Abstract
Objective
Peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α (PGC-1α) promotes hepatic gluconeogenesis by activating HNF4α and FoxO1. PGC-1α expression in the liver is highly elevated in obese and diabetic conditions, leading to increased hepatic glucose production. We previously showed that the spliced form of X-box binding protein 1 (XBP1s) suppresses FoxO1 activity and hepatic gluconeogenesis. The shared role of PGC-1α and XBP1s in regulating FoxO1 activity and gluconeogenesis led us to investigate the probable interaction between PGC-1α and XBP1s and its role in glucose metabolism.
Methods
We investigated the biochemical interaction between PGC-1α and XBP1s and examined the role of their interaction in glucose homeostasis using animal models.
Results
We show that PGC-1α interacts with XBP1s, which plays an anti-gluconeogenic role in the liver by suppressing FoxO1 activity. The physical interaction between PGC-1α and XBP1s leads to suppression of XBP1s activity rather than its activation. Upregulating PGC-1α expression in the liver of lean mice lessens XBP1s protein levels, and reducing PGC-1α levels in obese and diabetic mouse liver restores XBP1s protein induction.
Conclusions
Our findings reveal a novel function of PGC-1α as a suppressor of XBP1s function, suggesting that hepatic PGC-1α promotes gluconeogenesis through multiple pathways as a co-activator for HNF4α and FoxO1 and also as a suppressor for anti-gluconeogenic transcription factor XBP1s.
Keywords: PGC-1α, XBP1s, Glucose homeostasis, ER stress, UPR, Insulin resistance
Highlights
-
•
XBP1s suppresses FoxO1 activity and hepatic gluconeogenesis.
-
•
PGC-1α physically interacts with XBP1s and functions as a co-suppressor of XBP1s in the liver.
-
•
The suppression of XBP1s activity by PGC-1α leads to impaired glucose homeostasis in obese mice.
-
•
Hepatic PGC-1α promotes gluconeogenesis as a co-activator and also as a co-suppressor.
1. Introduction
Obesity is one of the most serious health problems around the globe. In the United States, 69% of the adult population is overweight and 36% is obese [1]. Globally, over 600 million adults, approximately 13% of world adult population, are obese [2]. Being overweight and obese leads to many debilitating pathologies such as hypertension, cardiovascular diseases, cancer, and, most notably, insulin resistance and subsequent development of type 2 diabetes [3], [4]. However, underlying mechanisms for the development of type 2 diabetes in obesity is still poorly understood.
The endoplasmic reticulum (ER), a membrane-bound organelle in eukaryotic cells, is the major site of synthesis of secretory and membrane proteins. Newly synthesized proteins are folded into their three-dimensional native structure within the lumen of the ER, and a variety of ER resident proteins monitor and help proper folding of newly synthesized proteins. An increased amount of unfolded proteins, which exceeds the handling capacity of the ER, creates perturbations in ER system and leads to development of a condition termed “ER stress”. In response to ER stress, cells employ an adaptive mechanism, the unfolded protein response (UPR), which re-establishes ER homeostasis [5], [6], [7]. In metazoans, three signaling branches of the UPR have been characterized. These branches are initiated by following three ER-transmembrane proteins: PERK (protein kinase RNA-like ER kinase), IRE1α (inositol requiring protein-1α), and ATF6 (activating transcription factor-6) [5], [6], [7]. PERK suppresses general protein translation, thereby reducing protein load into the ER [8]. IRE1α splices pro-mRNA of XBP1 (X-box binding protein 1), which leads to production of functioning transcription factor, XBP1s (spliced XBP1) [9], [10]. Together with another UPR transcription factor ATF6, XBP1s increases the folding capacity of the ER by increasing the transcription of various ER resident proteins [10], [11].
We and others have previously documented that i) as obesity and diabetes progress, metabolically crucial organs such as the hypothalamus, liver, and adipose tissues experience elevated ER stress leading to leptin and insulin resistance [12], [13], [14]; ii) increased ER stress in the liver is partly due to impaired XBP1s function [15], [16]; iii) restoring XBP1s action in the liver of obese and diabetic mice improves systematic glucose homeostasis by also suppressing hepatic gluconeogenic gene expression through XBP1s-mediated FoxO1 protein degradation [16], [17], [18].
Peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α (PGC-1α), a crucial regulatory molecule in various metabolic processes, plays key roles in the development of obesity, insulin resistance, and type 2 diabetes [19], [20], [21] and is a co-activator of a variety of transcription factors including PPARs, HNF4α, ERRs, and FoxO1 [22], [23], [24], [25], [26], [27], [28]. In particular, PGC-1α promotes gluconeogenesis by activating HNF4α and FoxO1. Its expression in the liver is highly elevated in obese and diabetic conditions, resulting in increased hepatic glucose production and contributing to the development of type 2 diabetes [23], [24], [27]. Since its discovery, PGC-1α has been considered to be a co-activator for many different transcription factors, but, thus far, no reports show that PGC-1α can also function as a repressor or a suppressor for a transcription factor.
2. Materials and methods
2.1. Animals
Leptin deficient (ob/ob) and wild type mice (C57BL7/J) were obtained from Jackson Laboratory. Mice were fed normal chow and given free access to water. Eight-week old male mice were used in the experiments. All animal procedures used in this study were approved by the Animal Care and Use Committee at Boston Children's Hospital.
2.2. Glucose tolerance test (GTT)
Mice were fasted overnight (15 h) and received dextrose (0.5 g/kg for ob/ob mice) intraperitoneally. Blood glucose levels were measured from the tail before and 15, 30, 60, 90, and 120 min after dextrose administration.
2.3. Primary hepatocyte isolation
Culture plates for mouse primary hepatocytes were prepared by coating with rat tail collagen I (Thermo Fisher Scientific) in 0.02 mg/ml in H2O with 20 mM acetic acid for 3–16 h before hepatocyte isolation.
For primary hepatocyte isolation, 7–8 week-old male mice (C57BL7/J) were anesthetized with Ketamine/Xylazine (100/20 mg per kg), and blood was drained out by perfusion of Perfusion Solution 1 (1× HBSS, 0.5 mM EGTA, 5.5 mM glucose, 100 U/ml of penicillin, and 100 μg/ml of streptomycin) via vena cava. Connective tissue within the liver was digested by perfusion of Perfusion Solution 2 (1× HBSS, 1.5 mM CaCl2, 5.5 mM glucose, 46.9 U/ml of type 1 collagenase, 125 U/ml of penicillin, and 125 μg/ml of streptomycin). Dissociated hepatocytes were filtered through 70 μm cell strainer and collected by centrifugation at 1,500 rpm at 4 °C for 3 min. Hepatocytes were further isolated from other cells by Percoll gradient centrifugation. Isolated hepatocytes were seeded on collagen-coated plates in Williams' media E with 10% FBS, 2 mM sodium pyruvate, 100 U/ml of penicillin and 100 μg/ml of streptomycin, 1 μM dexamethasone and 100 nM insulin for 3 h. Hepatocytes were infected with adenoviruses in Williams' media E with 0.2% BSA, 2 mM sodium pyruvate, 100 U/ml of penicillin and 100 μg/ml of streptomycin, 0.1 μM dexamethasone, and 1 nM insulin for overnight.
2.4. Total protein extraction from cells and tissues
Cells were lysed in lysis buffer (25 mM Tris–HCl, pH 7.4; 100 mM NaF; 50 mM Na4P2O7; 10 mM Na3VO4; 10 mM EGTA; 10 mM EDTA; 1% NP-40) with use of protease and phosphatase inhibitors (Roche). After rotation at 4 °C for 20 min at a speed of 13,000 rpm, cell debris was removed by centrifugation at 4 °C for another 20 min. Supernatants were collected, and protein concentration was quantified with use of the Protein Assay Kit (Bio-Rad). Liver tissues were homogenized with a bench-top homogenizer (Polytron, PT2100) or TissuLyserII (Qiagen) in ice-cold tissue lysis buffer (25 mM Tris–HCl, pH 7.4; 100 mM NaF; 50 mM Na4P2O7; 10 mM Na3VO4; 10 mM EGTA; 10 mM EDTA; 1% NP-40), containing protease and phosphatase inhibitors. After homogenization, lysates were rotated at 4 °C for 1 h, then clear protein lysates were separated by centrifugation at 4 °C for 20 min at a speed of 13,000 rpm. Protein concentration was quantified with use of the Protein Assay Kit. Each sample, with an equivalent protein concentration, was prepared in Laemmli buffer. Protein concentrations were normalized with lysis buffer, such that each sample had equivalent amounts of protein and volume. Protein was denatured by boiling at 100 °C for 5 min in Laemmli buffer, and lysates were cooled to room temperature before loading for western blot analysis.
2.5. Nuclear protein extraction from liver tissues
The nuclear extraction kit from Thermo Fisher Scientific (Waltham, MA) was used to isolate nuclear and cytoplasmic fractions from liver tissues, according to the manufacturer's instructions. Tissues were cut into small pieces, washed with PBS, and separated from the PBS by centrifugation at 500 × g for 5 min. Collected tissues were resuspended by company-supplied CER I buffer, homogenized with a Dounce homogenizer, vortexed, and incubated on ice for 10 min. CER II buffer was added, tissues were vortexed for 5 s, incubated for 1 min on ice, vortexed again, and then centrifuged for 5 min at maximum speed in a microcentrifuge. The supernatant (cytoplasmic fraction) was saved for later analysis, while pellets were resuspended with the supplied NER buffer, and underwent multiple cycles of vortexing (15 s) and incubation on ice (10 min), for a total of 40 min. After a 10-min centrifugation, the supernatant (containing the nuclear fraction) was collected. Protein concentrations from cytoplasmic and nuclear lysates were quantified with a Protein Assay Kit, and protein samples were prepared in Laemmli buffer and analyzed with immunoblotting.
2.6. Co-immunoprecipitation/immunoprecipitation
For co-immunoprecipitation, cells or tissues were lysed in co-immunoprecipitation lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 10 mM NaF, 0.5% NP-40), containing protease and phosphatase inhibitors. Equal amounts of protein from the lysates were mixed with specific antibodies to immunoprecipitate the target protein together with protein A-agarose or anti-flag M2 affinity gel and then incubated for 3 h. Immunoprecipitates were washed three times with the same co-immunoprecipitation buffer and eluted with sample buffer without reducing agent. Eluted samples were mixed with 2-mercaptoethanol and analyzed by western blotting. For immunoprecipitation, cells were lysed with radioimmuno-precipitation assay (RIPA) buffer (50 mM Tris HCl pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS), plus protease and phosphatase inhibitors. Equal amounts of proteins from the lysates were incubated overnight at 4 °C with antibodies specific for each target protein; samples were then further incubated with protein A-agarose beads (for rabbit antibody) or protein G-agarose beads (for mouse antibody) for an additional 2 h. Immunoprecipitates were washed three times with RIPA buffer and eluted with sample buffer with reducing agent.
2.7. Cycloheximide chase assay
MEF cells were infected with Ad-Xbp1s plus Ad-LacZ or Ad-PGC-1α. HEK293 cells were transfected with plasmids expressing Xbp1s constructs, with or without plasmid encoding PGC-1α. After overnight infection or transfection (16–24 h), cells were treated for various times with cycloheximide (50 μg/ml, Sigma), a translation initiation inhibitor. Samples were prepared as described for western blot analysis, and target protein levels were determined via immunoblotting. ImageJ was used to quantify three different western blots of the same experiment for Figure 2, Figure 4.
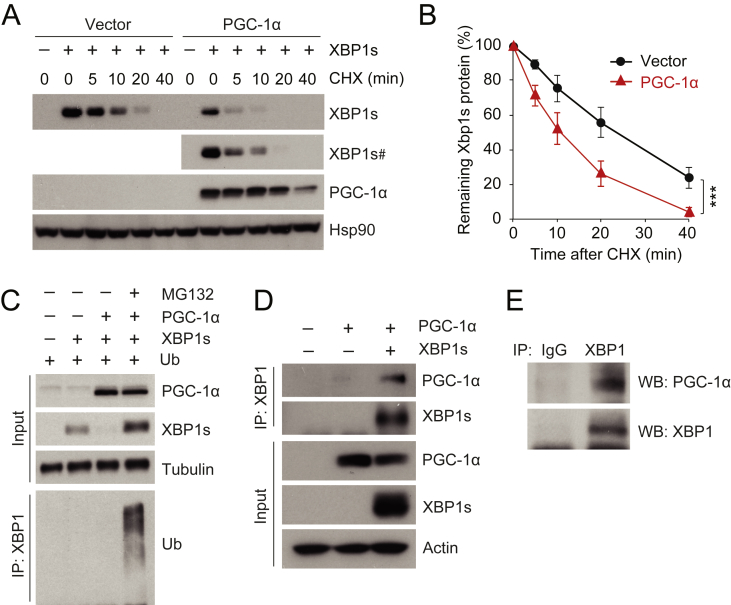
Figure 2.
PGC-1α physically interacts with XBP1s and promotes ubiquitin-mediated XBP1s protein degradation. (A, B) HEK293 cells, transfected with a plasmid expressing XBP1s plus empty vector or a plasmid expressing PGC-1α, were treated with cycloheximide (CHX, 50 μg/ml) for the indicated time. (A) Levels of XBP1s, PGC-1α, and Hsp90 protein levels. # indicates longer exposed film. (B) The quantified ratio of XBP1s to Hsp90 protein in A before and after treatment with CHX. (C) XBP1s, PGC-1α, tubulin, and ubiquitinated XBP1s protein levels. HEK293 cells expressing ubiquitin and XBP1s with or without PGC-1α were treated with DMSO or MG132 (10 μM) for 1 h. Ubiquitinated XBP1s was analyzed from immunoprecipitates pulled down with antibody specific to XBP1 (anti-XBP1), by immunoblotting with anti-ubiquitin. (D) Co-immunoprecipitation of XBP1s and PGC-1α. HEK293 cells were transfected with plasmid expressing PGC-1α, with or without the plasmid that encodes XBP1s. Cells were lysed and immunoprecipitated with anti-XBP1. (E) Co-immunoprecipitation of endogenous XBP1s and PGC-1α. Eight week-old male mice were fasted for 24 h, then refed for 90 min. Liver protein lysates were immunoprecipitated with either rabbit IgG or anti-XBP1, and co-immunoprecipitates were analyzed by western blotting. Each experiment was independently reproduced at least three times. Values are means ± s.e.m. Significance was determined by two-way analysis of variance (ANOVA), with Bonferroni multiple-comparison analysis (B). ***P < 0.001.
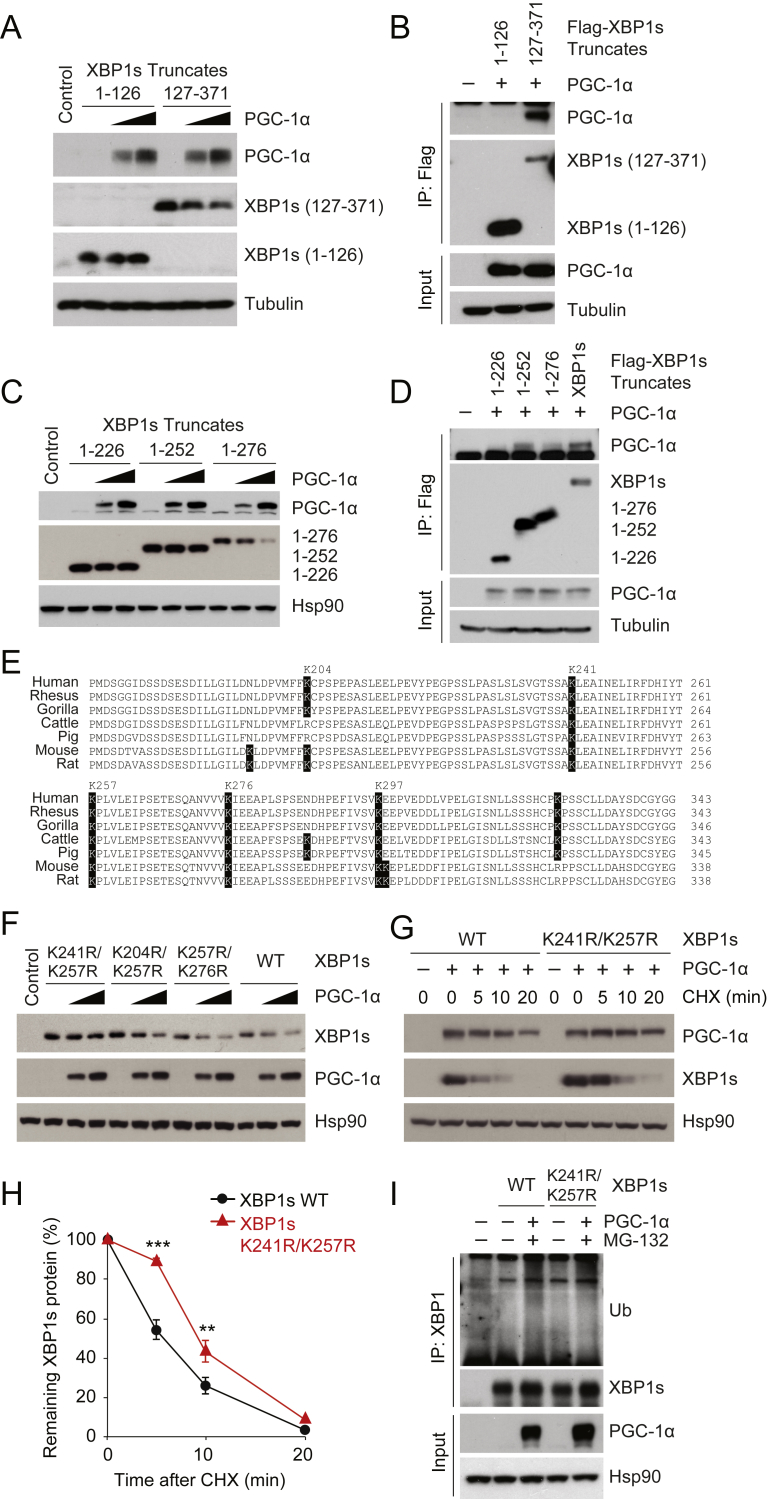
Figure 4.
Molecular interaction between the activation domain of XBP1s and PGC-1α. (A) Levels of flag-tagged XBP1s (1-126) and (127-371), in the presence of increasing amounts of PGC-1α. (B) Co-immunoprecipitation of flag-tagged XBP1s (1-126) or (127-371), and PGC-1α. Different XBP1s constructs were immunoprecipitated with anti-flag antibody conjugated with agarose beads. Immunoprecipitates were analyzed with indicated antibodies and anti-flag in western blot. (C) Levels of XBP1s (1-226), (1-252), and (1-276) protein, when co-expressed with increasing amounts of PGC-1α. (D) Co-immunoprecipitation of flag-tagged full-length XBP1s, XBP1s (1-226), (1-252), or (1-276), and PGC-1α. (E) Protein sequence alignment of the XBP1s activation domains from various mammalian species. The sequence location of lysines are based on mouse Xbp1s. (F) WT XBP1s, XBP1s with K204R/K257R, K241R/K257R, or K257R/K276R, PGC-1α and Hsp90 protein amounts, visualized by western blotting. Indicated XBP1s constructs were co-expressed with increasing doses of PGC-1α. (G,H) XBP1s (WT or K246R/K257R) was co-expressed with PGC-1α, and then treated with CHX (50 μg/ml) for the indicated time. (G) XBP1s (WT or K241R/K257R), PGC-1α and Hsp90 protein levels. (H) Quantitation of the ratio between levels of XBP1s and Hsp90 protein, before and after treatment with CHX. (I) Levels of XBP1s (WT or K241R/K257R), PGC-1α, Hsp90, and ubiquitinated Xbp1s proteins. HEK293 cells were transfected overnight with plasmids expressing ubiquitin and XBP1s (WT or K241R/K257R), with or without PGC-1α, then treated with DMSO or MG132 (10 μM) for 1 h. Ubiquitinated XBP1s was analyzed from immunoprecipitates with anti-XBP1 by immunoblotting with anti-ubiquitin. Each experiment was independently reproduced at least two times. Values are means ± s.e.m. Significance was determined by two-way ANOVA with Bonferroni multiple-comparison analysis (H). **P < 0.01, ***P < 0.001.
2.8. Statistical analysis
Data are presented as means ± s.e.m. Statistical significance was calculated by Student's t test (two-tailed) or by analysis of variance (ANOVA), using Prism (GraphPad). Significance was accepted at *P < 0.05, **P < 0.01 or ***P < 0.001. Numbers of cohorts and n values for each experiment were indicated in figure legends. Statistical outliers (judged by Grubb's outlier test) or samples not responding for known stimuli were excluded in the final analysis. No blinding or randomization was used. No statistical method was used to predetermine sample size and sample size was determined based on previous experiments and literatures. The variance was similar in the groups being compared.
3. Results
3.1. PGC-1α down-regulates XBP1s protein levels and activity
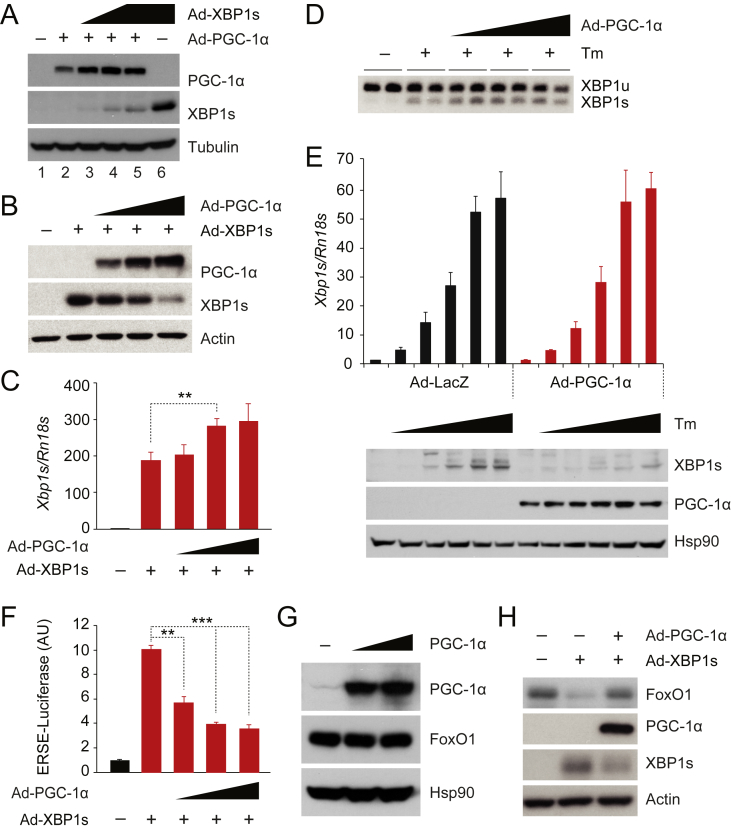
Given that PGC-1α is a co-activator of FoxO1 and promotes hepatic gluconeogenesis [23], we asked whether XBP1s also inhibits PGC-1α, as a mechanism for lowering hepatic glucose production, and whether it suppresses FoxO1 activity also through this alternative pathway. We infected mouse embryonic fibroblast (MEF) cells with a constant amount of adenovirus that expresses PGC-1α (Ad-PGC-1α), together with increasing doses of an adenovirus that encodes murine XBP1s (Ad-XBP1s) (Figure 1A); as a comparison, we also infected the MEF cells (in lane 6) with the same amount of Ad-XBP1s (as in lane 5), but without co-expressing PGC-1α (Figure 1A). Increased expression of XBP1s did not change the level of PGC-1α protein (Figure 1A); however, the total level of XBP1s protein was substantially decreased in the presence of PGC-1α (Figure 1A).
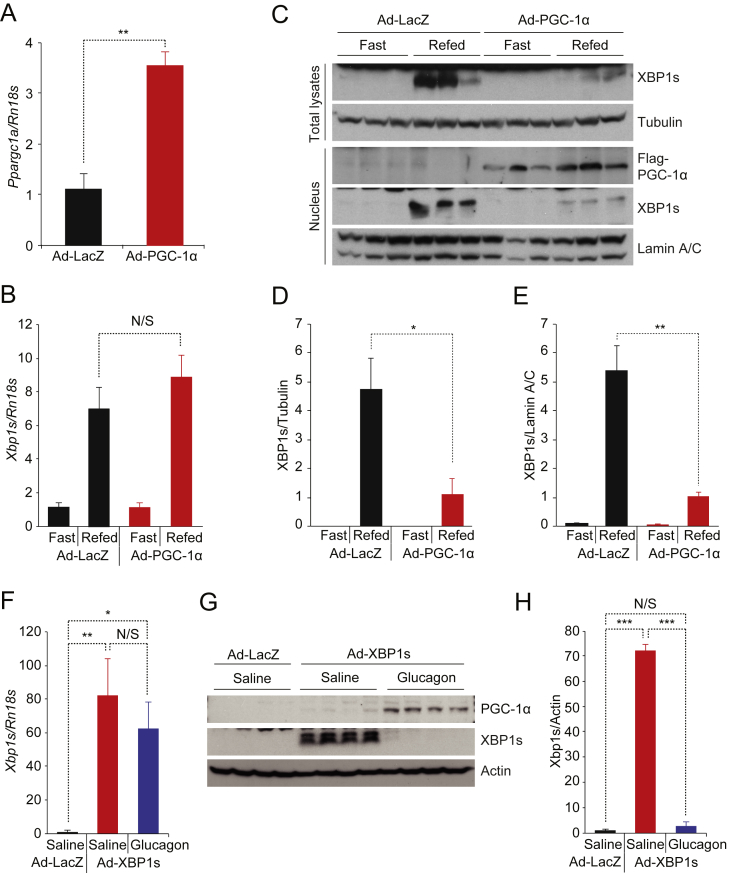
Figure 1.
PGC-1α decreases XBP1s protein levels and suppresses XBP1s activity. (A) XBP1s, PGC-1α, and tubulin protein levels in MEFs infected with equal doses of Ad-PGC-1α plus increasing doses of Ad-XBP1s. The same dose of Ad-XBP1s was used in lanes 5 and 6, except that Ad-LacZ was used in lane 6 instead of Ad-PGC-1α for co-expression. (B, C) MEFs were infected with the same amount of Ad-XBP1s, plus increasing amount of Ad-PGC-1α. (B) XBP1s, PGC-1α and actin protein levels in MEFs. (C) XBP1s mRNA levels measured by real time PCR (qPCR) (n = 5 for each group). (D) Levels of unspliced and spliced XBP1 mRNAs analyzed by reverse transcription PCR. MEFs were infected with increasing doses of Ad-PGC-1α, and treated with tunicamycin (Tm; 3 μg/ml, 90 min). (E) Levels of endogenous Xbp1s mRNA and protein in MEFs that were infected overnight with Ad-LacZ or Ad-PGC-1α then treated with increasing doses (0, 0.125, 0.25, 0.5, 1, 2 μM) of tunicamycin for 5 h. (F) ERSE-luciferase assay in MEFs infected with a fixed amount of Ad-XBP1s, plus increasing doses of Ad-PGC-1α, as in B and C. Renilla luciferase activity was used for normalization (n = 3 for each group). (G) Levels of endogenous FoxO1 protein in Fao cells infected with increasing doses of Ad-PGC-1α. (H) Levels of endogenous FoxO1 protein in Fao cells infected with equal amounts of Ad-XBP1s and Ad-LacZ or Ad-PGC-1α. We infected cells with the equal amount of virus by co-infecting with Ad-LacZ, and each experiment was repeated at least two times. Values are means ± s.e.m. Significance was determined by two-way analysis of variance (ANOVA), with Bonferroni multiple-comparison analysis (E) or Student's t test (C,F). AU, arbitrary units. **P < 0.01, ***P < 0.001.
The above result led us to hypothesize that PGC-1α negatively affects XBP1s protein expression levels and subsequently suppresses XBP1s activity. To test whether PGC-1α reduces the levels of XBP1s protein, MEFs were infected with Ad-XBP1s, along with Ad-LacZ or increasing amounts of Ad-PGC-1α. Elevated expression of PGC-1α indeed reduced the amount of XBP1s protein (Figure 1B), without lowering XBP1s mRNA levels (Figure 1C). PGC-1α protein also lessened both murine and human XBP1s protein in other cell lines, including rat hepatoma Fao cells (Supplementary Figure 1A–D). Endogenous XBP1s mRNA is generated from IRE1α-mediated splicing of full-length XBP1 mRNA in ER stress conditions [9], [10]. We thus investigated whether PGC-1α modulates XBP1 mRNA splicing. For this, we first infected MEFs with increasing doses of Ad-PGC-1α, and, after 16 h, we treated the cells with tunicamycin (3 μg/ml) for 90 min; PGC-1α expression did not alter the tunicamycin-induced splicing of XBP1 mRNA (Figure 1D).
We next explored whether PGC-1α also reduces the endogenous XBP1s protein amount. To express detectable levels of endogenous XBP1s, we treated Ad-PGC-1α- or Ad-LacZ-infected MEFs with increasing doses of tunicamycin for 5 h; this greatly increased XBP1s mRNA levels, despite the overexpression of PGC-1α (Figure 1E). Control cells infected with Ad-LacZ showed robust production of endogenous XBP1s protein as a result of tunicamycin treatment; however, this response was strongly diminished in PGC-1α-expressing MEFs, relative to that in LacZ-expressing MEFs. Together, these results indicate that PGC-1α specifically reduces the amount of XBP1s protein without affecting the XBP1s mRNA levels or the IRE1α endoribonuclease activity.
PGC-1α is considered to be a co-activator for various transcription factors [19], [20], [21], and the amount of transcription factor protein may not necessarily correlate with its activity. We thus asked whether the PGC-1α-mediated reduction in XBP1s protein also reduces the activity of XBP1s, that is, whether PGC-1α suppresses the transcriptional activity of XBP1s. To address this issue, we used an ER stress response element (ERSE)-driven luciferase system to record XBP1s transcriptional activity in the presence or absence of PGC-1α. MEFs expressing ERSE-driven luciferase and Renilla-luciferase were infected with Ad-XBP1s alone, or with Ad-XBP1s plus increasing doses of Ad-PGC-1α. XBP1s expression significantly upregulated the ERSE promoter-driven luciferase production (Figure 1F), but XBP1s activity was strongly ameliorated when PGC-1α was co-expressed with the XBP1s (Figure 1F).
Next, we tested whether PGC-1α expression suppresses XBP1s activity in mouse hepatocytes. Isolated mouse hepatocytes were infected with Ad-XBP1s alone or together with Ad-PGC-1α. XBP1s expression in hepatocytes increased its target gene expression. However, co-expression of PGC-1α markedly suppressed the increase of XBP1s target gene expression (Supplementary Figure 1E–H)
We have previously documented that XBP1s has a strong effect towards FoxO1 [17]. In the absence of XBP1s, PGC-1α expression itself was not able to alter endogenous FoxO1 protein levels in Fao cells (Figure 1G). We thus infected Fao cells with Ad-XBP1s along with Ad-LacZ or Ad-PGC-1α. XBP1s expression decreased the levels of endogenous FoxO1 protein, whereas co-expression of PGC-1α lessened the amount of XBP1s protein, and in turn restored levels of FoxO1 protein (Figure 1H). Collectively, our data suggest that PGC-1α negatively regulates the levels of XBP1s protein as well as its function.
3.2. PGC-1α physically interacts with XBP1s and promotes its degradation
To unravel the mechanistic underpinnings of the interaction between PGC-1α and XBP1s, we next inquired whether PGC-1α reduces levels of XBP1s protein by decreasing its stability. For this purpose, we treated cells expressing XBP1s with or without PGC-1α, with the translational inhibitor cycloheximide (50 μg/ml) for the indicated time periods. Western blotting was used to determine XBP1s protein levels before and after the addition of cycloheximide (Figure 2A). We also compared the rate of degradation of XBP1s protein in the presence or absence of PGC-1α (Figure 2B). Blots derived from cells co-expressing XBP1s plus PGC-1α were exposed for a longer time, and blots prepared from cells expressing XBP1s alone (and no PGC-1α) were exposed for a shorter time, in order to achieve the same level of XBP1s signal at the zero-time point for both groups (Figure 2A). As shown in Figure 2A, B, XBP1s protein was degraded faster in the presence of PGC-1α.
It has been suggested that XBP1s protein is down-regulated by ubiquitin-mediated proteasomal degradation [10], [29]. To test whether PGC-1α facilitates XBP1s ubiquitination as a means of degradation, ubiquitin-expressing HEK293 cells were transfected with XBP1s plasmid alone or together with a plasmid expressing PGC-1α; after 16 h, the HEK293 cells were treated with DMSO or MG-132 (10 μM) for 1 h, and lysates were immunoprecipitated with an antibody specific for XBP1 protein. Ubiquitination of immunoprecipitated XBP1s was analyzed by immunoblotting with an antibody against ubiquitin. The presence of PGC-1α diminished XBP1s protein amounts. However, this reduction was blocked by inhibition of 26S proteasome-mediated protein degradation with MG-132 treatment. The XBP1s was highly ubiquitinated in the presence of PGC-1α (Figure 2C), suggesting that PGC-1α promotes the degradation of the XBP1s protein via ubiquitination.
To examine whether PGC-1α and XBP1s interact physically to mediate the degradation of XBP1s protein, we transfected HEK293 cells with PGC-1α plasmid alone, or together with a plasmid for XBP1s. The cells were then lysed and co-immunoprecipitated with an anti-XBP1 antibody, and the immunoprecipitates were analyzed with western blotting. The anti-XBP1 antibody did not immunoprecipitate the PGC-1α protein in the absence of XBP1s (Figure 2D), documenting that the anti-XBP1 antibody did not cross-react with the PGC-1α protein. When PGC-1α and XBP1s were co-expressed and cell lysates immunoprecipitated with anti-XBP1, PGC-1α was detected in immunoprecipitates alongside XBP1s (Figure 2D); conversely, when PGC-1α was immunoprecipitated, XBP1s was detected in the immunoprecipitates (Supplementary Figure 2A).
To further investigate whether endogenous PGC-1α and endogenous XBP1s also bind to each other in the liver in vivo, total hepatic protein lysates were collected from mice, which had been refed for 90 min after 24 h fasting, to induce endogenous XBP1s expression. We detected PGC-1α in immunoprecipitates pulled down with anti-XBP1s from mouse liver (Figure 2E), while control IgG was not able to pull-down XBP1s and PGC-1α; this shows that endogenous XBP1s also interacts physically with PGC-1α in vivo. In sum, PGC-1α interacts with XBP1s in vitro and in vivo and it facilitates XBP1s protein degradation via ubiquitin-proteasome pathway instead of activating XBP1s activity.
3.3. The molecular interaction between the activation domain of PGC-1α and XBP1s promotes its degradation
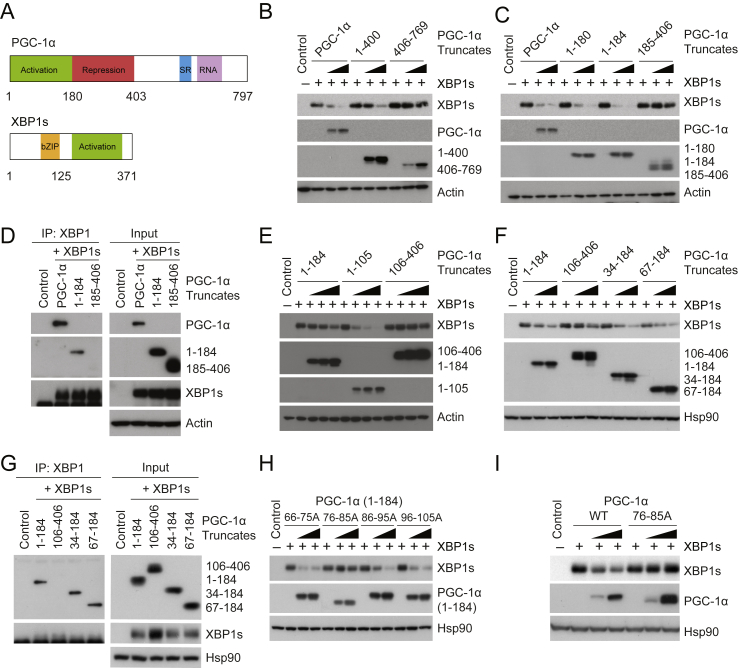
PGC-1α consists of an N-terminal region that contains activation and repression domains, a C-terminal region with a Ser/Arg-rich domain (SR domain), and an RNA binding domain (Figure 3A) [19], [20], [21], [22]. The C-terminal region interacts with MEF-2 and FoxO1, while the N-terminal region – including the activation domain and repression domains – binds to various nuclear receptors and NRF-1 [19], [20], [21], [28], [30], [31]. The activation domain of PGC-1α is necessary for promoting transcriptional activity of target transcription factors [32]. To understand which area (within PGC-1α) is involved in the physical interaction with XBP1s and how it functions as an XBP1s suppressor, we first expressed XBP1s in HEK293 cells – along with increasing doses of full-length PGC-1α, or of only the N-terminal half of PGC-1α (1-400), including the activation and repression domains, or of only the C-terminal half of PGC-1α (407-769). N-terminal PGC-1α (1-400) facilitated degradation of XBP1s protein, in a manner comparable to full length PGC-1α (Figure 3B), while the C-terminal of PGC-1α (407-769) was unable to reduce levels of XBP1s protein (Figure 3B).
Figure 3.
Molecular interaction between XBP1s and the activation domain of PGC-1α. (A) The protein domains of mouse PGC-1α and XBP1s. (B) XBP1s, PGC-1α, PGC-1α (1-400), PGC-1α (406-769), and actin protein levels. Plasmid expressing XBP1s was transfected to HEK293 cells, along with increasing doses of plasmid producing full-length PGC-1α, PGC-1α (1-400) or (406-769). (C) Protein amounts of XBP1s, PGC-1α, PGC-1α (1-180), (1-184), (185-406), and actin. XBP1s plasmid was transfected into HEK293 cells, along with increasing amounts of plasmid expressing full-length PGC-1α, PGC-1α (1-180), (1-184), or (185-406). (D) Co-immunoprecipitation of XBP1s and different regions of PGC-1α. HEK293 cells were transfected with plasmids expressing XBP1s, together with flag-tagged full-length PGC-1α, PGC-1α (1-184), or (185-406). Co-immunoprecipitation was performed using anti-XBP1, and immunoprecipitates were analyzed with indicated antibodies and flag antibody (anti-flag) for PGC-1α (1-184) and (185-406) in immunoblot. (E) Levels of XBP1s, PGC-1α (1-184), (1-105), (106-406), and actin proteins in HEK293 cells transfected to express XBP1s plus increasing doses of indicated PGC-1α truncates. (F) XBP1s, PGC-1α (106-406), (34-184), (67-184), and Hsp90 protein levels. (G) Co-immunoprecipitation of XBP1s and different PGC-1α truncates. HEK293 cells expressing XBP1s, along with indicated flag-tagged PGC-1α constructs, were lysed and used for immunoprecipitation with anti-XBP1. (H) Immunoblot of XBP1s, of PGC-1α (1-184) constructs with indicated amino acids mutated to alanine, and of Hsp90 protein. HEK293 cells were transfected with XBP1s plasmid plus increasing doses of indicated PGC-1α (1-184) constructs. (I) Protein levels of XBP1s, wild-type (WT) and mutant PGC-1α (76-85A), and Hsp90. HEK293 cells were transfected with XBP1s plasmid plus increasing doses of plasmids encoding wild-type PGC-1α or mutated PGC-1α (76-85A). Each experiment was repeated at least two times.
We next expressed XBP1s in HEK293 cells along with increasing doses of full-length PGC-1α, with the activation domains of PGC-1α (1-180, or 1-184) or with the repression domain of PGC-1α (185-406). XBP1s was robustly down-regulated by the activation domains (PGC-1α 1-180 and 1-184) as well as by full-length PGC-1α (Figure 3C); however, the repression domain (185-406) failed to promote XBP1s degradation (Figure 3C). Co-immunoprecipitation (to establish whether the activation domain of PGC-1α mediates XBP1s degradation through physical interaction with XBP1s) revealed that full-length PGC-1α, and also its activation domain (1-184), interact physically with XBP1s, but the repression domain (185-406) of PGC-1α does not (Figure 3D). By using PGC-1α that was further truncated within the activation domain, we found that residues 1-105 of PGC-1α, or the activation domain (1-184), were both able to diminish XBP1s protein levels, while amino acids 106-406 of PGC-1α failed to reduce the amount of XBP1s protein (Figure 3E). Moreover, residues 34-184 and 67-184 of PGC-1α also retained their ability to down-regulate XBP1s protein (Figure 3F). Co-immunoprecipitation also revealed a physical interaction between residues 34-184 or 67-184 of PGC-1α and XBP1s, while not from residues 106-406 of PGC-1α, (Figure 3G), suggesting that the area within 67–105 of PGC-1α is responsible for the degradation of XBP1s protein.
We then made a series of mutations within the PGC-1α activation domain (1-184) by changing each set of ten amino acids (within 67–105) to alanine, thereby generating constructs with amino acid residues between 66 and 75 (66-75A), 76 and 85 (76-85A), 86 and 95 (86-95A), or 96 to 105 (96-105A) changed to alanine. When these mutated activation domains of PGC-1α were co-expressed with XBP1s, only the activation domain of PGC-1α with residues 76 to 85 mutated (76-85A) lost its ability to reduce XBP1s protein, while the other mutations still retained their ability to degrade XBP1s (Figure 3H); and the activation domain of PGC-1α (76-85A) also had no ability to bind to XBP1s (Supplementary Figure 2B). Next, we introduced the 76-85A mutation into full-length PGC-1α and co-expressed the mutated protein with XBP1s. While wild-type PGC-1α promotes XBP1s protein degradation, PGC-1α (76-85A) could not facilitate such degradation (Figure 3I), reinforcing the conclusion that the amino acids 76-85 within the activation of domain of PGC-1α are critical for the down-regulation of XBP1s protein.
3.4. The activation domain of XBP1s interacts with PGC-1α
We also sought to uncover the parts of XBP1s that are involved in interacting with PGC-1α, starting with N-terminal XBP1s (1-126), which includes a basic leucine-zipper (bZIP) DNA binding domain, and with the C-terminal activation domain (127-371) (Figure 3A). When we expressed either the N-terminal (1-126) or the C-terminal (127-371) of XBP1s along with increasing doses of full-length PGC-1α in HEK293 cells, the levels of N-terminal XBP1s (1-126) protein remained stable, whereas those of C-terminal XBP1s (127-371) protein were strongly diminished in the presence of PGC-1α (Figure 4A). Additionally, we tested whether the binding of PGC-1α to the C-terminal activation domain (127-371) could mediate the protein degradation of XBP1s. When flag-tagged XBP1s 1-126 or 127-371 was co-expressed with PGC-1α and then immunoprecipitated with a flag antibody, we observed that N-terminal XBP1s exhibited no interaction with PGC-1α, while the C-terminal activation domain of XBP1s could bind to PGC-1α even when the amount of the activation domain protein was robustly reduced due to the co-expression of PGC-1α (Figure 4B).
We further examined the protein interaction of XBP1s and PGC-1α within the activation domain of XBP1s. First, we created various truncated forms of XBP1s, by adding a stop codon at different locations in the activation domain; we then tested the function of these truncated forms in vitro, by co-expressing them with PGC-1α. When XBP1s (1-226), (1-252), or (1-276) was co-expressed with increasing doses of PGC-1α in HEK 293 cells, the levels of XBP1s (1-226) and (1-252) were unchanged; however, the amount of XBP1s (1-276) protein was robustly diminished by the co-expression of PGC-1α (Figure 4C). Next, we examined the protein interaction between PGC-1α and the following flag-tagged XBP1s truncates: full-length XBP1s, XBP1s (1-226), (1-252) or (1-276); this was achieved by co-expressing the truncated forms of XBP1s with PGC-1α. As shown in Figure 4D, XBP1s (1-276), as well as full-length XBP1s, interacted physically with PGC-1α, while XBP1s (1-226) failed to show such binding. These observations again suggest that a physical interaction between the two proteins is required for PGC-1α-mediated down-regulation of XBP1s. Interestingly, XBP1s (1-252) displayed levels of protein interaction with PGC-1α that were comparable with those of other XBP1s constructs – such as XBP1s (1-276) - even though PGC-1α was not able to reduce the levels of XBP1s (1-252) protein (Figure 4C, D).
The above results suggest that amino acid residues between 227 and 252 within the activation domain of XBP1s also participate in physical interaction with PGC-1α, whereas residues 253–276 of XBP1s are still required for subsequent processes – possibly for ubiquitin ligase recruitment and/or ubiquitination – that, in turn, lead to XBP1s down-regulation.
Therefore, we next explored sites within the activation domain of XBP1s, where ubiquitination may be stimulated by PGC-1α. Ubiquitins are covalently conjugated to lysine residues of substrate proteins by ubiquitin ligases [33]. Interestingly, there are several lysine residues located within the activation domain of XBP1s such as K241, K257, K276, and K297, which are highly conserved among various mammalian XBP1s (Figure 4E). We thus mutated each of single lysine to arginine at the 241, 257, 276, or 297 positions of XBP1s, expressed these mutated XBP1s along with PGC-1α and found that XBP1s with K257R was more stable compared to the other mutated XBP1s, when co-expressed with PGC-1α (Supplementary Figure 2C). However, subsequent co-expression of XBP1s K257R with higher levels of PGC-1α, compared to the levels in Supplementary Figure 2C, still led to degradation of XBP1s K257R (Supplementary Figure 2D). When we made double mutant XBP1s, in which two lysine residues were changed to alanine, and expressed these in the presence of PGC-1α, we found that XBP1s with K241R and K257R were much more resistant to PGC-1α-mediated degradation, relative to other double-mutated XBP1s (Figure 4F).
Next, we determined whether the half-life of XBP1s is increased when ubiquitination at K241 and K257 is prevented in the presence of PGC-1α. To achieve this, we expressed either wild-type or XBP1s with K241R and K257R along with PGC-1α, blocked further protein synthesis by adding cycloheximide, and then followed the half-life of XBP1s (Figure 4G,H). The stability and half-life of XBP1s K241R and K257R were significantly increased compared to those of wild-type XBP1s when PGC-1α was co-expressed (Figure 4G, H); however, in the absence of PGC-1α, the half-lives of wild-type XBP1s and of XBP1s K241R/K257R were not significantly different (Supplementary Figure 2E).
Furthermore, when either wild-type XBP1s or XBP1s with a double mutation at K241R and K257R was co-expressed with PGC-1α, we noted that PGC-1α-induced ubiquitination was strongly diminished in the double-mutated XBP1s (K241R and K257R), suggesting that each of these lysine residues is a primary ubiquitination site, which is stimulated by the interaction with PGC-1α (Figure 4I). Previous proteomic analysis has also identified K262 of human XBP1s (equivalent to K257 of murine XBP1s) as a possible ubiquitination site [34].
The above observations in vitro led us to conclude that (i) the activation domains of PGC-1α and XBP1s interact physically; (ii) the interaction between these two proteins curbs the activity of XBP1s as an UPR transcription factor and as a FoxO1 suppressor, rather than promoting the transcriptional activity of XBP1s; and (iii) such suppression mediated by PGC-1α occurs via increased ubiquitination and protein degradation of XBP1s. Thus, when the physical interaction between two proteins is prevented by deletion studies or mutations, PGC-1α loses its ability to suppress XBP1s activity.
3.5. Increased hepatic PGC-1α expression down-regulates XBP1s protein levels
Next we investigated whether the interaction between PGC-1α and XBP1s also leads to a reduction in XBP1s protein levels in vivo. Given that PGC-1α expression is highly elevated and XBP1s function is impaired in the livers of obese and diabetic mice [15], [16], [24], [35], we examined whether levels of XBP1s protein are diminished in the livers of lean mice when PGC-1α expression is increased. The only known physiological way to induce endogenous XBP1s in the liver in vivo is feeding [15]; thus, we analyzed XBP1s protein amount during refeeding after fasting with or without PGC-1α overexpression.
We overexpressed PGC-1α in the liver of 8-week-old lean male mice by tail vein injection of adenovirus expressing PGC-1α (Ad-PGC-1α) or control adenovirus (Ad-LacZ); five days post injection, mice were fasted for 24 h, then refed for 1 h. Ad-PGC-1α injection successfully increased hepatic PGC-1α expression, both at the transcript and protein levels (Figure 5A,C and Supplementary Figure 3A). Control mice exhibited increased hepatic XBP1s expression (in both transcript and protein levels) and nuclear translocation upon refeeding (Figure 5B–E). Refeeding generated a comparable amount of spliced XBP1 mRNA (Xbp1s) in the livers of mice injected with Ad-PGC-1α compared to those of Ad-LacZ controls (Figure 5B). However, total XBP1s protein amount and, accordingly, nuclear translocation of XBP1s during refeeding were significantly diminished in the livers of mice with increased expression of PGC-1α (Figure 5C–E).
Figure 5.
Increasing PGC-1α expression in the liver down-regulates endogenous hepatic XBP1s activity. (A–E) Eight week-old lean male mice were infected with Ad-LacZ (n = 4) or Ad-PGC-1α (1.2 × 108 PFU/g, n = 4) via tail-vein injection; 5 days later, the mice were fasted for 24 h, then re-fed for 1 h. (A) PGC-1α mRNA (Ppargc1a) levels and (B) Xbp1s mRNA levels in the liver determined by qPCR. (C) Levels of endogenous XBP1s and tubulin protein in total liver extracts, and levels of nuclear XBP1s, flag-PGC-1α, and lamin A/C protein levels in nuclear extracts from the liver. (D, E) Quantitation of XBP1s protein levels in C. (D) XBP1s protein amounts in total liver lysates normalized with tubulin and (E) XBP1s protein amounts in the nuclear fraction normalized with Lamin A/C. (F–H) Eight week-old lean male mice were infected with Ad-LacZ or Ad-XBP1s (4 × 107 PFU/g) via tail-vein injection. 5 days later, 4 h-fasted mice received either saline or glucagon (500 μg/kg) by intraperitoneal injection. The liver was collected 2 h after the treatment. (F) Xbp1s mRNA levels in the liver (n = 5 for each). (G) Levels of XBP1s, PGC-1α, and actin proteins in the liver. (H) Quantified XBP1s protein levels in G. Each experiment was independently repeated in three independent cohorts (A–H). Values are means ± s.e.m. Significance was determined by Student's t test or one-way ANOVA with Bonferroni post hoc test (F, H). *P < 0.05, **P < 0.01, ***P < 0.001. N/S – not significant.
Next, we expressed XBP1s in the liver of lean mice via injection of Ad-XBP1s through the tail vein and then induced endogenous PGC-1α expression by administering glucagon (500 μg/kg, intraperitoneal injection). Ad-XBP1s injection increased both transcript and protein levels of XBP1s (Figure 5F–H). Glucagon treatment successfully increased endogenous PGC-1α expression in the liver (Figure 5G) without altering mRNA level of XBP1s (Figure 5F). However, glucagon treatment significantly down-regulated XBP1s protein levels (Figure 5G,H). Collectively, our data suggest that PGC-1α protein also reduces XBP1s protein levels in vivo.
3.6. Reducing hepatic PGC-1α expression increases XBP1s activity in obese mice
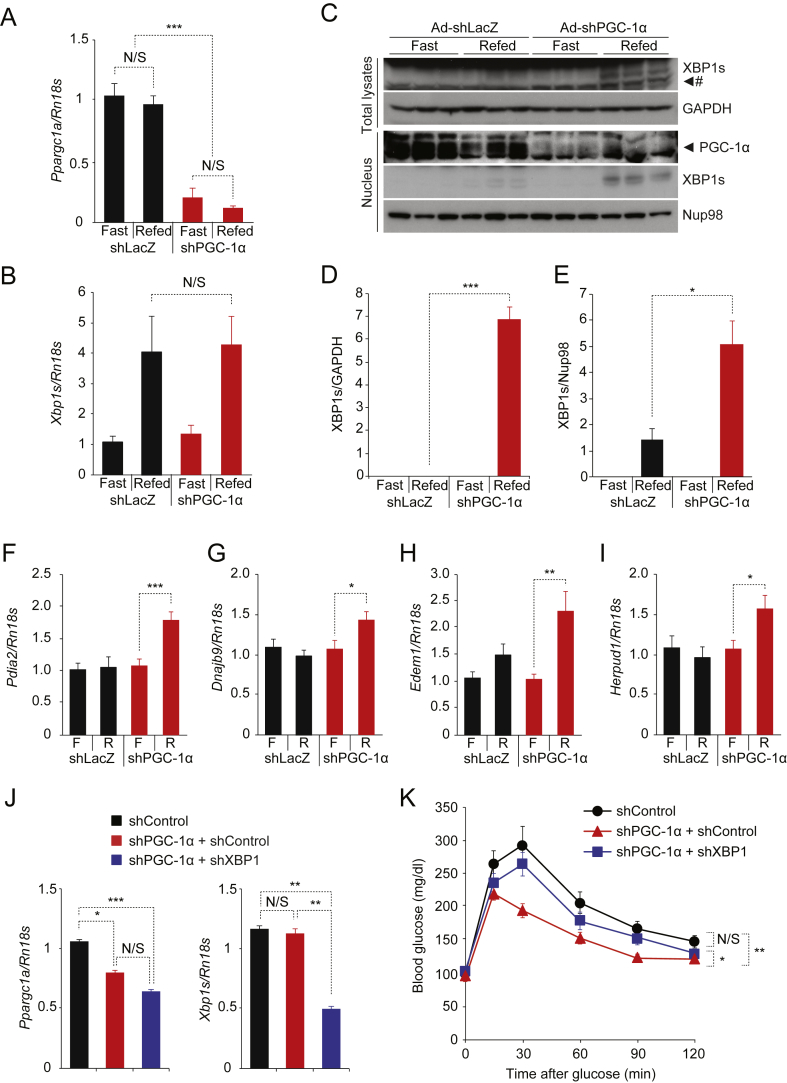
We have previously shown that one of the pathologies leading to development of glucose intolerance in obesity is reduced XBP1s activity [15], [16]. Considering the fact that PGC1α leads to degradation of XBP1s, and that PGC1α expression and activity are increased in the livers of obese mice [24], we sought to investigate whether reducing PGC-1α expression in the liver of obese mice will increase XBP1s protein levels and subsequently contribute to improvement of glucose homeostasis. Eight-week-old ob/ob male mice were injected via the tail vein with adenovirus expressing shRNA against PGC-1α while control ob/ob mice were injected with shLacZ-producing adenovirus. Glucose tolerance test (GTT) demonstrated a marked improvement of glucose clearance in obese mice expressing shPGC-1α compared to control ob/ob mice (Supplementary Figure 3B, C). Both fed and fast blood glucose were significantly reduced in Ad-shPGC-1α-infected ob/ob mice (Supplementary Figure 3D, E). ShPGC-1α significantly lowered PGC-1α gene expression (Ppargc1a) in the liver (Supplementary Figure 3F) and also down-regulated other gluconeogenic gene expression including G6pc and Pck1 (Supplementary Figure 3F). Reduced PGC-1α expression in obese mouse liver did not lead to any significant change in Xbp1s mRNA levels (Supplementary Figure 3G); however, nuclear XBP1s levels were significantly increased in mouse liver expressing shPGC-1α (1.1 ± 0.2 vs. 12.0 ± 3.9, shLacZ vs. shPGC-1α, p = 0.03) (Supplementary Figure 3H, I). In line with elevated XBP1s protein levels in the nucleus, total FoxO1 protein levels were reduced (Supplementary Figure 3J, K), and the expression of XBP1s target genes was increased in the liver (Supplementary Figure 3L), suggesting that decreased PGC-1α activity up-regulates XBP1s function.
We further investigated whether reducing PGC-1α protein levels restores XBP1s protein induction and activity during refeeding in obese mouse liver. Ob/ob mice were injected with adenovirus expressing shPGC-1α or shLacZ as control, and, six days after virus injection, mice were fasted for 24 h and refed for 1 h (Figure 6A–E) or 3 h (Figure 6F–I). ShRNA against PGC-1α effectively reduced the PGC-1α mRNA levels in livers of ob/ob mice (Figure 6A). Reduction of PGC-1α expression again did not alter levels of Xbp1s mRNA generated during refeeding, which were comparable to those in control mouse liver (Figure 6B). ShLacZ-expressing ob/ob mice exhibited that low amount of XBP1s proteins were generated and translocated into the nucleus in the liver during refeeding (Figure 6C–E), however ob/ob mice with shPGC-1α demonstrated a significant increase in total protein induction and nuclear translocation of XBP1s during refeeding (Figure 6C–E). Accordingly, when we measured XBP1s target gene expression during refeeding, we found that refeeding failed to induce Protein Disulfide Isomerase Family A, Member 2 (Pdia2), DnaJ (Hsp40) Homolog, Subfamily B, Member 9 (Dnajb9), ER Degradation Enhancer, Mannosidase Alpha-Like 1 (Edem1) and Homocysteine-Inducible, Endoplasmic Reticulum Stress-Inducible, Ubiquitin-Like Domain Member 1 (Herpud1) gene expression in control obese mouse liver as we documented previously [15] (Figure 6F–I); however, depletion of PGC-1α restored feeding-induced XBP1s target gene expression in the liver (Figure 6F–I).
Figure 6.
Reducing PGC-1α expression in the obese mouse liver restores endogenous XBP1s activity. (A–I) Eight week-old ob/ob mice received Ad-shLacZ (n = 4) or Ad-shPGC-1α (6 × 107 PFU/g, n = 4) via tail-vein injection, and mice were fasted for 24 h and then re-fed for 1 h (A–E) or 3 h (F–I) on day 7 after virus injection. (A) Ppargc1a mRNA levels and (B) Xbp1s mRNA levels in the liver after 24 h fast and 1 h refeeding. (C) Endogenous XBP1s and GAPDH protein levels in total liver extracts, and XBP1s, PGC-1α and NUP98 protein levels in nuclear extracts from the liver. (D,E) Quantitation of XBP1s protein levels (in J) in total liver lysates (D) and in the nuclear fraction (E). Each experiment in A-E was independently repeated in two different cohorts. (F) Pdia2, (G) Dnajb9, (H) Edem1, and (I) Herpud1 mRNA levels in the liver after 24 h fast and 3 h refeeding. Results in F-I are average of three independent cohorts (total mice numbers are 12 for each group). (J, K) Eight week-old ob/ob mice received adenoviruses expressing shControl, shPGC-1α (shPGC-1α + shControl), or shPGC-1α together with shXBP1 (shPGC-1α + shXBP1) via tail-vein injection. (J) mRNA levels of Ppargc1a (Left) and Xbp1s (Right) in the liver measured by qPCR. (K) Glucose tolerance test (GTT, glucose 0.5 g/kg). Results in J and K are average of four independent cohorts (total n = 24 for shControl, n = 27 for shPGC-1α + shControl, n = 26 for shPGC-1α + shXBP1). See Supplementary Experimental Procedures for the details. Values are means ± s.e.m. Significance was determined by one-way ANOVA with Bonferroni post hoc test (J), two-way ANOVA (K) or by Student's t test. *P < 0.05, **P < 0.01, ***P < 0.001. N/S – not significant. # Non-specific bands.
Next, we investigated whether the interaction between PGC-1α and XBP1s contributes to glucose homeostasis in vivo. We injected ob/ob mice with adenoviruses expressing shControl, shPGC-1α (shPGC-1α + shControl), or shPGC-1α with shXBP1 (shPGC-1α + shXBP1) via tail vein injection. In order to deliver both shPGC-1α and shXBP1 to mice without creating toxicity, we used a lower dose of adenovirus for shPGC-1α (see Supplementary Experimental Procedures for the detail) compared to other experiments in Figure 6A–I and Supplementary Figure 3. The reduced dose we used for shPGC-1α (shPGC-1α + shControl and shPGC-1α + shXBP1) still significantly down-regulated endogenous PGC-1α levels in the liver compared with shControl (Figure 6J). ShXBP1 (shPGC-1α + shXBP1) also produced marked down-regulation of hepatic XBP1s levels (Figure 6J). There was no difference in food intake and body weight among groups (Supplementary Figure 4). Ob/ob mice expressing shPGC-1α alone (shPGC-1α + shControl) showed improved glucose tolerance compared to shControl group (Figure 6K). However, simultaneous down-regulation of XBP1s along with PGC-1α in the liver (shPGC-1α + shXBP1) reversed such an improvement in glucose tolerance created in shPGC-1α + shControl group (Figure 6K). There was no difference in glucose tolerance between shControl and shPGC-1α + shXBP1 groups (Figure 6K).
Collectively, our data in vivo demonstrated the existence of molecular interaction between endogenous PGC-1α and endogenous XBP1s in the liver and documented that increased PGC-1α contributes to reduced XBP1s activity in obesity, and reducing PGC-1α in obese mouse liver significantly improves XBP1s action that contributes to the reduction of hepatic gluconeogenesis.
4. Discussion
As obesity surges to an epidemic level globally, obesity-associated pathologies such as type 2 diabetes and cardiovascular diseases have become serious health, economic, and social concerns; but while the development of therapeutics to address these disorders remains a central goal in the field, success has been elusive. A more detailed understanding of the mechanisms underlying diabetes and other obesity-related diseases may lead to new clinical treatment options.
Recent efforts including ours have demonstrated that ER stress plays a crucial role in the development of obesity-linked metabolic disorders and can be promising therapeutic target to treat them [5], [6]. Increased ER stress was documented in the hypothalamus, liver, and adipose tissues both from obese animal models and human subjects [12], [13], [14], [36]. Furthermore, reducing ER stress by administrating chemical chaperones such as 4-phenylbutyrate (4-PBA), tauroursodeoxycholic acid (TUDCA), Celastrol, and Withaferin A has shown to improve leptin sensitivity and systematic glucose homeostasis not just in animal models but also in obese and insulin resistant human patients [13], [37], [38], [39], [40], [41].
In recent years, we have identified several mechanistic pathologies that lead to development of ER stress in obesity conditions [15], [16], [42]. One of the most important mechanisms that we identified was that the activity of XBP1s, the master regulator of ER homeostasis, was reduced in obesity conditions [15], [16] and that this reduction not only plays a crucial role for development of ER stress in obesity but also directly leads to up-regulation of hepatic glucose production [17]. Under normal conditions, XBP1s interacts with FoxO1 leading to FoxO1 degradation in post-prandial conditions, which ultimately reduces the hepatic gluconeogenic gene expression [17], whereas reduced effectiveness of this mechanism in obesity increases hepatic glucose output and hyperglycemia [17].
PGC-1α is a central regulator of cellular pathways involved in metabolic homeostasis in various organs such as the brain, muscle, brown and white adipose tissues, pancreatic β cells, and liver [22], [43], [44], [45], [46] and is recognized as a co-activator of a variety of transcription factors. PGC-1α is a powerful inducer of hepatic glucose production by its ability to increase the activity of gluconeogenic transcription factors such as FoxO1 and HNF4α during fasting conditions. As detailed above, XBP1s in the liver plays an important role in down-regulation of hepatic gluconeogenesis during post-prandial state in healthy conditions [15], [16], [17], [47].
Our current work shows that PGC-1α interacts with XBP1s both in vitro and in vivo, but, unexpectedly, their physical interaction leads to the suppression – rather than the promotion – of XBP1s activity by facilitating ubiquitination and protein degradation of XBP1s via the 26S proteasome. Our findings are the first to report a negative role for PGC-1α on activation of a transcription factor through the physical interaction. This unexpected negative input from PGC-1α to XBP1s activity probably evolved to ensure proper functioning of PGC-1α-FoxO1 axis when the organism is shifting to fasting from a fed state. Hepatic glucose production is one of the most crucial processes that is necessary to keep the blood glucose level within a very narrow range. Hepatic XBP1s protein amount is high in fed states and this has a powerful effect on blockade of FoxO1 activity. However, when an organism starts to shift to a fasting state, hepatic glucose production has to be increased to prevent hypoglycemia. In this state, PGC-1α levels and activity start to increase to activate the transcription factors such as HNF4α and FoxO1 that are involved in up-regulating the hepatic glucose production. Our results indicate that PGC-1α, while activating the gluconeogenic transcription factors, also inhibits XBP1s to prevent XBP1s's effect towards FoxO1, to increase the efficiency of FoxO1 (Supplementary Figure 5). Indeed, our results document that PGC-1α has dominance over XBP1s-FoxO1 axis; in the presence of PGC-1α, XBP1s loses its ability to inhibit FoxO1.
While in healthy conditions this mechanism is beneficial for regulation of blood glucose levels, in obesity, hepatic PGC-1α expression is highly increased, and this up-regulation has previously shown to contribute to the increased hepatic glucose production and consequently to the hyperglycemia [24], [27], [35]. Probably, one of the most challenging times for keeping blood glucose levels of type 2 diabetic patients under control is the post-prandial state. Our results indicate that increased hepatic PGC-1α in obesity and a consequent decrease in XBP1s activity lead to a higher level of hepatic glucose production through up-regulation of FoxO1 activity. Indeed, elevated hepatic PGC-1α expression in otherwise healthy lean mice is associated with reduced amount and nuclear translocation of XBP1s protein during post-prandial state; however, attenuation of PGC-1α expression in obese mouse liver leads to up-regulation of XBP1s protein during refeeding.
5. Conclusions
In summary, our results both in vitro and in vivo collectively point to a new function of PGC-1α in obesity and document that PGC-1α, through interacting with XBP1s and subsequently directing it to 26S proteasome-mediated degradation, leads to suppression of XBP1s activity. These results indicate that PGC-1α not only is a co-activator but also leads to suppression of the activity of its binding transcription factors. Moreover, PGC-1α′s function as a co-suppressor of XBP1s, in addition to as a co-activator of FoxO1 and HNF4α, leads to impaired glucose homeostasis in obese and diabetic mice. Thus, disruption of the interaction between PGC-1α and XBP1s might provide a potential therapeutic avenue for restoring glucose metabolism in obese and diabetic patients.
Acknowledgments
This work was supported by the funds provided to U.O. from the Department of Medicine, Boston Children's Hospital, and by grants to U.O. from the National Institutes of Health (R01DK098496, R01DK081009 and R56DK089111) and the American Diabetes Association Career Development grant (#7-09-CD-10).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.molmet.2017.10.010.
Contributor Information
Jaemin Lee, Email: jaeminlee@dgist.ac.kr.
Umut Ozcan, Email: umut.ozcan@childrens.harvard.edu.
Conflict of interest
U.O. is a scientific founder, shareholder, and scientific advisory board and board of directors member of ERX Pharmaceuticals Inc.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Ogden C.L., Carroll M.D., Kit B.K., Flegal K.M. Prevalence of childhood and adult obesity in the United States, 2011-2012. The Journal of the American Medical Association. 2014;311:806–814. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng M., Fleming T., Robinson M., Thomson B., Graetz N., Margono C. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014;384:766–781. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olshansky S.J., Passaro D.J., Hershow R.C., Layden J., Carnes B.A., Brody J. A potential decline in life expectancy in the United States in the 21st century. The New England Journal of Medicine. 2005;352:1138–1145. doi: 10.1056/NEJMsr043743. [DOI] [PubMed] [Google Scholar]
- 4.Berrington de Gonzalez A., Hartge P., Cerhan J.R., Flint A.J., Hannan L., MacInnis R.J. Body-mass index and mortality among 1.46 million white adults. The New England Journal of Medicine. 2010;363:2211–2219. doi: 10.1056/NEJMoa1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee J., Ozcan U. Unfolded protein response signaling and metabolic diseases. The Journal of Biological Chemistry. 2014;289:1203–1211. doi: 10.1074/jbc.R113.534743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park S.W., Ozcan U. Potential for therapeutic manipulation of the UPR in disease. Seminars in Immunopathology. 2013;35:351–373. doi: 10.1007/s00281-013-0370-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nature Reviews Molecular Cell Biology. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 8.Harding H.P., Zhang Y., Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 9.Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 11.Haze K., Yoshida H., Yanagi H., Yura T., Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Molecular Biology of the Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozcan U., Cao Q., Yilmaz E., Lee A.H., Iwakoshi N.N., Ozdelen E. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 13.Ozcan L., Ergin A.S., Lu A., Chung J., Sarkar S., Nie D. Endoplasmic reticulum stress plays a central role in development of leptin resistance. Cell Metabolism. 2009;9:35–51. doi: 10.1016/j.cmet.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Won J.C., Jang P.G., Namkoong C., Koh E.H., Kim S.K., Park J.Y. Central administration of an endoplasmic reticulum stress inducer inhibits the anorexigenic effects of leptin and insulin. Obesity (Silver Spring) 2009;17:1861–1865. doi: 10.1038/oby.2009.194. [DOI] [PubMed] [Google Scholar]
- 15.Park S.W., Zhou Y., Lee J., Lu A., Sun C., Chung J. The regulatory subunits of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nature Medicine. 2010;16:429–437. doi: 10.1038/nm.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J., Sun C., Zhou Y., Lee J., Gokalp D., Herrema H. p38 MAPK-mediated regulation of Xbp1s is crucial for glucose homeostasis. Nature Medicine. 2011;17:1251–1260. doi: 10.1038/nm.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y., Lee J., Reno C.M., Sun C., Park S.W., Chung J. Regulation of glucose homeostasis through a XBP-1-FoxO1 interaction. Nature Medicine. 2011;17:356–365. doi: 10.1038/nm.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park S.W., Herrema H., Salazar M., Cakir I., Cabi S., Basibuyuk Sahin F. BRD7 regulates XBP1s' activity and glucose homeostasis through its interaction with the regulatory subunits of PI3K. Cell Metabolism. 2014;20:73–84. doi: 10.1016/j.cmet.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finck B.N., Kelly D.P. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. The Journal of Clinical Investigation. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J., Handschin C., Spiegelman B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metabolism. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Rodgers J.T., Lerin C., Gerhart-Hines Z., Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Letters. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puigserver P., Wu Z., Park C.W., Graves R., Wright M., Spiegelman B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 23.Puigserver P., Rhee J., Donovan J., Walkey C.J., Yoon J.C., Oriente F. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 24.Yoon J.C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 25.Huss J.M., Kopp R.P., Kelly D.P. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. The Journal of Biological Chemistry. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 26.Vega R.B., Huss J.M., Kelly D.P. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Molecular and Cellular Biology. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhee J., Inoue Y., Yoon J.C., Puigserver P., Fan M., Gonzalez F.J. Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knutti D., Kaul A., Kralli A. A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Molecular and Cellular Biology. 2000;20:2411–2422. doi: 10.1128/mcb.20.7.2411-2422.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee A.H., Iwakoshi N.N., Anderson K.C., Glimcher L.H. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 31.Michael L.F., Wu Z., Cheatham R.B., Puigserver P., Adelmant G., Lehman J.J. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puigserver P., Adelmant G., Wu Z., Fan M., Xu J., O'Malley B. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 33.Weissman A.M. Themes and variations on ubiquitylation. Nature Reviews Molecular Cell Biology. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 34.Higgins R., Gendron J.M., Rising L., Mak R., Webb K., Kaiser S.E. The unfolded protein response triggers site-specific regulatory ubiquitylation of 40S ribosomal proteins. Molecular Cell. 2015;59:35–49. doi: 10.1016/j.molcel.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herzig S., Long F., Jhala U.S., Hedrick S., Quinn R., Bauer A. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 36.Boden G., Duan X., Homko C., Molina E.J., Song W., Perez O. Increase in endoplasmic reticulum stress-related proteins and genes in adipose tissue of obese, insulin-resistant individuals. Diabetes. 2008;57:2438–2444. doi: 10.2337/db08-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozcan U., Yilmaz E., Ozcan L., Furuhashi M., Vaillancourt E., Smith R.O. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J., Lee J., Salazar Hernandez M.A., Mazitschek R., Ozcan U. Treatment of obesity with celastrol. Cell. 2015;161:999–1011. doi: 10.1016/j.cell.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kars M., Yang L., Gregor M.F., Mohammed B.S., Pietka T.A., Finck B.N. Tauroursodeoxycholic Acid may improve liver and muscle but not adipose tissue insulin sensitivity in obese men and women. Diabetes. 2010;59:1899–1905. doi: 10.2337/db10-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao C., Giacca A., Lewis G.F. Sodium phenylbutyrate, a drug with known capacity to reduce endoplasmic reticulum stress, partially alleviates lipid-induced insulin resistance and beta-cell dysfunction in humans. Diabetes. 2011;60:918–924. doi: 10.2337/db10-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J., Liu J., Feng X., Salazar Hernandez M.A., Mucka P., Ibi D. Withaferin A is a leptin sensitizer with strong antidiabetic properties in mice. Nature Medicine. 2016;22:1023–1032. doi: 10.1038/nm.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park S.W., Zhou Y., Lee J., Lee J., Ozcan U. Sarco(endo)plasmic reticulum Ca2+-ATPase 2b is a major regulator of endoplasmic reticulum stress and glucose homeostasis in obesity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19320–19325. doi: 10.1073/pnas.1012044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin J., Wu H., Tarr P.T., Zhang C.Y., Wu Z., Boss O. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 44.Ma D., Li S., Lucas E.K., Cowell R.M., Lin J.D. Neuronal inactivation of peroxisome proliferator-activated receptor gamma coactivator 1alpha (PGC-1alpha) protects mice from diet-induced obesity and leads to degenerative lesions. The Journal of Biological Chemistry. 2010;285:39087–39095. doi: 10.1074/jbc.M110.151688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kleiner S., Mepani R.J., Laznik D., Ye L., Jurczak M.J., Jornayvaz F.R. Development of insulin resistance in mice lacking PGC-1alpha in adipose tissues. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9635–9640. doi: 10.1073/pnas.1207287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon J.C., Xu G., Deeney J.T., Yang S.N., Rhee J., Puigserver P. Suppression of beta cell energy metabolism and insulin release by PGC-1alpha. Developmental Cell. 2003;5:73–83. doi: 10.1016/s1534-5807(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 47.Deng Y., Wang Z.V., Tao C., Gao N., Holland W.L., Ferdous A. The Xbp1s/GalE axis links ER stress to postprandial hepatic metabolism. The Journal of Clinical Investigation. 2013;123:455–468. doi: 10.1172/JCI62819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.