Abstract
A Klebsiella pneumoniae clinical strain, named SCKP83, was isolated and found to be resistant to colistin thanks to the presence plasmid-borne colistin resistant gene mcr-1. The strain was subjected to whole genome sequencing and conjugation experiments. The subsequent analysis indicated that the strain belongs to ST15 and the capsular type K41. In SCKP83, mcr-1 was carried by a 97.4-kb non-self-transmissible plasmid, a 90.9-kb region of which was predicted as an intact phage. This phage was 47.79% GC content, encoded 105 proteins and contained three tRNAs. mcr-1 was located downstream of two copies of the insertion sequence ISApl1 (one complete and one truncated) and was inserted in the ant1 gene, which encodes a putative antirepressor for antagonizing C1 repression, in this phage. The phage is highly similar to phage P7 (77% coverage and 98% identity) from Escherichia coli. Several similar mcr-1-carrying plasmids have been found in E. coli at various locations in China, suggesting that these phage-like plasmids have circulated in China. The findings in this study suggest that the P7 phage-like plasmids are not restricted to E. coli and may represent new vehicles to mediate the inter-species spread of mcr-1.
Keywords: colistin, resistance, phagemid, plasmids, Klebsiella pneumoniae
Introduction
Klebsiella pneumoniae is a major pathogen causing a variety of infections in humans. Colistin is the last resort antimicrobial agent to treat infections caused by K. pneumoniae including those with resistance to carbapenems. However, colistin-resistant K. pneumoniae have emerged worldwide (Olaitan et al., 2014a). A few mechanisms including both chromosomal and plasmid-borne ones have been identified to be responsible for resistance to colistin in K. pneumoniae (Olaitan et al., 2014b). Plasmid-borne colistin resistance genes including mcr-1 (Liu et al., 2016), mcr-2 (Xavier et al., 2016), and mcr-3 (Yin et al., 2017) have been found recently. In particular, mcr-1 has been identified in various species of the Enterobacteriaceae in many countries (Poirel et al., 2017).
Bacteriophages (phages) are viruses able to infect and replicate within bacteria. Phages mediate the transfer of genetic components between bacteria via transduction. Phages may have a lytic cycle or a lysogenic cycle or both. In the lytic cycle, phage genomes are replicated and are assembled into particles, which cause cell lysis and are then released. In the lysogenic cycle, phage genomes integrate into the chromosome of host bacterial cells to exist in a latent or dormant state without causing cell lysis (Feiner et al., 2015). The structure of phages typically consists of a protein head that encapsulates a DNA or RNA genome and a tail that attacks the bacterial host (Wurtz, 1992). Phage genomes vary remarkbly in form and size but usually encode products for host takeover, replication, virion assembly, or lysis (Black and Thomas, 2012). Some phages may integrate into plasmids and can therefore be transferred by the host plasmid (Oliver et al., 2005; Shin and Ko, 2015).
mcr-1 is commonly carried by plasmids of the IncI2 or IncX4 replicon type and has also been found on IncF, IncHI2, or IncP plasmids (Poirel et al., 2017). We have found a plasmid carrying mcr-1 and phage P7-like sequences, which is reported here.
Methods
Bacterial strain
K. pneumoniae strain SCKP83 was recovered from a sputum sample of a 90-year-old male patient with severe pneumonia in February 2017 in China, who did not receive colistin before. Species identification was performed using Vitek II (bioMérieux, Marcy-l′Étoile, France) and MALTI-TOF (Bruker, Billerica, MA, USA). In vitro susceptibility of colistin was performed using the broth dilution method of the Clinical Laboratory Standards Institute (CLSI) (CLSI, 2017) and breakpoints of colistin defined by EUCAST (http://www.eucast.org/) were applied. The presence of plasmid-borne colistin resistant genes mcr-1, mcr-2, and mcr-3 was screened by PCR as described previously (Xavier et al., 2016; Zhao and Zong, 2016; Yin et al., 2017).
Whole genome sequencing and analysis
The strain was subjected to whole genome sequencing. Genomic DNA was prepared using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) and whole genome sequencing was performed using the HiSeq X10 Sequencer (Illumina, San Diego, CA). The coverage was approximately 300 × coverage, which was calculated based on the estimated genome size and the average output of the sequencer. Reads were trimmed using Trimmomatic (version 0.36) (Bolger et al., 2014) and were then assembled to contigs using SPAdes (version 3.11) (Bankevich et al., 2012) with careful mode turned on. Sequence type and capsular type were determined using the genomic sequence to query the multi-locus sequence typing and wzi allele databases of K. pneumoniae available at http://bigsdb.pasteur.fr/klebsiella/klebsiella.html. Antimicrobial resistance genes were identified from genome sequences using the ABRicate program (https://github.com/tseemann/abricate) and ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/). The plasmid carrying mcr-1, designated pMCR_SCKP-LL83, was circularized using PCR and Sanger sequencing to fill in gaps between contigs. Plasmid replicon was determined using the PlasmidFinder tool at http://genomicepidemiology.org/. Similar plasmids were retrieved from the GenBank and pairwise comparisons were preformed using BLASTn alignment (Altschul et al., 1990) and BRIG (Alikhan et al., 2011). The presence of phages was screened using PHASTER (http://phaster.ca/) (Arndt et al., 2016). tRNAs were screened using tRNA-SE (http://lowelab.ucsc.edu/tRNAscan-SE/) (Lowe and Chan, 2016).
Nucleotide sequence accession numbers
Draft whole-genome sequence of strain SCKP83 has been deposited into GenBank under the accession number NOKM00000000. Short reads of the whole-genome sequence of strain SCKP83 has been deposited into Short Reads Achieve under the accession number SRP099296. The complete sequences of pMCR_SCKP-LL83 has been deposited into GenBank under the accession numbery MF510496.
Conjugation and transformation experiments
Conjugation experiments were performed using both broth- and filter-based methods as described previously (Coque et al., 2002; Novais et al., 2006; Valenzuela et al., 2007). The azide-resistant Escherichia coli strain J53 was used as the recipient and 2 μg/ml colistin plus 150 μg/ml sodium azide were used for selecting transconjugants. Plasmids were prepared from strain SCKP83 using alkaline lysis (Sambrook and Russell, 2001) and were used for electroporation. Electroporation was conducted using a Gene Pulser (Bio-Rad, Hercules, CA, USA) with an electrical pulse of 25 μF capacitance, 2.5 kV and 200 Ω sample resistance. E. coli strain DH5α and a colistin-susceptible K. pneumoniae strain 020018 were used as recipient strains. Potential transformants were selected on agar plates containing 2 μg/ml colistin.
Induction of bacteriophage
To determine the nature of pMCR_SCKP-LL83, we performed the induction assay using ultraviolet ray and mitomycin C as described previously (Mitsui et al., 1973; Raya and H'bert, 2009). Briefly, for UV induction, 1 ml culture of strain SCKP-LL83 in the exponential phase was harvested and resuspended in 0.05 M phosphate buffer (pH 6.8). The suspension was adjusted to the 0.5 McFarland turbidity. Six aliquots of 150 μl were spotted on a 9 cm Petri dish and irradiated by a germicidal UV lamp at a distance of 100 cm. The drops were collected at 10, 20, 30, 60, 90, and 120 s serially, each of which was then incubated with 1 ml LB broth under 37°C in dark for 3–4 h. Lysis was observed by naked eyes. For mitomycin C induction, 100 ml cultures of strain SCKP-LL83 were added with mitomycin C to a final concentration of 0.1, 1, 10, 20, and 40 μg/ml and were incubated under 37°C with shaking. Aliquots (1 ml) were sampled at 2, 4, 12, and 24 h. The cultures were filtrated through 0.22 μm polyethersulfone membranes (Merck Millipore, Billerica, MA, USA) and the membranes were used for the plaque formation test, which was carried out via the agar overlay method (Kropinski et al., 2009). All of the tests were performed in triplicate.
Assay for replication module
The replication initiation protein-encoding gene repB and its replication origin sequence (ori) of pMCR_SCKP-LL83 were amplified with self-designed primers OriF (CGGAATTCGAAATGGGATCAACATTGACTATACG) and OriR (CGGAATTCATCAATACCACTGCTTGATGAGA; EcoRI sites are underlined). The amplicons were cloned onto the vector pKC1139, which has a temperature sensitive origin oriT and cannot replicate at temperatures higher than 30°C. The ligated vectors were transformed into E. coli DH5α and the transformants were screened by apramycin (100 μg/ml) at 37°C. The presence of repB and ori in transformants were confirmed by PCR with M13 (-21) Forward and M13-R primers binding to the clone vector and Sanger sequencing.
Results and discussion
Strain SCKP83 was resistant to colistin (MIC, 8 μg/ml) and had mcr-1 but no mcr-2 and mcr-3 genes. Whole genome sequencing of strain SCKP83 generated 5,247,124 clean reads, which were then assembled to 119 contigs (89 >1,000 bp) with a 50.38% GC content. Strain SCKP83 belonged to ST15, which is a relative common type of K. pneumoniae seen in China (Zhang et al., 2017b). The capsular type of strain SCKP83 was K41.
mcr-1 was carried by a 97.4 kb plasmid, pMCR_SCKP-LL83, which did not carry any additional known antimicrobial resistance genes. Despite repeated attempts, no colistin resistant transconjugants were obtained, suggesting that pMCR_SCKP-LL83 is not self-transmissible. In addition, the transformation of this plasmid into E. coli strain DH5α and a colistin-susceptible K. pneumoniae strain was unsuccessful. This suggests that this plasmid may be strain-specific or its transformation occurs at a low frequency, which could not be detected in our experiments. pMCR_SCKP-LL83 had a single pO111 plasmid replicon. Transformants containing repB and its ori were obtained. The presence of repB and ori allows the temperature sensitive vector pKC1139 to replicate at 37°C, suggesting that the replication module of pMCR_SCKP-LL83 indeed leads to the replication of this plasmid.
On pMCR_SCKP-LL83, mcr-1 was located downstream of a complete insertion sequence ISApl1. The phosphoesterase-encoding pho gene that is always located downstream of mcr-1 was truncated at its 3′-end with only 38 bp out of the 747-bp gene remaining. Surprisingly, immediate upstream of the complete ISApl1 (1,070 bp in length) lies another ISApl1 that is truncated at its 5′-end with the presence of 223 bp including an intact right-hand inverted repeat (IRR) (Figure 1). When we artificially subtract the ISApl1Δ-ISApl1-mcr-1-phoΔ region from pMCR_SCKP-LL83, the remaining artificially-joining sequence perfectly matched the ant1 gene, which encodes a putative antirepressor for antagonizing C1 repression by formation of Ant1/Ant2/C1 complex. It therefore becomes evident that the ISApl1Δ-ISApl1-mcr-1-phoΔ structure is inserted into ant1. It has been found that a single copy of ISApl1 is able to mobilize mcr-1 and pho together with itself (Li et al., 2017; Zhao et al., 2017). The insertion of ISApl1 would generate 2-bp direct target repeats (DR). However, no 2-bp DRs were present flanking the ISApl1Δ-ISApl1-mcr-1-phoΔ structure, suggesting that the formation of such a complex structure was not directly due to the insertion mediated by ISApl1. The mechanism responsible for generating the ISApl1Δ-ISApl1-mcr-1-phoΔ structure remains unclear but might have involved recombination.
Figure 1.
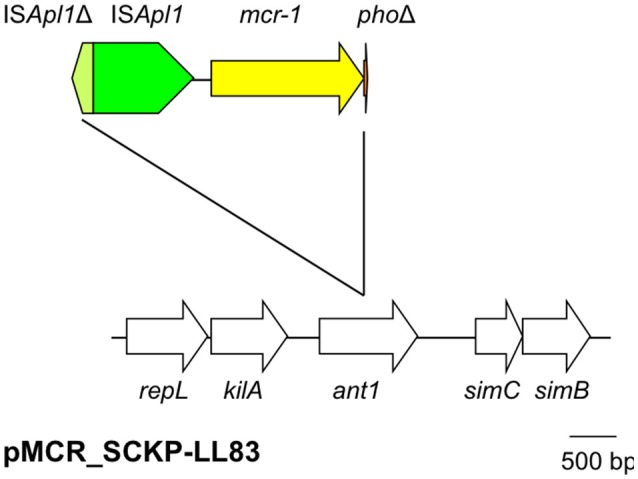
The genetic context of mcr-1 on pMCR_SCKP-LL83. The ISApl1Δ-ISApl1-mcr-1-phoΔ structure is inserted into ant1 but without the 2-bp DR characteristic of the insertion of ISApl1. The two ISApl1 are at contrary oppositions. Δ refers to truncated genes or elements. ant1 encodes a putative antirepressor. The phage genes surrounding ant1 include repL (encoding replication protein), kliA (encoding a putative host killing protein), simB and simC (both encoding proteins for host immunity).
A 90.9-kb region of the 97.4-kb pMCR_SCKP-LL83 was predicted as an intact phage. Neither the appearance of lysis nor the formation of plaques was observed in the UV induction. In mitomycin C induction, no plaques were formed at the tested concentrations and intervals. These results suggest that pMCR_SCKP-LL83 was indeed a plasmid. Nonetheless, the phage region on pMCR_SCKP-LL83 had 47.79% GC content, encoded 105 proteins and contained three tRNAs, i.e., tRNA-Asn, tRNA-Thr, and tRNA-Met (Table 1). pMCR_SCKP-LL83 is highly similar (72% coverage and 98% identity) to the 101.7-kb Enterobacteria phage P7 (GenBank accession no. AF503408). Phage P7 (previously called as φamp) was isolated from E. coli of human fecal flora (Smith, 1972) and exists as a nonintegrated autonomous circular plasmid that constitutes a unique compatibility group (Hedges et al., 1975). Compared with P7, pMCR_SCKP-LL83 did not have the blaTEM−1-carrying transposon Tn3, the type I restriction-modification system EcoP7, a 4-kb invertible C-segment and a few genes, most of which encode proteins of unknown function (Table S1 in the Supplementary file and Figure 2). C-segment contains several genes encoding phage tail fibers and also determines the host specificity of the phage (Iida, 1984). In contrast, pMCR_SCKP-LL83 had a few extra genes including an unnamed type I restriction-modification system, mcr-1 and a 5-kb putative invertible C-segment (Table S1), which is highly similar (92% coverage and 99% identity) to the multiple DNA inversion region min on plasmid p15B of E. coli 15T (Sandmeier et al., 1991).
Table 1.
Features of pMCR_SCKP-LL83.
| Featurea | Position (start–end) | Function |
|---|---|---|
| 0001 | 356–1912 | Type I restriction-modification system subunit M |
| 0002 | 1909–3114 | Restriction endonuclease subunit S |
| 0003 | 3235–6351 | Type I restriction enzyme EcoR124II R protein |
| 0004 | 6616–7122 | 3′-Phosphatase, 5′-polynucleotide kinase |
| 0005/pmgS | 7195–8457 | Putative morphogenetic protein |
| 0006 | 8459–8677 | Hypothetical protein |
| 0007 | 8759–9460 | Hypothetical protein |
| 0008/pphA | 9457–10134 | Serine/Threonine protein phosphatase |
| 0009/pmgP | 10131–10757 | Putative morphogenetic protein |
| 0010 | 11259–11414 | Hypothetical protein |
| 0011/pmgM | 11481–12059 | Putative morphogenetic function protein |
| 0012 | 12062–12307 | Putative morphogenetic protein |
| 0013 | 12571–12831 | Baseplate protein |
| 0014 | 12841–14058 | Tail protein |
| 0015 | 14062–14790 | Tail protein |
| 0016 | 14777–15562 | Hypothetical protein |
| 0017 | 15564–16580 | Tail length tape measure protein |
| 0018 | 16573–17205 | Putative baseplate protein |
| 0019 | 17252–18250 | Hypothetical protein |
| 0018/dnaB | 18250–19614 | Replicative DNA helicase |
| 0021 | 19900–19975 | tRNA-Met |
| 0024/tciA | 20250–20675 | Putative tellurite or colicin resistance protein |
| 0025 | 21187–21360 | Hypothetical protein |
| 0026 | 21603–21678 | tRNA-Thr |
| 0027 | 21681–21756 | tRNA-Asn |
| 0028/dmt | 22429–24693 | DNA adenine methylase family protein |
| 0029/rdgC | 24690–25595 | Recombination-associated protein RdgC |
| 0030 | 25588–25872 | Hypothetical protein |
| 0031 | 25857–26096 | Hypothetical protein |
| 0032 | 26335–27123 | Hypothetical protein |
| 0033 | 27163–27585 | Outer membrane lytic protein |
| 0034/upfB | 27763–28155 | Hypothetical protein |
| 0035 | 28048–28311 | Hypothetical protein |
| 0036/repA | 28491–29375 | Initiator replication family protein of pO111-like replicon |
| 0037 | 29668–30477 | Helicase |
| IS1294 | 32106–32205 | Insertion sequence |
| 0040/parA | 32334–33530 | Plasmid partition protein A |
| 0041/parB | 33547–34548 | Plasmid partition protein B |
| 0042 | 34774–36480 | Putative baseplate protein |
| 0043 | 36541–38130 | Hypothetical protein |
| 0044 | 38140–38955 | Tail tube protein |
| 0045/pmgG | 38991–39572 | Putative morphogenetic protein |
| 0046/bplB | 39584–40093 | Putative baseplate structural protein |
| 0047 | 40217–40423 | Hypothetical protein |
| 0048 | 40547–40792 | Hypothetical protein |
| 0049/repL | 40843–41652 | Replication protein |
| 0050/kilA | 41718–42518 | Putative host killing protein |
| 0051 | 42682–43587 | Hypothetical protein |
| 0052/mcr-1 | 43541–45166 | Colistin resistance |
| ISApl1 | 45353–46422 | Insertion sequence |
| ISApl1Δ | 46423–46645 | Insertion sequence, truncated |
| 0055 | 46580–46915 | Antirepressor protein |
| 0056 | 46912–47133 | Hypothetical protein |
| 0057/simB | 47561–48031 | Superimmunity linked function |
| 0058/simC | 48039–48818 | Superimmunity linked function |
| 0059/pmgC | 49028–49594 | putative morphogenetic protein |
| 0060/tubB | 49605–50216 | Major tail tube protein |
| 0061/pmgB | 50231–51112 | Putative morphogenetic protein |
| 0062 | 51194–54586 | Transglycosylase SLT domain protein |
| 0063/pmgA | 54586–54942 | Putative morphogenetic protein |
| 0064 | 54939–56372 | Putative baseplate structural protein |
| 0065 | 56372–57208 | Putative tail tube protein |
| 0066 | 57287–57721 | Putative tail fiber structure or assembly protein |
| 0067 | 57733–59214 | Hypothetical protein |
| 0068 | 59483–59728 | Hypothetical protein |
| 0069 | 59769–60206 | Hypothetical protein |
| 0070 | 60217–60645 | Hypothetical protein |
| 0071 | 60686–61159 | Hypothetical protein |
| 0072 | 61188–61646 | Hypothetical protein |
| 0073/tfaE | 62160–62771 | Prophage tail fiber assembly protein TfaE |
| 0074 | 62771–63229 | Hypothetical protein |
| 0075 | 63240–63683 | Hypothetical protein |
| 0076/pin | 63773–64345 | Site-specific recombinase |
| 0077 | 64781–65044 | Hypothetical protein |
| 0078/lydA | 65119–65448 | Lysis determining protein |
| 0079 | 65445–65888 | Lysis determining protein |
| 0080 | 65875–66477 | Hypothetical protein |
| 0081/darA | 66479–68398 | Hypothetical protein |
| 0082/ddrA | 68395–68760 | Hypothetical protein |
| 0083 | 68797–71760 | Hypothetical protein |
| 0084/hxr | 71750–72061 | Putative repressor protein Hxr |
| 0085/ompD | 72804–73916 | Outer membrane porin protein OmpD |
| 0086/ssb | 74150–74638 | Single-stranded DNA-binding protein |
| 0087/lys | 74808–75365 | Lysozyme |
| 0088 | 75657–76676 | Putative head processing protein |
| 0089 | 76669–78378 | Putative portal protein |
| 0090 | 78454–85221 | Putative DNA adenine methyltransferase |
| 0091 | 85255–85695 | Hypothetical protein |
| 0092 | 85692–85940 | Modulator protein |
| 0093 | 85982–87286 | Hypothetical protein |
| 0094 | 87343–87984 | Maturation control protein |
| 0095/ref | 88173–88733 | Recombination enhancement function protein |
| 0096 | 88981–89190 | Putative lysogeny establishment protein |
| 0097/cre | 89343–90374 | GST-loxP-cre recombinase fusion protein |
| 0098/cra | 90382–90603 | Putative Cre-associated regulatory protein |
| 0099 | 91208–91417 | C1 repressor inactivator |
| 0100 | 91528–92379 | Primary repressor of lytic function |
| 0101 | 92405–93889 | Putative large terminase protein |
| 102/pacA | 93889–95082 | Terminase A protein |
| 0103/lpa | 95169–95621 | Late promoter activating protein |
| 0104 | 95710–96753 | Hypothetical protein |
| 0105 | 96781–96960 | Hypothetical protein |
| 0106/doc | 96965–97345 | Toxin Doc |
| 0001 | 356–1912 | Type I restriction-modification system subunit M |
| 0002 | 1909–3114 | Restriction endonuclease subunit S |
| 0003 | 3235–6351 | Type I restriction enzyme EcoR124II R protein |
| 0004 | 6616–7122 | 3′-Phosphatase, 5′-polynucleotide kinase |
| 0005/pmgS | 7195–8457 | Putative morphogenetic protein |
| 0006 | 8459–8677 | Hypothetical protein |
| 0007 | 8759–9460 | Hypothetical protein |
| 0008/pphA | 9457–10134 | Serine/Threonine protein phosphatase |
| 0009/pmgP | 10131–10757 | Putative morphogenetic protein |
| 0010 | 11259–11414 | Hypothetical protein |
| 0011/pmgM | 11481–12059 | Putative morphogenetic function protein |
| 0012 | 12062–12307 | Putative morphogenetic protein |
| 0013 | 12571–12831 | Baseplate protein |
| 0014 | 12841–14058 | Tail protein |
| 0015 | 14062–14790 | Tail protein |
| 0016 | 14777–15562 | Hypothetical protein |
| 0017 | 15564–16580 | Tail length tape measure protein |
| 0018 | 16573–17205 | Putative baseplate protein |
| 0019 | 17252–18250 | Hypothetical protein |
| 0018/dnaB | 18250–19614 | Replicative DNA helicase |
| 0021 | 19900–19975 | tRNA-Met |
| 0024/tciA | 20250–20675 | Putative tellurite or colicin resistance protein |
| 0025 | 21187–21360 | Hypothetical protein |
| 0026 | 21603–21678 | tRNA-Thr |
| 0027 | 21681–21756 | tRNA-Asn |
| 0028/dmt | 22429–24693 | DNA adenine methylase family protein |
| 0029/rdgC | 24690–25595 | Recombination-associated protein RdgC |
| 0030 | 25588–25872 | Hypothetical protein |
| 0031 | 25857–26096 | Hypothetical protein |
| 0032 | 26335–27123 | Hypothetical protein |
| 0033 | 27163–27585 | Outer membrane lytic protein |
| 0034/upfB | 27763–28155 | Hypothetical protein |
| 0035 | 28048–28311 | Hypothetical protein |
| 0036/repA | 28491–29375 | Initiator replication family protein of pO111-like replicon |
| 0037 | 29668–30477 | Helicase |
| IS1294 | 32106–32205 | Insertion sequence |
| 0040/parA | 32334–33530 | Plasmid partition protein A |
| 0041/parB | 33547–34548 | Plasmid partition protein B |
| 0042 | 34774–36480 | Putative baseplate protein |
| 0043 | 36541–38130 | Hypothetical protein |
| 0044 | 38140–38955 | Tail tube protein |
| 0045/pmgG | 38991–39572 | Putative morphogenetic protein |
| 0046/bplB | 39584–40093 | Putative baseplate structural protein |
| 0047 | 40217–40423 | Hypothetical protein |
| 0048 | 40547–40792 | Hypothetical protein |
| 0049/repL | 40843–41652 | Replication protein |
| 0050/kilA | 41718–42518 | Putative host killing protein |
| 0051 | 42682–43587 | Hypothetical protein |
| 0052/mcr-1 | 43541–45166 | Colistin resistance |
| ISApl1 | 45353–46422 | Insertion sequence |
| ISApl1Δ | 46423–46645 | Insertion sequence, truncate |
| 0055 | 46580–46915 | Antirepressor protein |
| 0056 | 46912–47133 | Hypothetical protein |
| 0057/simB | 47561–48031 | Superimmunity linked function |
| 0058/simC | 48039–48818 | Superimmunity linked function |
| 0059/pmgC | 49028–49594 | Putative morphogenetic protein |
| 0060/tubB | 49605–50216 | Major tail tube protein |
| 0061/pmgB | 50231–51112 | Putative morphogenetic protein |
| 0062 | 51194–54586 | Transglycosylase SLT domain protein |
| 0063/pmgA | 54586–54942 | putative morphogenetic protein |
| 0064 | 54939–56372 | putative baseplate structural protein |
| 0065 | 56372–57208 | Putative tail tube protein |
| 0066 | 57287–57721 | Putative tail fiber structure or assembly protein |
| 0067 | 57733–59214 | Hypothetical protein |
| 0068 | 59483–59728 | Hypothetical protein |
| 0069 | 59769–60206 | Hypothetical protein |
| 0070 | 60217–60645 | Hypothetical protein |
| 0071 | 60686–61159 | Hypothetical protein |
| 0072 | 61188–61646 | Hypothetical protein |
| 0073/tfaE | 62160–62771 | Prophage tail fiber assembly protein TfaE |
| 0074 | 62771–63229 | Hypothetical protein |
| 0075 | 63240–63683 | Hypothetical protein |
| 0076/pin | 63773–64345 | Site-specific recombinase |
| 0077 | 64781–65044 | Hypothetical protein |
| 0078/lydA | 65119–65448 | Lysis determining protein |
| 0079 | 65445–65888 | Lysis determining protein |
| 0080 | 65875–66477 | Hypothetical protein |
| 0081/darA | 66479–68398 | Hypothetical protein |
| 0082/ddrA | 68395–68760 | Hypothetical protein |
| 0083 | 68797–71760 | Hypothetical protein |
| 0084/hxr | 71750–72061 | Putative repressor protein Hxr |
| 0085/ompD | 72804–73916 | Outer membrane porin protein OmpD |
| 0086/ssb | 74150–74638 | Single-stranded DNA-binding protein |
| 0087/lys | 74808–75365 | Lysozyme |
| 0088 | 75657–76676 | Putative head processing protein |
| 0089 | 76669–78378 | Putative portal protein |
| 0090 | 78454–85221 | Putative DNA adenine methyltransferase |
| 0091 | 85255–85695 | Hypothetical protein |
| 0092 | 85692–85940 | Modulator protein |
| 0093 | 85982–87286 | Hypothetical protein |
| 0094 | 87343–87984 | Maturation control protein |
| 0095/ref | 88173–88733 | Recombination enhancement function protein |
| 0096 | 88981–89190 | Putative lysogeny establishment protein |
| 0097/cre | 89343–90374 | GST-loxP-cre recombinase fusion protein |
| 0098/cra | 90382–90603 | Putative Cre-associated regulatory protein |
| 0099 | 91208–91417 | C1 repressor inactivator |
| 0100 | 91528–92379 | Primary repressor of lytic function |
| 0101 | 92405–93889 | Putative large terminase protein |
| 102/pacA | 93889–95082 | Terminase A protein |
| 0103/lpa | 95169–95621 | Late promoter activating protein |
| 0104 | 95710–96753 | Hypothetical protein |
| 0105 | 96781–96960 | Hypothetical protein |
| 0106/doc | 96965–97345 | Toxin Doc |
Features: genes, mobile genetic elements or C-segments. The allele numbers of genes present on pMCR_SCKP-LL83 are shown.
Figure 2.
Comparison of pMCR_SCKP-LL83 with phage P7 (GenBank accession no. AF503408). Similar regions are indicated with the degree of nucleotide identity being shown in gray scales. Mobile genetic elements, type I restriction-modification (RM) systems and C-segments are shown in green, yellow, and blue, respectively.
It is well known that phages can transfer genetic components between bacterial isolates, but the role of phages in disseminating antimicrobial resistance genes is still a matter of debate (Colavecchio et al., 2017; Enault et al., 2017). Nonetheless, some studies have found that phages are able to transfer genes conferring resistance to aminoglycosides (aadA, aphA1, strA, strB), β-lactams (blaCMY−2, blaCTX−M−9, blaOXA−2, blaOXA−20, blaPSE−1, blaTEM), chloramphenicol (floR), or tetracycline (tet(A), tet(B), tetG, tetO, tetW) via transduction (Zhang and LeJeune, 2008; Colomer-Lluch et al., 2014; Bearson and Brunelle, 2015; Ross and Topp, 2015; Shousha et al., 2015; Anand et al., 2016). In addition, a recent study has identified that two E. coli phages could promote the transformation of plasmids carrying antimicrobial resistance genes (Keen et al., 2017).
During the process of this work, mcr-1 in either complete or interrupted version has been found on plasmids containing similar phage sequences including pHYEC7-mcr1 (GenBank accession no. KX518745), pSLK172-1 (GenBank accession no. CP017632) (Bai et al., 2017), and pMCR-1-P3 (GenBank accession no. KX880944) (Zhang et al., 2017a). All of these plasmids have been recovered from E. coli at various locations of China and are highly similar (75–79% coverage, 97–99% identity, identified by BLAST; Figure 3) to pMCR_SCKP-LL83. This suggests that the phage sequence-containing plasmids represent new vehicles, which may have circulated in China, to mediate the spread of mcr-1 in addition to plasmids of IncI2, X4, F, HI2, and P types. The identification of pMCR_SCKP-LL83 from a K. pneumoniae is worrisome, suggesting that the P7 phage-like plasmids are not restricted to E. coli and may involve in the inter-species spread of mcr-1. The various locations of mcr-1 on these plasmids suggest that these plasmids may have acquired mcr-1 independently.
Figure 3.
Comparison of phage P7 and similar mcr-1-carrying plasmids. The comparison is a pairwise BLASTn alignment performed using BRIG (Alikhan et al., 2011). Plasmids are pMCR_SCKP-LL83 (this study), pHYEC7-mcr1 (GenBank accession no. KX518745) and pSLK172-1 (GenBank accession no. CP017632) and pMCR-1-P3 (GenBank accession no. KX880944). Coding sequences (CDS) of phage P7 (GenBank accession no. AF503408) are indicated. CDS of phage P7 absent from pMCR_SCKP-LL83 or vice-verse are listed in Table S1.
In the previous study on the ability of E. coli phages to promote the transformation of plasmids carrying antimicrobial resistance gene, phages, and plasmids are separate entities (Keen et al., 2017), which are different from the phage-like plasmid in the present study. As mentioned above, the conjugation and transformation of pMCR_SCKP-LL83 were unsuccessful. Among phage-like plasmids carrying mcr-1, pMCR-1-P3 was not self-transmissible and there are no data about whether it can be transferred by transformation (Zhang et al., 2017a), while pSLK172-1 was self-transmissible (Bai et al., 2017). This suggests that some phage-like plasmids may have lost the conjugative module and are therefore not self-transmissible. It is possible that these plasmids acquire genes encoding the conjugative module to become self-transmissible.
In conclusion, we identified and characterized a mcr-1-carrying P7 phage-like plasmid from a K. pneumoniae clinical isolate. Such phage-like plasmids may represent new types of vehicles to mediate the spread of mcr-1.
Author contributions
ZZ: designed the experiments, analyzed the data, and wrote the MS. LL: performed the experiments and analyzed the data. WZ and YF: analyzed the data.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (project no. 81222025, 81572030, and 81661130159).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00011/full#supplementary-material
References
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Anand T., Bera B. C., Vaid R. K., Barua S., Riyesh T., Virmani N., et al. (2016). Abundance of antibiotic resistance genes in environmental bacteriophages. J. Gen. Virol. 97, 3458–3466. 10.1099/jgv.0.000639 [DOI] [PubMed] [Google Scholar]
- Arndt D., Grant J. R., Marcu A., Sajed T., Pon A., Liang Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. 10.1093/nar/gkw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L., Wang J., Hurley D., Yu Z., Wang L., Chen Q., et al. (2017). A novel disrupted mcr-1 gene and a lysogenized phage P1-like sequence detected from a large conjugative plasmid, cultured from a human atypical enteropathogenic Escherichia coli (aEPEC) recovered in China. J. Antimicrob. Chemother. 72, 1531–1533. 10.1093/jac/dkw564 [DOI] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearson B. L., Brunelle B. W. (2015). Fluoroquinolone induction of phage-mediated gene transfer in multidrug-resistant Salmonella. Int. J. Antimicrob. Agents 46, 201–204. 10.1016/j.ijantimicag.2015.04.008 [DOI] [PubMed] [Google Scholar]
- Black L. W., Thomas J. A. (2012). Condensed genome structure. Adv. Exp. Med. Biol. 726, 469–487. 10.1007/978-1-4614-0980-9_21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2017). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. M100-S27. (Wayne, PA: Clinical and Laboratory Standards Institute; ). [Google Scholar]
- Colavecchio A., Cadieux B., Lo A., Goodridge L. D. (2017). Bacteriophages contribute to the spread of antibiotic resistance genes among foodborne pathogens of the enterobacteriaceae family - a review. Front. Microbiol. 8:1108. 10.3389/fmicb.2017.01108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomer-Lluch M., Calero-Cáceres W., Jebri S., Hmaied F., Muniesa M., Jofre J. (2014). Antibiotic resistance genes in bacterial and bacteriophage fractions of Tunisian and Spanish wastewaters as markers to compare the antibiotic resistance patterns in each population. Environ. Int. 73, 167–175. 10.1016/j.envint.2014.07.003 [DOI] [PubMed] [Google Scholar]
- Coque T. M., Oliver A., Perez-Diaz J. C., Baquero F., Canton R. (2002). Genes encoding TEM-4, SHV-2, and CTX-M-10 extended-spectrum β-lactamases are carried by multiple Klebsiella pneumoniae clones in a single hospital (Madrid, 1989 to 2000). Antimicrob. Agents Chemother. 46, 500–510. 10.1128/AAC.46.2.500-510.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enault F., Briet A., Bouteille L., Roux S., Sullivan M. B., Petit M. A. (2017). Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J. 11, 237–247. 10.1038/ismej.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiner R., Argov T., Rabinovich L., Sigal N., Borovok I., Herskovits A. A. (2015). A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 13, 641–650. 10.1038/nrmicro3527 [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Jacob A. E., Barth P. T., Grinter N. J. (1975). Compatibility properties of P1 and PhiAMP prophages. Mol. Gen. Genet. 141, 263–267. 10.1007/BF00341804 [DOI] [PubMed] [Google Scholar]
- Iida S. (1984). Bacteriophage P1 carries two related sets of genes determining its host range in the invertible C segment of its genome. Virology 134, 421–434. 10.1016/0042-6822(84)90309-X [DOI] [PubMed] [Google Scholar]
- Keen E. C., Bliskovsky V. V., Malagon F., Baker J. D., Prince J. S., Klaus J. S., et al. (2017). Novel “Superspreader” bacteriophages promote horizontal gene transfer by transformation. MBio 8:e02115-16. 10.1128/mBio.02115-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropinski A. M., Mazzocco A., Waddell T. E., Lingohr E., Johnson R. P. (2009). Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 501:69. 10.1007/978-1-60327-164-6_7 [DOI] [PubMed] [Google Scholar]
- Li R., Xie M., Zhang J., Yang Z., Liu L., Liu X., et al. (2017). Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 72, 393–401. 10.1093/jac/dkw411 [DOI] [PubMed] [Google Scholar]
- Liu Y.-Y., Wang Y., Walsh T. R., Yi L.-X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Lowe T. M., Chan P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. 10.1093/nar/gkw413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui N., Kiritani K., Nishida S. (1973). A Lysin(s) in lysates of Clostridium botulinum A190 induced by ultraviolet ray or Mitomycin C. Jpn. J. Microbiol. 17, 353–360. 10.1111/j.1348-0421.1973.tb00786.x [DOI] [PubMed] [Google Scholar]
- Novais A., Canton R., Valverde A., Machado E., Galan J. C., Peixe L., et al. (2006). Dissemination and persistence of blaCTX−M−9 are linked to class 1 integrons containing CR1 associated with defective transposon derivatives from Tn402 located in early antibiotic resistance plasmids of IncHI2, IncP1-α, and IncFI groups. Antimicrob. Agents Chemother. 50, 2741–2750. 10.1128/AAC.00274-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaitan A. O., Diene S. M., Kempf M., Berrazeg M., Bakour S., Gupta S. K., et al. (2014a). Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int. J. Antimicrob. Agents 44, 500–507. 10.1016/j.ijantimicag.2014.07.020 [DOI] [PubMed] [Google Scholar]
- Olaitan A. O., Morand S., Rolain J. M. (2014b). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. 10.3389/fmicb.2014.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver A., Coque T. M., Alonso D., Valverde A., Baquero F., Cantón R. (2005). CTX-M-10 linked to a phage-related element is widely disseminated among Enterobacteriaceae in a Spanish hospital. Antimicrob. Agents Chemother. 49, 1567–1571. 10.1128/AAC.49.4.1567-1571.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Jayol A., Nordmann P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. 10.1128/CMR.00064-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya R. R., H'bert E. M. (2009). Isolation of phage via Induction of Lysogens. Methods Mol. Biol. 501, 23–32. 10.1007/978-1-60327-164-6_3 [DOI] [PubMed] [Google Scholar]
- Ross J., Topp E. (2015). Abundance of antibiotic resistance genes in bacteriophage following soil fertilization with dairy manure or municipal biosolids, and evidence for potential transduction. Appl. Environ. Microbiol. 81, 7905–7913. 10.1128/AEM.02363-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2001). Molecular Cloning. A Laboratory Manual. Cold Spring Harbour, NY: Cold Spring Harbour Laboratory Press. [Google Scholar]
- Sandmeier H., Iida S., Hübner P., Hiestand-Nauer R., Arber W. (1991). Gene organization in the multiple DNA inversion region min of plasmid p15B of E. coli 15T-: assemblage of a variable gene. Nucleic Acids Res. 19, 5831–5838. 10.1093/nar/19.21.5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Ko K. S. (2015). A plasmid bearing the blaCTX−M−15 gene and phage P1-like sequences from a sequence type 11 Klebsiella pneumoniae isolate. Antimicrob. Agents Chemother. 59, 6608–6610. 10.1128/AAC.00265-15 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shousha A., Awaiwanont N., Sofka D., Smulders F. J., Paulsen P., Szostak M. P., et al. (2015). Bacteriophages isolated from chicken meat and the horizontal transfer of antimicrobial resistance genes. Appl. Environ. Microbiol. 81, 4600–4606. 10.1128/AEM.00872-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. W. (1972). Ampicillin resistance in Escherichia coli by phage infection. Nat. New Biol. 238, 205–206. 10.1038/newbio238205a0 [DOI] [PubMed] [Google Scholar]
- Valenzuela J. K., Thomas L., Partridge S. R., van der Reijden T., Dijkshoorn L., Iredell J. (2007). Horizontal gene transfer in a polyclonal outbreak of carbapenem-resistant Acinetobacter baumannii. J. Clin. Microbiol. 45, 453–460. 10.1128/JCM.01971-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz M. (1992). Bacteriophage structure. Electron Microsc. Rev. 5, 283–309. 10.1016/0892-0354(92)90013-G [DOI] [PubMed] [Google Scholar]
- Xavier B. B., Lammens C., Ruhal R., Kumar-Singh S., Butaye P., Goossens H., et al. (2016). Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 21:30280. 10.2807/1560-7917.ES.2016.21.27.30280 [DOI] [PubMed] [Google Scholar]
- Yin W., Li H., Shen Y., Liu Z., Wang S., Shen Z., et al. (2017). Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. MBio 8:e00543–e00517. 10.1128/mBio.00543-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Feng Y., Liu F., Jiang H., Qu Z., Lei M., et al. (2017a). A phage-like IncY plasmid carrying the mcr-1 gene in Escherichia coli from a pig farm in China. Antimicrob. Agents Chemother. 61:e02035–e02016. 10.1128/AAC.02035-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Liu L., Zhou H., Chan E. W., Li J., Fang Y., et al. (2017b). Nationwide surveillance of clinical carbapenem-resistant enterobacteriaceae (CRE) strains in China. EBioMed. 19, 98–106. 10.1016/j.ebiom.2017.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., LeJeune J. T. (2008). Transduction of blaCMY−2, tet(A), and tet(B) from Salmonella enterica subspecies enterica serovar Heidelberg to S. Typhimurium. Vet. Microbiol. 129, 418–425. 10.1016/j.vetmic.2007.11.032 [DOI] [PubMed] [Google Scholar]
- Zhao F., Zong Z. (2016). Kluyvera ascorbata carrying the mcr-1 colistin resistance gene from hospital sewage. Antimicrob. Agents Chemother. 60, 7498–7501. 10.1128/aac.01165-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Feng Y., Lü X., Mcnally A., Zong Z. (2017). Remarkable diversity of Escherichia coli carrying mcr-1 from hospital sewage with the identification of two new mcr-1 variants. Front. Microbiol. 8:2094. 10.3389/fmicb.2017.02094 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




