ABSTRACT
IQG-607 is a metal complex previously reported as a promising anti-tuberculosis (TB) drug against isoniazid (INH)-resistant strains of Mycobacterium tuberculosis. Unexpectedly, we found that INH-resistant clinical isolates were resistant to IQG-607. Spontaneous mutants resistant to IQG-607 were subjected to whole-genome sequencing, and all sequenced colonies carried alterations in the katG gene. The katG(S315T) mutation was sufficient to confer resistance to IQG-607 in both MIC assays and inside macrophages. Moreover, overexpression of the InhA(S94A) protein caused IQG-607's resistance.
KEYWORDS: antibiotic resistance, MIC, spontaneous mutants, tuberculosis, whole-genome sequencing
TEXT
The pentacyano(isoniazid)ferrate(II) complex (Na3[FeII(CN)5(INH)] · 3H2O), also known as IQG-607, is an isoniazid (INH) analog that contains a cyanoferrate moiety bound to the pyridinic nitrogen of the INH molecule (Fig. 1) (1). So far, two alternative hypotheses were proposed to explain its mechanism of action. Enzymatic inhibition studies demonstrated that IQG-607 inhibits the wild-type NADH-dependent enoyl-[acyl carrier protein] reductase (InhA) enzyme (Enzyme Commission no. EC 1.3.1.9) and its structural mutants (I21V, S94A, and I47T) in a time-dependent manner (2, 3). Interestingly, this inhibition was observed in the absence of NAD+ or NADH and without requiring activation by the catalase-peroxidase KatG (EC 1.11.1.21), suggesting that IQG-607 could be used against INH-resistant Mycobacterium tuberculosis strains whose mechanism of resistance involves mutations in the katG (Rv1908c) gene (2, 3). Another possible explanation involves an intramolecular electron transference mediated by the metal center that would self-activate the INH moiety from IQG-607 inside the host macrophages, thus forming a hypothetical adduct with NADH (4). Despite the extensive characterization of IQG-607 interaction with the InhA enzyme, its activity in mice (5), and its favorable toxicological profile in rats (6) and mini pigs (7), it is still unknown if the compound is active against M. tuberculosis strains carrying mutations in the katG gene, hence our work focused on elucidating the mechanism of resistance of the M. tuberculosis to the compound IQG-607 by employing a genetic approach with a set of different INH-resistant strains.
FIG 1.
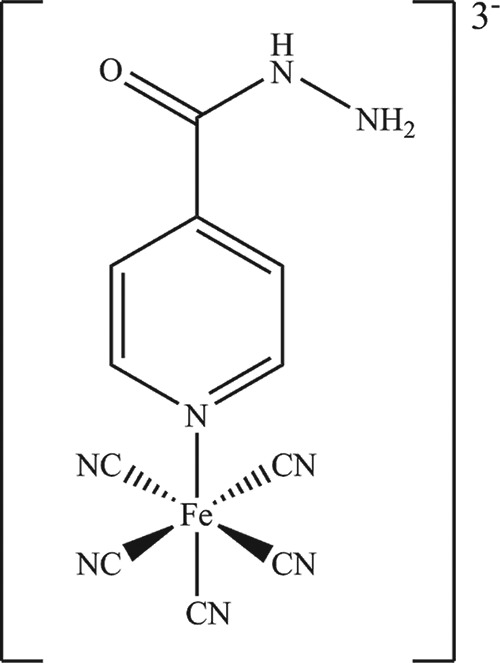
Chemical structure of IQG-607 (Na3[FeII(CN)5(INH)] · 3H2O). This inorganic complex contains a pentacyanoferrate(II) moiety bound to the nitrogen atom of the heterocyclic ring of isoniazid. This moiety is thought to activate the isoniazid molecule.
We first tested the compound activity against several multidrug-resistant tuberculosis (MDR-TB) clinical isolates by determining MICs on Middlebrook 7H9 liquid medium as described previously (8). Eight MDR-TB clinical isolates carried the most common mutation found in the katG gene (Ser315Thr) (9), while one INH-susceptible clinical isolate carried the wild-type gene. As shown in Table 1, all strains were resistant to both INH (MIC > 25 mg/liter) and IQG-607 (MIC > 100 mg/liter), except for the CDCT-28 isolate. This result was unexpected, since it suggests an association between the presence of the katG(S315T) mutation and an increase of at least 64× in the MIC values of IQG-607.
TABLE 1.
Activity of IQG-607 and several anti-TB compounds against nine clinical isolatesa
| Isolatec | katG mutation | inhA promoter genotype | rpoB mutation | MIC (mg/liter) forb |
|||
|---|---|---|---|---|---|---|---|
| INH | IQG-607 | RIF | MFX | ||||
| H37Rv | WT | WT | WT | 0.39 | 1.56 | <0.2 | <0.2 |
| CDCT-1 | S315T | WT | H526D | >100 | >100 | >100 | <0.2 |
| CDCT-2 | S315T | C(−15)T | H526D | 100 | >100 | >100 | <0.2 |
| CDCT-3 | S315T | WT | D516V | 25 | >100 | 6.25 | <0.2 |
| CDCT-4 | S315T | WT | S531L | >100 | >100 | >100 | <0.2 |
| CDCT-5 | S315T | WT | D516V | 50 | >100 | >100 | <0.2 |
| CDCT-10 | S315T | WT | H526D | 100 | >100 | >100 | <0.2 |
| CDCT-16 | S315T | C(−15)T | D516V | 100 | >100 | >100 | <0.2 |
| CDCT-27 | S315T | WT | WT | 25 | >100 | <0.2 | <0.2 |
| CDCT-28 | WT | WT | D516A | 0.39 | 1.56 | <0.2 | <0.2 |
TB, tuberculosis; WT, wild type.
MIC values reported here were observed in two independent experiments or were the highest value found among three independent tests. INH, isoniazid; IQG-607, pentacyano(isoniazid)ferrate(II); RIF, rifampin; MFX, moxifloxacin.
Clinical isolate identification.
Next, spontaneous mutants resistant to IQG-607 were isolated on 7H10 plates using compound concentrations above the MIC (≥2 mg/liter). We confirmed the resistance phenotype by determining MICs of six colonies (named IQG1 to 6) selected from the highest compound concentration tested (16 mg/liter). The six selected mutants displayed cross-resistance only to INH (Table 2). We retested the resistance of one representative mutant (IQG1) by monitoring its growth rate, as previously described (10), in the presence of IQG-607 or INH above their MIC values, and indeed neither of the compounds prevented the growth of this mutant (see Fig. S1 in the supplemental material).
TABLE 2.
Activity of IQG-607, INH, ETH, and MFX against several laboratory strains of M. tuberculosis
| Strain | katG genotype | inhA promoter genotype | inhA genotype | MIC (mg/liter) fora |
|||
|---|---|---|---|---|---|---|---|
| INH | ETH | MFX | IQG-607 | ||||
| H37Rv | WT | WT | WT | 0.39 | 3.13 | 0.08 | 1.56 |
| IQG1b | 120del1 | WT | WT | >100 | 3.13 | 0.16 | >100 |
| IQG2 | 120del1 | WT | WT | >100 | 1.56 | 0.08 | >100 |
| IQG3 | T(271)C | WT | WT | >100 | 3.13 | 0.16 | >100 |
| IQG4 | 1298del1 | WT | WT | >100 | 1.56 | 0.08 | >100 |
| IQG5 | 120del1 | WT | WT | >100 | 1.56 | 0.02 | >100 |
| IQG6 | C(1249)G | WT | WT | >100 | 3.13 | 0.08 | >100 |
| pNIP::furA+katG(WT) | WT | WT | WT | <0.2 | 3.13 | <0.2 | 0.78 |
| pNIP::furA+katG(S315T) | G(944)Cc | WT | WT | >100 | 0.78 | <0.2 | >100 |
| pNIP::Ø | WT | WT | WT | 0.39 | 6.25 | <0.2 | 1.56 |
| pNIP::InhA(WT) | WT | WT | WT | 0.39 | 6.25 | <0.2 | 3.13 |
| pNIP::InhA(S94A) | WT | WT | T(280)Gd | 25 | >100 | <0.2 | >100 |
MIC values reported here were observed in two independent experiments or were the highest value found among three independent tests. ETH, ethionamide.
IQG1-6 refers to the IQG-607 spontaneous mutant strains.
This mutation changes the serine-315 residue by a threonine (S315T).
This strain still expresses the underlying level of the wild-type inhA gene but carries the mutant inhA gene [T(280)G], cloned into the integrative plasmid pNIP40/b. This mutation changes the serine-94 residue by an alanine (S94A).
We performed the whole-genome sequencing (by MiSeq platform) of three colonies (IQG1 to 3) to uncover the mutations associated with the resistance phenotype, and we found that all colonies carried mutations in the katG gene. This result was confirmed by target DNA sequencing of the six mutants (IQG1 to 6) using several primers described in Table S1 in the supplemental material. Indeed, all six mutants carried alterations in the katG gene (Table 2). IQG1, 2, and 5 had a single deletion of cytosine 120 (120del1), which changed the reading frame of katG and introduced a premature stop codon at position 45. The resultant defective protein lacks almost all the residues from the N-terminal domain, including the key active site residues (11) and the entire C-terminal domain. The deletion of adenine 1298 (1298del1) in the mutant IQG4 introduced a premature stop codon at position 467 and led to a truncated protein lacking only the C-terminal domain. These findings are in agreement with previous work using INH to select spontaneous mutants (12, 13). The W91R mutation found in IQG3 was already described in clinical isolates at a very low frequency (14–17), and it is expected to confer resistance to INH by a still unclear mechanism, possibly involving a disruption of an important H-bonding network in the active site of KatG (18). To the best of our knowledge, the H417D mutation found with IQG6 has never been described in either clinical isolates or in laboratory strains. Although the role of this particular histidine residue is not known, some studies have already uncovered the importance of the neighbor residue Arg-418 for catalysis and its involvement in INH resistance (18–20).
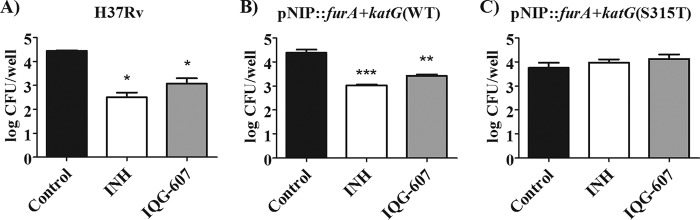
To establish a causal relationship between the clinically relevant mutation katG(S315T) and resistance to IQG-607, we knocked out the wild-type katG gene from M. tuberculosis H37Rv strain by using an allelic exchange approach (21) and complemented it with the wild-type, furA+katG(WT), or the mutated allele, furA+katG(S315T) (see Fig. S2 in the supplemental material). MIC for IQG-607 increased by more than 64-fold in the mutated strain (Table 2), clearly suggesting that the compound depends on KatG to be active. However, MIC assays lack the oxidizing environment of the macrophage, and therefore would not allow a route for the redox-mediated self-activation of IQG-607 previously proposed (4). To test this premise, we infected murine macrophages as described previously (22) with ΔfurA::katG laboratory strains carrying the wild-type gene or the S315T mutant and treated them with IQG-607. The mutant strain preserved its resistance phenotype toward IQG-607 inside macrophages (Fig. 2), indicating that the host intracellular environment is not sufficient to trigger the self-activation mechanism proposed for IQG-607, which reinforces the necessity of the KatG enzyme for the activity of IQG-607.
FIG 2.
Treatment of macrophages infected with (A) H37Rv, (B) pNIP::furA+katG(WT), or (C) pNIP::furA+katG(S315T) laboratory strains of M. tuberculosis with 50 μM (6.85 mg/liter) of INH (white bar) or 50 μM (23.2 mg/liter) of IQG-607 (gray bar). Data are expressed as mean numbers of the logarithms of CFU per well from triplicates. Two-way ANOVA with Bonferroni's post hoc test was employed to compare the treatments. Asterisks (*/**/***) represent P values below 0.05, 0.01, and 0.001, respectively, compared with the untreated control group (black bar).
Finally, we evaluated the antimicrobial activity of IQG-607 in mycobacterial cells constitutively overexpressing the S94A mutant InhA enzyme, a mutation that is known to cause INH resistance (23). The mutant inhA gene (Rv1484) was cloned into an integrative plasmid (pNIP40/b) under the control of the hsp60 promoter. In addition to the expected resistance to INH and also to ethionamide (ETH), the expression of the InhA(S94A) mutant enzyme (see Fig. S3 in the supplemental material) resulted in mycobacteria also resistant to IQG-607 (Table 2). This result genetically validated the InhA protein as the target of IQG-607. However, it also showed that the Ser-94 residue is essential for the compound's interaction with the enzyme, somehow contradicting the inhibition observed against the purified InhA(S94A) enzyme (3).
In conclusion, we demonstrated that IQG-607 does not activate itself inside macrophages or mycobacteria. As with INH, IQG-607 also requires the activity of KatG in order to be active against M. tuberculosis strains. These findings challenge its planned usage against INH-resistant strains. Nonetheless, the compound has already been reported to be less toxic than INH in a preclinical study using mice (5, 24). This is a remarkable feature, since INH was previously associated with hepatotoxicity in animal and humans due to its toxic metabolites, such as hydrazine (25). We performed a checkerboard assay as previously described (26) and found that IQG-607 has an indifferent effect when combined with other anti-TB drugs (see Table S2 in the supplemental material), showing it could be used in association. Further experiments will be required to understand its metabolism inside mycobacteria and to establish its utility as an INH surrogate with low toxicity.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of Science and Technology in Tuberculosis (INCT-TB), Brazil (grant Decit/SCTIE/MS-MCT-CNPq-FNDCT-CAPES), the National Council for Scientific and Technological Development of Brazil (CNPq) (grant 441720/2014-5 to A.D.V.), and the Brazilian Development Bank (BNDES) (grant 14.2.0914.1). B.L.A acknowledges the fellowship by Coordination for the Improvement of Higher Education Personnel (CAPES). L.A.B. (CNPq, grant 520182/99-5) and C.V.B are Research Career Awardees of CNPq.
We acknowledge Silvana Spindola de Miranda from the Federal University of Minas Gerais (Belo Horizonte, Brazil) for providing the clinical isolates used in this study, Mariane Rotta for providing purified InhA(WT) enzyme used in the Western blot assay, and Mary Jackson and Brigitte Gicquel for providing the pNIP40/b and pPR27xylE plasmids.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02222-17.
REFERENCES
- 1.Sousa EH, Basso LA, Santos DS, Diógenes IC, Longhinotti E, Lopes LG, Moreira IS. 2012. Isoniazid metal complex reactivity and insights for a novel anti-tuberculosis drug design. J Biol Inorg Chem 17:275–283. doi: 10.1007/s00775-011-0848-x. [DOI] [PubMed] [Google Scholar]
- 2.Oliveira JS, Sousa EH, Basso LA, Palaci M, Dietze R, Santos DS, Moreira IS. 2004. An inorganic iron complex that inhibits wild-type and an isoniazid-resistant mutant 2-trans-enoyl-ACP (CoA) reductase from Mycobacterium tuberculosis. Chem Commun (Camb.) 7:312–313. doi: 10.1039/b313592f. [DOI] [PubMed] [Google Scholar]
- 3.Vasconcelos IB, Meyer E, Sales FAM, Moreira IS, Basso LA, Santos DS. 2008. The mode of inhibition of Mycobacterium tuberculosis wild-type and isoniazid-resistant 2-trans-enoyl-ACP(CoA) reductase enzymes by an inorganic complex. Antiinfect Agents Med Chem 7:50–62. doi: 10.2174/187152108783329799. [DOI] [Google Scholar]
- 4.Sousa EH, de Mesquita Vieira FG, Butler JS, Basso LA, Santiago DS, Diógenes IC, Lopes LG, Sadler PJ. 2014. [Fe(CN)5(isoniazid)](3−): an iron isoniazid complex with redox behavior implicated in tuberculosis therapy. J Inorg Biochem 140:236–244. doi: 10.1016/j.jinorgbio.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Rodrigues-Junior VS, Dos Santos Junior A, Dos Santos AJ, Schneider CZ, Calixto JB, Sousa EH, de França Lopes LG, Souto AA, Basso LA, Santos DS, Campos MM. 2012. Activity of IQG-607, a new orally active compound, in a murine model of Mycobacterium tuberculosis infection. Int J Antimicrob Agents 40:182–185. doi: 10.1016/j.ijantimicag.2012.04.019. [DOI] [PubMed] [Google Scholar]
- 6.Rodrigues-Junior VS, Machado P, Calixto JB, Siqueira JM, Andrade EL, Bento AF, Campos MM, Basso LA, Santos DS. 2017. Preclinical safety evaluation of IQG-607 in rats: acute and repeated dose toxicity studies. Regul Toxicol Pharmacol 86:11–17. doi: 10.1016/j.yrtph.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 7.Rodrigues-Junior VS, Cintra L, Machado P, Dadda A, Basso LA, Mafra ACCN, Campos AH, Campos MM, Santos DS. 2017. Toxicological profile of IQG-607 after single and repeated oral administration in minipigs: an essential step towards phase I clinical trial. Regul Toxicol Pharmacol 90:78–86. doi: 10.1016/j.yrtph.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 8.Giacobbo BC, Pissinate K, Rodrigues-Junior V, Villela AD, Grams ES, Abbadi BL, Subtil FT, Sperotto N, Trindade RV, Back DF, Campos MM, Basso LA, Machado P, Santos DS. 2017. New insights into the SAR and drug combination synergy of 2-(quinolin-4-yloxy)acetamides against Mycobacterium tuberculosis. Eur J Med Chem 126:491–501. doi: 10.1016/j.ejmech.2016.11.048. [DOI] [PubMed] [Google Scholar]
- 9.Unissa AN, Subbian S, Hanna LE, Selvakumar N. 2016. Overview on mechanisms of isoniazid action and resistance in Mycobacterium tuberculosis. Infect Genet Evol 45:474–492. doi: 10.1016/j.meegid.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 10.McGrath M, Gey van Pittius NC, Sirgel FA, Van Helden PD, Warren RM. 2014. Moxifloxacin retains antimycobacterial activity in the presence of gyrA mutations. Antimicrob Agents Chemother 58:2912–2915. doi: 10.1128/AAC.02583-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertrand T, Eady NA, Jones JN, Jesmin Nagy JM, Jamart-Grégoire B, Raven EL, Brown KA. 2004. Crystal structure of Mycobacterium tuberculosis catalase-peroxidase. J Biol Chem 279:38991–38999. doi: 10.1074/jbc.M402382200. [DOI] [PubMed] [Google Scholar]
- 12.Bergval IL, Schuitema AR, Klatser PR, Anthony RM. 2009. Resistant mutants of Mycobacterium tuberculosis selected in vitro do not reflect the in vivo mechanism of isoniazid resistance. J Antimicrob Chemother 64:515–523. doi: 10.1093/jac/dkp237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brossier F, Cambau E, Tessier E, Jarlier V, Sougakoff W. 2016. The in vitro mechanisms of isoniazid and ethionamide resistance poorly reflect those in vivo in Mycobacterium tuberculosis. Tuberc 101:144–145. doi: 10.1016/j.tube.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 14.Vilchèze C, Jacobs WR. 2014. Resistance to isoniazid and ethionamide in Mycobacterium tuberculosis: genes, mutations, and causalities. Microbiol Spectr 2:MGM2-0014–2013. doi: 10.1128/microbiolspec.MGM2-0014-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cardoso RF, Cooksey RC, Morlock GP, Barco P, Cecon L, Forestiero F, Leite CQ, Sato DN, Shikama ML, Mamizuka EM, Hirata RD, Hirata MH. 2004. Screening and characterization of mutations in isoniazid-resistant Mycobacterium tuberculosis isolates obtained in Brazil. Antimicrob Agents Chemother 48:3373–3381. doi: 10.1128/AAC.48.9.3373-3381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meumann EM, Globan M, Fyfe JA, Leslie D, Porter JL, Seemann T, Denholm J, Stinear TP. 2015. Genome sequence comparisons of serial multi-drug-resistant Mycobacterium tuberculosis isolates over 21 years of infection in a single patient. Microb Genom 1:e000037. doi: 10.1099/mgen.0.000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramaswamy SV, Reich R, Dou SJ, Jasperse L, Pan X, Wanger A, Quitugua T, Graviss EA. 2003. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 47:1241–1250. doi: 10.1128/AAC.47.4.1241-1250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghiladi RA, Medzihradszky KF, Rusnak FM, Ortiz de Montellano PR. 2005. Correlation between isoniazid resistance and superoxide reactivity in Mycobacterium tuberculosis KatG. J Am Chem Soc 127:13428–13442. doi: 10.1021/ja054366t. [DOI] [PubMed] [Google Scholar]
- 19.Mo L, Zhang W, Wang J, Weng XH, Chen S, Shao LY, Pang MY, Chen ZW. 2004. Three-dimensional model and molecular mechanism of Mycobacterium tuberculosis catalase-peroxidase (KatG) and isoniazid-resistant KatG mutants. Microb Drug Resist 10:269–279. doi: 10.1089/mdr.2004.10.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cade CE, Dlouhy AC, Medzihradszky KF, Salas-Castillo SP, Ghiladi RA. 2010. Isoniazid-resistance conferring mutations in Mycobacterium tuberculosis KatG: catalase, peroxidase, and INH-NADH adduct formation activities. Protein Sci 19:458–474. doi: 10.1002/pro.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villela AD, Rodrigues VD, Pinto AF, Wink PL, Sánchez-Quitian ZA, Petersen GO, Campos MM, Basso LA, Santos DS. 2017. Characterisation of iunH gene knockout strain from Mycobacterium tuberculosis. Mem Inst Oswaldo Cruz 112:203–208. doi: 10.1590/0074-02760160462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodrigues-Junior VS, dos Santos Junior AA, Villela AD, Belardinelli JM, Morbidoni HR, Basso LA, Campos MM, Santos DS. 2014. IQG-607 abrogates the synthesis of mycolic acids and displays intracellular activity against Mycobacterium tuberculosis in infected macrophages. Int J Antimicrob Agents 43:82–85. doi: 10.1016/j.ijantimicag.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 23.Vilchèze C, Wang F, Arai M, Hazbón MH, Colangeli R, Kremer L, Weisbrod TR, Alland D, Sacchettini JC, Jacobs WR. 2006. Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat Med 12:1027–1029. doi: 10.1038/nm1466. [DOI] [PubMed] [Google Scholar]
- 24.Basso LA, Schneider CZ, Anderson JAB, dos Santos AA Jr, AA, Campos MM, Souto AA, Santos DS. 2010. An inorganic complex that inhibits Mycobacterium tuberculosis enoyl reductase as a prototype of a new class of chemotherapeutic agents to treat tuberculosis. J Braz Chem Soc 21:1384–1389. doi: 10.1590/S0103-50532010000700026. [DOI] [Google Scholar]
- 25.Hassan HM, Guo HL, Yousef BA, Luyong Z, Zhenzhou J. 2015. Hepatotoxicity mechanisms of isoniazid: a mini-review. J Appl Toxicol 35:1427–1432. doi: 10.1002/jat.3175. [DOI] [PubMed] [Google Scholar]
- 26.Reddy VM, Einck L, Andries K, Nacy CA. 2010. In vitro interactions between new antitubercular drug candidates SQ109 and TMC207. Antimicrob Agents Chemother 54:2840–2846. doi: 10.1128/AAC.01601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.