ABSTRACT
Staphylococcus schleiferi is a beta-hemolytic, coagulase-variable colonizer of small animals that can cause opportunistic infections in humans. In veterinary isolates, the rate of mecA-mediated oxacillin resistance is significant, with reported resistance rates of >39%. The goal of this study was to evaluate oxacillin and cefoxitin disk diffusion (DD) and MIC breakpoints for detection of mecA-mediated oxacillin resistance in 52 human and 38 veterinary isolates of S. schleiferi. Isolates were tested on multiple brands of commercial media and according to Clinical and Laboratory Standards Institute (CLSI) methods. Zone diameters and MIC values were interpreted using CLSI breakpoints (CLSI, Performance Standards for Antimicrobial Susceptibility Testing. M100-S27, 2017) for Staphylococcus aureus/Staphylococcus lugdunensis, coagulase-negative staphylococci (CoNS), and Staphylococcus pseudintermedius. Results were compared to those of mecA PCR. Twenty-nine of 90 (32%) isolates were mecA positive. Oxacillin inhibition zone sizes and MICs interpreted by S. pseudintermedius breakpoints reliably differentiated mecA-positive and mecA-negative isolates, with a categorical agreement (CA) of 100% and no very major errors (VMEs) or major errors (MEs) for all media. For cefoxitin DD results interpreted using S. aureus/S. lugdunensis and CoNS breakpoints, CA values were 85% and 75%, respectively, and there were 72% and 64% VMEs, respectively, and 0 MEs. For cefoxitin MICs interpreted using S. aureus/S. lugdunensis breakpoints, CA was 81%, and there were 60% VMEs and no MEs. Our data demonstrate that oxacillin DD or MIC testing methods using the current S. pseudintermedius breakpoints reliably identify mecA-mediated oxacillin resistance in S. schleiferi, while cefoxitin DD and MIC testing methods perform poorly.
KEYWORDS: breakpoints, cefoxitin, mecA, oxacillin, PBP2a, Staphylococcus schleiferi
INTRODUCTION
Staphylococcus schleiferi is an emerging zoonotic pathogen that colonizes the skin and mucosal surfaces of small animals (1–3). Isolates form medium to large, nonpigmented, beta-hemolytic colonies on 5% sheep blood agar (4, 5). The species is further divided into S. schleiferi subsp. schleiferi and S. schleiferi subsp. coagulans. Staphylococcus schleiferi subsp. schleiferi was first isolated from human specimens, in 1988 (4), while S. schleiferi subsp. coagulans was first isolated from dogs with otitis externa, in 1990 (5). The subspecies have high levels of DNA homology but differ in several phenotypic characteristics, including clumping factor, tube coagulase, and urease production. S. schleiferi subsp. schleiferi is clumping factor positive, tube coagulase negative, and urease negative, while S. schleiferi subsp. coagulans is clumping factor negative, tube coagulase positive, and urease positive (4, 5). Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) can reliably identify S. schleiferi to the species level, but further biochemical testing is needed to differentiate the two subspecies (6).
In small animals, S. schleiferi most frequently colonizes the skin, nares, ears, and rectum of dogs, where it can cause inflammatory skin disease, otitis externa, and otitis media (2, 3, 5, 7–14). Though found at a lower prevalence than that in dogs, S. schleiferi has also been isolated from healthy cats (1), cats with inflammatory skin disease (1), and parrots (2). Oxacillin resistance rates vary but can be high, with some studies reporting oxacillin resistance for 39 to 73% of veterinary isolates (7–9, 15, 16). Staphylococcal cassette chromosome mec type IV (SCCmec IV) has been identified in S. schleiferi subsp. coagulans (16). S. schleiferi has also been reported to carry SCCmec types I and IV (17, 18). In addition to oxacillin resistance, reduced susceptibility to clindamycin, erythromycin, and fluoroquinolones has been reported (13, 19, 20). In a small study, S. schleiferi isolates were negative for beta-lactamase production (21).
Though it is primarily a veterinary pathogen, S. schleiferi can also cause opportunistic infections in humans. Cases of endophthalmitis (22), endocarditis (23, 24), bacteremia (25), osteomyelitis (26), and wound (27), surgical site (27), and pacemaker (28) infections have been reported. S. schleiferi was implicated in an outbreak of surgical site infections and was originally misidentified as Staphylococcus aureus (29). Interestingly, while both subspecies can cause infection, S. schleiferi subsp. schleiferi is more prevalent in causing human infections (27, 29).
S. schleiferi has been reported to give false-positive results in latex agglutination tests for S. aureus identification, at rates of 25 to 75% (30). Use of MALDI-TOF MS will likely increase the number of S. schleiferi isolates identified, and the high oxacillin resistance rates in this species are concerning. Many clinical laboratories use cefoxitin disk diffusion (DD) testing for detection of oxacillin resistance in staphylococci other than S. pseudintermedius, as outlined by the Clinical and Laboratory Standards Institute (CLSI) M100-S27 document (31, 32). Previous data supporting the use of cefoxitin as a surrogate agent for detection of oxacillin resistance in coagulase-negative staphylococci (CoNS) were largely derived from Staphylococcus epidermidis isolates (33). However, the most accurate methods for detecting mecA-mediated oxacillin resistance in other CoNS species have yet to be determined. Some studies have shown that cefoxitin disk testing has low sensitivity for detecting oxacillin resistance in S. schleiferi veterinary isolates and S. pseudintermedius (15, 33, 34).
The goal of the present study was to evaluate oxacillin and cefoxitin DD and broth microdilution (BMD) MIC testing for detection of mecA-mediated oxacillin resistance in 52 human and 38 veterinary isolates of S. schleiferi. Oxacillin-resistant staphylococci are resistant to all beta-lactam antibiotics except for new anti-methicillin-resistant S. aureus (anti-MRSA) cephalosporins. We demonstrate that oxacillin DD results interpreted by the CLSI M100-S27 breakpoints for S. pseudintermedius reliably detect mecA-positive and mecA-negative S. schleiferi isolates, while cefoxitin is an unreliable surrogate agent.
(The results of this study were presented to the CLSI Antimicrobial Susceptibility Testing Subcommittee in June 2017, leading to the addition of specific breakpoints for oxacillin disk diffusion and MIC testing of S. schleiferi for the forthcoming 28th edition of the M100 document.)
MATERIALS AND METHODS
Specimens.
A total of 90 S. schleiferi isolates were included in this study (Table 1). Human isolates (n = 52) were submitted by Becton Dickinson and Company (BD) (n = 13), JMI Laboratories (n = 22), and Weill Cornell Medicine (n = 17). Canine and other small animal isolates (n = 38) were obtained from the Texas A&M University College of Veterinary Medicine (n = 12), the University of Tennessee College of Veterinary Medicine (n = 25), and Weill Cornell Medicine (n = 1). Isolates were identified to the species or subspecies level at each institution, using the corresponding standard operating procedures. Subspecies identifications were confirmed by urease testing at the Ronald Reagan UCLA Medical Center, Los Angeles, CA. Urease testing on urea agar slants (BD) was performed according to the manufacturer's protocol, with the slants incubated at 35°C in ambient air for 24 h.
TABLE 1.
Summary of study isolates submitted by various institutions
| Institute | No. of isolates | Source | Specimen source(s) | No. of isolates |
|
|---|---|---|---|---|---|
| S. schleiferi subsp. schleiferi | S. schleiferi subsp. coagulans | ||||
| BD | 13 | Human | Unknown | 13 | 0 |
| JMI Laboratories | 22 | Human | Unknown | 9 | 13 |
| Weill Cornell Medicine | 17 | Human | Blood, catheter tip, scalp, skin, urine, unknown, wound | 11 | 6 |
| Texas A&M | 12 | Canine | Ear, lung, urine | 0 | 12 |
| University of Tennessee | 25 | Canine | Bone, ear, skin | 0 | 25 |
| Weill Cornell Medicine | 1 | Small animal | Ear | 0 | 1 |
| Total | 90 | 33 | 57 | ||
mecA PCR.
Fifty-four isolates with known mecA genotypes were submitted. For the remaining isolates, colony PCR was performed on 18- to 24-h isolates grown on 5% sheep blood agar plates (SBAP). The following primers were used to amplify a 533-bp product from the mecA gene: 5′-AAAATCGATGGTAAAGGTTGGC-3′ and 5′-AGTTCTGCAGTACCGGATTTGC-3′ (35). A pipette tip was used to transfer a pinpoint amount of an isolated colony to a 25-μl PCR mixture containing 12.5 μl AmpliTaq Gold 360 master mix (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA), 0.5 μM (each) mecA primers, and 11.25 μl water. Reaction mixtures were incubated at 95°C for 10 min, followed by 35 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s. The final extension was performed at 72°C for 7 min. As a positive control, colony PCR was performed on S. aureus ATCC 43300, a mecA-positive strain, and 1 μl purified genomic DNA from the same strain. PCR products were visualized on a 4% precast agarose gel (Lonza, Basel, Switzerland).
Antimicrobial susceptibility testing (AST).
Isolates were stored as described previously (34) and subcultured twice on 5% SBAP before testing. DD and BMD tests were performed as described by the CLSI (31, 36). DD was evaluated on Mueller-Hinton agar (MHA) plates obtained from 3 vendors: Remel (Lenexa, KS), Hardy Diagnostics (Santa Maria, CA), and BD. Disks containing 1 μg oxacillin and 30 μg cefoxitin (BBL, BD) were used. BMD was performed by the CLSI reference method, using frozen-form panels containing cation-adjusted Mueller-Hinton broth (CA-MHB) with cefoxitin (unsupplemented) or oxacillin supplemented with 2% NaCl. BMD panels were made by Thermo Fisher. MIC tests using CA-MHB from 3 different manufacturers (Difco, BD, and Oxoid) were evaluated on a single panel. Oxacillin and cefoxitin were tested in 2-fold dilutions at concentrations ranging from 0.015 μg/ml to 32 μg/ml. Isolated colonies grown overnight on SBAP at 35 to 37°C in ambient air were resuspended in 0.85% saline to obtain a 0.5 McFarland standard. The suspensions were used to inoculate all DD and MIC plates per CLSI recommendations (31, 36). DD test plates were incubated at 35°C in ambient air, and zones of inhibition were measured at 16 to 18 h for oxacillin and 24 h for cefoxitin. During preliminary studies, zones of inhibition were read with both transmitted and reflected light, and there was no difference noted between the two methods. Therefore, for the final study, both oxacillin and cefoxitin zones of inhibition were read using reflected light. BMD test plates were incubated at 35°C in ambient air and read at 16 to 20 h for cefoxitin and 24 h for oxacillin. S. aureus ATCC 25923 and S. aureus ATCC 29213 were used as quality control strains for DD and BMD tests, respectively.
Data analysis.
Zone diameters and MIC values were interpreted using breakpoints for the following organisms, obtained from the CLSI M100-S27 document: (i) S. aureus/S. lugdunensis; (ii) CoNS, excluding S. lugdunensis and S. pseudintermedius; and/or (iii) S. pseudintermedius (Table 2) (32). Results were compared to the results of mecA PCR, which was considered the gold standard for oxacillin resistance. Categorical agreement (CA), very major errors (VMEs), and major errors (MEs) were calculated as previously described (37). CA was determined using mecA PCR results as the reference to define isolates as resistant or susceptible. VMEs were counted as identifications of isolates that were mecA positive but oxacillin or cefoxitin susceptible. MEs were defined as identifications of isolates that were mecA negative but oxacillin or cefoxitin resistant.
TABLE 2.
Breakpoints used for prediction of S. schleiferi mecA-mediated oxacillin resistance in this studya
| Organism | Oxacillin breakpoint |
Cefoxitin breakpoint |
||||||
|---|---|---|---|---|---|---|---|---|
| DD inhibition zone (mm) |
MIC (μg/ml) |
DD inhibition zone (mm) |
MIC (μg/ml) |
|||||
| S | R | S | R | S | R | S | R | |
| S. aureus/S. lugdunensis | NA | NA | ≤2 | ≥4 | ≥22 | ≤21 | ≤4 | ≥8 |
| Coagulase-negative staphylococci, except S. lugdunensis and S. pseudintermedius | NA | NA | ≤0.25 | ≥0.5 | ≥25 | ≤24 | NA | NA |
| S. pseudintermediusb | ≥18 | ≤17 | ≤0.25 | ≥0.5 | NA | NA | NA | NA |
From the CLSI M100-S27 document (32). NA, not applicable; S, susceptible; R, resistant.
In the 28th edition of the CLSI M100 document, the guidance for S. pseudintermedius will also apply to S. schleiferi.
Discrepancy analysis.
For any isolates with oxacillin results that were discordant with mecA PCR results, testing was repeated by both AST and PCR. If the error resolved, it was excluded as an error.
PBP2a testing.
Penicillin binding protein 2a (PBP2a) testing was performed on 54 S. schleiferi isolates by use of Alere PBP2a SA culture colony test kits (Alere Inc., Scarborough, ME) and Oxoid PBP2′ latex agglutination test kits (Thermo Fisher Scientific, Waltham, MA). Only 54 isolates were tested due to a limited number of available testing kits. Isolates were chosen based on mecA PCR results to give an almost even distribution of mecA-positive and mecA-negative isolates (28 and 26, respectively). Forty-three isolates were S. schleiferi subsp. coagulans, and 11 were S. schleiferi subsp. schleiferi. Thirty-eight isolates were from animals (all S. schleiferi subsp. coagulans), and 16 were from humans (5 S. schleiferi subsp. coagulans isolates and 11 S. schleiferi subsp. schleiferi isolates). Colonies from SBAP used for DD and BMD inoculum preparation were tested for noninduced PBP2a expression according to the manufacturers' instructions for S. aureus. S. aureus ATCC 43300 and S. aureus 25923 were used as positive and negative controls, respectively.
RESULTS
Isolates.
Ninety isolates were tested in this study (Table 1). Fifty-two (58%) were isolated from human specimens (blood, wounds, urine, ears, catheters, skin, and the scalp). Thirty-eight (42%) isolates were urine, bone, lung, pyoderma (of the ear), or skin isolates from canines or other small animals. Thirty-three (37%) isolates were identified as S. schleiferi subsp. schleiferi, all of which were isolated from human specimens. Fifty-seven (63%) isolates were identified as S. schleiferi subsp. coagulans; 19 (33%) of these were isolated from human specimens, and 38 (67%) were isolated from animals.
mecA PCR.
Twenty-nine (32%) isolates were mecA positive. All of the mecA-positive isolates were S. schleiferi subsp. coagulans, and four of these were isolated from human specimens. Sixty-one (68%) isolates were mecA negative. Of these, 33 (54%) were S. schleiferi subsp. schleiferi and 28 (46%) were S. schleiferi subsp. coagulans.
Cefoxitin DD and BMD testing.
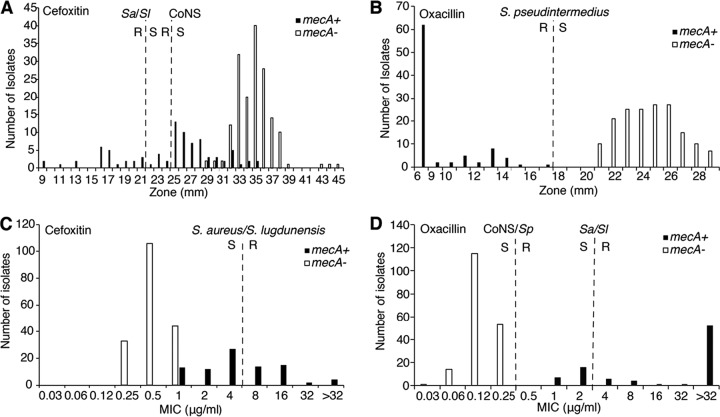
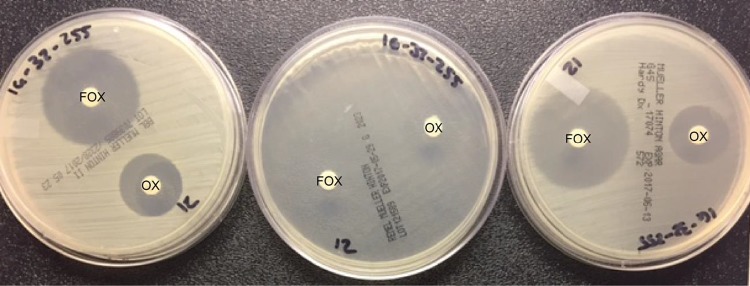
Results from the cefoxitin DD and BMD tests are summarized in Table 3 and Fig. 1. For cefoxitin tests, neither the zones of inhibition nor the MICs showed clear divisions between mecA-positive and mecA-negative isolates for any medium (either MHA or CA-MHB) brand tested (Fig. 1; see Fig. S1 and S2 in the supplemental material). For DD testing, 17 isolates showed only faint growth on Remel MHA medium (Fig. 2), and thus zones of inhibition could not be measured. Therefore, a total of 253 data points were collected for all media (Fig. 1A).
TABLE 3.
Performances of cefoxitin DD and BMD testing for detection of mecA-mediated oxacillin resistance in S. schleiferi
| Breakpoints | Summary |
BD medium |
Hardy medium |
Remel medium |
Difco CA-MHB |
BD CA-MHB |
Oxoid CA-MHB |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CA (%) | No. of errors/no. of isolates (%) |
CA (%) | No. of errors/no. of isolates (%) |
CA (%) | No. of errors/no. of isolates (%) |
CA (%)a | No. of errors/no. of isolates (%) |
CA (%) | No. of errors/no. of isolates (%) |
CA (%) | No. of errors/no. of isolates (%) |
CA (%) | No. of errors/no. of isolates (%) |
||||||||
| VMEs | MEs | VMEs | MEs | VMEs | MEs | VMEs | MEsa | VMEs | MEs | VMEs | MEs | VMEs | MEs | ||||||||
| CLSI M100-S27 DD breakpoints | |||||||||||||||||||||
| S. aureus/S. lugdunensis | 85 | 63/87 (72) | 0/166 (0) | 78 | 20/29 (69) | 0/61 (0) | 76 | 22/29 (76) | 0/61 (0) | 71 | 21/29 (72) | 0/44 (0) | |||||||||
| CoNS staphylococci (except for S. lugdunensis and S. pseudintermedius) | 75 | 56/87 (64) | 0/166 (0) | 81 | 17/29 (59) | 0/61 (0) | 77 | 21/29 (72) | 0/61 (0) | 75 | 18/29 (62) | 0/44 (0) | |||||||||
| CLSI M100-S27 MIC breakpoints | |||||||||||||||||||||
| S. aureus/S. lugdunensis | 81 | 52/87 (60) | 0/183 (0) | 81 | 12/29 (41) | 0/61 (0) | 81 | 12/29 (41) | 0/61 (0) | 80 | 11/29 (38) | 0/61 (0) | |||||||||
We were unable to read zones for 17 mecA-negative isolates due to poor growth; these were not included in the denominator.
FIG 1.
Distributions of cefoxitin and oxacillin growth inhibition zone diameters and MICs as determined by DD and BMD testing on all media tested. M100-S27 DD and MIC breakpoints are shown for S. aureus/S. lugdunensis (Sa/Sl), coagulase-negative staphylococci (excluding S. lugdunensis and S. pseudintermedius) (CoNS), and S. pseudintermedius. R, resistant; S, susceptible. (A) Cefoxitin DD testing (for BD, Hardy, and Remel media, n = 253). (B) Oxacillin DD testing (for BD, Hardy, and Remel media, n = 253). (C) Cefoxitin MICs (for Difco, BD, and Oxoid media, n = 270). (D) Oxacillin MICs (for Difco, BD, and Oxoid media, n = 270).
FIG 2.
Growth of a single strain of S. schleiferi on BD (left), Remel (middle), and Hardy (right) MHA plates with oxacillin (OX) and cefoxitin (FOX) disks.
DD and MIC results were interpreted using the cefoxitin breakpoints listed in Table 2. On applying the CLSI M100-S27 S. aureus/S. lugdunensis breakpoints for DD testing using cefoxitin, the CA values for BD, Hardy, and Remel media were 78%, 76%, and 71%, respectively. For cefoxitin, there were 20/29 (69%), 22/29 (76%), and 21/29 (72%) VMEs for the BD, Hardy, and Remel media, respectively. There were no MEs because all mecA-negative isolates were susceptible by DD testing on all media tested, with zone sizes ranging from 29 to 45 mm (Fig. 1). For BMD, all brands performed similarly with cefoxitin. The CA values and numbers of VMEs for BD, Difco, and Remel CA-MHB were 81% and 12/29 (41%), 81% and 12/29 (41%), and 80% and 11/29 (38%), respectively. There were no MEs for any of the CA-MHB medium brands tested. On applying the CoNS breakpoints for DD testing using cefoxitin, the CA value and percentage of VMEs were 81% and 59% for BD medium, 77% and 72% for Hardy medium, and 75% and 62% for Remel medium (Table 3). There were no MEs with application of the CoNS breakpoints.
Overall, for cefoxitin DD results interpreted using S. aureus/S. lugdunensis and CoNS breakpoints, CA was 85% and 75%, respectively, and there were 63/87 (72%) and 56/87 (64%) VMEs, respectively. Cefoxitin MICs interpreted using S. aureus/S. lugdunensis breakpoints yielded an overall CA of 81% and 52/87 (60%) VMEs. There were no MEs for either DD or MIC results interpreted using any breakpoints.
Oxacillin DD and BMD testing.
Results from the oxacillin DD and BMD tests are summarized in Table 4 and Fig. 1. In contrast to the cefoxitin results, there was a clear division between mecA-positive and mecA-negative isolates for both zones of inhibition and MICs for all medium brands tested (Fig. 1; Fig. S3 and S4). On applying the S. pseudintermedius breakpoints for DD testing, CA was 100% for all medium brands. There were no VMEs or MEs. On Remel medium, 16 isolates did not grow well enough for reading of the oxacillin DD zones, so the denominator (n) was 45.
TABLE 4.
Performances of oxacillin DD and BMD testing for detection of mecA-mediated oxacillin resistance in S. schleiferi
| Breakpoints | Summary |
BD medium |
Hardy medium |
Remel medium |
Difco CA-MHB |
BD CA-MHB |
Oxoid CA-MHB |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CA (%)a | No. of errors/no. of isolates (%) |
CA (%) | No. of errors/no. of isolates (%) |
CA (%) | No. of errors/no. of isolates (%) |
CA (%)a | No. of errors/no. of isolates (%) |
CA (%) | No. of errors/no. of isolates (%) |
CA (%) | No. of errors/no. of isolates (%) |
CA (%) | No. of errors/no. of isolates (%) |
||||||||
| VMEs | MEsa | VMEs | MEs | VMEs | MEs | VMEs | MEsa | VMEs | MEs | VMEs | MEs | VMEs | MEs | ||||||||
| CLSI M100-S27 DD breakpoints | |||||||||||||||||||||
| S. pseudintermedius | 100 | 0/87 (0) | 0/167 (0) | 100 | 0/29 (0) | 0/61 (0) | 100 | 0/29 (0) | 0/61 (0) | 100 | 0/29 (0) | 0/45 (0) | |||||||||
| CLSI M100-S27 MIC breakpoints | |||||||||||||||||||||
| S. aureus/S. lugdunensis | 91 | 23/87 (26) | 0/183 (0) | 98 | 6/29 (21) | 0/61 (0) | 92 | 7/29 (24) | 0/61 (0) | 89 | 10/29 (35) | 0/61 (0) | |||||||||
| Coagulase-negative staphylococci (except for S. lugdunensis and S. pseudintermedius) | 100 | 0/87 (0) | 0/183 (0) | 100 | 0/29 (0) | 0/61 (0) | 100 | 0/29 (0) | 0/61 (0) | 100 | 0/29 (0) | 0/61 (0) | |||||||||
| S. pseudintermediusb | 100 | 0/87 (0) | 0/183 (0) | 100 | 0/29 (0) | 0/61 (0) | 100 | 0/29 (0) | 0/61 (0) | 100 | 0/29 (0) | 0/61 (0) | |||||||||
We were unable to read zones for 16 mecA-negative isolates; these were not included in the denominator.
In the 28th edition of the CLSI M100 document, the guidance for S. pseudintermedius will also apply to S. schleiferi.
For oxacillin BMD, CA values with the S. aureus/S. lugdunensis breakpoints were 98%, 92%, and 89% for Difco, BD, and Oxoid CA-MHB, respectively. The numbers of VMEs for Difco, BD, and Oxoid CA-MHB were 6/29 (21%), 7/29 (24%), and 10/29 (35%), respectively. There were no MEs for the three medium brands tested. The oxacillin MIC breakpoints for CoNS and S. pseudintermedius are the same. CA was 100%, and there were no VMEs or MEs for the three medium brands.
Overall, for oxacillin DD results interpreted using the S. pseudintermedius breakpoints, overall CA was 100%, and there were no VMEs or MEs. Oxacillin MICs interpreted using the S. aureus/S. lugdunensis breakpoints yielded an overall CA of 91%, and there were 23/87 (26%) isolates with VMEs and no MEs. Overall CA for oxacillin MICs interpreted using CoNS and S. pseudintermedius breakpoints was 100%, with no VMEs or MEs.
Discrepant analysis.
One isolate tested mecA PCR negative but was PBP2a positive and oxacillin resistant by DD and BMD testing. When all tests were repeated, the mecA PCR was positive, so this error was excluded from our analyses.
PBP2a testing.
Compared to mecA PCR as the gold standard, PBP2a results showed 100% CA for all 54 S. schleiferi isolates tested, using both Alere PBP2a SA culture colony test kits and Oxoid PBP2′ latex agglutination test kits.
DISCUSSION
The widespread adoption of MALDI-TOF MS for bacterial identification has allowed clinical laboratories to better identify staphylococci to the species level (6, 38). Rapid adoption of this technology, combined with an increasing immunocompromised population and close contact between humans and companion animals, has increased the number and diversity of clinically significant CoNS isolates identified (39). This increase leads to the issue of determining which methods are best for detecting mecA-mediated oxacillin resistance in CoNS, which has not been critically evaluated for many species.
Here we present oxacillin and cefoxitin DD and BMD data for detection of mecA-mediated oxacillin resistance in 90 human and veterinary isolates of S. schleiferi. PCR for mecA was used to define oxacillin resistance. The correlation between oxacillin resistance and mecA detection has previously been reported to be 93 to 95% (12, 16). In previous studies, the mechanism of oxacillin resistance in mecA-negative isolates was not determined (12, 16).
While a similar study has been performed on veterinary isolates (15), our study included 52 human isolates, MIC testing, MHA manufactured by BD, Hardy, and Remel, and CA-MHB manufactured by Difco, BD, and Oxoid. The 3 brands of CA-MHB performed similarly for both oxacillin and cefoxitin MIC testing (Tables 3 and 4). Some isolates did not grow satisfactorily on Remel MHA to produce a readable zone of inhibition (Fig. 2). However, for those isolates that grew well, major differences in performance between brands were not noted (Fig. 1; Tables 3 and 4). Nonetheless, laboratories should be cognizant of medium-to-medium variability for commercial MHAs.
For DD testing of staphylococci other than S. pseudintermedius, the 2017 guidance for the CLSI reference method uses cefoxitin as a surrogate agent for detecting mecA-mediated oxacillin resistance (31, 32). Our data show that cefoxitin DD testing does not accurately predict the presence of mecA in S. schleiferi by use of the M100-S27 breakpoints for CoNS. Although cefoxitin DD testing accurately identified mecA-negative isolates, VME rates were unacceptably high for S. schleiferi on all media. Our findings are similar to those of previous studies that have shown species-dependent results for cefoxitin disk diffusion testing of CoNS. Compared to PBP2a testing results, the overall sensitivity of cefoxitin DD testing was only 25% for 150 isolates of S. intermedius and S. schleiferi (15). In a study of 170 isolates of mecA-positive CoNS, cefoxitin DD testing failed to identify five mecA-positive isolates of S. simulans (33). For S. saprophyticus, cefoxitin DD testing was 100% sensitive but only 56% specific compared to mecA PCR (40). Finally, the VME rate for cefoxitin DD testing for S. pseudintermedius was 29.7% compared to the mecA PCR results (34). The findings from our study may be problematic for laboratories that identify CoNS by use of phenotypic methods alone because S. schleiferi can be misidentified as S. aureus, leading to erroneous oxacillin susceptibility results.
As a result of this study, along with the data demonstrated previously for S. pseudintermedius (28), the CLSI recently removed cefoxitin DD testing as an option for confirming that non-S. epidermidis CoNS isolates from serious infections with oxacillin MICs in the 0.5- to 2.0-μg/ml range are truly oxacillin resistant. Laboratories should confirm susceptibility for such isolates by mecA or PBP2a tests. At present, the Alere PBP2a SA culture colony test is FDA cleared for PBP2a testing in S. aureus, while the Oxoid PBP2′ latex agglutination test kit is FDA cleared for testing PBP2a in S. aureus and induced CoNS. In this study, PBP2a testing was 100% sensitive and 100% specific for identifying mecA-positive and mecA-negative isolates when colonies were tested without induction. For S. schleiferi, the Oxoid PBP2′ latex agglutination test kit has shown 85 to 100% CA between PBP2a expression and oxacillin resistance (3, 15), while the Alere PBP2a SA culture colony test has shown 100% CA (41). Additionally, the CLSI Staphylococcus Ad Hoc Working Group will continue to systematically evaluate the performance of current recommended phenotypic testing options for predicting mecA-mediated oxacillin resistance in the genus Staphylococcus.
Oxacillin DD and BMD testing performed most reliably in detecting mecA-mediated oxacillin resistance in S. schleiferi. Our findings are similar to those of Bemis et al. (15), who tested 43 S. schleiferi canine isolates for PBP2a and correlated the results with those of oxacillin and cefoxitin DD testing. Using breakpoints for animal isolates, they found 100% CA between PBP2a testing and oxacillin DD testing for both subspecies but only 0% and 46% CA between PBP2a testing and cefoxitin DD testing for S. schleiferi subsp. schleiferi and S. schleiferi subsp. coagulans, respectively (15). Additionally, Wu et al. (34) found that oxacillin DD testing performed better than cefoxitin DD testing for detecting mecA in S. pseudintermedius, a common colonizer of dogs and cats. One limitation to our study is that all 29 mecA-positive isolates were S. schleiferi subsp. coagulans because we were unable to obtain mecA-positive S. schleiferi subsp. schleiferi isolates. There may be differences in mecA-positive S. schleiferi subsp. schleiferi DD and BMD testing that could not be examined in this study.
In summary, we have shown that oxacillin DD and BMD testing methods using the current S. pseudintermedius breakpoints accurately identify mecA-mediated oxacillin resistance in S. schleiferi. Cefoxitin DD and BMD were unreliable for identifying oxacillin resistance due to the large number of false-susceptible results observed. The results from this study were presented to the CLSI Antimicrobial Susceptibility Testing Subcommittee in June 2017, leading to specific breakpoints for oxacillin DD and MIC testing of S. schleiferi.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge those at the Ronald Reagan UCLA Medical Center, Weill Cornell Medicine, the Texas A&M University College of Veterinary Medicine, the University of Tennessee College of Veterinary Medicine, BD, and JMI Laboratories who contributed isolates and/or participated in the testing of isolates. We also thank Thermo Fisher Scientific for making the BMD panels used in this study and Alere for providing PBP2a kits.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01653-17.
REFERENCES
- 1.Abraham JL, Morris DO, Griffeth GC, Shofer FS, Rankin SC. 2007. Surveillance of healthy cats and cats with inflammatory skin disease for colonization of the skin by methicillin-resistant coagulase-positive staphylococci and Staphylococcus schleiferi ssp. schleiferi. Vet Dermatol 18:252–259. doi: 10.1111/j.1365-3164.2007.00604.x. [DOI] [PubMed] [Google Scholar]
- 2.Briscoe JA, Morris DO, Rosenthal KL, Shofer FS, Rankin SC. 2009. Evaluation of mucosal and seborrheic sites for staphylococci in two populations of captive psittacines. J Am Vet Med Assoc 234:901–905. doi: 10.2460/javma.234.7.901. [DOI] [PubMed] [Google Scholar]
- 3.Griffeth GC, Morris DO, Abraham JL, Shofer FS, Rankin SC. 2008. Screening for skin carriage of methicillin-resistant coagulase-positive staphylococci and Staphylococcus schleiferi in dogs with healthy and inflamed skin. Vet Dermatol 19:142–149. doi: 10.1111/j.1365-3164.2008.00663.x. [DOI] [PubMed] [Google Scholar]
- 4.Freney J, Brun Y, Bes M, Meugnier H, Grimont F, Grimont PAD, Nervi C, Fleurette J. 1988. Staphylococcus lugdunensis sp. nov. and Staphylococcus schleiferi sp. nov., two species from human clinical specimens. Int J Syst Evol Microbiol 38:168–172. [Google Scholar]
- 5.Igimi S, Takahashi E, Mitsuoka T. 1990. Staphylococcus schleiferi subsp. coagulans subsp. nov., isolated from the external auditory meatus of dogs with external ear otitis. Int J Syst Bacteriol 40:409–411. doi: 10.1099/00207713-40-4-409. [DOI] [PubMed] [Google Scholar]
- 6.Argemi X, Riegel P, Lavigne T, Lefebvre N, Grandpre N, Hansmann Y, Jaulhac B, Prevost G, Schramm F. 2015. Implementation of matrix-assisted laser desorption ionization–time of flight mass spectrometry in routine clinical laboratories improves identification of coagulase-negative staphylococci and reveals the pathogenic role of Staphylococcus lugdunensis. J Clin Microbiol 53:2030–2036. doi: 10.1128/JCM.00177-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck KM, Waisglass SE, Dick HL, Weese JS. 2012. Prevalence of meticillin-resistant Staphylococcus pseudintermedius (MRSP) from skin and carriage sites of dogs after treatment of their meticillin-resistant or meticillin-sensitive staphylococcal pyoderma. Vet Dermatol 23:369–375, e66–e67. doi: 10.1111/j.1365-3164.2012.01035.x. [DOI] [PubMed] [Google Scholar]
- 8.Cain CL, Morris DO, O'Shea K, Rankin SC. 2011. Genotypic relatedness and phenotypic characterization of Staphylococcus schleiferi subspecies in clinical samples from dogs. Am J Vet Res 72:96–102. doi: 10.2460/ajvr.72.1.96. [DOI] [PubMed] [Google Scholar]
- 9.Frank LA, Kania SA, Hnilica KA, Wilkes RP, Bemis DA. 2003. Isolation of Staphylococcus schleiferi from dogs with pyoderma. J Am Vet Med Assoc 222:451–454. doi: 10.2460/javma.2003.222.451. [DOI] [PubMed] [Google Scholar]
- 10.Kania SA, Williamson NL, Frank LA, Wilkes RP, Jones RD, Bemis DA. 2004. Methicillin resistance of staphylococci isolated from the skin of dogs with pyoderma. Am J Vet Res 65:1265–1268. doi: 10.2460/ajvr.2004.65.1265. [DOI] [PubMed] [Google Scholar]
- 11.May ER, Kinyon JM, Noxon JO. 2012. Nasal carriage of Staphylococcus schleiferi from healthy dogs and dogs with otitis, pyoderma or both. Vet Microbiol 160:443–448. doi: 10.1016/j.vetmic.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 12.Penna B, Mendes W, Rabello R, Lilenbaum W. 2013. Carriage of methicillin susceptible and resistant Staphylococcus schleiferi among dog with or without topic infections. Vet Microbiol 162:298–299. doi: 10.1016/j.vetmic.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 13.Vanni M, Tognetti R, Pretti C, Crema F, Soldani G, Meucci V, Intorre L. 2009. Antimicrobial susceptibility of Staphylococcus intermedius and Staphylococcus schleiferi isolated from dogs. Res Vet Sci 87:192–195. doi: 10.1016/j.rvsc.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Yamashita K, Shimizu A, Kawano J, Uchida E, Haruna A, Igimi S. 2005. Isolation and characterization of staphylococci from external auditory meatus of dogs with or without otitis externa with special reference to Staphylococcus schleiferi subsp. coagulans isolates. J Vet Med Sci 67:263–268. doi: 10.1292/jvms.67.263. [DOI] [PubMed] [Google Scholar]
- 15.Bemis DA, Jones RD, Hiatt LE, Ofori ED, Rohrbach BW, Frank LA, Kania SA. 2006. Comparison of tests to detect oxacillin resistance in Staphylococcus intermedius, Staphylococcus schleiferi, and Staphylococcus aureus isolates from canine hosts. J Clin Microbiol 44:3374–3376. doi: 10.1128/JCM.01336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts S, O'Shea K, Morris D, Robb A, Morrison D, Rankin S. 2005. A real-time PCR assay to detect the Panton Valentine leukocidin toxin in staphylococci: screening Staphylococcus schleiferi subspecies coagulans strains from companion animals. Vet Microbiol 107:139–144. doi: 10.1016/j.vetmic.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh A, Singh Y, Kapil A, Dhawan B. 2016. Staphylococcal cassette chromosome mec (SCCmec) typing of clinical isolates of coagulase-negative staphylococci (CoNS) from a tertiary care hospital in New Delhi, India. Indian J Med Res 143:365–370. doi: 10.4103/0971-5916.182629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki T, Tsubakishita S, Kuwahara-Arai K, Matsuo M, Lu YJ, Tanaka Y, Hiramatsu K. 2015. Complete genome sequence of methicillin-resistant Staphylococcus schleiferi strain TSCC54 of canine origin. Genome Announc 3:e01268-15. doi: 10.1128/genomeA.01268-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cain CL, Morris DO, Rankin SC. 2011. Clinical characterization of Staphylococcus schleiferi infections and identification of risk factors for acquisition of oxacillin-resistant strains in dogs: 225 cases (2003–2009). J Am Vet Med Assoc 239:1566–1573. doi: 10.2460/javma.239.12.1566. [DOI] [PubMed] [Google Scholar]
- 20.Intorre L, Vanni M, Di Bello D, Pretti C, Meucci V, Tognetti R, Soldani G, Cardini G, Jousson O. 2007. Antimicrobial susceptibility and mechanism of resistance to fluoroquinolones in Staphylococcus intermedius and Staphylococcus schleiferi. J Vet Pharmacol Ther 30:464–469. doi: 10.1111/j.1365-2885.2007.00896.x. [DOI] [PubMed] [Google Scholar]
- 21.Hebert GA. 1990. Hemolysins and other characteristics that help differentiate and biotype Staphylococcus lugdunensis and Staphylococcus schleiferi. J Clin Microbiol 28:2425–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzamalis A, Chalvatzis N, Anastasopoulos E, Tzetzi D, Dimitrakos S. 2013. Acute postoperative Staphylococcus schleiferi endophthalmitis following uncomplicated cataract surgery: first report in the literature. Eur J Ophthalmol 23:427–430. doi: 10.5301/ejo.5000254. [DOI] [PubMed] [Google Scholar]
- 23.Kumar D, Cawley JJ, Irizarry-Alvarado JM, Alvarez A, Alvarez S. 2007. Case of Staphylococcus schleiferi subspecies coagulans endocarditis and metastatic infection in an immune compromised host. Transpl Infect Dis 9:336–338. doi: 10.1111/j.1399-3062.2007.00222.x. [DOI] [PubMed] [Google Scholar]
- 24.Leung MJ, Nuttall N, Mazur M, Taddei TL, McComish M, Pearman JW. 1999. Case of Staphylococcus schleiferi endocarditis and a simple scheme to identify clumping factor-positive staphylococci. J Clin Microbiol 37:3353–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martiny D, Potvliege C, Jonckheer J. 2010. Fatal bacteremia due to Staphylococcus schleiferi subsp. schleiferi. Clin Microbiol Newsl 32:85–86. doi: 10.1016/j.clinmicnews.2010.05.002. [DOI] [Google Scholar]
- 26.Calvo J, Hernandez JL, Farinas MC, Garcia-Palomo D, Aguero J. 2000. Osteomyelitis caused by Staphylococcus schleiferi and evidence of misidentification of this Staphylococcus species by an automated bacterial identification system. J Clin Microbiol 38:3887–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez JL, Calvo J, Sota R, Aguero J, Garcia-Palomo JD, Farinas MC. 2001. Clinical and microbiological characteristics of 28 patients with Staphylococcus schleiferi infection. Eur J Clin Microbiol Infect Dis 20:153–158. [DOI] [PubMed] [Google Scholar]
- 28.Celard M, Vandenesch F, Darbas H, Grando J, Jean-Pierre H, Kirkorian G, Etienne J. 1997. Pacemaker infection caused by Staphylococcus schleiferi, a member of the human preaxillary flora: four case reports. Clin Infect Dis 24:1014–1015. doi: 10.1093/clinids/24.5.1014. [DOI] [PubMed] [Google Scholar]
- 29.Kluytmans J, Berg H, Steegh P, Vandenesch F, Etienne J, van Belkum A. 1998. Outbreak of Staphylococcus schleiferi wound infections: strain characterization by randomly amplified polymorphic DNA analysis, PCR ribotyping, conventional ribotyping, and pulsed-field gel electrophoresis. J Clin Microbiol 36:2214–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Griethuysen A, Bes M, Etienne J, Zbinden R, Kluytmans J. 2001. International multicenter evaluation of latex agglutination tests for identification of Staphylococcus aureus. J Clin Microbiol 39:86–89. doi: 10.1128/JCM.39.1.86-89.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.CLSI. 2015. Performance standards for antimicrobial disk susceptibility tests. M012-A12. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 32.CLSI. 2017. Performance standards for antimicrobial susceptibility testing. M100-S27. Clinical and Laboratory Standards Institute, Wayne, PA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swenson JM, Tenover FC, Cefoxitin Disk Study Group. 2005. Results of disk diffusion testing with cefoxitin correlate with presence of mecA in Staphylococcus spp. J Clin Microbiol 43:3818–3823. doi: 10.1128/JCM.43.8.3818-3823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu MT, Burnham CA, Westblade LF, Dien Bard J, Lawhon SD, Wallace MA, Stanley T, Burd E, Hindler J, Humphries RM. 2016. Evaluation of oxacillin and cefoxitin disk and MIC breakpoints for prediction of methicillin resistance in human and veterinary isolates of Staphylococcus intermedius group. J Clin Microbiol 54:535–542. doi: 10.1128/JCM.02864-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. 1991. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol 29:2240–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.CLSI. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 37.Clark RB, Lewinski MA, Loeffelholz MJ, Tibbetts RJ. 2009. Cumitech 31A, Verification and validation of procedures in the clinical microbiology laboratory. Coordinating ed, Sharp SE. ASM Press, Washington, DC. [Google Scholar]
- 38.Dupont C, Sivadon-Tardy V, Bille E, Dauphin B, Beretti JL, Alvarez AS, Degand N, Ferroni A, Rottman M, Herrmann JL, Nassif X, Ronco E, Carbonnelle E. 2010. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and two automated systems. Clin Microbiol Infect 16:998–1004. doi: 10.1111/j.1469-0691.2009.03036.x. [DOI] [PubMed] [Google Scholar]
- 39.Elamin WF, Ball D, Millar M. 2015. Unbiased species-level identification of clinical isolates of coagulase-negative staphylococci: does it change the perspective on Staphylococcus lugdunensis? J Clin Microbiol 53:292–294. doi: 10.1128/JCM.02932-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson KN, Andreacchio K, Edelstein PH. 2014. Detection of methicillin-resistant coagulase-negative staphylococci by the Vitek 2 system. J Clin Microbiol 52:3196–3199. doi: 10.1128/JCM.01162-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnold AR, Burnham CA, Ford BA, Lawhon SD, McAllister SK, Lonsway D, Albrecht V, Jerris RC, Rasheed JK, Limbago B, Burd EM, Westblade LF. 2016. Evaluation of an immunochromatographic assay for rapid detection of penicillin-binding protein 2a in human and animal Staphylococcus intermedius group, Staphylococcus lugdunensis, and Staphylococcus schleiferi clinical isolates. J Clin Microbiol 54:745–748. doi: 10.1128/JCM.02869-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.