Summary
Ethylene response factor 1 (ERF1) is an essential integrator of the jasmonate and ethylene signalling pathways coordinating a large number of genes involved in plant defences. Its orthologue in Hevea brasiliensis, HbERF‐IXc5, has been assumed to play a major role in laticifer metabolism and tolerance to harvesting stress for better latex production. This study sets out to establish and characterize rubber transgenic lines overexpressing HbERF‐IXc5. Overexpression of HbERF‐IXc5 dramatically enhanced plant growth and enabled plants to maintain some ecophysiological parameters in response to abiotic stress such as water deficit, cold and salt treatments. This study revealed that HbERF‐IXc5 has rubber‐specific functions compared to Arabidopsis ERF1 as transgenic plants overexpressing HbERF‐IXc5 accumulated more starch and differentiated more latex cells at the histological level. The role of HbERF‐IXc5 in driving the expression of some target genes involved in laticifer differentiation is discussed.
Keywords: genetic modification, latex, plant hormone, rubber, transcription factor
Introduction
The plant hormone ethylene plays a crucial role in regulating plant growth, development and responses to biotic and abiotic stress (Abeles et al., 1992). Ethylene binds to endoplasmic reticulum‐localized receptors that function as negative regulators of ethylene signalling in the absence of the hormone (Chen et al., 2002). Ethylene response factors (ERFs) are the last known actors in the ethylene transduction pathway and regulate downstream ethylene‐responsive genes. Members of the ERF transcription factor gene family have at least one conserved AP2 domain of about 60 amino acids. ERFs are trans‐acting factors that bind to GCC, or GCC box, or dehydration‐responsive element (DRE)/C‐repeat motifs (DRE/CRT), cis‐acting elements in the promoter region of target genes. Following Nakano's classification, the ERF family was subdivided into ten functional groups (Nakano et al., 2006). Group IX is known to have several members involved in ethylene, jasmonate and pathogen responses. Two members of this group, ERF1 and ORA59, are essential integrators of the jasmonate and ethylene signal transduction pathways (Lorenzo et al., 2003; Pré et al., 2008). Constitutive expression of ERF1 in Arabidopsis confers resistance to pathogens and several necrotrophic fungi (Berrocal‐Lobo and Molina, 2004; Berrocal‐Lobo et al., 2002; Onate‐Sanchez and Singh, 2002). The ERF1 gene is used for applications in several cultivated species, notably to improve tolerance to abiotic stress. A patent has been filed by the University of Pennsylvania on this gene and its use in plants (Ecker and Solano, 2002). Constitutive expression of ERF1 activates the transcription of effector genes encoding basic chitinase (ChiB), thionin 2.1 (Thi2.1) and DEFENSIN 1.2 (PDF1.2) (Brown et al., 2003; Manners et al., 1998; Solano et al., 1998). ERF1 is also a major regulatory factor, independent of ABA and of the proline biosynthesis enzyme P5CS1, which is one of the essential factors for tolerance to water stress (Szabados and Savoure, 2010).
Ethylene and jasmonate play a crucial role in controlling natural rubber production by regulating responses to harvesting stress (Pirrello et al., 2014; Putranto et al., 2015a,c). Hevea brasiliensis is the main commercial source of natural rubber. Natural rubber is produced in laticifers, which constitute an articulated network of latex cells. These cells are differentiated from the cambium and are inserted into the phloem tissue. The latex is harvested by tapping soft bark, and stimulation with ethephon, an ethylene releaser, is used to improve latex flow and latex regeneration between two tappings. Jasmonates and wounding are also factors that induce laticifer differentiation (Hao and Wu, 2000; Tian et al., 2015). The ERF family has been identified and classified in H. brasiliensis (Duan et al., 2013). Two orthologues to Arabidopsis ERF1 have been predicted in the rubber tree (Putranto et al., 2015a). These genes, HbERF‐IXc4 and HbERF‐IXc5, are highly expressed in bark and latex tissues (Piyatrakul et al., 2014). They are also activated in the latex from tapped or ethephon‐stimulated trees, and in leaves of plants grown under abiotic stress (Putranto et al., 2015a). A mix of ethylene (in the form of ethylene gas or ethephon), and methyl jasmonate (MeJA) or wounding stress (such as tapping or mechanical wounding known to induce the production of JA) trigger a very strong expression of these genes in Hevea, as has been found for the ERF1 orthologue in cotton (Champion et al., 2009). Moreover, subcellular localization and transactivation experiments showed that HbERF‐IXc4 and HbERF‐IXc5 act as transcriptional activators in the nucleus (Putranto et al., 2015a). These results suggest that HbERF‐IXc4 and HbERF‐IXc5 play an essential role in latex stress signalling. No major regulation of these genes occurs at the onset of the physiological syndrome known as tapping panel dryness (TPD), which affects latex production (Putranto et al., 2015b).
This study sets out to characterize the function of one of the ERF1 orthologue namely HbERF‐IXc5 gene in a nonmodel species. An efficient Agrobacterium tumefaciens‐mediated genetic modification procedure opens up opportunities for further functional analysis in the rubber tree (Blanc et al., 2006; Leclercq et al., 2010). Although a Hevea gene was first inserted in transgenic rubber plants in 2003 (Jayashree et al., 2003), the first complete functional analysis of a gene encoding a Hevea copper zinc superoxide dismutase was recently carried out in transgenic rubber (Leclercq et al., 2012). Based on this effective procedure, the study sets out to produce and characterize transgenic rubber plants overexpressing the HbERF‐IXc5 gene under the transcriptional control of two promoters, CaMV 35S and Hev2.1 (Montoro et al., 2008). Sixteen independent transgenic callus lines were established. Transgenic plants were regenerated and cultured for 1 year after acclimatization. The plants were characterized in terms of growth, anatomy and biochemical compounds. Some ecophysiological parameters were also monitored in response to abiotic stress such as water deficit, cold and salt treatments. These data revealed better tolerance to abiotic stress, suggesting that HbERF‐IXc5 may be involved in plant defence mechanisms. HbERF‐IXc5 is assumed to be an integrator of the ethylene and jasmonate signalling pathways. This study also provided new insight into developmental processes, such as latex cell differentiation, which is induced by the plant stress hormone jasmonate.
Results
Establishment, plant regeneration and molecular analyses of transgenic callus lines
Callus from embryogenic and friable line CI07060 was inoculated with A. tumefaciens strain EHA105 harbouring binary vectors pCamway‐35S::HbERF‐IXc5 and pCamway‐HEV2.1::HbERF‐IXc5. After a 5‐day coculture, calli were transferred as small aggregates to the decontamination medium, DM. After a selection phase of 10 subcultures of 3 weeks on DM medium supplemented with paromomycin, 5 and 11 paromomycin‐resistant lines were established from about 300 callus aggregates initially cultured for each construct, respectively (Table 1, Data S1). Promoters directing the HbERF‐IXc5 gene had an important effect on the number of established lines. The regeneration of somatic embryos and their conversion into plantlets were attempted for each transgenic line and the wild‐type line CI07060. All lines regenerated somatic embryos. The plant regeneration process of stable transgenic lines is described in Figure 1. Of the 16 transgenic lines, 10 were able to produce well‐shaped embryos but only six transgenic lines were able to produce plantlets. The embryo to plantlet conversion rate was very variable between transgenic lines. Lines TS19A90, TS20A69 and TS20A75 showed higher embryogenic capacity than WT. TS20A75 produced the largest number of total somatic embryos (223 per g of callus). On the other hand, TS20A69 achieved the highest production of well‐shaped embryos.
Table 1.
Plant regeneration ability of wild‐type and HbERF‐IXc5 transgenic lines. The somatic embryos consisted of two classes: well‐shaped embryos and abnormal embryos. Only well‐shaped embryos were transferred for conversion into plantlets. The conversion percentage was calculated as follows: number of plants/number of well‐shaped somatic embryos
| Replication | Total embryos | Well‐shaped embryo | Plant | ||
|---|---|---|---|---|---|
| Construct | Line | No RITA | No g−1 FM | No g−1 FM | No g−1 FM |
| None | CI07060 | 20 | 58c | 22b | 9abc |
| 35S::HbERF‐IXc5 | TS19A46 | 12 | 32c | 7b | 5bc |
| TS19A59 | 5 | 13c | 4b | 1c | |
| TS19A90 | 14 | 176ab | 56a | 19ab | |
| TS19A99 | 13 | 102bc | 11b | 0c | |
| TS19A101 | 7 | 5c | 0b | 0c | |
| HEV2.1::HbERF‐IXc5 | TS20A29 | 7 | 12c | 5b | 0c |
| TS20A38 | 8 | 4c | 0b | 0c | |
| TS20A44 | 6 | 17c | 0b | 0c | |
| TS20A45 | 6 | 10c | 0b | 0c | |
| TS20A47 | 7 | 12c | 2b | 1c | |
| TS20A53 | 8 | 24c | 4b | 0c | |
| TS20A69 | 14 | 176ab | 82a | 16abc | |
| TS20A75 | 15 | 223a | 76a | 24a | |
| TS20A78 | 5 | 5c | 1b | 0c | |
| TS20A82 | 10 | 4c | 0b | 0c | |
| TS20A90 | 6 | 2c | 0b | 0c |
Statistical analysis was performed with an ANOVA followed by the Tukey test. Values with the same letter were not significantly different at the 0.05 probability level.
Figure 1.

Description of plant regeneration from friable callus lines of wild‐type and HbERF‐IXc5 transgenic lines. Callus of wild‐type (a), and transgenic lines TS19A90 (b), TS20A75 (c). View of RITA system with brown callus and somatic embryo of wild‐type (d), and transgenic lines TS19A90 (e), TS20A75 (f); 12‐month‐old plant of wild‐type (g), and transgenic lines TS19A90 (h), TS20A75 (i). Root system of wild‐type (j), and transgenic lines TS19A90 (k), TS20A75 (l)
The four lines producing a sufficient number of plants for further experiments (TS19A46, TS19A90, TS20A69 and TS20A75) had only one copy of T‐DNA according to Southern blot hybridization (Figure 2). A gene expression analysis by real‐time RT‐PCR revealed a significant higher relative transcript abundance in all the transgenic lines compared to WT, which expressed only the native HbERF‐IXc5 gene (Table 2, Data S2). This confirmed overexpression of the HbERF‐IXc5 gene in the transgenic lines.
Figure 2.
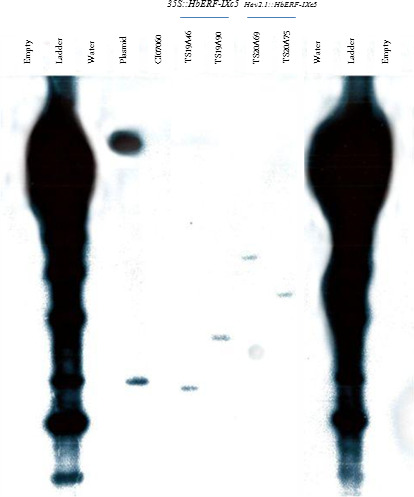
Determination of copy number by Southern blot hybridization. Genomic DNA of leaves were digested with Eco RI. The blot was hybridized with a 32P radio‐labelled probe corresponding to NPTII gene
Table 2.
Relative transcript abundance for the HbERF‐IXc5 gene in leaves of wild‐type and HbERF‐IXc5 transgenic lines. The reference gene used was HbRh2b (Piyatrakul et al. 2014). Each data are the mean of four biological replicates
| Construct | Line | Relative transcript abundance |
|---|---|---|
| None | WT | 1.15 E − 04c |
| 35S::HbERF‐IXc5 | TS19A46 | 1.98 E − 02b |
| 35S::HbERF‐IXc5 | TS19A90 | 2.77 E + 00a |
| Hev2.1::HbERF‐IXc5 | TS20A69 | 1.23 E + 00a |
| Hev2.1::HbERF‐IXc5 | TS20A75 | 3.66 E − 03b |
Statistical analysis was performed with an ANOVA followed by the Tukey test. Values with the same letter were not significantly different at the 0.05 probability level.
Monitoring growth and ecophysiological parameters in 1‐year‐old plants
Several parameters were measured on WT and transgenic plants to quantify plant growth (Table 3, Data S3–S4). All the transgenic lines were significantly more vigorous than the WT line, except line TS19A90. The plants of these lines (TS19A46, TS20A69 and TS20A745) were taller (+15 to 34%) and heavier (+170 to 190%), and had more leaves than the WT plants. Although all parts of the plant were heavier, the root system was comparatively more developed in the transgenic lines (1.6–2.7 times) than in the WT line. A comparison of photosynthetic nitrogen use efficiency (PNUE) revealed that the plants from line TS20A75 maintained significantly greater photosynthetic activity (19.22 μmol CO2 mol N−1) than the WT plants (5.11 μmol CO2 mol N−1) under optimum culture conditions (Figure 3, Data S5).
Table 3.
Growth analysis of 1‐year‐old plants from wild‐type and HbERF‐IXc5 transgenic lines. Each data item is the mean of 40 replications for morphological and eight replications for weight measurements. Dry matter was measured on eight plants for leaf and stem, and statistical analysis was performed with an ANOVA followed by the Newman–Keuls test
| Construct | Line | Number of leaves | Stem diameter (mm) | Stem height (mm) | Dry matter (g/plant) | |||
|---|---|---|---|---|---|---|---|---|
| Total | Leaves | Stem | Root | |||||
| None | CI07060 | 20.98c | 6.87c | 570c | 32.335c | 9.114c | 9.330b | 13.861c |
| 35S::HbERF‐IXc5 | TS19A46 | 28.73b | 9.55a | 742a | 87.291a | 19.794ab | 30.491a | 37.006a |
| 35S::HbERF‐IXc5 | TS19A90 | 19.44c | 6.48c | 478d | 56.584b | 15.539b | 19.174b | 21.871bc |
| HEV2.1::HbERF‐IXc5 | TS20A69 | 32.84a | 9.03a | 761a | 87.803a | 24.619a | 32.631a | 30.553ab |
| HEV2.1::HbERF‐IXc5 | TS20A75 | 26.68b | 8.19b | 657b | 93.831a | 23.314a | 32.834a | 37.682a |
Values with the same letter were not significantly different at the 0.05 probability level.
Figure 3.
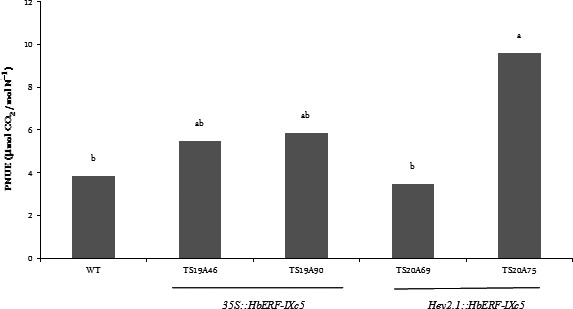
Comparison of photosynthetic nitrogen use efficiency (PNUE, A400/Na) between wild‐type and several transgenic lines under optimum growth conditions without water stress. Statistical analysis was performed with an ANOVA followed by the Newman and Keuls test. Values with the same letter were not significantly different at the 0.05 probability level
Quantitative and qualitative analyses of some histological parameters
A histological analysis did not reveal any structural changes between WT and transgenic plants (Data S6–S9). No significant differences were noted for leaves (width of cuticle, upper epidermis, palisade parenchyma, lower epidermis and stomata), stem (width of bark, cambium, xylem, pith) and taproot (width of bark, xylem and pith), except for the Hev2.1::HbERF‐IXc5 lines (TS20A69 and TS20A75) with the wider cambium (73–75 μm) compared to WT (38 μm) in the taproot. However, two striking observations were made. Firstly, a substantial accumulation of starch reserves was especially observed in the lignified part of the stem (Figure 4). Secondly, latex cells were more abundant in the leaf and stem (Table 4).
Figure 4.
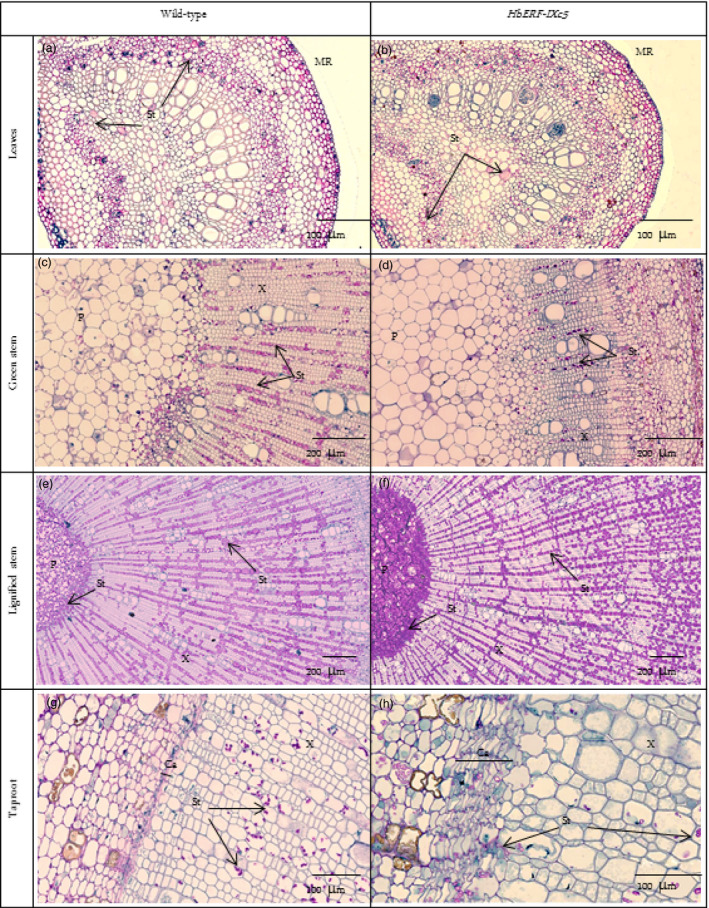
Histological analysis revealing the accumulation of starch reserves. (a) Leaves of wild‐type, (b) Leaves of HbERF‐IXc5 transgenic lines, (c) Green stem of wild type, (d) Green stem of HbERF‐IXc5 transgenic lines, (e) Lignified stem of wild type, (f) Lignified stem of HbERF‐IXc5 transgenic lines, (g) Taproot of wild type, (h) Taproot of HbERF‐IXc5 transgenic lines. The histological sections were stained with Schiff Naphthol Blue Black. Histological sections were annotated by arrows as follows: MR, midrib; P, pith; X, xylem; Ca, cambium; St, starch
Table 4.
Qualitative and quantitative analysis on histological slides of primary and secondary latex cells in leaves, green and lignified stems, and taproot of 1‐year‐old plants from wild‐type and transgenic lines. The abundance of primary latex cells (PCL) and secondary latex cells (SLC) was estimated as follows: (−): absence; (+): rare; (++): 5–10; (+++): 10–50; (++++): >50
| Construct | Line | Primary latex cell | Secondary latex cell | |||||
|---|---|---|---|---|---|---|---|---|
| Leaf | Green stem | Lignified stem | Root | Green stem | Lignified stem | |||
| Midrib | Lamina | |||||||
| None | CI07060 | 50a | 4a | + | − | + | + | +++ |
| 35S::HbERF‐IXc5 | TS19A46 | 157c | 5a | +++ | − | + | +++ | ++++ |
| 35S::HbERF‐IXc5 | TS19A90 | 37a | 3a | + | − | + | + | ++ |
| HEV2.1::HbERF‐IXc5 | TS20A69 | 113b | 12a | + | − | + | + | + |
| HEV2.1::HbERF‐IXc5 | TS20A75 | 84b | 3a | ++ | − | + | + | ++ |
Statistical analysis was performed with an ANOVA followed by the Tukey test. Values with the same letter were not significantly different at the 0.05 probability level.
Primary latex cells could be counted on 2 mm of leaves. Lines TS19A46, TS20A69 and TS20A75 had significantly much more primary latex cells in the midrib of leaves (157, 113 and 84) compared to the WT line (50). A qualitative analysis of primary and secondary latex cells in green and lignified stems revealed a clear abundance of these types of cells in the plants of line TS19A46 (Table 4, Figure 5). Numerous secondary latex cells were randomly distributed, either as isolated cells or in a group not yet forming anastomoses. Laticifer cells had a thick cell wall, nontransparent, elastic cytoplasm and were stained in pink‐red (Figure 5).
Figure 5.
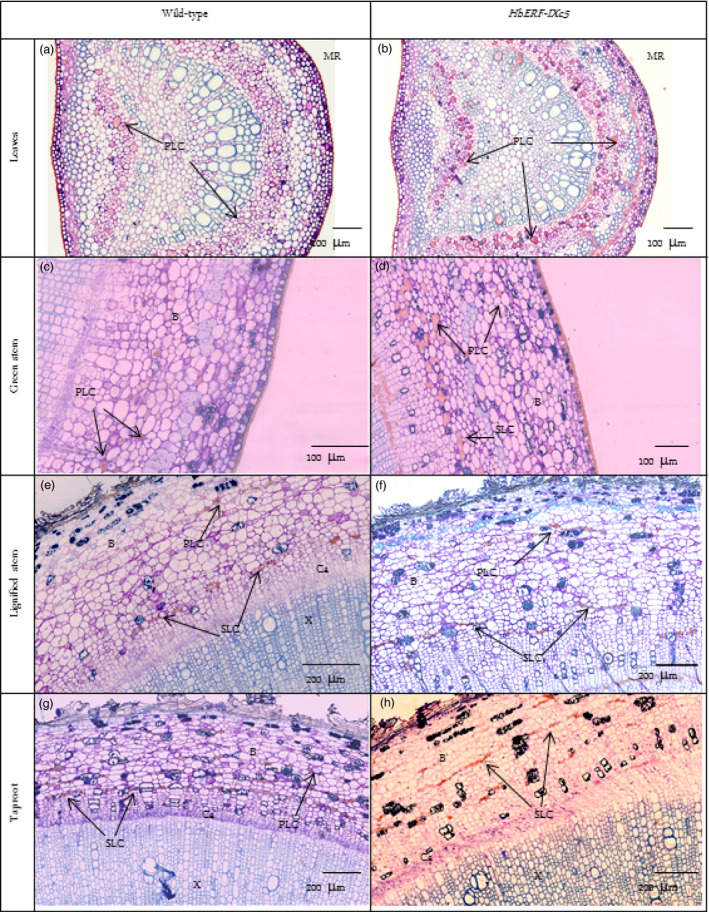
Presence of latex cells in leaves, stem and root of 1‐year‐old plants from wild‐type and HbERF‐IXc5 transgenic lines. (a) Latex cell in leaves of wild type; (b) Latex cell in leaves of HbERF‐IXc5 transgenic lines; (c) Latex cell in green stem of wild type; (d) Latex cell in green stem of HbERF‐IXc5 transgenic lines; (e) Latex cell in lignified stem of wild‐type; (f) Latex cell in lignified stem of HbERF‐IXc5 transgenic lines; (g) Latex cell in taproot of wild type; (h) Latex cell in taproot of HbERF‐IXc5 transgenic lines. The histological sections were stained with Oil Red O. Histological sections were annotated by arrows as follows: B, bark; Ca, cambium; PLC, primary latex cells; SLC, secondary latex cell; X, xylem
Carbohydrate and antioxidant contents
In an attempt to confirm an accumulation of starch reserves, several carbohydrate compounds were quantified in 1‐year‐old plants growing under standard conditions (Table 5, Data S10). Starch was observed as pink‐red coloured grains stained by periodic acid–Schiff. The starch content increased dramatically in the bark and pith of both green and lignified stems of the transgenic lines compared to WT, but the variability did not lead to significant differences. By contrast, there were no large differences in contents for soluble sugar (glucose, fructose and sucrose). Interestingly, we found that sucrose was the most important carbohydrate in stem bark and in leaves, while the starch content was the highest compound in stem pith. Starch reserves were more abundant in the bark of lignified stems from the transgenic lines compared to the wild type.
Table 5.
Quantification of starch accumulation in the pith of lignified stem of 1‐year‐old plants from wild‐type and HbERF‐IXc5 transgenic lines. All values are expressed in mg of each carbohydrate per gram of dry matter except starch content, which was converted into equivalent glucose
| Tissue | Construct | Line | Starch | Glucose | Fructose | Sucrose | Soluble sugar |
|---|---|---|---|---|---|---|---|
| Bark lignified stem | None | CI07060 | 17b | 12b | 6b | 127cde | 145bcd |
| 35S::HbERF‐IXc5 | TS19A90 | 41b | 6b | 3b | 131cde | 140bcd | |
| HEV2.1::HbERF‐IXc5 | TS20A69 | 79b | 8b | 4b | 140cde | 152bcd | |
| HEV2.1::HbERF‐IXc5 | TS20A75 | 99b | 7b | 4b | 128cde | 139bcd | |
| Pith lignified stem | None | CI07060 | 147ab | 2b | 1b | 36de | 39e |
| 35S::HbERF‐IXc5 | TS19A90 | 68b | 1b | 0b | 33e | 34e | |
| HEV2.1::HbERF‐IXc5 | TS20A69 | 266a | 1b | 0b | 47de | 48de | |
| HEV2.1::HbERF‐IXc5 | TS20A75 | 125b | 1b | 1b | 32e | 34e | |
| Bark green stem | None | CI07060 | 8b | 31b | 3b | 148bcde | 182bc |
| 35S::HbERF‐IXc5 | TS19A90 | 21b | 23b | 2b | 128cde | 153bcd | |
| HEV2.1::HbERF‐IXc5 | TS20A69 | 35b | 19b | 7b | 152bcde | 178bc | |
| HEV2.1::HbERF‐IXc5 | TS20A75 | 73b | 22b | 3b | 159bcd | 184b | |
| Pith green stem | None | CI07060 | 23b | 11b | 11b | 48de | 70 |
| 35S::HbERF‐IXc5 | TS19A90 | 74b | 13b | 0b | 80de | 93cde | |
| HEV2.1::HbERF‐IXc5 | TS20A69 | 159ab | 8b | 8b | 51de | 67de | |
| HEV2.1::HbERF‐IXc5 | TS20A75 | 108b | 10b | 11b | 61de | 82de | |
| Leaves | None | CI07060 | 7b | 82a | 18ab | 249ab | 349a |
| 35S::HbERF‐IXc5 | TS19A90 | 9b | 78a | 18ab | 285a | 381a | |
| HEV2.1::HbERF‐IXc5 | TS20A69 | 3b | 62ab | 24ab | 227abc | 313a | |
| HEV2.1::HbERF‐IXc5 | TS20A75 | 19b | 99a | 47a | 211abc | 357a |
Analysis was performed with an ANOVA followed by the Newman–Keuls test. Values with the same letter were not significantly different at the 0.05 probability level.
HbERF‐IXc5 is assumed to control the expression of genes involved in ROS‐scavenging systems. For that reason, two main antioxidant compounds were quantified in leaves of 1‐year‐old plants growing under standard conditions (Figure 6, Data S11). Total glutathione contents ranged from 1150 to 1760 nmoles/gDM. Two transgenic lines TS19A90 and TS20A75 had a significant greater content than the control (Figure 6a). The ascorbic acid content was about 400 nmoles/gDM in WT and in lines TS19A90 and TS20A69, and peaked at 783 nmoles/gDM for line TS20A75 (Figure 6b).
Figure 6.
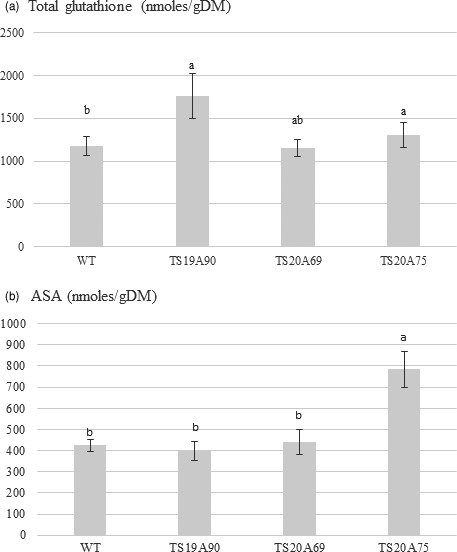
Antioxidant contents in leaves of 1‐year‐old plants from wild‐type (WT) and transgenic lines. (a) Total glutathione. (b) Ascorbic acid content (ASA)
Effect of abiotic stress
In order to test tolerance to abiotic stress, plants were subjected to water deficit, salt and cold treatments. Up to 8 days after water deficit, plants had turgescent leaves (Figure 7a,b). Fourteen days after no watering, both WT and transgenic plants displayed the same leaf wilting symptoms (Figure 7c). In the absence of an obvious phenotype, the water deficit effect was evaluated by monitoring the fraction of transpirable soil water (FTSW) (Data S12, Figure 7d). Moderate and severe physiological stress statuses were considered for FTSW below 0.4 and 0.2, respectively. Moderate and severe stress status was reached for plants of all lines within 8 and 9 days after no watering, respectively, revealing that moderate stress status was very transient except for line TS19A90, which maintained this status for 4 days.
Figure 7.
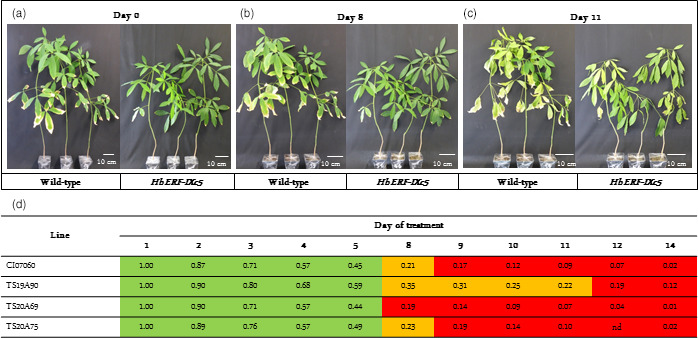
Effect of water deficit on 8‐month‐old plants from wild‐type and transgenic lines and their FTSW. (a) Plants before water stress on day 0 (FTSW about 1). (b) Plants after 8 days of water deficit (FTSW ≥ 0.2). (c) Plants after 14 days of water deficit (FTSW < 0.2). (d) Daily change in FTSW value after water deficit treatment. Highlighting in green (1 > FTSW > 0.4), in orange (0.4 > FTSW > 0.2), in red (FTSW < 0.2). nd: nondetermined
The Fv/Fm and PI values decreased dramatically during the 4 days of cold treatment and then increased for all lines except TS1A90 (Figure 8a,c). Overall, TS20A75 showed the highest average Fv/Fm values, while TS20A69 and TS20A75 showed the highest average PI values. Higher Fv/Fm and PI values indicated the high tolerance to cold stress (Kadir et al., 2007). By contrast, the chlorophyll content (SPAD) decreased slightly during and after treatment (Figure 8e). All lines showed a higher SPAD value compared to the wild type.
Figure 8.
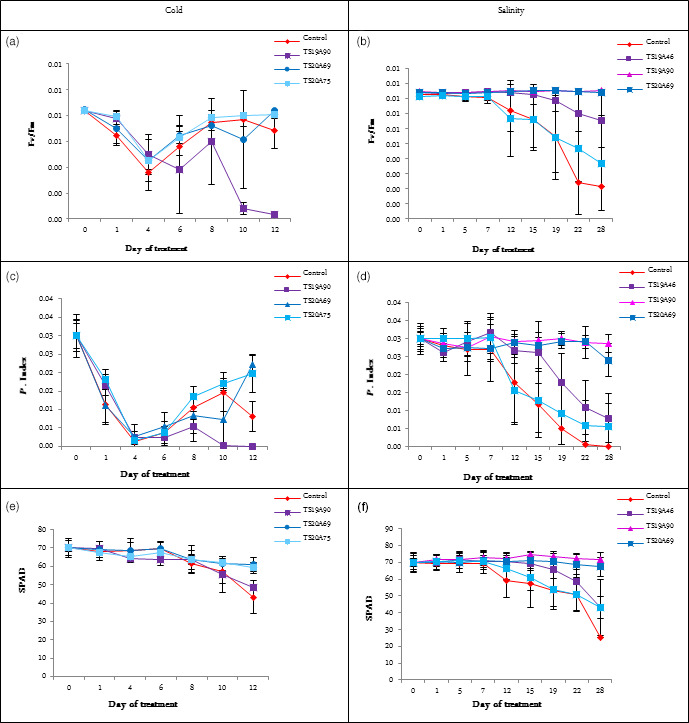
Effect of cold and salinity stress on various ecophysiological parameters for wild‐type and several transgenic lines. (a) Effect of cold stress on Fv/Fm value. (b) Effect of salinity stress on Fv/Fm value. (c) Effect of cold stress on P. Index value. (d) Effect of salinity stress on P. Index value. (e) Effect of cold stress on SPAD value. (f) Effect of salinity stress on SPAD value. Statistical analysis was performed with an ANOVA followed by the Tukey test. Values with the same letter were not significantly different at the 0.05 probability level
As regards the salinity treatment, the Fv/Fm and PI values showed that WT plants, presenting the lowest values, were more affected than transgenic lines after 28 days of treatments (Figure 8b,d). This indicates that the photosystem II capacity was degraded in WT plants. At the same time, the chlorophyll content decreased particularly in WT plants (Figure 8f). TS19A90 and TS20A69 were found to perform better and had a more stable chlorophyll content. Both lines also gave the highest SPAD value during the treatment. This suggests that transgenic plants may absorb light and accumulate energy better and thus avoid oxidative damage. The PI values peaked for lines TS19A90 and TS20A69. This result suggests that these transgenic lines might delay leaf senescence under high salinity concentrations, which is considered to be an important and favourable trait, known as ‘stay‐green’.
Discussion
HbERF‐IXc5 belongs to group IX of the ERF transcription factor family (Duan et al., 2013). This gene shares several common functional features of its putative orthologous gene ERF1 (Duan et al., 2013; Putranto et al., 2015a). Interestingly, this study revealed several new characteristics of rubber transgenic lines overexpressing HbERF‐IXc5 in terms of somatic embryogenesis, growth, starch reserves and laticifer differentiation, as well as better tolerance to abiotic stress.
Overexpression of HbERF‐IXc5 affects the somatic embryogenesis process
Overexpression of the HbERF‐IXc5 gene induced greater variability in callus proliferation, production of somatic embryos and plant regeneration capacity (Table 1, Figure 1). Callus browning was correlated with somatic embryogenesis induction (Lardet et al., 2009), but the HbERF‐IXc5 callus lines turned very rapidly brown, almost necrotic, in response to the low growth regulator content in the EXP medium. This feature had a negative impact on the production of somatic embryos for several lines. By contrast, a small number of lines could produce a larger number of embryos and plantlets compared to the wild‐type line. This observation led to the conclusion that there is no deleterious effect of this gene whatever the stages of development. Use of the HEV2.1 promoter to direct the HbERF‐IXc5 gene reduced the negative effect of the transgene with a larger number of established transgenic lines. ROS‐scavenging system modified transgenic lines remained yellow throughout the somatic embryogenesis process, and they mostly lost their ability to regenerate somatic embryos and plantlets (Leclercq et al., 2012). Indeed, ROS detoxification enzymes and ethylene play an important role in somatic embryogenesis of woody species (Gomez‐Garay et al., 2013; Jo et al., 2014). This suggests that HbERF‐IXc5 might control several target genes triggering a metabolic response related to the oxidation of polyphenols and did not produce a sufficient amount of antioxidants to protect cells.
Stress in plant tissue cultures induces ethylene production, and consequent negative effects in terms of tissue browning. But it can also promote growth and cell proliferation depending on the type of tissue, stage of development and environmental factors (Kepczynska and Zielinska, 2011; Pierik et al., 2006). ERFs coordinate ethylene response. Among the differentially expressed ERF genes at different stages of the Hevea somatic embryogenesis process (Piyatrakul et al., 2012), the HbERF‐IXc4 gene was highly expressed in callus during embryogenesis induction in highly embryogenic lines and down‐regulated in nonembryogenic lines. By contrast, its paralogue HbERF‐IXc5 was not regulated at all during the somatic embryogenesis process. These data suggest that an overexpression of the HbERF‐IXc5 gene might induce, among others, HbERF‐IXc4 target genes involved in somatic embryogenesis. Nevertheless, variability in the response of Hevea tissue could indicate that this factor is not the main one and may well cooperate with others in fine‐tuning this biological process.
Overexpression of HbERF‐IXc5 improves the vigour of plant material and influences photosynthetic activity
Transgenic plants from the HbERF‐IXc5 lines had better growth than the wild‐type plants except line TS19A90 (Table 3). The better performance of the HbERF‐IXc5 transgenic lines can be attributed to several features. Firstly, the root system was well developed and could contribute to better nutrient and water uptake. Secondly, these transgenic plants had more leaves and this well‐developed canopy may have contributed to a strong photosynthetic capacity. This is especially the case for line TS20A75 harbouring HEV2.1::HbERF‐IXc5, which had a twofold intrinsic photosynthetic nitrogen use efficiency (PNUE) compared to the WT plants (Figure 3). Thirdly, all the transgenic lines tended to accumulate more starch reserves and, for line TS20A75, more ascorbic acid and total glutathione. The presence of starch could be an indirect indicator of photosynthetic activity (Geigenberger et al., 2011). Rubber trees adjust the amount of carbohydrate reserves stored to the level of metabolic demand for latex regeneration at the expense of growth (Silpi et al., 2007).
The performance of the HbERF‐IXc5 lines was quite singular. ERF1‐overexpressing plants showed an extreme dwarf phenotype similar to that of the constitutive ethylene response mutant ctr1 and EIN3/EIL1‐overexpressing transgenic plants. However, this effect on growth can differ in some other reports. Interaction between the environment and the ethylene signalling pathway may affect growth regulation (Vahala et al., 2013). Moreover, root growth inhibition by ethylene was reported in overexpressing ERF1 lines (Mao et al., 2016). This analysis also suggested that HbERF‐IXc5 may have an effect other than cell elongation and development when compared to ERF1. Plants from line TS19A90 cannot be considered as dwarf plants but simply smaller plants than control. The expression of HbERF‐IXc5 in leaves was much greater in transgenic lines than in the control line and peaked for line TS19A90. Position effect of transgene into the genome and promoters may have a significant impact on transgene expression. Expression of HbERF‐IXc5 gene is under the control of CaMV 35S promoter in line TS19A90. Hevein2.1 is a major latex protein having laticifer‐specific expression in roots and stems, so the promoter of this gene can be considered as a strong promoter. Finally, this promoter is also induced by light and consequently can drive transgene expression in leaves (Montoro et al., 2008). For that reason, these two promoters can drive strong gene expression in leaves. Transgene dose effect could be observed in nonphotosynthetic tissues such as callus (impact on somatic embryogenesis) and bark (impact on laticifer differentiation).
Overexpression of HbERF‐IXc5 improves tolerance to abiotic stress
The HbERF‐IXc5 lines were screened for their tolerance to water deficit, cold and salinity treatments. HbERF‐IXc5‐overexpressing plants did not show any obvious morphological characteristics of tolerance to abiotic stress within the duration of the treatments, but several ecophysiological parameters revealed better stability of leaf physiological status. Similarly to Arabidopsis ERF1, HbERF‐IXc5 may be involved in controlling responses to abiotic stress. ERF1 up‐regulates specific target genes in response to biotic and abiotic stress by stress‐specific binding to GCC or DRE/CRT cis‐acting elements (Cheng et al., 2013). ERF1 positively regulates Arabidopsis resistance to the pathogens Botrytis cinerea and Plectosphaerella cucumerina by binding GCC boxes in defence‐related genes (Berrocal‐Lobo et al., 2002; Solano et al., 1998). Conversely, ERF1 binds to DRE motifs but not GCC boxes in response to abiotic stress. Transactivation experiments revealed that HbERF‐IXc5 can activate a GCC box, but no information is available on its action on DRE (Putranto et al., 2015a).
Upstream, expression of the HbERF‐IXc5 gene was induced by dehydration and down‐regulated by cold in rubber juvenile plants (Putranto et al., 2015a). In Hevea transgenic lines, overexpression of the HbERF‐IXc5 transgene ultimately gave the same effect for both the salinity and cold treatments, even though the native gene was down‐regulated by the cold treatment. Plant defence mechanisms usually include a strengthening of ROS‐scavenging systems. This study showed an increase in antioxidant contents in some HbERF‐IXc5‐overexpressing lines. Line TS20A75 accumulated in plant leaves both more total glutathione and ascorbic acid than other lines. By contrast, a strong browning of transgenic callus lines was noted after somatic embryogenesis induction. This suggests that HbERF‐IXc5 gene induction by stress may not lead to a sufficient activation of genes involved in ROS‐scavenging systems.
HbERF‐IXc4 and HbERF‐IXc5 are assumed to be involved in the crosstalk of the ethylene and jasmonate signalling pathways for several reasons. Firstly, HbERF‐IXc4 and HbERF‐IXc5 belong to ERF group IX and it is orthologous to Arabidopsis ERF1, which is an integrator of JA and ET signals in plant defence (Lorenzo et al., 2003). Secondly, HbERF‐IXc4 and HbERF‐IXc5 are controlled by multiple harvesting stresses, tapping and ethephon stimulation, related to jasmonate and ethylene production, respectively (Putranto et al., 2015a). Lastly, expression of the HbERF‐IXc4 and HbERF‐IXc5 genes was induced by wounding, methyl jasmonate and ethylene, but it was dramatically enhanced by a combination of MeJA and ET like ERF1 (Putranto et al., 2015a). The involvement of JA and ET signalling was reported in salt stress induction (Cheng et al., 2013). Taken together, these data suggest that HbERF‐IXc5 contributes to the implementation of defence mechanisms in the rubber tree but not necessarily through the sensing of cell redox homoeostasis and the control of antioxidant production. Further characterization of transgenic HbERF‐IXc4 lines will allow to confirm its involvement in stress tolerance as HbERF‐IXc5.
Overexpression of HbERF‐IXc5 induces laticifer differentiation
HbERF‐IXc5‐overexpressing plants had significantly more primary latex cells in the midrib of leaves compared to WT. For line TS19A46, both primary and secondary latex cells were more abundant in green and lignified stems (Table 4). This suggests that HbERF‐IXc5 may contribute to the laticifer differentiation mechanism. Primary latex cells are isolated within parenchymatous tissues when secondary laticifers are differentiated from vascular cambium. Activation of cambium was found in the Hevea transgenic lines. ERFs are essential for ethylene‐stimulated cambial cell division (Etchells et al., 2012). Overexpression of some ERFs can stimulate diameter growth in Populus plants (Vahala et al., 2013). Young stems had primary latex cells and no secondary laticifers, with the latter appearing after development of five extension units (Hao and Wu, 1982). However, a large number of secondary laticifers were observed in young green stems of line TS19A46 revealing an earlier occurrence than in wild‐type plants. Tapping and mechanical wounding were first shown to activate laticifer differentiation (Hao and Wu, 1982). Then, jasmonates were identified as factors involved in laticifer differentiation induced by mechanical wounding (Hao and Wu, 2000; Tian et al., 2003; Wu et al., 2002). When applied to extending young green stems, jasmonates led to a significant increase in the number of primary laticifers but did not induce secondary laticifer differentiation (Hao and Wu, 2000). Secondary laticifer differentiation was obtained by jasmonate application to the extended young stems. Ethephon caused no obvious change in the number of laticifer rings when applied to trees without latex exploitation (Hao and Wu, 1984). This suggests that ethylene does not play a direct role in laticifer differentiation. By contrast, interactions between jasmonic acid (JA) and cytokinin or the stress signal pathway mediate wound‐induced secondary laticifer differentiation in rubber trees (Tian et al., 2015; Zhang and Tian, 2015; Zhang et al., 2015). Intensive studies on the identification of differentially expressed genes and signalling pathways should lead to a better understanding of secondary laticifer differentiation (Hong et al., 2015; Loh et al., 2016).
Several AP2/ERF genes (HbERF1, HbERF2, HbERF3 and RAV1) were induced by JA in bark during JA‐induced laticifer differentiation (Wu et al., 2010). HbERF1, HbERF2 and HbERF3 corresponded to HbERF‐VIIa3, HbERF‐VIIa17 and HbERF‐VIIa1 according to the Hevea AP2/ERF superfamily classification (Duan et al., 2013). The ERFs from group VII are known to regulate hypoxia‐responsive genes (Licausi et al., 2013). Although HbERF‐IXc5 was not identified in JA‐induced laticifer differentiation, HbERF‐IXc5 might be at the crosstalk of the ET and JA signalling pathways and control some target genes directly involved in this biological process.
Conclusions
This study is the first successful functional analysis of a transcription factor in transgenic lines from Hevea brasiliensis, a nonmodel perennial latex‐producing species. HbERF‐IXc5‐overexpressing lines turned rapidly brown after somatic embryogenesis induction. Of the sixteen independent transgenic callus lines, sufficient numbers of plants were regenerated from four lines for further characterization. Overexpression of HbERF‐IXc5 enhanced growth and tolerance to abiotic stress in rubber plants. HbERF‐IXc5 might control some target genes involved in carbohydrate metabolism, which is an essential feature for latex production. This study also provided new insight into environmentally controlled developmental processes such as laticifer differentiation. Through its role as an integrator of the ethylene and jasmonate signalling pathways, HbERF‐IXc5 might play a role in JA‐induced laticifer differentiation. Given that HbERF‐IXc5 should have pleiotropic effects, further characterization will be needed to identify its target genes associated with this biological process. Characterization in transgenic lines of its paralogue HbERF‐IXc4 should also improve our understanding of the specific role of ERF1 orthologues in Hevea.
Experimental procedures
Establishment of transgenic callus lines and plant regeneration
The HbERF‐IXc5 gene was cloned in two pCamway binary destination vectors harbouring the 35S CaMV and latex‐specific HEV2.1 promoters, respectively (Leclercq et al., 2015). These binary vectors were introduced into A. tumefaciens strain EHA105 by electroporation. For inoculation, bacteria were grown in liquid Lysogeny Broth medium (Duchefa, Haarlem, The Netherlands) supplemented with 50 mg L−1 kanamycin and 100 μm acetosyringone at 28 °C until OD600 nm = 0.6 (Blanc et al., 2006). Transgenic lines were established according to the procedure described by Blanc and Leclercq (Blanc et al., 2006; Leclercq et al., 2010). The production of embryos and their conversion into plantlets were carried out as described by Lardet and coll. (Lardet et al., 1999, 2007). Plantlets were then acclimatized in the greenhouse at 28 °C with 60% relative humidity.
Acclimatization and plant morphology measurements
Plantlets were transferred to 120‐mL paper pots (Jiffy pot, Ohio) and covered with a plastic box. After 2 months of acclimatization, plants were transferred to 2‐L plastic pots for 1 year of growth. Several parameters were measured on plants before acclimatization (length of taproot, diameter of taproot, height of stem, diameter of stem, number of leaves, number of leaflets and number of lateral roots, 2 and 6 months after acclimatization (height of plant, diameter of stem at the collar, number of leaves, and number of leaflets) and then 12 months after acclimatization (diameter of stem, height of stem, number of leaves and leaflets, fresh and dry weight of leaves, stem, total root and main root).
Molecular analyses
Isolation of genomic DNA and Southern blot hybridization was carried out as described by Leclercq and coll. (Leclercq et al., 2010). Hybridization was performed using random primed 32P radio‐labelled probes corresponding to NPTII genes amplified with the following primers: NPTII‐F: 5′‐CCGGCTACCTGCCCATTCGA‐3′ and NPTII‐R: 5′‐GCGATAGAAGGCGATGCG‐3′. The numbers of bands reflected the number of T‐DNA insertions.
RNA extraction using the caesium chloride cushion and gene expression analysis by real‐time RT‐PCR were carried out according to the methods described by Duan and coll. (Duan et al., 2010). This experiment followed recommendations specified by (Udvardi et al., 2008). HbRH2b was selected from eleven housekeeping genes as the best reference gene according to its stability in tissues from various treatments in juvenile plants and mature trees (Putranto et al., 2015b). Real‐time RT‐PCR analysis was carried out using a Light Cycler 480 (Roche, Basel, Switzerland). The transcript abundance level for each gene was relatively quantified by normalization with the transcript abundance of the reference HbRH2b gene. Gene HbERF‐IXc5 was amplified with primers HbERFCL13262‐F381 5′‐CAGTTGAAAGAGTGAAGGAATC‐3′ and HbERFCL13262‐R567 5′‐TCCAAGTAATCAGCACCCAAG‐3′. Relative transcript abundance took into account primer efficiencies. All the normalized ratios corresponding to transcript accumulation were calculated automatically by Light Cycler Software version 1.5.0 provided by the manufacturer using the following calculation: Normalized Ratio = Efficiency−Δ(Cp target‐Cp RH2b).
Histo‐cytological analysis
Leaf, green stem, lignified stem and taproot samples followed the same process described in Montoro and coll. (Montoro et al., 2008). Sections were stained with two staining treatments: Oil Red O‐Toluidine Blue staining to identify lipid compounds such as latex in red‐orange, lignins, sclerenchyma and polyphenols in blue‐green (Lillie and Ashburn, 1943), and periodic acid–Schiff/Naphthol Blue Black (NBB) to specifically stain walls and polysaccharide storage (starch) in violet, and proteins in blue (Fisher, 1968). To validate the detection of latex cells, stem samples were placed in an ethanol/acetic acid fixative buffer then treated with denaturant agent (iodine bromide) according to the Shi and Hu's method (Shi and Hu, 1965). Samples were directly cut by a microtome into thick section of 40 μm. Rubber inclusions in latex cells appeared in brown and red colours, respectively, for iodine bromide and oil red O stainings (Figure S1).
Quantification of carbohydrates
Leaves and both the bark and pith of green and lignified stems were collected from wild‐type and transgenic plants and placed quickly in liquid nitrogen. Samples were freeze‐dried and powdered with a laboratory mill to obtain a particle size >100 μm. Sugars were determined by the HPAEC‐PAD technique (ThermoFisher Scientific, Waltham, Massachusetts, USA) with a CarboPack PA1 column and isocratic conditions (150 mm sodium hydroxide) (Mialet‐Serra et al., 2005). Starch was quantified by enzymatic assay using a spectrophotometer (Mialet‐Serra et al., 2005). Glucose was quantified as described by Boehringer using hexokinase and glucose 6 phosphate dehydrogenase, followed by spectrophotometry of NADPH at 340 nm (Boehringer, 1984).
Colorimetric measurement of antioxidants
Half a gram of leaves was ground in liquid nitrogen and mixed in 5 mL of 3% sulphosalicylic acid at 4 °C. Samples were centrifuged twice at 11 000 g for 15 min at 4 °C. Determination of ascorbic acid (ASA) and total glutathione content in leaves was performed on the supernatant according to the method published by Masato and Queval, respectively (Masato, 1980; Queval and Noctor, 2007).
Application of environmental stress
For the water deficit treatment, 1‐year‐old plants were cut back and placed in pots with 250 g of mould substrate according to the method described by Sanier and coll. (Sanier et al., 2013b). Once the plants had developed two growth units, the pots were sealed to prevent soil evaporation and were not watered anymore. The fraction of transpirable soil water (FTSW) was measured and calculated as described by Luquet and coll. (Luquet et al., 2008). The FTSW threshold indicates the timing of stomatal closure in response to a soil water deficit (Ray et al., 2002; Sinclair and Ludlow, 1986). The experiment ended when the transpiration rate of each stress pot was less than 10% of that of the fully watered pots (Sinclair and Ludlow, 1986). FTSW was decreased from 1 (normal water status) to 0.4 (moderate stress) and 0.2 (severe stress) (Sanier et al., 2013a).
For the cold treatment, 1‐year‐old plants growing in 2‐L pots were transferred from the greenhouse at 28 °C to a climatic room at 10 °C for 96 h with a photoperiod of 8 h/16 h. After cold treatment, plants were placed in a room at 20 °C for 1 day, and then transferred to a greenhouse at 28 °C.
For the salinity treatment, plants were watered with 1 litre of a 500 mm sodium chloride solution. Each pot was watered once a week to prevent any increase in osmotic potential from salt.
Eco‐physiological measurements
For wild‐type and transgenic plants grown under standard conditions, leaf photosynthesis was measured on a mature leaf (stage D) with a portable photosynthesis system (GFS‐3000, Walz, Germany). The measurements were made in situ between 9:00 am and 11:00 am at a light intensity of 1200 μmol m2.s−1 of PAR, with a CO2 concentration maintained at 400 ppm, a controlled leaf temperature at 28 °C, a relative humidity in the cuvette set at 65% and a constant flow rate through the cuvette of 800 mL min−1. The measured variables were calculated according to von Caemmerer & Farquhar's method (von Caemmerer and Farquhar, 1981), and included leaf net CO2 assimilation (A), Gs (stomatal conductance), the transpiration rate (E) and the intercellular CO2 mole fraction (Ci). Photosynthetic nitrogen use efficiency was computed (PNUE), expressed as the ratio between net CO2 assimilation and leaf nitrogen content (A400/Na). The leaves used for gas exchange measurements were also used to determine nitrogen content in the leaves as a percentage of the dry weight (DW; Nm in mg g−1 DW), SLA, (cm2 g−1), leaf mass per area (LMA, the inverse of SLA in g.cm2) and, accordingly, nitrogen concentration on a leaf area basis multiplying Nm by LMA (Na in gN m2). Total leaf nitrogen was analysed based on the Dumas combustion method using a LECO FP‐528 nitrogen analyser, in the CIRAD plant analysis laboratory. Chlorophyll content was also measured on the same leaf using a SPAD‐502 (Minolta, Ltd., Tokyo, Japan).
For cold and salinity stress, chlorophyll fluorescence measurements were performed using a Handy‐PEA fluorometer (Hansatech Instrument, King's Lynn, Norfolk,VA) on control and stressed plants in the morning on mature leaves up to 12 days after treatment. The fluorescence transients were induced by 1‐s illumination with an array of six light‐emitting diodes providing a maximum light intensity of 3000 μmol (photons) m−2 s−1 and uniform irradiation over a 4‐mm‐diameter leaf area. Fast fluorescence kinetics (F0 to FM) were recorded from 10 μs to 1 s. The fluorescence intensity at 50 μs was considered as F0 (Strasserf et al., 1995). Readings were taken on the abaxial side of mature leaves, dark adapted with a lightweight plastic leaf clip for 30 min before measurement. Maximum photosystem II (PSII) photochemical efficiency F v /F m, the ratio of variable fluorescence (F v) to maximum fluorescence (F m) and the performance index (PI) plant vitality indicator (Strasserf et al., 1995; Strauss et al., 2006) were calculated automatically. As for photosynthesis measurement, the chlorophyll content value was measured on the same leaf using a SPAD‐502 (Minolta, Ltd., Tokyo Japan).
Database and statistical analysis
All the experiments comprised several biological replications which are specified in the tables of the results section. The data were normalized prior to statistical analysis using XLSTAT (Addinsoft, Paris, France). Raw data were analysed by ANOVA with various statistical tests. The significance of values was determined for a P‐value under 0.05.
Conflict of interest
Co‐authors declare they have no conflict of interest.
Supporting information
Figure S1 Comparison of latex cell‐specific staining using (a) iodine bromide treatment (Shi and Hu, 1965) and (b) oil red O (Montoro et al., 2008). Latex cells (LC) are specified by arrows and appear in brown and red for the two stainings, respectively.
Data S1 Number of GFP positive aggregates during the selection of transgenic lines.
Data S2 HbERF‐IXc5 gene expression.
Data S3 Growth raw data for each acclimatized plants.
Data S4 Plant weight raw data for each acclimatized plants.
Data S5 Ecophysiological raw data for each analysed plants.
Data S6 Quantitative and qualitative histological raw data for leaves of each analysed plants.
Data S7 Quantitative and qualitative histological raw data for green stem of each analysed plants.
Data S8 Quantitative and qualitative histological raw data for lignified stem of each analysed plant.
Data S9 Quantitative and qualitative histological raw data for taproot of each analysed plant.
Data S10 Carbohydrate content raw data for each analysed plants.
Data S11 Antioxidant content raw data for each analysed plants.
Data S12 Fraction of transpirable soil water (FTSW) raw data for each analysed plants.
Acknowledgements
This work was supported by CIRAD, the Institut Français du Caoutchouc, the Socfinco and SIPH companies, the Directorate General of Higher Education (DGHE) Ministry of National Education of Indonesia (Scholarship No. 386/E4.4/K/2012), French Ministry of Foreign Affairs and AGREENIUM. We are grateful to Marc Lartaud and Armelle Soutiras for technical help. The authors also thank Peter Biggins for his revision of the English.
References
- Abeles, F.B. , Morgan, P.W. and Saltveit, M.E. (1992) Ethylene in Plant Biology. San Diego, California: Academic Press Inc. [Google Scholar]
- Berrocal‐Lobo, M. and Molina, A. (2004) Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol. Plant Microbe Interact. 17, 763–770. [DOI] [PubMed] [Google Scholar]
- Berrocal‐Lobo, M. , Molina, A. and Solano, R. (2002) Constitutive expression of ETHYLENE‐RESPONSE‐FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32. [DOI] [PubMed] [Google Scholar]
- Blanc, G. , Baptiste, C. , Oliver, G. , Martin, F. and Montoro, P. (2006) Efficient Agrobacterium tumefaciens‐mediated transformation of embryogenic calli and regeneration of Hevea brasiliensis Mull Arg. plants. Plant Cell Rep. 24, 724–733. [DOI] [PubMed] [Google Scholar]
- Boehringer, S.A. (1984) Methods of Enzymatic Food Analysis using Single Reagents. Germany: Mannheim. [Google Scholar]
- Brown, R.L. , Kazan, K. , McGrath, K.C. , Maclean, D.J. and Manners, J.M. (2003) A role for the GCC‐box in jasmonate‐mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 132, 1020–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Caemmerer, S. and Farquhar, G.D. (1981) Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves. Planta, 153, 376–387. [DOI] [PubMed] [Google Scholar]
- Champion, A. , Hebrard, E. , Parra, B. , Bournaud, C. , Marmey, P. , Tranchant, C. and Nicole, M. (2009) Molecular diversity and gene expression of cotton ERF transcription factors reveal that group IXa members are responsive to jasmonate, ethylene and Xanthomonas. Mol. Plant Pathol. 10, 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.F. , Randlett, M.D. , Findell, J.L. and Schaller, G.E. (2002) Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J. Biol. Chem. 277, 19861–19866. [DOI] [PubMed] [Google Scholar]
- Cheng, M.C. , Liao, P.M. , Kuo, W.W. and Lin, T.P. (2013) The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress‐responsive gene expression by binding to different cis‐acting elements in response to different stress signals. Plant Physiol. 162, 1566–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, C. , Rio, M. , Leclercq, J. , Bonnot, F. , Oliver, G. and Montoro, P. (2010) Gene expression pattern in response to wounding, methyl jasmonate and ethylene in the bark of Hevea brasiliensis . Tree Physiol. 30, 1349–1359. [DOI] [PubMed] [Google Scholar]
- Duan, C. , Argout, X. , Gébelin, V. , Summo, M. , Dufayard, J.‐F. , Leclercq, J. , Kuswanhadi et al. (2013) Identification of the Hevea brasiliensis AP2/ERF superfamily by RNA sequencing. BMC Genom. 14, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker, J.R. and Solano, R. (2002) Ethylene‐response‐factor1 (erf1) in Plants. University of Pennsylvania, Philadelphia, Pennsylvania: Google Patents. [Google Scholar]
- Etchells, J.P. , Provost, C.M. and Turner, S.R. (2012) Plant vascular cell division is maintained by an interaction between PXY and ethylene signalling. PLoS Genet. 8, e1002997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, D.B. (1968) Protein staining of ribboned epon sections for light microscopy. Histochemie, 16, 92–96. [DOI] [PubMed] [Google Scholar]
- Geigenberger, P. , Tiessen, A. and Meurer, J. (2011) Use of non‐aqueous fractionation and metabolomics to study chloroplast function in Arabidopsis. Methods Mol. Biol. 775, 135–160. [DOI] [PubMed] [Google Scholar]
- Gomez‐Garay, A. , Lopez, J.A. , Camafeita, E. , Bueno, M.A. and Pintos, B. (2013) Proteomic perspective of Quercus suber somatic embryogenesis. J. Proteomics. 93, 314–325. [DOI] [PubMed] [Google Scholar]
- Hao, B.‐Z. and Wu, J.‐L. (1982) Effects of wound (tapping) on laticifer differentiation in Hevea brasiliensis . Acta Bot. Sin. 24, 388–391. [Google Scholar]
- Hao, B.‐Z. and Wu, J.‐L. (1984) Acceleration of laticifer differentiation in Hevea brasiliensis by latex drainage. Chinese J. Trop. Crops, 5, 19–23. [Google Scholar]
- Hao, B.‐Z. and Wu, J.‐L. (2000) Laticifer differentiation in Hevea brasiliensis: induction by Exogenous Jasmonic Acid and Linolenic Acid. Ann. Bot. 85, 37–43. [Google Scholar]
- Hong, H. , Xiao, H. , Yuan, H. , Zhai, J. and Huang, X. (2015) Cloning and characterisation of JAZ gene family in Hevea brasiliensis . Plant Biol. 17, 618–624. [DOI] [PubMed] [Google Scholar]
- Jayashree, R. , Rekha, K. , Venkatachalam, P. , Uratsu, S.L. , Dandekar, A.M. , Kumari Jayasree, P. , Kala, R.G. et al. (2003) Genetic transformation and regeneration of rubber tree (Hevea brasiliensis Muell. Arg) transgenic plants with a constitutive version of an anti‐oxidative stress superoxide dismutase gene. Plant Cell Rep. 22, 201–209. [DOI] [PubMed] [Google Scholar]
- Jo, L. , Dos Santos, A.L. , Bueno, C.A. , Barbosa, H.R. and Floh, E.I. (2014) Proteomic analysis and polyamines, ethylene and reactive oxygen species levels of Araucaria angustifolia (Brazilian pine) embryogenic cultures with different embryogenic potential. Tree Physiol. 34, 94–104. [DOI] [PubMed] [Google Scholar]
- Kadir, S. , Von Weihe, M. and Al‐Khatib, K. (2007) Photochemical efficiency and recovery of photosystem II in grapes after exposure to sudden and gradual heat stress. J. Am. Soc. Hortic. Sci. 132, 764–769. [Google Scholar]
- Kepczynska, E. and Zielinska, S. (2011) Disturbance of ethylene biosynthesis and perception during somatic embryogenesis in Medicago sativa L. reduces embryos’ ability to regenerate. Acta Physiol. Plant. 33, 1969–1980. [Google Scholar]
- Lardet, L. , Piombo, G. , Oriol, F. , Dechamp, E. and Carron, M.P. (1999) Relations between biochemical characteristics and conversion ability in Hevea brasiliensis zygotic and somatic embryos. Can. J. Bot. 77, 1168–1177. [Google Scholar]
- Lardet, L. , Martin, F. , Dessailly, F. , Carron, M.P. and Montoro, P. (2007) Effect of exogenous calcium on post‐thaw growth recovery and subsequent plant regeneration of cryopreserved embryogenic calli of Hevea brasiliensis (Mull. Arg.). Plant Cell Rep. 26, 559–569. [DOI] [PubMed] [Google Scholar]
- Lardet, L. , Dessailly, F. , Carron, M.P. , Rio, M.A. , Ferriere, N. and Montoro, P. (2009) Secondary somatic embryogenesis in Hevea brasiliensis (Mull. Arg.): an alternative process for long‐term somatic embryogenesis. J. Rubber Res. 12, 215–228. [Google Scholar]
- Leclercq, J. , Lardet, L. , Martin, F. , Chapuset, T. , Oliver, G. and Montoro, P. (2010) The green fluorescent protein as an efficient selection marker for Agrobacterium tumefaciens‐mediated transformation in Hevea brasiliensis (Mull. Arg). Plant Cell Rep. 29, 513–522. [DOI] [PubMed] [Google Scholar]
- Leclercq, J. , Martin, F. , Sanier, C. , Clement‐Vidal, A. , Fabre, D. , Oliver, G. , Lardet, L. et al. (2012) Over‐expression of a cytosolic isoform of the HbCuZnSOD gene in Hevea brasiliensis changes its response to a water deficit. Plant Mol. Biol. 80, 255–272. [DOI] [PubMed] [Google Scholar]
- Leclercq, J. , Szabolcs, T. , Martin, F. and Montoro, P. (2015) Development of a new pCAMBIA binary vector using Gateway(R) technology. Plasmid, 81, 50–54. [DOI] [PubMed] [Google Scholar]
- Licausi, F. , Pucciariello, C. and Perata, P. (2013) New role for an old rule: N‐end rule‐mediated degradation of ethylene responsive factor proteins governs low oxygen response in plants(F). J. Integr. Plant Biol. 55, 31–39. [DOI] [PubMed] [Google Scholar]
- Lillie, R.D. and Ashburn, L.L. (1943) Supersaturated solutions of fat stains in dilute isopropanol for demonstration of acute fatty degeneration not shown by Herxheimer's technique. Arch. Pathol. 36, 432–440. [Google Scholar]
- Loh, S.C. , Thottathil, G.P. and Othman, A.S. (2016) Identification of differentially expressed genes and signalling pathways in bark of Hevea brasiliensis seedlings associated with secondary laticifer differentiation using gene expression microarray. Plant Physiol. Biochem. 107, 45–55. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O. , Piqueras, R. , Sanchez‐Serrano, J.J. and Solano, R. (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell, 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luquet, D. , Clément‐Vidal, A. , Fabre, D. , This, D. and Sonderegger, N. (2008) Orchestration of transpiration, growth and carbohydrate dynamics in rice during a dry‐down cycle. Funct. Plant Biol. 35, 689–704. [DOI] [PubMed] [Google Scholar]
- Manners, J.M. , Penninckx, I.A. , Vermaere, K. , Kazan, K. , Brown, R.L. , Morgan, A. , Maclean, D.J. et al. (1998) The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol. Biol. 38, 1071–1080. [DOI] [PubMed] [Google Scholar]
- Mao, J.L. , Miao, Z.Q. , Wang, Z. , Yu, L.H. , Cai, X.T. and Xiang, C.B. (2016) Arabidopsis ERF1 mediates cross‐talk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PLoS Genet. 12, e1006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masato, O. (1980) An improved method for determination of l‐ascorbic acid and l‐dehydroascorbic acid in blood plasma. Clin. Chim. Acta, 103, 259–268. [DOI] [PubMed] [Google Scholar]
- Mialet‐Serra, I. , Clement, A. , Sonderegger, N. , Roupsard, O. , Jourdan, C. , Labouisse, J.P. and Dingkuhn, M. (2005) Assimilate storage in vegetative organs of coconuts (Cocos nucifera). Expl. Agric. 41, 161–174. [Google Scholar]
- Montoro, P. , Lagier, S. , Baptiste, C. , Marteaux, B. , Pujade‐Renaud, V. , Leclercq, J. and Alemanno, L. (2008) Expression of the HEV2.1 gene promoter in transgenic Hevea brasiliensis. Plant Cell, Tissue Organ Cult. 94, 55–63. [Google Scholar]
- Nakano, T. , Suzuki, K. , Fujimura, T. and Shinshi, H. (2006) Genome‐wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onate‐Sanchez, L. and Singh, K.B. (2002) Identification of Arabidopsis ethylene‐responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiol. 128, 1313–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik, R. , Tholen, D. , Poorter, H. , Visser, E. and Voesenek, L. (2006) The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci. 11, 176–183. [DOI] [PubMed] [Google Scholar]
- Pirrello, J. , Leclercq, J. , Dessailly, F. , Rio, M. , Piyatrakul, P. , Kuswanhadi, K. , Tang, C. et al. (2014) Transcriptional and post‐transcriptional regulation of the jasmonate signalling pathway in response to abiotic and harvesting stress in Hevea brasiliensis . BMC Plant Biol. 14, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piyatrakul, P. , Putranto, R.A. , Martin, F. , Rio, M. , Dessailly, F. , Leclercq, J. , Dufayard, J.F. et al. (2012) Some ethylene biosynthesis and AP2/ERF genes reveal a specific pattern of expression during somatic embryogenesis in Hevea brasiliensis . BMC Plant Biol. 12, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piyatrakul, P. , Yang, M. , Putranto, R.A. , Pirrello, J. , Dessailly, F. , Hu, S. , Summo, M. et al. (2014) Sequence and expression analyses of ethylene response factors highly expressed in latex cells from Hevea brasiliensis . PLoS One, 9, e99367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pré, M. , Atallah, M. , Champion, A. , De Vos, M. , Pieterse, C.M. and Memelink, J. (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putranto, R.A. , Duan, C. , Kuswanhadi, Chaidamsari, T. , Rio, M. , Piyatrakul, P. , Herlinawati, E. et al. (2015a) Ethylene response factors are controlled by multiple harvesting stresses in Hevea brasiliensis . PLoS One, 10, e0123618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putranto, R.A. , Herlinawati, E. , Rio, M. , Leclercq, J. , Piyatrakul, P. , Gohet, E. , Sanier, C. et al. (2015b) Involvement of ethylene in the latex metabolism and tapping panel dryness of Hevea brasiliensis . Int. J. Mol. Sci. 16, 17885–17908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putranto, R.A. , Herlinawati, E. , Rio, M. , Leclercq, J. , Piyatrakul, P. , Gohet, E. , Sanier, C. et al. (2015c) Involvement of ethylene in the latex metabolism and tapping panel dryness of Hevea brasiliensis . Int. J. Mol. Sci. 16, 17885–17908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval, G. and Noctor, G. (2007) A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Anal. Biochem. 363, 58–69. [DOI] [PubMed] [Google Scholar]
- Ray, J.D. , Gesch, R.W. , Sinclair, T.R. and Hartwell Allen, L. (2002) The effect of vapor pressure deficit on maize transpiration response to a drying soil. Plant Soil, 239, 113–121. [Google Scholar]
- Sanier, C. , Oliver, G. , Clement‐Vidal, A. , Fabre, D. , Lardet, L. and Montoro, P. (2013a) Influence of water deficit on the physiological and biochemical parameters of in vitro plants from Hevea brasiliensis Clone PB 260. J. Rubber Res. 16, 61–74. [Google Scholar]
- Sanier, C. , Oliver, G. , Clément Demange, A. , Fabre, D. , Lardet, L. and Montoro, P. (2013b) Influence of water deficit on physiological parameters and antioxidant system on in vitro plants from the Hevea brasiliensis clone PB260. J. Rubber Res. 16, 61–74. [Google Scholar]
- Shi, Z.Q. and Hu, Z.H. (1965) Method for making slide of rubber containing plant tissue. Acta Bot. Sin. 13, 179–183. [Google Scholar]
- Silpi, U. , Lacointe, A. , Kasempsap, P. , Thanysawanyangkura, S. , Chantuma, P. , Gohet, E. , Musigamart, N. et al. (2007) Carbohydrate reserves as a competing sink: evidence from tapping rubber trees. Tree Physiol. 27, 881–889. [DOI] [PubMed] [Google Scholar]
- Sinclair, T. and Ludlow, M. (1986) Influence of soil water supply on the plant water balance of four tropical grain legumes. Funct. Plant Biol. 13, 329–341. [Google Scholar]
- Solano, R. , Stepanova, A. , Chao, Q. and Ecker, J.R. (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE‐INSENSITIVE3 and ETHYLENE‐RESPONSE‐FACTOR1. Genes Dev. 12, 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasserf, R.J. , Srivastava, A. and Govindjee, G. (1995) Polyphasic chlorophyll a fluorescence transient in plants and Cyanobacteria. Photochem. Photobiol. 61, 32–42. [Google Scholar]
- Strauss, A.J. , Krüger, G.H.J. , Strasser, R.J. and Heerden, P.D.R.V. (2006) Ranking of dark chilling tolerance in soybean genotypes probed by the chlorophyll a fluorescence transient O‐J‐I‐P. Environ. Exp. Bot. 56, 147–157. [Google Scholar]
- Szabados, L. and Savoure, A. (2010) Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97. [DOI] [PubMed] [Google Scholar]
- Tian, W. , Shi, M. , Yu, F. , Wu, J. , Hao, B. and Cui, K. (2003) Localized effects of mechanical wounding and exogenous jasmonic acid on the induction of secondary laticifer differentiation in relation to the distribution of jasmonic acid in Hevea brasiliensis . Acta Bot. Sin. 45, 1366–1372. [Google Scholar]
- Tian, W.M. , Yang, S.G. , Shi, M.J. , Zhang, S.X. and Wu, J.L. (2015) Mechanical wounding‐induced laticifer differentiation in rubber tree: an indicative role of dehydration, hydrogen peroxide, and jasmonates. J. Plant Physiol. 182, 95–103. [DOI] [PubMed] [Google Scholar]
- Udvardi, M.K. , Czechowski, T. and Scheible, W.R. (2008) Eleven golden rules of quantitative RT‐PCR. Plant Cell, 20, 1736–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahala, J. , Felten, J. , Love, J. , Gorzsás, A. , Gerber, L. , Lamminmäki, A. , Kangasjärvi, J. et al. (2013) A genome‐wide screen for ethylene‐induced ethylene response factors (ERFs) in hybrid aspen stem identifies ERF genes that modify stem growth and wood properties. New Phytol. 200, 511–522. [DOI] [PubMed] [Google Scholar]
- Wu, J.‐L. , Hao, B.‐Z. and Tan, H.‐Y. (2002) Wound‐induced laticifer differentiation in Hevea brasiliensis shoots mediated by jasmonic acid. J. Rubber Res. 5, 53–63. [Google Scholar]
- Wu, H.L. , Yu, B. , Cheng, Q.Q. , Zeng, R.Z. , Duan, C.F. , Nie, Z.Y. and Li, Y. (2010) Cloning and Characterization of Jasmonic Acid‐Induced AP2/EREBP Genes in Laticifer from Rubber Tree (Hevea brasiliensis Muell. Arg.). Chin. Agric. Sci. Bull. 26, 287–293. [Google Scholar]
- Zhang, S.X. and Tian, W.M. (2015) Cross talk between cytokinin and jasmonates in regulating the secondary laticifer differentiation in rubber tree (Hevea brasiliensis Muell. Arg.). J. Rubber Res. 18, 38–48. [Google Scholar]
- Zhang, S.X. , Wu, S.H. , Chen, Y.Y. and Tian, W.M. (2015) Analysis of Differentially Expressed Genes Associated with coronatine‐induced laticifer differentiation in the rubber tree by subtractive hybridization suppression. PLoS One, 10, e0132070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Comparison of latex cell‐specific staining using (a) iodine bromide treatment (Shi and Hu, 1965) and (b) oil red O (Montoro et al., 2008). Latex cells (LC) are specified by arrows and appear in brown and red for the two stainings, respectively.
Data S1 Number of GFP positive aggregates during the selection of transgenic lines.
Data S2 HbERF‐IXc5 gene expression.
Data S3 Growth raw data for each acclimatized plants.
Data S4 Plant weight raw data for each acclimatized plants.
Data S5 Ecophysiological raw data for each analysed plants.
Data S6 Quantitative and qualitative histological raw data for leaves of each analysed plants.
Data S7 Quantitative and qualitative histological raw data for green stem of each analysed plants.
Data S8 Quantitative and qualitative histological raw data for lignified stem of each analysed plant.
Data S9 Quantitative and qualitative histological raw data for taproot of each analysed plant.
Data S10 Carbohydrate content raw data for each analysed plants.
Data S11 Antioxidant content raw data for each analysed plants.
Data S12 Fraction of transpirable soil water (FTSW) raw data for each analysed plants.


