ABSTRACT
Prion diseases are transmissible spongiform encephalopathies (TSEs) characterized by fatal, progressive neurologic diseases with prolonged incubation periods and an accumulation of infectious misfolded prion proteins. Antemortem diagnosis is often difficult due to a long asymptomatic incubation period, differences in the pathogenesis of different prions, and the presence of very low levels of infectious prion in easily accessible samples. Chronic wasting disease (CWD) is a TSE affecting both wild and captive populations of cervids, including mule deer, white-tailed deer, elk, moose, muntjac, and most recently, wild reindeer. This study represents a well-controlled evaluation of a newly developed real-time quaking-induced conversion (RT-QuIC) assay as a potential CWD diagnostic screening test using rectal biopsy sections from a depopulated elk herd. We evaluated 69 blinded samples of recto-anal mucosa-associated lymphoid tissue (RAMALT) obtained from USDA Veterinary Services. The results were later un-blinded and statistically compared to immunohistochemical (IHC) results from the USDA National Veterinary Services Laboratories (NVSL) for RAMALT, obex, and medial retropharyngeal lymph node (MRPLN). Comparison of RAMALT RT-QuIC assay results with the IHC results of RAMALT revealed 92% relative sensitivity (95% confidence limits: 61.52–99.8%) and 95% relative specificity (95% confidence limits: 85.13–99%). Collectively, our results show a potential utility of the RT-QuIC assay to advance the development of a rapid, sensitive, and specific prion diagnostic assay for CWD prions.
KEYWORDS: Ante-mortem test, chronic wasting disease (CWD), elk, immunohistochemistry (IHC), medial retropharyngeal lymph node (MRPLN), obex, prion, real-time quaking-induced conversion (RT-QuIC) assay, recto-anal mucosa-associated lymphoid tissue (RAMALT)
INTRODUCTION
Chronic wasting disease (CWD) is a type of transmissible spongiform encephalopathy (TSE) that affects a wide range of captive and wild cervids such as elk, white-tailed deer, mule deer, moose, muntjac, reindeer1-5 and can experimentally be transmitted to swine.6 CWD was first identified in Colorado and has since expanded to several states and territories of North America including Iowa, Wisconsin and Illinois.4,7 CWD has also been reported in South Korea, Canada and very recently in Norway.1 Conversion of normal cellular prion protein (PrPC) into its abnormally folded isoform, disease-associated prion protein (PrPD), results in protease resistance and the consequent accumulation of PrPD, which serves as the diagnostic marker to identify prion infectivity.8,9 PrPC is involved in various biological functions such as cell signaling, metal homeostasis, apoptosis and oxidative stress.10-13 CWD achieves efficient horizontal transmission to neighboring animals because PrPD becomes widely distributed among the lymph nodes, nervous tissue3,14,15 and blood,16 and it is also excreted in other body fluids where it persists in the environment.17-19 The presence of abnormal PrPD serves as an important marker to diagnose CWD. In white-tailed deer, PrPD accumulation in brain tissues is preceded by its accumulation in several lymphoid tissues like tonsils, lymph nodes, and gut-associated lymphoid tissue.20,21,22 Thus, given the potential for earlier detection, this peripheral tissue distribution was exploited in the utilization of rectal biopsies for antemortem screening of scrapie, thus helping surveillance programs in Canada and the US since 2006 and 2008, respectively.4,5 Due to the rapid and widespread nature of CWD in captive and free-ranging animals, the serious concern that needs to be addressed is how to detect the disease early enough in live animals from both captive and wild populations for intervention measures to halt the spread of the disease.19,23 To date, several diagnostic methods have been approved as official regulatory test methods in the US to detect PrPD, such as immunohistochemistry (IHC), Western blot, and ELISA. However, the utility of these assays is limited to postmortem tissues (such as obex and medial retropharyngeal lymph nodes) in the case of cervids with CWD.
New protein-misfolding amplification platforms such as real-time quaking-induced conversion (RT-QuIC) and serial protein-misfolding cyclic amplification (PMCA) assays have recently been evaluated for their utility in detecting PrPD, as well as for their sensitivity and specificity. However, the diagnostic utility of these techniques has not been rigorously tested for cervid biopsies. The RT-QuIC and PMCA assays show great promise for developing new tests that will enable us to detect prion disease early in live animals.24,25 Furthermore, the RT-QuIC assay is well documented for its flexibility and versatility utilizing a high-throughput format. To detect PrPD in CWD cases, most of the current diagnostic methods use brainstem, medial retropharyngeal lymph node (MRPLN), and palatine tonsils from postmortem animals. In white-tailed deer and elk, utilizing recto-anal mucosa-associated lymphoid tissue (RAMALT) and other lymphoid tissue biopsies provides promising sample tools for antemortem diagnosis of CWD. Also, collecting RAMALT from live animals is a relatively simple and reliable procedure that has already been adopted for use in experimental animals and sheep populations and could be translated to field conditions for both captive and wild cervid management.20,26-28,29,30 Despite these advancements in diagnosing prion diseases, several challenges still need to be addressed for sensitive and specific detection of CWD in early stages of infection.
In the present study, researchers from Iowa State University, USDA Wildlife Services, and USDA Veterinary Services employed a blinded study design to evaluate the ability of the RT-QuIC assay to detect CWD from post-mortem RAMALT samples in an elk herd of 69 animals depopulated due to CWD infection. The RAMALT RT-QuIC assay results correlated well with the IHC detection of PrPD from the RAMALT and with the severity of prion infection. Our study identifies RAMALT samples as a suitable candidate for the RT-QuIC assay and compares RT-QuIC with currently approved testing methods. Our results demonstrate that RAMALT could be adopted for the antemortem diagnosis of CWD. With the validation of RT-QuIC assay for RAMALT samples, CWD could be diagnosed from live elk, increasing the number of strategies available to prevent the spread of the disease.
RESULTS
Determination of Quality of Substrate Used for RT-QuIC Assay
The purity of substrate is a key determinant of protein misfolding kinetics in the RT-QuIC assay. Therefore, we first tested the purity of our in-house generated, Syrian Hamster recombinant prion protein (SHrPrP) (residues 90–231). The high-purity recombinant prion protein was generated using the bacterial expression system and isolated by affinity chromatography under denaturing conditions, and on-column refolding followed by elution of protein (as described in methods, Fig. 1A, top panel). Biochemical characterization of the SHrPrP using total protein stain (Ponceau) revealed that the protein was maximally pure (Fig. 1A, middle panel). We also performed Western blot analysis of the purified protein using a mouse monoclonal 6D11 antibody, which revealed a single band specific for PrP immunoreactivity at ∼16 kD molecular weight (Fig. 1A, bottom panel). Additionally, we used SHrPrP (residues 90–231) received from Dr. Caughey and colleagues from the NIH Rocky Mountain Laboratory31 as a positive control. This was loaded in lane 8 of the Western blot (Fig. 1A), corresponding to the same molecular weight as our lab-purified protein in lane 6 (center of the peak fraction during elution of protein), which was used in the RT-QuIC assay. Total protein stain and Western blot analysis of SHrPrP from our lab and the positive control from Caughey's lab are shown in Fig. 1B. Next, we performed biophysical characterization of the substrate using NMR spectroscopy. The spectra revealed the purity and homogeneity of the substrate and conformation of the alpha helical-rich protein (Fig. 1C). Furthermore, CD spectroscopy of the purified protein confirmed the high α-helical content of its secondary structure (Fig. 1D). For NMR and CD spectroscopy, we also plotted our lab-purified protein against positive controls from the Caughey lab. Lastly, mass spectroscopy indicated >99% purity of the rPrP (data not shown).
FIGURE 1.
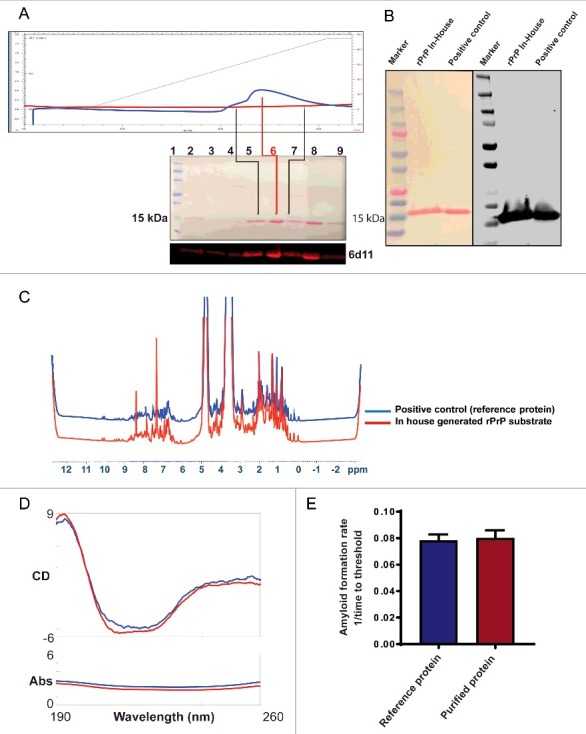
Biochemical and biophysical characterization of purified Syrian Hamster recombinant prion protein (SHrPrP) (90–231 residues). (A) The top panel is the elution of rPrP using FPLC. The middle panel consists of total protein stain using Ponceau S solution. The bottom panel is the Western blot analysis of eluted rPrP using 6D11 antibody. The numbers 1–9 in the middle panel show samples loaded in each well during Western blot. Lane 1 represents molecular weight standards. Lanes 2–7 represent the protein fractions collected at different steps of protein purification. Lanes 5 & 7 (black lines) represent the early and late peak fractions, respectively, during elution of protein. Lane 6 (red line) represents the middle of the peak fraction of the eluted protein, which is used as a substrate in the RT-QuIC assay. Lane 8 represents the positive control, which is also an SHrPrP (90–231), from Dr. Caughey and colleagues, used as a reference. (B) Total protein stain (Ponceau S) and Western blot analysis of pooled SHrPrP (90–231) of purified protein and a positive control with the 6D11 antibody. The first lane is molecular weight standards, the second lane is purified protein and the third lane is a positive control. (C & D) NMR and CD spectroscopy images of SHrPrP (90–231). The blue line represents the SHrPrP (90–231) positive control purified at the Caughey lab, and the red line represents SHrPrP (90–231) that we purified. (E) AFR in the RT-QuIC assay using reference protein and purified protein as substrates. The blue bar indicates reference protein, while the red bar indicates purified protein.
Along with other confirmatory tests, we also compared purified protein in the RT-QuIC assay, from which it is evident that both the reference protein and purified protein have similar amyloid formation rates (AFR) (Fig. 1E). AFR was derived by taking the inverse of the time to cross the threshold fluorescence, which was defined as the control mean fluorescence plus 5 standard deviations.32 Every purified batch of the rPrP90–231 was validated using total protein stain, Western blotting and test plates with known standards before using in the RT-QuIC assay.
Optimal Seed Concentration of Recto-Anal Mucosa-Associated Lymphoid Tissue (RAMALT) Samples for RT-QuIC Analysis
As in many assays, it is imperative to quantify the amount of seed needed for the assay. Too much or too little seed may reduce the sensitivity of the RT-QuIC assay. Therefore, to determine the optimal seed concentration of a RAMALT sample that works best in the RT-QuIC assay, we did a serial log dilution of a known positive control RAMALT sample. The sample was homogenized with phosphate-buffered saline (PBS) to 10% and later diluted (in 0.05% SDS/PBS) to generate concentrations (%) of 2, 2 × 10−1, 2 × 10−2, 2 × 10−3, 2 × 10−4 and so on until 2 × 10−7. From each serially diluted homogenate, 5 µl was added to 95 µl of the RT-QuIC reaction mixture (described in Methods), thus further diluting the seed in the well. Seed concentrations of 2 × 10−2 and 2 × 10−3 amplified optimally compared to all other dilutions, which amplified at later time points (Fig. 2A). When different seed concentrations from the serial dilution were plotted against the AFR, a linear relationship was observed for those seeded with a positive control. High AFR values indicate the sample is crossing the threshold at an early time point and has a low lag period, making it more suitable for testing. Threshold-crossing time for any sample depends on its seed concentration. No amyloid was formed when negative seeds were used (Fig. 2B). All RAMALT samples from 69 elk were tested using 2 × 10−2 dilution in the RT-QuIC assay (Fig. 2C & 2D). The RT-QuIC fluorescence amplification readout showed a clear increase in fluorescence intensity in CWD-positive samples as compared to negative controls.
FIGURE 2.
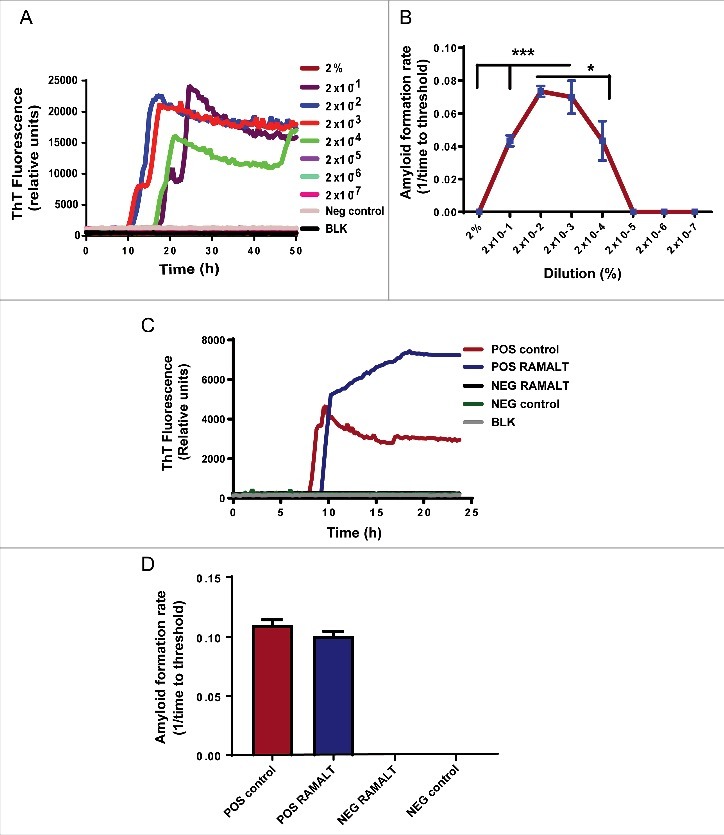
Choosing optimal working dilution and testing RAMALT samples in the RT-QuIC assay. (A) Different dilutions of a positive control on the RT-QuIC assay. Each dilution ran in triplicate. The 2 × 10−2 dilution was used for all tested samples. (B) AFR for different dilutions of a positive control. The 2 × 10−2 dilution has more AFR and is significantly different. Asterisks *p ≤ 0.05 and ***p < 0.001 added to show the sample concentrations that were significantly different. (C) Amplification kinetics of positive and negative elk RAMALT samples along with controls. Blue trace represents the RAMALT sample that tested positive in the RT-QuIC assay. (D) AFR of RAMALT sample along with a positive control. The blue bar indicates a representative trace of one RAMALT sample.
CWD Status of the Herd
Immunohistochemical (IHC) analysis of obex, MRPLN, and RAMALT (Fig. 3A and 3B) were independently performed in all 69 elk samples at the National Veterinary Services Laboratories, but the RT-QuIC analysis was done at Iowa State University and only for RAMALT samples. Out of 69 elk, 51 were MM, and 17 were ML genotypes at codon 132 in the Prnp gene; genotyping data were not available for one animal, so only 68 animals were used for data analysis (Table 1). Among the 51 MM animals, IHC analyses identified 14 animals that were CWD-positive in both the obex and MRPLN, one in obex sample only, and six in MRPLN only. For RAMALT samples, both IHC and the RT-QuIC assay identified 10 animals that were positive by IHC of obex and MRPLN. Of the 17 ML animals, IHC analyses identified 2 animals that were CWD-positive in IHC of both the obex and MRPLN, and two in obex only. However, IHC and RT-QuIC analyses of RAMALT samples only identified 2 animals as CWD-positive (Table 1). This discrepancy in PrPD detections when comparing RAMALT results to those of obex and MRPLN may be due to lack of PrPD in tested samples. Samples positive by IHC analysis of obex were scored as 0, 1, 2, 3 & 4 based on a previously reported methodology33 (Table 1). Collectively, 19/68 (28%) elk samples were CWD-positive by IHC of obex, and 22/68 (32%) were positive by IHC of MRPLN. Notably, out of these positive animals, IHC of RAMALT identified 12 positives, and RT-QuIC analysis of RAMALT identified 14 positives.
FIGURE 3.
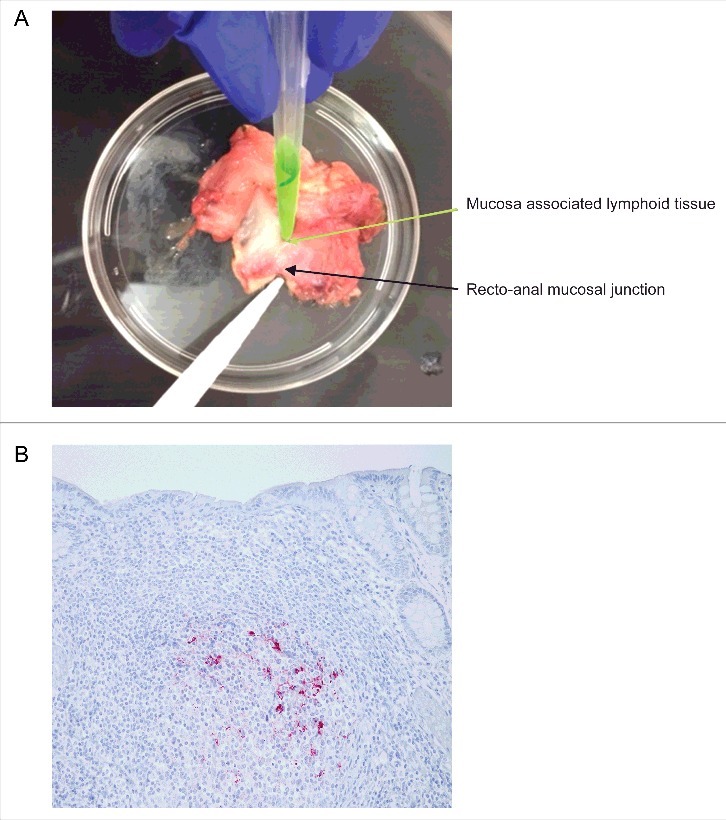
Isolation and immunohistochemical detection of PrPD from RAMALT sample. (A) Dissection of recto-anal mucosal (RA) junction to isolate mucosa-associated lymphoid tissue (MALT) for RAMALT sample. (B) Immunohistochemistry of RAMALT from elk (Cervus elaphus nelsoni). Note the proteinase-resistant prion staining (red chromogen) distribution within the lymphoid follicle located in the lamina propria. Staining was done using a mouse monoclonal antibody F99/97.6.1 against epitopes at residues 220–225 of prion protein and developed with streptavidin-alkaline phosphatase method. The slides were further counterstained with hematoxylin (blue stain).
TABLE 1.
Summary of CWD status by IHC and RT-QuIC assay results.
| Animal number | Genotype | IHC of obex | Obex score | IHC of MRPLN | IHC of RAMALT | CWD status | RT-QuIC of RAMALT |
|---|---|---|---|---|---|---|---|
| 1 | MM | P | 4 | P | P | P | P |
| 2 | MM | ND | ND | ND | ND | ND | |
| 3 | MM | ND | ND | ND | ND | ND | |
| 4 | MM | ND | ND | ND | ND | ND | |
| 5 | MM | ND | ND | ND | ND | ND | |
| 6 | ML | ND | ND | ND | ND | ND | |
| 7 | MM | ND | ND | ND | ND | ND | |
| 8 | ML | P | 1 | ND | P | P | P |
| 9 | MM | ND | ND | ND | ND | ND | |
| 10 | MM | P | 3 | P | P | P | P |
| 11 | MM | ND | ND | ND/I | ND | ND | |
| 12 | ML | P | 1 | P | ND | P | ND |
| 13 | MM | ND | ND | ND | ND | ND | |
| 14 | ML | ND | ND | ND | ND | ND | |
| 15 | MM | ND | ND | ND | ND | ND | |
| 16 | MM | ND | 0 | P | ND | P | P |
| 17 | MM | P | 1 | P | P | P | P |
| 18 | ML | ND | ND | ND | ND | ND | |
| 19 | MM | ND | ND | ND | ND | ND | |
| 20 | ML | ND | ND | ND | ND | ND | |
| 21 | MM | ND | ND | ND | ND | ND | |
| 22 | MM | ND | ND | ND | ND | ND | |
| 23 | MM | ND | 0 | P | ND | P | ND |
| 24 | ML | ND | ND | ND | ND | ND | |
| 25 | MM | ND | ND | ND | ND | ND | |
| 26 | MM | ND | ND | ND | ND | ND | |
| 27 | MM | ND | 0 | P | ND | P | ND |
| 28 | ML | ND | ND | ND | ND | ND | |
| 29 | MM | P | 3 | P | P | P | P |
| 30 | MM | P | 2 | P | P | P | P |
| 31 | MM | ND | ND | ND | ND | ND | |
| 32 | MM | P | 3 | P | P | P | P |
| 33 | MM | ND | ND | ND | ND | ND | |
| 34 | MM | ND | ND | ND | ND | ND | |
| 35 | ML | ND | ND | ND | ND | ND | |
| 36 | MM | ND | ND | ND | ND | ND | |
| 37 | MM | ND | ND | ND | ND | ND | |
| 38 | MM | ND | ND | ND | ND | ND | |
| 39 | ML | ND | ND | ND | ND | ND | |
| 40 | MM | ND | ND | ND | ND | ND | |
| 41 | MM | ND | ND | ND | ND | ND | |
| 42 | ML | ND | ND | ND | ND | ND | |
| 43 | ML | P | 1 | ND | ND | P | ND |
| 44 | ML | ND | ND | ND | ND | ND | |
| 45 | MM | ND | ND | ND | ND | ND | |
| 46 | MM | ND | 0 | P | ND | P | ND |
| 47 | MM | P | 2 | P | ND | P | ND |
| 48 | MM | P | 1 | ND | ND | P | ND |
| 49 | ML | ND | ND | ND | ND | ND | |
| 50 | MM | P | 2 | P | P | P | ND |
| 51 | MM | ND | 0 | P | ND | P | ND |
| 52 | ML | ND | ND | ND | ND | ND | |
| 53 | MM | P | 1 | P | ND | P | ND |
| 54 | MM | P | 2 | P | ND | P | ND |
| 55 | ML | ND | ND | ND | ND | ND | |
| 56 | MM | P | 3 | P | P | P | P |
| 57 | ML | P | 3 | P | P | P | P |
| 58 | MM | ND | ND | ND | ND | ND | |
| 59 | MM | ND | ND | ND | ND | ND | |
| 60 | MM | ND | ND | ND | ND | ND | |
| 61 | MM | P | 3 | P | P | P | P |
| 62 | MM | ND | ND | ND | ND | ND | |
| 63 | MM | P | 1 | P | P | P | P |
| 64 | MM | ND | ND | ND | ND | P | |
| 65 | MM | P | 1 | P | ND | P | ND |
| 66 | MM | ND | ND | ND | ND | P | |
| 67 | MM | ND | ND | ND | ND | ND | |
| 68 | MM | ND | 0 | P | ND | P | ND |
| Total | MM- 51 | P- 19 | 0- 6; | P- 22 | P- 12 | P- 25 | P- 14 |
| ML- 17 | ND- 49 | 1- 8; | ND- 46 | ND- 56 | ND- 43 | ND- 54 | |
| 2- 4; | |||||||
| 3- 6; | |||||||
| 4- 1 |
The number of animals testing positive and negative for CWD were tabulated. Out of 69 samples, genotyping was available for only 68 animals. IHC of MRPLN detected the highest number of positives followed by IHC of obex, RT-QuIC assay of RAMALT and IHC of RAMALT. Samples that were positive in IHC of obex were scored as 0, 1, 2, 3 & 4 based on the distribution and extent of PrPD in the brain. Obex score of zero represents elk that were MRPLN-positive but had no staining in the obex, and a score of one represents the minimal distribution of PrPD around vagal nuclei with higher scores indicating more extensive distributions. A score of 4 represents widespread distribution in the gray and white matter of the brainstem. P = positive; ND = not detected, ND/I = not detected, insufficient follicles.
RAMALT RT-QuIC and IHC Test Results
Based on genotyping and other diagnostic test results, CWD is more prevalent in animals homozygous for MM at the Prnp codon 132 since more positives were observed in MM animals compared to those with the ML genotype (Fig. 4A). This is consistent with the previous reports.34,35 To evaluate the RAMALT RT-QuIC results, we used the Receiver Operating Characteristics (ROC) curve, which plots sensitivity versus 1-specificty of a diagnostic test. The specificity of RT-QuIC versus a diagnostic test (e.g., IHC) is calculated as TN/(TN+FP), where TN (true negative) is the number of samples identified as negative (not infected) by both tests and FP (false positive) is the number of samples identified as positive (infected) by RT-QuIC but negative by the other test (e.g., IHC). The area under the curve (AUC) represents the accuracy of the test and offers a way to quantitatively compare two or more diagnostic tests. Here, we compared the RAMALT RT-QuIC results with IHCs of RAMALT, obex, and MRPLN samples. In general, an AUC >90% indicates high accuracy. IHC of RAMALT and RAMALT RT-QuIC assay (Fig. 4B) share 93.1% AUC. Thus, RAMALT RT-QuIC assay results were closely comparable to RAMALT IHC results. Overall, the correlation between IHC of RAMALT and those of RT-QuIC analyses was nearly 81% (Table 2). By comparing RAMALT samples, the relative sensitivity and relative specificity of the RT-QuIC assay and IHC were 92% and 95%, respectively. The sensitivity of the RT-QuIC assay for RAMALT samples with other tests like IHC's of obex and MRPLN is 50% along with 95% specificity.
FIGURE 4.

CWD status of the herd. (A) Stacked bar graph showing infectivity of different tissues from MM and ML genotypes at PrP codon 132. Red bars indicate the number of animals that are negative for CWD, while the blue bars indicate CWD-positive animals. (B) ROC curves of RAMALT RT-QuIC assay compared against other IHC tests. An AUC >90% indicates high accuracy. Among different IHC's, IHC of RAMALT has highest AUC which is 93.1%, indicating RAMALT RT-QuIC assay results were comparable to RAMALT IHC results.
TABLE 2.
Correlation between various tests.
| Variable | By variable | Correlation | Lower 95% | Upper 95% |
|---|---|---|---|---|
| RAMALT RT-QuIC | IHC of RAMALT | 0.8144 | 0.7156 | 0.8812 |
| IHC of obex | IHC of RAMALT | 0.7443 | 0.6162 | 0.8341 |
| IHC of obex | RAMALT RT-QuIC | 0.5764 | 0.3934 | 0.7155 |
| IHC of MRPLN | IHC of RAMALT | 0.5886 | 0.4089 | 0.7244 |
| IHC of MRPLN | RAMALT RT-QuIC | 0.5054 | 0.3053 | 0.6628 |
| IHC of MRPLN | IHC of obex | 0.6922 | 0.5447 | 0.7981 |
Correlation between different diagnostic tests with upper and lower 95% confidence intervals. When comparing all detection procedures, the outcome of the RT-QuIC analysis of RAMALT was highly correlated to those of IHC results demonstrating the efficacy of RT-QuIC assay for CWD detection in RAMALT samples.
Discussion
The lack of an antemortem diagnostic test for CWD is a concerning problem. CWD's widespread nature and very long incubation period warrants the pressing need for a robust antemortem test for early diagnosis, which would reduce much of the uncertainty plaguing modern herd management. Recently, the RT-QuIC assay has been experimentally used for diagnosing Creutzfeldt–Jakob disease (CJD),36 scrapie,37 and CWD.38 Very recently, we have reported that the RT-QuIC assay can be readily adopted for a rapid detection of prion infection using an ex vivo brain culture model.39 Though the RT-QuIC assay was developed to rapidly diagnose misfolded prions from human samples, the seed used has been mostly limited to post-mortem samples like brain tissue. Although attempts were made to detect PrPD in noninvasive samples from biofluids like blood or saliva, the presence of inhibitors that interfere with aggregation limits their utility as diagnostic samples in RT-QuIC assays.40-42 More recently, nasal brushings from human patients with CJD showed the promising diagnostic application in the antemortem diagnosis of prion disease using RT-QuIC43 and PMCA.44 However, elk nasal brushings did not form a good seed in the RT-QuIC assay for early cases since cervid olfactory sensory neurons are not infected until later stages of CWD.34 In elk, abnormal prions accumulate in the brain, lymph nodes in the head, and spread to peripheral lymphoid tissues like RAMALT.45 In the current study, we have evaluated a highly sensitive RT-QuIC assay to detect CWD using RAMALT samples in a blinded fashion. IHC of RAMALT, obex, and MRPLN were also performed. We noticed a slight increase in the number of positives found in animals with MM polymorphism at codon 132 when compared to the ML genotype. This is consistent with previous reports showing higher CWD prevalence rate within this genotype.34,35 Based on the RT-QuIC analysis of RAMALT samples, we observed a strong correlation between the RT-QuIC assay and IHC of RAMALT sample results, which agree with earlier reports.33 Therefore, the RT-QuIC assay has the potential to be a useful and complementary diagnostic tool, especially given the accessibility of RAMALT samples for CWD diagnosis. Advantages of the assay, including the need for only a minor amount of tissue, and simplified sample preparation and reading of results, are making it a widely used and versatile technique. The low tissue requirements for the RT-QuIC assay also minimize the amount of trauma experienced by the animal during sample collection.46
However, unlike the comparison with RAMALT IHC, we observed a relatively low (∼50%) sensitivity for RAMALT RT-QuIC assay results when compared with IHC testing of obex and MRPLN samples. The low sensitivity is likely due to the variability of PrPD tissue distribution in elk (compared to other animals) together with the fact that we only used a small quantity of RAMALT tissues in the RT-QuIC assay. Therefore, if a selected sample of RAMALT tissue did not contain any PrPD, the RT-QuIC test would be falsely negative for that animal. This could potentially be avoided by making homogenate from all the available RAMALT tissue and further preparing the seed from this homogenate. Nonetheless, the higher sensitivity of RAMALT RT-QuIC assay when compared with IHC of RAMALT results demonstrated that the RT-QuIC assay is useful for detecting CWD from RAMALT samples. The comparison of RAMALT RT-QuIC assay results with the obex grades demonstrates that animals with higher obex grades (3 and above) are more likely to be positive in the RT-QuIC assay than those with lower obex grades, indicating that disease state is highly variable between animals.
In summary, we evaluated the diagnostic performance of the RT-QuIC assay for RAMALT samples from captive elk in a blinded manner. Comparing standard IHC and RT-QuIC assays of RAMALT samples revealed that these two are well correlated.
Further improvements in the RT-QuIC seeding assays for cervids could enhance optimal screening to achieve greater sensitivity and specificity. This could be achieved using other species of rPrP, by optimizing assay conditions, using mouse bioassays and other diagnostic tests like PMCA as well. For example, transgenic mice expressing bank vole PrP are highly susceptible to a wide variety of prion diseases and rPrP-expressing bank vole PrP (BVrPrP) was able to detect many prion diseases including human prions in the RT-QuIC assay.47 Similarly, BVrPrP and cervid rPrP could be important alternate substrates for diagnosing CWD, but this still remains to be tested. We are progressing towards developing an efficient and sensitive RT-QuIC assay for ante-mortem samples to screen herds for CWD status, which will assist in deploying rapid control and/or preventive measures to better manage CWD.
MATERIALS AND METHODS
Purification of Syrian Hamster Recombinant Prion Protein (SHrPrP)
SHrPrP encoding a 90–231 amino acid sequence was transformed, expressed in E. coli Rosetta cells (EMD Millipore) and purified using previously published methods.39,48,49 In brief, the SHr PrP-expressing Rosetta strain DE2 of E. coli was inoculated into lysogeny broth (LB) along with auto-induction system1 (EMD Millipore) and selection antibiotics. After the cultures were induced overnight, the bacteria were harvested after reaching optical density 3 at 600 nm. Inclusion bodies were isolated from these cell pellets by treating with 1X Bug Buster (EMD Millipore) followed by 0.1X Bug Buster and centrifugation at 13000 × g for 15 minutes. Then the inclusion bodies were denatured in 8 M guanidine hydrochloride (GuHCl) in 100 mM Na Phosphate at pH 8 using OMNI International Tissue Homogenizer and rotated for 50 min at RT. Next, the solution with denatured protein was centrifuged at 8000 × g for 5 min and the supernatants were added to Ni-NTA superflow resin (Qiagen) that had been equilibrated in denaturing buffer [100 mM sodium phosphate (pH 8.0), 10 mM Tris, 6 M GuHCl]. To allow rPrP to bind to beads, Ni-NTA superflow resin along with denatured rPrP was incubated on a rotator for 40 min at RT. Later, the beads were packed in an Akta XK26 column at RT. Subsequent steps were carried out under refrigerated conditions for fast protein liquid chromatography using Bio-Rad Duoflow. Refolding of rPrP was attained on the column by using a gradient from denaturing buffer to refolding buffer (100 mM NaPO4, 10 mM Tris [pH 8.0]). After refolding, rPrP was eluted from the column using a gradient from refolding buffer to elution buffer (100 mM NaPO4, 10 mM Tris [pH 8.0], 500 mM imidazole [pH 5.5]) (Fig. 1A, Top panel). Fractions from the middle of the peak were collected and mixed with pre-chilled dialysis buffer (10 mM NaPO4 [pH 5.5]) and dialyzed with three sets of 3.6 L of dialysis buffer.
Tissue Processing and RT-QuIC Method
After collection, samples were stored in a −80°C freezer until being tested. RAMALT samples measuring 0.3–0.5 cm were dissected from larger rectal biopsies collected 1 cm anterior to the mucocutaneous junction (Fig. 3A). This tissue was first homogenized to 10% (w/v) in PBS with the help of a Bullet Blender (Next Advance) using 0.5-mm Zirconium oxide beads in 1.5-mL screw cap tubes. Conditions for homogenization are three 5-min cycles at a speed setting of 10. Next, the homogenates were diluted to 2% (w/v) in PBS. The samples were then diluted in RT-QuIC dilution buffer (0.05% SDS in PBS).32 Next, 5 µl of 0.02% rectal biopsy homogenate was added as a seed to 95 μl of RT-QuIC reaction buffer (1X PBS supplemented with 220 mM NaCl, 10 μM EDTA, 10 μM Thioflavin T (ThT), and 0.1 mg/ml rPrP), in each well of a 96-well plate. We evaluated each rectal biopsy sample (3 replicates per sample) against a blank, as well as positive and negative controls in a 96-well flat-bottomed (Nunc) plates. Later, the sealed plates were analyzed using a Cytation 3 multi-mode plate reader (BioTek) at 42°C with shaking (807 rpm, double orbital) and rest cycles that alternated every minute for 24 h. Fluorescence readings of ThT were bottom-read at 450-nm excitation and 480-nm emission every 15 min using a gain setting of 1200. A sample was judged to be positive based on the previously reported criterion32 of when the mean of replicate fluorescence readings crossed the predetermined threshold (mean fluorescence plus 10 times the standard deviation of known negative controls). Each sample ran in quad-druplicate with known positive and negative controls. After completing data analysis and reporting results, the samples were unblinded to reveal the true diagnostic status. Then we calculated the AFR, which is the inverse of the threshold-crossing time for each dilution. We used the dilution with the highest AFR to seed all samples.40
Biophysical and Biochemical Analyses of rPrP Substrate
To confirm the structure of expressed truncated rPrP, we performed Nuclear Magnetic Resonance (NMR) spectroscopy and Circular Dichroism (CD) according to published methods.50,51 In brief, the CD spectra of the purified protein were recorded on a Jasco J-710 Spectropolarimeter using a thermostatted cuvette (Helma) with 1-mm path length. Measurements were carried out using a protein diluted in sodium phosphate buffer at 14°C, pH 7, yielding a final protein concentration of 8 µM. NMR spectra were recorded using a Bruker Avance 800 spectrometer. The concentration of protein was 1 mM in sodium phosphate buffer along with 5% D2O. Samples were prepared in special NMR tubes (Catalog# 897240–0000, Kimble Chase).
In addition to the biophysical characterization, we tested the purity of rPrP substrate using SDS-polyacrylamide gel electrophoresis using previously published methods.39 To visualize total proteins, Coomassie staining of the gels and Ponceau S staining of membranes were used. Western blotting against prion protein used monoclonal antibody 6D11 (Catalog# SIG-399810, Covance). In brief, proteins were loaded from different fractions and separated on 15% SDS-PAGE gels. After transferring the proteins onto nitrocellulose membranes, blots were first evaluated with Ponceau S stain (Catalog# P7170, Sigma-Aldrich) to locate the protein since Ponceau S does not interfere with polypeptides. The stain was removed by washing membranes in distilled water until the background was clean. To block nonspecific binding sites, nitrocellulose membranes were blocked with blocking buffer (Li-Cor) for 1 h. Later, the membranes were probed with the primary antibody 6D11 for 20 min at RT and washed 5 times with PBS containing 0.05% Tween 20. Then, secondary goat anti-mouse (1:20000, Alexa flour) was used to detect PrPC with the Odyssey IR imaging system.
IHC on the Brainstem, RLN, and RAMALT
The brainstem at the level of the obex, the MRPLN and RAMALT were fixed in 10% buffered formalin, processed, sectioned and stained for IHC using the mouse monoclonal primary anti-prion antibody F99/97.6.1 (Anti-Prion 99, Ventana Medical Systems) on an automated staining system (Ventana Medical Systems). Positive and negative scrapie ovine control tissues were included in each staining run and individual tissues were diagnosed as positive or not detected based on the presence or absence of characteristic CWD chromogen deposits in the tissue sections. RAMALT samples were expected to have a minimum of six lymphoid follicles to be a representative sample of the rectal mucosa. A single RAMALT sample (animal 11) had fewer than six follicles and was scored as not detected/insufficient follicles (Fig. 3B).33
Obex scoring was used to estimate disease progression in animals whose IHC results for brainstem were positive following previously published scoring criteria.33,52 Briefly, an obex score of zero had no detectable positive staining in the obex. An obex score of one had limited staining within the vagal nuclei and progressively more staining in the obex equated to progressively higher scores. A maximum obex score of four was assigned when widespread positive staining occurred in both the gray and white matter throughout the entire section of the brainstem.
Statistical Analysis
Data analysis was performed using Prism 5.0 software (GraphPad). Raw data were analyzed using one-way ANOVA, and then Tukey's post-test was performed to compare different dilutions. Differences with p ≤ 0.05 were considered significantly different, and asterisks were assigned as follows: *p ≤ 0.05, **p < 0.01, and ***p < 0.001. ROC curves were drawn using R statistical software.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
A.G.K is a shareholder of PK Biosciences Corporation (Ames, IA), which is interested in developing diagnostic and therapeutic strategies for Parkinson's disease and other related neurodegenerative diseases. A.G.K. has no commercial interest in the present work. Other authors declare no potential conflicts of interest.
ACKNOWLEDGMENTS
We would like to thank Dr. Byron Caughey and his lab members for providing constructs and Dr. Brian Lee and CureFFI.org for help with rPrP purification. The W. Eugene and Linda Lloyd Endowed Chair are also acknowledged. We would also like to acknowledge Mr. Gary Zenitsky, Dr. Huajun Jin and Dr. Vellareddy Anantharam for help with manuscript preparation.
FUNDING
This work was supported by ISU Veterinary Diagnostic Lab, ISU Wildlife initiative, NIH grants ES019267 and ES026892.
REFERENCES
- 1.Benestad SL, Mitchell G, Simmons M, Ytrehus B, Vikoren T. First case of chronic wasting disease in Europe in a Norwegian free-ranging reindeer. Vet Res. 2016;47:88. doi: 10.1186/s13567-016-0375-4. PMID:27641251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baeten LA, Powers BE, Jewell JE, Spraker TR, Miller MW. A natural case of chronic wasting disease in a free-ranging moose (Alces alces shirasi). J Wildl Dis. 2007;43:309–14. doi: 10.7589/0090-3558-43.2.309. PMID:17495319. [DOI] [PubMed] [Google Scholar]
- 3.Williams ES. Chronic wasting disease. Vet Pathol. 2005;42:530–49. doi: 10.1354/vp.42-5-530. [DOI] [PubMed] [Google Scholar]
- 4.Williams ES, Young S. Chronic wasting disease of captive mule deer: A spongiform encephalopathy. J Wildl Dis. 1980;16:89–98. doi: 10.7589/0090-3558-16.1.89. [DOI] [PubMed] [Google Scholar]
- 5.Nalls AV, McNulty E, Powers J, Seelig DM, Hoover C, Haley NJ, Hayes-Klug J, Anderson K, Stewart P, Goldmann W, et al.. Mother to offspring transmission of chronic wasting disease in reeves' muntjac deer. PLoS One. 2013;8:e71844. doi: 10.1371/journal.pone.0071844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore SJ, West Greenlee MH, Kondru N, Manne S, Smith JD, Kunkle RA, Kanthasamy A, Greenlee JJ. Experimental transmission of the chronic wasting disease agent to swine after oral or intracranial inoculation. J Virol. 2017;91(19):e00926–17. doi: 10.1128/JVI.00926-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams ES, Young S. Spongiform encephalopathy of Rocky Mountain elk. J Wildl Dis. 1982;18:465–71. doi: 10.7589/0090-3558-18.4.465. [DOI] [PubMed] [Google Scholar]
- 8.Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–83. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Z, Xiao G. Conformational conversion of prion protein in prion diseases. Acta Biochim Biophys Sin (Shanghai). 2013;45:465–76. doi: 10.1093/abbs/gmt027. [DOI] [PubMed] [Google Scholar]
- 10.Choi CJ, Anantharam V, Saetveit NJ, Houk RS, Kanthasamy A, Kanthasamy AG. Normal cellular prion protein protects against manganese-induced oxidative stress and apoptotic cell death. Toxicol Sci. 2007;98:495–509. doi: 10.1093/toxsci/kfm099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi CJ, Anantharam V, Martin DP, Nicholson EM, Richt JA, Kanthasamy A, Kanthasamy AG. Manganese upregulates cellular prion protein and contributes to altered stabilization and proteolysis: Relevance to role of metals in pathogenesis of prion disease. Toxicol Sci. 2010;115:535–46. doi: 10.1093/toxsci/kfq049. PMID:20176619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harischandra DS, Kondru N, Martin DP, Kanthasamy A, Jin H, Anantharam V, Kanthasamy AG. Role of proteolytic activation of protein kinase Cdelta in the pathogenesis of prion disease. Prion. 2014;8:143–53. doi: 10.4161/pri.28369. PMID:24576946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anantharam V, Kanthasamy A, Choi CJ, Martin DP, Latchoumycandane C, Richt JA, Kanthasamy AG. Opposing roles of prion protein in oxidative stress- and ER stress-induced apoptotic signaling. Free Radic Biol Med. 2008;45:1530–41. doi: 10.1016/j.freeradbiomed.2008.08.028. PMID:18835352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams ES, Young S. Spongiform encephalopathies in Cervidae. Rev Sci Tech. 1992;11:551–67. doi: 10.20506/rst.11.2.611. PMID:1617203. [DOI] [PubMed] [Google Scholar]
- 15.Perrott MR, Sigurdson CJ, Mason GL, Hoover EA. Evidence for distinct chronic wasting disease (CWD) strains in experimental CWD in ferrets. J Gen Virol. 2012;93:212–21. doi: 10.1099/vir.0.035006-0. PMID:21918005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orru CD, Wilham JM, Raymond LD, Kuhn F, Schroeder B, Raeber AJ, Caughey B. Prion disease blood test using immunoprecipitation and improved quaking-induced conversion. MBio. 2011;2:e00078–11. doi: 10.1128/mBio.00078-11. PMID:21558432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson DM, Manca M, Haley NJ, Denkers ND, Nalls AV, Mathiason CK, Caughey B, Hoover EA. Rapid antemortem detection of CWD prions in deer saliva. PLoS One. 2013;8:e74377. doi: 10.1371/journal.pone.0074377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haley NJ, Mathiason CK, Zabel MD, Telling GC, Hoover EA. Detection of sub-clinical CWD infection in conventional test-negative deer long after oral exposure to urine and feces from CWD+ deer. PLoS One. 2009;4:e7990. doi: 10.1371/journal.pone.0007990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nichols TA, Pulford B, Wyckoff AC, Meyerett C, Michel B, Gertig K, Hoover EA, Jewell JE, Telling GC, Zabel MD. Detection of protease-resistant cervid prion protein in water from a CWD-endemic area. Prion. 2009;3:171–83. doi: 10.4161/pri.3.3.9819. PMID:19823039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keane D, Barr D, Osborn R, Langenberg J, O'Rourke K, Schneider D, Bochsler P. Validation of use of rectoanal mucosa-associated lymphoid tissue for immunohistochemical diagnosis of chronic wasting disease in white-tailed deer (Odocoileus virginianus). J Clin Microbiol. 2009;47:1412–7. doi: 10.1128/JCM.02209-08. PMID:19261781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keane DP, Barr DJ, Bochsler PN, Hall SM, Gidlewski T, O'Rourke KI, Spraker TR, Samuel MD. Chronic wasting disease in a Wisconsin white-tailed deer farm. J Vet Diagn Invest. 2008;20:698–703. doi: 10.1177/104063870802000534. [DOI] [PubMed] [Google Scholar]
- 22.Balachandran A, Harrington NP, Algire J, Soutyrine A, Spraker TR, Jeffrey M, González L, O'Rourke KI. Experimental oral transmission of chronic wasting disease to red deer (Cervus elaphus elaphus): Early detection and late stage distribution of protease-resistant prion protein. Can Vet J. 2010;51:169–78. PMID:20436863. [PMC free article] [PubMed] [Google Scholar]
- 23.Jennelle CS, Henaux V, Wasserberg G, Thiagarajan B, Rolley RE, Samuel MD. Transmission of chronic wasting disease in Wisconsin white-tailed deer: Implications for disease spread and management. PLoS One. 2014;9:e91043. doi: 10.1371/journal.pone.0091043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–3. doi: 10.1038/35081095. PMID:11459061. [DOI] [PubMed] [Google Scholar]
- 25.Atarashi R, Moore RA, Sim VL, Hughson AG, Dorward DW, Onwubiko HA, Priola SA, Caughey B. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat Methods. 2007;4:645–50. doi: 10.1038/nmeth1066. PMID:17643109. [DOI] [PubMed] [Google Scholar]
- 26.Wild MA, Spraker TR, Sigurdson CJ, O'Rourke KI, Miller MW. Preclinical diagnosis of chronic wasting disease in captive mule deer (Odocoileus hemionus) and white-tailed deer (Odocoileus virginianus) using tonsillar biopsy. J Gen Virol. 2002;83:2629–34. doi: 10.1099/0022-1317-83-10-2629. [DOI] [PubMed] [Google Scholar]
- 27.Wolfe LL, Spraker TR, Gonzalez L, Dagleish MP, Sirochman TM, Brown JC, Jeffrey M, Miller MW. PrPCWD in rectal lymphoid tissue of deer (Odocoileus spp.). J Gen Virol. 2007;88:2078–82. doi: 10.1099/vir.0.82342-0. PMID:17554043. [DOI] [PubMed] [Google Scholar]
- 28.Spraker TR, VerCauteren KC, Gidlewski T, Schneider DA, Munger R, Balachandran A, O'Rourke KI. Antemortem detection of PrPCWD in preclinical, ranch-raised Rocky Mountain elk (Cevvus elaphus nelsoni) by biopsy of the rectal mucosa. J Vet Diagn Invest. 2009;21:15–24 [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez L, Jeffrey M, Siso S, Martin S, Bellworthy SJ, Stack MJ, Chaplin MJ, Davis L, Dagleish MP, Reid HW. Diagnosis of preclinical scrapie in samples of rectal mucosa. Vet Rec. 2005;156:846–7. doi: 10.1136/vr.156.26.846-b. [DOI] [PubMed] [Google Scholar]
- 30.Espenes A, Press CM, Landsverk T, Tranulis MA, Aleksandersen M, Gunnes G, Benestad SL, Fuglestveit R, Ulvund MJ. Detection of PrP(Sc) in rectal biopsy and necropsy samples from sheep with experimental scrapie. J Comp Pathol. 2006;134:115–25. doi: 10.1016/j.jcpa.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Wilham JM, Orru CD, Bessen RA, Atarashi R, Sano K, Race B, Meade-White KD, Taubner LM, Timmes A, Caughey B. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 2010;6:e1001217. doi: 10.1371/journal.ppat.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haley NJ, Carver S, Hoon-Hanks LL, Henderson DM, Davenport KA, Bunting E, Gray S, Trindle B, Galeota J, LeVan I, et al.. Detection of chronic wasting disease in the lymph nodes of free-ranging cervids by real-time quaking-induced conversion. J Clin Microbiol. 2014;52:3237–43. doi: 10.1128/JCM.01258-14. PMID:24958799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomsen BV, Schneider DA, O'Rourke KI, Gidlewski T, McLane J, Allen RW, McIsaac AA, Mitchell GB, Keane DP, Spraker TR, et al.. Diagnostic accuracy of rectal mucosa biopsy testing for chronic wasting disease within white-tailed deer (Odocoileus virginianus) herds in North America: Effects of age, sex, polymorphism at PRNP codon 96, and disease progression. J Vet Diagn Invest. 2012;24:878–87. doi: 10.1177/1040638712453582. [DOI] [PubMed] [Google Scholar]
- 34.Haley NJ, Siepker C, Hoon-Hanks LL, Mitchell G, Walter WD, Manca M, Monello RJ, Powers JG, Wild MA, Hoover EA, et al.. Seeded amplification of chronic wasting disease prions in nasal brushings and recto-anal mucosa-associated lymphoid tissues from elk by real-time quaking-induced conversion. J Clin Microbiol. 2016;54:1117–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamir AN, Gidlewski T, Spraker TR, Miller JM, Creekmore L, Crocheck M, Cline T, O'Rourke KI. Preliminary observations of genetic susceptibility of elk (Cervus elaphus nelsoni) to chronic wasting disease by experimental oral inoculation. J Vet Diagn Invest. 2006;18:110–4. doi: 10.1177/104063870601800118. [DOI] [PubMed] [Google Scholar]
- 36.Orru CD, Groveman BR, Hughson AG, Zanusso G, Coulthart MB, Caughey B. Rapid and sensitive RT-QuIC detection of human Creutzfeldt-Jakob disease using cerebrospinal fluid. MBio. 2015;6(1):e02451–14. doi: 10.1128/mBio.02451-14. PMID:25604790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dassanayake RP, Orru CD, Hughson AG, Caughey B, Graca T, Zhuang D, Madsen-Bouterse SA, Knowles DP, Schneider DA. Sensitive and specific detection of classical scrapie prions in the brains of goats by real-time quaking-induced conversion. J Gen Virol. 2016;97:803–12. doi: 10.1099/jgv.0.000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng YC, Hannaoui S, John TR, Dudas S, Czub S, Gilch S. Early and non-invasive detection of chronic wasting disease prions in elk feces by real-time quaking induced conversion. PLoS One. 2016;11:e0166187. doi: 10.1371/journal.pone.0166187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kondru N, Manne S, Greenlee J, West Greenlee H, Anantharam V, Halbur P, Kanthasamy A, Kanthasamy A. Integrated organotypic slice cultures and RT-QuIC (OSCAR) assay: Implications for translational discovery in protein misfolding diseases. Sci Rep. 2017;7:43155. doi: 10.1038/srep43155. PMID:28233859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henderson DM, Davenport KA, Haley NJ, Denkers ND, Mathiason CK, Hoover EA. Quantitative assessment of prion infectivity in tissues and body fluids by real-time quaking-induced conversion. J Gen Virol. 2015;96:210–9. doi: 10.1099/vir.0.069906-0. PMID:25304654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vieira TC, Cordeiro Y, Caughey B, Silva JL. Heparin binding confers prion stability and impairs its aggregation. FASEB J. 2014;28:2667–76. doi: 10.1096/fj.13-246777. PMID:24648544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castilla J, Saá P, Soto C. Detection of prions in blood. Nat Med. 2005;11:982–5. PMID:16127436. [DOI] [PubMed] [Google Scholar]
- 43.Orru CD, Bongianni M, Tonoli G, Ferrari S, Hughson AG, Groveman BR, Fiorini M, Pocchiari M, Monaco S, Caughey B, et al.. A test for Creutzfeldt-Jakob disease using nasal brushings. N Engl J Med. 2014;371:519–29. doi: 10.1056/NEJMoa1315200. PMID:25099576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moda F, Gambetti P, Notari S, Concha-Marambio L, Catania M, Park KW, Maderna E, Suardi S, Haïk S, Brandel JP, et al.. Prions in the urine of patients with variant Creutzfeldt-Jakob disease. N Engl J Med. 2014;371:530–9. doi: 10.1056/NEJMoa1404401. PMID:25099577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sigurdson CJ, Aguzzi A. Chronic wasting disease. Biochim Biophys Acta. 2007;1772:610–8. PMID:17223321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monello RJ, Powers JG, Hobbs NT, Spraker TR, O'Rourke KI, Wild MA. Efficacy of antemortem rectal biopsies to diagnose and estimate prevalence of chronic wasting disease in free-ranging cow elk (Cervus elaphus nelsoni). J Wildl Dis. 2013;49:270–8. doi: 10.7589/2011-12-362. PMID:23568902. [DOI] [PubMed] [Google Scholar]
- 47.Orru CD, Groveman BR, Raymond LD, Hughson AG, Nonno R, Zou W, Ghetti B, Gambetti P, Caughey B. Bank vole prion protein as an apparently universal substrate for RT-QuIC-based detection and discrimination of prion strains. PLoS Pathog. 2015;11:e1004983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilham JM, Orrú CD, Bessen RA, Atarashi R, Sano K, Race B, Meade-White KD, Taubner LM, Timmes A, Caughey B. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 2010;6:e1001217. doi: 10.1371/journal.ppat.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.West Greenlee MH, Lind M, Kokemuller R, Mammadova N, Kondru N, Manne S, Smith J, Kanthasamy A, Greenlee J. Temporal resolution of misfolded prion protein transport, accumulation, glial activation, and neuronal death in the retinas of mice inoculated with scrapie. Am J Pathol. 2016;186:2302–9. doi: 10.1016/j.ajpath.2016.05.018. PMID:27521336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Donne DG, Viles JH, Groth D, Mehlhorn I, James TL, Cohen FE, Prusiner SB, Wright PE, Dyson HJ.. Structure of the recombinant full-length hamster prion protein PrP(29–231): The N terminus is highly flexible. Proc Natl Acad Sci U S A. 1997;94:13452–7. doi: 10.1073/pnas.94.25.13452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zahn R, von Schroetter C, Wüthrich K. Human prion proteins expressed in Escherichia coli and purified by high-affinity column refolding. FEBS Lett. 1997;417:400–4. doi: 10.1016/S0014-5793(97)01330-6. PMID:9409760. [DOI] [PubMed] [Google Scholar]
- 52.Spraker TR, Balachandran A, Zhuang D, O'Rourke KI. Variable patterns of distribution of PrP(CWD) in the obex and cranial lymphoid tissues of Rocky Mountain elk (Cervus elaphus nelsoni) with subclinical chronic wasting disease. Vet Rec. 2004;155:295–302. doi: 10.1136/vr.155.10.295. PMID:15478500. [DOI] [PubMed] [Google Scholar]


