Abstract
Over the past few decades, as gene discovery methods and sequencing technologies have evolved, many genetic variations that significantly increase the risk of or cause neurodegenerative diseases have been identified. However, knowledge of those pathogenic mutations and subsequent mechanism-focused studies has rarely yielded effective treatments, warranting alternative strategies for refining rational therapeutic targets. Nevertheless, with the evolution of gene targeting methods, it has been increasingly recognized that the disease-causing gene itself is the best therapeutic target even when we do not have a full understanding of its biological functions. Considering this, CRISPR/Cas gene editing technology offers the promise of permanently silencing or correcting the disease-causing mutations, potentially overcoming key limitations of RNA-targeting approaches. The versatile CRISPR/Cas-based strategies have the potential to become treatment options for challenging disorders such as neurodegenerative diseases. Here, we summarize recent reports of preclinical applications of CRISPR/Cas in models of neurodegenerative disorders to provide perspectives on therapeutic gene editing for diseases of the nervous system.
Keywords: allele-specific, CRISPR/Cas, gRNA, neurodegenerative disease, PAM, precision medicine
Introduction
Neurodegenerative diseases collectively represent disease conditions involving loss of neurons in the nervous systems. Progressive neurodegeneration processes affect movement, balancing, breathing, cognition and behavior. Thus, neurodegenerative diseases gradually increase one’s dependency over time, resulting in devastation for affected individuals and their family members. Despite numerous implicated underlying mechanisms and even known causal genetic mutations,1–5 most neurodegenerative diseases have been defying the development of effective treatments, reflecting the difficulty in defining rational drug targets through mechanism-focused approaches. However, recent development of site-specific gene editing technologies such as CRISPR (clustered regularly interspaced short palindromic repeats)/Cas (CRISPR-associated systems) offers new hope for the development of effective treatments for neurodegenerative diseases with known causative mutations.
In the literature, applications of CRISPR/Cas focused on the investigation of gene function, disease modeling and preclinical studies.6–13 Of note, preclinical investigation or clinical use of CRISPR/Cas for disease treatment may offer several advantages over RNA-lowering approaches.14 For example, CRISPR/Cas strategies can overcome key limitations of RNAi and/or antisense oligonucleotide (ASO) methods, such as a potentially high off-target activity14 and requirement of repeated treatments,15 which may increase the risk of complications in individuals compromised by chronic progressive neurodegenerative diseases. By contrast, most CRISPR/Cas approaches generate irreversible changes (e.g. inactivation or correction) in the DNA, and therefore, if a cell is correctly targeted, the cell does not need repeated treatments. In addition, demonstration of the feasibility of genome editing in post-mitotic neurons and the mammalian brain7,12 support the potential of CRISPR/Cas strategies as a therapeutic route for neurodegenerative disorders. Also, CRISPR/Cas is highly flexible and thus applicable to neurodegenerative diseases due to loss-of-function mutations, providing advantages over RNA-lowering methods, which is therapeutically meaningful to gain-of-function mutations. In contrast to other gene editing techniques, site-specificity for CRISPR/Cas is mediated by an interaction between guide RNA (gRNA) and target DNA. Thus, protein engineering is not required for CRISPR/Cas, making this approach highly feasible and affordable.16,17
The field has quickly applied various CRISPR/Cas strategies to models of neurodegenerative diseases, and demonstrated therapeutic potential in preclinical studies. CRISPR/Cas approaches for neurodegenerative diseases can be broadly grouped into (1) correction of disease-causing mutations; (2) inactivation of gain-of-function mutations; and (3) modulation of transcription. In this review article, we summarize recent applications of CRISPR/Cas9 to neurodegenerative diseases, focusing on describing intervention strategies based on CRISPR/Cas gene editing.
Background on the CRISPR/Cas gene editing system
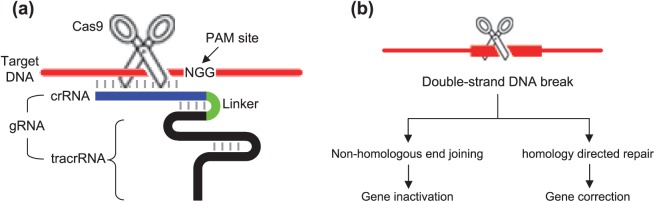
The CRISPR/Cas system is a bacterial defense mechanism that inactivates foreign genetic material.16–20 However, its recent modification, fusing CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA) into a chimeric single gRNA (Figure 1(a))21,22 has facilitated site-specific gene editing in mammalian cells, leading to widespread applications in biomedical research. Among three types of CRISPR, the type II system (i.e. CRISPR/Cas9 from Streptococcus pyogenes) is the most commonly utilized in the laboratories. In type II, the CRISPR locus is transcribed and processed, resulting in crRNA–tracrRNA dsRNA (double-stranded RNA) formation to direct Cas nuclease to the target sequence16,17 and to generate double-strand breaks (DSBs).21,23,24 DSBs are repaired by non-homologous end joining (NHEJ) repair mechanisms or homology-directed repair (HDR).25 Although canonical NHEJ appears to result in conservative DSB repair,26 NHEJ repair involved in CRISPR/Cas usually leads to frameshift mutations and thus is likely to produce downstream premature stop codons, resulting in inactivation of the target gene through a nonsense-mediated decay pathway16,17 (Figure 1(b)). In the following sections, we describe how CRISPR/Cas strategies have been used to address various genetic mutations that cause neurodegenerative diseases.
Figure 1.
Components and mechanisms of CRISPR/Cas-mediated gene editing.
(a) CRISPR/Cas gene editing requires target DNA (red horizontal line), gRNA (crRNA and tracrRNA fusion), Cas endonuclease (scissors) and a PAM site. Cas9 and NGG PAM site of S. pyogenes are shown in this illustration.
(b) A double-strand break induced by CRISP/Cas is processed by two distinct pathways. Non-homologous end joining and homology-directed repair lead to gene inactivation and correction, respectively.
Correction of disease-causing DNA repeats and single-nucleotide variants
Many forms of neurodegenerative diseases are due to genetic mutations,1–5,27 and attempts have been made to correct disease-causing mutations using CRISPR/Cas strategies in various model systems.
Huntington’s disease
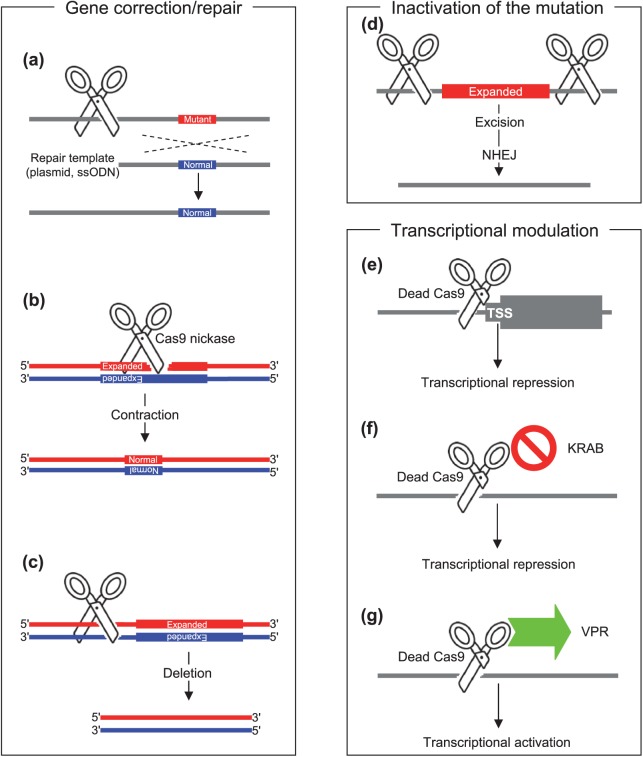
Many neurodegenerative diseases [e.g. Huntington’s disease (HD), spinocerebellar ataxias] are caused by expansions of unstable nucleotide repeats in certain genes.28 Reducing the sizes of disease-causing expanded repeats using CRISPR/Cas is therefore hypothesized to ameliorate associated disease pathogenesis. HD is a dominantly inherited neurodegenerative disease, caused by an expansion of CAG trinucleotide repeat in the first exon of huntingtin gene (HTT).3,29 HTT CAG repeat is highly polymorphic in the normal population30; once their lengths become greater than 35, various characteristic neurological symptoms occur.29 The first attempt to reduce the size of the disease-generating CAG repeat in induced pluripotent stem cells (iPSCs) derived from an individual with HD (carrying 72 and 19 CAGs) was based on homologous recombination using a repair template of bacterial artificial chromosome (BAC) containing the entire HTT with a normal CAG repeat (21 CAGs).31 Expression profiling analysis and apoptosis assays showed that genetically corrected iPSC clonal lines showed normalization of various cellular pathogenic signaling pathways (e.g. cadherin, TGF-β, BDNF) and disease phenotypes (e.g. susceptibility to cell death),31 supporting its therapeutic benefits. Subsequently, CRISPR/Cas9 was used to improve the efficiency of homologous recombination (Figure 2(a)) when making isogenic allelic series of HD cell models.32 More recently, an expanded HTT CAG repeat in HD patient-derived iPSC line was corrected using CRISPR/Cas9 in combination with a piggyBac transposon-based selection system through HDR33; neural rosette formation deficit and increased cell death following growth factor withdrawal were reversed in corrected HD iPSC lines.34 Contracting expanded CAG/CTG repeats using CRISPR/Cas9 D10A nickase without using a repair template was also reported. DSBs generated by CRISPR/Cas9 directly targeting the repeat sequence induced both expansion and contraction of the repeat. However, when single-strand breaks were produced by Cas9 D10A nickase, the repeat in the reporter vector tended to contract due to the activation of distinct DNA repair mechanisms (Figure 2(b)).35 CRISPR/Cas approaches directly targeting disease-causing repeats have to be thoroughly evaluated for their target gene specificities to avoid permanent modification of other repeat-containing genes.
Figure 2.
CRISPR/Cas strategies for neurodegenerative diseases.
Broadly, three CRISPR/Cas strategies were applied to model systems of neurodegenerative diseases: gene correction; inactivation of mutation; and transcriptional modulation. Depending on the objective of the gene editing, Cas9 endonuclease ((a), (c), (d)), Cas9 nickase (b), or dead Cas9 ((e), (f), (g)) were used to correct the genetic defects ((a), (b), (c)), inactivate the gain-of-function mutation ((d)), or modulate transcription of disease-related genes ((e), (f), (g)).
Amyotrophic lateral sclerosis/frontotemporal dementia
An expansion of GGGGCC hexanucleotide (G4C2) repeats in the C9orf72 gene is the most common cause of familial amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD).27,36 An expanded G4C2 repeat in an iPSC line from a subject with C9orf72 mutation was corrected via homologous recombination using CRISPR/Cas9 and a plasmid DNA donor template (Figure 2(a)); motor neurons differentiated from gene-corrected iPSC lines showed amelioration of disease-associated cellular phenotypes such as abnormal protein aggregation and stress granule formation.37 Applications of CRISPR/Cas to correct pathogenic single-nucleotide variants causing neurodegenerative diseases (Figure 2(a)) are also described in the literature. Mutations in several genes including SOD1 and FUS are found in familial cases of ALS,4,38–40 and targeted gene correction methods using CRISPR/Cas9 were able to convert pathogenic alleles such as SOD A272C and FUS G1566A into non-pathogenic normal alleles in iPSCs. The repair template was provided by either (1) a donor plasmid containing wild-type allele and homology arm or (2) a linear single-stranded oligodeoxynucleotide (ssODN).41 Corrected iPSC clones maintained pluripotency markers and differentiation capability.
Frontotemporal dementia and Alzheimer disease
Similar CRISPR/Cas-mediated correction approaches (Figure 2(a)) were applied to cell models of FTD and Alzheimer’s disease in an effort to generate an isogenic cell panel of disease models. Briefly, R406W and P301L mutations in microtubule associated protein tau (MAPT) were corrected using ssODN-mediated CRISPR/Cas9 in iPSCs.42–44 Similarly, A79V and L150P mutations in presenilin 1 (PSEN1) were corrected using CRISPR/Cas and ssODN repair templates.45,46
Fragile X-associated tremor ataxia syndrome
A neurodegenerative disease, Fragile X-associated tremor ataxia syndrome (FXTAS) is caused by an expansion of CGG repeat in the 5′-untranslated region of the fragile X mental retardation gene (FMR1). The size of expanded repeats determines the type of disease; a full mutation (>200 repeats) and pre-mutation (55–200 repeats) result in Fragile X syndrome (FXS) and FXTAS, respectively.47–50 Application of a CRISPR/Cas9 strategy aiming at targeting the end of the upstream sequence of the CGG repeat of FMR1 could cause large deletions of abnormally expanded repeats (Figure 2(c)), and reverse FMR1 methylation in patient-derived iPSC and pluripotent stem cells.51
Inactivation of gain-of-function mutation
Allele-specific CRISPR/Cas approaches to (selectively) inactivate the disease allele (Figure 2(d)) have been tested in model systems of a dominantly inherited neurodegenerative disorder.
Huntington’s disease
HD results from a CAG expansion mutation in HTT through gain-of-function mechanisms. Complete lack of Htt leads to embryo lethality in mice,52–54 and compound heterozygous nullifying variants in HTT are associated with a neurodevelopmental disorder in humans.55 Whether or not huntingtin is dispensable in the adult brain is controversial.56,57 However, loss of one copy of the gene due to balanced translocation does not cause HD,58 supporting therapeutic CRISPR/Cas gene silencing. These reinforce that therapeutic CRISPR/Cas silencing strategy for HD needs to be mutant allele-specific to avoid adverse outcomes due to collateral silencing of the normal allele. Recently, mutant allele-specific CRISPR/Cas strategies using SNPs that create or eliminate PAM sites were developed.59 By mapping all CRISPR PAM sites on HTT gene haplotypes and comparing haplotypes in a pair-wise manner, the mutant HTT gene haplotype-specific CRISPR PAM sites for a given diplotype were identified. In a proof-of-principle experiment, two mutant allele-specific gRNAs whose targets encompass (1) important regions for gene transcription including promoter region and transcription start site and (2) the expanded CAG repeat in the first exon, were used to selectively inactivate the mutant allele. The simultaneous use of two mutant allele-specific gRNAs eliminated an approximately 44 Kb DNA segment including the promoter region, transcription start site and expanded CAG repeats from the mutant allele without affecting the normal HTT allele in patient-derived fibroblast, iPS and NPC cells. This excision ultimately prevented the generation of mutant HTT mRNA and protein, indicating complete and permanent inactivation of the HD chromosome.
Subsequently, a similar gene silencing approach to inactivate the mutant allele by excising the first exon of HTT in an allele-specific manner was reported.60 Using one allele-specific and one non-allele-specific gRNA simultaneously, the first exon of Htt including the CAG expansion mutation was excised, leading to reduced expression levels of the mutant Htt in a population of cells. Although moderate, the normal HTT expression levels were also reduced by a dual gRNA CRISPR approach possibly due to the interference of transcription of normal allele by non-allele-specific gRNA. Nevertheless, the delivery of dual gRNAs and Cas9 using a rAAV shuttle vector system into BACHD transgenic mice significantly lowered the mutant HTT mRNA levels, demonstrating the effectiveness of CRISPR/Cas in vivo for the first time.
Novel concepts such as (1) allele specificity using PAM-altering SNPs, (2) targeting the haplotype carrying the mutation, rather than the mutation itself and (3) preventing transcription of the mutant allele to inactivate the gene were integrated to permanently silence the mutant gene in a completely allele-specific manner in HD. Although strategies of transcription prevention by CRISPR/Cas may be less efficient compared to single gRNA CRISPR/Cas approaches because of (1) excision of a rather larger region and (2) requirement of two gRNAs, haplotype-targeting CRISPR/Cas silencing strategies are broadly applicable to other dominant diseases.
Transcriptional modulation of neurodegenerative disease-related genes
Targeting specific regions of a gene using (modified) CRISPR/Cas systems permits modulation of expression levels of genes that cause or are associated with neurodegenerative diseases.
Huntington’s disease
It has been recently demonstrated that the expression levels of HTT could be lowered by CRISPR/Cas approaches using a single gRNA, targeting non-coding regions of the gene.61 In mesenchymal stem cells extracted from the bone marrow of the YAC128 HD mouse model,62 single gRNA-mediated CRISPR/Cas9 DNA editing at the 5′ untranslated region (UTR) or exon1–intron1 junction of HTT resulted in reduced HTT mRNA and protein expression levels.61 A significant reduction of HTT mRNA expression levels was also achieved by targeting the transcription start site of HTT using a single gRNA and dead Cas9 (dCas9) (Figure 2(e)).63 If genetic variations that permit allele-specific CRISPR/Cas9 targeting are available in the region important for transcriptional and/or translational regulation of HTT, this approach may provide therapeutic benefits without producing significant side effects in HD.
Parkinson’s disease and Alzheimer’s disease
In addition, a mutant form of Cas9 and various fusions were used to modulate the expression levels of neurodegenerative disease-related genes. The use of dCas9 and a gRNA targeting the transcription start sites significantly reduced the expression levels of disease-associated genes such as SNCA, MAPT and APP.63 Also, mRNA and protein expression levels of SNCA could be precisely up- and downregulated in the human iPSC-derived neurons by CRISPR/Cas9 systems using dCas9-KRAB (Figure 2(f)) and dCas9-VPR effector domain fusion (Figure 2(g)).63
Challenges and opportunities
Various CRISPR/Cas strategies tested in the model systems of neurodegenerative disease demonstrated good target engagement and amelioration of disease-associated cellular phenotypes, supporting their therapeutic potential in clinical trials. Still, most reported CRISPR/Cas strategies in the scientific literature are not immediately applicable for treating humans with neurodegenerative diseases. Various technical hurdles and biological questions need to be addressed before therapeutic use of CRISPR/Cas in humans.
Off-targeting
Most CRISPR/Cas approaches aim at generating permanent changes in DNA, and therefore off-targeting is one of the major concerns of clinical applications of CRISPR/Cas. Cas9 requires extensive homology between sequences of target DNA and gRNA for cleavage.17,64 Still, Cas9 can bind with off-targets with mismatches,64 and such transient binding may result in variable off-target activities.65 Thus, levels of off-targeting may vary widely depending on the CRISPR/Cas strategies, and it may be very difficult to develop ones without any off-target activity. However, levels of off-target activities of CRISPR/Cas can be significantly reduced by using (1) optimized gRNA design methods,65 (2) truncated gRNA,66 (3) Cas9 with increased fidelity,67,68 (4) Cas9 nickase mutant,69,70 (5) low levels of Cas923 and (6) controllable Cas9.71,72 Combinatorial approaches may further lower off-target activity of CRISPR/Cas to the extent that therapeutic benefits exceed off-targeting-mediated side effects.
Delivery
The most promising CRISPR/Cas delivery vector for neurodegenerative disease is adeno-associated viruses (AAVs), as they are relatively safe and efficient in generating a gene of interest, without genome integration.73,74 Due to their limited transgene capacities, smaller endonucleases such as saCas975 or Cpf176–78 may be better suited for AAV vectors. Nevertheless, dual AAV vector systems for spCas9 and gRNA efficiently generated DNA modification in mouse brains.12,60 However, prolonged expression of high levels of Cas9 may lead to increased levels of off-target activity.23 Alternatively, Cas9 protein may be delivered for efficient DNA targeting with minimal off-targeting in the context of protein therapy.79,80
Homology-directed repair versus non-homologous end joining
CRISPR/Cas strategies for genetic correction through HDR are believed to be less efficient, and may instead induce NHEJ, potentially resulting in knocking-out the target gene. In the worst-case scenario, CRISPR/Cas strategies aimed at correcting the mutant allele may inactivate the normal allele. In preclinical studies, screening assays were performed to select genetically corrected cell clones for subsequent molecular characterization. Since selection is not desirable in therapeutic applications of CRISPR/Cas, development of genome editing strategies that induce high-fidelity HDR with low NHEJ is critical.23,81 The use of mutant allele-specific CRISPR/Cas strategies designed for HDR of the mutant allele will also contribute to minimizing the levels of NHEJ of the normal allele.
Requirement of personalization for allele-specific CRISPR/Cas
Allele-specific CRISPR/Cas strategies targeting genetic variations that are linked to the mutations require customized designs because each patient may have different combinations of mutant and normal allele haplotypes. Considering the cost associated with clinical trials, personalized allele-specific CRISPR/Cas strategies may sound unrealistic. However, certain disease subjects share the founder mutation,82 and therefore alleles on the ancestral haplotype with low allele frequency in the normal population may allow a single mutant allele-specific CRISPR/Cas gene editing to be applicable to most patients who share the founder mutation. In addition, when distributions of haplotypes of mutant allele and normal allele are quite distinct (e.g. HD),83 a set of a small number of mutant allele-specific CRISPR/Cas strategies may be developed for the majority of patients. Identification of broadly applicable allele-specific target sites will facilitate the efficacy testing of CRISPR/Cas strategies in humans.
Limitations of transcriptional modulation
The use of dead Cas9 would generate temporary effects due to the lack of DNA modification, and therefore it remains to be investigated whether CRISPR-mediated transcription modulation strategies using dCas9 (CRISPRi) provide advantages over conventional RNA-lowering approaches such as ASO. Also, the likelihood of allele-specific application would be variable, depending on the availability of human genetic variations near transcription start sites of target genes. Still, transcriptional activation using dCas9–VPR fusion indicates versatility of CRISPR approaches and supports its utility in treating human diseases due to haploinsufficiency.
Treatment window
Since the CRISPR field is actively developing highly efficient and gene-selective CRISPR genome editing methodologies and delivery mechanisms,12,67,68,72,84,85 many of the technical problems including off-targeting are expected to be solved in the future. With a promising forecast for technical aspects of CRISPR/Cas, integration of allele-specific strategies with efficient delivery methods is likely to open routes for intervening in neurodegenerative diseases in humans. However, important biological questions of neurodegenerative disorders still remain to be addressed. Considering the nature of the disease, primary objectives of treatments for neurodegenerative disease are to prevent the loss of neurons and their functions, and therefore maximum therapeutic benefits may be achieved when the treatments are applied reasonably early. Symptomatic interventions were able to attenuate related pathological markers and behavioral phenotypes in animal models of neurodegenerative diseases, but neuronal loss was not reverted.86–89 In support of stage-dependent therapeutic efficacy, disease was completely and partially reversed when the production of mutant ataxin-1 was halted at an early stage and at a later stage, respectively, in a model system of spinocerebellar ataxia type 1 (SCA1).90 When it comes to CRISPR/Cas therapeutics for chronic and progressive diseases of the nervous system, it is particularly important to choose an optimal treatment window to obtain maximum therapeutic benefits without generating unwanted side effects. For example, if CRISPR/Cas treatments are delivered too late, damaged neurons may not be able to restore their cellular integrity and functions; if gene silencing CRISPR/Cas treatments are applied too early, unwanted adverse consequences may arise due to unexpected compromised gene functions. Despite its importance, the optimal treatment window for each neurodegenerative disease is unclear due to the lack of preclinical CRISPR/Cas experiments on relevant animal models. Thus, generation of genetically faithful animal models with relevant human genetic variations that allow tests for allele-specific CRISPR/Cas strategies, and subsequent time-series analysis, will provide valuable insights into temporal aspects of CRISPR/Cas therapeutics, facilitating the clinical application of powerful CRISPR/Cas genome engineering technology in treating neurodegenerative diseases.
Conclusions
The field has witnessed promises and limitations of CRISPR/Cas as a therapeutic means for challenging health issues such as neurodegenerative disorders. Technical barriers and safety issues certainly must be solved before applying CRISPR/Cas strategies to treat human diseases. Nevertheless, conceptual foundations and key components necessary for the application of CRISPR/Cas to neurodegenerative diseases have been addressed in preclinical studies, and the potential of integrative approaches was also demonstrated. Now, personal genomics becomes more affordable than ever, facilitating the discovery of disease-causing genes and providing knowledge of targets for CRISPR/Cas. Conversely, development of broadly applicable and efficient therapeutic CRISPR/Cas genome engineering tools will permit targeting the root cause of the disease, facilitating genetic tests and the identification of disease-producing mutations through personal genomics. The synergistic interaction between two disciplines will eventually support widespread applications of CRISPR/Cas in precision medicine for neurodegenerative diseases and more, significantly contributing to understanding diseases and improving human health.
Footnotes
Funding: This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Conflict of interest statement: The authors declare that there is no conflict of interest.
Contributor Information
Jun Wan Shin, Molecular Neurogenetics Unit, Center for Genomic Medicine, Massachusetts General Hospital, 185 Cambridge Street, Boston, MA 02114, USA; Department of Neurology, Harvard Medical School, Boston, MA 02115, USA; Medical and Population Genetics Program, The Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Jong-Min Lee, Molecular Neurogenetics Unit, Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA, USA Department of Neurology, Harvard Medical School, Boston, MA, USA.
References
- 1. Deng HX, Hentati A, Tainer JA, et al. Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase. Science 1993; 261: 1047–1051. [DOI] [PubMed] [Google Scholar]
- 2. Goate A, Chartier-Harlin MC, Mullan M, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 1991; 349: 704–706. [DOI] [PubMed] [Google Scholar]
- 3. The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993; 72: 971–983. [DOI] [PubMed] [Google Scholar]
- 4. Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993; 362: 59–62. [DOI] [PubMed] [Google Scholar]
- 5. Tanzi RE, Vaula G, Romano DM, et al. Assessment of amyloid beta-protein precursor gene mutations in a large set of familial and sporadic Alzheimer disease cases. Am J Hum Genet 1992; 51: 273–282. [PMC free article] [PubMed] [Google Scholar]
- 6. Staahl BT, Benekareddy M, Coulon-Bainier C, et al. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat Biotechnol 2017; 35: 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Straub C, Granger AJ, Saulnier JL, et al. CRISPR/Cas9-mediated gene knock-down in post-mitotic neurons. PloS One 2014; 9: e105584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tabebordbar M, Zhu K, Cheng JK, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016; 351: 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nelson CE, Hakim CH, Ousterout DG, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016; 351: 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun 2015; 6: 7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shah AN, Davey CF, Whitebirch AC, et al. Rapid reverse genetic screening using CRISPR in zebrafish. Nat Methods 2015; 12: 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Swiech L, Heidenreich M, Banerjee A, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol 2015; 33: 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shen Z, Zhang X, Chai Y, et al. Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Dev Cell 2014; 30: 625–636. [DOI] [PubMed] [Google Scholar]
- 14. Heidenreich M, Zhang F. Applications of CRISPR-Cas systems in neuroscience. Nat Rev Neurosci 2016; 17: 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Southwell AL, Skotte NH, Kordasiewicz HB, et al. In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol Ther 2014; 22: 2093–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doudna JA, Charpentier E. Genome editing: the new frontier of genome engineering with CRISPR-Cas9. Science 2014; 346: 1258096. [DOI] [PubMed] [Google Scholar]
- 17. Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014; 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garneau JE, Dupuis ME, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010; 468: 67–71. [DOI] [PubMed] [Google Scholar]
- 19. Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010; 327: 167–170. [DOI] [PubMed] [Google Scholar]
- 20. Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, et al. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 2005; 60: 174–182. [DOI] [PubMed] [Google Scholar]
- 21. Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012; 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013; 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science 2013; 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fishman-Lobell J, Rudin N, Haber JE. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol 1992; 12: 1292–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Betermier M, Bertrand P, Lopez BS. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet 2014; 10: e1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011; 72: 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci 2007; 30: 575–621. [DOI] [PubMed] [Google Scholar]
- 29. Bates GP, Dorsey R, Gusella JF, et al. Huntington disease. Nat Rev Dis Primers. 2015; 1: 15005. [DOI] [PubMed] [Google Scholar]
- 30. Lee JM, Ramos EM, Lee JH, et al. CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology 2012; 78: 690–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. An MC, Zhang N, Scott G, et al. Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell 2012; 11: 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. An MC, O’Brien RN, Zhang N, et al. Polyglutamine disease modeling: epitope based screen for homologous recombination using CRISPR/Cas9 system. PLoS Curr 2014; 6 DOI: 10.1371/currents.hd.0242d2e7ad72225efa72f6964589369a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yusa K. Seamless genome editing in human pluripotent stem cells using custom endonuclease-based gene targeting and the piggyBac transposon. Nat Protoc 2013; 8: 2061–2078. [DOI] [PubMed] [Google Scholar]
- 34. Xu X, Tay Y, Sim B, et al. Reversal of phenotypic abnormalities by CRISPR/Cas9-mediated gene correction in Huntington disease patient-derived induced pluripotent stem cells. Stem Cell Reports 2017; 8: 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cinesi C, Aeschbach L, Yang B, et al. Contracting CAG/CTG repeats using the CRISPR-Cas9 nickase. Nat Commun 2016; 7: 13272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011; 72: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mutihac R, Ababneh N, Scaber J, et al. Modelling amyotrophic lateral sclerosis (ALS) using mutant and CAS9/CRISPR-corrected motor neurons from patients with C9ORF72 mutations reveals disease-specific cellular phenotypes. J Neurol Sci 2015; 357(Suppl. 1): e48. [Google Scholar]
- 38. Bruijn LI, Houseweart MK, Kato S, et al. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science 1998; 281: 1851–1854. [DOI] [PubMed] [Google Scholar]
- 39. Drepper C, Herrmann T, Wessig C, et al. C-terminal FUS/TLS mutations in familial and sporadic ALS in Germany. Neurobiol Aging 2011; 32: 548.e1–4. [DOI] [PubMed] [Google Scholar]
- 40. Vance C, Rogelj B, Hortobagyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 2009; 323: 1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang L, Yi F, Fu L, et al. CRISPR/Cas9-mediated targeted gene correction in amyotrophic lateral sclerosis patient iPSCs. Protein Cell 2017; 8: 365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nimsanor N, Kitiyanant N, Poulsen U, et al. Generation of an isogenic, gene-corrected iPSC line from a symptomatic 57-year-old female patient with frontotemporal dementia caused by a P301L mutation in the microtubule associated protein tau (MAPT) gene. Stem Cell Res. 2016; 17: 556–559. [DOI] [PubMed] [Google Scholar]
- 43. Nimsanor N, Poulsen U, Rasmussen MA, et al. Generation of an isogenic, gene-corrected iPSC line from a pre-symptomatic 28-year-old woman with an R406W mutation in the microtubule associated protein tau (MAPT) gene. Stem Cell Res 2016; 17: 600–602. [DOI] [PubMed] [Google Scholar]
- 44. Nimsanor N, Poulsen U, Rasmussen MA, et al. Generation of an isogenic, gene-corrected iPSC line from a symptomatic 59-year-old female patient with frontotemporal dementia caused by an R406W mutation in the microtubule associated protein tau (MAPT) gene. Stem Cell Res 2016; 17: 576–579. [DOI] [PubMed] [Google Scholar]
- 45. Pires C, Schmid B, Petraeus C, et al. Generation of a gene-corrected isogenic control cell line from an Alzheimer’s disease patient iPSC line carrying a A79V mutation in PSEN1. Stem Cell Res 2016; 17: 285–288. [DOI] [PubMed] [Google Scholar]
- 46. Poon A, Schmid B, Pires C, et al. Generation of a gene-corrected isogenic control hiPSC line derived from a familial Alzheimer’s disease patient carrying a L150P mutation in presenilin 1. Stem Cell Res 2016; 17: 466–469. [DOI] [PubMed] [Google Scholar]
- 47. Botta-Orfila T, Tartaglia GG, Michalon A. Molecular pathophysiology of fragile X-associated tremor/ataxia syndrome and perspectives for drug development. Cerebellum 2016; 15: 599–610. [DOI] [PubMed] [Google Scholar]
- 48. Cronister A, Teicher J, Rohlfs EM, et al. Prevalence and instability of fragile X alleles: implications for offering fragile X prenatal diagnosis. Obstet Gynecol 2008; 111: 596–601. [DOI] [PubMed] [Google Scholar]
- 49. Usdin K, Hayward BE, Kumari D, et al. Repeat-mediated genetic and epigenetic changes at the FMR1 locus in the fragile X-related disorders. Front Genet 2014; 5: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Usdin K, Kumari D. Repeat-mediated epigenetic dysregulation of the FMR1 gene in the fragile X-related disorders. Front Genet 2015; 6: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Park CY, Halevy T, Lee DR, et al. Reversion of FMR1 methylation and silencing by editing the triplet repeats in fragile X iPSC-derived neurons. Cell Rep 2015; 13: 234–241. [DOI] [PubMed] [Google Scholar]
- 52. White JK, Auerbach W, Duyao MP, et al. Huntingtin is required for neurogenesis and is not impaired by the Huntington’s disease CAG expansion. Nat Genet 1997; 17: 404–410. [DOI] [PubMed] [Google Scholar]
- 53. Zeitlin S, Liu JP, Chapman DL, et al. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat Genet 1995; 11: 155–163. [DOI] [PubMed] [Google Scholar]
- 54. Nasir J, Floresco SB, O’Kusky JR, et al. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell 1995; 81: 811–823. [DOI] [PubMed] [Google Scholar]
- 55. Rodan LH, Cohen J, Fatemi A, et al. A novel neurodevelopmental disorder associated with compound heterozygous variants in the huntingtin gene. Eur J Hum Genet 2016; 24: 1826–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet 2000; 26: 300–306. [DOI] [PubMed] [Google Scholar]
- 57. Wang G, Liu X, Gaertig MA, et al. Ablation of huntingtin in adult neurons is nondeleterious but its depletion in young mice causes acute pancreatitis. Proc Natl Acad Sci USA 2016; 113: 3359–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ambrose CM, Duyao MP, Barnes G, et al. Structure and expression of the Huntington’s disease gene: evidence against simple inactivation due to an expanded CAG repeat. Somat Cell Mol Genet 1994; 20: 27–38. [DOI] [PubMed] [Google Scholar]
- 59. Shin JW, Kim KH, Chao MJ, et al. Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet 2016; 25: 4566–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Monteys AM, Ebanks SA, Keiser MS, et al. CRISPR/Cas9 editing of the mutant Huntingtin allele in vitro and in vivo. Mol Ther 2017; 25: 12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kolli N, Lu M, Maiti P, et al. CRISPR-Cas9 mediated gene-silencing of the mutant Huntingtin gene in an in vitro model of Huntington’s disease. Int J Mol Sci 2017; 18: 754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Slow EJ, van Raamsdonk J, Rogers D, et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet 2003; 12: 1555–1567. [DOI] [PubMed] [Google Scholar]
- 63. Heman-Ackah SM, Bassett AR, Wood MJ. Precision modulation of neurodegenerative disease-related gene expression in human iPSC-derived neurons. Sci Rep 2016; 6: 28420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu X, Scott DA, Kriz AJ, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 2014; 32: 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 2015; 33: 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fu Y, Sander JD, Reyon D, et al. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 2014; 32: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016; 529: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Slaymaker IM, Gao L, Zetsche B, et al. Rationally engineered Cas9 nucleases with improved specificity. Science 2016; 351: 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013; 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cho SW, Kim S, Kim Y, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 2014; 24: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Konermann S, Brigham MD, Trevino A, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 2013; 500: 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Davis KM, Pattanayak V, Thompson DB, et al. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol 2015; 11: 316–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Burger C, Nash K, Mandel RJ. Recombinant adeno-associated viral vectors in the nervous system. Hum Gene Ther 2005; 16: 781–791. [DOI] [PubMed] [Google Scholar]
- 74. Taymans JM, Vandenberghe LH, Haute CV, et al. Comparative analysis of adeno-associated viral vector serotypes 1, 2, 5, 7, and 8 in mouse brain. Hum Gene Ther 2007; 18: 195–206. [DOI] [PubMed] [Google Scholar]
- 75. Ran FA, Cong L, Yan WX, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015; 520: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim D, Kim J, Hur JK, et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol 2016; 34: 863–868. [DOI] [PubMed] [Google Scholar]
- 77. Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015; 163: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kleinstiver BP, Tsai SQ, Prew MS, et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol 2016; 34: 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang M, Zuris JA, Meng F, et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc Natl Acad Sci USA 2016; 113: 2868–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zuris JA, Thompson DB, Shu Y, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol 2015; 33: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Miyaoka Y, Berman JR, Cooper SB, et al. Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Sci Rep 2016; 6: 23549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mok K, Traynor BJ, Schymick J, et al. Chromosome 9 ALS and FTD locus is probably derived from a single founder. Neurobiol Aging 2012; 33: 209.e3– e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chao MJ, Gillis T, Atwal RS, et al. Haplotype-based stratification of Huntington’s disease. Eur J Hum Genet 2017; 25: 1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Doench JG, Fusi N, Sullender M, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016; 34: 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wojtal D, Kemaladewi DU, Malam Z, et al. Spell checking nature: versatility of CRISPR/Cas9 for developing treatments for inherited disorders. Am J Hum Genet 2016; 98: 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell 2000; 101: 57–66. [DOI] [PubMed] [Google Scholar]
- 87. Regulier E, Trottier Y, Perrin V, et al. Early and reversible neuropathology induced by tetracycline-regulated lentiviral overexpression of mutant huntingtin in rat striatum. Hum Mol Genet 2003; 12: 2827–2836. [DOI] [PubMed] [Google Scholar]
- 88. Diaz-Hernandez M, Torres-Peraza J, Salvatori-Abarca A, et al. Full motor recovery despite striatal neuron loss and formation of irreversible amyloid-like inclusions in a conditional mouse model of Huntington’s disease. J Neurosci 2005; 25: 9773–9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. DiFiglia M, Sena-Esteves M, Chase K, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci USA 2007; 104: 17204–17209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zu T, Duvick LA, Kaytor MD, et al. Recovery from polyglutamine-induced neurodegeneration in conditional SCA1 transgenic mice. J Neurosci 2004; 24: 8853–8861. [DOI] [PMC free article] [PubMed] [Google Scholar]