Abstract
IMPORTANCE
Conjunctival melanoma (CM) is a highly aggressive ocular cancer for which treatment options are limited; the molecular pathogenesis is poorly understood.
OBJECTIVE
To identify the molecular characteristics of CM using next-generation whole-exome sequencing (WES).
DESIGN, SETTING, AND PARTICIPANTS
Whole-exome sequencing was performed on tumor DNA extracted from the archived specimens of 5 patients with CM who had been treated with surgical excision between 2006 and 2011. These samples were analyzed at a tertiary academic ocular oncology referral center using a customized bioinformatic pipeline.
MAIN OUTCOMES AND MEASURES
Sample analyses were designed to detect driver mutations, chromosome copy number aberrations, and mutation signatures.
RESULTS
The study’s 5 patients ranged in age from 51 to 77 years. Four of the 5 were female, and all were white. Mutations were detected in known oncogenes, including BRAF, NRAS, NF1, EGFR, ALK, TERT, and APC. None of the mutations associated with uveal melanoma were found. All samples demonstrated a C→T mutation signature typical of UV-induced DNA damage. The most common CNA was a gain in chromosome 6p.
CONCLUSIONS AND RELEVANCE
In these 5 patients, WES allowed identification of mutations that can be targeted with therapy and supported the role of UV light in CM pathogenesis. These findings indicate a need for larger studies to evaluate the diagnostic, prognostic, and therapeutic value of WES for CM.
Conjunctival melanoma (CM) is a rare but potentially deadly ocular malignant condition, with a 10-year disease-specific mortality of 9% to 35%.1 Primary treatment of CM consists of local surgical excision with wide margins and adjuvant therapy (cryotherapy, brachytherapy, and/or topical application of mitomycin C). However, regional and systemic metastasis occurs in approximately 30% of patients within 3 years, and there are no effective treatments for metastatic disease.1 Conjunctival melanoma appears to be a distinct entity compared with other mucosal melanomas. In contrast to these malignant conditions, CM incidence is often associated with UV sunlight exposure. Conjunctival melanoma is also associated with a higher 5-year survival rate (86%) compared with melanomas of the gastrointestinal tract (4%–33%), urogenital tract (7%–22%), and respiratory mucosal tissues (0%–31%); this difference is possibly related to earlier detection or differences in the innate aggressiveness of the tumor.2
The molecular attributes of CM remain poorly characterized, which is a problem that has hindered the development of novel therapies. One study3 reported mutations in BRAF and NRAS in 29% and 18% of CMs, respectively, but the technology used in this study did not allow for a comprehensive assessment of driver mutations, chromosome copy number aberrations (CNAs), and mutational signatures. In the present study, whole-exome sequencing (WES) permits more comprehensive characterization of the molecular biology of CM.
Methods
Five formalin-fixed, paraffin-embedded, archival CM specimens were selected based on the availability of sufficient tissue for testing. Tumor DNA was extracted from all 5 and prepared for WES. Ethical approval was obtained from the University of Miami institutional review board for this study, which adheres to the tenets of the Declaration of Helsinki and is compliant with the Health Insurance Portability and Accountability Act of 1996 (HIPAA). Per institutional review board approval, the use of archival samples obviated the need for the informed consent of the included patients.
The University of Miami Sequencing Core facility conducted the WES using the customized bioinformatic pipeline of the present study, with a mean exome coverage target of ×60. Sequences were aligned to the human reference genome GRCh37/hg19 using NovoAlign (Novocraft Technologies Sdn Bhd). Since matched blood samples were not available, we used a panel of normal tissue samples (n = 117), a high-coverage blood sample, and somatic single-nucleotide and insertion and deletion (indel) variant-caller MuTect2 to call out tumor variants and filter out likely silent germline polymorphisms.4 To minimize artifacts introduced by the specimen archiving process, we filtered out all genetic variants present in less than 20% of sequencing reads. Additionally, variants were excluded if they were outside of coding or splicing regions, had fewer than 3 alternate reads, were present in greater than 0.5% of the population, or were predicted to be non-damaging using ANNOVAR (Children’s Hospital of Philadelphia, Pennsylvania). Chromosome copy number aberrations were assessed using CNVkit (University of California, San Francisco) and Genomic Identification of Significant Targets in Cancer (GISTIC) version 2.0 (Broad Institute), whereas mutation signatures were analyzed using pmsignature (the University of Chicago) in R (R Development Core Team). Additional details are available in the Supplement in the eAppendix.
Results
All patients were white, and none had a history of cutaneous melanoma or uveal melanoma. Two of the 5 patients (40%) had CM that arose from primary acquired melanosis; all lesions arose on sun-exposed areas. Four of the 5 patients had bulbar CM (located within the conjunctiva covering the globe) and the remaining individual had palpebral CM (located within the tarsal conjunctiva of the eyelid). None of the 5 patients had had treatment of their disease prior to receiving care at the center where the archiving of their samples occurred. Clinical features are further summarized in the Table.
Table.
Demographic and Clinical Features of Study Population
| Patient No./Sex/Age, y | Tumor Location | Tumor Characteristics | Tumor Area, mm2 | Adjuvant Therapy | Recurrence After Primary Excision | Time to Recurrence | Treatment of Recurrence |
|---|---|---|---|---|---|---|---|
| 1/M/68 | Temporal bulbar, right eye | Separate focus of PAM; diffuse disease | 24 | Cryo plus MMC | Local recurrence | 3 y | MMC or IFN plus surgery |
| 2/F/65a | Plica and inferior palpebral, right eye; caruncle, right eye | Diffuse growth into tarsus/caruncle + large circumscribed palpebral lesion | 78 | Cryo plus IFN | Local recurrence | 10 mo | Exenteration |
| 3/F/52 | Temporal bulbar, right eye | Limbus, overlying cornea | 24.5 | Cryo | No | NA | NA |
| 4/F/51 | Nasal bulbar, left eye | Limbus, overlying cornea | 15 | Cryo plus MMC | Local recurrence | 3 mo | MMC or IFN plus surgery |
| 5/F/77 | Inferotemporal bulbar, left eye | Arising from PAM, diffuse along bulbar conjunctiva | 3 | Cryo | No | NA | NA |
Abbreviations: cryo, cryotherapy; IFN, interferon-alfa-2b; F, female; M, male; MMC, mitomycin C; NA, not applicable; PAM, primary acquired melanosis.
Patient died.
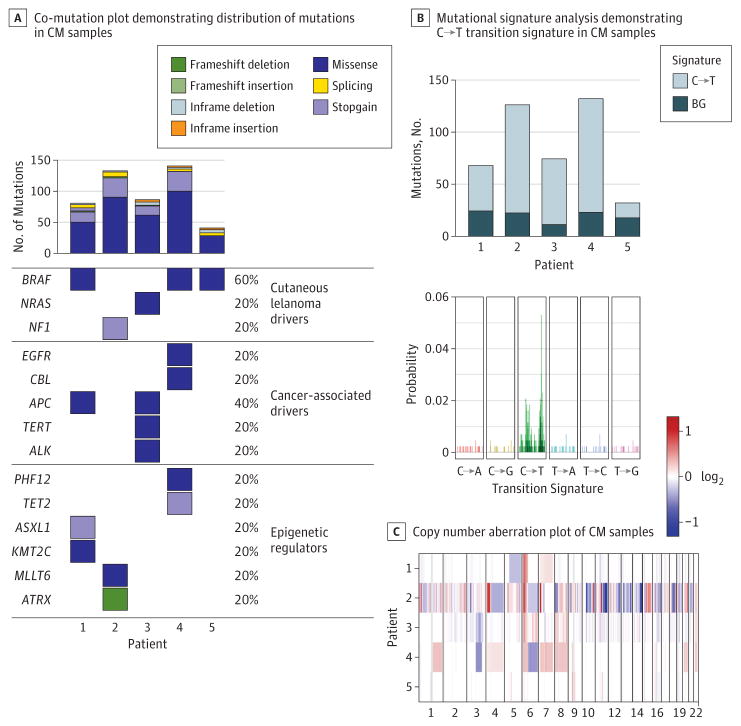
Deleterious mutations were identified in all 5 CM samples (Figure). These mutations included BRAF V600E, BRAF V600K, and NRAS Q61R, which have been reported previously in CM and cutaneous melanoma.5,6 One sample (patient 2) harbored a mutation in the tumor suppressor NF1 (without systemic manifestations), which previously has been reported in CM only in association with neurofibromatosis.7 Mutations previously unreported in CM occurred in other cancer-associated genes, including APC (n = 2), EGFR (n = 1), CBL (n = 1), and ALK (n = 1). In cutaneous melanoma as well as CM,8 TERT mutations have been found to occur in the promoter sequence, there by altering messenger RNA expression, whereas the TERT mutation in the present case was a missense alteration within the coding sequence and was there fore predicted to alter protein function. Mutations were also found in epigenetic regulators, including TET2 (n = 1), ATRX (n = 1), and ASXL1 (n = 1). The gene ASXL1 encodes a protein-binding partner of BAP1, a gene that is commonly mutated in uveal melanoma9; however, no samples contained mutations in BAP1 or other genes commonly mutated in uveal melanoma.10
Figure. Co-Mutation Plot, Mutational Signature Analysis, and Copy Number Aberration Plot of Conjunctival Melanoma (CM) Tissue Samples.
A, Top bar plot demonstrates number of predicted detrimental mutations in coding and splicing regions; mutations were categorized into clusters associated with cutaneous melanoma, cancer-driving mutations, and epigenetic mutations. Mutations in genes associated with uveal melanoma were absent. B, Bottom plot shows probability of mutations in single nucleotides. Top plot shows frequency of C→T transition signature in each patient in study sample. C, Patients 2 and 4, whose samples demonstrated the largest number of copy number aberrations and mutations, experienced rapid recurrence of disease following primary tumor excision.
Discussion
Several of these mutations could nominate new therapeutic options. The mutations in BRAF, NRAS, and NF1 activate the mitogen-activated protein kinase (MAPK) pathway, which can be pharmacologically inhibited with mitogen-activated protein kinase kinase (MEK) inhibitors.11 The mutations in APC, EGFR, and ALK can be inhibited with other targeted molecular agents.12
The role of UV light in the pathogenesis of CM has been the subject of discussion.13 However, consistent with a recent report,14 we observed in all 5 cases a prominent C→T mutation signature associated with UV light-induced DNA damage (Figure), which strongly implicates UV light as a mutagen in CM. The C→T mutations are important in UV-driven malignant conditions. Notably, non–UV-exposed conjunctival melanomas with KIT mutations have also been described, particularly in Chinese populations.2
The only CNA identified in all 5 samples was a chromosome 6p gain, which is also common in cutaneous melanoma and uveal melanoma. However, other recurrent CNAs were observed (Figure; eTables 1–4 in the Supplement). Samples from patients 1, 3, and 5 appeared to have a lower mutational burden than those found inmucosal melanomas in other locations, as previously described.15 Interestingly, the 2 samples from individuals who experienced rapid recurrences within a year (patient 2 and patient 4) had the greatest number of mutations (Figure) and the largest amount of CNAs (Figure), suggesting that WES data may be valuable in predicting clinical outcome. This is an interesting observation that deserves further evaluation.
Limitations
Limitations of the present study include the small number of samples. In addition, 2 of 5 cases (40%) arose from primary acquired melanosis, which is thought to serve as the origin of malignant transformation in 75% of CM cases.1 It is possible that the mutational signatures of de novo CM observed in this study may differ from those arising from primary acquired melanosis. In addition, the location of the tumor (bulbar vs palpebral) may carry distinct signatures as well, given that the areas experience differences in exposure to UV light. All of the CM samples in this series occurred in sun-exposed areas. Larger studies would be appropriate for further evaluation of these potential distinctions.
Conclusions
Whole-exome sequencing is a genome-wide comprehensive approach for identifying mutations and chromosomal alterations. This study demonstrates the potential value of WES for understanding the pathogenesis of CM and potentially providing precision medicine in the future for patients with this disease.
Supplementary Material
Key Points.
Question
What mutations can be identified with whole-exome sequencing (WES) of conjunctival melanoma (CM)?
Findings
With WES, CM was found to harbor mutations in BRAF, NRAS, and NF1; previously unreported mutations in EGFR, APC, TERT and other cancer-associated genes; and the C→T mutation signature consistent with UV-induced DNA damage. The most common chromosomal alteration was 6p gain.
Meaning
Whole-exome sequencing might enable the detection of molecular mutations targetable by cancer therapies and provide insight into the pathogenesis of CM.
Acknowledgments
Funding/Support: Financial support for this study includes the following sources: Dr Galor has received support from the Department of Veterans Affairs and Veterans Health Administration; Dr Harbour has received support from the Office of Research and Development via Clinical Sciences Research grants EPID-006-15S and R01 CA125970, as well as a generous gift from Mark J. Daily, MD; Dr Field has received support from the Office of Research and Development via Clinical Sciences Research grant F30 CA206430; and Dr Dubovy has received support from the Florida Lions Eye Bank. Dr Karp has received support from the Ronald and Alicia Lepke Grant, the Lee and Claire Hager Grant, the Jimmy and Gaye Bryan Grant, the Richard Azar Family Grant, the Robert Baer Family Grant, the Emilyn Page and Mark Feldberg Grant, and the Gordon Charitable Trust. Further grants specific to the project include National Institutes of Health Clinical Sciences Research grant R01EY026174, National Institutes of Health Center Core Grant P30EY014801, Department of Defense Grant W81XWH-13-1-0048, an unrestricted grant from Research to Prevent Blindness, and a Richard and Kathy Lesser grant.
Footnotes
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. No disclosures were reported.
Role of the Funder/Sponsor: All funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Author Contributions: Dr Karp had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Wang, Galor, Harbour, Karp.
Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: Swaminathan, Wang, Galor, Dubovy, Harbour.
Critical revision of the manuscript for important intellectual content: Swaminathan, Field, Sant, Galor, Harbour, Karp.
Statistical analysis: Swaminathan, Field, Sant.
Obtained funding: Galor.
Administrative, technical, or material support: Field, Dubovy, Karp.
Study supervision: Wang, Harbour, Karp.
Other: Field.
References
- 1.Wong JR, Nanji AA, Galor A, Karp CL. Management of conjunctival malignant melanoma: a review and update. Expert Rev Ophthalmol. 2014;9(3):185–204. doi: 10.1586/17469899.2014.921119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mikkelsen LH, Larsen AC, von Buchwald C, Drzewiecki KT, Prause JU, Heegaard S. Mucosal malignant melanoma—a clinical, oncological, pathological and genetic survey. APMIS. 2016;124(6):475–486. doi: 10.1111/apm.12529. [DOI] [PubMed] [Google Scholar]
- 3.Griewank KG, Westekemper H, Murali R, et al. Conjunctival melanomas harbor BRAF and NRAS mutations and copy number changes similar to cutaneous and mucosal melanomas. Clin Cancer Res. 2013;19(12):3143–3152. doi: 10.1158/1078-0432.CCR-13-0163. [DOI] [PubMed] [Google Scholar]
- 4.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kakavand H, Walker E, Lum T, et al. BRAF(V600E) and NRAS(Q61L/Q61R) mutation analysis in metastatic melanoma using immunohistochemistry: a study of 754 cases highlighting potential pitfalls and guidelines for interpretation and reporting. Histopathology. 2016;69(4):680–686. doi: 10.1111/his.12992. [DOI] [PubMed] [Google Scholar]
- 6.Larsen AC, Dahl C, Dahmcke CM, et al. BRAF mutations in conjunctival melanoma: investigation of incidence, clinicopathological features, prognosis and paired premalignant lesions. Acta Ophthalmol. 2016;94(5):463–470. doi: 10.1111/aos.13007. [DOI] [PubMed] [Google Scholar]
- 7.Rubinstein TJ, Plesec TP, Singh AD. Desmoplastic melanoma of the eyelid and conjunctival melanoma in neurofibromatosis type 1: a clinical pathological correlation. Surv Ophthalmol. 2015;60(1):72–77. doi: 10.1016/j.survophthal.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Koopmans AE, Ober K, Dubbink HJ, et al. Rotterdam Ocular Melanoma Study Group. Prevalence and implications of TERT promoter mutation in uveal and conjunctival melanoma and in benign and premalignant conjunctival melanocytic lesions. Invest Ophthalmol Vis Sci. 2014;55(9):6024–6030. doi: 10.1167/iovs.14-14901. [DOI] [PubMed] [Google Scholar]
- 9.Decatur CL, Ong E, Garg N, et al. Driver mutations in uveal melanoma: associations with gene expression profile and patient outcomes. JAMA Ophthalmol. 2016;134(7):728–733. doi: 10.1001/jamaophthalmol.2016.0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330(6009):1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HS, Jung M, Kang HN, et al. Oncogenic BRAF fusions in mucosal melanomas activate the MAPK pathway and are sensitive to MEK/PI3K inhibition or MEK/CDK4/6 inhibition. Oncogene. 2017;36(23):3334–3345. doi: 10.1038/onc.2016.486. [DOI] [PubMed] [Google Scholar]
- 12.Drilon A, Siena S, Ou SI, et al. Safety and antitumor activity of the multitargeted pan-TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase i trials (ALKA-372-001 and STARTRK-1) Cancer Discov. 2017;7(4):400–409. doi: 10.1158/2159-8290.CD-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu GP, Hu DN, McCormick SA. Latitude and incidence of ocular melanoma. Photochem Photobiol. 2006;82(6):1621–1626. doi: 10.1562/2006-07-17-RA-970. [DOI] [PubMed] [Google Scholar]
- 14.Rivolta C, Royer-Bertrand B, Rimoldi D, et al. UV light signature in conjunctival melanoma; not only skin should be protected from solar radiation. J Hum Genet. 2016;61(4):361–362. doi: 10.1038/jhg.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furney SJ, Turajlic S, Stamp G, et al. Genome sequencing of mucosal melanomas reveals that they are driven by distinct mechanisms from cutaneous melanoma. J Pathol. 2013;230(3):261–269. doi: 10.1002/path.4204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.