ABSTRACT
With poliovirus eradication nearing, few pockets of active wild poliovirus (WPV) transmission remain in the world. Intratypic differentiation (ITD) plays a crucial part in laboratory surveillance as the molecular detection method that can identify and distinguish wild and vaccine-like polioviruses isolated from acute flaccid paralysis cases or environmental sources. The need to detect new variants of WPV serotype 1 (WPV1) and the containment of all serotype 2 polioviruses (PV2) in 2015 required changes to the previous version of the method. The ITD version 5.0 is a set of six real-time reverse transcription-PCR (rRT-PCR) assays that serve as accurate diagnostic tools to easily detect and differentiate PV serotypes and genotypes. We describe the creation and properties of quantitation standards, including 16 control RNA transcripts and nine plaque-isolated viruses. All ITD rRT-PCR assays were validated using these standards, and the limits of detection were determined for each assay. We designed and pilot tested two new assays targeting recently circulating WPV1 genotypes and all PV2 viruses. The WPV1 assay had 99.1% specificity and 100% sensitivity, and the PV2 assay had 97.7% specificity and 92% sensitivity. Before proceeding to the next step in the global poliovirus eradication program, we needed to gain a better understanding of the performance of the ITD 5.0 suite of molecular assays and their limits of detection and specificities. The findings and conclusions in this evaluation serve as building blocks for future development work.
KEYWORDS: diagnostic assay, molecular typing, polio eradication, poliovirus, rRT-PCR, real-time reverse transcription-PCR, serotype identification
INTRODUCTION
The Global Polio Eradication Initiative (GPEI) has reached a critical juncture with very little room for error (1). The detection of polioviruses from surveillance stool samples of acute flaccid paralysis (AFP) cases will remain crucial to monitoring progress of poliovirus eradication. Molecular detection methods must be able to identify poliovirus from AFP surveillance or environmental sources and function as the frontline defense in case of accidental release (2, 3). Wild poliovirus (WPV) continues to circulate in West Africa and South Asia. In 2016, there were 37 cases of WPV type 1 (WPV1) detected in Afghanistan (n = 13), Pakistan (n = 20), and Nigeria (n = 4) (4). The continual circulation of WPV in Nigeria was unexpected, but the reduction in WPV1 cases and the continuing absence of WPV3 since 2012 have been encouraging.
In the current polio laboratory surveillance algorithm, poliovirus is detected by virus isolation in cell culture. Cultures exhibiting viral cytopathic effect (CPE) are tested by real-time reverse-transcription PCR (rRT-PCR) to identify programmatically important viruses (WPV, vaccine-derived poliovirus [VDPV], and serotype 2 poliovirus [PV2]). This process is termed intratypic differentiation (ITD) and is performed in conjunction with an evolving algorithm (5–8). Previous versions of the ITD kit comprised a set of diagnostic rRT-PCR assays containing primer and probe mixes with in-house buffers and PCR template positive controls to be used with commercially available reverse transcriptase and Taq polymerase enzymes (9). The method accommodated highly degenerate primers and probes, but run conditions were complex, and the method required more than 3 h to complete (9). To avoid the long run times and complex reagent preparation required by older methods (10), we replaced the previous buffer system with commercially available rRT-PCR enzyme master mix and simplified the thermocycler run method. In 2014, continued genetic drift of the WPV1 South Asia genotype caused a decrease in the sensitivity of the WPV1 molecular assays for the drifted WPV1 variants. Although poliovirus detection was not impacted, it added to laboratory workload due to the increased number of isolates that were referred for sequencing (8, 11). As a transitional step to improve assay performance, the ITD version 4.1 kit was introduced in 2015. It included a modified WPV1 assay to account for evolution of circulating WPV1 and an updated WPV3 assay to reduce background signal (CDC, unpublished data).
Wild poliovirus type 2 (WPV2) was last detected in 1999, and global certification of WPV2 eradication was announced in September 2015 (12). Vaccine-derived polioviruses (VDPVs) may arise by genetic reversion of vaccine strains: as long as oral polio vaccine (OPV) is used, there is a risk of VDPV emergence. VDPVs are phenotypically equivalent to WPV and may cause paralysis and circulate in areas with immunity gaps (13, 14). Type 2 VDPVs have emerged and caused outbreaks in multiple countries. In individuals with primary immune deficiency disorders, VDPVs may replicate for prolonged periods and are termed immunodeficiency-related VDPVs (iVDPVs) (15). In April 2016, poliovirus type 2 was removed from the OPV formulation, and withdrawal of trivalent OPV was coordinated globally in a switch to bivalent OPV, containing only serotypes 1 and 3. A monovalent OPV2 (mOPV2) formulation is kept in a global stockpile to support outbreak response, and it has already been used to respond to VDPV circulation in several countries. After WPV2 eradication and the withdrawal of OPV2 from routine use, detection of any live PV2 constitutes a potential emergency that requires rapid investigation and response. New molecular methods were necessary to detect all PV2s, inclusive of VPDV2, WPV2, and wild seed strains used in production of inactivated polio vaccine; as a result, the Global Polio Laboratory Network (GPLN) incorporated a PV2 assay into the current suite of ITD assays (ITD 5.0), and the assays were released to the GPLN laboratories in 2016. We now report on the validation of the ITD 5.0 suite of assays, including assay sensitivity and specificity, and results of pilot testing in three GPLN laboratories.
MATERIALS AND METHODS
Reference virus panel.
To determine specificity, all six ITD assays were tested against a reference virus panel (n = 200) representing all PV serotypes as well as nonpolio enteroviruses and CPE-negative culture supernatants (Table 1). Viruses were isolated in cell cultures of L20B cells (recombinant murine cells that express human poliovirus receptor [PVR]) and human rhabdomyosarcoma cells (RD; ATCC CCL-136) inoculated with stools from AFP or contact cases that were collected from 1999 through 2016 by laboratories in the GPLN (5, 7, 16, 17). For PV2 assay development, 63 viral protein 1 (VP1) sequences in CDC's internal poliovirus database of iVDPV2 isolates from 1990 to 2015 (WHO Eastern Mediterranean Region, n = 53; WHO Americas Region, n = 9), as well as the PV2 strain used for IPV production (MEF-1; accession no. AY238473.1) and a Sabin 2 representative (accession no. AY184220), were aligned in BioEdit (18). A maximum composite likelihood phylogenetic tree was constructed in MEGA version 6 (19), and representative iVDPV2 viruses were selected for PV2 assay development (data not shown).
TABLE 1.
Summary of reference poliovirus isolates by serotype and genotype
| Serotypea | Genotypeb | No. of specimens (n = 200) |
|---|---|---|
| PV1 | Sabin | 10 |
| VDPV1 | 11 | |
| WEAF-B1 | 15 | |
| SOAS | 22 | |
| PV2 | Sabin | 15 |
| VDPV | 16 | |
| Wild | 10 | |
| iVDPV2 | 16 | |
| PV3 | Sabin | 12 |
| VDPV | 14 | |
| WPV AFR | 13 | |
| WPV SOAS | 20 | |
| Negative | NPEVc | 15 |
| Negative | 11 |
PV1 to -3, poliovirus serotypes 1 to 3.
VDPV, vaccine-derived poliovirus; WEAF-B1, West Africa B1 genotype; SOAS, South Asia genotype; iVDPV2, immunodeficient vaccine-derived poliovirus (16 iVDPV2 virus isolates were used for assaying PV2 rRT-PCR only); WPV AFR, wild poliovirus African genotype; WPV SOAS, wild poliovirus South Asia genotype. IPV isolates of serotype 1 (Mahoney) and serotype 3 (Saukett) were positive in both EV and PanPV assays. The Mahoney strain was positive in the Sabin 1 assay and Saukett in the Sabin 3 assay (data not shown).
NPEV, nonpolio enterovirus, including coxsackievirus A4 (CV-A4), CV-A9, CV-A21, CV-B4, and CV-B5, echovirus 6 (E-6), E-9, E-11 (n = 2), and E-12, enterovirus A71 (EV-A71), EV-D68, and EV/rhinovirus, rhinovirus A47, and unknown NPEV (n = 1).
Plaque purification for quantification standards.
Polioviruses representing each of the three serotypes were plaque purified using an adaptation of previously published methods (20, 21). First HeLa cell monolayers were seeded into separate 10-cm culture dishes and inoculated with virus dilutions from 10−2 to 10−8. The infected cell monolayers were overlaid with 0.9% agarose in 2× complete minimal essential medium (MEM; Thermo Fisher Scientific, Waltham, MA). The solidified agarose-cell monolayers were incubated at 37°C for 48 to 72 h. From each virus, 20 to 30 plaques were harvested into 0.5 ml MEM and subjected to three freeze-thaw cycles. Virus clones were propagated in RD cells (highly susceptible for poliovirus) at 37°C for 24 h, by which time 100% CPE was observed. Supernatants were harvested and stored at −80°C until further use. To verify the identity of the plaque-purified viruses, full-genome sequencing was performed as adapted from previously described methods (22). Viral RNA was extracted from virus clones with the QIAamp viral RNA minikit following the manufacturer's instructions (Qiagen, Germantown, MD) and sequenced from two overlapping amplicons. First, two overlapping amplicons (3.5 and 4 kbp) were separately synthesized from viral RNA by using a two-step RT-PCR and SuperScript II reverse transcriptase (Thermo Fisher Scientific) (23). cDNAs were synthesized with reverse-sense primers 7500A and Q8, and the remaining single-stranded RNA was digested with RNase H at 37°C for 30 min. The two cDNA fragments were amplified using a combination of primers 28S and Q8 (5′ amplicon) or Y7 and 7500A (3′ amplicon) (see Table S4 in the supplemental material) (24–26). PCR amplification using Q5 high-fidelity polymerase was run at 98°C for 30 s, followed by 35 cycles of 98°C for 10 s, 50°C for 30 s, and 72°C for 3 min, with a final incubation at 72°C for 5 min (New England BioLabs, Ipswich, MA). Amplicons were gel extracted, purified, and bidirectionally sequenced using the primers listed in Table S4 (22). Terminal sequences were determined by using a 2nd generation 5′→3′ random amplification of cDNA ends (RACE) kit according to the manufacturer's instructions (Roche, Indianapolis, IN).
Development of synthetic poliovirus quantification standards.
Synthetic poliovirus VP1 transcripts were used to evaluate assay performance. Constructs were synthesized comprising a concatenated VP1 region (≈900 nucleotides [nt]) of the respective target sequence (flanking sequences) and 163 nt from the 5′ untranscribed region (UTR) of a Sabin type 1 virus isolate (see Table S3 in the supplemental material) (Genscript, Piscataway, NY). We generated 16 RNA transcripts derived from sequences of the Sabin vaccine strains (n = 3), WPV serotypes 1 (n = 4) and 3 (n = 4), including both West Africa (WEAF-B1) and South Asia (SOAS) genotypes, VDPV2 (n = 3), and the WPV2 strain (MEF-1) used in inactivated polio vaccine (IPV) production. The concatenated VP1 and 5′-UTR sequences were inserted into a pUC19 vector with a T7 promoter to allow synthesis of positive-sense RNA transcripts from the plasmid insert. RNA transcripts were synthesized by in vitro transcription of linearized plasmid DNA by using the HiScribe T7 high-yield RNA synthesis kit with RNase inhibitor (New England BioLabs). DNA was enzymatically removed from the transcription reaction by DNase I treatment (New England BioLabs) and cleaned up with Select-a-Size DNA Clean, selecting material of >300 bp following the manufacturer's instructions (Zymo Research, Irvine, CA). RNA transcripts were concentrated with RNA Clean & Concentrator-100, including a secondary in-column DNase I treatment step (DNase I set; Zymo Research). RNA transcripts were quantified using the Qubit RNA BR assay kit (Thermo Fisher Scientific). All standards were diluted to 106 to 108 copies·μl−1 and stored in single-use aliquots at −80°C until further use.
LOD for poliovirus intratypic differentiation assays.
Each ITD PCR mixture consists of 10 μl of qScript XLT one-step RT-quantitative PCR (qPCR) ToughMix (Quanta Biosciences, Beverly, MA), 1 μl of primer-probe mixture (contained in the ITD 5.0 kit; CDC, Atlanta, GA), 8 μl RNase-free water, and 1 μl of template (cell culture supernatant or extracted viral RNA). The thermocycling conditions are 30 min at 50°C for the RT step and 1 min of incubation at 95°C, followed by 40 cycles of 95°C for 15 s, 50°C for 1 min, and 72°C for 5 s. (A reduced ramp rate of 25% between annealing and elongation was applied on the Applied Biosystems 7500 real-time PCR system [Thermo Fisher Scientific].) This minimized dissociation of primers and probes in the PanPV rRT-PCR assay containing multiple degeneracies and inosine as a base analog at several positions (27). The limit of detection (LOD) for each assay was determined by testing limiting dilutions of the 16 different RNA transcripts and nine different plaque-purified viruses. Six replicates of each concentration of RNA transcripts (104 to 10−2 copies·μl−1) and serial dilutions of nine viruses (107 to 100 50% cell culture infective doses [CCID50]·ml−1) were tested in triplicate for each of the dilution steps. The 95% LOD was determined by running 20 replicates of the last dilution step with 100% positivity and the next two lower concentrations.
Data analysis.
To analyze 95% LOD data collected from rRT-PCR assay runs, cycle threshold (CT) values were recorded for all individual replicates for each target assay. Cycle thresholds were used only for qualitative determination: (i) positive (CT between 1 and 40) or (ii) negative (no CT). The probability of samples residing within the 95% confidence interval (CI) was calculated from the number of positive samples out of 20 replicates for all templates used in the assays. The results were grouped by ≥95% or <95% sensitivity for each dilution step.
Statistical analysis.
The statistical significance of the probability to detect a template was evaluated comparing the number of positive replicates for each concentration out of the total number tested (n = 20) using a binominal test in R (28). Clinical sensitivity and specificity were assessed in two-by-two contingency tables. Fisher's exact test was used to determine whether template type was independent of RNA concentration (i.e., no association between template type and LOD) with the gmodels package in R (28). To determine which of the templates was significantly less sensitive, a post hoc test for comparing multiple proportions, the Marascuilo method, was applied (29).
Ethical considerations.
CDC's Internal Program for Research Determination deemed that this study is categorized as public health nonresearch and that human subject regulations did not apply.
Accession number(s).
Full-length nucleotide sequences have been submitted to the GenBank sequence database under accession no. KY941931 to KY941935 (see Table S5 in the supplemental material).
RESULTS
Design and performance of updated WPV1 assay.
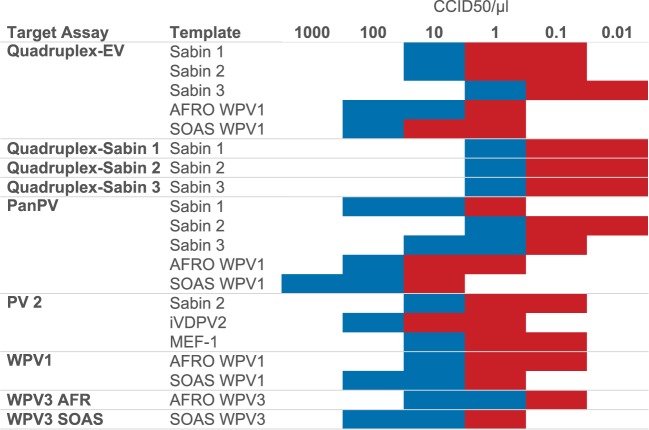
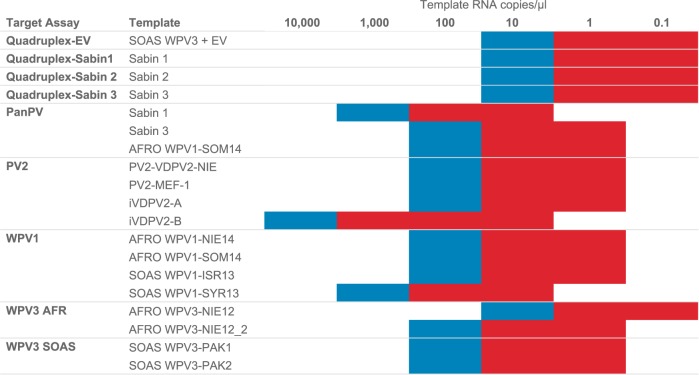
Complete WPV1 VP1 region sequences were retrieved from GenBank and from the internal CDC poliovirus database. A total of 1,433 sequences from 2012 to 2016 were analyzed using R for nucleotide heterogeneity at position 163, which is within the forward primer target site, a recently identified site of sequence heterogeneity (see Fig. S1 in the supplemental material). Sequences were grouped by genetic clusters with viruses that share ≥95% nucleotide identity in the VP1 region. Viruses from 2012 to 2013 contained a thymidine at position 163, whereas isolates of clusters R4B5A, R4B5C, and R4B7 detected in 2014 to 2016 had a cytosine; in viruses from 2016, the majority of viruses sequenced carried the thymidine. To increase diagnostic sensitivity and to simplify interpretation, a new WPV1 rRT-PCR assay was designed to detect all recent variants that have been found in Africa (genotype WEAF-B1) and in South Asia (genotype SOAS). Initially, multiple primers and probes were tested with 58 viruses from a reference virus panel (Table 1), and the best-performing primer-probe combinations were selected for pilot studies (Table 2). In collaboration with GPLN partners, the WPV1 assay was validated with 327 poliovirus isolates at the National Institute for Communicable Diseases (NICD), Johannesburg, South Africa (n = 67), the Enterovirus Research Centre (ERC), Mumbai, India (n = 90), and the National Institute of Health, Islamabad, Pakistan (n = 170). The specificity of the WPV1 assay was 99.1% (334 out of 337 isolates) and the sensitivity was 100% compared to the previous version of the WPV1 assay (Table 3). The limit of detection (LOD) for the WPV1 rRT-PCR assay was 10 CCID50·μl−1 with WPV1-SOAS and WPV1-WEAF virus templates (Fig. 1; see Table S1 in the supplemental material). WPV1 templates WPV1-WEAF NIE14, WPV1-WEAF SOM14, WPV1-SOAS ISR13, and WPV1-SOAS SYR13 were not significantly different from each other (P > 0.05). The LOD with RNA transcripts ranged between 100 and 1,000 RNA copies·μl−1 (Fig. 2; see Fig. S2 and Table S2 in the supplemental material).
TABLE 2.
List of primers and probes used in the ITD 5.0 rRT-PCR assays by assay, primer and probe sequences, and binding regions
| Assay type and target | Primer or probe (dye)a | Oligonucleotide sequence (5′→3′) | nt positionb |
|---|---|---|---|
| Quadruplex | |||
| Sabin 1 | Sabin 1 2S | AGG TCA GAT GCT TGA AAG C | 2505–2523 |
| Sabin 1 probe A4 (Cy5) | CGC CCC CAC CGT TTC ACG GA | 2540–2559 | |
| Sabin 1 1A | CCA CTG GCT TCA GTG TTT | 2600–2583 | |
| Sabin 2 | Sabin 2 2S | CCG TTG AAG GGA TTA CTA AA | 2525–2544 |
| Sabin 2 probe (FAM) | ATT GGT TCC CCC GAC TTC CAC CAA T | 2550–2572 | |
| Sabin 2 1A | CGG CTT TGT GTC AGG CA | 2595–2579 | |
| Sabin 3 | Sabin 3 2S | AGG GCG CCC TAA CTT T | 2537–2552 |
| Sabin 3 probe (ROX) | TCA CTC CCG AAG CAA CAG | 2554–2571 | |
| Sabin 3 1A | TTA GTA TCA GGT AAG CTA TC | 2591–2572 | |
| PanEV (any EV) | PAN-EV S | GGC CCC TGA ATG CGG CTA ATC C | 458–480 |
| PAN-EV probe (VIC) | CCG ACT ACT TTG GGW GTC CGT GT | 546–568 | |
| PAN-EV A | GCG ATT GTC ACC ATW AGC AGY CA | 603–581 | |
| PanPV (any poliovirus) | PanPV/PCR-S1 | TTG GAG TTC TTC ACI TAI TCI MGI TTY GAY ATG | 2832–2864 |
| PanPV PCR-probe 1A (FAM) | TGR TTN ARI GCR TGI CCR TTR TT | 2926–2904 | |
| PanPV PCR-1A | GGA GCT CCG GGT GGG AYR TAC ATI ATY TGR TAI AC | 2962–2928 | |
| WPV1 (wild PV1) | WEAF WPV1 S | GTA CAA ACC AGT CAY GTN AT | 2661–2680 |
| SOAS WPV1 S | CGT ACA GAC TAG RCA YGT NAT | 2660–2680 | |
| WPV1 probe S (FAM) | CAT WAT GGT TAC RCA MGC ACC T | 2729–2750 | |
| WPV1 A | GAG AAT AAY TTG TCY TTK GAY GT | 2800–2778 | |
| PV2 (any serotype 2) | PV Type 2 S | GAT GCA AAY AAC GGI CAT GC | 2911–2930 |
| PV Type 2 probe S (FAM) | ATG ACT ATA CGT GGC AGA C | 2993–3011 | |
| PV Type 2 probe 1D S (FAM) | CRC CKA TIC CTG GYA | 2972–2986 | |
| PV Type 2 A | TCA TAA AAG TGG GAR TAC GCR TT | 3110–3088 | |
| PV Type 2 A 1C | TCG TAA AAA TGA GAA TAT GCA TT | 3110–3088 | |
| AFR WPV3 | |||
| WPV3-I | SOAS WPV3 S | CAG GGA GTA GAT GAY CTN AT | 2443–2462 |
| WEAF genotype | WEAF WPV3 S | CAG GGG GTT GAT GAY TTR AT | 2443–2462 |
| WPV3 probe S (Cy5) | CNC ARA ACA GYC TTC CGG ATA CC | 2504–2526 | |
| WPV3 A | ACK GTG TCT GAY GGN AC | 2623–2607 | |
| SOAS WPV3 | |||
| WPV3-II | SOAS 6S | GTY RTA CAR CGR CGY AGY AGR A | 2671–2692 |
| SOAS genotype | SOAS WPV3 probe S (FAM) | TTC TTY GCA AGI GGR GCR TGY GT | 2713–2735 |
| SOAS 5A | TCY TTR TAI GTR ATG CGC CAA G | 2816–2795 |
Probes labeled with 6-carboxyfluorescein (FAM) were quenched with black hole quencher 1 (BHQ1), those labeled with ROX were quenched with BHQ2, those labeled with Cy5 were quenched with BHQ3, and those labeled with VIC were quenched with 6-carboxytetramethylrhodamine (TAMRA).
All nucleotide positions are relative to those described by Toyoda et al. (34), except PanPV GenBank accession no. AY184219.1 and SOAS WPV3 GenBank accession no. AJ293918.1.
TABLE 3.
WPV1 assay performance compared to the previous rRT-PCR assay, ITD 4.0 WPV1a
| Result for WPV1 ITD 5.0b | No. of isolates with expected result from WPV1 ITD 4.0 |
|
|---|---|---|
| Positive | Negative | |
| Positive | 130 | 3 |
| Negative | 0 | 334 |
Results from a total of 467 isolates are shown.
Sensitivity, 100%; specificity, 99.1%; positive predictive value, 97.7% (95% CI, 93.6 to 99.2%); negative predictive value, 100% (95% CI, 98.9 to 100%).
FIG 1.
Sensitivity of detection of plaque-purified polioviruses (CCID50 per microliter) in the ITD 5.0 assays by target assay and template. The color code indicates sensitivity of ≥95% (blue) or <95% (red) (calculated as a percentage of positive samples per dilution; n = 20).
FIG 2.
Sensitivity of detection of synthetic RNA transcripts (RNA copy number per microliter) in the ITD 5.0 assays by target assay and template. The color code indicates sensitivity of ≥95% (blue) or <95% (red) (calculated as a percentage of positive samples per dilution; n = 20).
Development and performance of PV2 rRT-PCR assay.
Because WPV2 has been eradicated and Sabin 2 removed from OPV, there is increased priority to detect type 2 polioviruses. Primer and probe sets that detected iVDPV reference viruses, Sabin 2, VDPV2, and MEF-1 (inactivated polio vaccine [IPV] seed strain) were designed based on sequence alignments of relevant virus sequences, and the concentrations were optimized (Table 2). The best-performing primer and probe combinations were tested with a panel of 184 reference virus isolates (Table 1). The PV2 assay was also assessed with 16 iVDPV2s (for a total of 200), isolated from stools between 2002 and 2015 in Asia, North America, and Africa. Additionally, we tested the PV2 PCR assay with four RNA transcripts derived from Sabin 2, MEF-1, an iVDPV strain (iVDPV2-A) with 6 nucleotide differences (4 amino acids), and an iVDPV (iVDPV2-B) with 112 nucleotide changes (20 amino acids) compared to Sabin 2. Six out of 69 PV2 isolates were not detected, resulting in 92% assay sensitivity compared to VP1 sequencing results (Table 4). Four of these WPV2 isolates were collected between 1998 and 1999 and have not been detected since (12). The new PV2 assay detected all 16 iVDPV2s from previously VP1-sequenced isolates.
TABLE 4.
PV2 assay performance compared to the previous method, VP1 sequencinga
| Result for PV2b | No. of isolates with expected result by VP1 sequencing |
|
|---|---|---|
| Positive | Negative | |
| Positive | 69 | 7 |
| Negative | 6 | 294 |
Results from a total of 376 isolates are shown.
Sensitivity, 92%; specificity, 97.7%; positive predictive value, 90.8% (95% CI, 82.2 to 95.5%); negative predictive value, 98% (95% CI, 95.7 to 99.1%).
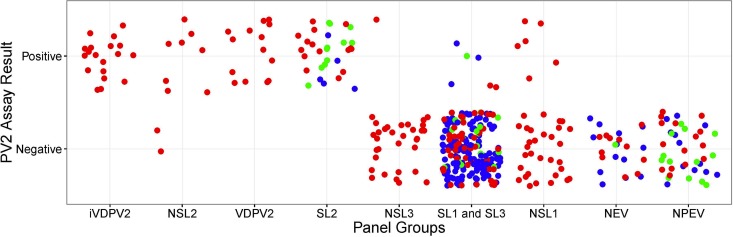
The PV2 assay was pilot tested with 149 virus isolates at ERC and 27 virus isolates at NICD, in addition to the reference virus panel tested at CDC (n = 200; Table 1). The combined PV2 rRT-PCR assay specificity was 97.7%, with 294 rRT-PCR negative from 300 expected negatives determined before by ITD screening and VP1 sequencing (Fig. 3 and Table 4). The LOD of the PV2 assay was lower with the Sabin 2 and MEF-1 virus templates (10 CCID50·μl−1) than with an iVDPV2 virus template (100 CCID50·μl−1) (Fig. 1; see Fig. S3 and Table S1 in the supplemental material). The PV2 assay was the least sensitive with the RNA transcript iVDPV2-B compared to the templates PV2-Sabin, PV2-MEF-1, and iVDPV2-A (P < 0.05), with a 95% LOD of 10,000 RNA copies (P < 0.05) (Fig. S3 and Table S1). This iVDPV2-B RNA transcript was highly divergent with 112 nucleotides different (20 amino acid changes) from Sabin 2's VP1 region and had several mismatches with the PV2 primers and probes (data not shown).
FIG 3.
PV2 assay performance by participating pilot testing laboratory and reference viruses tested. Each dot represents a single virus tested against the PV2 assay. Colors indicate the participating laboratory: Centers for Disease Control and Prevention, United States (red; n = 200), Enterovirus Research Centre, India (blue; n = 149), and National Institute for Communicable Diseases (NICD), South Africa (green; n = 40).
ITD assays and improved performance with new run conditions.
Each of the six rRT-PCR assay identifies between one and four different targets and follows a specific algorithm (see Fig. S4 in the supplemental material): The quadruplex EV-Sabin assay includes primers and probes for the detection of any enterovirus (EV) and the oral vaccine strains (Sabin 1, 2, and 3) (Table 2). The other five assays detect any poliovirus (PanPV), WPV1, PV2, and WPV3 (two separate assays, with high sensitivity for the WEAF and SOAS genotypes, respectively). To reduce false-positive results and decrease turnaround time, a new run method was adopted for all ITD and VDPV primer sets. From 2014 onwards, a simpler method was introduced that used 42°C annealing temperatures to accommodate the inosine-containing primers and probes (8). We further shortened cycling conditions, increased the annealing temperature to 50°C to achieve higher specificity, and reduced the overall estimated run time to 132 min (in the Applied Biosystems 7500 system) with both the ITD and VDPV assays.
LOD of ITD rRT-PCR assays.
The 95% LOD was determined with 16 RNA transcripts and 9 plaque-purified poliovirus isolates representing all three poliovirus serotypes, including two wild genotypes each for types 1 and 3 (West Africa, South Asia), OPV and IPV seed strains. The sensitivity of the quadruplex assay for Sabin 1, 2, and 3 was identical for all three virus strains, at 1 CCID50·μl−1 (95% confidence interval [CI], 83 to 100%) (Fig. 1). When RNA transcripts were tested in the same assay, Sabin 1, 2, and 3 templates were detected 100% of the time at 10 RNA copies·μl−1 (95% CI, 83 to 100%) (Fig. 2). The sensitivity of the EV component of the quadruplex assay ranged from 1 CCID50·μl−1 when tested with a Sabin 3 template to 100 CCID50·μl−1 when tested with a WPV1-SOAS template (Table S1). The EV assay was significantly more sensitive for Sabin 3 than for WPV1-SOAS templates (P < 0.05). When assaying RNA transcripts in the EV assay, the 95% LOD was 10 copies·μl−1 across transcripts. The sensitivity of the PanPV rRT-PCR assay (detecting all polioviruses) ranged between 1 CCID50·μl−1 for Sabin 2 and 100 CCID50·μl−1 for WPV1 SOAS (Fig. 1). Sabin 2 and WPV1-SOAS virus templates sensitivities differed significantly from each other (P < 0.005) in the PanPV assay. The WPV3 assay sensitivities ranged from 1 CCID50·μl−1 for the WPV3 AFR assay to 10 CCID50·μl−1 for the WPV3 SOAS assay (Fig. 1; Table S1). Sensitivity was 100 RNA copies·μl−1 for one of the two AFRO WPV3 transcripts and both SOAS WPV3 transcripts with their corresponding WPV3 assay (Fig. 2; Table S2).
DISCUSSION
Having improved molecular diagnostic methods is critical for the polio endgame, especially to document disappearance of OPV2 and to assist in detecting components of complex mixtures (8). ITD has been an essential component of polio surveillance in the GPLN for >25 years, from probe hybridization to conventional RT-PCR and multiple iterations of rRT-PCR (9, 10, 27, 30–32). Methods and assays are periodically updated to incorporate new technologies, to improve their sensitivity and specificity, to account for continued poliovirus evolution, and to respond to changing needs of the GPEI. We developed a new ITD algorithm and rRT-PCR assays that detect all serotype 2 polioviruses and both circulating WPV1 genotypes, West Africa (WEAF-B1) and South Asia (SOAS). The use of commercial one-step RT-PCR master mix reagents simplifies kit manufacture and potentially reduces lot-to-lot variation.
We describe the validation and performance data of an additional four rRT-PCR assays targeting all three poliovirus serotypes and WPV3 genotypes, with simplified run conditions, including a multiplex rRT-PCR assay detecting four targets in a single tube. Assay specificities were assessed with 174 virus isolates covering all PV serotypes and genotypes, 15 nonpolio enteroviruses, and 11 nonenteroviruses. Assays were pilot tested in GPLN reference laboratories in Africa and South Asia that serve the regions in which poliovirus was most recently found to be endemic. Multisite validation of ITD 5.0 in GPLN consistently showed results identical to or better than those from previous ITD versions. The WPV1 rRT-PCR assay displayed high sensitivity and specificity in identifying distinct genetic clusters that have circulated from 2014 to the present. We observed fewer false-negative results and could detect both WPV1 genotypes in a single duplex assay. The new run method reduced false-positive results because of a greater stringency, with higher annealing and extension temperatures, and decreased the overall turnaround time for all rRT-PCR assays, with shorter cycle times. Simultaneously running all ITD assays under the same cycling conditions facilitates higher laboratory throughput and a standardized approach to performing the complex set of rRT-PCRs that comprise the ITD algorithm (Fig. S4).
The PV2 assay detected both OPV and IPV seed strains (i.e., Sabin 2 and MEF-1), cVDPV2, and iVDPV2. The 10-fold reduction in sensitivity between vaccine strains and iVDPV2 strains is caused by high sequence diversity and mismatches in the primer and probe annealing sites; however, iVDPV sequences are not predictable, so some degree of sensitivity loss is expected. With the introduction of the PV2 assay, the testing algorithm was changed and the VDPV2 assay became obsolete, since all type 2 viruses must now be referred for sequencing (Fig. S4) (9, 24). The design of the ITD 5.0 algorithm incorporates redundancies to detect all polioviruses with multiple assays to protect against genetic drift or user error. Any virus with discordant results (e.g., PanPV positive but negative for all other polio assays) should be referred to an accredited WHO poliovirus sequencing laboratory.
For the purposes of validation, proficiency testing, and distribution of molecular detection assays, RNA transcripts continue to be extremely valuable, given the containment requirements for live PV2. The least sensitive ITD assay was PanPV with either RNA transcripts or virus isolates, probably due to the high degeneracy of the primer and probes (10). Regardless of its reduced sensitivity, the PanPV assay is a cornerstone of the ITD 5.0 algorithm, since it captures all poliovirus serotypes and genotypes. Following the ITD 5.0 algorithm, these viruses would still be detected as indeterminate polioviruses (positive for PanPV and PanEV but negative in all other assays or positive for PanEV, negative for PanPV, but positive for one or more of the other assays) and would be referred for VP1 sequencing (24).
Challenges that remain to be addressed include improving PanPV performance, identifying iVDPV2 from highly diverged polioviruses from immunocompromised persons (15), and increasing the sensitivity of the assays for the eventual detection of viral genome in RNA extracted directly from stool or concentrated sewage. A process control increasingly gains importance in future iterations of the ITD assays to control for RNA extraction performance as well as PCR inhibition. Continual monitoring of poliovirus sequences is crucial to account for genetic drift and to revise existing ITD assays for sensitive poliovirus detection supporting AFP and environmental surveillance (4, 24).
Future GPLN testing strategies will eliminate the cell culture isolation algorithm from poliovirus diagnostics. ITD 5.0 lays the groundwork for further improvements such as direct detection in stool extracts and sewage concentrates. This shift will have major advantages such as a reduction in turnaround time and elimination of the need to maintain multiple cell lines (i.e., RD and L20B). The direct detection method must be as sensitive as virus isolation in cell culture, and it must be robust, inexpensive, and easy to implement in all GPLN laboratories. That method must align with the requirements established by the WHO Global Action Plan referred to as GAPIII (33). A better understanding of the currently applied molecular assay capabilities and limitations is needed before proceeding to the next step in the global poliovirus eradication program.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Naomi Dybdahl-Sissoko and Hong Pang for assistance in growing and handling reference virus isolates. We acknowledge the contributions of Hongmei Liu for assistance with transfection assays and sequencing, Ling Wei for expertise with plaque purification of viruses, and Elizabeth Henderson and Jane Iber for help with the specimen and VP1 sequencing databases. We thank David R. Kilpatrick for contributions to the early development of several of the real-time RT-PCR assays. We appreciate helpful discussions with colleagues in the Global Polio Laboratory Network and WHO headquarters and regional offices. We thank Laila Bassioni (WHO Regional Reference Polio Laboratory, VACSERA, Cairo, Egypt) and Berhane Beyene (National Polio & Measles Laboratory, Ethiopian Health and Nutrition Research Institute, Addis Ababa, Ethiopia) for permitting the use of sequence data.
IHRC, Inc., is a contracting agency to the Division of Viral Diseases, Centers for Diseases Control and Prevention, Atlanta, Georgia, USA.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. The use of trade names is for identification only and does not imply endorsement by the CDC or the U.S. government.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01624-17.
REFERENCES
- 1.WHO. 2013. The Global Polio Eradication Initiative. WHO, Geneva, Switzerland: http://www.polioeradication.org/Resourcelibrary/Strategyandwork.aspx Accessed 10 April 2017. [Google Scholar]
- 2.Duizer E, Rutjes S, de Roda Husman AM, Schijven J. 2016. Risk assessment, risk management and risk-based monitoring following a reported accidental release of poliovirus in Belgium, September to November 2014. Euro Surveill 21:30169. doi: 10.2807/1560-7917.ES.2016.21.11.30169. [DOI] [PubMed] [Google Scholar]
- 3.Shulman LM, Gavrilin E, Jorba J, Martin J, Burns CC, Manor Y, Moran-Gilad J, Sofer D, Hindiyeh MY, Gamzu R, Mendelson E, Grotto I, Genotype-Phenotype Identification (GPI) Group. 2014. Molecular epidemiology of silent introduction and sustained transmission of wild poliovirus type 1, Israel, 2013. Euro Surveill 19:20709. doi: 10.2807/1560-7917.ES2014.19.7.20709. [DOI] [PubMed] [Google Scholar]
- 4.WHO. 15 February 2017. Polio this week. WHO, Geneva, Switzerland: http://www.polioeradication.org/Dataandmonitoring/Poliothisweek.aspx Accessed 17 February 2017. [Google Scholar]
- 5.Wood DJ, Hull B. 1999. L20B cells simplify culture of polioviruses from clinical samples. J Med Virol 58:188–192. doi:. [DOI] [PubMed] [Google Scholar]
- 6.WHO. 2004. Polio laboratory manual, 4th ed, vol WHO/IVB/04.10 WHO, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/68762/1/WHO_IVB_04.10.pdf. [Google Scholar]
- 7.WHO. 2007. S1. Supplement to the WHO Polio Laboratory Manual: an alternative test algorithm for poliovirus isolation and characterization. WHO, Geneva, Switzerland: http://polioeradication.org/wp-content/uploads/2017/05/NewAlgorithmForPoliovirusIsolationSupplement1.pdf. [Google Scholar]
- 8.WHO. 2015. The 21st Informal Consultation on the Global Polio Laboratory Network 25. WHO, Geneva, Switzerland: http://polioeradication.org/wp-content/uploads/2016/07/GPLN_Meeting_recommendations_2015.pdf Accessed 25 May 2017. [Google Scholar]
- 9.Kilpatrick DR, Ching K, Iber J, Chen Q, Yang SJ, De L, Williams AJ, Mandelbaum M, Sun H, Oberste MS, Kew OM. 2014. Identification of vaccine-derived polioviruses using dual-stage real-time RT-PCR. J Virol Methods 197:25–28. doi: 10.1016/j.jviromet.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kilpatrick DR, Yang C-F, Ching K, Vincent A, Iber J, Campagnoli R, Mandelbaum M, De L, Yang S-J, Nix A, Kew OM. 2009. Rapid group-, serotype-, and vaccine strain-specific identification of poliovirus isolates by real-time reverse transcription-PCR using degenerate primers and probes containing deoxyinosine residues. J Clin Microbiol 47:1939–1941. doi: 10.1128/JCM.00702-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jorba J. 2016. Phylogenetic analysis of poliovirus sequences, p 227–237. In Martín J. (ed), Poliovirus: methods and protocols. Springer, New York, NY. doi: 10.1007/978-1-4939-3292-4_11. [DOI] [PubMed] [Google Scholar]
- 12.WHO. 20 September 2015. Global eradication of wild poliovirus type 2 declared. WHO, Geneva, Switzerland: http://polioeradication.org/news-post/global-eradication-of-wild-poliovirus-type-2-declared/ Accessed 13 November 2016. [Google Scholar]
- 13.Kew O, Morris-Glasgow V, Landaverde M, Burns C, Shaw J, Garib Z, Andre J, Blackman E, Freeman CJ, Jorba J, Sutter R, Tambini G, Venczel L, Pedreira C, Laender F, Shimizu H, Yoneyama T, Miyamura T, van Der Avoort H, Oberste MS, Kilpatrick D, Cochi S, Pallansch M, de Quadros C. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356–359. doi: 10.1126/science.1068284. [DOI] [PubMed] [Google Scholar]
- 14.WHO. March 2017. Fact sheet: vaccine-derived poliovirus. WHO, Geneva, Switzerland: http://polioeradication.org/wp-content/uploads/2016/07/GPEI-cVDPV-factsheet_March-2017.pdf. [Google Scholar]
- 15.Dunn G, Klapsa D, Wilton T, Stone L, Minor PD, Martin J. 2015. Twenty-eight years of poliovirus replication in an immunodeficient individual: impact on the Global Polio Eradication Initiative. PLoS Pathog 11:e1005114. doi: 10.1371/journal.ppat.1005114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McAllister RM, Melnyk J, Finklestein JZ, Adams EC, Gardner MB. 1969. Cultivation in vitro of cells derived from a human rhabdomyosarcoma. Cancer 24:520–526. doi:. [DOI] [PubMed] [Google Scholar]
- 17.Mendelsohn CL, Wimmer E, Racaniello VR. 1989. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 18.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 19.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shulman LM, Manor Y, Hindiyeh M, Sofer D, Mendelson E. 2016. Molecular characterization of polio from environmental samples: ISSP, The Israeli Sewage Surveillance Protocol, p 55–107. In Martín J. (ed), Poliovirus: methods and protocols. Springer, New York, NY. doi: 10.1007/978-1-4939-3292-4_5. [DOI] [PubMed] [Google Scholar]
- 21.Burns CC, Shaw J, Campagnoli R, Jorba J, Vincent A, Quay J, Kew O. 2006. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. J Virol 80:3259–3272. doi: 10.1128/JVI.80.7.3259-3272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu HM, Zheng DP, Zhang LB, Oberste MS, Pallansch MA, Kew OM. 2000. Molecular evolution of a type 1 wild-vaccine poliovirus recombinant during widespread circulation in China. J Virol 74:11153–11161. doi: 10.1128/JVI.74.23.11153-11161.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilpatrick DR, Iber JC, Chen Q, Ching K, Yang SJ, De L, Mandelbaum MD, Emery B, Campagnoli R, Burns CC, Kew O. 2011. Poliovirus serotype-specific VP1 sequencing primers. J Virol Methods 174:128–130. doi: 10.1016/j.jviromet.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 24.Burns CC, Kilpatrick DR, Iber JC, Chen Q, Kew OM. 2016. Molecular properties of poliovirus isolates: nucleotide sequence analysis, typing by PCR and real-time RT-PCR, p 177–212. In Martín J. (ed), Poliovirus: methods and protocols. Springer, New York, NY. doi: 10.1007/978-1-4939-3292-4_9. [DOI] [PubMed] [Google Scholar]
- 25.Jorba J, Campagnoli R, De L, Kew O. 2008. Calibration of multiple poliovirus molecular clocks covering an extended evolutionary range. J Virol 82:4429–4440. doi: 10.1128/JVI.02354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Zhu S, Yan D, Liu G, Bai R, Wang D, Chen L, Zhu H, An H, Kew O, Xu W. 2010. Natural type 3/type 2 intertypic vaccine-related poliovirus recombinants with the first crossover sites within the VP1 capsid coding region. PLoS One 5:e15300. doi: 10.1371/journal.pone.0015300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kilpatrick DR, Nottay B, Yang CF, Yang SJ, Mulders MN, Holloway BP, Pallansch MA, Kew OM. 1996. Group-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residue at positions of codon degeneracy. J Clin Microbiol 34:2990–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Core Team. 2016. R: a language and environment for statistical computing, v3.3.1. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org. [Google Scholar]
- 29.Marascuilo LA, Serlin RC. 1988. Statistical methods for the social and behavioral sciences. W. H. Freeman, New York, NY. [Google Scholar]
- 30.WHO. 1990. Manual for the virological investigation of poliomyelitis, p 104 WHO, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/62186/1/WHO_EPI_CDS_POLIO_90.1.pdf. [Google Scholar]
- 31.De L, Nottay B, Yang CF, Holloway BP, Pallansch M, Kew O. 1995. Identification of vaccine-related polioviruses by hybridization with specific RNA probes. J Clin Microbiol 33:562–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kilpatrick DR, Nottay B, Yang CF, Yang SJ, Da Silva E, Penaranda S, Pallansch M, Kew O. 1998. Serotype-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residues at positions of codon degeneracy. J Clin Microbiol 36:352–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.WHO. 2015. WHO Global Action Plan to minimize poliovirus facility-associated risk after type-specific eradication of wild polioviruses and sequential cessation of oral polio vaccine use. WHO, Geneva, Switzerland: http://polioeradication.org/wp-content/uploads/2016/09/GAPIII_2014.pdf. [Google Scholar]
- 34.Toyoda H, Kohara M, Kataoka Y, Suganuma T, Omata T, Imura N, Nomoto A. 1984. Complete nucleotide sequences of all three poliovirus serotype genomes. J Mol Biol 174:561–585. doi: 10.1016/0022-2836(84)90084-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.