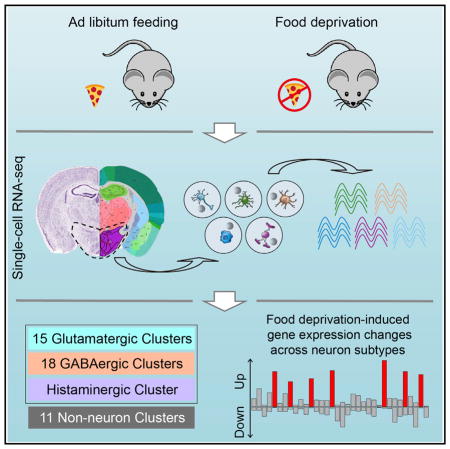
SUMMARY
The hypothalamus is one of the most complex brain structures involved in homeostatic regulation. Defining cell composition and identifying cell-type-specific transcriptional features of the hypothalamus is essential for understanding its functions and related disorders. Here, we report single-cell RNA sequencing results of adult mouse hypothalamus, which defines 11 non-neuronal and 34 neuronal cell clusters with distinct transcriptional signatures. Analyses of cell-type-specific transcriptomes reveal gene expression dynamics underlying oligodendrocyte differentiation and tanycyte subtypes. Additionally, data analysis provides a comprehensive view of neuropeptide expression across hypothalamic neuronal subtypes and uncover Crabp1+ and Pax6+ neuronal populations in specific hypothalamic subregions. Furthermore, we found food deprivation exhibited differential transcriptional effects among the different neuronal subtypes, suggesting functional specification of various neuronal subtypes. Thus, the work provides a comprehensive transcriptional perspective of adult hypothalamus, which serves as a valuable resource for dissecting cell-type-specific functions of this complex brain region.
In Brief
Chen et al. perform single-cell RNA sequencing analysis of the adult mouse hypothalamus to probe the rich cell diversity of this complex brain region. They also identify neuronal subtype-specific transcriptional responses to food deprivation.
INTRODUCTION
The hypothalamus is one of the most complex brain regions, essential for regulating physiological and behavioral homeostasis. Numerous studies have revealed its role in orchestrating a wide range of animal behaviors (Denton et al., 1996; Elmquist et al., 1999; Navarro and Tena-Sempere, 2011). Commensurate with its functional diversity, is its highly complex anatomical and cellular composition (Puelles and Rubenstein, 2015; Shimogori et al., 2010). Works in the last few decades have identified various cell types in hypothalamus based on different properties (Brown et al., 2013; Lee et al., 2015; Mathew, 2008; Wu et al., 2014). However, a comprehensive cell-type classification of hypothalamus has not been achieved.
Although different methodologies have been used for classifying cell types in the nervous system (Greig et al., 2013; Jiang et al., 2015), the most direct and unambiguous method to define a cell type is its transcriptional feature, as it underlies other cell features such as morphology, connectivity, and function (Greig et al., 2013; Toledo-Rodriguez et al., 2004). In addition, gene expression-based cell classification can be reliably and conveniently adapted by the entire research community (Gong et al., 2003), making data comparison among different groups possible. Indeed, a systematic in situ hybridization (ISH) database has revealed extensive cell-type heterogeneity in brain (Lein et al., 2007). However, the limitation of ISH on assessing co-expression of multiple genes prevents a definitive cell-type classification.
Recent advances in single-cell RNA sequencing (scRNA-seq) have facilitated the transcriptional cataloguing of cell types in many tissues, including those in the nervous system (Gokce et al., 2016; Macosko et al., 2015; Tasic et al., 2016; Zeisel et al., 2015). While cell diversity in the cerebral cortex (Lake et al., 2016; Tasic et al., 2016; Zeisel et al., 2015), hippocampus (Zeisel et al., 2015), and striatum (Gokce et al., 2016) has been cataloged to an unprecedented level, the cost and effort of profiling large numbers of single cells by conventional scRNA-seq methods prevent its broader application to highly complex brain regions, such as the hypothalamus. To overcome this challenge, cost-efficient scRNA-seq methods have been developed to achieve high-throughput parallel analysis (Klein et al., 2015; Macosko et al., 2015), making scRNA-seq analysis of complex brain regions possible.
Here, we applied high-throughput Drop-seq method (Macosko et al., 2015) to profile single cells dissociated from the adult mouse hypothalamus. Through clustering analysis, we identified 11 non-neuronal and 34 neuronal cell types. Data analysis revealed the transcriptional dynamics underlying the oligodendrocyte differentiation, as well as the transcriptional heterogeneity of tanycytes, a hypothalamus-specific non-neuronal cell type whose function remains poorly characterized. Additionally, single-cell transcriptome analysis revealed not only highly divergent expression patterns of neuropeptides and receptors across neuron subtypes, but also Crabp1+ and Pax6+ neuronal populations at specific hypothalamic regions. Furthermore, cell-type-specific transcription responses to food deprivation among various cell types were also revealed. Thus, our study provides a comprehensive gene expression map across divergent cell types in the hypothalamus, which will facilitate broader functional understanding of this complex brain region.
RESULTS
Overview of the Cell Types in Hypothalamus Identified by Single-Cell RNA-Seq
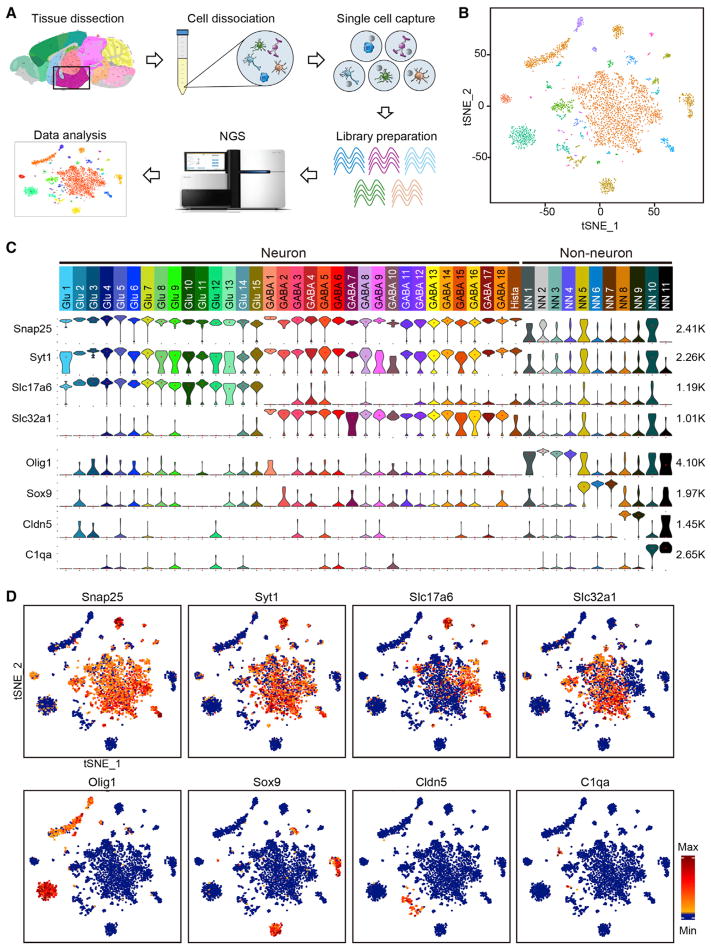
To characterize cellular heterogeneity in the hypothalamus, we performed scRNA-seq using cells dissociated from adult mouse hypothalamus (Figures 1A and S1A). In five independent experiments, we sequenced more than 14,000 single cells derived from dissociated hypothalamus tissues of seven mice. To assess the cell-type-specific transcriptional response to food-deprivation, four normal fed mice and three food-deprived (24 hr) mice were used. To increase cell number, and considering that 24 hr food-deprivation is a relatively mild treatment that unlikely to cause changes in cell identify, we combined the cells from both control and food-deprived mice for clustering analysis. From the 14,000 cells analyzed, 3,319 cells have more than 2,000 genes detectable in a single cell (Figure S1B). Semi-supervised clustering analysis (unsupervised clustering analysis [Macosko et al., 2015] followed by manually filtering [see the Experimental Procedures]) of the 3,319 cells (Figures S1C and S1D) identified 45 cell clusters with distinct gene expression signatures (Figures 1B and 1C). We applied the SC3 method to reclassify the same 3,319 cells (Kiselev et al., 2016) and found that the results were largely consistent (Figure S1E), demonstrating the reliability of our clustering results.
Figure 1. Identification of 45 Cell Types in Adult Mouse Hypothalamus by scRNA-Seq.
(A) Workflow of single-cell RNA-seq of mouse hypothalamus. Hypothalamic tissues were dissected from adult mouse brain and dissociated into single-cell suspension. Single cells and barcoded beads were captured into droplets followed by cDNA synthesis, amplification, and library preparation. After next generation sequencing, cells were classified based on their transcriptomes.
(B) tSNE plot showing the overall gene expression relationship among the 3,319 single cells with more than 2,000 genes detected in each cell. Different cell clusters are color-coded.
(C) Violin plot showing the expression of pan marker genes across the 45 cell clusters. Each cluster is color-coded. The mRNA level is shown on linear scale and adjusted for different genes. The maximum TPM value of each pan marker gene is presented on the right. Snap25 and Syt1, pan-neuronal markers; Slc17a6, pan-glutamatergic marker; Slc32a1, pan-GABAergic marker; Olig1, Sox9, Cldn5, and C1qa each marks multiple non-neuronal clusters. Glu1–Glu15, glutamatergic neuron cluster 1–15; GABA1–GABA18, GABAergic neuron cluster 1–18; Hista, histaminergic neuron; NN1–NN11, non-neuron cluster 1–11.
(D) tSNE plots showing expression of pan marker genes in distinct cell clusters. The gene expression level is color-coded. See also Figure S1.
Based on the expression of the pan neuronal makers Snap25 and Syt1, the 45 cell clusters were divided into 34 neuronal (Snap25/Syt1-high) and 11 non-neuronal clusters (Snap25/Syt1-negative or low) (Figures 1C and 1D). The 34 neuronal clusters could be further divided into 15 glutamatergic (Glu1–Glu15) and 18 GABAergic (GABA1–GABA18) subtypes based on their differential expression of Slc17a6 and Slc32a1 (Figures 1C and 1D). However, the “Hista” cluster did not belong to either of the two categories as neither Slc17a6 nor Slc32a1 was expressed in this cluster (Figure 1C). Among the non-neuronal clusters (NN1–NN11), Oligo1, Sox9, Cldn5, and C1qa were highly expressed in NN1–NN4, NN5–NN7, NN8–NN9, and NN10–NN11, respectively (Figures 1C and 1D).
Based on the above clustering results, we re-assigned each of the 14,437 single cells (≥800 transcripts detected) to the 45 cell clusters. We found that the cell number increased for each cluster (Table S1). Notably, more cells were categorized into non-neuronal clusters than neuronal clusters (Table S1), consistent with the fact that neurons express more genes than non-neuronal cells (Zeisel et al., 2015). Importantly, most of the 3,319 informative cells (≥2,000 genes detected) were correctly assigned to the original clusters, and the pooled transcriptome of each cluster before and after adding the cells with <2,000 genes detected had a high Pearson’s correlation coefficiency (Table S1), indicating that our clustering results based on the 3,319 cells could be reliably extended to the ~14,000 cells sequenced. Although some animals did not contribute to all of the 45 clusters, this is likely due to variations in tissue dissection, because each cluster includes cells from multiple animals and different treatments (Figure S1F), indicating that neither food deprivation nor different animals affects cell clustering.
Classification of Non-neuronal Cell Types in Hypothalamus
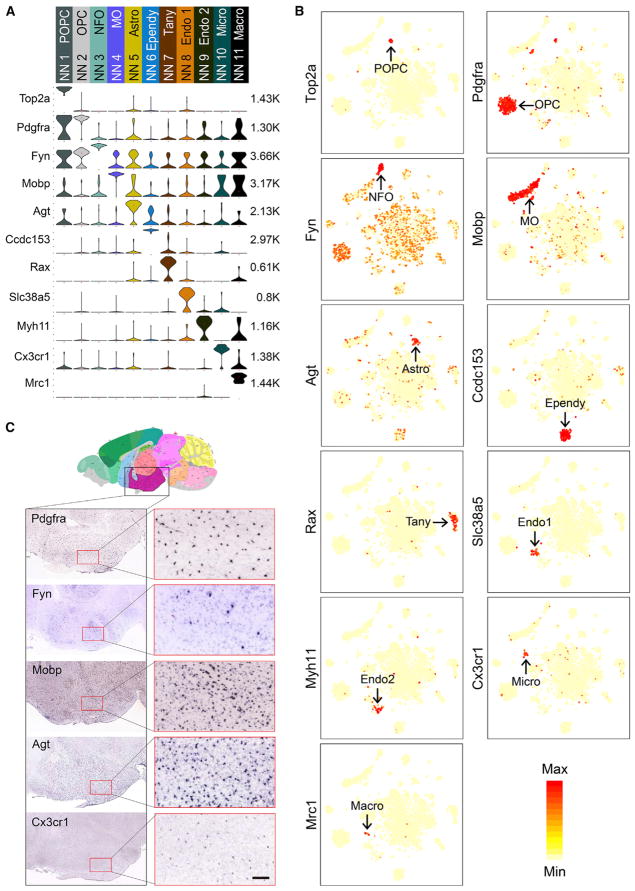
We generated gene expression heatmaps and identified marker genes for each of the non-neuronal clusters (Figures 2A and S2). The four Oligo1+ oligodendrocyte clusters could be further distinguished from each other by the expression of Top2a (proliferating oligodendrocyte precursor cell [POPC]), Pdgfra (oligodendrocyte precursor cell [OPC]), Fyn (newly formed oligodendrocyte [NFO]), and Mobp (myelinating oligodendrocyte [MO]) (Figures 2A and 2B). These subtypes reflect distinct stages of oligodendrocyte differentiation (Emery and Lu, 2015). The three Sox9+ cell clusters could also be distinguished from one another by subtype markers Agt (astrocytes [Astro]), Ccdc153 (ependymocyte [Ependy]), and Rax (tanycyte [Tany]) (Figures 2A and 2B). The two Cldn5+ endothelial cell subtypes (Endo 1 and Endo 2) were marked by Slc38a5 and Myh11, respectively (Figures 2A and 2B). The two C1qa+ groups expressed either Cx3cr1 or Mrc1 (Figures 2A and 2B), representing microglia (Micro) and macrophages (Macro). The expression of some non-neuronal subtype markers in mouse hypothalamus is confirmed by ISH data from the Allen Brain Atlas (Figure 2C). Although the non-neuronal cell types identified here are similar to those found in other brain regions (Tasic et al., 2016; Zeisel et al., 2015), tanycytes, localizing in the ventral walls of the third ventricle (3V), were only identified in our dataset, demonstrating the power of our method in capturing region-specific cell types.
Figure 2. Overview of the 11 Non-neuronal Cell Clusters in Hypothalamus.
(A) Violin plot showing the expression profile of representative marker genes in the 11 non-neuronal cell clusters. Different clusters are color-coded. The mRNA level is shown on a linear scale and adjusted for different genes. The maximum TPM value of each gene is presented on the right. POPC, proliferating oligodendrocyte progenitor cell; OPC, oligodendrocyte progenitor cell; NFO, newly formed oligodendrocytes; MO, myelinating oligodendrocyte; Astro, astrocyte; Ependy, ependymocyte; Tany, tanycyte; Endo, endothelial cell; Micro, microglia; Macro, macrophage.
(B) tSNE plots showing the expression of representative marker genes are restricted to specific non-neuronal clusters among all of the cells. The expression level is color-coded.
(C) In situ hybridization (ISH) data from Allen Brain Atlas showing the expression of non-neuronal subtype markers Pdgfra, Fyn, Mobp, Agt, and Cx3cr1 in hypothalamus. Left: the coronal sections of the entire hypothalamic region. Right: enlarged images of the regions in red squares. Scale bar, 100 μm.
See also Figure S2.
Oligodendrocyte Differentiation Is Associated with Dynamic Transcriptional Changes
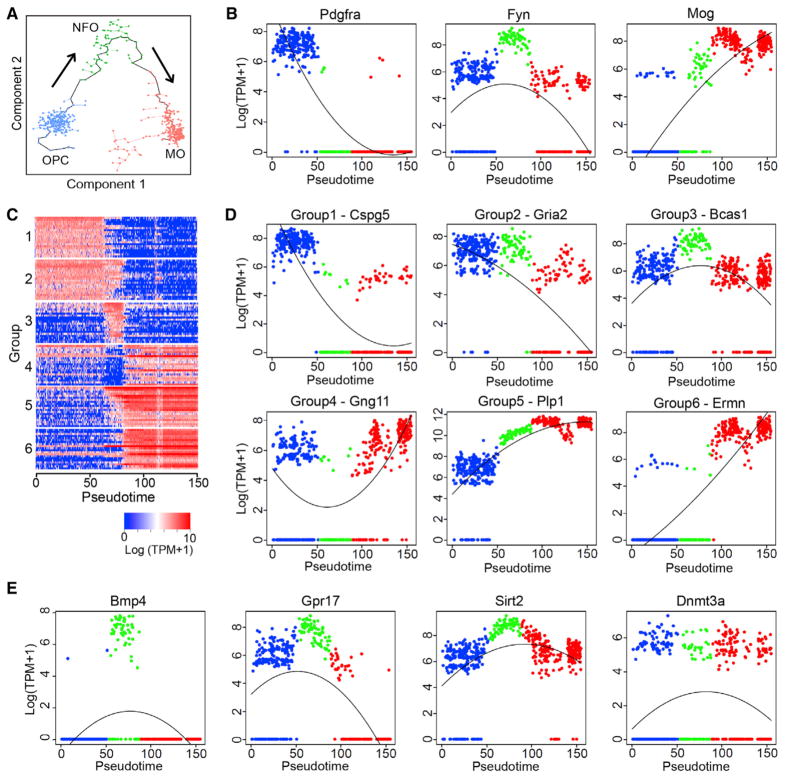
Identifying OPC, NFO, and MO cell clusters in our dataset indicated that oligodendrocyte differentiation and axon myelination are still taking place in adult hypothalamus. Additionally, the single-cell transcriptomes of oligodendrocytes at different maturation stages provide an opportunity for understanding the transcriptional program of oligodendrocyte differentiation in vivo. To this end, we performed an unsupervised pseudotime analysis with Monocle (Trapnell et al., 2014), in which OPCs, NFOs, and MOs were linked according to their gene expression profile (Figure 3A). Based on the established differentiation direction of oligodendrocyte (Emery and Lu, 2015), we chose the direction of OPCs to NFOs and then to MOs (Figure 3A). Sequential expression of Pdgfra, Fyn, and Mog matches this direction (Figure 3B), indicating that the pseudotime axis mimics the oligodendrocyte maturation process. To further characterize the transcriptional program underlying oligodendrocyte differentiation, we identified six groups of genes with distinct expression patterns along the differentiation process (Figures 3C and 3D; Table S2). Gene ontology (GO) analysis revealed that different biological processes were enriched in different gene groups (Table S3). For example, the genes in groups 1 and 2, which are highly expressed in OPCs but repressed during oligodendrocyte maturation, are enriched for regulation of development and differentiation, while genes in groups 5 and 6 with a low-to-high trend are enriched for axon ensheathment and myelination. Notably, genes in group 3 are high in NFOs, suggesting that these genes might be important for OPCs to MOs transition. Consistently, several genes of this group have been found to play a role in oligodendrocyte differentiation, such as Bmp4 (Samanta and Kessler, 2004) and Gpr17 (Chen et al., 2009) (Figure 3E). Interestingly, some epigenetic factors, such as Sirt2 and Dnmt3a, are also highly transcribed in NFO (Figure 3E), indicating that chromatin remodeling and epigenetic regulation may play an important role during oligodendrocyte maturation.
Figure 3. Transcriptional Dynamics during Oligodendrocyte Maturation.
(A) Unsupervised ordering of OPCs (blue), NFOs (green), and MOs (red) based on their gene expression profiles. Minimal spanning tree is shown in black. Arrows indicate the direction of differentiation.
(B) Scatterplots showing the transcriptional dynamics of Pdgfra, Fyn, and Mog along the pseudotime. X axis represents the pseudotime axis, y axis shows gene expression level on log scale. Blue, green, and red dots represent OPCs, NFOs, and MOs, respectively.
(C) Heatmap showing six groups of genes with distinct expression dynamics during oligodendrocyte maturation. Columns are individual cells organized along the pseudotime and rows represent individual genes. Twenty of the most representative genes from each group are plotted. Expression level is color-coded. (D) Scatterplots showing the transcriptional dynamics of representative genes belong to groups 1 to 6 in Figure 2C along the maturation of oligodendrocyte. (E) Scatterplots showing the expression of NFO-specific genes Bmp4, Grp17, Sirt2, and Dnmt3a along the pseudo-timeline.
See also Figure S3.
A very recent study also analyzed mouse oligodendrocyte subtypes and differentiation using scRNA-seq (Marques et al., 2016). Using Drop-seq, a different scRNA-seq technique, we successfully captured the OPC to MO differentiation process in adult mouse hypothalamus through pseudotime analysis. Importantly, the transcriptional dynamics of stage-specific genes along the developmental axis revealed in our study is highly similar to the other study (Marques et al., 2016) (Figure S3), thus validating our approach.
ScRNA-Seq Reveals Transcriptional Features of Tanycyte and Tanycyte Subtypes
Tanycyte is a hypothalamus-specific non-neuronal cell type. The cell bodies of tanycytes occupy the floor and ventrolateral walls of the 3V and project laterally into adjacent hypothalamic regions, including dorsomedial hypothalamic nucleus (DMH), ventromedial hypothalamic nucleus (VMH), arcuate hypothalamic nucleus (ARH), and median eminence (ME) (Bolborea and Dale, 2013). This morphological feature distinguishes tanycytes from neighboring ependymocytes that occupy dorsal walls of the 3V (Goodman and Hajihosseini, 2015) (Figure 4A). Previous studies have revealed important functions of tanycytes in regulating energy homeostasis through diverse mechanisms (Goodman and Hajihosseini, 2015), but the underlying molecular mechanism remains largely unknown.
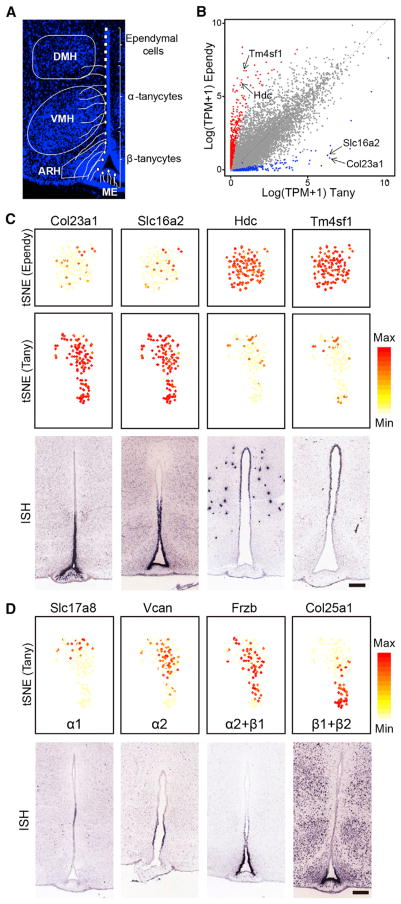
Figure 4. Gene Expression Features of Tanycyte and Tanycyte Subtypes.
(A) Schematic diagram showing the spatial distribution and morphology of tanycytes, which can be further divided into α and β subtypes. ARH, arcuate hypothalamic nucleus; VHM, ventromedial hypothalamic nucleus; DMH, dorsomedial hypothalamic nucleus; ME, median eminence.
(B) Scatterplot comparing the expression profiles of tanycytes and ependymocytes. X axis and y axis represent the average expression level of certain genes among all tanycytes and all ependymal cells, respectively. Each dot represents a single gene, blue dots represent tanycyte-enriched genes and red dots represent ependymocyte-enriched genes. The tanycyte-specific genes Col23a1 and Slc16a2, as well as the ependymocyte-specific genes Hdc and Tm4sf1 are indicated by arrows.
(C) Expression patterns of selected tanycyte- and ependymocyte-specific genes. tSNE plots (upper and middle panels) showing the selective expression of Col23a1 and Slc16a2 in tanycytes (Tany), Hdc and Tm4sf1 in ependymocytes (Ependy). Gene expression level is color-coded. ISH data (lower panels, from Allen Brain Atlas) show the distribution of corresponding genes along the 3V walls. Scale bar, 200 μm.
(D) Potential tanycyte subtype markers identified by scRNA-seq. tSNE plots (upper panels) showing the expression of selected genes enriched in subsets of tanycytes. The genes are ordered according to their expression level along the vertical axis of the tSNE map. For each gene, potential tanycyte subtype(s) that express the marker gene are listed. Gene expression level is color-coded. ISH data (lower panels, from Allen Brain Atlas) indicate a dorsal-to-ventral distribution of corresponding genes along the 3V walls. Scale bar, 200 μm. See also Figure S4.
Our clustering analysis identified a Rax+ tanycyte cell cluster (Miranda-Angulo et al., 2014), which is distinct from the Ccdc153+ ependymocyte cluster (Figures 2A and 2B), indicating that tanycytes are transcriptionally distinct from ependymocytes and other cell types (Figure S4A). Consistent with previous studies (Lee et al., 2012; Robins et al., 2013), we found that the radial glia makers Nestin (Nes) and Vimentin (Vim) are highly transcribed in tanycytes (Figure S4B), suggesting that these cells may originate from embryonic radial glia and function as neural stem cells in adult hypothalamus (Lee et al., 2012). However, both Nes and Vim are also highly expressed in ependymal cells (Figure S4B) and as such cannot serve as tanycyte-specific markers. To characterize the molecular features of tanycytes and their neighboring ependymal cells, we compared the transcription profiles of these two cell clusters (Figure 4B), which revealed many genes that are differentially expressed in tanycytes and ependymocytes (Table S4). GO analysis of the tanycyte-specific genes identified terms that include signal transduction, GPCR signaling pathway, and modulation of synaptic transmission (Figure S4C), consistent with the known function of tanycytes in transmission of metabolic signals to neurons in regulating homeostasis (Goodman and Hajihosseini, 2015). The tanycyte-enriched genes include Col23a1, Slc16a2, Lhx2, and Ptn (Figures 4C and S4D), some of which have been linked to tanycyte development and function, such as Lhx2 (Salvatierra et al., 2014) and Slc16a2 (Mayerl et al., 2014). To test the potential of these differentially expressed genes to serve as tanycyte- and ependymocyte-specific markers, we examined their expression pattern in mouse brain with the ISH data from Allen Brain Atlas. Data shown in Figures 4C and S4D confirmed that these genes marked different cell populations along the dorsal-ventral axis of the 3V walls, which is consistent with the known location of tanycytes and ependymal cells (Figure 4A).
In addition to identifying tanycyte-specific markers, we further analyzed transcriptional heterogeneity among the tanycyte subtypes. Currently, tanycytes are separated into different subpopulations based on their physical location: dorsal tanycytes projecting to DMH and VMH are named α tanycytes, which are further divided into more dorsal α1 subtype and more ventral α2 subtype, the cells distributed at the ventral 3V walls that are in contact with ARH are termed β1 tanycytes, and the cells located at the bottom of the 3V walls that are in contact with ME are regarded as β2 tanycytes (Mathew, 2008). The lack of reliable molecular markers of tanycyte subtypes is a major hurdle preventing understanding tanycyte subtype-specific function. To overcome this hurdle, we attempted to identify tanycyte subtype markers by analyzing the scRNA-seq data of the tanycytes. We first identified highly variable genes within tanycyte cluster through principle components analysis (PCA). By plotting these genes on the tSNE map and comparing these with the public ISH dataset, we found a good correlation between the distribution pattern of the candidate marker genes on tSNE map and their in situ expression pattern along the ventral-dorsal axis (Figure 4D). For example, the Slc17a8 and Col25a1 located in the two most distal parts of the tSNE map are consistent with the ISH data showing that they are expressed in the most dorsal-and ventral-tanycyte cell populations, respectively (Figure 4D). Based on the tSNE map and ISH data, and considering the spatial distribution of tanycyte subtypes (Mathew, 2008), we believe that Slc17a8 and Col25a1 can serve as potential markers for α1 and β subtypes, respectively. Similarly, the expression patterns of other tanycyte subtype-specific genes can be predicted based on tSNE map (Figures 4D and S4E) and some of these are supported by ISH data (Figure 4D). Notably, although specific marker genes (or combinations of marker genes) can be used to roughly separate tanycyte subtypes, many genes exhibited a gradient, rather than a clear-cut distribution across tanycyte subpopulations (Figures 4D and S4E), consistent with the notion that tanycytes may be composed of continuous cell trajectory with transition zones between different subtypes (Mathew, 2008).
The Hypothalamus Harbors Multiple Transcriptionally Distinct Neuron Subtypes
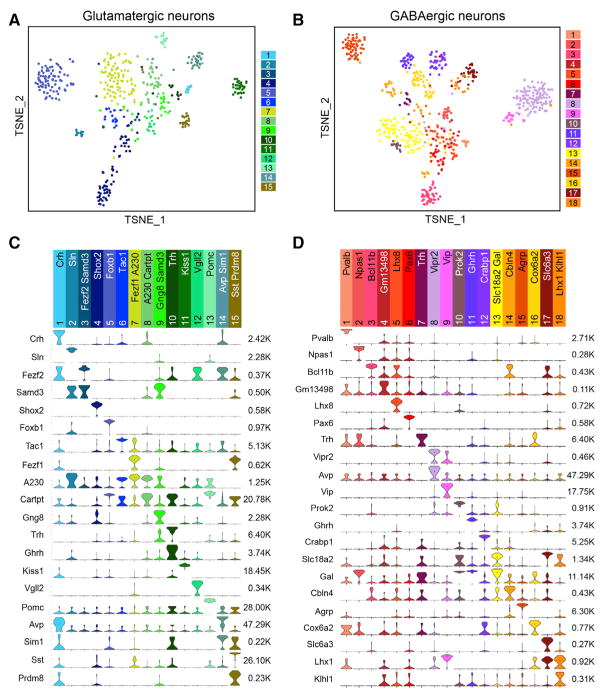
Our clustering analyses identified 15 glutamatergic neuron subtypes (Glu1–GLu15), 18 GABAergic neuron subtypes (GABA1–GABA18), and 1 histaminergic neuron cluster (Hista) expressing high levels of Hdc but negligible Slc17a6 or Slc32a1 (Figure 1C). We generated single-cell transcriptome heatmaps (Figure S5A) and visualized the glutamatergic and GABAergic neuron subtypes by tSNE (Figures 5A and 5B). In addition, potential subtype-specific marker genes for each of the 34 neuronal clusters were identified (Table S5). The majority of the 34 neuronal clusters contain subtype-specific genes that are unique to that cluster, and in some cases, a neuron cluster could be defined by the combinatorial expression of marker genes (Figures 5C and 5D). As expected, a number of neuron subtypes are distinguished by the expression of specific neuropeptides. For example, Kiss1 and Pomc could represent Glu11 and Glu13 cell clusters, respectively (Figure 5C), while Vip and Agrp could represent GABA9 and GABA15 cell clusters, respectively (Figure 5D). Additionally, many transcription factors (e.g., Foxb1 for Glu5, Npas1 for GABA2, Lhx8 for GABA5) exhibited neuron subtype-specific expression pattern (Figures 5C and 5D) consistent with their role in controlling neuron differentiation and identity.
Figure 5. Glutamatergic and GABAergic Neuron Subtypes in Hypothalamus.
(A) tSNE plot showing the 15 glutamatergic neuron subtypes identified in hypothalamus. Differentially expressed genes among all subtypes are used for dimension reduction. Different neuron subtypes are color-coded.
(B) tSNE plot showing the 18 GABAergic neuron subtypes in hypothalamus.
(C) Violin plots showing the expression of subtype markers across the 15 glutamatergic neuron subtypes. Columns represent different neuron clusters which are color-coded. Marker genes for each cluster are indicated. The gene expression level is shown on linear scale and adjusted for different genes. The maximum TPM value for each marker gene is presented on right. A230 is A230065H16Rik. (D) Violin plots showing the expression of subtype markers across the 18 GABAergic neuron subtypes. Columns represent different neuron clusters which are color-coded.
See also Figure S5.
Based on expression of the marker genes, many neuronal subtypes identified here could be assigned to neuron subtypes already described in the hypothalamus. For example, in glutamatergic clusters, Glu5 represents neurons in the mammillary body (MM) that express Foxb1 (Figure S5B), while Glu11 and Glu13 represent Kiss1+ and Pomc+ neurons in ARH. On the other hand, the GABA8 and GABA9 clusters represent Avp+ and Vip+ neurons in the suprachiasmatic nucleus (SCH), while GABA11 and GABA15 clusters represent Ghrh+ and Agrp+ neurons in ARH. In addition to confirming known hypothalamic neuron subtypes, our dataset also revealed gene expression features in specific neuron clusters. For example, Cartpt and Cck are actively transcribed in the Glu5 cluster (Foxb1+ neuron in MM), while Prph and Wif1 were co-expressed with Hdc in the Hista cluster (histaminergic neurons in tuberomammillary nucleus), both of which were verified by ISH (Figures S5B and S5C). Additionally, we found that the combined expression of Sst and Prdm8 defines the Sst+ neuroendocrine cells (Figure 5C, Glu15) located in PVpo (periventricular hypothalamic nucleus, preoptic part) with known function in regulating growth hormone secretion (Murray et al., 2015), but without a definitive molecular marker. We confirmed the co-expression of Sst and Prdm8 in PVpo, but not in cortex, by immunostaining (Figure S5D). Collectively, these results demonstrate that our unbiased scRNA-seq analyses are able to reveal cell types as well as cell-type-specific transcriptional features in the hypothalamus.
Relationship among Neuron Subtypes Assessed by Transcriptome
Currently, the developmental relationship among the different neuron subtypes in the hypothalamus is largely unknown. Taking advantage of the scRNA-seq dataset, we performed pairwise comparison and hierarchical clustering analyses on the glutamatergic and GABAergic neuron populations to assess their relationship based on transcriptome (Figure S5E). Interestingly, we found six glutamatergic clusters (Glu10 to Glu15), which represented different neurosecretory cells, are clustered together (Figure S5E). The separation of these neurons from the other glutamatergic subtypes indicates that these neurons possess distinct transcriptional programs that may reflect their distinct developmental process and functional properties. On the other hand, among GABAergic clusters, the SCH Vipr2+ (GABA8) and Vip+ (GABA9) neurons clustered together (Figure S5E), consistent with their similar developmental origin (VanDunk et al., 2011) and function (regulating circadian rhythm) (Dibner et al., 2010). These observations indicate that our cell-type-specific transcriptomic dataset has the potential to reveal lineage relationship and functional relevance among the various hypothalamic neurons.
Divergent Expression Patterns of Neuropeptides and Receptors across Neuronal Subtypes
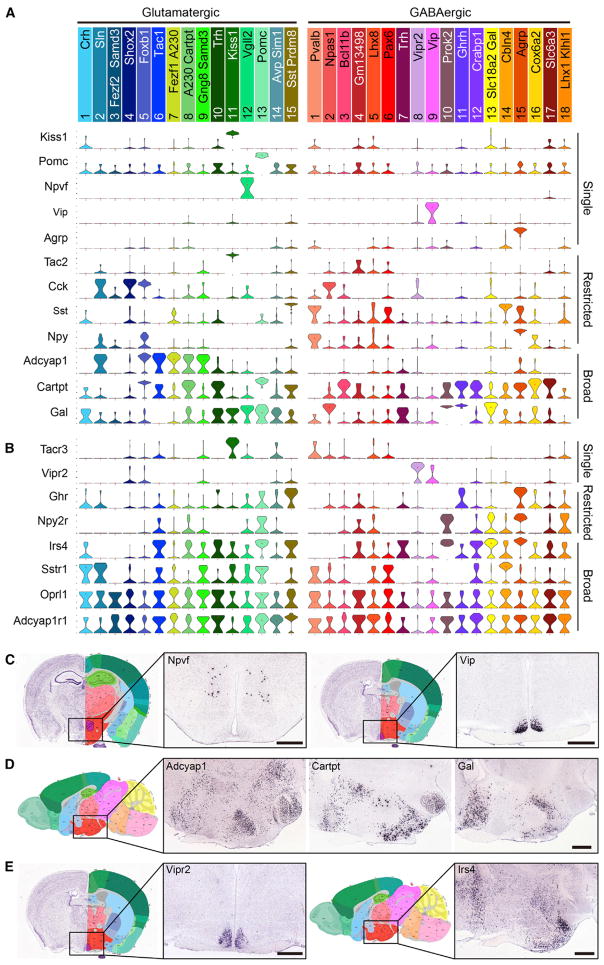
The expression and function of various neuropeptides in the hypothalamus has been widely studied. However, a comprehensive expression profile of neuropeptide and receptors in the hypothalamic neurons is still unavailable. Thus, we assessed the expression profile of different neuropeptides and receptors in different hypothalamic neuron subtypes and found a highly divergent expression pattern among the various neuronal subtypes (Figures 6A and 6B and S6).
Figure 6. Divergent Expression Pattern of Neuropeptides and Receptors among the Hypothalamic Neuron Subtypes.
(A) Violin plots showing the expression of selected neuropeptides among the neuron subtypes in hypothalamus. Gene expression level is presented on linear scale and adjusted for different genes.
(B) Violin plots showing the expression of selected neuropeptide receptors among the neuron subtypes in hypothalamus.
(C) Coronal sections showing that Npvf and Vip are selectively expressed in different hypothalamic regions. ISH data are from Allen Brain Atlas. The boxed regions in the left panels were enlarged and shown in right panels.
(D) Sagittal sections showing the broad distribution of Adcyap1, Cartpt, and Gal in hypothalamus.
(E) ISH showing that Vipr2 is selectively expressed in superchiasmatic nucleus while Irs4 is widely distributed in hypothalamus. Scale bars, 500 μm. See also Figure S6.
First, we found that the expression of the majority of neuropeptide genes is restricted to one or a few neuron subtypes. For example, Kiss1, Pomc, Npvf, Vip, and Agrp are each expressed in only one neuron subtype, while Tac2, Cck, Sst, and Npy are each expressed in two to four neuron subtypes with distinct patterns (Figures 6A and 6C). However, some neuropeptide genes, such as Adcyap1, Cartpt, and Gal, are expressed in multiple neuron subtypes (Figure 6A), consistent with their broad distribution in hypothalamus (Figure 6D). Although previous studies have used ISH or immunostaining to assess the co-expression of different neuropeptides, these methods can only profile a limited number of neuropeptides and neuron subtypes. Taking the advantage of our scRNA-seq dataset, we analyzed the gene expression profile of all detectable neuropeptides in the hypothalamic neuron subtypes. We found that co-expression of multiple neuropeptides is a common feature of many hypothalamic neurons. For instance, Npy is also expressed in Agrp+ neurons (GABA15) and Cartpt is also expressed in Pomc+ neurons (Glu13) (Figure 6A), which are consistent with previous findings (Schwartz et al., 2000). In addition, we found that Sst+ neurons (GABA1) express Npy, while Foxb1+ neurons (Glu5) co-express Cck, Adcyap1, and Cartpt (Figure 6A). Interestingly, Ghrh co-expresses with Trh in a glutamatergic population (Glu10) (Figure 5C), but also co-localizes with Gal in a GABAergic neuron subtype (GABA11) (Figure 5D). The extensive co-expression of multiple neuropeptide genes in a neuron subtype suggests complex crosstalk among different peptide signaling pathways.
Expression of a specific neuropeptide in certain neuron subtypes determines a specific signal that can be sent from these cells. Conversely, expression of a specific neuropeptide receptor indicates that these cells can receive a specific signal. Thus, we also analyzed the gene expression profiles of neuropeptide receptors among the various hypothalamic neuron subtypes. We found that like the expression profile of neuropeptides, the expression profile of neuropeptide receptors mRNA is also very diverse (Figures 6B and S6B). Some receptor genes are expressed in only one or a few neuron subtypes, such as Vipr2 (GABA8) and Ghr (Glu15 and GABA15) (Figures 6B and 6E), which are consistent with previous studies (Harmar et al., 2002; Murray et al., 2015). In contrast, several peptide receptor genes, such as Irs4, Sstr1, OPrl1, and Adcyap1r1, are broadly expressed in multiple neuron subtypes (Figures 6B and 6E), suggesting extensive regulatory roles of these signaling pathways in the hypothalamus. Notably, a number of neuropeptide receptors genes are co-expressed with their ligands in some neuron subtypes, such as Tacr3 in Tac2+ neurons (Glu11), Sstr1 in Sst+ neurons (GABA1 and GABA14), and Npy2r in Npy+ neurons (GABA15) (Figures 6A and 6B), indicating that a feedback mechanism may be used to regulate the corresponding neuropeptide secretion in these neuron subtypes.
Earlier studies on peptidergic neuron function in hypothalamus have paid less attention to the role of fast synaptic transmission mediated by conventional neurotransmitters. However, the potential interactions between slow-acting peptidergic signals and fast ionotropic signals are supported by some recent studies, which suggest an essential role of GABA/glutamate-mediated synaptic transmission in controlling animal behaviors in certain peptidergic neurons (Krashes et al., 2014; Tong et al., 2008). Consistently, our dataset shows that all hypothalamic peptidergic neurons can be categorized into either glutamatergic (Slc17a6+) or GABAergic (Slc32a1+) (Figure 6A), indicating that most, if not all, peptidergic neurons could communicate with downstream targets through fast synaptic transmission. Notably, the neurotransmitter and neuropeptide present in the same neuron do not necessarily function simultaneously because they are packed into different types of vesicles and respond to distinct neuronal activity (Ludwig and Leng, 2006).
Collectively, our data provide a comprehensive neuropeptide and receptor expression profile of hypothalamic neuron subtypes, which suggests complex crosstalk among the different neuropeptide signaling pathways, between neuropeptides and neurotransmitters, and between different peptidergic neuron populations.
Neuronal Subtypes Revealed by Single Cell RNA-Seq
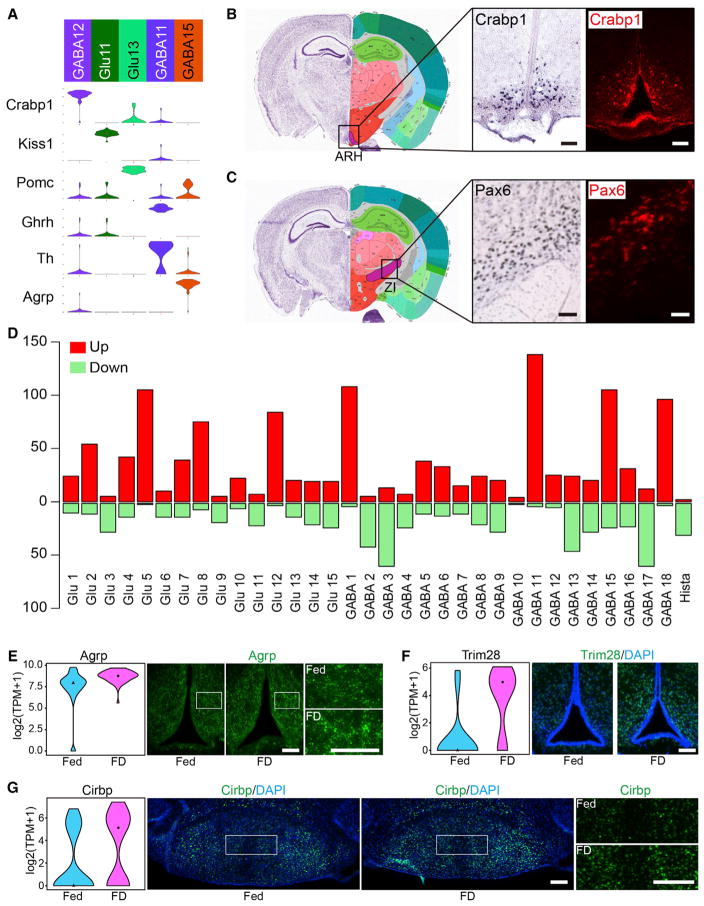
Unbiased scRNA-seq is capable of de novo discovery of cell types with distinct transcriptional features. Indeed, we identified several neuronal subtypes with specific molecular markers from our dataset. For instance, we found that Crabp1, which encodes a retinoic acid binding protein, is highly expressed in the GABA12 cluster (Figure 7A). ISH and immunostaining showed that this gene is restricted to the ARH across the hypothalamus (Figure 7B), suggesting that it marks a distinct neuronal subtype in this region. To exclude the possibility that Crabp1 is expressed from other known cell populations located in ARH, we queried the expression of Kiss1, Pomc, Ghrh, Th, and Agrp in Crabp1+ neurons. All of them are highly expressed in specific cell types in ARH (Figure 7A). Our data showed that none of these genes are expressed in Crabp1+ neurons (Figure 7A), supporting the notion that the Crabp1+ neurons represent a cell type in ARH. Another neuron subtype identified in our study is the Pax6+ GABAergic neuron (GABA6). Pax6 has an established function in neural development (Ypsilanti and Rubenstein, 2016), but its expression profile and function in the adult brain is largely unknown. A recent study has identified a Pax6+ interneuron population in layer I of cerebral cortex (Zeisel et al., 2015). Here, we found that Pax6 is specifically expressed in the GABA6 cluster (Figure 5D). Both ISH and antibody staining support the localization of Pax6+ neurons in the zona incerta (ZI) (Figure 7C), a hypothalamic region whose cell composition and function are poorly understood. Identifying Crabp1+ and Pax6+ neuronal subtypes in the hypothalamus has set the stage for dissecting their specific functions in the hypothalamus.
Figure 7. Neuron Types and Assessment of Food Deprivation-Induced Transcriptional Changes in Hypothalamus.
(A) Violin plot showing the expression of selected genes in neuron subtypes located in ARH. Gene expression level is shown on linear scale and adjusted for different genes.
(B) ISH and immunostaining on coronal hypothalamus sections showing that the Crabp1+ neurons are restricted in ARH. ISH data are from Allen Brain Atlas. Scale bars, 100 μm.
(C) ISH and immunostaining showing presence of Pax6+ neurons in ZI. Scale bars, 100 μm.
(D) Bar graphs showing the number of genes affected by food deprivation across hypothalamic neuron subtypes. Red and green bars represent up- and downregulated genes. See also Table S6.
(E) Violin plots and immunostaining showing that Agrp is upregulated by food deprivation. Fed, n = 62; FD, n = 50. Fold change = 1.52, p = 0.00016. Fed, normal feeding; FD, food deprivation. Scale bars, 100 μm.
(F) Violin plots and immunostaining showing that Trim28 is upregulated in ARH by food deprivation. Fed, n = 16; FD, n = 19. Fold change = 2.76, p = 0.00081. Scale bar, 100 μm.
(G) Violin plot and immunostaining showing that Cirbp in MM neurons is upregulated by food deprivation. Fed, n = 103; FD, n = 97. Fold change = 1.97, p = 0.00099. Scale bars, 200 μm.
See also Figure S7.
Cell-Type-Specific Transcriptional Response to Food Deprivation Revealed by scRNA-Seq
Hypothalamus is an important brain region controlling feeding and metabolism (Schwartz et al., 2000). Classification of the various neuronal subtypes allows us to identify the neuron subtypes involved in regulating feeding behavior. To this end, we examined the transcriptional response to food deprivation across the hypothalamic neuronal subtypes by comparing single-cell gene expression profiles between ad libitum fed and food-deprived mice. The results revealed that food deprivation has differential effects on gene expression of the various hypothalamic neuron subtypes. The differentially expressed genes (DEGs) are mainly enriched in Glu5, Glu8, Glu12, GABA1, GABA11, GABA15, and GABA18 clusters (Figures 7D and S7A; Table S6), indicating that these neuronal subtypes are the main cells responding to food deprivation. Identification of the Agrp+ (GABA15), Ghrh+ (GABA11), and Npvf+ (Glu12) neurons as food deprivation-responding cell types are consistent with previous studies (Henry et al., 2015; Murray et al., 2015; Wahab et al., 2015) and thus validate our approach. Importantly, scRNA-seq uncovered cell types, such as MM neurons (Glu5), that were not previously linked to energy homeostasis, exemplifying the ability of our unbiased approach in revealing neuron subtype-specific functions. GO analyses showed that DEGs identified in different neuronal clusters are enriched for genes of distinct functions (Figure S7B; Table S7) indicating that different neuronal subtypes are functionally different. To confirm the gene expression changes revealed by scRNA-seq, we first focused on Agrp+ neurons (GABA15), which are known for controlling feeding and energy expedition. Immunostaining showed that the Agrp level was significantly increased upon food deprivation (Figure 7E), consistent with our scRNA-seq result and previous knowledge (Henry et al., 2015). Additionally, immunostaining confirmed upregulation of Trim28 in Ghrh+ neurons (GABA11) in the ARH and increased expression of Cirbp in MM neurons (Glu5) upon food deprivation (Figures 7F and 7G).
DISCUSSION
Comprehensive Cell-Type Classification in Hypothalamus Can Facilitate Its Functional Study
Understanding the cell composition and cell-type-specific transcription features of the hypothalamus is essential for understanding the function of this important brain region. Several previous studies have attempted to characterize the cell types in the hypothalamus based on cell location, morphology, connection, function, and marker gene expression (Brown et al., 2013; Lee et al., 2015; Mathew, 2008; Wu et al., 2014). However, a systematic hypothalamic cell map of transcriptomic features is still lacking. Using the Drop-seq technique (Macosko et al., 2015), we profiled transcriptomes of more than 14,000 single cells and identified 45 transcriptionally distinct cell subtypes in the adult mouse hypothalamus. Most of the non-neuronal cell types identified in this study are similar to those found in the cerebral cortex (Zeisel et al., 2015), suggesting that most non-neuronal cells are widely distributed across various brain regions. In contrast, the neuronal cell types are largely hypothalamus-specific, indicating that different neuron composition underlies distinct functions of different brain regions. Our work, therefore, provides a comprehensive overview of hypothalamic cell types based on their transcriptional features, which will greatly facilitate functional studies of hypothalamus.
First, the cell-type-specific markers identified here provide an unambiguous cell-type definition, which can help to unify studies from different laboratories to resolve confusion generated due to the use of different criteria in defining cell types. Second, the information of unique and combinatorial markers for different cell types enable the development of genetic or viral tools to achieve cell-type-specific labeling and manipulation, which is essential for dissecting cell-type-specific function in such a complex brain region. Third, by assessing the relationship of different hypothalamic cell types based on transcriptional profiles, hypotheses regarding the development and function of different cell subtypes can be generated and tested, which in turn can advance our understanding of hypothalamus. Fourth, identifying Crabp1+ neurons in ARH and Pax6+ neurons in ZI indicates that uncharacterized neuron subtypes in hypothalamus still remain, even within a relatively well-studied region such as the ARH.
Divergent Expression Pattern of Neuropeptides and Receptors in the Hypothalamus
Understanding the expression and function of various neuropeptides has long been an interest of the hypothalamus scientific community. Our scRNA-seq datasets provide a comprehensive expression profile of neuropeptides and their receptors across hypothalamic neuron subtypes. Analysis of this expression profile has revealed several important features. First, neuropeptides feature a very divergent expression pattern and that cellular heterogeneity can be largely resolved based on the expression of a specific neuropeptide or a combination of neuropeptides. Second, co-expression of multiple neuropeptides can be analyzed in our dataset, thus facilitating the study of crosstalk among different neuropeptide signals. Third, by examining the distribution of neuropeptides and corresponding receptors, the potential regulatory relationship within individual neuron subtypes or among different neuron populations can be analyzed, revealing complex intra- or inter-population regulation. Fourth, our analysis has uncovered extensive overlap between neuropeptide and conventional neurotransmitters across hypothalamic neuron subtypes, indicating a functional interaction between the two neuronal signaling systems, which begs for further studies on the crosstalk between the slow peptidergic signaling and the fast ionotropic signaling.
Gene Expression Features of Tanycyte and Tanycyte Subtypes
Tanycyte is a hypothalamus-specific, non-neuronal cell type. Accumulating evidence suggests its diverse physiological functions in neuroendocrine, metabolism, and neurogenesis (Goodman and Hajihosseini, 2015). However, the molecular features underlying tanycyte heterogeneity and diverse function have been elusive. Our scRNA-seq dataset not only confirm tanycyte as a distinct cell type, but also reveal tanycyte-specific markers that can be used to distinguish them from ependymal cells. More importantly, by analyzing single-cell transcriptomes of tanycytes, we identified tanycyte subtypes with distinct transcription profiles. The tanycyte- and tanycyte-subtype-specific marker genes identified will allow the development of genetic tools for achieving cell-(sub)type-specific manipulation for dissection the function of tanycyte and tanycyte subtypes.
Revealing Cell-Type-Specific Transcriptional Response to Food Deprivation
In addition to cell-type classification, scRNA-seq can be applied to dissect cell-type-specific transcriptional dynamics in complex tissues under different physiological and pathological conditions. This is particularly important for the nervous system that has great cell heterogeneity and cell-subtype-specific functions (Knight et al., 2012), but conventional RNA-seq cannot resolve such level of cell-type heterogeneity. As a proof-of-principle study, we compared the transcriptional program between normal-fed and food-deprived animals across hypothalamic neuronal subtypes, which revealed 7 out of the 34 subtypes exhibit significant transcriptome changes. The analysis not only revealed the specific neuronal clusters that respond to food deprivation, but also uncovered cell types that have not been previously linked to feeding and energy homeostasis, thus highlighting the capability of unbiased single-cell profiling in revealing biological insight into brain functions.
EXPERIMENTAL PROCEDURES
Animals
All animal experiments followed the guidelines of the Institutional Animal Care and Use Committee at Harvard Medical School. Young adult female (8- to 10-week-old) B6D2F1 mice (C57B6 female × DBA2 male) were used. One day before the experiments, each animal was separated into individual fresh cages. For 24 hr food deprivation treatment, only water was provided.
Tissue Dissection, Single-Cell Dissociation, and Library Construction
The hypothalamus tissue was dissected from acute brain slices of adult (8- to 10-week-old) mice. The tissue was dissociated into single-cell suspension using a papain-based dissociation protocol (Brewer and Torricelli, 2007) with some modifications (see the Supplemental Experimental Procedures). The single cells and barcoded-beads were captured into nanolitersized droplets with microfluidic device, followed by library construction as previously described (Macosko et al., 2015).
Cell Clustering
The 3,319 cells with at least 2,000 genes detected in each single cell were used for clustering analysis. To classify the cells, the R package Seurat was used (Macosko et al., 2015). The highly variable genes were identified from these cells using Seurat with the default setting. Then these highly variable genes were used for principle component analysis (PCA). The statistically significant PCs were used for two-dimension t-distributed stochastic neighbor embedding (tSNE). Based on the tSNE map, density-based clustering (DBSCAN) was used to cluster cells based on their proximity, resulting in 40 clusters. The largest neuronal cluster containing 1,574 cells was extracted for further clustering using the same strategy described above. The same analysis was repeated for another two rounds. In total, 73 cell clusters were identified. Following this unsupervised clustering analysis, we further filtered out clusters representing double droplets, clusters from non-hypothalamic tissues, as well as clusters with less than ten cells (see the Supplemental Experimental Procedures). After filtering, a total of 45 cell clusters were identified.
Supplementary Material
Highlights.
Transcriptome-based cell-type classification in mouse adult hypothalamus
Transcriptional features of oligodendrocyte differentiation and tanycyte subtypes
Neuropeptide and receptor expression patterns of hypothalamic neuronal subtypes
Cell-type-specific transcriptional responses to food deprivation
Acknowledgments
We thank Drs. Li Shen and Hao Wu for their initial help in establishing the Drop-seq platform, Dr. Sarah E. Ross for the Prdm8 antibody, and Dr. Luis M. Tuesta for critical reading of the manuscript. This project was partly supported by NIH U01 MH105960 and HHMI. Y.Z. is an investigator of the Howard Hughes Medical Institute.
Footnotes
ACCESSION NUMBERS
The accession number for the sequencing data reported in this work is GEO: GSE87544.
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2017.03.004.
AUTHOR CONTRIBUTIONS
Y.Z. and R.C. conceived the project. Y.Z. and R.C. designed the study. R.C. and X.W. performed the experiments. L.J. and X.W. analyzed the data. R.C., X.W., L.J., and Y.Z. interpreted the data. R.C. and Y.Z. wrote the manuscript.
References
- Bolborea M, Dale N. Hypothalamic tanycytes: potential roles in the control of feeding and energy balance. Trends Neurosci. 2013;36:91–100. doi: 10.1016/j.tins.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR. Isolation and culture of adult neurons and neurospheres. Nat Protoc. 2007;2:1490–1498. doi: 10.1038/nprot.2007.207. [DOI] [PubMed] [Google Scholar]
- Brown CH, Bains JS, Ludwig M, Stern JE. Physiological regulation of magnocellular neurosecretory cell activity: integration of intrinsic, local and afferent mechanisms. J Neuroendocrinol. 2013;25:678–710. doi: 10.1111/jne.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wu H, Wang S, Koito H, Li J, Ye F, Hoang J, Escobar SS, Gow A, Arnett HA, et al. The oligodendrocyte-specific G protein-coupled receptor GPR17 is a cell-intrinsic timer of myelination. Nat Neurosci. 2009;12:1398–1406. doi: 10.1038/nn.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton DA, McKinley MJ, Weisinger RS. Hypothalamic integration of body fluid regulation. Proc Natl Acad Sci USA. 1996;93:7397–7404. doi: 10.1073/pnas.93.14.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Elmquist JK, Elias CF, Saper CB. From lesions to leptin: hypothalamic control of food intake and body weight. Neuron. 1999;22:221–232. doi: 10.1016/s0896-6273(00)81084-3. [DOI] [PubMed] [Google Scholar]
- Emery B, Lu QR. Transcriptional and epigenetic regulation of oligodendrocyte development and myelination in the central nervous system. Cold Spring Harb Perspect Biol. 2015;7:a020461. doi: 10.1101/cshperspect.a020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokce O, Stanley GM, Treutlein B, Neff NF, Camp JG, Malenka RC, Rothwell PE, Fuccillo MV, Südhof TC, Quake SR. Cellular taxonomy of the mouse striatum as revealed by single-cell RNA-seq. Cell Rep. 2016;16:1126–1137. doi: 10.1016/j.celrep.2016.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Goodman T, Hajihosseini MK. Hypothalamic tanycytes-masters and servants of metabolic, neuroendocrine, and neurogenic functions. Front Neurosci. 2015;9:387. doi: 10.3389/fnins.2015.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, et al. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- Henry FE, Sugino K, Tozer A, Branco T, Sternson SM. Cell type-specific transcriptomics of hypothalamic energy-sensing neuron responses to weight-loss. eLife. 2015;4 doi: 10.7554/eLife.09800. http://dx.doi.org/10.7554/eLife.09800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Shen S, Cadwell CR, Berens P, Sinz F, Ecker AS, Patel S, Tolias AS. Principles of connectivity among morphologically defined cell types in adult neocortex. Science. 2015;350:aac9462. doi: 10.1126/science.aac9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev VY, Kirschner K, Schaub MT, Andrews T, Yiu A, Chandra T, Natarajan KN, Reik W, Barahona M, Green AR, Hemberg M. SC3 - consensus clustering of single-cell RNA-seq data. bioRxiv. 2016;036558 doi: 10.1038/nmeth.4236. http://dx.doi.org/10.1101/036558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight ZA, Tan K, Birsoy K, Schmidt S, Garrison JL, Wysocki RW, Emiliano A, Ekstrand MI, Friedman JM. Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell. 2012;151:1126–1137. doi: 10.1016/j.cell.2012.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes MJ, Shah BP, Madara JC, Olson DP, Strochlic DE, Garfield AS, Vong L, Pei H, Watabe-Uchida M, Uchida N, et al. An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature. 2014;507:238–242. doi: 10.1038/nature12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DA, Bedont JL, Pak T, Wang H, Song J, Miranda-Angulo A, Takiar V, Charubhumi V, Balordi F, Takebayashi H, et al. Tanycytes of the hypothalamic median eminence form a diet-responsive neurogenic niche. Nat Neurosci. 2012;15:700–702. doi: 10.1038/nn.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IT, Chang AS, Manandhar M, Shan Y, Fan J, Izumo M, Ikeda Y, Motoike T, Dixon S, Seinfeld JE, et al. Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron. 2015;85:1086–1102. doi: 10.1016/j.neuron.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Ludwig M, Leng G. Dendritic peptide release and peptide-dependent behaviours. Nat Rev Neurosci. 2006;7:126–136. doi: 10.1038/nrn1845. [DOI] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcao A, Xiao L, Li H, Häring M, Hochgerner H, Romanov RA, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352:1326–1329. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew TC. Regional analysis of the ependyma of the third ventricle of rat by light and electron microscopy. Anat Histol Embryol. 2008;37:9–18. doi: 10.1111/j.1439-0264.2007.00786.x. [DOI] [PubMed] [Google Scholar]
- Mayerl S, Müller J, Bauer R, Richert S, Kassmann CM, Darras VM, Buder K, Boelen A, Visser TJ, Heuer H. Transporters MCT8 and OATP1C1 maintain murine brain thyroid hormone homeostasis. J Clin Invest. 2014;124:1987–1999. doi: 10.1172/JCI70324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Angulo AL, Byerly MS, Mesa J, Wang H, Blackshaw S. Rax regulates hypothalamic tanycyte differentiation and barrier function in mice. J Comp Neurol. 2014;522:876–899. doi: 10.1002/cne.23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PG, Higham CE, Clayton PE. 60 YEARS OF NEUROENDOCRINOLOGY: The hypothalamo-GH axis: the past 60 years. J Endocrinol. 2015;226:T123–T140. doi: 10.1530/JOE-15-0120. [DOI] [PubMed] [Google Scholar]
- Navarro VM, Tena-Sempere M. Neuroendocrine control by kisspeptins: role in metabolic regulation of fertility. Nat Rev Endocrinol. 2011;8:40–53. doi: 10.1038/nrendo.2011.147. [DOI] [PubMed] [Google Scholar]
- Puelles L, Rubenstein JL. A new scenario of hypothalamic organization: rationale of new hypotheses introduced in the updated prosomeric model. Front Neuroanat. 2015;9:27. doi: 10.3389/fnana.2015.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins SC, Stewart I, McNay DE, Taylor V, Giachino C, Goetz M, Ninkovic J, Briancon N, Maratos-Flier E, Flier JS, et al. α-Tanycytes of the adult hypothalamic third ventricle include distinct populations of FGF-responsive neural progenitors. Nat Commun. 2013;4:2049. doi: 10.1038/ncomms3049. [DOI] [PubMed] [Google Scholar]
- Salvatierra J, Lee DA, Zibetti C, Duran-Moreno M, Yoo S, Newman EA, Wang H, Bedont JL, de Melo J, Miranda-Angulo AL, et al. The LIM homeodomain factor Lhx2 is required for hypothalamic tanycyte specification and differentiation. J Neurosci. 2014;34:16809–16820. doi: 10.1523/JNEUROSCI.1711-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- Schwartz MW, Woods SC, Porte D, Jr, Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000;404:661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, et al. A genomic atlas of mouse hypothalamic development. Nat Neurosci. 2010;13:767–775. doi: 10.1038/nn.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Rodriguez M, Blumenfeld B, Wu C, Luo J, Attali B, Goodman P, Markram H. Correlation maps allow neuronal electrical properties to be predicted from single-cell gene expression profiles in rat neocortex. Cereb Cortex. 2004;14:1310–1327. doi: 10.1093/cercor/bhh092. [DOI] [PubMed] [Google Scholar]
- Tong Q, Ye CP, Jones JE, Elmquist JK, Lowell BB. Synaptic release of GABA by AgRP neurons is required for normal regulation of energy balance. Nat Neurosci. 2008;11:998–1000. doi: 10.1038/nn.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDunk C, Hunter LA, Gray PA. Development, maturation, and necessity of transcription factors in the mouse suprachiasmatic nucleus. J Neurosci. 2011;31:6457–6467. doi: 10.1523/JNEUROSCI.5385-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahab F, Shahab M, Behr R. The involvement of gonadotropin inhibitory hormone and kisspeptin in the metabolic regulation of reproduction. J Endocrinol. 2015;225:R49–R66. doi: 10.1530/JOE-14-0688. [DOI] [PubMed] [Google Scholar]
- Wu Z, Autry AE, Bergan JF, Watabe-Uchida M, Dulac CG. Galanin neurons in the medial preoptic area govern parental behaviour. Nature. 2014;509:325–330. doi: 10.1038/nature13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ypsilanti AR, Rubenstein JL. Transcriptional and epigenetic mechanisms of early cortical development: An examination of how Pax6 coordinates cortical development. J Comp Neurol. 2016;524:609–629. doi: 10.1002/cne.23866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.