Abstract
Background
Pathogens often secrete molecules that mimic those present in the plant host. Recent studies indicate that some of these molecules mimic plant hormones required for development and immunity.
Scope and Conclusion
This Viewpoint reviews the literature on microbial molecules produced by plant pathogens that functionally mimic molecules present in the plant host. This article includes examples from nematodes, bacteria and fungi with emphasis on RaxX, a microbial protein produced by the bacterial pathogen Xanthomonas oryzae pv. oryzae. RaxX mimics a plant peptide hormone, PSY (plant peptide containing sulphated tyrosine). The rice immune receptor XA21 detects sulphated RaxX but not the endogenous peptide PSY. Studies of the RaxX/XA21 system have provided insight into both host and pathogen biology and offered a framework for future work directed at understanding how XA21 and the PSY receptor(s) can be differentially activated by RaxX and endogenous PSY peptides.
Keywords: Molecular mimicry, plant pathogen, microbial mimic, RaxX, XA21, PSY, engineering receptor
INTRODUCTION: WHAT IS BIOLOGICAL MIMICRY?
Biological mimicry and molecular mimicry
In classical mimicry, species that are attractive to predators as food have evolved mechanisms that facilitate resemblance to inedible species that are dangerous or unpleasant to eat. A well-known example is Henry Bates’s work on Amazonian butterflies. Bates theorized that palatable species occasionally produced mutant forms with visual characteristics in their wings similar to toxic or unpalatable species, making them less likely to be chosen by birds for food (Bates, 1862). Like this case described by Bates, early studies of biological mimicry focused on the visual resemblance between species. For example, some orchid species produce a flower that mimics bee females to attract males for pollination (Pasteur, 1982). Another example is an early barnyard grass, a weed that resembles rice, and thus farmers do not pull it out from rice fields (Barrett, 1983).
In 1964, researchers discovered examples of molecular mimicry: structural, functional or immunological similarities of molecules that are shared between infectious pathogens and the hosts that they infect. Molecular mimicry has been observed in diverse species of pathogens that infect both plants and animals. Because establishment and maintenance of pathogenesis depend on the passing and receiving of signals between host and pathogen, some of the molecules produced by pathogens mimic host components to gain evolutionary advantages (Mitchum et al., 2012; Eves-Van Den Akker et al., 2016). Such microbial molecules functionally or structurally resemble host factors that are required for host survival, such as ligands of host receptors, substrates of host enzymes, or host proteins themselves (Knodler et al., 2001; Nesic et al., 2010). Some of these microbially produced molecules mimic plant hormones that control growth, development, and regulation of innate immunity.
In this Viewpoint, we highlight recent examples of molecular mimics produced by bacteria, nematodes and fungal pathogens.
CORONATINE
A well-studied case of hormone mimicry in plants is the production of coronatine by the gram-negative biotrophic bacterium Pseudomonas syringae (Weiler et al., 1994). Coronatine is a structural and functional mimic of jasmonoyl-l-isoleucine (JA-Ile), a bioactive form of the plant hormone jasmonic acid (JA) (Weiler et al., 1994; Fonseca et al., 2009). Thus, coronatine targets a JA receptor, COI-1 (CORONATINE INSENSITIVE1)/JAZ (JASMONATE ZIM DOMAIN) co-receptor, and activates JA signalling (Katsir et al., 2008; Melotto et al., 2008) (Fig. 1). JA signalling by coronatine suppresses salicylic acid (SA)-dependent defence through antagonistic cross-talk (Robert-Seilaniantz et al., 2011). SA plays a central role in regulating plant defences against biotrophic and hemibiotrophic pathogens, including P. syringae (Robert-Seilaniantz et al., 2011). These studies indicate that coronatine produced during P. syringae infection mimics JA action, suppressing the host defence response to facilitate infection (Geng et al., 2012).
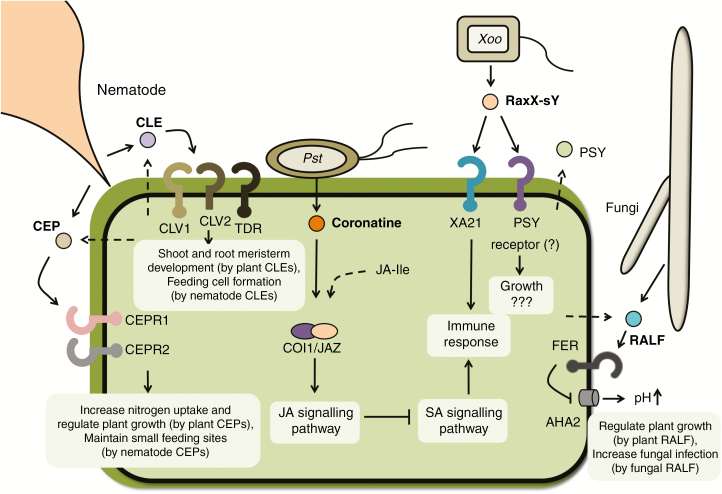
Fig. 1.
Plant pathogens produce molecular mimics that modulate plant signalling pathways. Pseudomonas syringae pv. tomato (Pst) produces coronatine, a structural and functional mimic of jasmonoyl-l-isoleucine (JA-Ile). Coronatine binds the plant JA receptor, COI1/JAZ, to activate the JA signalling pathway, which suppresses salicylic acid (SA)-mediated signalling and inhibits the immune response. Nematodes secrete a mimic of a plant peptide called CLAVATA3/ENDOSPERM SURROUNDING REGION-related (CLE), which is perceived by the plant CLAVATA1 (CLV1)/CLV2 heterodimer or a tracheary element differentiation inhibitory factor (TDIF) receptor (TDR). It is hypothesized that nematode CLE peptides subvert plant CLE-mediated shoot and root meristem development to instead produce feeding cells for the nematode. Another class of nematode effectors mimics the plant C-TERMINALLY ENCODED PEPTIDEs (CEPs). Plant CEPs are produced in the roots under nitrogen starvation and then move through xylem vessels to shoots, where they are recognized by two receptors, CEPR1 and CEPR2. Activation of CEPRs induces nitrogen-demand signals, which increase expression of nitrogen transporters, inhibit primary root elongation and initiate lateral root development to take up nitrogen. The benefit to the nematode is that nematode CEPs induce more nitrogen uptake and keep the size of the feeding site small for biotrophic interaction with plants. Nematodes need to maintain small feeding sites to prevent excessive nutrient drainage and allow host plants to survive. The fungal pathogen Fusarium oxysporum secretes a mimic of the plant rapid alkalinization factor (RALF) peptide. Plant RALF targets the FERONIA (FER) receptor to activate a plasma membrane H(+)-ATPase 2 (AHA2) and thus alkalinizes the extracellular space in planta. RALF-induced extracellular alkalinization regulates the plant cell expansion required for plant growth and development. Fungal RALF-induced alkalinization in the plant apoplast is beneficial to fungal infection and multiplication but the underlying mechanism remains unclear. The sulphated RaxX (RaxX-sY) peptide from Xanthomonas oryzae pv. oryzae (Xoo) mimics the plant peptide hormone PSY (plant peptide containing sulphated tyrosine). RaxX-sY activates PSY signalling and promotes plant growth. Rice XA21 recognizes and responds specifically to microbial RaxX to activate the immune response. Straight lines indicate the secretion of pathogen molecules. Dashed lines indicate products of the endogenous factor from the plant. Question marks indicate pathways that have not yet been fully elucidated.
CLAVATA3/ENDOSPERM SURROUNDING REGION-RELATED PEPTIDES
Plant-parasitic nematodes also produce mimics of endogenous plant hormones. All sedentary plant-parasitic nematodes produce peptides similar to plant CLAVATA3/ENDOSPERM SURROUNDING REGION-related (CLE) peptides, which regulate shoot meristem differentiation, root growth and vascular development (Chen et al., 2015; Guo et al., 2017). A-type plant CLEs suppress shoot and root apical meristem activity and promote cell differentiation, while B-type plant CLEs function in the vascular meristem to suppress differentiation of tracheary elements and promote procambial cell division (Katsir et al., 2011). Nematodes produce both A-type and B-type CLEs and secrete those precursor proteins into plant tissues, where they undergo post-translational modifications and proteolytic processing to become bioactive CLE peptides resembling plant CLEs (Chen et al., 2015). The mature nematode CLEs, which are 12-amino-acid arabinosylated glycopeptide, strongly interact with plant CLE receptor complexes (Wang et al., 2005; Mitchum et al., 2008; Yamaguchi et al., 2016; Guo et al., 2017). A-type CLEs bind CLAVATA1 (CLV1)/CLV2 heterodimer, while B-type CLEs bind a tracheary element differentiation inhibitory factor (TDIF)-receptor (TDR) (Kucukoglu and Nilsson, 2015). The plant requires synergistic interaction between A- and B-type CLEs, spatial regulation and negative feedback for fine-tuning of vascular development (Whitford et al., 2008; Guo et al., 2017). Unlike plant CLE-mediated signalling, nematodes secrete A- and B-type CLEs directly into procambial cells in the roots, which induces massive cell proliferation and formation of feeding cells, where nematodes obtain nutrients (Fig. 1) (Guo et al., 2017).
C-TERMINALLY ENCODED PEPTIDES
C-TERMINALLY ENCODED PEPTIDEs (CEPs) are a large and diverse family of effector peptides produced by some sedentary plant-parasitic nematodes (Bobay et al., 2013; Ogilvie et al., 2014; Bird et al., 2015; Eves-Van Den Akker et al., 2016). CEPs are plant peptide hormones that are 15 amino acids in length with hydroxylation modification on prolines (Ohyama et al., 2008). Plant CEP genes are highly upregulated in the portion of roots undergoing nitrogen starvation (Imin et al., 2013). Root-derived plant CEP peptides move through xylem vessels to the shoots, where they interact with two leucine-rich repeat receptors, CEPR1 and CEPR2 (Tabata and Sawa, 2014). The CEP/CEPR interaction induces shoot-derived polypeptides named CEP downstream 1 (CEPD1) and CEPD2 (Ohkubo et al., 2017). CEPD1 and CEPD2 in turn travel to the roots (specifically to those roots in nitrogen rich soils) and then upregulate Arabidopsis nitrogen transporter NRT2.1 to take up nitrogen in the roots (Ohkubo et al., 2017) (Fig. 1). Over-expression of CEP genes or exogenous application of CEPs suppresses root cell proliferation and thus reduces primary root elongation and accelerates lateral root development. Taken together, these observations show that root-derived CEPs under nitrogen starvation induce systemic N-demand signals for nitrate uptake from nitrogen-rich soil, while suppressing root growth where nitrogen is limiting. Nematode CEPs also upregulate NRT2.1 expression and reduce primary root length. Nematode CEPs limit expansion of feeding sites, which they rely on for nutrient acquisition for several weeks. It is hypothesized that nematodes keep the size of feeding sites small, to their own benefit because over-sized feeding sites may drain excessive nutrient from plants and kill host plants ( Eves-Van Den Akker et al., 2016). Altogether, nematodes mimic plant CEPs to sustain their biotrophic interaction with plants ( Eves-Van Den Akker et al., 2016).
RAPID ALKALINIZATION FACTOR
The root-infecting fungus Fusarium oxysporum secretes a functional mimic of the plant regulatory peptide RALF (rapid alkalinization factor) (Masachis et al., 2016). Plant RALF targets the receptor FERONIA (FER), which inactivates plasma membrane H(+)-ATPase 2 (AHA2) and thus inhibits proton transport (Murphy and De Smet, 2014). This plant RALF/FER interaction induces extracellular alkalinization and impacts cell expansion during plant growth and development (Murphy and De Smet, 2014). The root-infecting fungus F. oxysporum secretes fungal RALF (F-RALF) to induce FER-mediated extracellular alkalinization in the host tissue in a similar manner to plant RALFs (Masachis et al., 2016). Extracellular alkalinization has been reported to contribute to fungal infection and indeed F-RALF-induced alkalinization increases fungal pathogenicity (Murphy and De Smet, 2014; Masachis et al., 2016) (Fig. 1). The underlying mechanisms remain to be elucidated. Stegmann et al. (2017) recently found that RALF peptides inhibit the FERONIA-mediated formation of active immune receptor complexes. Thus, fungal RALFs may suppress the plant immune response by inhibiting the assembly of receptor kinase complexes. Taken together, these findings show that fungal pathogens use F-RALFs to increase fungal infection and to suppress host immunity.
Together, these studies indicate that molecular mimics can suppress host immune responses, facilitate infection and/or enhance plant health, maintenance of which is critically important for propagation and survival of biotrophic pathogens.
THE XA21 IMMUNE RECEPTOR
Here, we describe the discoveries that led to the identification and characterization of a small sulphated protein produced by a plant pathogen that triggers an immune response in rice plants expressing the XA21 immune receptor and that induces root growth. These findings were presented at the Annals of Botany Lecture in Melbourne Australia, and are detailed in two publications (Pruitt et al., 2015, 2017).
Discovery of XA21
In 1905, British geneticist and plant breeder Rowland Biffen demonstrated that it is possible to generate wheat varieties with resistance to a devastating disease by genetic manipulation. He cross-pollinated a resistant wheat variety with a susceptible wheat variety and showed that the resulting seed carried the resistance of the parent. Today, more than 100 years after Biffen’s discovery, plant breeders have introduced ‘resistance genes’ into virtually every crop plant that we consume. However, for many years the identity of these genes and the functional basis of resistance remained elusive.
An explosion of research in the 1990s led to the isolation of several resistance genes from diverse plant species (Martin et al., 1993; Bent et al., 1994; Jones et al., 1994; Whitham et al., 1994; Lawrence et al., 1995; Song et al., 1995). Many of the resistance genes encoded intracellular proteins carrying a nucleotide binding site (NBS), leucine-rich repeats (LRRs), coiled coil and Toll /IL-1 receptor (TIR) domains that recognize specific races of a microbial species. Some of the resistance genes encoded cell surface receptors and were later named pattern recognition receptors (PRRs). PRRs recognize molecules that are conserved across a large class of microbes.
In 1995, we isolated the Xa21 gene, which encodes a PRR carrying both an extracellular receptor domain and an intracellular kinase domain (Song et al., 1995). Rice plants carrying Xa21 confer broad-spectrum resistance to the bacterial pathogen Xanthomonas oryzae pv. oryzae (Xoo) (Wang et al., 1996), which normally caused a devastating disease of rice in Asia and Africa (Ikeda et al., 1990).
Discovery of rax genes (required for activation of xa21-mediated immunity)
Efforts to identify the microbial molecule that activates the XA21-mediated immune response led to the identification of a number of Xoo genes that are required for activation of XA21 (rax genes). These genes encode a tyrosine sulphotransferase, RaxST, and three components of a predicted type 1 secretion system (T1SS): a membrane fusion protein, RaxA; an adenosine triphosphate (ATP)-binding cassette (ABC) transporter, RaxB; and an outer membrane protein, RaxC. raxST, raxA and raxB are located in a single operon (raxSTAB) (da Silva et al., 2004). RaxB falls into a clade of ABC transporters that carry a proteolytic peptidase domain in their N-termini, termed C39, which recognizes and cleaves a conserved N-terminal leader sequence (Dirix et al., 2004, Havarstein et al., 1995). On the basis of these findings, we hypothesized that the activator of XA21-mediated immunity is tyrosine-sulphated by RaxST, a peptide processed and secreted via RaxABC T1SS (da Silva et al., 2004).
Discovery of RaxX
In 2015, we reported the discovery of RaxX, a small protein with a tyrosine sulphation site and a predicted N-terminal leader sequence. The RaxX sequence is highly conserved in many Xanthomonas species. This conservation suggested that RaxX serves a key biological function. Xoo strains that lack RaxX, or carry mutations in the single RaxX tyrosine residue (Y41), are able to evade XA21-mediated immunity (Pruitt et al., 2015). Y41 of RaxX is sulphated by the prokaryotic tyrosine sulphotransferase RaxST. Sulphated, but not non-sulphated, RaxX triggers hallmarks of the plant immune response in an XA21-dependent manner. A sulphated, 21-amino-acid synthetic RaxX peptide (RaxX21-sY) is sufficient for this activity (Pruitt et al., 2015).
As a result of the explosion of studies on PRRs in both plant and animal systems, it has now become clear that the molecules recognized by PRRs can be conserved quite widely across genera (e.g. flagellin) or more narrowly within a genus (e.g. Pep13) (Brunner et al., 2002) and, further, that sequence variation and posttranslational modifications can modulate PRR-dependent pathogen recognition (Takeuchi et al., 2003; Sun et al., 2006). Like microbial molecules recognized by other PRRs, the RaxX protein is conserved within a class of microbes (RaxX is conserved in most, but not all, Xanthomonas species) and is posttranslationally modified (McDonald et al., 2017).
These results indicate that the molecules recognized by NBS-LRR proteins and those recognized by PRR proteins share overlapping characteristics. Furthermore, the transcriptomic patterns triggered by different types of resistance proteins are overlapping (Tao et al., 2003; Tsuda et al., 2008). Although the many terms used to describe plant–pathogen interactions in the past [avirulence gene, effector-triggered immunity (ETI), pathogen associated molecular patterns (PAMPs)-triggered immunity (PTI) etc. (Jones and Dangl, 2006)] were very useful in advancing our understanding of plant–pathogen interactions, the discovery of diverse receptors and their ligands in the last 10 years has blurred the distinction between NBS-LRR and PRR proteins. Thus, broad generalizations that place these receptors and their corresponding microbial determinants into specific classes are less useful today than previously. For this reason, we avoid using the older terms.
RaxX shares similarity to a plant peptide hormone
Although RaxX shares no homology with microbial proteins of known function, sequence analysis revealed that RaxX residues 40–52 are similar to the plant peptide hormone PSY (plant peptide-containing sulphated tyrosine) (Amano et al., 2007, Pruitt et al., 2015, 2017). Alignment of PSY peptides from different plants revealed a highly conserved 13-amino-acid region, which corresponds precisely to the region of sequence similarity between RaxX and Arabidopsis PSY1 (AtPSY1) (Pruitt et al., 2015, 2017). AtPSY1 is the best-characterized member of the plant PSY peptide family. AtPSY1 is an 18-amino-acid glycopeptide with a single sulphotyrosine residue that is secreted and processed and promotes root elongation primarily through regulation of cell size (Amano et al., 2007). AtPSY1 is widely expressed in Arabidopsis tissues (Amano et al., 2007). AtPSY1 promotes acidification of the apoplastic space through activation of membrane proton pumps (Fuglsang et al., 2014). This acidification is thought to activate pH-dependent expansins and cell wall remodelling enzymes that loosen the cellulose network (Cosgrove, 2000; Hager, 2003). Concomitant water uptake by the cell leads to cellular expansion.
The sequence similarity suggests the possibility that RaxX functionally mimics PSY1. We recently demonstrated that RaxX peptides derived from diverse Xanthomonas species promote root growth, mimicking the growth-promoting activities of PSY peptides (Pruitt et al., 2017). We also showed that, unlike RaxX, PSY peptides do not activate XA21-mediated immunity (Pruitt et al., 2017). Thus, XA21 is a highly selective immune receptor capable of specifically recognizing the bacterial mimic (Fig. 1).
What is the biological function of RaxX?
In a classical evolutionary arms race, both the pathogen and host develop and deploy an arsenal of strategies to infect or resist their partner (Jones and Dangl, 2006). For example, many pathogens secrete an array of molecular factors designed to manipulate host biology and suppress the immune response. In turn, plants have developed a set of immune receptors that recognize these molecules or their activities and launch mechanisms to destroy the pathogen, which the pathogen then tries to counter. Our studies of the RaxX/XA21 interaction provide another example. Based on our findings, we propose a model whereby, in the absence of XA21, Xoo and other xanthomonads use sulphated RaxX to mimic PSY1-like peptides to suppress host defence responses, facilitate infection or enhance plant health, maintenance of which is critically important for propagation and survival of biotrophic pathogens (Amano et al., 2007; Mosher et al., 2013; Mosher and Kemmerling, 2013). We recently showed that a Xoo raxX mutant is less virulent than wild-type Xoo on rice leaves in the absence of XA21 (Pruitt et al., 2017). This result supports the ideas that that RaxX is required for the full virulence of Xoo to infect rice leaves and that XA21 later evolved to recognize and respond specifically to RaxX. We have not yet identified the molecular mechanism utilized by RaxX to enhance virulence.
The robust immune response conferred by XA21 is likely to cause a strong selective pressure on the raxX gene in Xoo. Indeed, we have identified natural variants of RaxX21 that are able to evade the activation of XA21 immunity by altering one or two amino acids (e.g. P44, A46 and/or P48) (Pruitt et al., 2015). Variation of RaxX P44 and P48 completely abolishes the immunogenic activity of RaxX in XA21 rice (Pruitt et al., 2015). The amino acid change of A46 has a partial effect. However, these RaxX variants retain the ability to mimic the growth-stimulating properties of PSY (Pruitt et al., 2017). We do not know whether the amino acid changes in these RaxX variants arose in response to the presence of XA21 or were pre-existing in the Xoo population. Epidemiological studies with documentation of disease occurrence over time and space are needed to further investigate the evolution of raxX.
It is also possible that RaxX evolved to mimic specific host PSY peptides. Indeed, three residues in RaxX21 from Xoo (strain PXO99) are identical to those in OsPSY1a (Pruitt et al., 2017). Similarly, the amino acids of RaxX21 from X. oryzae pv. oryzicola (strain BSL256) are similar to those in OsPSY2 (Pruitt et al., 2017). If these two peptides evolved to mimic different PSY peptides, it would indicate that there are multiple PSY receptors in rice, which differentially recognize diverse PSY peptides. Unlike RaxXs from Xoo and Xoc, comparative genetic analysis of other sequenced RaxX and PSY peptides did not reveal strict correlation between the sequences of RaxX from the pathogen and PSYs from a corresponding compatible host (Pruitt et al., 2017). However, alignment of the plant PSY sequences highlights variations at positions 5, 7 and 9 that correspond to RaxX amino acids 44, 46 and 48 (Pruitt et al., 2017). The variation in both plant and pathogen RaxX shares several common amino acids, which suggests that the change is not random. These observations have led us to additional questions about this system: Do specific amino acids affect the ability of peptides to activate specific PSY receptor(s)? Is the PSY receptor(s) able to recognize diverse amino acid variants?
Tyrosine sulphation is critical for RaxX activity
Our results indicate that tyrosine sulphation is critical for RaxX activity. The presence or absence of sulphation and the residues near Y41 are decisive for the ability of RaxX to trigger XA21-mediated immunity and to activate PSY signalling. We have identified RaxX in at least nine Xanthomonas species that infect maize, cassava, sugar cane, tomatoes, peppers, wheat, alfalfa, onions, banana and citrus (Pruitt et al., 2015, 2017). In all of these strains, raxX is encoded upstream of raxST, raxA and raxB and carries a conserved tyrosine residue (Pruitt et al., 2015, 2017). The co-localization of the rax genes suggests that they function in a common biological process and that the RaxX proteins in these species are also substrates for RaxST-mediated tyrosine sulphation.
Although tyrosine sulphation has not been demonstrated in other prokaryotic species, it is a common posttranslational modification of eukaryotic proteins and plays important roles in regulating plant development and in mediating the interactions of plants and animals with microbes. For example, in humans, sulphation of the C-C chemokine receptor type 5 (CCR5) is critical for binding of the envelope glycoprotein gp120 by HIV (Farzan et al., 1999). In addition to PSY, plants produce at least three other classes of tyrosine sulphated peptides: phytosulphokine (PSK) (Matsubayashi and Sakagami, 1996), root meristem growth factor (RGF) (Matsuzaki et al., 2010) and Casparian strip integrity factor (CIF) (Doblas et al., 2017; Nakayama et al., 2017). PSK, RGF and CIF are also processed, secreted, and play a role in the regulation of growth and development in the root.
Further directions: anticipatory breeding and receptor engineering
The robust immunity conferred by XA21 on most Xoo strains is a highly valued agronomic trait. For this reason, XA21 has been introgressed into diverse rice varieties grown by rice farmers (Chen et al., 2000; Toenniessen et al., 2003; Sundaram et al., 2008; Gopalakrishnan et al., 2008; Huang et al., 2012; Win et al., 2012). The discovery of raxX allelic variants that can evade XA21-mediated recognition can be used to screen Xoo field populations for the presence of potentially virulent variants. Such an anticipatory breeding approach would alert farmers to the need to plant different varieties or mixtures before strains that overcome XA21-mediated immunity cause crop damage.
In some cases, knowledge of plant receptor–ligand interaction has been used to engineer resistance to abiotic and biotic stress. For example, Park et al. (2015) described engineering of the abscisic acid (ABA) receptor, PYR1 (Pyrabactin Resistance 1), to activate ABA-mediated drought tolerance. Park et al. (2015) established a set of PYR1 mutants that contains all possible single amino acid substitutions in the ABA binding pocket. The mutant library was then screened for receptor mutants that could respond to mandipropamid (MD). This is a fungicide that controls severely pathogenic oomycetes. The authors identified several mutated receptors that weakly responded to MD. Based on the screening results, the effective mutated residues were combined and extra mutations were added in the engineered PYR1 to optimize the MD response. These experiments led to the generation of a new receptor, PYR1MANDI, which is activated by nanomolar levels of MD. PYR1MANDI can give a solution to a few problems associated with the natural response by ABA/PYR, which is moderate and is activated too late to protect crops from drought stress. Synthesized ABA is too expensive to spray on crop fields. Application of PYR1MANDI to crop fields will allow MD to efficiently activate ABA-mediated drought tolerance (Park et al., 2015; Rodriguez and Lozano-Juste, 2015).
Over-expression of PRRs has been used to improve the resistance of plants to infection by bacterial pathogens. However, in some cases the engineered plants do not have sufficient resistance to suppress disease. For example, a chitin receptor conferred moderate resistance to Magnaporthe oryzae, the causal agent of rice blast disease (Kishimoto et al., 2010). To create plants with enhanced resistance, Kishimoto et al. (2010) constructed a chimeric receptor that contains the extracellular domain of chitin elicitor binding protein (CEBiP) and the intracellular domain of XA21. The new receptor has high affinity to chitin, and rice plants expressing the chimeric receptor exhibited enhanced resistance to M. oryzae (Kishimoto et al., 2010). These results indicate that engineering receptors can be an effective strategy for enhancing disease resistance.
These reports of plant receptors engineered for tolerance to biotic and abiotic stress suggest that knowledge of receptors that recognize microbial mimics, which often serve as virulence factors, may also lead to novel strategies for disease prevention (Park et al., 2015; Rodriguez and Lozano-Juste, 2015). Recent studies of the JA receptor support this idea. The endogenous JA receptor is sensitive to both JA-Ile and the mimic coronatine. By making a structure-guided point mutation of a single amino acid, Zhang et al. (2015) generated a modified JA receptor that has reduced sensitivity to coronatine but retains endogenous JA-Ile recognition. Arabidopsis with the modified JA receptor displayed enhanced resistance to coronatine-producing Pseudomonas strains, and has a normal phenotype in the absence of infection (Zhang et al., 2015). The Zhang et al. study demonstrates how an understanding of bacterial mimicry of host factors can be used to engineer plants with enhanced resistance to bacterial pathogens.
CONCLUSIONS
Studies of the RaxX/XA21 system have provided insight into both host and pathogen biology and offered a framework for future work directed at understanding how XA21 and the PSY receptor(s) can be differentially activated by RaxX and endogenous PSY peptides. These studies also suggest mechanisms for engineering plants for enhanced resistance. For example, XA21 variants engineered for resistance to strains that can evade XA21-mediated immunity would be of use to farmers. Knowledge of the RaxX genetic variants in virulent Xoo strains makes this strategy theoretically possible. Future availability of a XA21/RaxX co-crystal structure would allow researchers to identify amino acids of XA21 that are required for recognition and response to RaxX. With this knowledge, substitutions in the identified key residues could be tested for their ability to respond to RaxX genetic variants. If successful, these results would set the stage for engineering rice plants with new receptors that recognize the RaxX variants. Furthermore, because Xanthomonas species affect virtually all crop plants, similar approaches could be used to engineer resistance to non-rice pathogens. For example, X. axonopodis pv. citrumelo, X. euvesicatoria and X. oryzae pv. oryzicola, which cause serious diseases on citrus, tomato and rice, carry alternative raxX alleles.
LITERATURE CITED
- Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y. 2007. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proceedings of the National Academy of Sciences of the USA 104: 18333–18338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett S. 1983. Mimicry in plants. Scientific American 257: 76–83. [Google Scholar]
- Bates HW. 1862. XXXII. Contributions to an insect fauna of the Amazon valley. Lepidoptera: Heliconidæ. Transactions of the Linnean Society of London 23: 495–566. [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D et al. 1994. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265: 1856–1860. [DOI] [PubMed] [Google Scholar]
- Bird DM, Jones JT, Opperman CH, Kikuchi T, Danchin EG. 2015. Signatures of adaptation to plant parasitism in nematode genomes. Parasitology 142Suppl 1: S71–S84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay BG, DiGennaro P, Scholl E, Imin N, Djordjevic MA, Bird DM. 2013. Solution NMR studies of the plant peptide hormone CEP inform function. FEBS Letters 587: 3979–3985. [DOI] [PubMed] [Google Scholar]
- Brunner F, Rosahl S, Lee J et al. 2002. Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. EMBO Journal 21: 6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, LX H., Xu CG, Zhang Q. 2000. Improvement of bacterial blight resistance of ‘Minghui 63’, an elite restorer line of hybrid rice, by molecular marker-assisted selection. Crop Science 40: 239–244. [Google Scholar]
- Chen S, Lang P, Chronis D et al. 2015. In planta processing and glycosylation of a nematode CLAVATA3/ENDOSPERM SURROUNDING REGION-like effector and its interaction with a host CLAVATA2-like receptor to promote parasitism. Plant Physiology 167: 262–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. 2000. Expansive growth of plant cell walls. Plant Physiology and Biochemistry 38: 109–124. [DOI] [PubMed] [Google Scholar]
- Dirix G, Monsieurs P, Dombrecht B et al. 2004. Peptide signal molecules and bacteriocins in Gram-negative bacteria: a genome-wide in silico screening for peptides containing a double-glycine leader sequence and their cognate transporters. Peptides 25: 1425–1440. [DOI] [PubMed] [Google Scholar]
- Doblas VG, Smakowska-Luzan E, Fujita S et al. 2017. Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355: 280–284. [DOI] [PubMed] [Google Scholar]
- Eves-Van Den Akker S, Lilley CJ, Yusup HB, Jones JT, Urwin PE. 2016. Functional C-TERMINALLY ENCODED PEPTIDE (CEP) plant hormone domains evolved de novo in the plant parasite Rotylenchulus reniformis. Molecular Plant Pathology 17: 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan M, Mirzabekov T, Kolchinsky P et al. 1999. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96: 667–676. [DOI] [PubMed] [Google Scholar]
- Fonseca S, Chini A, Hamberg M et al. 2009. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nature Chemical Biology 5: 344–350. [DOI] [PubMed] [Google Scholar]
- Fuglsang AT, Kristensen A, Cuin TA et al. 2014. Receptor kinase-mediated control of primary active proton pumping at the plasma membrane. Plant Journal 80: 951–964. [DOI] [PubMed] [Google Scholar]
- Geng X, Cheng J, Gangadharan A, Mackey D. 2012. The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of arabidopsis defense. Plant Cell 24: 4763–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan S, Sharma RK, Anand Rajkumar K et al. 2008. Integrating marker assisted background analysis with foreground selection for identification of superior bacterial blight resistant recombinants in Basmati rice. Plant Breeding 127: 131–139. [Google Scholar]
- Guo X, Wang J, Gardner M et al. 2017. Identification of cyst nematode B-type CLE peptides and modulation of the vascular stem cell pathway for feeding cell formation. PLoS Pathogens 13: e1006142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager A. 2003. Role of the plasma membrane H+-ATPase in auxin-induced elongation growth: historical and new aspects. Journal of Plant Research 116: 483–505. [DOI] [PubMed] [Google Scholar]
- Havarstein LS, Diep DB, Nes IF. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Molecular Microbiology 16: 229–240. [DOI] [PubMed] [Google Scholar]
- Huang B, Xu JY, Hou MS, Ali J, Mou TM. 2012. Introgression of bacterial blight resistance genes Xa7, Xa21, Xa22 and Xa23 into hybrid rice restorer lines by molecular marker-assisted selection. Euphytica 187: 449–459. [Google Scholar]
- Ikeda R, S KG, Tabien RE, Zhang H. 1990. A new gene for resistance to bacterial blight from O. longistaminata. Japanese Journal of Breeding 40: 280–281. [Google Scholar]
- Imin N, Mohd-Radzman NA, Ogilvie HA, Djordjevic MA. 2013. The peptide encoding MtCEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. Journal of Experimental Botany 64: 5395–5409. [DOI] [PubMed] [Google Scholar]
- Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JD. 1994. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266: 789–793. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Katsir L, Davies KA, Bergmann DC, Laux T. 2011. Peptide signaling in plant development. Current Biology 21: R356–R364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. 2008. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proceedings of the National Academy of Sciences of the USA 105: 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto K, Kouzai Y, Kaku H, Shibuya N, Minami E, Nishizawa Y. 2010. Perception of the chitin oligosaccharides contributes to disease resistance to blast fungus Magnaporthe oryzae in rice. Plant Journal 64: 343–354. [DOI] [PubMed] [Google Scholar]
- Knodler LA, Celli J, Finlay BB. 2001. Pathogenic trickery: deception of host cell processes. Nature Reviews Molecular Cell Biology 2: 578–588. [DOI] [PubMed] [Google Scholar]
- Kucukoglu M, Nilsson O. 2015. CLE peptide signaling in plants – the power of moving around. Physiologia Plantarum 155: 74–87. [DOI] [PubMed] [Google Scholar]
- Lawrence GJ, Finnegan EJ, Ayliffe MA, Ellis JG. 1995. The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell 7: 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GB, Brommonschenkel SH, Chunwongse J et al. 1993. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262: 1432–1436. [DOI] [PubMed] [Google Scholar]
- Masachis S, Segorbe D, Turrà D et al. 2016. A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nature Microbiology 1: 16043. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Sakagami Y. 1996. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proceedings of the National Academy of Sciences of the USA 93: 7623–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. 2010. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329: 1065–1067. [DOI] [PubMed] [Google Scholar]
- McDonald M, Schwessinger B, Stewart V, Ronald P. 2017. Distribution and inheritance of a gene cluster encoding a sulfated tyrosine peptide in Xanthomonas spp. bioRxiv: 149930. [Google Scholar]
- Melotto M, Mecey C, Niu Y et al. 2008. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interaction with the COI1 F-box protein. Plant Journal 55: 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum MG, Wang X, Davis EL. 2008. Diverse and conserved roles of CLE peptides. Current Opinion in Plant Biology 11: 75–81. [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Wang X, Wang J, Davis EL. 2012. Role of nematode peptides and other small molecules in plant parasitism. Annual Review of Phytopathology 50: 175–195. [DOI] [PubMed] [Google Scholar]
- Mosher S, Kemmerling B. 2013. PSKR1 and PSY1R-mediated regulation of plant defense responses. Plant Signaling & Behavior 8: e24119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher S, Seybold H, Rodriguez P et al. 2013. The tyrosine-sulfated peptide receptors PSKR1 and PSY1R modify the immunity of Arabidopsis to biotrophic and necrotrophic pathogens in an antagonistic manner. Plant Journal 73: 469–482. [DOI] [PubMed] [Google Scholar]
- Murphy E, De Smet I. 2014. Understanding the RALF family: a tale of many species. Trends in Plant Science 19: 664–671. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Shinohara H, Tanaka M, Baba K, Ogawa-Ohnishi M, Matsubayashi Y. 2017. A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355: 284. [DOI] [PubMed] [Google Scholar]
- Nesic D, Miller MC, Quinkert ZT, Stein M, Chait BT, Stebbins CE. 2010. Helicobacter pylori CagA inhibits PAR1-MARK family kinases by mimicking host substrates. Nature Structural & Molecular Biology 17: 130–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie HA, Imin N, Djordjevic MA. 2014. Diversification of the C-TERMINALLY ENCODED PEPTIDE (CEP) gene family in angiosperms, and evolution of plant-family specific CEP genes. BMC Genomics 15: 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo Y, Tanaka M, Tabata R, Ogawa-Ohnishi M, Matsubayashi Y. 2017. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nature Plants 3: 17029. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Ogawa M, Matsubayashi Y. 2008. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant Journal 55: 152–160. [DOI] [PubMed] [Google Scholar]
- Park SY, Peterson FC, Mosquna A, Yao J, Volkman BF, Cutler SR. 2015. Agrochemical control of plant water use using engineered abscisic acid receptors. Nature 520: 545–548. [DOI] [PubMed] [Google Scholar]
- Pasteur G. 1982. A classificatory review of mimicry systems. Annual Review of Ecology and Systematics 13: 169–199. [Google Scholar]
- Pruitt RN, Schwessinger B, Joe A et al. 2015. The rice immune receptor XA21 recognizes a tyrosine-sulfated peptide from a Gram-negative bacterium. Science Advances 1: e1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RN, Joe A, Zhang W et al. 2017. A microbially derived tyrosine sulfated peptide mimics a plant peptide hormone. New Phytologist 215: 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. 2011. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annual Review of Phytopathology 49: 317–343. [DOI] [PubMed] [Google Scholar]
- Rodriguez PL, Lozano-Juste J. 2015. Unnatural agrochemical ligands for engineered abscisic acid receptors. Trends in Plant Science 20: 330–332. [DOI] [PubMed] [Google Scholar]
- da Silva FG, Shen Y, Dardick C et al. 2004. Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Molecular Plant–Microbe Interactions 17: 593–601. [DOI] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL et al. 1995. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806. [DOI] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E et al. 2017. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289. [DOI] [PubMed] [Google Scholar]
- Sun W, Dunning FM, Pfund C, Weingarten R, Bent AF. 2006. Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell 18: 764–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M, Vishnupriya MR, Biradar SK et al. 2008. Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 160: 411–422. [Google Scholar]
- Tabata R, Sawa S. 2014. Maturation processes and structures of small secreted peptides in plants. Frontiers in Plant Science 5: 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Taguchi F, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y. 2003. Flagellin glycosylation island in Pseudomonas syringae pv. glycinea and its role in host specificity. Journal of Bacteriology 185: 6658–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W et al. 2003. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toenniessen GH, O’Toole JC, DeVries J. 2003. Advances in plant biotechnology and its adoption in developing countries. Current Opinion in Plant Biology 6: 191–198. [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F. 2008. Interplay between MAMP-triggered and SA-mediated defense responses. Plant Journal 53: 763–775. [DOI] [PubMed] [Google Scholar]
- Wang GL, Song WY, Ruan DL, Sideris S, Ronald PC. 1996. The cloned gene, Xa21, confers resistance to multiple Xanthomonas oryzae pv. oryzae isolates in transgenic plants. Molecular Plant–Microbe Interactions 9: 850–855. [DOI] [PubMed] [Google Scholar]
- Wang X, Mitchum MG, Gao B et al. 2005. A parasitism gene from a plant-parasitic nematode with function similar to CLAVATA3/ESR (CLE) of Arabidopsis thaliana. Molecular Plant Pathology 6: 187–191. [DOI] [PubMed] [Google Scholar]
- Weiler EW, Kutchan TM, Gorba T, Brodschelm W, Niesel U, Bublitz F. 1994. The Pseudomonas phytotoxin coronatine mimics octadecanoid signalling molecules of higher plants. FEBS Letters 345: 9–13. [DOI] [PubMed] [Google Scholar]
- Whitford R, Fernandez A, De Groodt R, Ortega E, Hilson P. 2008. Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proceedings of the National Academy of Sciences of the USA 105: 18625–18630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B. 1994. The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78: 1101–1115. [DOI] [PubMed] [Google Scholar]
- Win KM, Korinsak S, Jantaboon J, Siangliw M, Siangliw JL, Sirithunya P. 2012. Breeding the Thai jasmine rice variety kdml105 for non-age-related broad-spectrum resistance to bacterial blight disease based on combined marker-assisted and phenotypic selection. Field Crops Research 137: 186–194. [Google Scholar]
- Yamaguchi YL, Ishida T, Sawa S. 2016. CLE peptides and their signaling pathways in plant development. Journal of Experimental Botany 67: 4813–4826. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yao J, Withers J et al. 2015. Host target modification as a strategy to counter pathogen hijacking of the jasmonate hormone receptor. Proceedings of the National Academy of Sciences of the USA 112: 14354–14359. [DOI] [PMC free article] [PubMed] [Google Scholar]