ABSTRACT
Molecular mechanisms underlying the negative health effects of shift work are poorly understood, which remains a barrier to developing intervention strategies to protect the long-term health of shift workers. We evaluated genome-wide differences in DNA methylation (measured in blood) between 111 actively employed female nightshift and 86 actively employed female dayshift workers from the Seattle metropolitan area. We also explored the effect of chronotype (i.e., measure of preference for activity earlier or later in the day) on DNA methylation among 110 of the female nightshift workers and an additional group of 131 male nightshift workers. Methylation data were generated using the Illumina Infinium HumanMethylation450 BeadChip (450K) Array. After applying the latest methylation data processing methods, we compared methylation levels at 361,210 CpG loci between the groups using linear regression models adjusted for potential confounders and applied the false-discovery rate (FDR) ≤ 0.05 to account for multiple comparisons. No statistically significant associations at the genome-wide level were observed with shift work or chronotype, though based on raw P values and absolute effect sizes, there were suggestive associations in genes that have been previously linked with cancer (e.g., BACH2, JRK, RPS6KA2) and type-2 diabetes (e.g., KCNQ1). Given that our study was underpowered to detect moderate effects, examining these suggestive results in well-powered independent studies or in pooled data sets may improve our understanding of the pathways underlying the negative health effects of shift work and the influence of personal factors such as chronotype. Such an approach may help identify potential interventions that can be used to protect the long-term health of shift workers.
KEYWORDS: Shift work, chronotype, DNA methylation
Introduction
Shift work is a feature of our 24-hour economy, with 18% of workers in the developed world engaged in it fulltime.1 This high prevalence, coupled with growing evidence for its impact on chronic disease, including cardiovascular disease, diabetes and cancer,2–5 makes shift work a pressing public health concern. An understanding of the mechanisms behind shift work's relationship with chronic disease could aid strategies to mitigate its negative health effects; however, the molecular mechanisms underlying the negative health effects of shift work are poorly understood. Several promising mechanisms have been proposed.6,7 Among these, changes in DNA methylation are compelling for 3 reasons: 1) Differential DNA methylation associates with transcriptional programming;8,9 2) Changes in DNA methylation are inducible by exogenous factors;10 and 3) Changes in DNA methylation have been observed in the early stages of development of various chronic diseases.11–13
In a previous study, we evaluated DNA methylation, as measured in blood, across ∼473,800 CpG loci among 65 dayshift and 59 nightshift workers.6 Significant differences, at the genome-wide level, between dayshift and nightshift workers were observed at 16,135 loci. We now expand on that study by comparing DNA methylation between 86 dayshift and 111 nightshift workers from another previous study, originally conducted to examine the effects of nightshift work on circulating melatonin and reproductive hormones in women,14 while applying the latest methylation data processing methods, which have advanced rapidly since our earlier study.
Chronotype (i.e., preference for engaging in activity earlier or later in the day) may impact a person's ability to adapt to shift work schedules.15,16 While evening-type individuals are thought to be less disrupted by shift work, and some studies have observed decreased risks of cancer among evening-type compared with morning-type shift workers,17,18 other studies have observed increased risks of cancer among evening-type compared with morning-type shift workers.19,20 To further our understanding of the effects of chronotype in the context of shift work, we also explore the impact of a measure of chronotype on DNA methylation among 110 of the female nightshift workers and an additional 131 male nightshift workers from a previous study of nightshift work in association with circulating melatonin and cortisol levels in men.21
Results
Shift work and DNA methylation
For the 197 female subjects included in the analysis of shift work and DNA methylation (86 dayshift, 111 nightshift), the distribution of selected variables is presented by shift status in Table 1. Nightshift workers were slightly younger and had a slightly higher body mass index (BMI) than dayshift workers. The proportion of nightshift workers who are non-white were also greater than in dayshift workers. On average, dayshift workers were engaged in their current shift schedule longer than nightshift workers. While the fraction of smokers was the same for night and dayshift workers, alcoholic beverage consumption was greater among dayshift workers. The mean proportions of cell types in blood for dayshift and nightshift workers, inferred from the DNA methylation data, are also provided in Table 1. No major differences in cell-type proportions were apparent between dayshift and nightshift workers.
Table 1.
Distribution of select factors, by shift status, among female participants included in the analysis of shift work in association with genome-wide DNA methylation as measured in blood.
| Dayshiftn (%) | Nightshiftn (%) | |
|---|---|---|
| Agea,b | ||
| 22–29 | 19 (22) | 36 (33) |
| 30–36 | 20 (23) | 27 (24) |
| 37–42 | 28 (33) | 27 (24) |
| 43–48 | 19 (22) | 21 (19) |
| Body Mass Index [(lbs/in2)×703]a,c | ||
| 17.9–21.5 | 27 (31) | 23 (21) |
| >21.5–23.4 | 22 (26) | 27 (24) |
| >23.4–25.5 | 17 (20) | 32 (29) |
| >25.5–29.4 | 20 (23) | 29 (26) |
| Race | ||
| White | 72 (84) | 81 (73) |
| Other | 14 (16) | 30 (27) |
| Smoking | ||
| No | 80 (93) | 104 (94) |
| Yes | 6 ( 7 ) | 7 ( 6 ) |
| Alcohol | ||
| No | 58 (67) | 103 (93) |
| Yes | 28 (33) | 8 ( 7 ) |
| Months engaged in current shift schedule (months)d | ||
| 6-<12 | 13 (15) | 21 (19) |
| 12-<24 | 26 (30) | 33 (30) |
| 24-<54 | 21 (25) | 34 (31) |
| ≥ 54 | 26 (30) | 23 (20) |
| Proportion of blood cell types (means) | ||
| B-cells | 0.04 | 0.05 |
| CD8 T-cells | 0.10 | 0.10 |
| CD4 T-cells | 0.13 | 0.14 |
| Natural Killer cells | 0.04 | 0.04 |
| Granulocytes | 0.62 | 0.65 |
| Monocytes | 0.10 | 0.11 |
Age and BMI categories based on quartiles among all 197 participants included in shift work analysis.
Mean (Standard Deviation) for Age: Dayshift = 35.9 (7.0); Nightshift = 34.4 (7.4).
Mean (Standard Deviation) for Body Mass Index: Dayshift = 23.0 (2.6) ; Nightshift = 23.8 (2.7).
Mean (Standard Deviation) for Months engaged in current shift schedule: Dayshift = 46.4 (47.9); Nightshift = 36.8 (36.1).
M-values were calculated as measures of methylation for each CpG dinucleotide represented on the 450K Array. The correlation coefficients between M-values of the replicate samples were ≥0.95, indicating suitable precision of the assay. After applying our bioinformatics processing pipeline (see Materials and Methods) 361,210 CpG loci remained for analysis. In linear regression models adjusted for age, BMI, race, alcohol consumption, smoking, and leukocyte cell mixture, and after accounting for multiple comparisons using the false-discovery rate (FDR), shift work was not statistically significantly associated with methylation differences at any of the loci that we analyzed (q-value >0.99) (Supplementary Table 1). The top 5 associations based on P values and top 5 associations based on differences in M-values are highlighted in a volcano plot of the overall results (Fig. 1).
Figure 1.
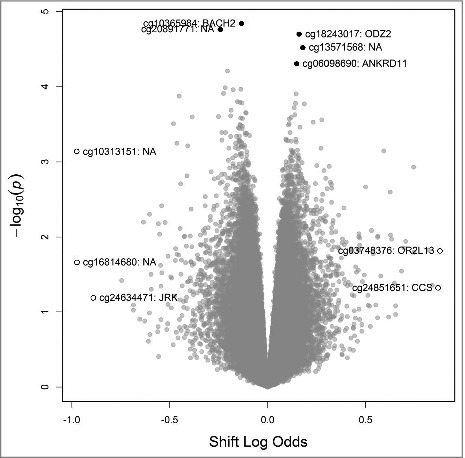
Volcano plot of results from genome-wide analysis of DNA methylation and shift work status, highlighting the 5 loci with the smallest unadjusted P values (•) and the 5 loci with largest absolute differences in effect size (○).
Chronotype and DNA methylation among nightshift workers
One female nightshift worker was missing data with which to calculate chronoscore, leaving a total of 241 nightshift workers (110 female and 131 male) for inclusion in our analysis. Based on our previous study of chronotype and its impacts on melatonin suppression in the same population, we considered 2 chronoscore categorization schemes.22 In the first scheme, chronoscore was dichotomized at the midpoint of the scale (evening-types, 13–33; morning-types, 34–55). In the second scheme, evening-types are those with scores of 28 or less, intermediate-types are those with scores from 29–39 and morning-types are those with scores of 40 or higher. When examining the dichotomized variable, 51% of the nightshift workers were of evening chronotypes, and this distribution was similar when considering men and women separately (results not shown). The distributions of selected variables are presented by the dichotomized chronoscore variable in Table 2. Those with lower chronoscores tended to be younger but were similar to those with higher chronoscores across all other factors. After accounting for multiple comparisons, only one locus had a q-value <0.99 (Supplementary Table 2). cg24873410 in the body of the Ribosomal Protein S6 Kinase A2 (RPS6KA2) gene, which encodes a protein kinase, was hypermethylated among those shift workers with lower chronoscores (i.e., eveningness) compared with those with higher chronoscores (q-value = 0.21). The top 5 associations based on raw P values and top 5 associations based on differences in M-values are highlighted in a volcano plot of the overall results (Fig. 2). When using the 3 category chronoscore variable, such that individuals with scores of 28 or less were compared with those with scores of 40 or higher (individuals with scores of 29 to 39 were excluded), we did not detect any significant associations. The q-value for the CpG locus in RPS6KA2 did change from 0.21 to 0.99, though the effect estimate was only marginally altered (results not shown).
Table 2.
Distribution of select factors, by chronoscore, among nightshift workers included in the analysis of chronoscore in association with genome-wide DNA methylation as measured in blood
| Chronoscorea |
||
|---|---|---|
| Higher | Lower | |
| n (%) | n (%) | |
| Gender | ||
| Female | 53 (45) | 57 (47) |
| Male | 66 (55) | 65 (53) |
| Ageb,c | ||
| 22–28 | 31 (26) | 46 (38) |
| 29–34 | 24 (20) | 23 (19) |
| 35–41 | 27 (23) | 30 (24) |
| 40–55 | 37 (31) | 23 (19) |
| Body Mass Index [(lbs/in2)×703]b,d | ||
| 17.9–22.6 | 31 (26) | 30 (25) |
| >22.6–24.9 | 26 (22) | 34 (28) |
| >24.9–27.4 | 30 (25) | 30 (24) |
| >27.4–31.2 | 32 (27) | 28 (23) |
| Race | ||
| White | 83 (70) | 84 (70) |
| Other | 36 (30) | 37 (30) |
| Smoking | ||
| No | 115 (97) | 112 (92) |
| Yes | 4 ( 3 ) | 10 ( 8 ) |
| Alcohol | ||
| No | 106 (89) | 112 (92) |
| Yes | 13 (11) | 10 ( 8 ) |
| Months engaged in current shift schedule (months)e | ||
| 6-<12 | 20 (17) | 16 (13) |
| 12-<24 | 28 (23) | 36 (30) |
| 24-<54 | 28 (32) | 44 (36) |
| ≥ 54 | 33 (28) | 26 (21) |
| Proportion of blood cell types (means) | ||
| B-cells | 0.04 | 0.05 |
| CD8 T-cells | 0.09 | 0.09 |
| CD4 T-cells | 0.14 | 0.15 |
| Natural Killer cells | 0.06 | 0.04 |
| Granulocytes | 0.59 | 0.59 |
| Monocytes | 0.12 | 0.10 |
Score from the Composite Scale of Morningness was dichotomized at the midpoint of the scale; higher score (≥ 34) = mornigness; lower score (≤ 33) = eveningness
Age and BMI categories based on quartiles among all 241 participants included in chronoscore analysis.
Mean (Standard Deviation) for Age: Higher chronoscore = 36.1 (8.1); Lower chronoscore = 33.7 (8.4).
Mean (Standard Deviation) for Body Mass Index: Higher chronoscore = 25.0 (3.0); Lower chronoscore = 24.8 (2.8).
Mean (Standard Deviation) for Months engaged in current shift schedule: Higher chronoscore = 43.1 (43.7); Lower chronoscore = 40.1 (41.8).
Figure 2.
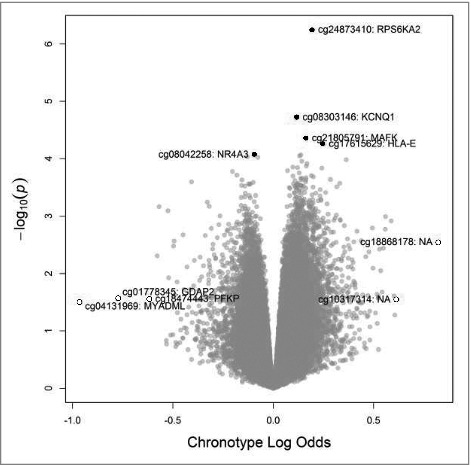
Volcano plot of results from genome-wide analysis of DNA methylation and chronoscore, highlighting the 5 loci with the smallest unadjusted P values (•) and the 5 loci with largest absolute differences in effect size (○).
Discussion
In the analysis of shift work and genome-wide DNA methylation, we observed no statistically significant associations after accounting for multiple comparisons. However, our exploratory results, which are provided in their entirety (Supplementary Table 1) may be worth examining in larger, independent studies or pooled analyses of multiple smaller independent data sets examining the impacts of shift work on DNA methylation. For example, when considering raw P values, the top association consisted of decreased methylation among nightshift workers at cg10365984, a locus in a CpG island at the 5’UTR of the BTB Domain and CNC Homolog 2 (BACH2) gene, which encodes a transcriptional regulator. Decreased methylation at the 5’UTR may suggest increased expression of this gene among nightshift workers.23 BACH2 expression has been previously associated with immunosuppression within tumors.24
As another example, when considering absolute effect sizes, one of the largest associations observed was for cg24634471, a locus in a CpG island near the transcription start site of the JRK gene. The locus was hypomethylated in nightshift compared with dayshift workers, possibly indicating increased expression of the Jrk Helix-Turn-Helix Protein (JRK) gene,23 which is a DNA binding protein. JRK has been observed as overexpressed in colorectal, breast and ovarian cancers.25
There were also no statistically significant associations observed in the chronoscore analysis (Supplementary Table 2). When considering raw P values, the most significant association was observed for a locus in the body of the RPS6KA2 (RSK) gene (cg24873410), which was hypermethylated among those with a later diurnal preference. Since CpG methylation of the gene body has been positively associated with expression,23 RPS6KA2 may be expressed at higher levels in individuals tending toward eveningness. RPS6KA2 is a downstream signaler of the mitogen-activated protein kinase pathway and was identified as a putative tumor suppressor gene for ovarian cancer.26 In a previous study from our group, there was suggestive evidence that the risk of ovarian cancer among evening-type shift workers was lower than it was among morning-type shift workers.18
Based on raw P values, another interesting association with chronoscore that may be worth exploring in future studies is at a CpG locus (cg08303146) in the first exon of the Potassium Voltage-Gated Channel Subfamily Q Member 1 (KCNQ1) gene, which encodes a voltage-gated potassium channel. The locus was hypermethylated among shift workers with a later diurnal preference, suggesting decreased expression of the gene among these individuals.23 SNPs in this gene have been associated with type-2 diabetes across multiple study populations.27 Recently, these polymorphisms were linked to differential DNA methylation in a study of type-2 diabetes, suggesting that differential methylation may be on the causal pathway linking the SNPs to an increased risk of type-2 diabetes.28 An analysis in the Nurses’ Health Cohort demonstrated that nurses working out of synch with their chronotype (e.g., morning-type nightshift workers, evening-type dayshift workers) were at significantly increased risk of developing type-2 diabetes.29
Results of the current study differ from those we observed in our previous study of DNA methylation.6 In that study of 65 dayshift and 59 nightshift workers, we observed 16,135 loci that were significantly differentially methylated between night and dayshift workers after FDR correction. In the few years since that study was published, the field has made rapid advances in understanding the limitations of array-based measurement of DNA methylation and in the pre-processing of Illumina 450K data. Unlike the previous study, our current study excluded 124,302 CpG sites due to quality control concerns and incorporated improved normalization techniques, which may partly explain differences in findings. Further, our current statistical analysis is based on applying the FDR to the results of individual linear regression models and is more conservative than the statistical analysis of microarrays (SAM) approach that we previously used.30 Differences in blood cell composition between the current and previous study may also account for differing results. In the previous study, DNA was extracted from isolated lymphocytes, leaving few granulocytes to contribute DNA (∼4%). In the current study, granulocytes, by far, were the largest contributor of DNA among dayshift and nightshift workers (>60%). Even though we adjusted for cell profile in both studies, this adjustment may not have been adequate to normalize such large differences in cell proportion between the 2 studies.
Our findings also did not replicate those from the previous study of methylation and shift work by Zhu et al. (2011). This may be attributable to differences in the study populations and/or differences in the methylation data processing. Zhu et al. (2011) evaluated female participants with a cumulative history of ≥10 y of nightshift work (did not have to be consecutive years and did not have to be currently engaged in shift work), whereas our participants were actively engaged in nightshift work for no less than 6 consecutive months. They found hypomethylation in the promoter region of CLOCK and hypermethylation in the promoter region of CRY,31 which we did not observe at the genome-wide level.
We performed sensitivity analyses to determine if the ability to detect statistically significant effects of nightshift work on DNA methylation would be improved if analyses excluded dayshift and nightshift workers that reported less than 12 months on their current shift schedule. Q-values remained unchanged (>0.99; results not shown).
Relative to the large number of loci that were examined, our sample size was small, limiting our power to detect moderate effects. Another limitation of our study is the lack of data on long-term cigarette smoking or alcohol consumption. Long-term cigarette smoking and alcohol consumption have been associated with differential DNA methylation,32,33 and differences in cigarette smoking and alcohol consumption have been reported between dayshift and nightshift workers.34,35 Instead, we had data on whether or not cigarettes had been used or alcohol had been consumed in the 24-hour periods over which urine specimens were collected, which may or may not have coincided with collection of the blood sample that was ultimately assayed for genome-wide DNA methylation.
While actigraphy-based sleep quality data were available for the majority of participants, we did not include sleep quality data in the analyses because sleep quality would most likely be a mediator of the effect of shift work on DNA methylation and carcinogenesis. Mediation analyses were beyond the scope of this manuscript but may be explored in the future to improve understanding of the specific aspects of shift work that may be impacting DNA methylation levels.
Exploring the impacts of shift work on DNA methylation in blood is a reasonable strategy since shiftwork has been associated with cancer across multiple tissues types and sites,36–40 including hematopoietic cancers. In addition, previous studies have shown that methylation in blood can serve as a systemic marker of methylation in other tissues.41,42
Our study did not identify any statistically significant differences in genome-wide DNA methylation by shift status or chronoscore. Because our study was underpowered to detect moderate effects, examination of some of the suggestive associations we observed in well-powered, independent studies or in pooling efforts including multiple independent data sets may help improve our understanding of mechanisms underlying the negative health effects of shift work. Insights afforded by such studies may inform the development of much needed interventions to protect the long-term health of shift workers.
Materials and methods
Study subjects
The study was approved by the Institutional Review Board of the Fred Hutchinson Cancer Research Center. Study subjects were drawn from participants of 2 previous studies of shift work conducted among healthcare workers from the Seattle metropolitan area: the Female Shift Worker Study (FSWS)14 and the Male Shift Worker Study (MSWS).21 Subjects in both studies were recruited through advertisements at local area hospitals, direct mailing to Washington State Department of Health licensed and certified health care workers, and referrals from eligible and ineligible participants. To be eligible for these studies, participants had to be actively engaged in dayshift or nightshift work at the time of recruitment and during data collection. In the FSWS (recruitment and data collection from November 2003 to August 2007) and MSWS (recruitment and data collection from October 2007 to May 2011), nightshift workers were required to work at least 20 h per week (at least 8 h per shift, stopping work no earlier than 06:00) and to sleep at night during days off. Dayshift workers were required to work at least 20 h per week and work exclusively during the dayshift (i.e., work at least 8 h per shift, beginning work no earlier than 06:00).
For the FSWS, participants were required to be women aged 20 to 49 y. Additional eligibility criteria included: regular menstrual periods; no personal history of breast cancer, chemotherapy, or tamoxifen therapy; no pregnancy or breast feeding within the past year; no use of supplements containing phytoestrogens or isoflavones; and consumption of no more than 5 servings per week of soy-based foods. For the MSWS, participants were required to be men aged 20 to 55 y. They could not be using medications or supplements to treat benign prostate conditions within 30 d of participation, could not have a personal history of prostate cancer or chemotherapy, and could not have undergone general anesthesia or major surgery at least 8 weeks before enrollment. For both the FSWS and MSWS, participants were required to have a BMI [weight in kilograms (kg) divided by the square of height in meters (m2)] between 18 and 30 kg/m2, and could not have been using hormones or supplements containing melatonin during the 30 d before enrollment.
Two-hundred and 88 participants from the FSWS (129 dayshift and 159 nightshift) and 208 participants from the MSWS (all nightshift) had available buffy coat samples for DNA extraction or previously extracted buffy coat DNA. Blood samples for DNA extraction were not collected from any dayshift workers in the MSWS. For the current study, we restricted to those participants who worked their respective shift schedules (day or night) for at least 6 consecutive months as we were interested in examining longer term impacts of shift work on DNA methylation. In total, 88 dayshift and 113 nightshift workers from the FSWS and 135 nightshift workers from the MSWS were selected.
Data and biospecimen collection
For eligible participants across the 2 studies, informed consent was obtained during either a home or work visit by trained study interviewers/phlebotomists. Structured interviews were conducted to collect information about current work and sleep schedules, work history, and physical activity. Height and weight measurements along with blood samples (10 mL EDTA vacutainer tubes) were also obtained during these interviews. For dayshift (nightshift) workers, data on caffeine, alcohol, tobacco, medication, and supplement intake were self-reported for the 24 h periods preceding the night (day) sleep period during which urine samples were collected to measure circulating melatonin levels (melatonin data not included in this study).
To assess chronotype, all participants completed the Composite Scale of Morningness, which is a self-reported questionnaire developed to assess chronotype among shift workers.43 The scale is used to assign a total chronoscore ranging from 13 to 55 points. Lower chronoscores are indicative of a later diurnal preference (i.e., evenigness) and higher scores are indicative of an earlier diurnal preference (i.e., morningness).43 Based on the student population in which this scale was originally developed, scores of 22 and less were classified as evening-types, 23 to 43 as intermediate-types and 44 and above as morning-types (14). However, as we previously pointed out in our analysis of the impact of chronotype on melatonin suppression among shift workers,22 these cut-points are arbitrary as they are based on the 10th and 90th percentiles of the distribution of chronoscores in the original student study population, and in our population with its much different age distribution, these cut-points lead to very few individuals being classified as morning and evening-types. Therefore, in our previous study of melatonin suppression,22 we evaluated alternative cut-points including simple dichotomization at the midpoint of the chronoscore scale (evening-types 33 or less and morning-types 34 or higher) and classification of individuals that are ±5 points from the exact midpoint of the scale as intermediate-types such that evening-types are those with scores of 28 or less, intermediate-types are those with scores from 29–39 and morning-types are those with scores of 40 or higher.
Specimen processing
Buffy coats were isolated from the whole blood samples and subsequently stored at −70°C. For the FSWS, buffy coat samples were stored from 7 to 11 y before DNA was extracted and used for the methylation assay. For the MSWS, buffy coat samples were stored up to 4 y before DNA extraction. The extracted DNA from the MSWS was subsequently stored for up to 8 years, at −20°C, before being used for the methylation assay. DNA extraction for both studies was performed on the buffy coats using a salt precipitation method.44 Extracted DNA (500 ng) for each participant was treated with sodium bisulfite using EZ DNA Methylation-DirectKit (Zymo Research, Irvine, CA). Treated DNA specimens were stored at −80°C, and the methylation assay was performed within 2 weeks of treatment.
Methylation assay
The Infinium HumanMethylation450 Bead Array (450K; Illumina, San Diego, CA) was used to quantify DNA methylation at 485,577 CpG loci. The array measures methylation across 21,154 genes, each with an average of 17 CpGs. CpGs span various gene regions, including those located 1500 bp and 200 bp upstream of transcription start sites (TSS1500 and TSS200 respectively) and those located within the 5' untranslated region, first exon, gene body, and 3' untranslated region. CpGs could be positioned within or near CpG islands.
The DNA methylation assay was performed as described previously.6 Briefly, 4 ml of bisulfite treated DNA was denatured and neutralized to prepare it for overnight isothermal whole-genome amplification. Next, the DNA was enzymatically fragmented for 60 min at 37°C and then precipitated with isopropanol and allowed to air dry. DNA was then resuspended in hybridization buffer. Samples were then applied to the methylation assay BeadChips, which were then incubated in a hybridization oven at 48°C for 16–24 h. After washing, the chip underwent extension and staining in capillary flow-through chambers. BeadChips were then scanned using the iScan+ (Illumina, San Diego, CA). Laboratory personnel were blinded to all study subject information, and specimens were identified by study ID number only. For 10% of subjects, a duplicate DNA sample was randomly included among the study samples (within and across batches) for quality control assurance.
Data processing
Based on a plot of log median probe intensities, we excluded 8 samples [4 samples from the FSWS (2 dayshift and 2 nightshift) and 4 samples from the MSWS] that clustered together with lower intensities in both the methylated and unmethylated channels compared with all other samples.45,46 These samples also had bisulfite conversion I and II control probe levels that were outliers compared with all other samples.
For the remaining samples, each CpG dinucleotide represented on the array was associated with an M-value, calculated as the log2 ratio of the intensities of the methylated and unmethylated probes. We used the “noob” method in the minfi Bioconductor package47 to resolve bias in the M-values associated with differences in background fluorescence.48 Functional normalization was used to rescale the M-values, which corrected any bias from differences in distributions of M-values from the 2 types of probes used in the array and removed technical variation while allowing for biologic differences in global methylation.49
We excluded non-CpG (i.e., CH) sites and CpG sites on the sex chromosomes. We also excluded CpG sites measured by poorly performing probes, which included CpG loci measured by probes with bead counts <3 in at least 10% of samples and CpG sites for which the detection P value was ≥0.01 in at least 10% of samples.47 Since the presence of SNPs at or near a CpG locus can affect probe hybridization,50 we excluded all CpG loci containing any SNP and those CpG loci within 10 base pairs of at least one SNP with a minor allele frequency >1% in any population based on 1000 Genomes data.51 We also excluded cross-reactive probes (i.e., probes that cross-hybridize to sites other than those for which they were designed).52,53 After implementing these processing procedures, 361,210 CpG loci remained for analysis.
Statistical analysis
The M-values for each CpG site were modeled as dependent variables using linear regression (R version 3.4.0). For the analysis of shift work and its association with DNA methylation, only data from the FSWS were included because there were no data for dayshift workers from the MSWS. All models included a variable for shift work status and adjustment variables for potential confounders: age (continuous), BMI (continuous), race (White or Non-White), alcohol consumption (yes or no), smoking (yes or no), and the mixture of leukocytes that contributed DNA to each participant's sample. Leukocyte cell mixture for 6 major leukocyte subsets (CD8+ T-cells, CD4+ T-cells, natural killer cells, B-cells, monocytes, and granulocytes) was inferred based on the method by Houseman et al. (2012).54 Variables indicating the proportion of these cell types (excluding monocytes to avoid collinearity) were included in all statistical models to adjust for cell type. Multiple testing was accounted for with FDR using the Benjamini-Hochberg (BH) method.55 Resulting q-values ≤0.05 were considered statistically significant.
For the analysis of chronotype association with DNA methylation, data for nightshift workers from both the FSWS and MSWS were included. Methods were similar to those described for the shift work analysis; however, instead of a variable for shift work, the model included a dichotomous variable for chronoscore. We also conducted an analysis comparing the highest to lowest categories of the 3 category chronotype variable described earlier. Along with variables for age, BMI, race, alcohol consumption, smoking, and cell mixture; a variable for gender was included in these models.
Disclosure of interests
The authors have no conflicts of interest to declare.
Funding details
The study was funded by grant R21 ES022863 from the US National Institute of Environmental Health Sciences, grants R01 CA097090 and R01 CA116400 from the US National Cancer Institute and an award from the Safeway Foundation. Charleen Adams is supported by Cancer Research UK (CRUK) [C18281/A19169] and works in the Medical Research Council Integrative Epidemiology Unit at the University of Bristol, which is supported by the Medical Research Council and the University of Bristol [MC_UU_12013/2].
References
- 1.Rajaratnam SM, Arendt J. Health in a 24-h society. Lancet. 2001;358:999-1005. doi: 10.1016/S0140-6736(01)06108-6. PMID:11583769 [DOI] [PubMed] [Google Scholar]
- 2.Lin X, Chen W, Wei F, Ying M, Wei W, Xie X. Night-shift work increases morbidity of breast cancer and all-cause mortality: a meta-analysis of 16 prospective cohort studies. Sleep Med. 2015;16:1381-7. doi: 10.1016/j.sleep.2015.02.543. PMID:26498240 [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Ji A, Zhu Y, Liang Z, Wu J, Li S, Meng S, Zheng X, Xie L. A meta-analysis including dose-response relationship between night shift work and the risk of colorectal cancer. Oncotarget. 2015;6:25046-60. doi: 10.18632/oncotarget.4502. PMID:26208480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jørgensen JT, Karlsen S, Stayner L, Andersen J, Andersen ZJ. Shift work and overall and cause-specific mortality in the Danish nurse cohort. Scand J Work Environ Health. 2017;43:117-26. doi: 10.5271/sjweh.3612. PMID:28245504 [DOI] [PubMed] [Google Scholar]
- 5.Vetter C, Devore EE, Wegrzyn LR, Massa J, Speizer FE, Kawachi I, Rosner B, Stampfer MJ, Schernhammer ES. Association Between Rotating Night Shift Work and Risk of Coronary Heart Disease Among Women. JAMA. 2016;315:1726-34. doi: 10.1001/jama.2016.4454. PMID:27115377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatti P, Zhang Y, Song X, Makar KW, Sather CL, Kelsey KT, Houseman EA, Wang P. Nightshift work and genome-wide DNA methylation. Chronobiol Int. 2015;32:103-12. doi: 10.3109/07420528.2014.956362. PMID:25187986 [DOI] [PubMed] [Google Scholar]
- 7.Fritschi L, Glass DC, Heyworth JS, Aronson K, Girschik J, Boyle T, Grundy A, Erren TC. Hypotheses for mechanisms linking shiftwork and cancer. Med Hypotheses. 2011;77:430-6. doi: 10.1016/j.mehy.2011.06.002. PMID:21723672 [DOI] [PubMed] [Google Scholar]
- 8.Bell JT, Pai A a, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y, Pritchard JK. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:1-13. doi: 10.1186/gb-2011-12-6-405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szyf M. DNA methylation signatures for breast cancer classification and prognosis. Genome Med. 2012;4:26. doi: 10.1186/gm325. PMID:22494847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore LE, Huang W-Y, Chung J, Hayes RB. Epidemiologic considerations to assess altered DNA methylation from environmental exposures in cancer. Ann N Y Acad Sci. 2003;983:181-96. doi: 10.1111/j.1749-6632.2003.tb05973.x. PMID:12724223 [DOI] [PubMed] [Google Scholar]
- 11.Dong Y, Zhao H, Li H, Li X, Yang S. DNA methylation as an early diagnostic marker of cancer (Review). Biomed Rep. 2014;2:326-30. PMID:24748968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baccarelli A, Rienstra M, Benjamin EJ. Cardiovascular Epigenetics: Basic Concepts and Results from Animal and Human Studies. Circ Cardiovasc Genet. 2010;3:567-73. doi: 10.1161/CIRCGENETICS.110.958744. PMID:21156932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillberg L, Ling C. The Potential Use of DNA Methylation Biomarkers to Identify Risk and Progression of Type 2 Diabetes. Front Endocrinol [Internet]. 2015;1–6 Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4378313/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis S, Mirick DK, Chen C, Stanczyk FZ. Night shift work and hormone levels in women. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2012;21:609-18. doi: 10.1158/1055-9965.EPI-11-1128 [DOI] [PubMed] [Google Scholar]
- 15.Erren TC. Shift work and cancer research: can chronotype predict susceptibility in night-shift and rotating-shift workers? Occup Env Med. 2013;70:283-4. doi: 10.1136/oemed-2012-100984 [DOI] [PubMed] [Google Scholar]
- 16.Saksvik IB, Bjorvatn B, Hetland H, Sandal GM, Pallesen S. Individual differences in tolerance to shift work - A systematic review. Sleep Med Rev. 2011;15:221-35. doi: 10.1016/j.smrv.2010.07.002. PMID:20851006 [DOI] [PubMed] [Google Scholar]
- 17.Hansen J, Lassen CF. Nested case-control study of night shift work and breast cancer risk among women in the Danish military. Occup Environ Med. 2012;69:551-6. doi: 10.1136/oemed-2011-100240. PMID:22645325 [DOI] [PubMed] [Google Scholar]
- 18.Bhatti P, Cushing-Haugen KL, Wicklund KG, a Doherty J, Rossing MA. Nightshift work and risk of ovarian cancer. Occup Env Med. 2013;70:231-7. doi: 10.1136/oemed-2012-101146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papantoniou K, Castaño-Vinyals G, Espinosa A, Aragonés N, Pérez-Gómez B, Ardanaz E, Altzibar JM, Sanchez VM, Gómez-Acebo I, Llorca J, et al.. Breast cancer risk and night shift work in a case-control study in a Spanish population. Eur J Epidemiol. 2016;9:867-78. doi: 10.1007/s10654-015-0073-y. PMID:26205167. [DOI] [PubMed] [Google Scholar]
- 20.Dickerman BA, Markt SC, Koskenvuo M, Hublin C, Pukkala E, Mucci LA, Kaprio J. Sleep disruption, chronotype, shift work, and prostate cancer risk and mortality: a 30-year prospective cohort study of Finnish twins. Cancer Causes Control CCC. 2016;27:1361-70. doi: 10.1007/s10552-016-0815-5. PMID:27734240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirick DK, Bhatti P, Chen C, Nordt F, Stanczyk FZ, Davis S. Night shift work and levels of 6-sulfatoxymelatonin and cortisol in men. Cancer Epidemiol Biomark Prev. 2013;22:1079-87. doi: 10.1158/1055-9965.EPI-12-1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhatti P, Mirick DK, Davis S. The impact of chronotype on melatonin levels among shift workers. Occup Environ Med. 2014;71:195-200. doi: 10.1136/oemed-2013-101730. PMID:24399070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484-92. doi: 10.1038/nrg3230. PMID:22641018 [DOI] [PubMed] [Google Scholar]
- 24.Roychoudhuri R, Eil RL, Clever D, Klebanoff CA, Sukumar M, Grant FM, Yu Z, Mehta G, Liu H, Jin P, et al.. The transcription factor BACH2 promotes tumor immunosuppression. J Clin Invest. 2016;126:599-604. doi: 10.1172/JCI82884. PMID:26731475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pangon L, Ng I, Giry-Laterriere M, Currey N, Morgan A, Benthani F, Tran PN, Al-Sohaily S, Segelov E, Parker BL, et al.. JRK is a positive regulator of β-catenin transcriptional activity commonly overexpressed in colon, breast and ovarian cancer. Oncogene. 2016;35:2834-41. doi: 10.1038/onc.2015.347. PMID:26455321 [DOI] [PubMed] [Google Scholar]
- 26.Bignone PA, Lee KY, Liu Y, Emilion G, Finch J, Soosay AER, Charnock FML, Beck S, Dunham I, Mungall AJ, et al.. RPS6KA2, a putative tumour suppressor gene at 6q27 in sporadic epithelial ovarian cancer. Oncogene. 2006;26:683-700. doi: 10.1038/sj.onc.1209827. PMID:16878154 [DOI] [PubMed] [Google Scholar]
- 27.Sun Q, Song K, Shen X, Cai Y. The Association between KCNQ1 Gene Polymorphism and Type 2 Diabetes Risk: A Meta-Analysis. PLOS ONE. 2012;7:e48578. doi: 10.1371/journal.pone.0048578. PMID:23133642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elliott HR, Shihab HA, Lockett GA, Holloway JW, McRae AF, Smith GD, Ring SM, Gaunt TR, Relton CL. The Role of DNA Methylation in Type 2 Diabetes Aetiology - Using Genotype as a Causal Anchor. Diabetes. 2017;. doi: 10.2337/db16-0874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vetter C, Devore EE, Ramin CA, Speizer FE, Willett WC, Schernhammer ES. Mismatch of Sleep and Work Timing and Risk of Type 2 Diabetes. Diabetes Care. 2015;38:1707-13. doi: 10.2337/dc15-0302. PMID:26109502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116-21. doi: 10.1073/pnas.091062498. PMID:11309499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobs DI, Hansen J, Fu A, Stevens RG, Tjonneland A, Vogel UB, Zheng T, Zhu Y. Methylation alterations at imprinted genes detected among long-term shiftworkers. Env Mol Mutagen. 2013;54:141-6. doi: 10.1002/em.21752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88:450-7. doi: 10.1016/j.ajhg.2011.03.003. PMID:21457905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou FC, Balaraman Y, Teng M, Liu Y, Singh R, Nephew KP. Alcohol alters DNA methylation patterns and inhibits neural stem cell differentiation. Alcohol Clin Exp Res. 2011;35:735-46. doi: 10.1111/j.1530-0277.2010.01391.x. PMID:21223309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Amelsvoort LGPM Jansen NWH, Kant I. Smoking among shift workers: More than a confounding factor. Chronobiol Int. 2006;23:1105-13. doi: 10.1080/07420520601089539. PMID:17190698 [DOI] [PubMed] [Google Scholar]
- 35.Dorrian J, Skinner N. Alcohol consumption patterns of shiftworkers compared with dayworkers. Chronobiol Int. 2012;29:610-8. doi: 10.3109/07420528.2012.675848. PMID:22621358 [DOI] [PubMed] [Google Scholar]
- 36.Rao D, Yu H, Bai Y, Zheng X, Xie L. Does night-shift work increase the risk of prostate cancer? A systematic review and meta-analysis. OncoTargets Ther. 2015;8:2817-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang F, Yeung KL, Chan WC, Kwok CC, Leung SL, Wu C, Chan EY, Yu IT, Yang XR, Tse LA. A meta-analysis on dose-response relationship between night shift work and the risk of colorectal cancer. Oncotarget. 2015;6:25046-60. doi: 10.18632/oncotarget.4502. PMID:26208480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin X, Chen W, Wei F, Ying M, Wei W, Xie X. Night-shift work increases morbidity of breast cancer and all-cause mortality: A meta-analysis of 16 prospective cohort studies. Sleep Med. 2015;16:1381-7. doi: 10.1016/j.sleep.2015.02.543. PMID:26498240 [DOI] [PubMed] [Google Scholar]
- 39.Ijaz S, Verbeek J, Seidler A, Lindbohm M-L, Ojajarvi A, Orsini N, Costa G, Neuvonen K. Night-shift work and breast cancer - a systematic review and meta-analysis. Scand J Work Env Health. 2013;39:431-47. doi: 10.5271/sjweh.3371 [DOI] [PubMed] [Google Scholar]
- 40.Jia Y, Lu Y, Wu K, Lin Q, Shen W, Zhu M, Huang S, Chen J. Does night work increase the risk of breast cancer? A systematic review and meta-analysis of epidemiological studies. Cancer Epidemiol. 2013;37:197-206. doi: 10.1016/j.canep.2013.01.005. PMID:23403128 [DOI] [PubMed] [Google Scholar]
- 41.Ma B, Wilker EH, Willis-Owen SA, Byun HM, Wong KC, Motta V, Baccarelli AA, Schwartz J, Cookson WO, Khabbaz K, et al.. Predicting DNA methylation level across human tissues. Nucleic Acids Res. 2014;42:3515-28. doi: 10.1093/nar/gkt1380. PMID:24445802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barault L, Ellsworth RE, Harris HR, Valente AL, Shriver CD, Michels KB. Leukocyte DNA as surrogate for the evaluation of imprinted Loci methylation in mammary tissue DNA. PLoS One. 2013;8:e55896. doi: 10.1371/journal.pone.0055896. PMID:23409079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith CS, Reilly C, Midkiff K. Evaluation of three circadian rhythm questionnaires with suggestions for an improved measure of morningness. J Appl Psychol 1989;74:728-38. doi: 10.1037/0021-9010.74.5.728. PMID:2793773 [DOI] [PubMed] [Google Scholar]
- 44.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988;16:1215. doi: 10.1093/nar/16.3.1215. PMID:3344216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fortin J-P, Hansen KD. Analysis of 450k data using minfi [Internet]. 2015. [cited 2017 Mar 14]; Available from: https://www.bioconductor.org/help/course-materials/2015/BioC2015/methylation450k.html#qc-plot.
- 46.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363-9. doi: 10.1093/bioinformatics/btu049. PMID:24478339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris TJ, Beck S. Analysis pipelines and packages for Infinium HumanMethylation450 BeadChip (450k) data. Methods. 2015;72:3-8. doi: 10.1016/j.ymeth.2014.08.011. PMID:25233806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Triche TJ, Weisenberger DJ, Van Den Berg D Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013;41:1-11. doi: 10.1093/nar/gkt090. PMID:23143271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilhelm-Benartzi CS, Koestler DC, Karagas MR, Flanagan JM, Christensen BC, Kelsey KT, Marsit CJ, a Houseman E, Brown R. Review of processing and analysis methods for DNA methylation array data. Br J Cancer. 2013;109:1394-402. doi: 10.1038/bjc.2013.496. PMID:23982603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morris N, Elston RC, Barnholtz-Sloan JS, Sun X, Comuzzie AG. Novel approaches to the analysis of family data in genetic epidemiology. Front Genet. 2015;6:27. doi: 10.3389/fgene.2015.00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butcher L. Illumina450ProbeVariants.db: Annotation Package combining variant data from 1000 Genomes Project for Illumina HumanMethylation450 Bead Chip probes. [Internet]. 2013. Available from: https://bioconductor.org/packages/release/data/experiment/html/Illumina450ProbeVariants.db.html. [Google Scholar]
- 52.Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203-9. doi: 10.4161/epi.23470. PMID:23314698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nordlund J, Bäcklin CL, Wahlberg P, Busche S, Berglund EC, Eloranta M-L, Flaegstad T, Forestier E, Frost B-M, Harila-Saari A, et al.. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013;14:r105. doi: 10.1186/gb-2013-14-9-r105. PMID:24063430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. PMID:22568884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289-300. [Google Scholar]


