Abstract
Hepatitis B is one of the most common infectious diseases, which leads to public health problems in the world, especially in Asian counties. In recent years, extensive human genetic association studies have been carried out to identify susceptible genes and genetic polymorphisms to understand the genetic contributions to the disease progression of HBV infection. HLA-DQ gene variations have been reported to be associated with HBV infection/clearance, disease progression and the development of hepatitis B-related complications, including liver cirrhosis (LC) and hepatocellular carcinoma (HCC). However, the results are either inconclusive or controversial. Therefore, to derive a more precise estimation of the association, a meta-analysis was performed. Our data revealed that the HLA-DQ alleles rs2856718-G, rs7453920-A and rs9275319-G were significantly associated with decreased risk of HBV infection and HBV natural clearance. Logistic regression analyses showed that HLA-DQ alleles rs9275572-A significantly increased HBV infection clearance, and decreased HBV natural clearance. However, rs2856718-G and rs9275572-A were not associated with development of cirrhosis. The HLA-DQ polymorphisms (rs2856718 and rs9275572) were associated with a decreased HBV-related HCC risk in all genetic models, but rs9272105-A increased the risk of HBV-related HCC. In addition, no significant association was observed between HLA-DQ rs9275319-G polymorphism and HBV-related HCC. These stratified analyses were limited due to relatively modest size of correlational studies. In future, further investigation on a large population and different ethnicities are warranted. Our findings contribute to the personalized care and prognosis in hepatitis B.
Keywords: HLA-DQ, Hepatitis B virus, Polymorphism, liver cirrhosis, hepatocellular carcinoma, Immunology
INTRODUCTION
Hepatitis B is an infectious disease caused by hepatitis B virus (HBV), which leads to the serious public health problems worldwide, especially in Asian counties. Nowadays, there are more than 240 million HBV carriers [1], among which 0.5-1.2 million died of chronic HBV infection each year [2]. As is known to all, the HBV infection is usually complex and variable [3], and can result in different clinical outcomes. Several progressive stages are confirmed for chronic HBV infection,including chronic hepatitis B (CHB), liver cirrhosis (LC), as well as hepatocellular carcinoma (HCC) [4]. Chronic HBV infection can progress into CHB, while about 10%-30% will progress to liver cirrhosis and HCC [5]. Thus, the degree of chronic HBV infection varies enormously among individuals, which represents a complex biological process where the cellular mechanisms and genetic contributions of pathogenesis remain unknown [6, 7]. These facts contribute to the development of more personalized therapy, diagnosis or prognosis, which then reduce the health disparity among the victims.
Viral factors (genotype and mutations) [8], host factors and environmental factors [9] are considered to involve in the disease progression of HBV infection, from HBV clearance to chronic infection that may progress into liver cirrhosis and HCC [10-12]. To date, several host factors are available, including age of infection, gender, volume of alcohol intake, obesity, smoking, diabetes, chemical exposure and chemical exposure [13, 14]. In addition, results from twin studies and candidate gene approaches demonstrated that host genetic factors may be closely associated with the outcome of HBV infection and progression [15-17]. Single nucleotide polymorphisms (SNPs), representing the most common type of genetic variation in human beings, may change the structure and biological function of the encoded protein [18]. Recently, genetic polymorphisms have attracted more attention due to their etiological roles in defining the disease progression of HBV infection. Recent studies indicated that variants in some host genes, including interleukin-4 (IL-4) gene -2590C/T (rs2243250) and -233C/T (rs20708742590) [19], tumor necrosis factor-α (TNF-α) gene -308 G/A [20], toll-like receptor 3 (TLR3) gene (rs1879026 and rs3775290) [21], vascular endothelial growth factor (VEGF) gene 634 G/C (rs2010963) [22] and cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) gene +49A/G [23], were associated with persistent HBV infection and natural clearance.
In the past few years, several genome-wide association studies (GWAS) have identified that SNPs proximate to the HLA-DP, HLA-DQ, and HLA-DR loci are significantly correlated with HBV infection outcomes [24-26]. Additionally, several studies on different populations have focused on the roles of HLA-DQ gene polymorphism in the pathogenesis of HBV infection. However, these findings are still controversial. Furthermore, a single-center study may have an inadequate sample size and lack of statistical power to obtain reliable conclusions. In this study, a comprehensive meta-analysis was utilized to precisely evaluate the correlation between HLA-DQ gene polymorphism and HBV infection complications (e.g. CHB, LC, and HCC).
RESULTS
Study characteristics
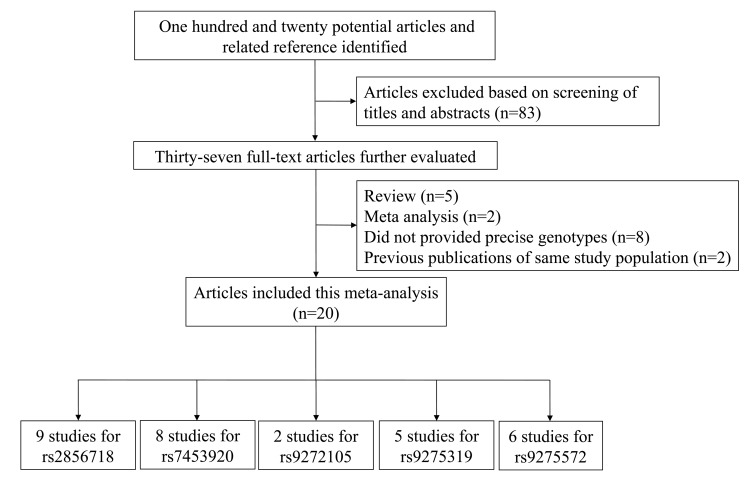
According to our search strategy, 120 publications were identified through the initial search after excluding 83 articles. A flow diagram of the detail selection and exclusion process was displayed in Figure 1. After full review, 37 studies were then excluded based on the following aspects: duplicate data, review articles, meta-analyses and case-only studies. Finally, 20 studies (28347 cases and 37329 controls) were chosen, and the data were extracted. Among these publications, there were 9 studies for rs2856718 [24, 25, 28, 31-33, 40, 43, 44], 8 for rs7453920 [24, 25, 28, 31, 34. 39, 44, 45], 2 for rs9272105 [29, 38], 5 for rs9275319 [33, 36-38, 40] and 6 for rs9275572 [30-32, 35, 41, 43]. The main features of each eligible study were summarized in Table 1 and Figure 2, respectively.
Figure 1. The flow charts of literature search and study selection.
Table1. Characteristics of the studies included in the meta-analysis.
| Study | Year | Ethnicity | Subgroup | Genotyping method | Case | Control | No. of cases | No. of controls | Polymorphisms | NOS |
|---|---|---|---|---|---|---|---|---|---|---|
| Mbarek H [24] | 2011 | Japanese | GWAS | GeneChip | CHB | non-HBV | 458 | 2056 | rs2856718; rs7453920 |
6 |
| 2011 | Japanese | First replication | Invader assay | CHB | non-HBV | 606 | 2023 | |||
| 2011 | Japanese | Second replication | TaqMan | CHB | non-HBV | 379 | 1539 | |||
| 2011 | Japanese | Third replication | TaqMan | CHB | non-HBV | 1226 | 879 | |||
| Hu LM [28] | 2012 | Chinese | TaqMan | HBV Carriers; HBV-HCC |
HBV clearance | 2644 | 1344 | rs2856718; rs7453920 |
7 | |
| Li SP [29] | 2012 | Chinese | GWAS Southern | Gene Chip | HCC | HBV positive | 1075 | 990 | rs9272105 | 7 |
| 2012 | Chinese | GWAS Central | Gene Chip | HCC | HBV positive | 500 | 500 | |||
| 2012 | Chinese | Validation 1 | iPLEX/TaqMan | HCC | HBV-positive | 2112 | 2208 | |||
| 2012 | Chinese | Validation 2 | iPLEX/TaqMan | HCC | HBV-positive | 1021 | 1491 | |||
| 2012 | Chinese | Replication | iPLEX/TaqMan | HCC | HBV-positive | 1298 | 1026 | |||
| Hu ZB [25] | 2013 | Chinese | GWAS | GeneChip | HBV carriers | HBV clearance | 951 | 937 | rs7453920 rs2856718 |
6 |
| 2013 | Chinese | Replication Ia | iPLEX | HBV carriers | HBV clearance | 1248 | 1248 | rs7453920 rs2856718 |
||
| 2013 | Chinese | Replication Ib | TaqMan | HBV carriers | HBV clearance | 1000 | 1803 | rs7453920 rs2856718 |
||
| 2013 | Chinese | Replication IIa | iPLEX | HBV carriers | HBV clearance | 981 | 1417 | rs7453920 | ||
| 2013 | Chinese | Replication IIb | TaqMan | HBV carriers | HBV clearance | 1001 | 1205 | rs7453920 | ||
| Chen KM [30] | 2013 | Chinese | TaqMan | HCC | CHB | 506 | 772 | rs9275572 | 8 | |
| Al-Qahtani AA [31] | 2014 | Saudi Arabian | PCR-based genotyping/ TaqMan |
HBV carriers (AsC, LC, HCC) |
Healthy controls, HBV clearance |
781 | 302, 587 | rs2856718; rs7453920; rs9275572 |
7 | |
| Zhang X [32] | 2014 | Chinese | Flight mass spectrometry | HBV carriers (CHB, LC, HCC); |
Healthy controls, HBV clearance |
792 | 507, 350 | rs2856718; rs9275572 |
8 | |
| Ji XW [33] | 2014 | Chinese | Real-time PCR | HBV carriers (CHB, ASCs, LC) |
Healthy controls; HBV clearance |
2489 | 1342; 327 | rs2856718; rs9275319 |
8 | |
| Liao Y [34] | 2014 | Chinese | HRM | chronic HBV carriers; HCC |
Healthy controls; HBV clearance |
677 | 237, 398 | rs7453920 | 8 | |
| Hou SH [35] | 2015 | Chinese | TaqMan | HBV carriers (CHB, LC, HCC) |
Healthy controls; HBV clearance |
310 | 316, 295 | rs9275572 | 8 | |
| Hou SH [36] | 2015 | Chinese | TaqMan | HBV carriers (CHB, LC, HCC) |
Healthy controls; HBV clearance |
310 | 316, 295 | rs9275319 | 8 | |
| Kim LH [37] | 2015 | Korean | TaqMan | CHB; HCC | Population control samples | 958 | 2880 | rs9275319 | 7 | |
| Wen J [38] | 2015 | Chinese | TaqMan | HCC | HBV persistent carriers | 1507 | 1560 | rs9272105; rs9275319 |
6 | |
| Liao Y [39] | 2015 | Chinese | Tibetans | HRM | HBV carriers | HBV clearance | 422 | 486 | rs7453920 | 7 |
| Chinese | Uygurs | HRM | HBV carriers | HBV clearance | 195 | 235 | ||||
| Jiang DK [40] | 2015 | Chinese | Shanghai | MassARRAY/TaqMan | LC | CHB | 440 | 1265 | rs9275319 | 6 |
| Chinese | Beijing | LC | CHB | 272 | 1336 | |||||
| Liu WX [41] | 2016 | Chinese | Flight mass spectrometry | HBV carriers | Healthy controls; HBV clearance |
396 | 254, 175 | rs2856718; rs9275572 |
8 | |
| Fan JH [42] | 2016 | Chinese | MassARRAY | HBV carriers | Healthy controls; HBV clearance |
397 | 238, 434 | rs9275319 | 8 | |
| Gao X [43] | 2016 | Chinese | Flight mass spectrometry | HBV carriers (CHB, LC, HCC) |
Healthy controls | 784 | 507 | rs2856718; rs9275572 |
8 | |
| Trinks [44] | 2017 | Argentinean | Central areas | TaqMan | HBV carriers | Healthy controls; HBV clearance |
201 | 207, 318 | rs2856718; rs7453920 |
8 |
| 2017 | Argentinean | North-western areas | TaqMan | HBV carriers | 200 | 201, 313 | ||||
| Pereira VRZB [45] | 2017 | Brazilian | TaqMan | CHB | Healthy controls | 210 | 210 | rs7453920 | 8 |
CHB: Chronic Hepatitis B; HBV: Hepatitis B Virus; HCC: Hepatocellular Carcinoma; LC: Liver Cirrhosis; AsC: Asymptomatic Carriers; NOS: Newcastle-Ottawa Scale.
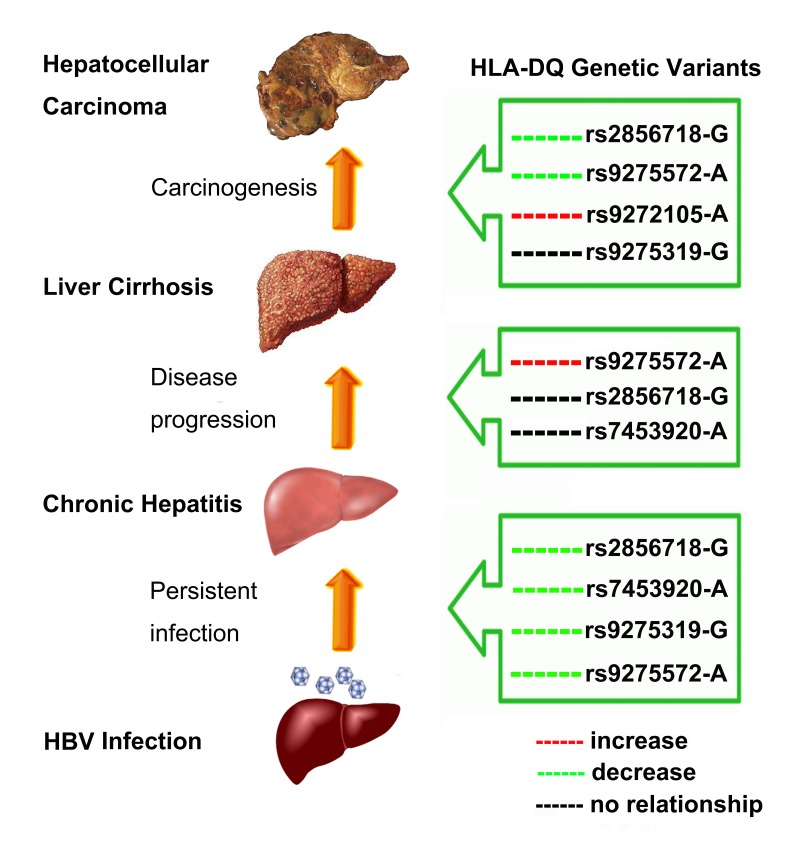
Figure 2. Host HLA-DQ region polymorphisms influencing infection outcomes.
Association between HLA-DQ rs2856718 polymorphism and outcome of HBV infection
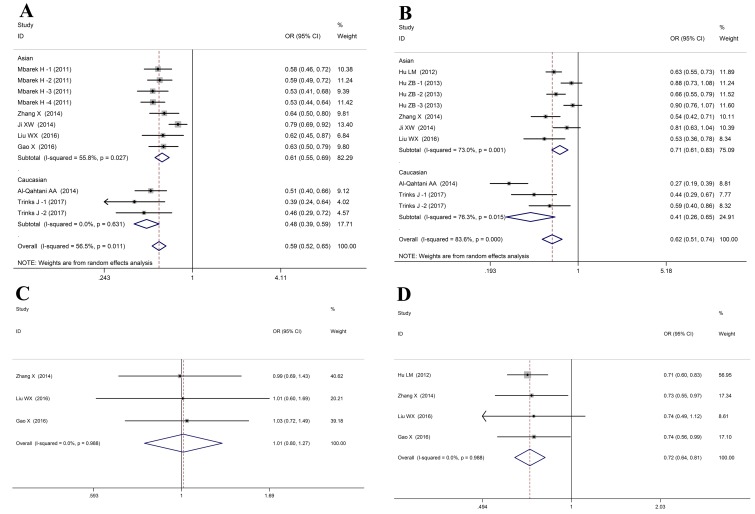
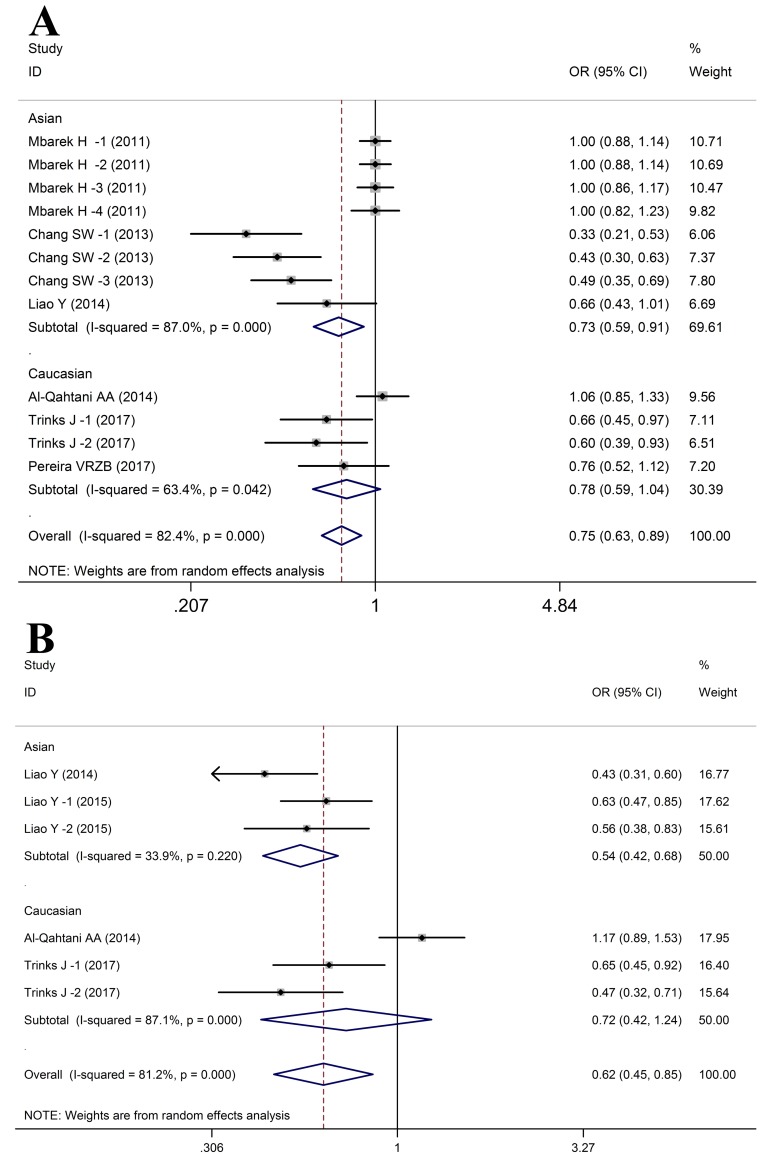
In the meta-analysis, 9 studies including 14155 cases and 17219 controls were included to investigate the associations between HLA-DQ rs2856718 polymorphism and HBV infection outcomes (Table 2). These results indicated that HLA-DQ rs2856718 was considered to be associated with a decrease of HBV infection risk (HBV infection vs. Control: allele: OR= 0.66, 95%CI: 0.60-0.73, PZ < 0.01; heterozygous: OR= 0.66, 95%CI: 0.62-0.71, PZ < 0.01; homozygous: OR= 0.46, 95%CI: 0.37-0.55, PZ < 0.01; recessive: OR= 0.60, 95%CI: 0.50-0.72, PZ < 0.01; dominant: OR= 0.59, 95%CI: 0.52-0.65, PZ < 0.01, Figure 3A). Whereas, in the Caucasian populations, no association was noticed in the recessive model (GG vs. AG+AA: OR = 0.63, 95%CI: 0.38-1.06, PZ = 0.08). Meanwhile, HLA-DQ rs2856718 polymorphism showed significant association with HBV clearance (HBV infection vs. SC: allele: OR= 0.74, 95%CI: 0.67-0.82, PZ < 0.01; heterozygous: OR= 0.63, 95%CI: 0.51-0.79, PZ < 0.01; homozygous: OR= 0.74, 95%CI: 0.70-0.78, PZ < 0.01; recessive: OR= 0.74, 95%CI: 0.63-0.87, PZ < 0.01; dominant: OR= 0.62, 95%CI: 0.51-0.74, PZ < 0.01, Figure 3B). Moreover, the HLA-DQ rs2856718 polymorphism was correlated with a decrease of HBV-related HCC risk in four genetic models (HCC vs. LC+CHB: allele: OR = 0.80, 95%CI: 0.76-0.90, PZ < 0.01; heterozygous: OR = 0.71, 95%CI: 0.63-0.81, PZ < 0.01; homozygous: OR = 0.74, 95%CI: 0.62-0.88, PZ < 0.01; dominant: OR = 0.72, 95%CI: 0.64-0.81, PZ < 0.01, Figure 3D). However, no association was noticed between HLA-DQ rs2856718 polymorphism and LC development from CHB in all genetic models (LC vs. CHB: allele: OR= 0.99, 95%CI: 0.84-1.17, PZ = 0.88; heterozygous: OR= 1.03, 95%CI: 0.81-1.32, PZ = 0.81; homozygous: OR = 0.96, 95%CI: 0.67-1.34, PZ = 0.78; recessive: OR= 0.94, 95%CI: 0.68-1.28, PZ = 0.69; dominant: OR= 1.01, 95%CI: 0.80-1.27, PZ = 0.93, Figure 3C).
Table 2. Main results of the meta-analysis of the association between HLA-DQ rs2856718 polymorphism and HBV infection outcomes.
| Comparison | Subgroup | Allele model (G vs. A) |
Heterozygous model (AG vs. AA) |
Homozygous model (GG vs. AA) |
Recessive model (GG vs. AG+AA) |
Dominant model (AG+GG vs. AA) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | ||
| HBV infection vs. Control | Overall | 0.66 (0.60-0.73) | <0.01 | <0.01 | 0.66 (0.62-0.71) | 0.04 | <0.01 | 0.46 (0.37-0.55) | <0.01 | <0.01 | 0.60 (0.50-0.72) | <0.01 | <0.01 | 0.59 (0.52-0.65) | 0.01 | <0.01 |
| Asian | 0.67 (0.60-0.75) | <0.01 | <0.01 | 0.70 (0.64-0.75) | 0.49 | <0.01 | 0.46 (0.37-0.58) | <0.01 | <0.01 | 0.58 (0.48-0.71) | <0.01 | <0.01 | 0.61 (0.55-0.69) | 0.03 | <0.01 | |
| Caucasian | 0.63 (0.50-0.78) | 0.08 | <0.01 | 0.46 (0.37-0.58) | 0.70 | <0.01 | 0.41 (0.24-0.70) | 0.02 | <0.01 | 0.63 (0.38-1.06) | <0.01 | 0.08 | 0.48 (0.39-0.59) | 0.63 | <0.01 | |
| HBV infection vs. NC | Overall | 0.74 (0.67-0.82) | <0.01 | <0.01 | 0.63 (0.51-0.79) | <0.01 | <0.01 | 0.74 (0.70-0.78) | <0.01 | <0.01 | 0.74 (0.63-0.87) | 0.04 | <0.01 | 0.62 (0.51-0.74) | <0.01 | <0.01 |
| Asian | 0.78 (0.70-0.87) | <0.01 | <0.01 | 0.75 (0.66-0.87) | 0.01 | <0.01 | 0.75 (0.71-0.80) | <0.01 | <0.01 | 0.74 (0.64-0.84) | <0.01 | <0.01 | 0.71 (0.61-0.83) | <0.01 | <0.01 | |
| Caucasian | 0.64 (0.56-0.73) | 0.64 | <0.01 | 0.41 (0.18-0.90) | <0.01 | 0.03 | 0.65 (0.57-0.70) | 0.56 | <0.01 | 0.72 (0.38-1.38) | <0.01 | 0.33 | 0.41 (0.26-0.65) | 0.02 | <0.01 | |
| LC vs. CHB | Chinese | 0.99 (0.84-1.17) | 0.99 | 0.88 | 1.03 (0.81-1.32) | 0.99 | 0.81 | 0.96 (0.67-1.34) | 0.99 | 0.78 | 0.94 (0.68-1.28) | 0.99 | 0.69 | 1.01 (0.80-1.27) | 0.99 | 0.93 |
| HCC vs. LC+CHB | Chinese | 0.80 (0.76-0.90) | 0.90 | <0.01 | 0.71 (0.63-0.81) | 0.85 | <0.01 | 0.74 (0.62-0.88) | 0.83 | <0.01 | 0.90 (0.77-1.06) | 0.52 | 0.20 | 0.72 (0.64-0.81) | 0.99 | <0.01 |
-*Because there was only one study with this genotype of rs2856718, the value could not be calculated.
Figure 3. Forest plots for HLA-DQ rs2856718 polymorphism and HBV infection outcomes.
A. HBV infection vs. Control (AA vs. AG+GG); B. HBV infection vs. NC (AA vs. AG+GG); C. LC vs. CHB (AA vs. AG+GG); D. HCC vs. LC+CHB (AA vs. AG+GG).
Meta-analysis for HLA-DQ rs9275572 polymorphism with HBV infection outcomes
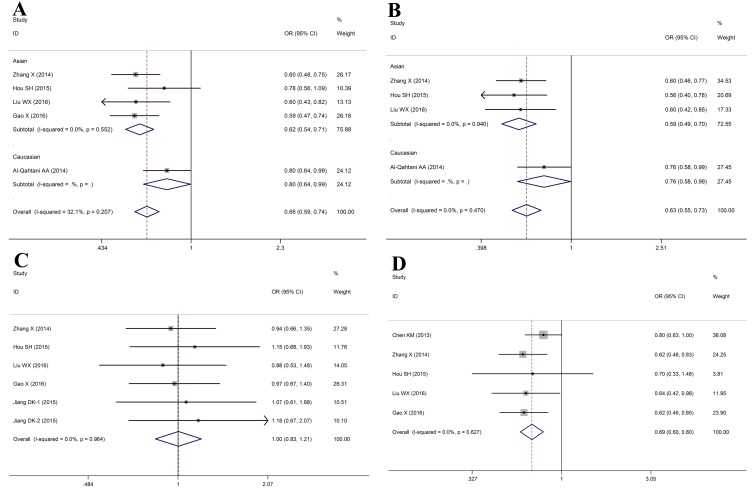
Finally, 6 studies including 3569 cases and 4065 controls were subject to analysis using fixed-effects or random-effects model (Table 3). Pooled analysis demonstrated that HLA-DQ rs9275572 polymorphism was correlated with a significantly increased risk of HBV infection in total population (HBV infection vs. Control: A vs. G: OR = 0.68, 95%CI: 0.62-0.74, PZ < 0.01; AG vs. GG: OR = 0.73, 95%CI: 0.65-0.82, PZ < 0.01; AA vs. GG: OR = 0.45, 95%CI: 0.37-0.56, PZ < 0.01; AA vs. AG+GG: OR = 0.51, 95%CI: 0.42-0.62, PZ < 0.01; AA+AG vs. GG: OR = 0.66, 95%CI: 0.59-0.74, PZ < 0.01, Figure 4A). With regards to the HBV clearance, our data indicated that subjects with the HLA-DQ rs9275572-A allele showed a significantly lower incidence of spontaneous clearance after HBV infection (HBV infection vs. NC: allele: OR = 0.70, 95%CI: 0.62-0.79, PZ < 0.01; heterozygous: OR = 0.65, 95%CI: 0.56-0.76, PZ < 0.01; homozygous: OR = 0.57, 95%CI: 0.44-0.76, PZ < 0.01; recessive: OR = 0.67, 95%CI: 0.52-0.87, PZ < 0.01; dominant: OR = 0.63, 95%CI: 0.55-0.73, PZ < 0.01, Figure 4B). Among LC and CHB patients, we found a significant relationship between the A allele and decreased risk of CHB to LC with an OR of 1.34 (95%CI: 1.14-1.56) for HLA-DQ rs9275572, but there was no significant correlation in the heterozygous model (OR = 0.97, 95%CI: 0.80-1.18, PZ = 0.76), homozygous model (OR = 1.22, 95%CI: 0.83-1.80, PZ = 0.31), recessive model (OR = 1.25, 95%CI: 0.85-1.82, PZ = 0.25) and dominant model (OR = 1.00, 95%CI: 0.83-1.21, PZ = 0.97, Figure 4C). These results revealed that a significant correlation might be presented between the HLA-DQ rs9275572 polymorphism and HBV-related HCC in all gene model (HCC vs. LC+CHB: allele: OR = 0.71, 95%CI: 0.63-0.80, PZ < 0.01; heterozygous: OR = 0.73, 95%CI: 0.64-0.84, PZ < 0.01; homozygous: OR = 0.49, 95%CI: 0.35-0.68, PZ < 0.01; recessive: OR = 0.54, 95%CI: 0.39-0.76, PZ < 0.01; dominant: OR = 0.69, 95%CI: 0.60-0.80, PZ < 0.01, Figure 4D).
Table 3. Main results of the meta-analysis of the association between HLA-DQ rs9275572 polymorphism and HBV infection outcomes.
| Comparison | Subgroup | Allele model (A vs. G) |
Heterozygous model (AG vs. GG) |
Homozygous model (AA vs. GG) |
Recessive model (AA vs. AG+GG) |
Dominant model (AA+AG vs. GG) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | ||
| HBV infection vs. Control | Overall | 0.68 (0.62-0.74) | 0.11 | <0.01 | 0.73 (0.65-0.82) | 0.34 | <0.01 | 0.45 (0.37-0.56) | 0.23 | <0.01 | 0.51 (0.42-0.62) | 0.41 | <0.01 | 0.66 (0.59-0.74) | 0.21 | <0.01 |
| Asian | 0.64 (0.54-0.71) | 0.34 | <0.01 | 0.68 (0.59-0.78) | 0.89 | <0.01 | 0.40 (0.31-0.52) | 0.35 | <0.01 | 0.47 (0.37-0.60) | 0.46 | <0.01 | 0.62 (0.54-0.71) | 0.55 | <0.01 | |
| Caucasian | 0.78 (0.66-0.91) | -* | <0.01 | 0.89 (0.71-1.13) | -* | 0.33 | 0.56 (0.40-0.78) | -* | <0.01 | 0.59 (0.43-0.81) | -* | <0.01 | 0.80 (0.64-0.99) | -* | 0.04 | |
| HBV infection vs. NC | Overall | 0.70 (0.62-0.79) | 0.61 | <0.01 | 0.65 (0.56-0.76) | 0.30 | <0.01 | 0.57 (0.44-0.76) | 0.99 | <0.01 | 0.67 (0.52-0.87) | 0.99 | <0.01 | 0.63 (0.55-0.73) | 0.47 | <0.01 |
| Asian | 0.66 (0.57-0.76) | 0.91 | <0.01 | 0.59 (0.49-0.71) | 0.92 | <0.01 | 0.57 (0.39-0.82) | 0.99 | <0.01 | 0.70 (0.49-1.00) | 0.99 | 0.05 | 0.59 (0.49-0.70) | 0.94 | <0.01 | |
| Caucasian | 0.78 (0.64-0.94) | -* | 0.01 | 0.82 (0.61-1.09) | -* | 0.17 | 0.58 (0.38-0.88) | -* | 0.01 | 0.65 (0.44-0.95) | -* | 0.03 | 0.76 (0.58-0.99) | -* | 0.05 | |
| LC vs. CHB | Chinese | 1.34 (1.14-1.56) | 0.99 | <0.01 | 0.97 (0.80-1.18) | 0.82 | 0.76 | 1.22 (0.83-1.80) | 0.82 | 0.31 | 1.25 (0.85-1.82) | 0.73 | 0.25 | 1.00 (0.83-1.21) | 0.96 | 0.97 |
| HCC vs. LC+CHB | Chinese | 0.71 (0.63-0.80) | 0.79 | <0.01 | 073 (0.64-0.84) | 0.55 | <0.01 | 0.49 (0.35-0.68) | 1.00 | <0.01 | 0.54 (0.39-0.76) | 0.99 | <0.01 | 0.69 (0.60-0.80) | 0.63 | <0.01 |
-*Because there was only one study with this genotype of rs9275572, the value could not be calculated.
Figure 4. Forest plots for HLA-DQ rs9275572 polymorphism and HBV infection outcomes.
A. HBV infection vs. Control (AA vs. AG+GG); B. HBV infection vs. NC (AA vs. AG+GG); C. LC vs. CHB (AA vs. AG+GG); D. HCC vs. LC+CHB (AA vs. AG+GG)
Association between HLA-DQ rs7453920 polymorphism and HBV infection outcome
In this meta-analysis, HLA-DQ rs7453920 polymorphism was confirmed to be significantly associated with HBV infection in the following genetic models (HBV infection vs. Control: A vs. G: OR = 0.72, 95%CI: 0.62-0.82, PZ < 0.01; AG vs. GG: OR = 0.76, 95%CI: 0.64-0.89, PZ < 0.01; AA+AG vs. GG: OR = 0.75, 95%CI: 0.63-0.89, PZ < 0.01, Figure 5A). In contrast, no significant correlation was identified between HLA-DQ rs7453920 polymorphism and HBV infection outcome in the Homozygous model (OR = 0.96, 95%CI: 0.82-1.12, PZ = 0.58) and Recessive model (OR = 0.99, 95%CI: 0.85-1.15, PZ = 0.90) (Table 4). Meanwhile, we confirmed that HLA-DQ rs7453920 polymorphism was associated with HBV clearance in total population (HBV infection vs. NC: allele: OR = 0.64, 95%CI: 0.40-1.02, PZ < 0.01; heterozygous: OR = 0.62, 95%CI: 0.45-0.85, PZ < 0.01; dominant: OR = 0.62, 95%CI: 0.45-0.85, PZ < 0.01, Figure 5B).
Figure 5. Forest plots for HLA-DQ rs7453920 polymorphism and HBV infection outcomes.
A. HBV infection vs. Control (AA vs. AG+GG); B. HBV infection vs. NC (AA vs. AG+GG).
Table 4. Main results of the meta-analysis of the association between HLA-DQ rs7453920 polymorphism and HBV infection outcomes.
| Comparison | Subgroup | Allele model (A vs. G) |
Heterozygous model (AG vs. GG) |
Homozygous model (AA vs. GG) |
Recessive model (AA vs. AG+GG) |
Dominant model (AA+AG vs. GG) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | ||
| HBV infection vs. Control | Overall | 0.72 (0.62-0.82) | <0.01 | <0.01 | 0.76 (0.64-0.89) | <0.01 | <0.01 | 0.96 (0.82-1.12) | 0.12 | 0.58 | 0.99 (0.85-1.15) | 0.22 | 0.90 | 0.75 (0.63-0.89) | <0.01 | <0.01 |
| Asian | 0.71 (0.59-0.85) | <0.01 | <0.01 | 0.75 (0.61-0.92) | <0.01 | <0.01 | 0.91 (0.75-1.11) | 0.13 | 0.34 | 0.91 (0.75-1.11) | 0.19 | 0.38 | 0.73 (0.59-0.91) | <0.01 | <0.01 | |
| Caucasian | 0.72 (0.60-0.87) | <0.01 | <0.01 | 0.77 (0.60-1.00) | 0.13 | 0.05 | 1.04 (0.81-1.35) | 0.19 | 0.75 | 1.11 (0.88-1.44) | 0.38 | 0.39 | 0.78 (0.59-1.04) | 0.04 | 0.09 | |
| HBV infection vs. NC | Overall | 0.64 (0.40-1.02) | <0.01 | 0.06 | 0.62 (0.47-0.81) | <0.01 | <0.01 | 0.69 (0.35-1.37) | <0.01 | 0.29 | 0.79 (0.43-1.47) | <0.01 | 0.46 | 0.62 (0.45-0.85) | <0.01 | <0.01 |
| Asian | 0.55 (0.46-0.65) | 0.42 | <0.01 | 0.54 (0.40-0.73) | 0.11 | <0.01 | 0.51 (0.20-1.30) | 0.16 | 0.16 | 0.59 (0.24-1.46) | 0.17 | 0.26 | 0.54 (0.42-0.68) | 0.22 | <0.01 | |
| Caucasian | 0.76 (0.33-1.73) | <0.01 | 0.51 | 0.70 (0.45-1.08) | 0.01 | 0.10 | 0.83 (0.33-2.08) | <0.01 | 0.70 | 0.96 (0.44-2.07) | 0.01 | 0.92 | 0.72 (0.42-1.24) | <0.01 | 0.24 | |
Meta-analysis for HLA-DQ rs9275319 polymorphism with HBV infection outcomes
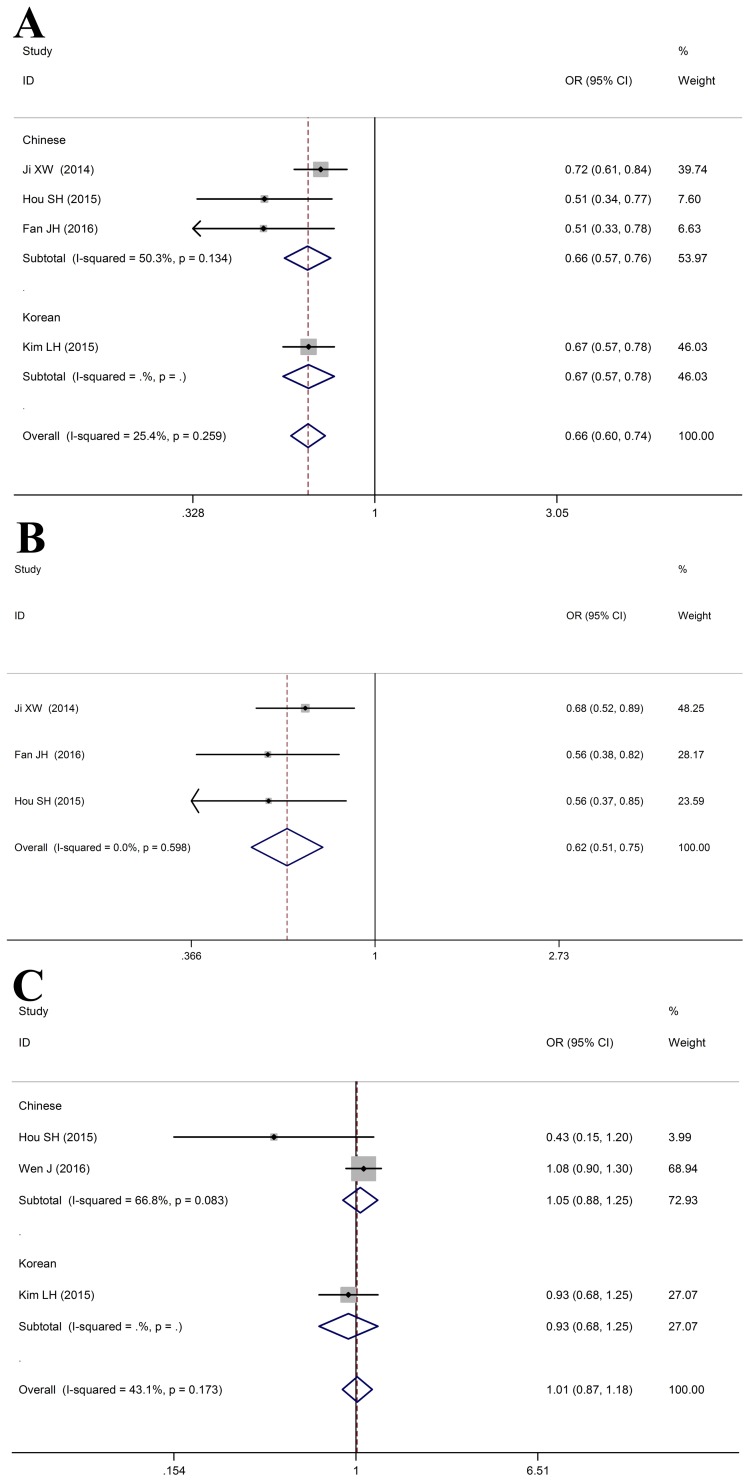
As shown in Table 5, Logistic regression analysis revealed a significant correlation between HLA-DQ rs9275319 polymorphism and a reduced risk of HBV infection in the HBV infection group (HBV infection vs. Control: allele: OR = 0.68, 95%CI: 0.62-0.74, PZ < 0.01; heterozygous: OR = 0.69, 95%CI: 0.62-0.77, PZ < 0.01; homozygous: OR = 0.50, 95%CI: 0.38-0.65, PZ < 0.01; recessive: OR = 0.55, 95%CI: 0.42-0.72, PZ < 0.01; dominant: OR = 0.66, 95%CI: 0.60-0.74, PZ < 0.01), as compared to healthy controls (Figure 6A). Meanwhile, HLA-DQ rs9275319 polymorphism was significantly associated with HBV clearance (HBV infection vs. NC: G vs. A: OR = 0.6, 95%CI: 0.54-0.76, PZ < 0.01; AG vs. AA: OR = 0.63, 95%CI: 0.51-0.77, PZ < 0.01; GG vs. AA: OR = 0.52, 95%CI: 0.28-0.95, PZ = 0.03; AG+GG vs. AA: OR = 0.62, 95%CI: 0.51-0.75, PZ < 0.01) (Figure 6B). However, no association was observed between HLA-DQ rs9275319 polymorphism and HBV-related HCC (HCC vs. LC+CHB: allele: OR = 0.99, 95%CI: 0.86-1.14, PZ = 0.92; heterozygous: OR = 1.04, 95%CI: 0.44-1.21, PZ = 0.67; homozygous: OR = 0.81, 95%CI: 0.50-1.32, PZ = 0.40; recessive: OR = 0.82, 95%CI: 0.51-1.32, PZ = 0.88; dominant: OR = 1.01, 95%CI: 0.87-1.18, PZ = 0.87, Figure 6C).
Table 5. Main results of the meta-analysis of the association between HLA-DQ rs9275319 polymorphism and HBV infection outcomes.
| Comparison | Subgroup | Allele model (G vs. A) |
Heterozygous model (AG vs. AA) |
Homozygous model (GG vs. AA) |
Recessive model (GG vs. AG+AA) |
Dominant model (AG+GG vs. AA) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | ||
| HBV infection vs. Control | Overall | 0.68 (0.62-0.74) | 0.12 | <0.01 | 0.69 (0.62-0.77) | 0.35 | <0.01 | 0.50 (0.38-0.65) | 0.16 | <0.01 | 0.55 (0.42-0.72) | 0.18 | <0.01 | 0.66 (0.60-0.74) | 0.26 | <0.01 |
| Chinese | 0.69 (0.61-0.79) | 0.06 | <0.01 | 0.66 (0.57-0.77) | 0.27 | <0.01 | 0.66 (0.42-1.03) | 0.20 | 0.07 | 0.71 (0.46-1.12) | 0.23 | 0.140 | 0.66 (0.57-0.76) | 0.13 | <0.01 | |
| Korean | 0.66 (0.8-0.76) | -* | <0.01 | 0.73 (0.62-0.86) | -* | <0.01 | 0.44 (0.31-0.62) | -* | <0.01 | 0.49 (0.35-0.68) | -* | <0.01 | 0.67 (0.57-0.78) | -* | <0.01 | |
| HBV infection vs. NC | Chinese | 0.64 (0.54-0.76) | 0.39 | <0.01 | 0.63 (0.51-0.77) | 0.78 | <0.01 | 0.52 (0.28-0.95) | 0.45 | 0.03 | 0.57 (0.37-1.04) | 0.45 | 0.07 | 0.62 (0.51-0.75) | 0.60 | <0.01 |
| HCC vs. LC+CHB | Overall | 0.99 (0.86-1.14) | 0.19 | 0.92 | 1.04 (0.44-1.21) | 0.18 | 0.67 | 0.81 (0.50-1.32) | 0.90 | 0.40 | 0.82 (0.51-1.32) | 0.88 | 0.41 | 1.01 (0.87-1.18) | 0.17 | 0.87 |
| Chinese | 1.02 (0.86-1.20) | 0.09 | 0.84 | 1.07 (0.89-1.29) | 0.09 | 0.45 | 0.73 (0.37-1.43) | 0.90 | 0.36 | 0.72 (0.37-1.42) | 0.95 | 0.35 | 1.05 (0.88-1.25) | 0.08 | 0.62 | |
| Korean | 0.94 (0.72-1.21) | -* | 0.61 | 0.93 (0.67-1.28) | -* | 0.65 | 0.91 (0.46-1.82) | -* | 0.80 | 0.93 (0.47-1.85) | -* | 0.84 | 0.93 (0.68-1.25) | -* | 0.62 | |
-*Because there was only one study with this genotype of rs9275319, the value could not be calculated.
Figure 6. Forest plots for HLA-DQ rs9275319 polymorphism and HBV infection outcomes.
A. HBV infection vs. Control (AA vs. AG+GG); B. HBV infection vs. NC (AA vs. AG+GG); C. HCC vs. LC+CHB (AA vs. AG+GG).
Meta-analysis for HLA-DQ rs9272105 polymorphism with HBV infection outcomes
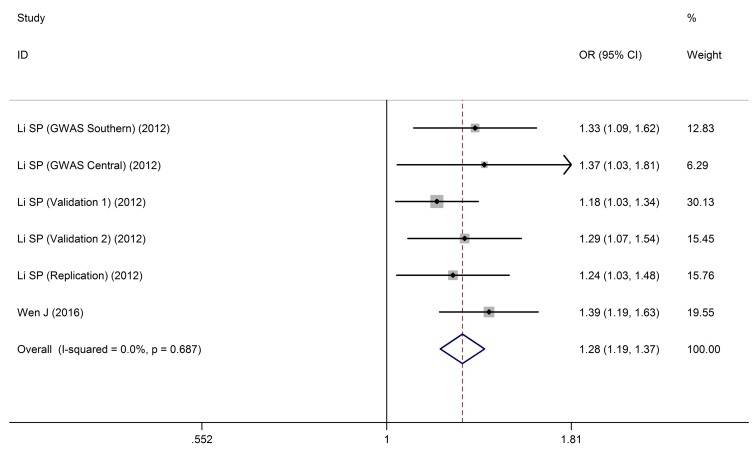
As shown in Table 6, HLA-DQ rs9272105 polymorphism was significantly associated with HBV-related HCC in all gene models (HCC vs. (LC+CHB): allele: OR = 1.31, 95%CI: 1.25-1.37, PZ < 0.01; heterozygous: OR = 1.11, 95%CI: 1.03-1.20, PZ < 0.01; homozygous: OR = 1.70, 95%CI: 1.56-1.86, PZ < 0.01; recessive: OR = 1.59, 95%CI: 1.48-1.72, PZ < 0.01; dominant: OR = 1.28, 95%CI: 1.19-1.37, PZ < 0.01, Figure 7).
Table 6. Main results of the meta-analysis of the association between HLA-DQ rs9272105 polymorphism and HBV infection outcomes.
| Comparison | Allele model (A vs. G) |
Heterozygous model (AG vs. GG) |
Homozygous model (AA vs. GG) |
Recessive model (AA vs. AG+GG) |
Dominant model (AA+AG vs. GG) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | OR (95%CI) | PH | PZ | |
| HCC vs. CHB | 1.31 (1.25-1.37) | 0.54 | <0.01 | 1.11 (1.03-1.20) | 0.55 | 0.01 | 1.70 (1.56-1.86) | 0.49 | <0.01 | 1.59 (1.48-1.72) | 0.20 | <0.01 | 1.28 (1.19-1.37) | 0.69 | <0.01 |
Figure 7. Forest plots for HLA-DQ rs9272105 polymorphism and HBV infection outcomes.
HCC vs. LC+CHB (AA vs. AG+GG).
Evaluation of sensitivity analysis
Sensitivity analysis was performed to assess the effects of an individual study on the overall OR. Meanwhile, the corresponding pooled ORs were not materially altered (Supplementary Figures 1-5).
Publication bias
Egger’s test was utilized to evaluate the publication bias of the included articles. The data showed that no evidence of publication bias was observed in HLA-DQ region polymorphisms (Supplementary Figures 6-10).
DISCUSSION
Epidemiologic studies have firmly confirmed that HBV infection plays a pivotal role in the chronic liver disease. To date, immune response has been considered to implicated in HBV clearance and HBV infection [46]. HLAs are members of the major histocompatibility complex (MHC) genes localized on chromosome 6p21, which play important roles in viral infectious diseases [47-49]. In a previous study, Zeng revealed that the proliferative responses of CD4+ T cells in patients with acute HBV infection were more severe than those with persistent HBV infection, suggesting MHC class II polymorphisms may affect the susceptibility of subjects to persistent infection [5]. Nowadays, three isotypes of HLA class II molecules have been identified including HLA-DR, HLA-DP, and HLA-DQ, which constitute a heterodimer formed by alpha- and beta-chains on the surface of antigen presenting cells (APCs) including macrophages, dendritic cells (DCs), and B lymphocytes [14]. HLA-DQ proteins, a group of heterodimeric molecules consisted of alpha- and beta-chains encoded by HLA-DQA1 and HLA-DQB1 genes [31], were implicated in immune-mediated diseases, including liver diseases and cancer [50, 51]. For example, several SNPs were considered to be associated with persistent HBV infection including HLA-DQA1*0302 [52], -DQB1*0301 [53], and -DQA1*0501 [54].
It is known to all, host genetic factors maybe closely involved in determination of the HBV infection outcome. HLA-DQ gene variations, such as HLA-DQ rs7453920, rs2856718, rs927210, rs9275319 and rs9275572, have been regarded to involve in HBV infection or clearance, as well as the disease progression and the development of hepatitis B associated complications (e.g. LC and HCC) [55]. In line with the previous study in Chinese population [28], HLA-DQ rs7453920 and rs2856718 SNPs haplotypes showed protective effects in a Japanese population-based study [24]. Hu et al found that HLA-DQ rs7453920 and rs2856718 were correlated with increased HBV clearance and decrease of HCC incidence in Han Chinese [28]. Zhang et al study demonstrated that HLA-DQ rs9275572A and rs2856718G polymorphism were significantly associated with decrease of HBV infection risk and HBV natural clearance. Additionally, rs9275572A was also related to the development of cirrhosis and HCC [32]. Interestingly, Al-Qahtani et al results showed that three SNPs (i.e. rs2856718, rs7453920, and rs9275572) of the HLA-DQ region contributed to the susceptibility to HBV infection in the Saudi Arabian population [31]. HLA-DQ rs9275319 was considered as an HBV-HCC susceptible SNP in a GWAS based on the Chinese populations [46], which was different from a previous study [33] in which rs9275319 variant genotypes were reported to be inversely related to HBV persistence and significantly related to HBV natural clearance [33]. Meanwhile, Li et al revealed that the rs9272105 variant allele was a risk factor for the HCC progression (OR = 1.30) [29]. To date, despite the fact that a large number of publications have focused on the association between HLA-DQ region polymorphisms and HBV infection outcomes, the results are still controversial. In this study, we conducted a meta-analysis to evaluate the relationship between the HLA-DQ region polymorphisms and HBV infection outcomes.
Compared to a single study, meta-analysis can provide sufficient results especially in analyzing unexplained studies [56]. In our previous meta-analysis including 11 case-control studies, we demonstrated that HLA-DQ rs2856718-G polymorphism showed protective effects against HBV infection, and rs2856718-A was a risk factor for chronic HBV infection [57]. Subsequently, Meta-analysis by Lv et al showed that rs2856718 and rs9275572 in HLA-DQ significantly decreased HBV-related HCC in total population, especially in Chinese other than in Saudi Arabian [58]. Whereas, in the analysis stratified by SNPs, only three SNPs (rs2856718, rs7453920, and rs9275572) for HBV infection and/or HBV-related HCC were included, with no study focusing on the rs9272105 and rs9275319. To our best knowledge, this is the first systematic and comprehensive meta-analysis exploring the associations between HLA-DQ region polymorphisms (rs2856718, rs7453920, rs9272105, rs9275319 and rs9275572) and HBV infection outcomes (including HBV infection, CHB, liver cirrhosis, and HBV-related HCC).
Indeed, there are some inherent limitations in this meta-analysis. Firstly, our results were obtained from unadjusted estimates due to lacking of raw data including age, gender, drinking, smoking, lifestyle, as well as environmental factors, which may lead to a confounding bias. Secondly, the number of studies was not large enough for a comprehensive meta-analysis. Thirdly, the gene-gene and of gene-environment interaction has not been evaluated in this study due to absence of original datasets. Finally, a lacking of the original data hampered our further evaluation on the potential interactions between clinical outcomes and viral backgrounds. Therefore, in future, further studies are needed to obtain more reliable results.
In summary, there are really variations between human populations. On this basis, a common SNP allele in a certain geographical or ethnic group may not be commonly observed in another geographical location or population. Our meta-analysis revealed that the HLA-DQ rs2856718-G and rs9275319-G were significantly associated with decreased risk of HBV infection and HBV natural clearance, but rs7453920-A was inconsistent in different populations. Because of the small sample size in Saudi Arabian population in this analysis, our findings need to be validated in future through a population-based study. Logistic regression analysis indicated that HLA-DQ allele rs9275572-A contributed to the significant increase of HBV infection clearance, and decreased HBV natural clearance. However, HLA-DQ alleles rs2856718-G and rs9275572-A were not associated with development of cirrhosis. The HLA-DQ (rs2856718 and rs9275572) polymorphisms were associated with a decreased HBV-related HCC risk in all genetic models, but HLA-DQ rs9272105 increased the risk of HBV-related HCC, which suggested that CHB patients with HLA-DQ rs9272105 should be monitored frequently for development of HCC. In addition, no association was observed between HLA-DQ rs9275319 polymorphism and HBV-related HCC. These findings contribute to the construction of a personalized hepatitis B therapy or prognosis in the near future.
MATERIALS AND METHODS
Literature search strategy
Literature search was performed from PubMed, EMBASE, China National Knowledge Infrastructure (CNKI) and Chinese WanFang databases, using the following keywords: “HLA-DQ”, “hepatitis B virus” or “HBV”, “HBV clearance” or “HBV natural clearance” or “NC”, “chronic hepatitis B” or “CHB”, “liver cirrhosis” or “LC” or “cirrhosis”, “Hepatocellular carcinoma” or “HCC” or “liver cancer”, “polymorphism” or “Single Nucleotide Polymorphism” or “SNP”, and “rs2856718” or “rs7453920” or “rs9272105” or “rs9275319” or “rs9275572”. Only the literatures published before June 21, 2017 were included, and were reviewed by two independent investigators (Tao Xu and Anyou Zhu). The search focused only on full articles for the meta-analysis. No language restriction was applied in the search process.
Inclusion and exclusion criteria
Eligible studies should meet the inclusion criteria as follows: (1) case-control studies; (2) studies with sufficient data for the estimation of an odds ratio (OR) with 95% confidence interval (CI); (3) studies reporting the genotype frequencies; (4) in cases of the same group of patients reported in multiple studies, only the most informative study was used to avoid duplication. The exclusion criteria were as follows: (1) duplicate data; (2) review articles; (3) case-only studies; (4) lacking of genotype frequency data; (5) with no full text available.
Quality assessment
Newcastle-Ottawa Scale (NOS) was applied to assess the quality of each included study [27]. The quality of studies was scored based on the following criteria: selection of cases, comparability of populations, and ascertainment of exposure to risks. Studies with a score of ≥ 6 were considered to be of high quality. In cases of any disagreement on the assigned grade, studies were fully reassessed until a consensus was achieved.
Data extraction
For the data extraction, the following data were independently extracted from the eligible studies: first author, publication date, ethnicity, genotyping method, cases stratified as HBV-related HCC, LC, and/or CHB; controls including the healthy controls and HBV clearance controls, total numbers of cases and controls. Two investigators (Tao Xu and Anyou Zhu) checked the data extraction results, and an open discussion or consultation was held in the presence of any disagreements.
Statistical analysis
SNP data were divided into four groups: HBV infection vs. healthy controls; HBV infection vs. NC; LC vs. CHB; HCC vs. (CHB and/or LC). The significance for five genetic models (allele model, heterozygous model, homozygous model, recessive model, and dominant model) was evaluated for each study, respectively. Statistical analysis was performed using STATA software (version 12.0; Stata Corporation, College Station, Texas, USA). Hardy-Weinberg equilibrium test (HWE) was evaluated for controls in each study by using the χ2-test, and P <0.05 was considered as departure from HWE. All the associations were estimated by ORs and 95% CIs. The significance of the pooled ORs was determined by Z-test and P <0.05 was considered statistically significant. Potential heterogeneity was evaluated using a χ2-based Q-test. PH ≥ 0.05 indicated a lack of heterogeneity among studies, and a fixed-effect model was used. Otherwise, a random-effects model was applied. Sensitivity analysis was performed by omitting each study in turn to determine the effects on the test of heterogeneity. Publication bias of literatures was assessed by Begg’s funnel plot.
SUPPLEMENTARY MATERIALS FIGURES
ACKNOWLEDGMENTS AND FUNDING
This work was supported by Anhui Provincial Natural Science Research Project of University (KJ2013A188, KJ2014A164 and KJ2016A472) and International Science and Technology Cooperation Project-Key Research and Development Program of Anhui Province (1604b0602026), and National Natural Science Foundation of China (81570011).
Footnotes
CONFLICTS OF INTEREST
The authors declare no competing financial interests.
REFERENCES
- 1.Gerlich WH. Medical virology of hepatitis B: how it began and where we are now. Virol J. 2013;10:239. doi: 10.1186/1743-422X-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liaw YF, Chu CM. Hepatitis B virus infection. Lancet. 2009;373:582–592. doi: 10.1016/S0140-6736(09)60207-5. [DOI] [PubMed] [Google Scholar]
- 3.Matsuura K, Isogawa M, Tanaka Y. Host genetic variants influencing the clinical course of hepatitis B virus infection. J Med Virol. 2016;88:371–379. doi: 10.1002/jmv.24350. [DOI] [PubMed] [Google Scholar]
- 4.Yin J, Xie J, Liu S, Zhang H, Han L, Lu W, Shen Q, Xu G, Dong H, Shen J, Zhang J, Han J, Wang L, et al. Association between the various mutations in viral core promoter region to different stages of hepatitis B, ranging of asymptomatic carrier state to hepatocellular carcinoma. Am J Gastroenterol. 2011;106:81–92. doi: 10.1038/ajg.2010.399. [DOI] [PubMed] [Google Scholar]
- 5.Zeng Z. Human genes involved in hepatitis B virus infection. World J Gastroenterol. 2014;20:7696–7706. doi: 10.3748/wjg.v20.i24.7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu Y, Wu Z, Peng Q, Ma L, Zhang X, Zhao J, Qin X, Li S. Role of IL-4 gene polymorphisms in HBV-related hepatocellular carcinoma in a Chinese population. PloS one. 2014;9:e110061. doi: 10.1371/journal.pone.0110061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang L, Liu C, Deng Y, Liu Y, Zhao J, Huang X, Tang W, Sun Y, Qin X, Li S. Association of Hypoxia-Inducible Factor-2 Alpha Gene Polymorphisms with the Risk of Hepatitis B Virus-Related Liver Disease in Guangxi Chinese: A Case-Control Study. PLoS One. 2016;11:e0158241. doi: 10.1371/journal.pone.0158241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim DW, Lee SA, Hwang ES, Kook YH, Kim BJ. Naturally occurring precore/core region mutations of hepatitis B virus genotype C related to hepatocellular carcinoma. PloS one. 2012;7:e47372. doi: 10.1371/journal.pone.0047372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thursz M. Genetic susceptibility in infectious diseases. Biotechnol Genet Eng Rev. 2000;17:253–264. [PubMed] [Google Scholar]
- 10.Yim HJ, Lok AS. Natural history of chronic hepatitis B virus infection: what we knew in 1981 and what we know in 2005. Hepatology. 2006;43:S173–S181. doi: 10.1002/hep.20956. [DOI] [PubMed] [Google Scholar]
- 11.Jiang X, Ma Y, Cui W, Li MD. Association of variants in HLA-DP on chromosome 6 with chronic hepatitis B virus infection and related phenotypes. Amino Acids. 2014;46:1819–1826. doi: 10.1007/s00726-014-1767-2. [DOI] [PubMed] [Google Scholar]
- 12.Ochi Y, Hashimoto S, Kawabe N, Murao M, Nakano T. HLA-DQ gene polymorphisms are associated with hepatocellular carcinoma and hepatitis B surface antigen in chronic hepatitis B virus infection. Hepatol Res. 2016 doi: 10.1111/hepr.12812. [DOI] [PubMed] [Google Scholar]
- 13.McCormack VA, Boffetta P. Today’s lifestyles, tomorrow’s cancers: trends in lifestyle risk factors for cancer in low- and middle-income countries. Ann. Oncol. 2011;22:2349–2357. doi: 10.1093/annonc/mdq763. [DOI] [PubMed] [Google Scholar]
- 14.Matsuura K, Isogawa M, Tanaka Y. Host genetic variants influencing the clinical course of hepatitis B virus infection. J Med Virol. 2016;88:371–379. doi: 10.1002/jmv.24350. [DOI] [PubMed] [Google Scholar]
- 15.Lin TM, Chen CJ, Wu MM, Yang CS, Chen JS, Lin CC, Kwang TY, Hsu ST, Lin SY, Hsu LC. Hepatitis B virus markers in Chinese twins. Anticancer Res. 1989;9:737–741. [PubMed] [Google Scholar]
- 16.Thursz MR, Kwiatkowski D, Allsopp CE, Greenwood BM, Thomas HC, Hill AV. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med. 1995;332:1065–1069. doi: 10.1056/NEJM199504203321604. [DOI] [PubMed] [Google Scholar]
- 17.Frodsham AJ. Host genetics and the outcome of hepatitis B viral infection. Transpl Immunol. 2005;14:183–186. doi: 10.1016/j.trim.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Tunçbilek S. Relationship between cytokine gene polymorphisms and chronic hepatitis B virus infection. World J Gastroenterol. 2014;20:6226–6235. doi: 10.3748/wjg.v20.i20.6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Y, Wu Z, Peng Q, Ma L, Zhang X, Zhao J, Qin X, Li S. Role of IL-4 gene polymorphisms in HBV-related hepatocellular carcinoma in a Chinese population. PLoS One. 2014;9:e110061. doi: 10.1371/journal.pone.0110061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azar SS, Mansoori M, Attar M, Shahbazi M. Tumor Necrosis Factor Alpha 308 G/A Single Nucleotide Polymorphism and Susceptibility to Hepatocellular Carcinoma Via Hepatitis B Infection. Asian Pac J Cancer Prev. 2016;17:3381338–4. [PubMed] [Google Scholar]
- 21.Huang X, Li H, Wang J, Huang C, Lu Y, Qin X, Li S. Genetic polymorphisms in Toll-like receptor 3 gene are associated with the risk of hepatitis B virus-related liver diseases in a Chinese population. Gene. 2015;569:218–224. doi: 10.1016/j.gene.2015.05.054. [DOI] [PubMed] [Google Scholar]
- 22.Ratnasari N, Nurdjanah S, Sadewa AH, Hakimi M. The role of vascular endothelial growth factor -634 G/C and its soluble receptor on chronic liver disease and hepatocellular carcinoma. Arab J Gastroenterol. 2016;17:61–66. doi: 10.1016/j.ajg.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Huang R, Hao Y, Fan Y, Yang C, Wu K, Cao S, Wu C. Association between cytotoxic T-lymphocyte-associated antigen 4 +49A/G polymorphism and persistent hepatitis B virus infection in the Asian population: evidence from the current studies. Genet Test Mol Biomarkers. 2013;17:601–606. doi: 10.1089/gtmb.2013.0069. [DOI] [PubMed] [Google Scholar]
- 24.Mbarek H, Ochi H, Urabe Y, Kumar V, Kubo M, Hosono N, Takahashi A, Kamatani Y, Miki D, Abe H, Tsunoda T, Kamatani N, Chayama K, et al. A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Genet. 2011;20:3884–3892. doi: 10.1093/hmg/ddr301. [DOI] [PubMed] [Google Scholar]
- 25.Hu Z, Liu Y, Zhai X, Dai J, Jin G, Wang L, Zhu L, Yang Y, Liu J, Chu M, Wen J, Xie K, Du G, et al. New loci associated with chronic hepatitis B virus infection in Han Chinese. Nat Genet. 2013;45:1499–1503. doi: 10.1038/ng.2809. [DOI] [PubMed] [Google Scholar]
- 26.Tan W, Xia J, Dan Y, Li M, Lin S, Pan X, Wang H, Tang Y, Liu N, Tan S, Liu M, He W, Zhang W, et al. Genome-wide association study identifies HLA-DR variants conferring risk of HBV-related acute-on-chronic liver failure. Gut. 2017 doi: 10.1136/gutjnl-2016-313035. [DOI] [PubMed] [Google Scholar]
- 27.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 28.Hu L, Zhai X, Liu J, Chu M, Pan S, Jiang J, Zhang Y, Wang H, Chen J, Shen H, Hu Z. Genetic variants in human leukocyte antigen/DP-DQ influence both hepatitis B virus clearance and hepatocellular carcinoma development. Hepatology. 2012;55:1426–1431. doi: 10.1002/hep.24799. [DOI] [PubMed] [Google Scholar]
- 29.Li S, Qian J, Yang Y, Zhao W, Dai J, Bei JX, Foo JN, McLaren PJ, Li Z, Yang J, Shen F, Liu L, Yang J, et al. GWAS identifies novel susceptibility loci on 6p21.32 and 21q21.3 for hepatocellular carcinoma in chronic hepatitis B virus carriers. PLoS Genet. 2012;8:e1002791. doi: 10.1371/journal.pgen.1002791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen K, Shi W, Xin Z, Wang H, Zhu X, Wu X, Li Z, Li H, Liu Y. Replication of genome wide association studies on hepatocellular carcinoma susceptibility loci in a Chinese population. PLoS One. 2013;8:e77315. doi: 10.1371/journal.pone.0077315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Qahtani AA, Al-Anazi MR, Abdo AA, Sanai FM, Al-Hamoudi W, Alswat KA, Al-Ashgar HI, Khalaf NZ, Eldali AM, Viswan NA, Al-Ahdal MN. Association between HLA variations and chronic hepatitis B virus infection in Saudi Arabian patients. PLoS One. 2014;9:e80445. doi: 10.1371/journal.pone.0080445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Jia J, Dong J, Yu F, Ma N, Li M, Liu X, Liu W, Li T, Liu D. HLA-DQ polymorphisms with HBV infection: different outcomes upon infection and prognosis to lamivudine therapy. J Viral Hepat. 2014;21:491–498. doi: 10.1111/jvh.12159. [DOI] [PubMed] [Google Scholar]
- 33.Ji X, Zhang Q, Li B, Du Y, Yin J, Liu W, Zhang H, Cao G. Impacts of human leukocyte antigen DQ genetic polymorphisms and their interactions with hepatitis B virus mutations on the risks of viral persistence, liver cirrhosis, and hepatocellular carcinoma. Infect Genet Evol. 2014;28:201–209. doi: 10.1016/j.meegid.2014.09.032. [DOI] [PubMed] [Google Scholar]
- 34.Liao Y, Cai B, Li Y, Chen J, Tao C, Huang H, Wang L. Association of HLA-DP/DQ and STAT4 polymorphisms with HBV infection outcomes and a mini meta-analysis. PLoS One. 2014;9:e111677. doi: 10.1371/journal.pone.0111677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou SH, Hu J, Zhang Y, Li QL, Guo JJ. Effects of interaction between genetic variants in human leukocyte antigen DQ and granulysin genes in Chinese Han subjects infected with hepatitis B virus. Microbiol Immunol. 2015;59:209–218. doi: 10.1111/1348-0421.12239. [DOI] [PubMed] [Google Scholar]
- 36.Hou SH, Li QL, Hu J, Peng H, Guo JJ. Association of HLA-DQ gene polymorphisms with the outcomes of hepatitis B virus infection. Basic & Clinical Medicine. 2015;35:873–878. (Chinese) [Google Scholar]
- 37.Kim LH, Cheong HS, Namgoong S, Kim JO, Kim JH, Park BL, Cho SW, Park NH, Cheong JY, Koh I, Shin HD, Kim YJ. Replication of genome wide association studies on hepatocellular carcinoma susceptibility loci of STAT4 and HLA-DQ in a Korean population. Infect Genet Evol. 2015;33:72–76. doi: 10.1016/j.meegid.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 38.Wen J, Song C, Jiang D, Jin T, Dai J, Zhu L, An J, Liu Y, Ma S, Qin N, Liang C, Chen J, Jiang Y, et al. Hepatitis B virus genotype, mutations, human leukocyte antigen polymorphisms and their interactions in hepatocellular carcinoma: a multi-centre case-control study. Sci Rep. 2015;5:16489. doi: 10.1038/srep16489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao Y, Cai B, Li Y, Chen J, Ying B, Tao C, Zhao M, Ba Z, Zhang Z, Wang L. Association of HLA-DP/DQ, STAT4 and IL-28B variants with HBV viral clearance in Tibetans and Uygurs in China. Liver Int. 2015;35:886–896. doi: 10.1111/liv.12643. [DOI] [PubMed] [Google Scholar]
- 40.Jiang DK, Ma XP, Wu X, Peng L, Yin J, Dan Y, Huang HX, Ding DL, Zhang LY, Shi Z, Zhang P, Yu H, Sun J, et al. Genetic variations in STAT4, C2, HLA-DRB1 and HLA-DQ associated with risk of hepatitis B virus-related liver cirrhosis. Sci Rep. 2015;5:16278. doi: 10.1038/srep16278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu WX, Zhang XL, Gao X, Yang L, Li BX, Liu DW. Association between HLA-DQ gene polymorphisms and different outcomes of hepatitis B virus infection. Chinese Journal of Epidemiology. 2016;37:384–388. doi: 10.3760/cma.j.issn.0254-6450.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 42.Fan JH, Hou SH, Qing-Ling L, Hu J, Peng H, Guo JJ. Association of HLA-DQ and IFNL4 polymorphisms with susceptibility to hepatitis B virus infection and clearance. Ann Hepatol. 2016;15:532–539. [PubMed] [Google Scholar]
- 43.Gao X, Liu W, Zhang X, Tang L, Wang L, Yan L, Yang H, Li T, Yang L, Ma N, Liu D. Genetic polymorphism of HLA-DQ confers susceptibility to hepatitis B virus-related hepatocellular carcinoma: a case-control study in Han population in China. Tumour Biol. 2016;37:12103–12111. doi: 10.1007/s13277-016-5077-z. [DOI] [PubMed] [Google Scholar]
- 44.Trinks J, Nishida N, Hulaniuk ML, Caputo M, Tsuchiura T, Marciano S, Haddad L, Blejer J, Bartoli S, Ameigeiras B, Frías SE, Vistarini C, Heinrich F, et al. Role of HLA-DP and HLA-DQ on the clearance of hepatitis B virus and the risk of chronic infection in a multiethnic population. Liver Int. 2017 doi: 10.1111/liv.13405. [DOI] [PubMed] [Google Scholar]
- 45.Pereira VRZB, Wolf JM, Stumm GZ, Boeira TR, Galvan J, Simon D, Lunge VR. Lack of association between human leukocyte antigen polymorphisms rs9277535 and rs7453920 and chronic hepatitis B in a Brazilian population. Genet Mol Res. 2017;16 doi: 10.4238/gmr16029565. [DOI] [PubMed] [Google Scholar]
- 46.Chisari FV. Rous-Whipple award lecture. Viruses, immunity, and cancer: Lessons from hepatitis B. Am J Pathol. 2000;156:1117–1132. doi: 10.1016/s0002-9440(10)64980-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang DK, Sun J, Cao G, Liu Y, Lin D, Gao YZ, Ren WH, Long XD, Zhang H, Ma XP, Wang Z, Jiang W, Chen TY, et al. Genetic variants in stat4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet. 2013;45:72–105. doi: 10.1038/ng.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishida N, Sawai H, Matsuura K, Sugiyama M, Ahn SH, Park JY, Hige S, Kang JH, Suzuki K, Kurosaki M, Asahina Y, Mochida S, Watanabe M, et al. Genome-wide association study confirming association of HLA-DQ with protection against chronic hepatitis B and viral clearance in Japanese and Korean. PLoS One. 2012;7:e39175. doi: 10.1371/journal.pone.0039175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blackwell JM, Jamieson SE, Burgner D. HLA and infectious diseases. Clin Microbiol Rev. 2009;22:370–385. doi: 10.1128/CMR.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Handunnetthi L, Ramagopalan SV, Ebers GC, Knight JC. Regulation of major histocompatibility complex class II gene expression, genetic variation and disease. Genes Immun. 2010;11:99–112. doi: 10.1038/gene.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi NM, Majumder P, Boss JM. Regulation of major histocompatibility complex class II genes. Curr Opin Immunol. 2011;23:81–87. doi: 10.1016/j.coi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu LP, Li XW, Liu Y, Sun GC, Chen ZH, Zhu XL, Hu QY, Li H. Association of haplotype formed on HLA-DRB1 and HLA-DQA1 alleles with outcomes of hepatitis B virus infection. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2006;23:427–430. [PubMed] [Google Scholar]
- 53.Thio CL, Carrington M, Marti D, O’Brien SJ, Vlahov D, Nelson KE, Astemborski J, Thomas DL. Class II HLA alleles and hepatitis B virus persistence in African Americans. J Infect Dis. 1999;179:1004–1006. doi: 10.1086/314684. [DOI] [PubMed] [Google Scholar]
- 54.Jiang YG, Wang YM, Liu TH, Liu J. Association between HLA class II gene and susceptibility or resistance to chronic hepatitis B. World J Gastroenterol. 2003;9:2221–2225. doi: 10.3748/wjg.v9.i10.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Zou ZQ, Wang K. Clinical Relevance of HLA Gene Variants in HBV Infection. J Immunol Res. 2016;2016:9069375. doi: 10.1155/2016/9069375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munafo MR, Clark TG, Flint J. Assessing publication bias in genetic association studies: evidence from a recent metaanalysis. Psychiatry Res. 2004;129:39–44. doi: 10.1016/j.psychres.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 57.Xu T, Sun M, Wang H. Relationship between HLA-DQ Gene Polymorphism and Hepatitis B Virus Infection. Biomed Res Int. 2017;2017:9679843. doi: 10.1155/2017/9679843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lv J, Xu T, Qian Z, Wang H. Association between HLA-DQ gene polymorphisms and HBV-related hepatocellular carcinoma. Gastroenterology Research and Practice. 2017;2017:7150386. doi: 10.1155/2017/7150386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.