SUMMARY
The eukaryotic translation initiation factor 5B (eIF5B) is a homolog of IF2, an ancient translation factor that enables initiator methionine-tRNAiMet (met-tRNAiMet) loading on prokaryotic ribosomes. While it can be traced back to the last universal common ancestor, eIF5B is curiously dispensable in modern aerobic yeast and mammalian cells. Here, we show that eIF5B is an essential element of the cellular hypoxic cap-dependent protein synthesis machinery. System-wide interrogation of dynamic translation machineries by MATRIX (mass spectrometry analysis of active translation factors using ribosome density fractionation and isotopic labeling experiments) demonstrated augmented eIF5B activity in hypoxic translating ribosomes. Global translatome studies revealed central carbon metabolism, cellular hypoxic adaptation, and ATF4-mediated stress response as major eIF5B-dependent pathways. These primordial processes rely on eIF5B even in the presence of oxygen and active eIF2, the canonical recruiter of met-tRNAiMet in eukaryotes. We suggest that aerobic eukarya retained eIF5B/IF2 to remodel anaerobic pathways during episodes of oxygen deficiency.
Graphical abstract
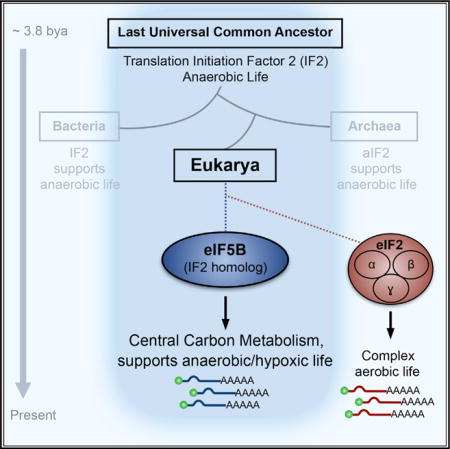
INTRODUCTION
The first living cells appeared on Earth ~4 billion years ago in an oxygen-deficient environment (Weiss et al., 2016). To sustain anaerobic life, primeval cells generated energy (ATP) through early forms of glycolysis, a highly conserved core component of central carbon metabolism that exists in virtually all extant species (Koonin, 2003). The polypeptides required to catalyze these primordial metabolic reactions were synthesized by two core ancient translation factors: initiation factor 1 (IF1) and IF2 (Koonin, 2003). After 1.5 billion years of anaerobic evolution, the increase in atmospheric oxygen catalyzed an upsurge in cellular energy production and biochemical complexity exempli fied by contemporary eukaryotes (Falkowski, 2006; Raymond and Segrè, 2006). The eukaryotic protein synthesis machinery is a paragon of this expansion of complexity. For initiation alone, eukaryotic cells employ at least 13 core eukaryotic-specific translation initiation factors (eIFs) composed of 33 known subunits (Jackson et al., 2010), highlighting the significance of aerobic protein synthesis to modern eukaryotic life. Yet the ancestral IF2, which can be traced back to the last universal common ancestor (LUCA), remains conserved in eukaryotes as eIF5B (Choi et al., 1998; Lee et al., 1999), just as it is in bacteria (IF2) and archaea (aIF2) (Dever, 2002). Despite such strong evolutionary conservation, however, eIF5B is not essential in eukaryotes under basal conditions, exhibiting minimal effects on global translation and cell viability (Choi et al., 1998; Thakor and Holcik, 2012). These observations raise the intriguing possibility that eIF5B/IF2, which first evolved in the absence of oxygen, was retained during aerobic eukaryotic evolution as a mechanism to sustain protein synthesis during oxygen deficiency.
In prokaryotes, IF2 facilitates initiator formylated met-tRNAfMet delivery to ribosomes (Laursen et al., 2005; Yatime et al., 2004). In contrast, this task is typically accomplished in eukaryotes by the textbook eIF2 that delivers met-tRNAiMet to the 40S ribosomal subunit (Jackson et al., 2010; Sonenberg and Hinnebusch, 2009). eIF2 function is tightly regulated by the integrated stress response (ISR), a ubiquitous eukaryotic program activated by various environmental stresses, including hypoxia (Harding et al., 2000a; Pakos-Zebrucka et al., 2016; Wouters and Koritzinsky, 2008). During hypoxic episodes, ISR-inducible kinases phosphorylate the eIF2α subunit and inhibit eIF2 activity (Liu et al., 2006). Although ISR-mediated eIF2 inhibition should theoretically abolish translation initiation, hypoxic cells are capable of engaging in robust cap-dependent protein synthesis by eIF4FH, the functional counterpart of the normoxic eIF4F (Ho et al., 2016; Liu et al., 2006; Uniacke et al., 2012). These data suggest that another translation initiation factor may complement, or assist, in the delivery of met-tRNAiMet for eIF4FH-directed translation during periods of hypoxic eIF2 inactivation.
In this study, we report that hypoxic cells exhibit increased dependence on eIF5B/IF2 for translation initiation and cell survival, with central carbon metabolism (CCM) being a major eIF5B-dependent target. A number of these targets rely preferentially on eIF5B, regardless of eIF2 activity. We suggest that during aerobic eukaryotic evolution, metabolic pathways that first evolved under anaerobic conditions retained their reliance on eIF5B/IF2, which remains active during oxygen deprivation.
RESULTS
MATRIX Identifies eIF5B as a Hypoxia-Enriched Translation Factor
The ancient eIF5B/IF2 likely evolved in an anaerobic environment. We hypothesized that eIF5B retains the ability to operate in eukaryotic cells exposed to low oxygen (Figure S1A), a stress that inactivates the canonical eIF2. To test this hypothesis impartially, we developed a method to assess global translation factor activity in living cells, termed MATRIX (mass spectrometry analysis of active translation factors using ribosome density fractionation and isotopic labeling experiments) (Figure 1A). Integrating metabolic pulse labeling, ribosome density fractionation, and high-throughput mass spectrometry, MATRIX offers the capability to generate an architectural blueprint of biologically active cellular translational machineries. Ribosome density fractionation effectively separates cellular assets based on translational activity. Specifically, factors actively engaged in protein synthesis are preferentially enriched in polysome fractions, while those disengaged from active translation are relatively consigned to the free fractions. We also analyzed ribosomal 40/60/80S fractions, which allow us to assess factors involved in translation initiation more specifically. Oligosome (mild translation) fractions were excluded from analysis because of the ambiguous or transitional state of protein synthesis that they represent. To minimize the confounding presence of newly synthesized peptides and proteins, cells grown in light stable isotope labeling by amino acids in cell culture (SILAC) media were pulsed with heavy SILAC media before protein harvest. This allows us to identify and exclude from further analysis peptides derived from ongoing translation.
Figure 1. MATRIX Identifies eIF5B as a Hypoxia-Enriched Translation Factor.
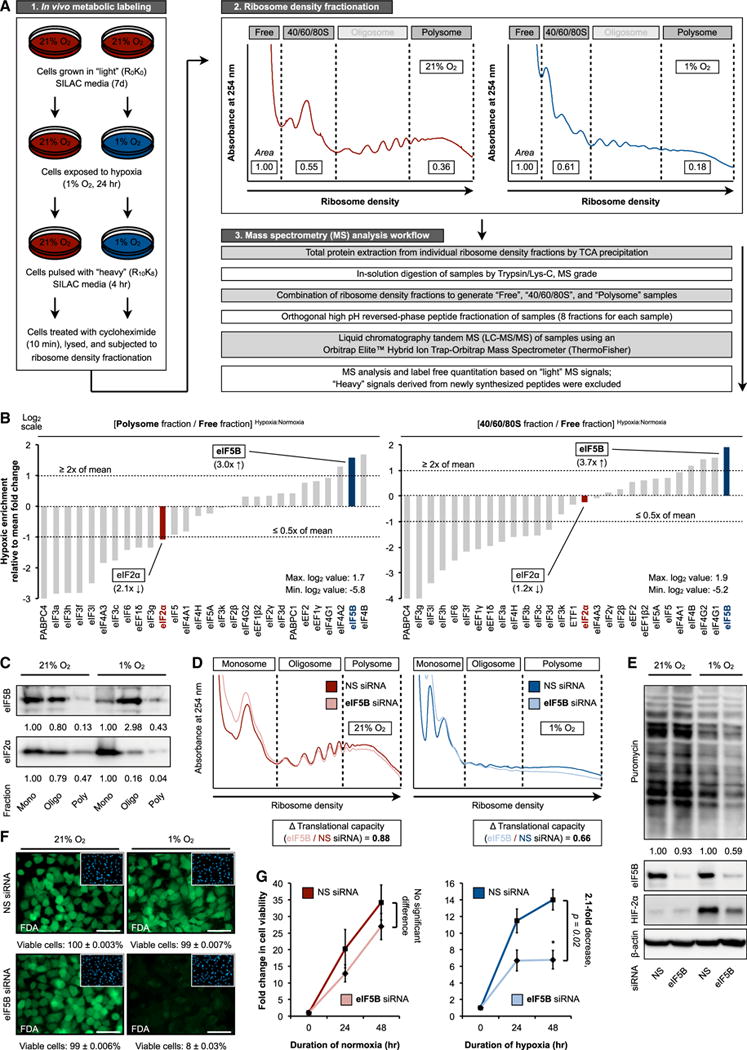
(A) MATRIX workflow.
(B) MATRIX analysis of eukaryotic translation factor activity in hypoxic versus normoxic protein synthesis in U87MG. Ratios of peptide/protein abundance in polysome to free fractions (left panel) and 40/60/80S to free fractions (right panel) were calculated.
(C) MATRIX validation by immunoblot in normoxic and hypoxic U87MG. Representative immunoblots are shown. Mono, monosome (40/60/80S); oligo, oligosome; poly, polysome.
(D and E) Ribosome density profiling (D) and representative immunoblots (E) of normoxic and hypoxic U87MG treated with control non-silencing (NS) or eIF5B-specific small interfering RNA (siRNA).
(F and G) Fluorescein diacetate (FDA) staining (F, green) with DAPI counterstaining (F, inset, blue) and cell viability measurements (G) were performed in normoxic and hypoxic U87MG treated with NS or eIF5B-specific siRNA.
Error bars represent SEM. *p < 0.05. See also Figure S1.
We performed MATRIX on U87MG glioblastoma cells subjected to normoxia (21% O2) and hypoxia (1% O2), a concentration that simultaneously activates eIF4FH and inhibits eIF4F (Figure 1B) (Ho et al., 2016; Uniacke et al., 2012). The ratio of peptide/protein abundance in polysome fractions (active, intense translation) to free fractions (translationally disengaged) was used as the primary readout (Figure 1B, left panel). The abundance ratio of proteins engaged in translation initiation (40/60/80S fractions) to free fractions served as a secondary readout (Figure 1B, right panel). Overall, MATRIX detected 2,728 well-represented (≥5 unique peptides) cellular factors (Table S1). We identified 30 translation factors with high confidence (i.e., detected across all measured fractions, ≥25% protein coverage). Based on the previously mentioned readouts, and an enrichment criterion of ≥2× mean fold change after outlier removal, MATRIX identified eIF5B (3× enrichment) (Figure 1B, blue highlight) as one of two translation factors (the other being eIF4B) that exhibited augmented activity under hypoxic conditions. Reassuringly, this impartial analysis confirmed the well-established hypoxic inhibition of eIF2α activity (2× decrease) (Figure 1B, red highlight). These findings were validated by immunoblot analysis of ribosome density fractions (Figure 1C). Thus, MATRIX analysis revealed that eIF5B concentrates in hypoxic translating ribosomes.
Involvement of eIF5B in Hypoxic Protein Synthesis
Next, we examined the effect of eIF5B depletion on global protein synthesis in various human cell lines exposed to normoxia or hypoxia using ribosome density profiling (Figures 1D and S1B). Normoxic eIF5B-depleted cells exhibited a modified ribosome density profile but maintained a largely similar translational intensity, as determined by quantitative area under curve measurements (Figures 1D, left panel, and S1C, top panel). In contrast, eIF5B-silencing in hypoxic cells resulted in a ~35% loss in translating ribosomes and decreased translational capacity (Figures 1D, right panel, and S1C, bottom panel). Similarly, puromycin incorporation showed that eIF5B silencing produced little discernable effect on normoxic translational capacity across multiple cell lines, while global translation was reproducibly decreased by ~40% in eIF5B-depleted hypoxic cells (Figures 1E and S1D). These findings agree with published observations that eIF5B is basally non-essential. Normoxic eIF5B depletion resulted only in a modest suppression of cell growth and viability (Figures 1F and 1G). In contrast, hypoxic cell viability was significantly decreased by eIF5B silencing (Figures 1F and 1G).
eIF5B Facilitates met-tRNAiMet Delivery during Hypoxia
eIF2 is the paradigmatic translation initiation factor that delivers met-tRNAiMet to eukaryotic ribosomes. As the affinity of eIF5B for met-tRNAiMet is considerably lower than that of eIF2, several studies have suggested a role for this translation factor in 80S ribosomal subunit joining (Pestova et al., 2000; Terenin et al., 2008). Confirming the observation that eIF5B was functionally enriched in hypoxic translating ribosomes (Figure 1B), we observed increased association between eIF5B and tRNAiMet in hypoxic versus normoxic conditions (Figures 2A and S2A). Likewise, eIF5B silencing caused a decrease in tRNAiMet levels in hypoxic ribosomes compared to eIF5B-replete controls (Figure 2B), unlike elongating tRNAs, such as tRNAArg, which remained unaffected (Figure S2B). This hypoxic augmentation of eIF5B activity may be accounted for, at least partly, by increased met-tRNAiMet availability as a result of eIF2 inhibition. Hypoxia-induced eIF2α subunit phosphorylation and eIF2 inactivation correlate positively with the induction of both ATF4, a master transcription factor of ISR induced regardless of initiating stress (Pakos-Zebrucka et al., 2016), and NDRG1, a hypoxia-inducible protein (Ho et al., 2016) (Figure 2C). This suggested that the synthesis of hypoxia-inducible proteins is eIF2 independent, instead relying on alternative factors to deliver met-tRNAiMet. eIF5B silencing severely attenuated the hypoxic induction of ATF4 and NDRG1 (Figures 2D and S2C, left panels) due to reduced translation efficiency (TE) (Figure 2E), but not steady-state levels of mRNAs (Figure S2D). Such hypoxic induction remained eIF5B dependent and eIF2 independent even following treatment with salubrinal, an ISR-prolonging eIF2 phosphatase inhibitor (Boyce et al., 2005) (Figures 2D and S2C, middle panels). Tunicamycin-induced endoplasmic reticulum (ER) stress, which leads to rapid eIF2α phosphorylation or eIF2 inactivation (Harding et al., 2000b), resulted in robust eIF5B-independent accumulation of the pan-stress-inducible ATF4, but not the hypoxia-induced protein NDRG1 (Figures 2D and S2C, right panels). Furthermore, cellular translational dependence on eIF5B was unaffected by heat shock (Figure S2E). These data suggested that eIF5B is preferentially regulated by hypoxic stress.
Figure 2. eIF5B Facilitates eIF2-Independent met-tRNAiMet Delivery.
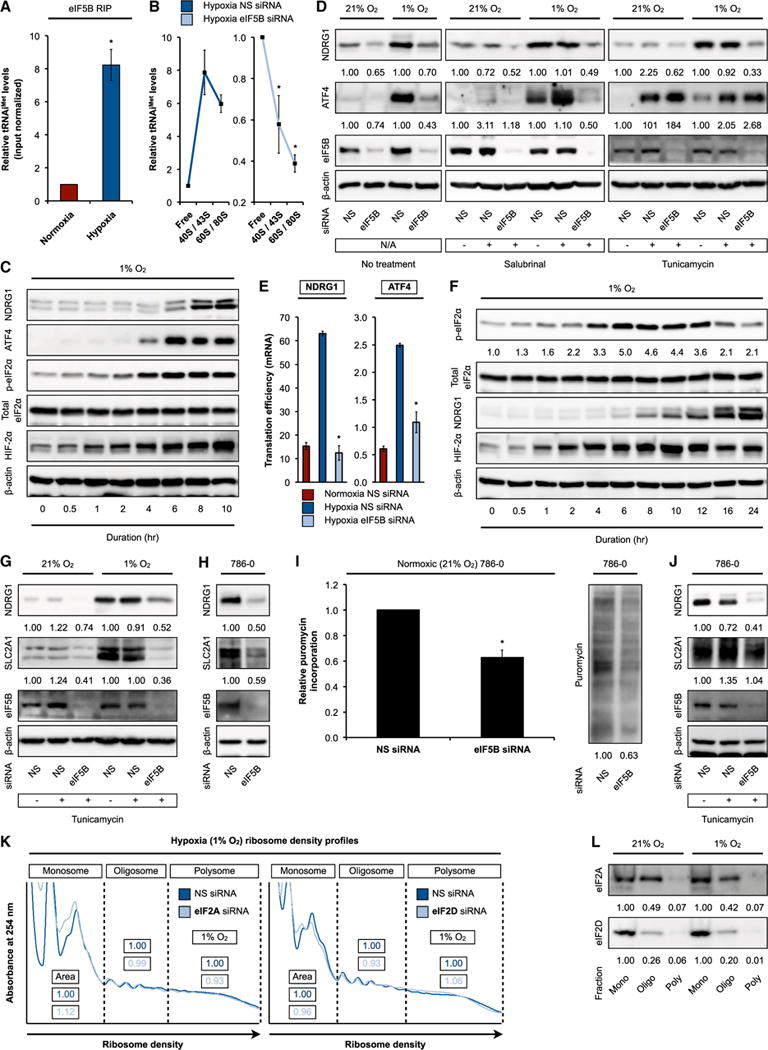
(A) eIF5B RNA immunoprecipitations (RIPs) followed by qRT-PCR measurements of input-normalized tRNAiMet levels from normoxic and hypoxic U87MG.
(B) Normoxic and hypoxic U87MG treated with control non-silencing (NS) or eIF5B-specific siRNA were subjected to ribosome density fractionation, followed by qRT-PCR measurements of tRNAiMet levels in the indicated fractions. (C and D) Representative immunoblots of hypoxic U87MG (C) and normoxic and hypoxic (1% O2, 10 hr) U87MG (D) treated with NS or eIF5B-specific siRNA (left panel) and with salubrinal (middle panel) or tunicamycin (right panel).
(E) Normoxic and hypoxic U87MG treated with NS or eIF5B-specific siRNA were subjected to ribosome density fractionation, followed by mRNA level measurements by qRT-PCR. Translation efficiency was defined as the ratio of polysome to monosome abundance.
(F–H) Representative immunoblots of hypoxic U87MG (F), normoxic and hypoxic U87MG treated with NS or eIF5B-specific siRNA and tunicamycin (G), and normoxic 786-0 treated with NS or eIF5B-specific siRNA (H).
(I) Puromycin incorporation measurements (left panel) and representative immunoblot (right panel) in normoxic 786-0 treated with NS or eIF5B-specific siRNA.
(J) Representative immunoblots of normoxic 786-0 treated with NS or eIF5B-specific siRNA and tunicamycin.
(K) Ribosome density profiling of hypoxic U87MG treated with NS, eIF2A-specific (left panel), or eIF2D-specific (right panel) siRNA. Area under curve measurements (area) are shown.
(L) Representative immunoblots of normoxic and hypoxic U87MG subjected to ribosome density fractionation.
Mono, monosome (40/60/80S); oligo, oligosome; poly, polysome. Error bars represent SEM. *p < 0.05. See also Figure S2.
We next examined the relationship between eIF5B-dependent met-tRNAiMet delivery and eIF2 activity status. In certain cellular systems, prolonged hypoxia is associated with ISR recovery and eIF2 reactivation (via eIF2α dephosphorylation) (Figures 2F and S2F) (Koritzinsky et al., 2006). Yet the induction of classic hypoxia-inducible proteins, such as NDRG1 and glucose transporter GLUT1/SLC2A1, under prolonged hypoxia remained eIF5B dependent but eIF2 independent, even with tunicamycin- or salubrinal-induced augmentation of eIF2 inhibition (Figures 2G, S2G, and S2H). We were mindful of the possibility that residual eIF2α phosphorylation under prolonged hypoxia could preclude full reactivation of eIF2 activity (Figure 2F). To address this issue, we employed the 786-0 clear cell renal cell carcinoma cell model, which behaves as a pseudo-hypoxic system under basal, normoxic conditions due to the genetic loss of von Hippel-Lindau (VHL). As such, normoxic 786-0 engages simultaneously in both normoxic and hypoxic protein synthesis (Ho et al., 2016). This represents an ideal model system to examine basal, normoxic eIF2 and eIF5B activities without the need for hypoxic stimulation. Results indicated that eIF5B silencing markedly reduced eIF5B-dependent NDRG1 and GLUT1 protein levels (Figures 2H and S2I), as well as global protein synthesis in normoxic 786-0 (Figure 2I). Again, eIF5B-dependent targets demonstrated eIF2 independence even in normoxic 786-0 (Figures 2J and S2J). These data suggest that eIF5B may act as the hypoxic surrogate of eIF2 through direct met-tRNAiMet delivery to initiating ribosomes and/or through ribosomal subunit joining. The relative contributions of various eIF5B activities to its role in hypoxic translation, e.g., met-tRNAiMet recruitment, GTPase activity, and ribosomal subunit joining (Shin et al., 2002), remain to be determined in future studies. The translation factors eIF2A and eIF2D can assemble with met-tRNAiMet under specific conditions (Sendoel et al., 2017). However, eIF2A and eIF2D silencing did not significantly alter ribosome density profiles (Figures 2K and S2K) or global protein synthesis (Figure S2L), and we observed no accumulation of these factors in actively translating hypoxic ribosomes (Figure 2L).
Global Interrogation of eIF5B-Dependent Translatome Remodeling
Next, we sought to determine the eIF5B-dependent target population in hypoxic cells using an unbiased, integrative approach that combines high-throughput RNomic and proteomic analyses, enabling the assessment of steady-state RNA and protein levels and translational output (TE/protein output) from the same samples, respectively (Figure 3A). Consistent with our biochemical assays (Figures 1D and 1E), RNomic analysis revealed a global decrease in TE (Figure S3A) for more than 70% of detected mRNAs in eIF5B-depleted cells compared to controls (Figure 3B, top panel), with minimal correlation with RNA level changes (Figure 3B, bottom panel). The high sensitivity of this assay allowed us to detect many low-abundance mRNAs whose TE changes were likely masked by global-scale assessments (Figures 1D and 1E). Complementary to RNA-based translatome analysis (Floor and Doudna, 2016; Ho et al., 2016; Ingolia et al., 2009), live-cell metabolic labeling by pulse-SILAC (pSILAC)-mass spectrometry (MS) represents a reliable, direct readout for global translational output (Schwanhäusser et al., 2011). Our pSILAC-MS analysis identified 480 proteins that exhibited reduced protein synthesis in eIF5B-depleted hypoxic cells (Figures 3C and S3B; Table S2). Translational output of these proteins exhibited significant positive correlation with steady-state protein levels (Figure 3C), and most (85%) demonstrated corresponding decreases in TE, but not RNA levels (Figures 3D and S3B).
Figure 3. Global Interrogation of eIF5B-Dependent Targets.
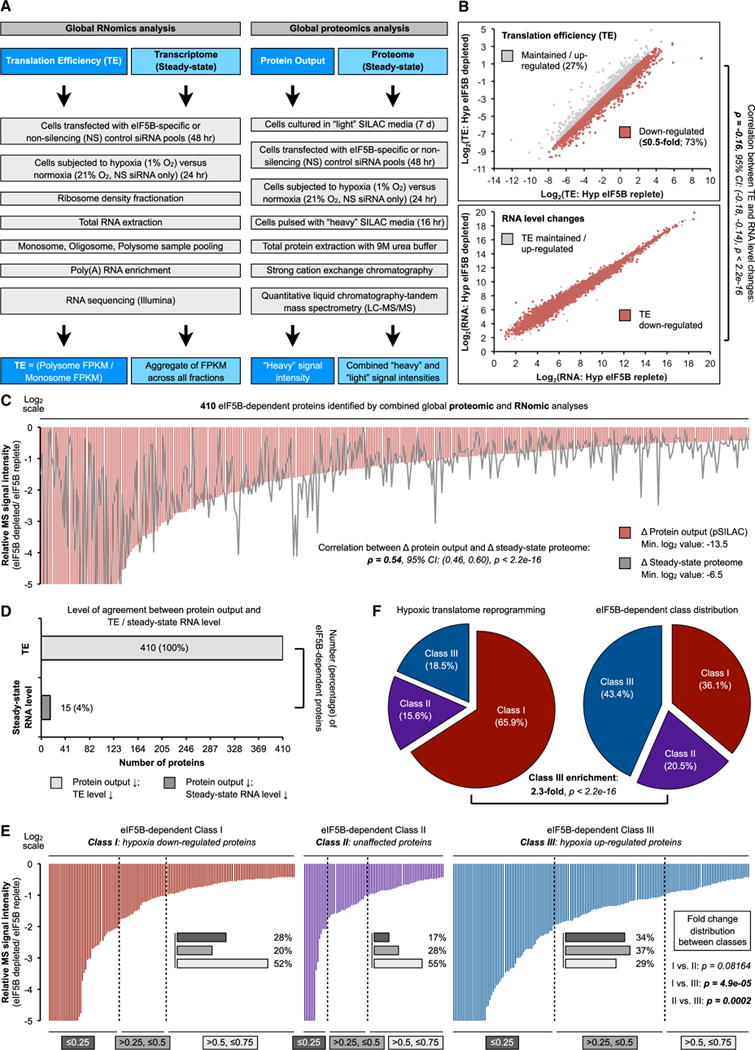
(A) Workflow of RNomic and proteomic analyses.
(B) RNomic analysis of translation efficiency (TE) and steady-state RNA levels. Transcripts exhibiting decreased TE (≤0.5-fold) are highlighted in red.
(C) eIF5B-dependent targets that exhibit decreased TE, protein output, and steady-state protein levels (≤0.75-fold).
(D) Agreement between TE, steady-state RNA level and protein output.
(E) Effect of eIF5B depletion on magnitude of TE decrease across hypoxia-responsive mRNA classes.
(F) Proportions of the three hypoxia-responsive mRNA classes under eIF5B-replete and eIF5B-depleted (by siRNA) conditions.
See also Figure S3.
We recently defined a class of eIF4FH-dependent cellular mRNAs that are translationally enhanced under hypoxic conditions, termed class III (Ho et al., 2016). Class III members were more profoundly affected by eIF5B in terms of both magnitude (Figure 3E) and proportion (2× enrichment) (Figure 3F) compared to class I (translationally downregulated during hypoxia) and class II (translation unaffected by oxygen levels). Protein output in eIF5B-depleted cells showed significant positive correlation with steady-state protein levels across all three classes (Figure S3C). Altogether, these studies identified a sizable eIF5B-dependent target population and provide evidence for eIF5B as an essential component of the hypoxic protein synthesis machinery.
Central Carbon Metabolism Is a Major eIF5B-Dependent Target
Pathway enrichment analysis revealed the ancient pathways of central carbon metabolism and cellular hypoxic response as some of the most prominent eIF5B-dependent systems (Figures 4A, S4A, and S4B). Multi-dimensional validation confirms concordant changes in TE, protein output, and steady-state protein levels for each identified eIF5B target (Figures 4A, bottom panel; 4B; and S4B, bottom panel). The observation that most eIF5B-dependent proteins are class III members (Figure S4C) highlights the critical involvement of eIF5B in adaptive hypoxic translatome reprogramming. The number of eIF5B-dependent enzymes and proteins involved in central carbon metabolism and fructolysis demonstrates the vital eIF5B dependence of these pathways (Figures 4C and S4D). Unlike proteins involved in hypoxic adaptation (Figure 4D), proteins implicated in other cellular stresses, e.g., heat shock (Hsp27, Hsp70, and Hsp90), appear to be eIF5B independent, at least in our system (Figures S4E–S4G). In addition, hypoxia-inducible factor (HIF) alpha subunits HIF-1α and HIF-2α, which orchestrate the transcriptional (Wang et al., 1995) and translational (Uniacke et al., 2012) arms of the conserved cellular hypoxic response, exhibited notable eIF5B dependence (Figures 1E, 4A, 4D, S1D, S2C, and S2E–S2G). These findings underscore the unique role of eIF5B in hypoxic adaptation. Central carbon metabolism is a vital pathway that operates at a minimal basal level under normoxic conditions. Quantification of normoxic immunoblots revealed the eIF5B dependence of these pathways (e.g., GLUT1 and NDRG1) even under standard, normoxic conditions (Figures 2D and 2G), including in normoxic 786-0 (Figures 2H and 2J). Finally, we confirmed the normoxic dependence of central carbon metabolism on eIF5B using a global pSILAC assessment of protein output in eIF5B-depleted, normoxic 786-0 (Figure 4E).
Figure 4. Central Carbon Metabolism Is a Major eIF5B-Dependent Pathway.
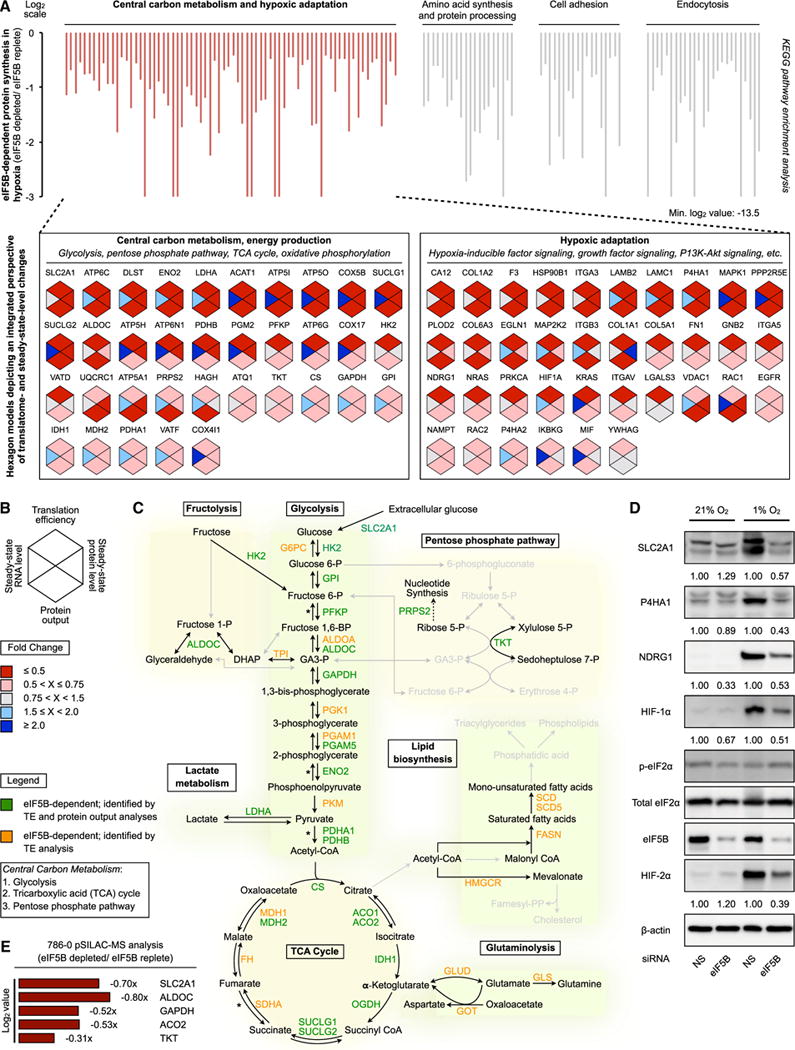
(A) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of eIF5B-dependent cellular processes (top panel). Hexagon models depicting TE, steady-state RNA level, protein output, and steady-state protein level for each identified protein involved in central carbon metabolism and hypoxic adaptation are shown (bottom panel).
(B) Legend for models in Figures 4A and S4A.
(C) eIF5B-dependent enzymes and proteins of central carbon metabolism and fructolysis identified by protein output and/or TE analyses. The asterisk indicates additional identified enzymes and/or isoforms (Figure S4D).
(D) Representative immunoblots of eIF5B-dependent proteins from normoxic and hypoxic U87MG treated with non-silencing (NS) control or eIF5B-specific siRNA.
(E) pSILAC analysis of protein output in normoxic 786-0 treated with NS or eIF5B-specific siRNA.
See also Figure S4.
DISCUSSION
In this report, we provide evidence that eIF5B is a key element of the cellular hypoxic protein synthesis machinery. Systemic translatome analyses revealed that eIF5B is principally involved in the translation of proteins involved in the ancient central carbon metabolism and cellular hypoxic response pathways. In addition, eIF5B is required for hypoxic cells to mount an ATF4-mediated stress response. It is tempting to speculate that eIF5B/IF2 was retained throughout the aerobic eukaryotic lineage to synthesize proteins that ensure cellular survival in oxygen-deficient environments, thus explaining their conservation to LUCA and across all domains of life. In addition, translatome analysis suggests that the HIF-2α and eIF4FH-dependent class III mRNAs (Ho et al., 2016; Uniacke et al., 2012) rely mostly on eIF5B, even when eIF2 is active. These data suggest that eIF5B facilitates met-tRNAiMet delivery for eIF4FH-mediated hypoxic cap-dependent translation.
From a broader perspective, our findings provide further evidence for the emerging paradigm of alternative, stress-specific translation machineries, such as the hypoxic cap-binding complex eIF4FH (Ho and Lee, 2016; Ho et al., 2016; Landon et al., 2014; Uniacke et al., 2012). Our current study expands upon this concept by demonstrating the hypoxic activation of eIF5B that may operate like eIF2A under certain eIF2-inactivating conditions (Sendoel et al., 2017).
Hypoxic augmentation of eIF5B activity was revealed by MATRIX, a global, unbiased approach that we developed to assess translation factor usage in living cells. Biological activity is a cornerstone of MATRIX, which prioritizes the ability to distinguish actively engaged translation factors from their disengaged or underused counterparts, especially across conditions. This feature offers MATRIX the innovative capability to reveal the lineup of active translation factors. This contrasts with studies that provide the roster or catalog of cellular factors, such as RNA-binding proteins (Castello et al., 2016) and ribosome-associated proteins (Simsek et al., 2017). Synergistic utilization of these complementary approaches could yield deeper insight into fundamental cellular processes, such as translation. We focused our current MATRIX analysis on known translation factors. Beyond these, MATRIX identified >70 ribosomal proteins, >40 recognized RNA-binding proteins, and many additional factors. Future studies are required to examine the biological roles of these factors.
Our studies demonstrate that central carbon metabolism is intricately dependent on eIF5B during normoxia and especially hypoxia. Oxygen-dependent oxidative phosphorylation is the primary mechanism of cellular energy generation in aerobic eukaryotes. Because hypoxia inhibits this process, hypoxic cells engage in a complex, coordinated effort to augment central carbon metabolism (glucose intake, glycolysis, and trichloroacetic acid [TCA] cycle) to meet energy demands. In contrast, basal central carbon metabolism functions at a lower level under standard, normoxic conditions. This difference in central carbon metabolism activity or requirement may explain the increased eIF5B dependence of translation initiation and cell viability during hypoxia, as well as the non-essentiality of eIF5B under basal normoxic conditions (Choi et al., 1998; Thakor and Holcik, 2012). Our data suggest that eIF5B enables the hypoxia-induced glycolytic switch (Nakazawa et al., 2016). Therapeutically, enhancing normoxic eIF5B activity to augment aerobic glycolysis may benefit Leigh syndrome patients who suffer from defective oxidative phosphorylation. Conversely, eIF5B inhibition represents a potential anti-cancer therapy given that aerobic glycolysis, or the Warburg effect, is a hallmark of cancer (Hanahan and Weinberg, 2011).
Finally, there exist eukaryotic species that can subsist in low-oxygen conditions, e.g., naked mole rats (Park et al., 2017) and humans living at high altitude (e.g., Tibetan highlanders) (Yi et al., 2010). It will be interesting to determine in future studies any differences in eIF5B dependence or utilization between hypoxia-tolerant and hypoxia-intolerant species and/or populations.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
U87MG, MCF7, and A549 cells were obtained from ATCC and propagated in DMEM (HyClone) with 10% fetal bovine serum (FBS) (Omega Scientific) and 1% penicillin-streptomycin (Pen-Strep) (HyClone). Cells were maintained at 37 C in a 5% CO2 humidified incubator. Cells were subjected to hypoxia (1% O2, 24 hr unless otherwise stated) at 37 C in a 5% CO2, N2-balanced, humidified H35 HypOxystation (HypOxygen). Final treatment concentrations were cycloheximide (Amresco) at 0.2 mg/mL, 75 μM salubrinal (Sigma-Aldrich) for 4 hr, and 25 μM tunicamycin (Sigma-Aldrich) for 4 hr.
SILAC
Cells were grown in light (R0K0) SILAC media (AthenaES) for 7 days before treatment. For MATRIX analysis, cells were pulsed with heavy (R10K8) SILAC media (AthenaES) for 4 hr (MATRIX) or 16 hr (pSILAC) following treatment.
Ribosome Density Fractionation
Polyribosome fractionations were performed essentially as previously described (Ho et al., 2016). Samples loaded based on equal cell number or equal total RNA yielded similar results. Total RNA was isolated from each fraction by phenol or chloroform extraction and ethanol precipitation following proteinase K treatment. Total protein was isolated by TCA precipitation.
MS Analysis
Liquid chromatography-tandem MS was performed by the SPARC BioCentre (The Hospital for Sick Children, Toronto, Canada). More details are provided in Supplemental Information.
RNA Sequencing Analysis
Poly(A) RNA selection was performed before library preparation and RNA sequencing runs. More details are provided in Supplemental Information.
Statistical Analysis
All experiments were performed at least three independent times, unless otherwise stated. Appropriate statistical analyses were performed, including Student’s t tests, Pearson’s correlation coefficient (ρ) calculations with 95% confidence intervals and p values, and chi-square tests, to investigate proportional differences. Statistical significance was defined as p < 0.05.
Supplementary Material
Highlights.
MATRIX produces system-wide blueprints of active translation factors
eIF5B is an essential component of the hypoxic protein synthesis machinery
eIF5B is the hypoxic surrogate of eIF2 that facilitates met-tRNAiMet delivery
Central carbon metabolism proteins are principally reliant on eIF5B for translation
Acknowledgments
S.L. is funded by grants from the NIH (NIGMS, 1R01GM115342, and NCI, 1R01CA200676) and the Sylvester Comprehensive Cancer Center. J.J.D.H. is a recipient of a Canadian Institutes of Health Research (CIHR) Postdoctoral Fellowship. We thank Dr. Siôn Ll. Williams and Dr. Steven Chen from the Sylvester Comprehensive Cancer Center Oncogenomics Core Facility and Biostatistics and Bioinformatics Core Facility, respectively, for RNA sequencing and analysis services.
Footnotes
DATA AND SOFTWARE AVAILABILITY
The accession number for the MS datasets reported in this paper is ProteomeXchange: PXD006799. The accession number for the RNA sequencing datasets reported in this paper is SRA: SRP110475.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, four figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2017.12.031.
AUTHOR CONTRIBUTIONS
J.J.D.H. and S.L. conceptualized the study and MATRIX. J.J.D.H. and S.L. conceived the experiments. J.J.D.H., N.C.B., G.C., P.D.M., and J.R.K. performed the experiments. J.J.D.H., N.C.B., J.R.K., and S.L. analyzed the data. J.J.D.H. and S.L. wrote the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, Yuan J. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- Castello A, Fischer B, Frese CK, Horos R, Alleaume AM, Foehr S, Curk T, Krijgsveld J, Hentze MW. Comprehensive identification of RNA-binding domains in human cells. Mol Cell. 2016;63:696–710. doi: 10.1016/j.molcel.2016.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SK, Lee JH, Zoll WL, Merrick WC, Dever TE. Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science. 1998;280:1757–1760. doi: 10.1126/science.280.5370.1757. [DOI] [PubMed] [Google Scholar]
- Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- Falkowski PG. Evolution. Tracing oxygen’s imprint on Earth’s metabolic evolution. Science. 2006;311:1724–1725. doi: 10.1126/science.1125937. [DOI] [PubMed] [Google Scholar]
- Floor SN, Doudna JA. Tunable protein synthesis by transcript isoforms in human cells. eLife. 2016;5:e10921. doi: 10.7554/eLife.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000a;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000b;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Ho JJ, Lee S. A cap for every occasion: alternative eIF4F complexes. Trends Biochem Sci. 2016;41:821–823. doi: 10.1016/j.tibs.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JJD, Wang M, Audas TE, Kwon D, Carlsson SK, Timpano S, Evagelou SL, Brothers S, Gonzalgo ML, Krieger JR, et al. Systemic reprogramming of translation efficiencies on oxygen stimulus. Cell Rep. 2016;14:1293–1300. doi: 10.1016/j.celrep.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol. 2003;1:127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- Koritzinsky M, Magagnin MG, van den Beucken T, Seigneuric R, Savelkouls K, Dostie J, Pyronnet S, Kaufman RJ, Weppler SA, Voncken JW, et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J. 2006;25:1114–1125. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landon AL, Muniandy PA, Shetty AC, Lehrmann E, Volpon L, Houng S, Zhang Y, Dai B, Peroutka R, Mazan-Mamczarz K, et al. MNKs act as a regulatory switch for eIF4E1 and eIF4E3 driven mRNA translation in DLBCL. Nat Commun. 2014;5:5413. doi: 10.1038/ncomms6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen BS, Sørensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Choi SK, Roll-Mecak A, Burley SK, Dever TE. Universal conservation in translation initiation revealed by human and archaeal homologs of bacterial translation initiation factor IF2. Proc Natl Acad Sci USA. 1999;96:4342–4347. doi: 10.1073/pnas.96.8.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21:521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa MS, Keith B, Simon MC. Oxygen availability and metabolic adaptations. Nat Rev Cancer. 2016;16:663–673. doi: 10.1038/nrc.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Reznick J, Peterson BL, Blass G, Omerbašić D, Bennett NC, Kuich PHJL, Zasada C, Browe BM, Hamann W, et al. Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science. 2017;356:307–311. doi: 10.1126/science.aab3896. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- Raymond J, Segrè D. The effect of oxygen on biochemical networks and the evolution of complex life. Science. 2006;311:1764–1767. doi: 10.1126/science.1118439. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, Levorse J, Dill BD, Schramek D, Molina H, et al. Translation from unconventional 5′ start sites drives tumour initiation. Nature. 2017;541:494–499. doi: 10.1038/nature21036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin BS, Maag D, Roll-Mecak A, Arefin MS, Burley SK, Lorsch JR, Dever TE. Uncoupling of initiation factor eIF5B/IF2 GTPase and translational activities by mutations that lower ribosome affinity. Cell. 2002;111:1015–1025. doi: 10.1016/s0092-8674(02)01171-6. [DOI] [PubMed] [Google Scholar]
- Simsek D, Tiu GC, Flynn RA, Byeon GW, Leppek K, Xu AF, Chang HY, Barna M. The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell. 2017;169:1051–1065. doi: 10.1016/j.cell.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol. 2008;15:836–841. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- Thakor N, Holcik M. IRES-mediated translation of cellular messenger RNA operates in eIF2α-independent manner during stress. Nucleic Acids Res. 2012;40:541–552. doi: 10.1093/nar/gkr701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniacke J, Holterman CE, Lachance G, Franovic A, Jacob MD, Fabian MR, Payette J, Holcik M, Pause A, Lee S. An oxygen-regulated switch in the protein synthesis machinery. Nature. 2012;486:126–129. doi: 10.1038/nature11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MC, Sousa FL, Mrnjavac N, Neukirchen S, Roettger M, Nelson-Sathi S, Martin WF. The physiology and habitat of the last universal common ancestor. Nat Microbiol. 2016;1:16116. doi: 10.1038/nmicrobiol.2016.116. [DOI] [PubMed] [Google Scholar]
- Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008;8:851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- Yatime L, Schmitt E, Blanquet S, Mechulam Y. Functional molecular mapping of archaeal translation initiation factor 2. J Biol Chem. 2004;279:15984–15993. doi: 10.1074/jbc.M311561200. [DOI] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


