New results link redox modulation, viroporin formation, and genome replication by (+)RNA viruses.
Abstract
Positive-strand RNA viruses replicate their genomes in membrane-bounded cytoplasmic complexes. We show that endoplasmic reticulum (ER)–linked genomic RNA replication by brome mosaic virus (BMV), a well-studied member of the alphavirus superfamily, depends on the ER luminal thiol oxidase ERO1. We further show that BMV RNA replication protein 1a, a key protein for the formation and function of vesicular BMV RNA replication compartments on ER membranes, permeabilizes these membranes to release oxidizing potential from the ER lumen. Conserved amphipathic sequences in 1a are sufficient to permeabilize liposomes, and mutations in these sequences simultaneously block membrane permeabilization, formation of a disulfide-linked, oxidized 1a multimer, 1a’s RNA capping function, and productive genome replication. These results reveal new transmembrane complexities in positive-strand RNA virus replication, show that—as previously reported for certain picornaviruses and flaviviruses—some alphavirus superfamily members encode viroporins, identify roles for such viroporins in genome replication, and provide a potential new foundation for broad-spectrum antivirals.
INTRODUCTION
Positive-strand RNA [(+)RNA] viruses comprise more than one-third of known virus genera and include poliovirus, hepatitis C virus, chikungunya virus, Zika virus, and many other serious human pathogens. With few effective vaccines or antiviral drugs for this largest of the seven major virus classes, new control approaches are sorely needed. A highly attractive target for controlling these viruses is their genomic RNA replication, which proceeds through RNA forms with no DNA intermediates. A universal feature of such (+)RNA virus genome replication is its location on intracellular membranes, often in conjunction with single- and/or double-membrane vesicle formation or other membrane rearrangements (1, 2). Virus replication proteins play key roles in generating these genome replication compartments by multimerizing and interacting with specific lipids and/or membrane proteins in host cells (2, 3). Membrane compartmentalization of these genome replication “factories” protects replicating RNA from competing processes, such as RNA translation and encapsidation, and from host defenses against double-strand RNA replication intermediates.
Brome mosaic virus (BMV) is a member of the large alphavirus-like superfamily of human, animal, and plant (+)RNA viruses, and has been studied as a model for genome replication. BMV has three genomic RNAs that encode, among other factors, RNA replication proteins 1a and 2apol. 1a contains an N-proximal RNA capping domain and a C-terminal nucleoside triphosphatase/helicase domain (4–6). 2apol has a polymerase domain and an N-terminal domain that interacts with the 1a helicase domain (7, 8). 1a is the master organizer of BMV RNA replication complex assembly (fig. S1A): 1a localizes to the outer perinuclear endoplasmic reticulum (ER) membranes, induces 50- to 75-nm vesicular invaginations or spherules, and recruits 2apol and viral RNA replication templates into these compartments (9). In these replication compartments, negative-strand RNAs [(−)RNAs] are synthesized, retained, and used as templates to synthesize genomic and subgenomic (+)RNAs that are then 5′-capped and exported into the cytosol [(10) and fig. S1, A and B]. 1a and 2apol direct BMV RNA replication in Saccharomyces cerevisiae, duplicating all major features of BMV replication in plant cells [reviewed by den Boon and Ahlquist (1)]. S. cerevisiae has proven a valuable model for normal and disease processes in human and other cells, and its powerful genetics enables studies of essential host factors (11, 12).
1a is localized on the cytosolic surface of perinuclear ER membranes (13), and 1a-induced RNA replication compartments, which are vesicular invaginations of the ER membrane, maintain an open neck-like connection between their interior and the cytosol, sufficient in diameter for the import of ribonucleotide substrates and export of the product RNA (9). Nevertheless, a previous study by one of us (M.N.) identified an unusual disulfide-linked 1a complex in BMV-infected plant cells (14). With some exemptions, disulfides are usually formed within the oxidized ER lumen and mitochondrial intermembrane space rather than the reduced cytoplasm (15). In S. cerevisiae, the thiol oxidase Ero1p is the major driver of disulfide bond formation within the ER lumen and is essential for cell viability (16–18). Here, we integrated yeast genetics, viral reverse genetics, and biochemistry to build on these findings to elucidate the molecular mechanisms by which 1a forms disulfide-linked complexes in the reducing cytoplasm. The results revealed unexpected features of 1a-membrane interactions in RNA genome replication and suggest potential broad-spectrum targets for virus control.
RESULTS AND DISCUSSION
To examine whether Ero1p might be involved in BMV RNA replication, we used a temperature-sensitive ero1-1 mutant and launched BMV RNA replication from plasmids expressing 1a, 2apol, and genomic RNA3 replication template. The ero1-1 mutation inhibited RNA3 replication by about twofold and viral RNA–dependent production of subgenomic RNA4 by about threefold at a semipermissive temperature (Fig. 1A). In contrast, RNA3 replication and RNA4 transcription were enhanced twofold by overexpressing wild-type (WT) ERO1 (ERO1ox) in WT yeast (Fig. 1B). 1a and 2apol levels were not significantly altered by mutation or overexpression of Ero1p (Fig. 1, A and B), implying no role for Ero1p in 1a and 2apol translation and stability. Overexpressing WT Ero1p or Ero1p-FLAG increased the accumulation of (+)RNA3 throughout the period examined (Fig. 1C and fig. S2). Thus, the ER luminal thiol oxidase Ero1p is a rate-limiting factor for cytoplasmic BMV RNA replication.
Fig. 1. Ero1p is a rate-limiting factor for BMV RNA replication.
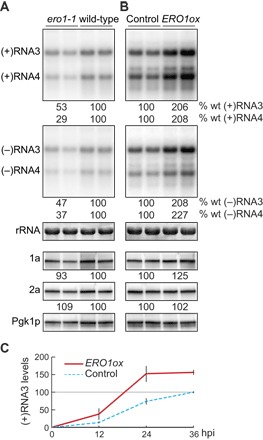
(A) Total RNA and protein were extracted from WT and ero1-1 strains at 18 hours after galactose induction at a semipermissive temperature (34°C). Accumulation of virus RNAs and proteins was analyzed by Northern and Western blotting. 25S ribosomal RNA (rRNA) and Pgk1p serve as loading controls. Note that (+)RNA3 signal is derived from DNA-dependent (+)RNA3 transcription, 1a-mediated (+)RNA3 stabilization, and 1a + 2apol–mediated (+)RNA3 replication (19). To allow visualization and measurement of the RNA bands, the blots showing negative-strand accumulation were exposed much longer than those for positive strands. (B) Total RNA and protein were isolated from control and WT ERO1-overexpressing (ERO1ox) cells grown under physiological conditions (30°C) and analyzed as described above. (C) Time-course analysis of (+)RNA3 accumulation in control and ERO1-overexpressing cells. The data are the mean values obtained in experiments performed with duplicate or triplicate samples, and each is representative of two or more independent experiments.
To gain further insight into Ero1p function in BMV RNA replication, we examined the effect of Ero1p overexpression on several fundamental steps in the BMV replication compartment assembly and function. In the absence of 2apol, 1a recruits RNA3 into genome replication compartments and markedly increases RNA3 stability and accumulation (9, 19). Ero1p overexpression had little effect on 1a-stimulated RNA3 accumulation (fig. S3), suggesting that Ero1p functions after the replication compartment assembly. Figure 1 shows that overexpressing Ero1p stimulates the accumulation of BMV RNA3 and RNA4 of both polarities. However, because positive-strand and negative-strand genomic RNA3 syntheses are co-dependent in the BMV replication cycle, specific activation of either step should increase the accumulation of RNA3 of both polarities. To circumvent this problem and test directly which step involves Ero1p, we used the RNA3 derivative 5′ Gal RNA3 [Fig. 2A and (20)], which lacks the cis-acting RNA signal for (+)RNA3 synthesis and functions only as a template for (−)RNA3 synthesis. Ero1p overexpression had little effect on the accumulation of 5′ Gal RNA3 of both polarities (Fig. 2B). In contrast, the accumulation of (+)RNA4 transcribed from (−)RNA3 was increased by Ero1p overexpression over fivefold (Fig. 2B). These results indicate that the primary effect of Ero1p is on BMV RNA template–directed (+)RNA synthesis and/or accumulation.
Fig. 2. Ero1p stimulates BMV (+)RNA accumulation by enhancing 1a’s disulfide-linked complex formation and capping function.
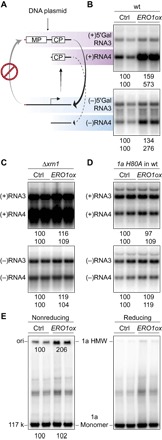
(A) Schematic representation of the limited BMV-directed RNA synthesis pathway for the RNA3 derivative 5′ Gal RNA3, which has the WT 5′ untranslated region replaced with that of yeast GAL1 mRNA (red line). MP (movement protein) and CP (capsid protein) designate RNA3 open reading frames. (B) Analysis of positive- and negative-strand 5′ Gal RNA3 and RNA4 from WT yeast cells expressing 1a, 2apol, and either vector (ctrl) or exogenous Ero1p (ERO1ox). (C) Analysis of RNA3 and RNA4 from Δxrn1 yeast cells expressing 1a, 2apol, and either vector or exogenous Ero1p. (D) Analysis of RNA3 and RNA4 from WT yeast cells expressing a capping-defective mutant 1a H80A, 2apol, and either vector or exogenous Ero1p. (E) Analysis of disulfide-linked 1a complexes from WT yeast cells used in Fig. 1B. 1a monomer and dithiothreitol (DTT)–sensitive, high–molecular weight (HMW) complex were analyzed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) using neutral 3 to 8% SDS-PAGE gels under both reducing and nonreducing conditions.
Xrn1p is a cytoplasmic 5′-3′ exonuclease that degrades uncapped mRNAs and, together with the 3′-5′ exonucleolytic exosome, controls cellular mRNA metabolism (21). Previously, Ahola et al. (22) identified Xrn1p as a major determinant of BMV (+)RNA accumulation: XRN1 deletion allows survival and accumulation of greatly increased levels of uncapped BMV (+)RNA, enhancing overall BMV (+)RNA3 and (+)RNA4 accumulation by two- to threefold and four- to sixfold, respectively. To test for a possible effect of Ero1p on 1a’s RNA capping, BMV replication assays were performed in the Δxrn1 mutant in the absence or presence of Ero1p overexpression. XRN1 deletion suppressed the ability of Ero1p to further stimulate (+)RNA accumulation, leading to equal accumulation of BMV (+)RNA with or without Ero1p overexpression (Fig. 2C). These results imply that, in WT cells, Ero1p stimulates BMV (+)RNA accumulation by enhancing 1a’s RNA capping function. Consistent with this, in WT cells, Ero1p overexpression did not stimulate the synthesis of uncapped (+)RNAs directed by capping-defective mutant 1a H80A and WT 2apol (Fig. 2D) (22).
As noted above, a previous study identified in BMV-infected plant cells a high–molecular weight 1a complex linked by oxidized disulfide bonds, which is active in covalent 1a guanylation for RNA capping (14). To examine whether Ero1p overexpression stimulates the formation of this disulfide-linked complex, 1a was extracted under nonreducing conditions from yeast cells used in the RNA replication assay (Fig. 1B). Overexpressing Ero1p enhanced the accumulation of 1a’s disulfide-linked complex by twofold (Fig. 2E), matching the twofold increase in RNA replication products induced by Ero1p overexpression (Fig. 1B). These results support a model in which Ero1p stimulates BMV (+)RNA accumulation by enhancing 1a’s disulfide-linked complex formation and RNA 5′ capping function.
How does the ER luminal thiol oxidase Ero1p influence cytoplasmic BMV RNA replication beyond the ER membrane? As a foundation for these studies, we confirmed that expressing 1a did not alter Ero1p-FLAG glycosylation and thus its location in the ER lumen (fig. S2). In principle, Ero1p oxidative events might indirectly affect BMV RNA replication via host factor(s), but this assumption is more challenging for several reasons: Cells tightly compartmentalize the ER lumen away from the cytosol, and neither a transmembrane electron/disulfide transfer system nor even a cytoplasmic–ER luminal shuttling protein has been observed for the ER. Another possibility is suggested by the fact that at least some members of the picornavirus and flavivirus superfamilies of (+)RNA viruses encode replication proteins that can induce membrane permeabilization, possibly by forming a pore (23, 24). Representative examples are the 2B proteins of poliovirus and coxsackievirus, members of the picornavirus superfamily. Mutations in the amphipathic helix that mediates coxsackievirus 2B “viroporin” activity alter 2B localization and inhibit viral genome replication and 2B’s effect on cellular secretory pathways (25–27). However, it is not known whether RNA replication protein(s) of any alphavirus superfamily members permeabilizes membranes.
Poliovirus 2B permeabilizes membranes and renders yeast and mammalian cells hypersensitive to hygromycin B, a membrane-impermeable translation inhibitor (28, 29). 1a phenocopied 2B by inducing similar hygromycin B sensitivity (Fig. 3A). Thus, BMV 1a and poliovirus 2B share a previously unrecognized common feature.
Fig. 3. BMV 1a permeabilizes ER membranes and releases oxidative equivalents from ER.
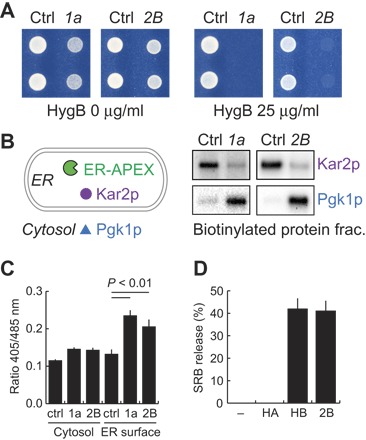
(A) Hygromycin B test. Yeast cells expressing indicated proteins are shown growing on defined solid media supplemented with the indicated hygromycin concentrations. (B) Assay for ER membrane integrity. Yeast cells expressing ER-APEX alone, or with BMV 1a or poliovirus 2B were subjected to biotin-phenol and H2O2 treatments to generate biotin-phenoxyl radicals in the ER lumen. Biotinylated protein fractions were then purified by using streptavidin-conjugated beads, electrophoresed, and visualized by Western blotting using antibodies against indicated proteins. 1a- or 2B-dependent leakage of biotin-phenoxyl radicals from the ER lumen was assessed by the biotinylation of Pgk1p, a cytosolic marker. (C) Redox status of the cytosol and cytosolic surface of the ER membrane. Two green fluorescent protein (GFP) reporters, roGFP2-Grx1 (Cytosol) and p450-roGFP2-Grx1 (ER surface), were used to monitor local glutathione redox statuses in control and 1a- and 2B-expressing yeast cells. Fluorescence intensities were measured with excitation at 405 nm for oxidized roGFP2 and at 485 nm for reduced roGFP2, and the observed fluorescence ratios were shown. (D) BMV 1a helix B and poliovirus 2B peptide-induced dye release from liposomes. Liposomes that encapsulated the self-quenching fluorescence dye SRB were incubated with indicated peptides (10 μM final). Dye release was monitored by fluorescence dequenching measurements.
APEX2 is an engineered ascorbate peroxidase that can be genetically targeted to a cellular region of interest, there catalyzing the oxidation of biotin-phenol to generate short-lived phenoxyl radicals upon H2O2 treatment and enabling in vivo proximity labeling (30–32). To examine whether 1a alters ER membrane permeability, we targeted APEX2 to the ER lumen (Fig. 3 and fig. S4) and observed protein biotinylation patterns of cytosolic and ER luminal markers such as Pgk1p and Kar2p, respectively. In control cells lacking 1a, ER-APEX selectively biotinylated Kar2p but not Pgk1p (Fig. 3B). However, in cells expressing either BMV 1a or poliovirus 2B, ER-APEX biotinylated both Kar2p and Pgk1p (Fig. 3B). The decline in the Kar2p biotinylation in cells expressing BMV 1a or poliovirus 2B is likely explained by reduction of ER-APEX–induced phenoxyl radicals in the ER lumen through their dispersal into the cytoplasm. Together, these results indicate that 1a induces the release of short-lived phenoxyl radicals and likely other small molecules from the ER lumen to the cytoplasm. In keeping with these results, 1a and 2B also induced a significantly more oxidized local environment at the cytoplasmic surface of the ER as monitored by the redox-sensitive reporter p450-roGFP2-Grx1, which was anchored on the outer face of the ER membrane but had much less oxidative effect in the general cytoplasm, as assayed by the cytosolic redox reporter roGFP2-Grx1 (Fig. 3C) (33).
A previous study identified in 1a an amphipathic helix, helix A, as a major ER-targeting region (34). Recent sequence analysis shows that the corresponding helix and another C-terminally adjacent amphipathic helix are conserved across the 1a-related replication proteins encoded throughout and beyond the alphavirus superfamily [(35) and fig. S5]. Similarly, poliovirus 2B contains two transmembrane α helices with both termini in the cytoplasm (23), and a peptide containing the first 2B amphipathic α helix is sufficient to permeabilize liposomes (36). To examine whether the putative amphipathic α-helical region in 1a is capable of membrane permeabilization, we used synthetic peptides and chemically defined liposomes encapsulating self-quenching fluorescence molecules. A peptide containing only the second amphipathic α-helical region in 1a (hereafter helix B), but not a peptide encoding only 1a helix A, released sulforhodamine B (SRB) from liposomes as efficiently as the 2B peptide (Fig. 3D).
Because Ero1p stimulates 1a’s disulfide-linked complex formation and RNA capping function, 1a viroporin mutations might inhibit these steps and show similarities to previously characterized 1a capping mutations (22). To determine whether and, if so, how 1a-mediated membrane permeabilization is necessary for BMV RNA replication, we used polymerase chain reaction (PCR) mutagenesis of several hydrophilic residues in critical membrane-permeabilizing helix B and suitable selection (see Materials and Methods) to isolate 1a mutants selectively lacking its viroporin function. These mutants, represented by 1a K424G and R426K, retain WT 1a’s early functions in recruiting and stabilizing (+)RNA3 templates in replication compartments in the absence of 2apol (Fig. 4C). However, in marked contrast to WT 1a, these mutants neither render yeast cells susceptible to hygromycin (Fig. 4A), disrupt the permeability barrier of the ER membrane (Fig. 4B), induce any ER-proximal cytoplasmic oxidizing environment (Fig. 4C), form any disulfide-linked 1a complex (Fig. 4E), nor support significant RNA-directed RNA3 or RNA4 accumulation in WT yeast (Fig. 4F). Consistent with their blocking 1a permeabilization of ER membranes in vivo (Fig. 4, B and C), these mutations also strongly interfered with helix B peptide–induced permeabilization of liposomes in vitro (Fig. 4D, upper panel). The simplified and more manipulatable in vitro liposome assay revealed that this last defect was concentration-dependent and could be largely overcome by increasing the peptide concentration 10-fold (Fig. 4D, lower panel). Because most viroporins permeabilize membranes by multimerizing to form pores (23), this suggests that the K424G and R426K mutations may shift the helix B pore assembly equilibrium toward a disassembled, inactive state, requiring 10-fold more peptide to achieve measurable pore assembly and activity. The in vivo RNA synthesis/accumulation defects of these viroporin mutants were partially rescued by XRN1 deletion (Fig. 4F). This partial complementation of the viroporin mutant phenotypes by XRN1 deletion both confirms the inferred crucial viroporin role in 1a’s RNA capping and suggests that 1a viroporin action may have additional important roles in other steps of BMV RNA replication.
Fig. 4. Linkage among 1a’s membrane permeabilization, disulfide bond formation, and capping functions.
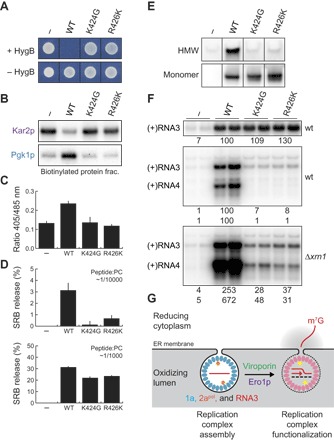
(A) Identification of viroporin-defective 1a mutants K424G and R426K, and (B to F) characterization of their phenotypes. (B) Assay for ER membrane integrity in yeast cells expressing WT 1a and 1a viroporin mutants by APEX-catalyzed protein biotinylation. (C) Redox statuses of the cytosolic surface of the ER membrane in yeast cells expressing WT 1a and 1a viroporin mutants. (D) Analysis of dye release from liposomes induced by WT, K424G, and R426K helix B peptides. Results are shown for two indicated molar ratios of peptide to phosphatidylcholine (PC); see Results and Discussion for further comments. (E) Analysis of disulfide-linked complexes in yeast cells expressing WT 1a and 1a viroporin mutants. 1a monomer serves as a loading control. (F) Effects of the viroporin mutations on 1a-mediated RNA3 recruitment in WT cells (upper panel) and 1a + 2apol–mediated RNA3 replication in WT and Δxrn1 cells (middle and lower panels). Note that a portion of the (+)RNA3 signals in the replication assay are derived from the recruitment and stabilization of RNA3 by WT 1a and 1a viroporin mutants. (G) Model for BMV RNA replication complex functionalization via membrane permeabilization and protein oxidation.
Thus, we show here that BMV requires the ER luminal thiol oxidase Ero1p for its cytoplasmic RNA replication. BMV replication protein 1a permeabilizes ER membranes to release Ero1p-generated oxidizing potential from the ER lumen to activate the cytoplasmic 1a protein’s RNA capping function via a new state of 1a multimerization involving oxidized linkages (Fig. 2E) and potentially 1a conformational changes (Fig. 4G). Several recent reports further imply an importance for oxidizing environments in genome replication by varied (+)RNA viruses: Knockdown or inactivation of respiratory burst oxidase homolog B of tobacco inhibits genome replication of two distinct plant viruses, BMV and red clover necrotic mosaic virus (37), and oxidizing agents can activate capping-related functions of alphavirus NSP1 and flavivirus NS5 (38). Similar to BMV 1a (14), the flavivirus NS5 forms a disulfide-linked dimer upon oxidation (38). In combination with previous results on picornaviruses and flaviviruses (23, 24), our findings reveal that genome replication proteins from three major superfamilies of (+)RNA viruses share membrane permeabilization as a previously unrecognized common feature. Therefore, these groups may also conserve an underlying molecular dependence on oxidation as shown here for BMV. Further study of the relevant mechanisms should be very informative. As noted above, our results show that 1a-induced membrane permeabilization is important for both viral RNA capping (Figs. 2, C to E, and 4, E and F) and apparently other aspects of BMV RNA replication (Fig. 4F). In keeping with this, the dependence of picornaviral replication on 2B viroporin activity must be independent of capping because picornaviral RNAs lack 5′ caps. Moreover, because picornavirus 2B(C) proteins increase cytosolic-free Ca2+ concentrations through viroporin functions (23, 39, 40), the potential influences on viral replication of luminal or extracellular calcium and other metal ions, as well as redox changes, should be investigated. Because protein channels are established, valuable therapeutic targets, further investigation and comparisons of relevant parallels among the three superfamilies could provide a new foundation for broad-spectrum antivirals.
MATERIALS AND METHODS
Yeast strain
Yeast strain YPH500 (MATα ura3-52 lys2-801 ada2-101 trp1-Δ63 his3-Δ200 leu2-Δ1) was used in all experiments (10). The Δxrn1 deletion mutant in the YPH500 genetic background was as described (41). The ero1-1 temperature-sensitive mutant in the YPH500 genetic background was generated by using a PCR-based strategy, as described (42), and showed extreme DTT sensitivity at a permissive temperature, as previously reported (16, 17).
Plasmids and plasmid construction
BMV 1a, 2apol, and genomic RNA3 were expressed from pB1YT3, pB2YT5, and pB3MS82 (22), or their derivatives with different selection markers. Viral and host factors were expressed under the control of the GAL1 promoter unless noted. In assays designed to lack 2apol-mediated, RNA-templated synthesis of (+)RNA3 (Fig. 2, A and B), pB3VG39 (20) was used to transcribe the RNA3 derivative 5′ Gal RNA3, in which the WT RNA3 5′ untranslated region was replaced with that of the GAL1 gene. Because BMV (+)RNAs both express and are strongly stabilized by the viral capsid protein, capsid protein expression can lead to positive feedback amplification effects on the levels of viral (+)RNAs, exaggerating differences in viral RNA replication. To avoid such possible effects, as in many previous BMV experiments, both full-length RNA3 from pB3MS82 and 5′ Gal RNA3 from pB3VG39 contain an early frameshift mutation in the BMV capsid protein gene.
To express Ero1p, the coding sequence was amplified from yeast genomic DNA by PCR using appropriate primers and inserted into a centromeric plasmid harboring the GAL1 promoter. For the expression of poliovirus 2B, yeast codon–optimized DNA fragments encoding 2Bc128 and 2Bc128*.3c (28) were chemically synthesized and inserted into centromeric plasmids containing the GAL1 promoter (Fig. 3, A and B). To express ER-APEX, a DNA fragment encoding the N-terminal signal sequence of Kar2p, APEX2 (31), FLAG, and HDEL retention signal was chemically synthesized and inserted in a centromeric plasmid harboring the GAL1 promoter. The plasmids were validated by Sanger sequencing.
RNA and protein analysis
Transformed yeast cells were precultured in selection media containing 2% raffinose to allow rapid GAL1 promoter induction upon galactose addition. Unless noted, the yeast cells were analyzed at 18 to 24 hours after galactose induction under either permissive or semipermissive conditions to minimize secondary effects of gene overexpression or mutation. RNA isolation and Northern blot analysis were as described (10). Northern blots were imaged on a Typhoon 9200 imager (GE Healthcare Life Sciences). Protein isolation, Western blot analysis, and antibodies against 1a, 2apol, FLAG, and Pgk1p were as described (11, 43). Anti-FLAG, anti-Kar2p, and anti-Pdi1p were purchased from MilliporeSigma, Santa Cruz Biotechnology, and Thermo Fisher Scientific, respectively. Endo H was purchased from New England Biolabs. Western blots were imaged on a ChemiDoc XRS system (Bio-Rad).
Isolation of 1a disulfide-linked complex
Two OD600 (optical density at 600 nm) units of yeast cells expressing WT 1a or 1a viroporin mutants were spheroplasted and then treated with 1 M sorbitol containing 10 mM N-ethylmaleimide (MilliporeSigma) for 10 min at room temperature. Spheroplasts were then lysed in 1 ml of 10% trichloroacetic acid (TCA), and total protein was collected by centrifugation for 5 min at 4°C at 15,000g. Protein pellets were washed with acetone twice. Dried pellets were suspended in 200 μl of 2× SDS loading buffer (Bio-Rad) containing 10 mM N-ethylmaleimide and vortexed for 10 min at 40°C. SDS-PAGE samples were divided into two, treated with or without DTT (final 100 mM), and vortexed for 10 min at 40°C. SDS-PAGE was performed using NuPAGE 3 to 8% tris-acetate gels (Thermo Fisher Scientific). After SDS-PAGE, the gels were soaked in a DTT-containing, alkaline transfer buffer (25 mM tris base, 190 mM glycine, 10 mM DTT) for 10 min at room temperature to break disulfide bonds of proteins and facilitate protein transfer to polyvinylidene difluoride (PVDF) membranes.
Hygromycin B test
Yeast cells were grown for 2 to 3 days on selection media containing 2% galactose in the presence and absence of hygromycin B (25 μg/ml; Thermo Fisher Scientific). A truncated, less-toxic 2B fragment, 2Bc128*.3c, was used in this assay (28).
APEX-catalyzed protein biotinylation
Protein biotinylation was performed essentially as described previously (30, 44). One OD600 unit of yeast cells was suspended in 0.1 M sorbitol containing 1 mM biotin-phenol for 30 min at 30°C and treated with 1 mM H2O2 for 1 min at room temperature. Phenoxyl radical reaction was then stopped by adding vitamin C and sodium azide (30). Total protein was immediately extracted from the yeast cells by a NaOH/TCA method (43). Resulting protein pellets were suspended in 100 μl of 1× SDS gel loading buffer (Bio-Rad) and vortexed for 10 min at 40°C. The total protein samples were then diluted 20 times with standard radioimmunoprecipitation assay (RIPA) buffer (MilliporeSigma) and mixed with streptavidin-coated Dynabeads (Thermo Fisher Scientific). Beads were washed four times with RIPA buffer before boiling in 1× SDS gel loading buffer and running samples in 4 to 15% SDS-PAGE gels (Bio-Rad). A longer 2B fragment, 2Bc128, was used as a positive control.
Redox monitoring
p415TEF cyto roGFP2-Grx1 was a gift from T. Dick (Addgene 65004) (33). An ER-targeting sequence corresponding to amino acids 1 to 34 from the N terminus of yeast NADP cytochrome P450 oxidoreductase (45) was fused to roGFP2-Grx1 (P450ss-roGFP2-Grx1). Fluorescent measurements were performed as described (46). A longer 2B fragment, 2Bc128, was used in this assay.
Liposome permeabilization assay
A poliovirus 2B peptide corresponding to the P3 peptide of Madan et al. (36) and two BMV 1a peptides corresponding to amino acids 388 to 422 (helix A) and 415 to 453 (helix B), respectively, were chemically synthesized by GenScript. Dulbecco’s phosphate-buffered saline was used as a solvent throughout the experiments. Lipid mixtures and SRB were purchased from MilliporeSigma. Liposome-encapsulated SRB was obtained by gel filtration using Sephadex G-25 resin (GE Healthcare Life Sciences). Liposomes containing ~1 mM phosphatidylcholine were incubated for 1 hour at room temperature with 0.1 or 1 μM peptide in Fig. 4C. In Fig. 3D, because the poliovirus 2B peptide required higher peptide-to-lipid molar ratios to permeabilize liposomes, the 2B and BMV peptides were each used at a concentration of 10 μM. SRB fluorescence was measured at an emission wavelength of 572 nm by using a Victor plate reader (PerkinElmer). The degree of SRB release was calculated as described (47).
Isolation of viroporin-defective mutants
Amino acid substitutions at each hydrophilic residue in helix B (H422, K424, R426, W428, or W429) were introduced by an overlap PCR method using primers harboring either ANN, CNN, or GNN mixed mutations at the corresponding codon. Separate libraries of random mutants at each of these five positions were constructed using homologous recombination of the resulting PCR products into pB1YT3L in YMI04, an RNA3-Ura3/RNA3-GUS reporter strain in the YPH500 genetic background (48). Transformed yeast cells were directly plated on galactose- and hygromycin-containing selection plates and grown for 5 to 6 days. Transformants that were not sensitive to hygromycin B but grew more slowly than control vector-transformed cells were selected to omit previously identified, faster-growing 1a mutants impaired in membrane targeting (34). 1a mutations responsible for the resulting selected phenotypes, and found capable of selective viral RNA template recruitment, were identified by Sanger sequencing of the selected transformed plasmids and were independently reintroduced into the 1a plasmid pB1YT3. K424G and R426K were identified through this screening, and their phenotypes were verified and further analyzed in our standard yeast strain YPH500.
Supplementary Material
Acknowledgments
We thank B. Sibert and J. den Boon for providing yeast cell lines and plasmids, M. Bracken and Z. Coleman for valuable technical assistance, other members of our laboratory for fruitful discussions, and K. Wessels and D. van de Velde for crucial administrative support. Funding: P.A. is an investigator of the Howard Hughes Medical Institute and the Morgridge Institute for Research, and gratefully acknowledges support from these institutes, the NIH, and the Rowe Family Virology Venture Fund. Author contributions: M.N. and P.A. conceived and designed the experiments. M.N. performed the experiments. M.N. and P.A. wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/1/eaap8258/DC1
fig. S1. Model for BMV RNA replication complex assembly and function.
fig. S2. Ero1p-FLAG stimulates BMV RNA replication and is glycosylated in control and 1a-expressing cells.
fig. S3. Effect of ERO1 overexpression on 1a-increased RNA3 accumulation.
fig. S4. Subcellular fractionation analysis of ER-APEX.
fig. S5. Evolutionarily conserved helix A and B sequences.
REFERENCES AND NOTES
- 1.den Boon J. A., Ahlquist P., Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 64, 241–256 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Ertel K. J., Benefield D., Castaño-Diez D., Pennington J. G., Horswill M., den Boon J. A., Otegui M. S., Ahlquist P., Cryo-electron tomography reveals novel features of a viral RNA replication compartment. eLife 6, e25940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altan-Bonnet N., Lipid tales of viral replication and transmission. Trends Cell Biol. 27, 201–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahola T., Ahlquist P., Putative RNA capping activities encoded by brome mosaic virus: Methylation and covalent binding of guanylate by replicase protein 1a. J. Virol. 73, 10061–10069 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X., Lee W.-M., Watanabe T., Schwartz M., Janda M., Ahlquist P., Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J. Virol. 79, 13747–13758 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishikiori M., Sugiyama S., Xiang H., Niiyama M., Ishibashi K., Inoue T., Ishikawa M., Matsumura H., Katoh E., Crystal structure of the superfamily 1 helicase from Tomato Mosaic Virus. J. Virol. 86, 7565–7576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kao C. C., Ahlquist P., Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J. Virol. 66, 7293–7302 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J., Ahlquist P., Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74, 4310–4318 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz M., Chen J., Janda M., Sullivan M., den Boon J., Ahlquist P., A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9, 505–514 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Janda M., Ahlquist P., RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72, 961–970 (1993). [DOI] [PubMed] [Google Scholar]
- 11.Gancarz B. L., Hao L., He Q., Newton M. A., Ahlquist P., Systematic identification of novel, essential host genes affecting bromovirus RNA replication. PLOS ONE 6, e23988 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kushner D. B., Lindenbach B. D., Grdzelishvili V. Z., Noueiry A. O., Paul S. M., Ahlquist P., Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. U.S.A. 100, 15764–15769 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.den Boon J. A., Chen J., Ahlquist P., Identification of sequences in Brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75, 12370–12381 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishikiori M., Meshi T., Ishikawa M., Guanylylation-competent replication proteins of Tomato mosaic virus are disulfide-linked. Virology 434, 118–128 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Riemer J., Bulleid N., Herrmann J. M., Disulfide formation in the ER and mitochondria: Two solutions to a common process. Science 324, 1284–1287 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Frand A. R., Kaiser C. A., The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell 1, 161–170 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Pollard M. G., Travers K. J., Weissman J. S., Ero1p: A novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell 1, 171–182 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Tu B. P., Weissman J. S., The FAD- and O2-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol. Cell 10, 983–994 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Janda M., Ahlquist P., Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. U.S.A. 95, 2227–2232 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grdzelishvili V. Z., Garcia-Ruiz H., Watanabe T., Ahlquist P., Mutual interference between genomic RNA replication and subgenomic mRNA transcription in brome mosaic virus. J. Virol. 79, 1438–1451 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parker R., RNA degradation in Saccharomyces cerevisae. Genetics 191, 671–702 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahola T., den Boon J. A., Ahlquist P., Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J. Virol. 74, 8803–8811 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nieva J. L., Madan V., Carrasco L., Viroporins: Structure and biological functions. Nat. Rev. Microbiol. 10, 563–574 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang Y.-S., Liao C.-L., Tsao C.-H., Chen M.-C., Liu C.-I., Chen L.-K., Lin Y.-L., Membrane permeabilization by small hydrophobic nonstructural proteins of Japanese encephalitis virus. J. Virol. 73, 6257–6264 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Kuppeveld F. J. M., Melchers W. J. G., Kirkegaard K., Doedens J. R., Structure-function analysis of coxsackie B3 virus protein 2B. Virology 227, 111–118 (1997). [DOI] [PubMed] [Google Scholar]
- 26.van Kuppeveld F. J. M., Galama J. M. D., Zoll J., van den Hurk P. J. J. C., Melchers W. J. G., Coxsackie B3 virus protein 2B contains cationic amphipathic helix that is required for viral RNA replication. J. Virol. 70, 3876–3886 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Jong A. S., Melchers W. J. G., Glaudemans D. H. R. F., Willems P. H. G. M., van Kuppeveld F. J. M., Mutational analysis of different regions in the coxsackievirus 2B protein: Requirements for homo-multimerization, membrane permeabilization, subcellular localization, and virus replication. J. Biol. Chem. 279, 19924–19935 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Barco A., Carrasco L., Identification of regions of poliovirus 2BC protein that are involved in cytotoxicity. J. Virol. 72, 3560–3570 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doedens J. R., Kirkegaard K., Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14, 894–907 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee H.-W., Zou P., Udeshi N. D., Martell J. D., Mootha V. K., Carr S. A., Ting A. Y., Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam S. S., Martell J. D., Kamer K. J., Deerinck T. J., Ellisman M. H., Mootha V. K., Ting A. Y., Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S.-Y., Kang M.-G., Park J.-S., Lee G., Ting A. Y., Rhee H.-W., APEX fingerprinting reveals the subcellular localization of proteins of interest. Cell Rep. 15, 1837–1847 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Morgan B., Ezeriņa D., Amoako T. N. E., Riemer J., Seedorf M., Dick T. P., Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nat. Chem. Biol. 9, 119–125 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Liu L., Westler W. M., den Boon J. A., Wang X., Diaz A., Steinberg H. A., Ahlquist P., An amphipathic α-helix controls multiple roles of brome mosaic virus protein 1a in RNA replication complex assembly and function. PLOS Pathog. 5, e1000351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahola T., Karlin D. G., Sequence analysis reveals a conserved extension in the capping enzyme of the alphavirus supergroup, and a homologous domain in nodaviruses. Biol. Direct 10, 16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madan V., Sánchez-Martínez S., Vedovato N., Rispoli G., Carrasco L., Nieva J. L., Plasma membrane-porating domain in poliovirus 2B protein. A short peptide mimics viroporin activity. J. Mol. Biol. 374, 951–964 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Hyodo K., Hashimoto K., Kuchitsu K., Suzuki N., Okuno T., Harnessing host ROS-generating machinery for the robust genome replication of a plant RNA virus. Proc. Natl. Acad. Sci. U.S.A. 114, E1282–E1290 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gullberg R. C., Jordan Steel J., Moon S. L., Soltani E., Geiss B. J., Oxidative stress influences positive strand RNA virus genome synthesis and capping. Virology 475, 219–229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Jong A. S., de Mattia F., Van Dommelen M. M., Lanke K., Melchers W. J. G., Willems P. H. G. M., van Kuppeveld F. J. M., Functional analysis of picornavirus 2B proteins: Effects on calcium homeostasis and intracellular protein trafficking. J. Virol. 82, 3782–3790 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Kuppeveld F. J. M., Hoenderop J. G. J., Smeets R. L. L., Willems P. H. G. M., Dijkman H. B. P. M., Galama J. M. D., Melchers W. J. G., Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 16, 3519–3532 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larimer F. W., Stevens A., Disruption of the gene XRN1, coding for a 5’----3’ exoribonuclease, restricts yeast cell growth. Gene 95, 85–90 (1990). [DOI] [PubMed] [Google Scholar]
- 42.Li Z., Vizeacoumar F. J., Bahr S., Li J., Warringer J., Vizeacoumar F. S., Min R., VanderSluis B., Bellay J., DeVit M., Fleming J. A., Stephens A., Haase J., Lin Z.-Y., Baryshnikova A., Lu H., Yan Z., Jin K., Barker S., Datti A., Giaever G., Nislow C., Bulawa C., Myers C. L., Costanzo M., Gingras A.-C., Zhang Z., Blomberg A., Bloom K., Andrews B., Boone C., Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat. Biotechnol. 29, 361–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horvath A., Riezman H., Rapid protein extraction from Saccharomyces cerevisiae. Yeast 10, 1305–1310 (1994). [DOI] [PubMed] [Google Scholar]
- 44.Hwang J., Espenshade P. J., Proximity-dependent biotin labelling in yeast using the engineered ascorbate peroxidase APEX2. Biochem. J. 473, 2463–2469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller D. J., Schwartz M. D., Dye B. T., Ahlquist P., Engineered retargeting of viral RNA replication complexes to an alternative intracellular membrane. J. Virol. 77, 12193–12202 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgan B., Sobotta M. C., Dick T. P., Measuring E(GSH) and H2O2 with roGFP2-based redox probes. Free Radic. Biol. Med. 51, 1943–1951 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Shukla A., Dey D., Banerjee K., Nain A., Banerjee M., The C-terminal region of the non-structural protein 2B from Hepatitis A Virus demonstrates lipid-specific viroporin-like activity. Sci. Rep. 5, 15884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishikawa M., Díez J., Restrepo-Hartwig M., Ahlquist P., Yeast mutations in multiple complementation groups inhibit brome mosaic virus RNA replication and transcription and perturb regulated expression of the viral polymerase-like gene. Proc. Natl. Acad. Sci. U.S.A. 94, 13810–13815 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian G., Xiang S., Noiva R., Lennarz W. J., Schindelin H., The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell 124, 61–73 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Frand A. R., Kaiser C. A., Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol. Cell 4, 469–477 (1999). [DOI] [PubMed] [Google Scholar]
- 51.Doh J. H., Lutz S., Curcio M. J., Co-translational localization of an LTR-retrotransposon RNA to the endoplasmic reticulum nucleates virus-like particle assembly sites. PLOS Genet. 10, e1004219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gautier R., Douguet D., Antonny B., Drin G., HELIQUEST: A web server to screen sequences with specific α-helical properties. Bioinformatics 24, 2101–2102 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/1/eaap8258/DC1
fig. S1. Model for BMV RNA replication complex assembly and function.
fig. S2. Ero1p-FLAG stimulates BMV RNA replication and is glycosylated in control and 1a-expressing cells.
fig. S3. Effect of ERO1 overexpression on 1a-increased RNA3 accumulation.
fig. S4. Subcellular fractionation analysis of ER-APEX.
fig. S5. Evolutionarily conserved helix A and B sequences.


