1016/j.ymben.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Paalman J.W.G., Verwaal R., Slofstra S.H., Verkleij A.J., Boonstra J., Verrips C.T. Trehalose and glycogen accumulation is related to the duration of the G1 phase of Saccharomyces cerevisiae. FEMS Yeast Res. 2003;3:261–268. doi: 10.1111/j.1567-1364.2003.tb00168.x. [DOI] [PubMed] [Google Scholar]
- Parrou J.L., Jules M., Beltran G., François J. Acid trehalase in yeasts and filamentous fungi: localization, regulation and physiological function. FEMS Yeast Res. 2005;5:503–511. doi: 10.1016/j.femsyr.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Parrou J.L., Teste M.A., Francois J. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: Genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology. 1997;143:1891–1900. doi: 10.1099/00221287-143-6-1891. [DOI] [PubMed] [Google Scholar]
- Peng Z.Y., Trumbly R.J., Reimann E.M. Purification and characterization of glycogen synthase from a glycogen-deficient strain of Saccharomyces cerevisiae. J. Biol. Chem. 1990;265:13871–13877. [PubMed] [Google Scholar]
- Pereira M., Eleutherio E., Panek A. Acquisition of tolerance against oxidative damage in Saccharomyces cerevisiae. BMC Microbiol. 2001;1:11. doi: 10.1186/1471-2180-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S.I., Kim J.E., Kim E.J., Chung S.K., Lee S.B. Catalytic reversibility of Pyrococcus horikoshii trehalose synthase: efficient synthesis of several nucleoside diphosphate glucoses with enzyme recycling. Process Biochem. 2011;46:128–134. [Google Scholar]
- Schick I., Haltrich D., Kulbe K.D. Trehalose phosphorylase from Pichia fermentans and its role in the metabolism of trehalose. Appl. Microbiol. Biotechnol. 1995;43:1088–1095. [Google Scholar]
- Schmidt K., Marx A., de Graaf A.A., Wiechert W., Sahm H., Nielsen J., Villadsen J. 13C tracer experiments and metabolite balancing for metabolic flux analysis: comparing two approaches. Biotechnol. Bioeng. 1998;58:254–257. doi: 10.1002/(sici)1097-0290(19980420)58:2/3<254::aid-bit19>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Schuetz R., Kuepfer L., Sauer U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol. Syst. Biol. 2007;3:119. doi: 10.1038/msb4100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifar R.M., Ras C., van Dam J.C., van Gulik W.M., Heijnen J.J., Van Winden W.A. Simultaneous quantification of free nucleotides in complex biological samples using ion pair reversed phase liquid chromatography isotope dilution tandem mass spectrometry. Anal. Biochem. 2009;388:213–219. doi: 10.1016/j.ab.2009.02.025. [DOI] [PubMed] [Google Scholar]
- Shi L., Sutter B.M., Ye X., Tu B.P. Trehalose is a key determinant of the quiescent metabolic state that fuels cell cycle progression upon return to growth. Mol. Biol. Cell. 2010;21:1982–1990. doi: 10.1091/mbc.E10-01-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambuk B.U., De Araujo P.S., Panek A.D., Serrano R. Kinetics and energetics of trehalose transport in Saccharomyces cerevisiae. Eur. J. Biochem. 1996;237:876–881. doi: 10.1111/j.1432-1033.1996.0876p.x. [DOI] [PubMed] [Google Scholar]
- Vallino J., Stephanopoulos G. Metabolic flux distributions in corynebacteríum glut icum during growth and lysine overproduction. Biotechnol. Bioeng. 1993;41:633–646. doi: 10.1002/bit.260410606. [DOI] [PubMed] [Google Scholar]
- van Dam J.C., Ras C., ten Pierick A. Analysis of glycolytic intermediates with ion chromatography- and gas chromatography-mass spectrometry. Methods Mol. Biol. 2011;708:131–146. doi: 10.1007/978-1-61737-985-7_7. [DOI] [PubMed] [Google Scholar]
- Van der Borght J., Chen C., Hoflack L., Van Renterghem L., Desmet T., Soetaert W. Enzymatic properties and substrate specificity of the trehalose phosphorylase from Caldanaerobacter subterraneus. Appl. Environ. Microbiol. 2011;77:6939–6944. doi: 10.1128/AEM.05190-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gulik W., Heijnen J. A metabolic network stoichiometry analysis of microbial growth and product formation. Biotechnol. Bioeng. 1995;48:681–698. doi: 10.1002/bit.260480617. [DOI] [PubMed] [Google Scholar]
- van Heerden J.H., Wortel M.T., Bruggeman F.J., Heijnen J.J., Bollen Y.J.M., Planqué R., Hulshof J., O’Toole T.G., Wahl S.A., Teusink B. Lost in transition: startup of glycolysis yields subpopulations of nongrowing cells. Science. 2014;343:1245114. doi: 10.1126/science.1245114. [DOI] [PubMed] [Google Scholar]
- Van Hoek P., Van Dijken J.P., Pronk J.T. Effect of specific growth rate on fermentative capacity of baker’s yeast. Appl. Environ. Microbiol. 1998;64:4226. doi: 10.1128/aem.64.11.4226-4233.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Winden W.A., Heijnen J.J., Verheijen P.J.T. Cumulative bondomers: a new concept in flux analysis from 2D [13C, 1H] COSY NMR data. Biotechnol. Bioeng. 2002;80:731–745. doi: 10.1002/bit.10429. [DOI] [PubMed] [Google Scholar]
- Van Winden W.A., Van Dam J.C., Ras C., Kleijn R.J., Vinke J.L., Van Gulik W.M., Heijnen J.J. Metabolic-flux analysis of Saccharomyces cerevisiae CEN.PK113-7D based on mass isotopomer measurements of 13C-labeled primary metabolites. FEMS Yeast Res. 2005;5:559–568. doi: 10.1016/j.femsyr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Vanrolleghem P.A., De Jong-Gubbels P., Van Gulik W.M., Pronk J.T., Van Dijken J.P., Heijnen S. Validation of a metabolic network for Saccharomyces cerevisiae using mixed substrate studies. Biotechnol. Prog. 1996;12:434–448. doi: 10.1021/bp960022i. [DOI] [PubMed] [Google Scholar]
- Varma A., Palsson B.O. Metabolic flux balancing: basic concepts, scientific and practical use. Nat. Biotechnol. 1994;12:994–998. [Google Scholar]
- Verheijen P. Data reconciliation and error detection. In: S. C., editor. The Metabolic Pathway Engineering Handbook. CRC Press; Boca Raton: 2010. [Google Scholar]
- Voit E.O. Biochemical and genomic regulation of the trehalose cycle in yeast: Review of observations and canonical model analysis. J. Theor. Biol. 2003;223:55–78. doi: 10.1016/s0022-5193(03)00072-9. [DOI] [PubMed] [Google Scholar]
- Wahl S., Nöh K., Wiechert W. 13C labeling experiments at metabolic nonstationary conditions: an exploratory study. BMC Bioinformatics. 2008;9:152. doi: 10.1186/1471-2105-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl S.A., Dauner M., Wiechert W. New tools for mass isotopomer data evaluation in 13C flux analysis: mass isotope correction, data consistency checking, and precursor relationships. Biotechnol. Bioeng. 2004;85:259–268. doi: 10.1002/bit.10909. [DOI] [PubMed] [Google Scholar]
- Wahl S.A., Seifar R., ten Pierick A., Ras C., van Dam J., Heijnen J., van Gulik W. Quantitative metabolomics using ID-MS. In: Krömer J.O., Nielsen L.K., Blank L.M., editors. vol. 1191. Springer; New York: 2014. pp. 91–105. (Metabolic Flux Analysis). [DOI] [PubMed] [Google Scholar]
- Wiechert W. 13C metabolic flux analysis. Metab. Eng. 2001;3:195–206. doi: 10.1006/mben.2001.0187. [DOI] [PubMed] [Google Scholar]
- Wiechert W., De Graaf A.A. Bidirectional reaction steps in metabolic networks: I. Modeling and simulation of carbon isotope labeling experiments. Biotechnol. Bioeng. 1997;55:101–117. doi: 10.1002/(SICI)1097-0290(19970705)55:1<101::AID-BIT12>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Wiechert W., Nöh K. Isotopically non-stationary metabolic flux analysis: complex yet highly informative. Curr. Opin. Biotechnol. 2013;24:979–986. doi: 10.1016/j.copbio.2013.03.024. [DOI] [PubMed] [Google Scholar]
- Willke T., Vorlop K.D. Industrial bioconversion of renewable resources as an alternative to conventional chemistry. Appl. Microbiol. Biotechnol. 2004;66:131–142. doi: 10.1007/s00253-004-1733-0. [DOI] [PubMed] [Google Scholar]
- Wilson W.A., Roach P.J., Montero M., Baroja-Fernández E., Munoz F.J., Eydallin G., Viale A.M., Pozueta-Romero J. Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol. Rev. 2010;34:952–985. doi: 10.1111/j.1574-6976.2010.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Mashego M.R., van Dam J.C., Proell A.M., Vinke J.L., Ras C., Van Winden W.A., van Gulik W.M., Heijnen J.J. Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly 13C-labeled cell extracts as internal standards. Anal. Biochem. 2005;336:164–171. doi: 10.1016/j.ab.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Young J.D., Shastri A.A., Stephanopoulos G., Morgan J.A. Mapping photoautotrophic metabolism with isotopically nonstationary 13C flux analysis. Metab. Eng. 2011;13:656–665. doi: 10.1016/j.ymben.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Kuijvenhoven K., Ras C., van Gulik W.M., Heijnen J.J., Verheijen P.J.T., van Winden W.A. Isotopic non-stationary 13C gluconate tracer method for accurate determination of the pentose phosphate pathway split-ratio in Penicillium chrysogenum. Metab. Eng. 2008;10:178–186. doi: 10.1016/j.ymben.2008.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material SM1. Metabolic network and system definitions
Supplementary material SM2. Metabolite data and quality check
Supplementary material SM3. Dynamics of mass isotopomers
Supplementary material SM4. Glucose tests
Supplementary material SM5. Calculated fluxes and flux maps
Supplementary material SM6. Thermodynamic analysis of the T6P-phosphatase reaction
Supplementary material SM7. Quantification of proteins-dehydrogenases