Abstract
Subtelomeric regions have several unusual characteristics, including complex repetitive structures, increased rates of evolution, and enrichment for genes involved in niche adaptation. The adaptive telomere failure hypothesis suggests that certain environmental stresses can induce a low level of telomere failure, potentially leading to elevated subtelomeric recombination that could result in adaptive mutational changes within subtelomeric genes. Here, we tested a key prediction of the adaptive telomere failure hypothesis—that telomere dysfunction mild enough to have little or no overall effect on cell fitness could still lead to substantial increases in the mutation rates of subtelomeric genes. Our results show that a mutant of Kluyveromyces lactis with stably short telomeres produced a large increase in the frequency of mutations affecting the native subtelomeric β-galactosidase (LAC4) gene. All lac4 mutants examined from strains with severe telomere dysfunction underwent terminal deletion/duplication events consistent with being due to break-induced replication. In contrast, although cells with mild telomere dysfunction also exhibited similar terminal deletion and duplication events, up to 50% of lac4 mutants from this background unexpectedly contained base changes within the LAC4 coding region. This mutational bias for producing base changes demonstrates that mild telomere dysfunction can be well suited as a force for altering the adaptive potential of subtelomeric genes.
Keywords: Subtelomere, break-induced replication, mild telomere dysfunction, β-galactosidase, yeast
TELOMERES are the nucleoprotein complexes found at the ends of linear chromosome that differentiate naturally occurring chromosomal ends from double-stranded breaks (DSBs) (De Lange et al. 2006; Wellinger and Zakian 2012). Telomeric DNA, composed of tandem arrays of a short repeat sequence, serves as a binding site for telomere proteins required for telomere capping and length regulation (Cech 2004; Wellinger and Zakian 2012). These repeats are also targets for telomerase, an enzyme that adds new telomeric repeats onto chromosome ends by copying the template region of its RNA subunit, counteracting the end replication problem (Greider and Blackburn 1989; Podlevsky and Chen 2012).
Mutations that alter telomere-binding protein activities or shorten telomeric repeat arrays can lead to improper telomere function (Garvik et al. 1995; Booth et al. 2001; Craven et al. 2002; Chan and Blackburn 2003; Hackett and Greider 2003; Mieczkowski et al. 2003). In cells where telomere capping function has become compromised, chromosome ends can resemble DNA DSBs (De Lange et al. 2006). DSBs are a severe form of DNA damage and are typically repaired by one of two pathways, nonhomologous end joining (NHEJ) or homologous recombination (HR) (Aylon and Kupiec 2004; Krejci et al. 2012; Chiruvella et al. 2013). NHEJ is a ligase IV-dependent reaction that joins two broken ends together and is active throughout the cell cycle (Lieber 2010; Waters et al. 2014). HR, on the other hand, utilizes homologous sequence, usually supplied by a sister chromatid, but sometimes by nonallelic sources of homology, as a template for repair (Pâques and Haber 1999; Mehta and Haber 2014).
Inappropriate repair of dysfunctional telomeres using NHEJ or HR can have varying effects on genomic stability. Dysfunctional telomeres repaired using NHEJ can result in telomere fusions and dicentric chromosomes, whereas repair facilitated through HR restores telomere function by means of less deleterious copying mechanisms. Specifically, repair through break-induced replication (BIR), a homology-based repair pathway that acts when only one DNA end is available (Teng and Zakian 1999), can restore critically shortened telomeres by copying a longer telomere (McEachern and Haber 2006). Both because it requires only a single end and because it is less likely to generate detrimental outcomes, the BIR pathway is predicted to be the preferable repair mechanism to act at dysfunctional telomeres in most circumstances (Ricchetti et al. 2003; Miller et al. 2011).
The effects of telomere dysfunction on overall genome stability have been well characterized (Blasco et al. 1997; Kim et al. 2001; Blackburn et al. 2015; Maciejowski and de Lange 2017) and can be particularly dramatic in the subtelomeric region found immediately adjacent to the telomere. Subtelomeres in many eukaryotes are characterized by low gene density, complex repetitive structure, rapid evolution, and dynamic epigenetic regulation (Gottschling et al. 1990; Pryde et al. 1997; Mefford and Trask 2002; Linardopoulou et al. 2005; Rudd et al. 2007). Increased rates of ectopic HR, deletions, translocations, and other rearrangement events within subtelomeres contribute to its dynamic nature and likely facilitate rapid sequence evolution between closely related species.
Driven by these elevated rates of mutation, gene families within subtelomeric regions are often larger and more rapidly evolving than those found at more internal chromosomal locations (Brown et al. 2010). Importantly, subtelomeres are enriched for “contingency genes,” genes critical for adaptation to novel or stressful environments (Moxon et al. 1994; Barry et al. 2003). Medically relevant examples include virulence genes found in eukaryotic pathogens such as trypanosomes and Plasmodium falciparum (Horn 2004; Horn and Barry 2005; Glover et al. 2011). Other well-characterized examples in yeasts include genes important in carbohydrate utilization, flocculation, and arsenic resistance (Bobrowicz et al. 1997; Verstrepen and Klis 2006; Brown et al. 2010).
The highly dynamic nature of subtelomeres makes them unsuitable locations for essential genes, but also makes these regions ideal locations for genes that can tolerate or even benefit from complex mutations. Sequence changes affecting subtelomeric contingency genes, including duplications, deletions, tandem repeat expansions, and chimeric gene formation, have the potential to confer fitness advantages in novel environments. Although subtelomeres appear to have great potential for facilitating rapid adaptation, the cellular mechanism responsible for such events are unclear.
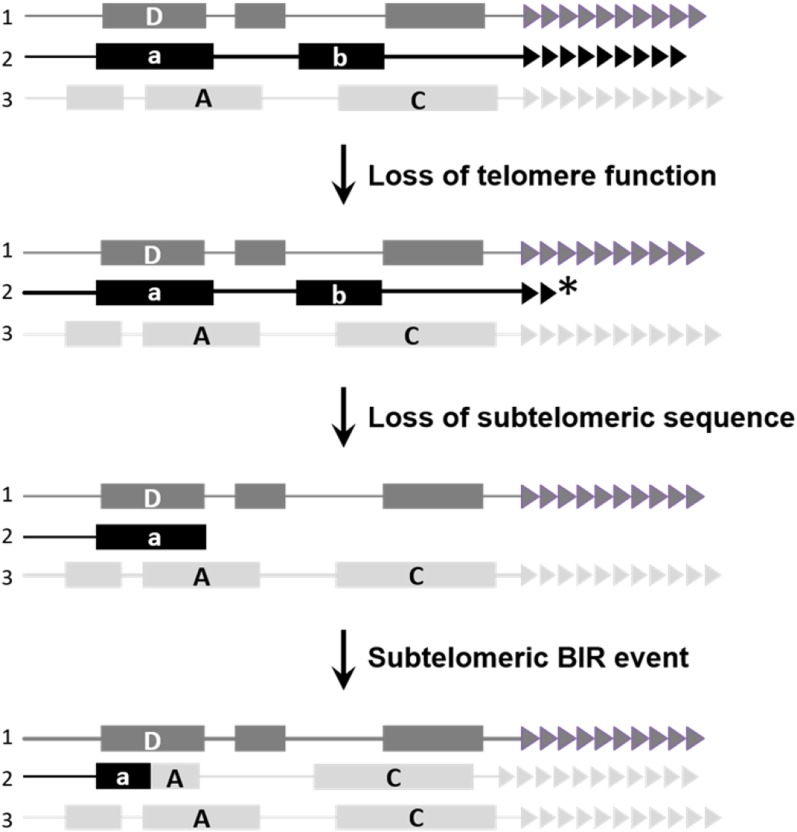
The adaptive telomere failure hypothesis proposed that telomeres can play a direct role in triggering subtelomeric evolution that can result in rapid adaptation to novel environments (McEachern 2008). This hypothesis suggests that mild telomere dysfunction, defined as telomere dysfunction that does not compromise cell growth nor cause an elevation of telomere–telomere fusions from NHEJ, can still lead to increased rates of HR at telomeres. Such events could potentially spread into the adjacent subtelomere, increasing incidences of subtelomeric BIR events (Figure 1). Mutational loss or duplication or modification of subtelomeric contingency genes could ultimately facilitate adaptation by generating novel mutants better able to survive environmental stress. This hypothesis suggests a previously unconsidered role for telomeres as an agent of adaptation through rapid evolution of subtelomeric regions and their associated genes.
Figure 1.
Adaptive telomere failure model. The adaptive telomere failure hypothesis suggests that limited telomere (triangles) failure can trigger BIR-events at subtelomeric homologous or homeologous sequence (a and A boxes), potentially leading to contingency gene loss (box b), duplication (box C), or chimeric gene product formation (box a/A). Asterisk indicates a dysfunctional telomere.
We have investigated the potential of mild telomere dysfunction to generate mutations in subtelomeric genes in the milk yeast Kluyveromyces lactis. Prior experiments have shown that both mild and severe telomere dysfunction can lead to highly elevated levels of recombination involving the immediately subtelomeric R element repeats of K. lactis (McEachern and Iyer 2001). In this work, we monitored a natural subtelomeric β-galactosidase gene (LAC4) for loss of function in the presence of different levels of telomere dysfunction. We found that although severe telomere dysfunction led to very high levels of BIR-like deletion/duplications, mild telomere dysfunction from constitutively short telomeres produced a near equal mix of BIR-like outcomes and LAC4 point mutations. Our results show that mild telomere dysfunction can trigger an unexpectedly diverse spectrum of mutations in subtelomeric genes, suggesting it is well suited to generate potentially adaptive mutations.
Materials and Methods
Yeast strains
All strains used are derivatives of the haploid K. lactis wild-type strain 7B520 (ura3-1 his2-2 trp1) (Wray et al. 1987). The ter1-∆ and ter1-28C(Taq) strains were all generated previously (McEachern and Blackburn 1995; Underwood et al. 2004; Wang et al. 2009) using a plasmid loop in, loop out procedure (McEachern and Blackburn 1995). Additionally the stn1-M1 and stn1-m1 ter1-∆ strains were made using standard yeast replacement procedures (Iyer et al. 2005).
White colony detection and quantification
The frequency at which white colonies arises from a blue parental colony was measured in all strain backgrounds. Independently derived isolates were examined from all strain backgrounds including 18 7B520, 27 ter1-∆, 61 ter1-Taq, 22 stn1-M1, and 18 stn1-m1 ter1-∆. Similarly sized colonies from each background were taken from standard rich medium (YPD) and were resuspended in water to a concentration of ∼1–2 × 104 cells/ml. A total of 25 μl of the cell suspension was spread on YPD plates containing 0.02% 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal). Two or three replicates were prepared simultaneously using the same cell suspension. Plates were grown at 30° for 3–4 days and scored for presence or absence of β-galactosidase activity as judged by colony color.
The frequency of white colony formation was calculated by dividing the total number of white colonies scored by total colonies counted. To calculate the median percentage of white colonies observed, the number of white colonies scored was averaged between replicates for each single isolate and the median was then calculated across these averages. White colony formation frequency in mutant lines were compared to the wild-type frequency using Pearson’s chi-squared test.
Growth on lactose was characterized in a subset of colonies, including 15 ter1-∆, 22 ter1-28C(Taq), 5 stn1-M1, and 7 stn1-m1 ter1-∆ white colonies, as well as a corresponding blue colony from the same plate. These isolates were grown on synthetically defined (SD) minimal media containing 2% lactose (purity ≥97%) with only uracil, histidine, and tryptophan supplemented into the medium.
Southern hybridizations
Genomic DNA from white and blue colonies was isolated from overnight cultures grown in liquid YPD at 30°. Digested genomic DNA was electrophoretically separated at 29 V for 15 hr on 1% agarose gels and transferred to Hybond N+ membranes. Presence or absence of sequence from the right arm of chromosome two (2R) was then assessed at intervals beginning at the telomere and extending inward toward the centromere. Oligonucleotides and primers used to generate DNA fragments for hybridization can be found in Supplemental Material, Table S1. All PCR products used for hybridizations were amplified from the wild-type strain 7B520.
Gene fragment probes were gel purified using the Qiagen Gel Extraction kit and were labeled using the Klenow fragment of DNA polymerase I. The telomere oligonucleotide probe was end labeled using the T4 polynucleotide kinase. Hybridizations were carried out overnight in 500 mM Na2HPO4 and 7% SDS at temperatures of 25° (oligo probe) and 60° (gene fragment probes). Membranes hybridized with the oligonucleotide probe were washed three times with 50 mM Na2HPO4 and 2% SDS, whereas membranes hybridized with gene fragment probes were washed with 100 mM Na2HPO4 and 2% SDS for 10 min.
LAC4-deficient mutant analysis
A 100-kb section of sequence from the end of the 2R chromosome was examined for homology shared with other subtelomeres using the Circoletto program (Darzentas 2010) set at the relaxed setting, with the E-value set to 0.1 for the BLAST run.
PCR primers were designed to amplify across the predicted chimeric regions generated in a repair event utilizing the 1R homology. Control reactions were carried out to amplify the proximal KLLA0B14751 gene through the native 2R region of homeology as well as the native 1R region of homology (Table S2). Additionally, primers were designed to amplify across the chimeric regions generated in a repair event utilizing the Z and X regions of mating locus. Control reactions amplified the nongenic region proximal to the cryptic mating locus on the 2R chromosome and the native Z and X regions flanking the left and right sides of the mating locus on 2R, respectively (Table S2). PCR products of the LAC4 gene and LAC4 promoter region from pale blue isolates from the ter1-28C(Taq) background were also examined (Table S2). Positive PCR products for predicted chimeric regions, as well as the LAC4 gene and LAC4 promoter region in pale blue ter1-28C(Taq) isolates, were sequenced by MacrogenUSA and analyzed using the genome analysis program Geneious.
Sequence analysis
The genomes from the K. marxianus strains DKMU3-1042 and K. lactis strain NRRL Y-1140 were downloaded from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/). Sequence visualization and analysis was carried out using CLC Workbench software (CLC Bio-Qiagen) and Geneious.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Strains are available upon request.
Results
K. lactis strains with telomere dysfunction have an increased frequency of loss of the subtelomerically encoded β-galactosidase activity
The adaptive telomere failure hypothesis proposes that telomeres in some organisms may have evolved to fail in response to certain environmental stresses (McEachern 2008). This could result in elevated recombination in subtelomeric regions and subsequent duplication, deletion, or recombination of the contingency genes enriched in these regions, potentially leading to increases in fitness (Figure 1). A prediction of the adaptive telomere failure hypothesis might be that even mild telomere dysfunction could be sufficient to produce elevated rates of recombination, affecting subtelomeric genes.
The study described here investigated sequence stability of the right arm of chromosome 2 (2R) in the dairy yeast K. lactis. The 2R subtelomere contains the naturally occurring β-galactosidase (LAC4) and lactose permease genes (LAC12) found 7 and 13 kb from the telomere, respectively (Figure 3A), both of which are required for lactose utilization (Kraepelin 1984). Hydrolysis of X-gal by β-galactosidase produces a visible blue color in K. lactis colonies, even in the presence of glucose (Dickson and Markin 1980), making the loss of blue color an effective screen for mutants with altered LAC4 function and a marker of subtelomeric sequence stability. To test the adaptive telomere failure hypothesis, subtelomeric LAC4 gene stability was examined in K. lactis mutants with either mild or severe telomere dysfunction, where all telomeres in the cell are subject to elevated levels of dysfunction.
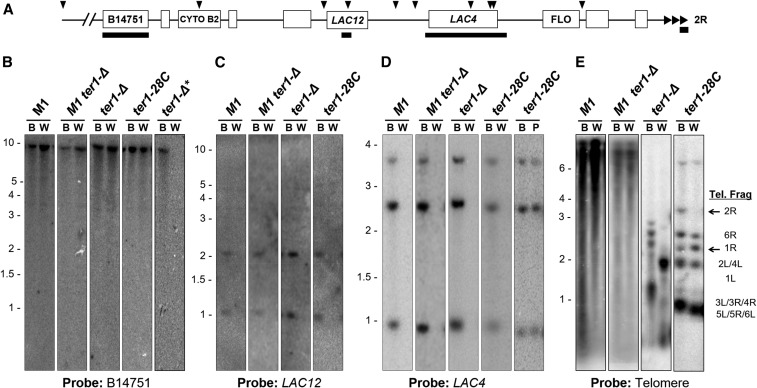
Figure 3.
Terminal deletions of the 2R chromosome end are present in most white colonies. (A) Map of the 2R chromosome end with telomere shown at right (triangles). Arrowheads indicate EcoRI sites, open boxes are open reading frames and solid bars are the probes used for hybridization. (B–E) Southern blots of EcoRI-digested genomic DNAs of white and blue isolates (B, W, and P indicates the blue, white, or pale colony color from which DNA was extracted) hybridized to probes, as indicated below the blots. * Shows a white ter1-Δ isolate [shown in (B) only] that is representative of isolates missing the KLLA0B14571 fragment. The identity of the telomere restriction fragments of wild-type and ter1-28C(Taq) are indicated to the right in (E). The 2R and 1R fragments are indicated by solid arrows. The faint band at ∼6 kb seen with the telomere probing of ter1-28C(Taq) is the ter1 gene.
White colony formation frequency was examined in K. lactis strains with severe or mild telomere dysfunction during growth on X-gal. For simplicity, we define severe telomere dysfunction as greatly perturbed telomere function accompanied by distinctly slowed cell growth and mild telomere dysfunction as the absence of either of those characteristics. Of the ∼58,000 colonies from the wild-type 7B520 background, all colonies examined on rich medium containing X-gal were blue in color, consistent with no detected loss of LAC4 (Figure 2 and Table 1). In contrast, strains with severely dysfunctional telomeres, including stn1-M1 and telomerase deletion mutants (ter1-Δ), exhibited significantly higher frequencies of white colonies (Figure 2 and Table 1). stn1-M1 and stn1-M1 ter1-Δ mutants produce chronic telomere dysfunction, leading to slower growing abnormal colonies and extremely long telomeres formed partly or entirely by recombination (Iyer et al. 2005; Xu and McEachern 2012). These mutants produced white colonies at the median frequencies of 1.3 and 2.4%, respectively (Table 1). The chronic disruption in the function of a telomere binding protein produced by the stn1-M1 mutation presumably underlies why that mutation is largely epistatic to the ter1-Δ mutation.
Figure 2.
White colony formation increases in strains with dysfunctional telomeres. Diluted cell suspensions of 7B520 and its derivatives, ter1-Δ, stn1-M1, stn1-M1 ter1-Δ, and ter1-28C(Taq) were spread on solid YPD medium containing 0.02% X-gal and scored for color. Shown are representative dilution plates with examples of white colonies in each strain, except 7B520, in which a white colony was never observed. Each panel shows an area on a respective plate that is 52 mm × 52 mm.
Table 1. Strains with telomere dysfunction have increased rates of white colony formation.
| TER1 allele | Telomere length | Total colonies observed | White colonies observed | Percentage of white colonies (%) | Median |
|---|---|---|---|---|---|
| Wild-type | 400–600 bp | 57,983 | 0 | <0.002 | 0.00% (18) |
| ter1-Δ | Variable length >100 bp at senescence | 32,484 | 1560 | 4.80 | 11.49% (27) |
| stn1-M1 | Very long and heterogenous | 55,272 | 173 | 0.31 | 1.30% (22) |
| stn1-M1 ter1Δ | Very long and heterogenous | 78,187 | 797 | 1.02 | 2.44% (18) |
| taq-28C | 75–150 bp | 134,375 | 90 | 0.07 | 0.02% (30) |
Dilution plates were examined for white colony formation in seven strains with varying levels of telomere function. The number of independent isolates used to generate the median frequency of white colonies is indicated in parentheses in the median column. Median frequency of white colonies formed was calculated from the average number of white colonies formed across the different independent isolates (see Materials and Methods for detailed description).
Telomerase deletion (ter1-Δ) mutants produce gradual telomere shortening accompanied by gradual, and eventually extreme growth senescence (McEachern and Blackburn 1995, 1996). ter1-Δ cells plated near the point of maximal senescence produced a median of 11.5% white colonies among the ∼32,500 postsenescence survivor colonies examined, a frequency at least 2700-fold higher than in wild-type cells. ter1-Δ cells at early stages of senescence or postsenescence survivors that have rebuilt telomeres using recombination showed no, or much lower frequencies of white colonies (data not shown). This is consistent with white colony formation being strongly linked to the telomere dysfunction of the highly senescent state rather than the lack of telomerase per se.
The mild telomere dysfunction strain used in these studies is the ter1-28C(Taq) mutant. This mutant has little or no effect on colony growth or morphology and has telomeres that are stable in length at 75–150 bp (about a quarter of wild-type size) that arise due to a base change in the Ter1 RNA template (Underwood et al. 2004). The ter1-28C(Taq) mutant produced 90 white colonies among ∼134,000 colonies inspected, at a median frequency of 0.02% (Table 1). This represents a white colony formation frequency at least 39-fold higher than in wild-type cells.
To confirm that the white colony formation in ter1-28C(Taq) was the result of telomere dysfunction and not due to a random mutation in some other gene acquired during its previous growth, six newly generated independent ter1-28C(Taq) mutants were examined for white colony formation. Of ∼15,000 total colonies examined, six (0.04%) were white (data not shown). This demonstrated that mild telomere dysfunction is responsible for the loss of function of a native subtelomeric gene in K. lactis.
We predicted that the inability to hydrolyze X-gal in the medium would correspond with the inability to metabolize lactose. To test this, 10 independent white isolates from each strain background were streaked on SD minimal medium containing 2% lactose as the sole carbon source. Although all blue isolate controls were able to grow on this medium, no white colonies were able to grow. Even after extensive passaging, none of the white clones were ever observed to revert back to blue, indicating the loss of β-galactosidase activity was not readily reversible and likely to be due to mutation rather than epigenetic change.
β-galactosidase is deleted in all LAC4-deficient isolates with severe telomere dysfunction but only in some isolates with mild telomere dysfunction
Southern hybridization studies were carried out to characterize the LAC4-deficient colonies isolated from dysfunctional telomere strains. All 30 independently derived white colonies from strains with severe telomere dysfunction (including 14 ter1-Δ, 5 stn1-M1, and 11 stn1-M1 ter1-Δ clones) showed no hybridization to the LAC4 probe (Figure 3D, and data not shown) indicating the entire gene was deleted. Additional hybridizations revealed that each white isolate was also deleted for LAC12 (encoding lactose permease), located ∼2 kb upstream from LAC4 (Figure 3C). Hybridization using a probe for the predicted gene KLLA0B14751, located ∼23 kb from the telomere, showed 14 of the 30 isolates (11 ter1-Δ and 3 stn1-M1 clones) were found to contain this fragment (Figure 3B). This suggested that the break point resulting in the loss of the 2R fragment containing the lactose utilization genes occurred between LAC12 and KLLA0B14751 gene in these 14 isolates. For the 16 other white isolates where the KLLA0B14751 was deleted, we concluded that the break point was more internally located. In stark contrast to the above results, the 30 blue isolates derived from the severe telomere dysfunction mutants never exhibited deletion of LAC4, LAC12, or KLLA0B14751 (Figure 3, B–D).
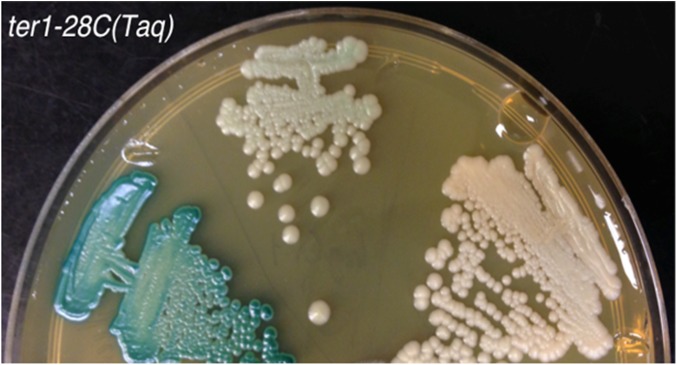
Southern hybridization was also used to characterize the white colonies derived from the ter1-28C(Taq) mutant with mild telomere dysfunction. Of the 23 LAC4 deficient white ter1-28C(Taq) mutants examined, 10 had experienced a deletion of both LAC4 and LAC12 (Figure 3, B–D and data not shown) but retained the more internal KLLA0B14751. This suggested that they may have experienced the same deletion event that predominated in severe telomere dysfunction mutants. Interestingly, the remaining 13 white ter1-28C(Taq) isolates were found to retain both LAC4 and LAC12 (Figure 3D and data not shown). Unlike the isolates lacking LAC4 and LAC12, these isolates were found to be pale blue in color once colonies had grown for a sufficient period of time on X-gal (Figure 4). However, like white isolates, these pale blue isolates were unable to grow on SD medium containing only lactose as a carbon source.
Figure 4.
ter1-28C(Taq) pale blue isolates were identified after extended growth on YPD plates with X-gal. Shown are comparisons between ter1-28C(Taq) clones producing blue colonies (left streak), a mutant producing pale blue colonies (middle streak), and a mutant producing white colonies (right streak). The YPD plate containing X-gal is shown after 5 days of growth.
Southern blotting was done to examine the terminal telomere fragments of the blue and white clones derived from the various mutants (Figure 3E and data not shown). EcoRI digestion of genomic DNA from the 7B520 parental strain separates the 12 telomeric restriction fragments into six different sized bands between ∼1 and 3.5 kb. The 2R EcoRI telomeric fragment is the largest band, making the loss of this fragment generally easy to detect. The highly variable and heterogeneous telomere restriction patterns in stn1-M1 and postsenescent ter1-Δ mutants made subtelomeric fragment identification from these strains infeasible. However, early passage ter1-28C(Taq) strains maintain a wild-type EcoRI restriction pattern and analysis of the two ter1-28C(Taq) white isolates (Figure 3E) showed that they lacked the 2R telomere fragment. This suggested that mutations responsible for the white colony phenotype are terminal deletions of the 2R chromosome end. Furthermore these terminal telomere patterns showed stronger than expected hybridization signal to the 1R telomere fragment (Figure 3E and data not shown). Quantification of the 1R band relative to other telomeric bands showed a 2.1-fold increase in signal compared to the 1R band of a blue ter1-28C(Taq) isolate used as a control, suggesting white isolates contained duplications that included the terminal EcoRI fragment of the 1R chromosome end.
Homeology between the 2R and 1R chromosome ends facilitated BIR-like repair in many 2R terminal deletion isolates
Sequence comparison of the 2R and 1R subtelomeric regions revealed an ∼1.1 kb shared region of homeology with 76% sequence identity (Figure S1). These subtelomeric regions fall ∼20 and 7.5 kb from the 2R and 1R chromosome end, respectively (Figure 5A). The homeologous region on the 2R chromosome end is found within an open reading frame with similarities to the Saccharomyces cerevisiae Cytochrome B2 (CYB2) gene required for lactate utilization (Alberti et al. 2000). The corresponding homeologous sequence on the 1R subtelomere is a divergent CYB2 pseudogene that is presumably no longer functional (Fairhead and Dujon 2006).
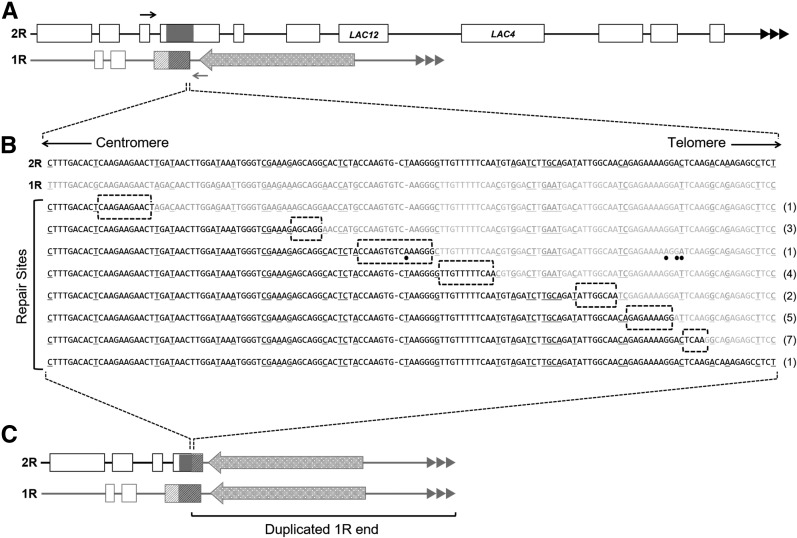
Figure 5.
Sequence junctions of 24 white isolates from strains with dysfunctional telomeres containing a chimeric 2R/1R gene fragment. (A) Structures of 2R and 1R chromosome ends. The shared 1.1 kb region of homology is shown as dark gray boxes. Positions of PCR forward primer and reverse primer are indicated by solid and shaded arrows, respectively. Open boxes represent open reading frames, the repeated triangles represent the telomeric repeat tracts, and the large shaded arrow represents the Tkl 1.1 transposable element that flanks the homologous region on 1R. (B) The top two sequences shown are the native 2R and 1R sequence in solid and shaded, respectively. The sequence reads from 24 2R/1R junctions are shown below. The underlined sequence represents the mismatched sequence between the 2R and 1R region. Numbers to the right of the sequence indicate the number of isolates that were repaired at the specific site of microhomology (highlighted by the dotted boxes). Dots indicate base changes that differ from both the 2R and 1R native sequence. (C) Predicted structures of the 1R and 2R ends after loss of sequence from 2R and duplication of the 1R terminal region.
In order to test whether this shared region of homeology was utilized in the repair of the 2R terminal deletion in isolates that have deleted LAC4 and LAC12 but retained KLLA0B14751, a PCR experiment was carried out. A forward primer was designed to the left flanking region of the CYB2-like homeology on the 2R chromosome and a reverse primer was designed to the right flanking region of the CYB2-like homeology on the 1R chromosome (Figure 5A). Using this primer set, PCR analysis was carried out on DNA from 14 white isolates with severe telomere dysfunction and 10 white isolates with mild telomere dysfunction. Amplification using this chimeric primer pair produced a PCR product consistent with the expected size of the 2R/1R hybrid in all 24 white isolates examined. As expected, control experiments using a primer pair designed to amplify the native CYB2 gene on the 2R chromosome failed to generate a PCR product in all white isolates examined. A PCR fragment of the expected size was amplified from the blue nondeleted control (data not shown). These results are consistent with the 2R deletions having recombination points with the 1R chromosome end within the CYB2-like sequences. Amplification across the native 1R CYB2-like homeology produced the expected PCR product size in all 24 white isolates as well as in the blue control suggesting that all white isolates still contained the native 1R subtelomeric region. This indicated that BIR repair duplicated the 1R end onto the truncated 2R end using the CYB2-like homeology to begin the copying reaction.
The 2R/1R chimeric PCR products were sequenced and aligned to the native 2R and 1R sequences to identify the point of recombination within the region of homeology. Although the homeologous region spans 1.1 kb, the recombination points in 23 of 24 isolates mapped to seven sites within a 150 bp region on the telomeric side of the homeologous region (Figure 5B). Furthermore, each break-point junction fell within a site of microhomology spanning 5–15 nucleotides in length. Sequencing analysis also detected four point mutations within the region duplicated during BIR in a single ter1-Δ isolate (Figure 5B).
A single isolate from the ter1-Δ background did not show a distinct switch from 2R sequence to 1R sequence when aligned to these two regions (Figure 5B). Instead, the entire length of the read was a perfect match to only the 2R region. Although the sequence information did not detect the exact point of repair, the PCR product that was sequenced was amplified using the 2R forward and 1R reverse primer set, which is specific only to the chimeric product. This suggests that this exceptional isolate uses an eighth site of microhomology for recombination that lies outside the specific region examined by our PCR assay.
Mating type loci on 2R and 3L chromosomes facilitate repair in some 2R terminal deletion isolates with severe telomere dysfunction
Southern analysis of strains with severe telomere dysfunction indicated 16 white isolates had deletions that extended internally as far as KLLA0B14751, but did not amplify the same 2R/1R junction fragment detected in the isolates described above. The 2R subtelomere was examined for additional regions of homology or homeology that could serve as alternative sites for BIR by comparing 100 kb of sequence from each chromosome end. Because seven predicted essential genes are found between ∼60 and 90 kb from the 2R telomere, terminal deletions of those sizes or larger are not likely to be survivable. This also predicts that any 2R homology used for repair would likely be located closer to the telomere.
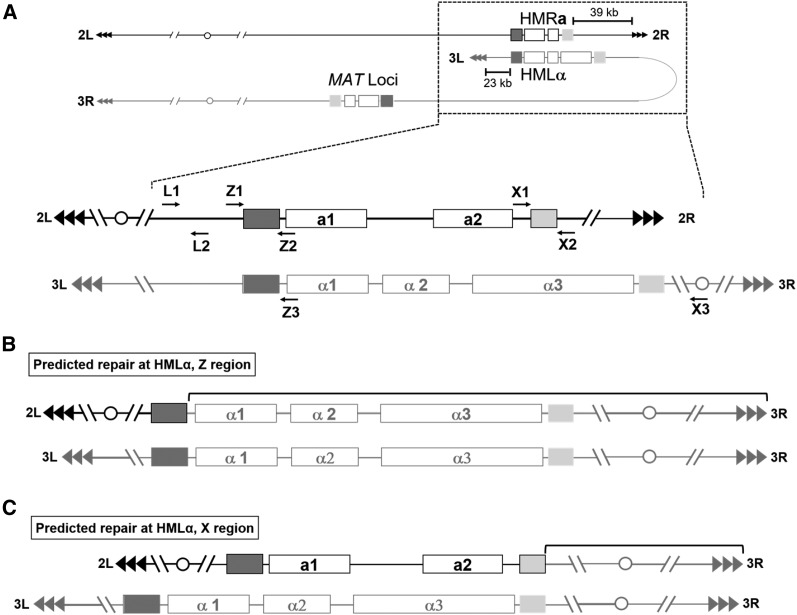
Two regions of homology shared between the transcriptionally silent cryptic mating loci located on chromosomes 2R (HMRa) and 3L (HMLα) as well as the active mating type locus (MAT) were identified ∼280 kb from the 3L telomere (Figure 6A). These two regions of perfect homology are the “Z” region, spanning 360 bp, and the “X” region, spanning 250 bp. The Z and X regions, which are not homologous to one another, are the left and right flanking sequences of all three mating loci and function to provide homologous blocks of sequence for recombination during mating type switching (Barsoum et al. 2010). It is important to note that, unlike the telomere proximal subtelomeric sequences, these regions on the 2R chromosome are in opposite orientations to the same regions on the 3L chromosome in respect to the telomere (Figure 6A).
Figure 6.
2R/3L chimeras at the common left and right flanking regions of the cryptic HMRa and HMLα mating loci were detected through PCR amplification across the left flanking region. (A) Overview of the mating type loci in 2L and 3R. Two blocks of perfect homology that are flanking the cryptic mating loci on the 2R and 3L subtelomeres are shown as dark and light shaded boxes. The left flanking region, or Z region (dark shaded) spans 360 bp, whereas the right flanking region, or X region (light shaded) spans 250 bp. The cryptic mating loci are found in opposite orientations with respect to their telomeres. Enlargement of cryptic mating type loci showing PCR primer locations (arrows) referred to in the text. Using these primers, PCR reactions designed to detect chimera formation across the left and right flanking regions was carried out in strains confirmed to have a terminal 2R deletion. The predicted outcome based on repair by homologous recombination is shown in (B), where recombination across the left flanking Z region results in the formation of a dicentric chromosome and gene conversion of the cryptic mating locus from a to α. The same type of PCR analysis was carried out to detect chimeras formed across the right flanking region, however no positive PCR reactions were identified. The predicted outcome based on homologous recombination across this right flanking region is shown in (C), also resulting in the formation of a dicentric chromosome.
To determine if repair of the 2R terminal deletion was initiated at the HMRa Z region, a PCR (using primers Z1 and Z3 in Figure 6A) was designed to amplify across the potential 2R/3L chimeric junction. Of the 16 uncharacterized white isolates from severe telomere dysfunction strains, five (including two ter1-Δ, two stn1-M1, and one stn1-M1 ter1-Δ mutant) were confirmed to produce a chimeric PCR product of the expected size spanning across the Z region (data not shown). Furthermore, PCR analysis across the native 2R Z region (using primers Z1 and Z2 in Figure 6A) failed to amplify the native sequence. Because this region of homology is a perfect match, the exact site of repair within the Z region could not be determined. Sequencing of these PCR products confirmed that the region on the left side of the Z homology was native 2R sequence, and the region on the right side of the homology was native 3L sequence in both the forward and reverse reads (Figure 6B). This indicated that a 3L Z region was used for repair, generating a 2R/3L junction.
The same strategy (using primers X1 and X3, Figure 6A) was carried out to amplify potential junctions generated from repair events that utilized the 250 bp X region of homology. However, PCR analysis was unable to amplify any chimeric fragments spanning this region from any of the remaining 11 uncharacterized white clones examined. For those 11 white clones (one ter1-Δ and 10 stn1-M1 ter1-Δ isolates), PCR reactions (using primers L1 and L2, Figure 6B) were successful at amplifying the region ∼1 kb upstream of HMRa on 2R. Although this is not direct evidence of repair at either the Z or X regions, it does strongly suggest that the terminal deletion of 2R does not extend internally past HMRa in any of the analyzed white clones. The complex nature of repair events at these internal chromosomal locations and the possibility that both the HMRa and the MAT loci are being utilized for repair make further PCR diagnostics of these repair events difficult.
ter1-28C(Taq) isolates with reduced LAC4 activity commonly contained base changes in the LAC4 gene
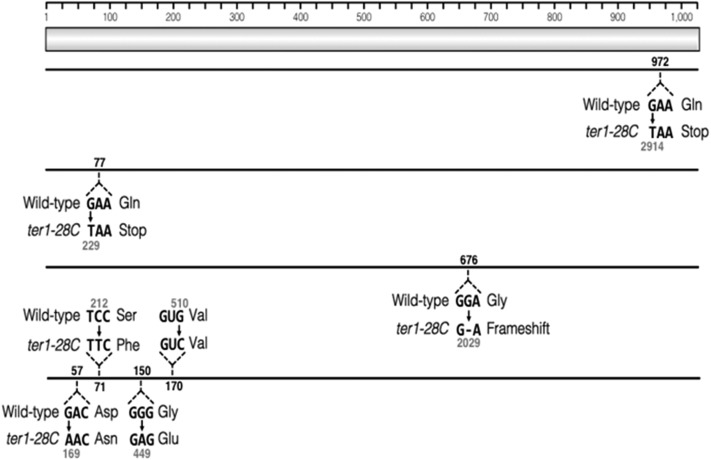
To determine whether the pale blue phenotype observed in some ter1-28C(Taq) isolates was due to mutation, we sequenced the LAC4 gene from six such isolates. The entire LAC4 locus, including 157 bp downstream of the gene and the ∼2.8 kb LAC4 regulatory region found between the LAC4 and LAC12, was sequenced. Four of the six isolates sequence contained at least one point mutation within the LAC4 open reading frame (Figure 7). Two of these isolates contained single-point mutations that resulted in premature stop codons. A single base pair deletion resulting in a frame-shift was detected in the third isolate. Four separate mutations were identified in the fourth isolate, three of which resulted in amino acid substitutions (Figure 7). No mutations were detected in two sequenced isolates and the basis of their phenotypes remains unclear. Because X-gal is passively transported into the cell, mutations within the LAC12 gene are not predicted to cause the light blue phenotype observed here. We conclude that the mild telomere dysfunction of ter1-28C(Taq) can lead to high levels of base changes within the subtelomere in the absence of any obvious BIR-like deletion/duplication.
Figure 7.
Some pale blue isolates contain point mutations within the LAC4 gene. Sanger sequencing of the LAC4 gene in six independent ter1-28C isolates was carried out. Sequence was compared to the wild-type LAC4 sequence, shown as the light shaded bar (amino acid sequence). Solid lines represent LAC4 sequence from the four isolates that contained mutations. The wild-type codon and mutated codon are show for each isolate with the resulting change in amino acid sequence. Black numbers represent the amino acid residue position affected by the mutation and gray number represent the nucleotide position of the mutation.
The K. lactis 2R telomere-adjacent region appears to be derived from K. marxianus
The LAC4 and LAC12 genes are not present in all strains of K. lactis and were previously predicted to have originated from the closely related species K. marxianus (Naumov 2005). This was based on the high sequence identity of the genes in the two species, and on the observation that a higher percentage of natural isolates of K. marxianus are able to assimilate lactose compared with K. lactis. To further address this issue, sequence comparisons of the 0.6–2 kb telomere-adjacent R elements (Nickles and McEachern 2004) of K. lactis and K. marxianus subtelomeres were carried out. Although 11 of the 12 K. lactis R elements share >85% sequence identity, the 2R telomere-adjacent region is unusual in sharing no more than 59% identity to the other R elements. Comparison of the 500 bp telomere proximal sequences from the divergent 2R R element revealed that it has much greater sequence identity (80–89%) to the telomere-adjacent regions of 15 out of 16 chromosome ends in K. marxianus than to the other R elements of K. lactis. High levels of sequence homology to K. marxianus, varying from 67% in the nongenic regions to as high as 98% in the coding regions, extend across the 2R terminal fragment, through the lactose permease gene. Homologous sequence was no longer detectable ∼100 bp upstream of LAC12. These results suggest that the entire ∼15 kb terminal region of the K. lactis 2R end, including the R element, may have been acquired from K. marxianus.
Discussion
Severely dysfunctional telomeres produce very high frequencies of lac4 mutations through BIR-like deletion/duplication events in subtelomeres.
By using a natural β-galactosidase gene (LAC4) as a marker of subtelomeric stability, we characterized subtelomeric mutations brought about by different levels of telomere dysfunction in the yeast K. lactis. We found that strains with high levels of telomere dysfunction (the ter1-Δ, stn1-M1, and stn1-M1 ter1-Δ mutants) displayed frequencies of lac4 mutant formation that were minimally two orders of magnitude greater than that seen in control cells. The frequency of LAC4 deletion appears to be related to the extent of telomere dysfunction in these mutants. The highest LAC4 deletion frequency (at least 2700-fold elevated) was seen in ter1-Δ cells at peak senescence, which have extremely dysfunctional telomeres and very poor growth. In contrast, ter1-Δ postsenescence survivors, which have longer telomeres and reduced telomere dysfunction, show vastly less LAC4 loss. The stn1-M1 and stn1-M1 ter1-Δ mutants, which exhibit chronic moderate telomere capping and growth defects reminiscent of moderately senescent ter1-Δ cells (Iyer et al. 2005; Xu and McEachern 2012), had LAC4 deletion frequencies that were minimally 180–500-fold higher than wild type, respectively. A modestly higher LAC4 loss of frequency in the stn1-M1 ter1-Δ strain is consistent with previous work that showed deletion of telomerase in the stn1-M1 ter1-Δ double mutant increased recombination rates within telomeric and R element sequences compared with the single stn1-M1 mutant (Xu and McEachern 2012).
The mutational events associated with LAC4 deletion in strains with severe telomere dysfunction were, without exception, due to terminal deletion of the 2R telomere. Two distinct classes of terminal deletions, occurring ∼20–∼40 kb from the 2R chromosome end, were found. The 20 kb deletions were all repaired by copying the 1R chromosome end using the first region of the 2R end internal from the LAC4 sequence that shares homeology to another subtelomeric region. Repair at this site replaced the 2R end through a nonreciprocal translocation duplicating the 1R end, as expected for a BIR event. Interestingly, repair junctions were isolated to small regions of microhomology clustered within the 3′ telomeric-facing end of the 1.1 kb homeologous sequence. Consistent with previous studies, this finding suggests that limited homology within the subtelomere can still act as substrates for subtelomeric BIR (Anand et al. 2014).
Repair of the ∼40 kb terminal deletions was found to have been carried out by recombination initiated at the homologous Z or X regions flanking the HMLα cryptic mating locus on 3L. Such BIR events are expected to lead to the formation of dicentric chromosomes, as the homologies found on 2R and 3L are found in opposite orientation to one another in respect to the telomere. Because of their instability, it is likely that any dicentric chromosomes that formed from the 2R/3L recombination would quickly have undergone further rearrangements to restore monocentric chromosomes. Alternatively, template switching of the invading 2R strand from the donor Z or X region duplex to another site of homology could bypass dicentric chromosome formation and result in the repair of the ∼40 kb terminal deletion with less deleterious genomic rearrangements (Smith et al. 2007; Ruiz et al. 2009; Sakofsky et al. 2015). Further analysis is required to fully understand the nature of repair of these larger terminal deletions.
Mild telomere dysfunction triggers a different spectrum of subtelomeric mutations than severe telomeric dysfunction
Our work also found that a strain with mild telomere dysfunction from stably shortened telomeres (ter1-28C(Taq)) gave rise to lac4 mutants at frequencies at least 39-fold higher than a wild-type strain. This demonstrates clearly that telomere dysfunction in K. lactis sufficiently mild to have no detectable detrimental effects on cell growth and viability can still induce substantially elevated rates of mutation that affect native subtelomeric genes. Characterization of lac4 isolates from ter1-28C(Taq) revealed that they also produced deletions of the 2R terminal fragment, as observed in strains with severe telomere dysfunction. Interestingly, however, repair of these terminal deletions in the ter1-28C(Taq) strain was only observed at the more terminally located 1R site of homeology and not at the more internal HMRa cryptic mating locus. This is consistent with a lower level of telomere dysfunction being less likely to lead to more extreme terminal deletions.
Our results further showed that ∼50% of lac4-deficient isolates generated in the ter1-28C(Taq) background had no obvious subtelomeric rearrangements. However, sequencing of the LAC4 gene from six such isolates revealed that four of them contained point mutations. This result indicates that mild-telomere dysfunction in K. lactis can produce a class of subtelomeric mutations (base changes unaccompanied by gross chromosomal rearrangements) that are not observed, or are at least are a much lower percentage of lac4 isolates, in strains with severely dysfunctional telomeres. Surprisingly, pale blue colony color, indicative of residual LAC4 function, was present in mutants containing premature stop codons within the LAC4 open reading frame. This suggests that LAC4 may have a weak alternative translational start point or that there is a detectable level of nonsense suppression occurring.
We have not examined whether the elevated rate of base changes from telomere dysfunction that we observed to occur in LAC4 is confined to subtelomeric regions. However, previous work showed that telomere dysfunction, even in severe form, did not lead to a detectable increase in HR occurring at a chromosome-internal site (McEachern and Iyer 2001). As it is expected that increased rates of both HR and base changes result from DNA ends being processed for repair, it is highly likely that elevated rates of base changes are largely confined to subtelomeres or regions able to undergo HR with subtelomeres.
How the point mutations that we observed in ter1-28C(Taq) lac4 mutants form is unknown. Past work in senescing S. cerevisiae cells lacking telomerase found that a 16-fold increase in base substitution and frameshift mutations at a subtelomeric location was dependent upon the Rev1 and Pol ζ translesion DNA polymerases (Meyer and Bailis 2007). More recently, it was shown that DNA synthesis during BIR is highly error prone, producing base changes at ∼1000 times the normal frequency (Deem et al. 2011). This is thought to be a consequence of the unusual, moving D-loop DNA synthesis that characterizes BIR (Saini et al. 2013). However, the base alterations in the lac4 gene formed in the ter1-28C(Taq) strain appear not to be due to classical BIR events, as there is no evidence that they are associated with nonreciprocal duplication of 2R sequences. We instead propose that they may be due to sister chromatid BIR events that occur from telomere dysfunction happening at just one of a pair of replicated sister telomeres. If one sister telomere becomes uncapped and is nucleolytically processed to lose some sequence, strand invasion of the shortened end into the intact sister may result in a BIR event that replaces the lost sequences exactly, except for base alterations introduced by the error-prone D-loop synthesis.
Why base changes are a far greater proportion of lac4 mutants in the mild telomere dysfunction ter1-28C(Taq) strain compared with the severe telomere dysfunction strains remains unclear. One possibility is that telomere uncapping occurring from severe dysfunction is more likely to persist through cell division and therefore act either before replication or affect both sister telomeres after replication. Consistent with this model, we note that the mild telomere dysfunction that characterizes the earliest stages of replicative senescence in human cells generally affects just one of a replicated pair of sister telomeres (Kaul et al. 2012). Another potential contributor to the different subtelomeric mutational profiles of the strains examined in our work could stem from severe telomere dysfunction tending to saturate one or more subprocesses involved in the repair of uncapped telomeres, and thereby perturb the outcomes of the repair events.
The strains used in this study are expected to lead to all telomeres in the cell having an equal chance of being dysfunctional. We therefore cannot conclude for certain that dysfunction of the 2R telomere was always the cause of mutation of 2R subtelomeric regions. However, dysfunctional telomeres are expected to act preferentially in cis with regard to causing mutations. This is because DNA ends that initiate HR typically act as recipients of genetic information donated or copied from another DNA molecule (Mehta and Haber 2014).
The potential of adaptive telomere failure to facilitate evolution of K. lactis subtelomeric DNA
Another major prediction of the adaptive telomere failure hypothesis, untested in the experiments presented in this work, is that many eukaryotic organisms, perhaps especially microbes, will generate occasional telomere dysfunction in certain stressful environments (McEachern 2008). This dysfunction might occur through many mechanisms, including targeted alteration or degradation of a telomere protein, or induction of telomere rapid deletion (Li and Lustig 1996; Bucholc et al. 2001; Bechard et al. 2011). Telomere uncapping could, in turn, sometimes stimulate recombination in subtelomeric genes and thereby occasionally produce beneficial mutations. If this type of telomere failure exists, it is unlikely to occur as simultaneous uncapping of all telomeres, but rather as failure affecting a single telomere in a cell at any given occurrence, with minimal effects on cell fitness. The mild telomere dysfunction of the ter1-28C(Taq) strain used here may therefore provide a useful model for understanding the nature of subtelomeric mutations that might be possible with environmentally induced telomere failure.
The potential benefits of elevated mutation rates (whether inducible or constitutive) in subtelomeric regions are likely related to the unusual types of genes found there. Subtelomeres are enriched in genes that exhibit copy number variation and/or function in adaptation to novel niches (Moxon et al. 1994; Barry et al. 2003). They are also common locations for genes that have introgressed from other related species (Naumov 2005; Naumova et al. 2011). We have shown here that the 2R telomere of K. lactis, including the β-galactosidase (LAC4) and lactose permease (LAC12) genes, has been acquired from the related species K. marxianus, a species previously shown be capable of mating with K. lactis (Steensma et al. 1988). Conceivably, the appearance of the human-created environments of dairies and creameries led to selection for lactose utilization and the LAC4 and LAC12 genes in K. lactis. Sugar utilization genes and gene families are common in yeast subtelomeric regions, and work on the Saccharomyces species has argued that some of them have evolved to have novel cleavage specificities (Brown et al. 2010).
Mild telomere dysfunction might increase the likelihood of beneficial mutations in subtelomeric genes through more than one mechanism. BIR-generated terminal duplications can increase the copy number of subtelomeric genes. Elevated subtelomeric recombination can also generate hybrid genes from dispersed family members. Our observation of BIR events repeatedly utilizing CYB2 sequences with only 76% sequence match to one another is an example of the potential of such processes. Increased base change rates in the absence of classical BIR deletion/duplication outcomes could obviously directly modify the sequence of subtelomeric genes. It is likely that subtelomeric genes that had accumulated mutations due to rare use, or that had been introgressed from another species, would be more likely to achieve fitness benefits from base changes than would normally be the case for other genes.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300607/-/DC1.
Acknowledgments
This work was supported in part by a National Institutes of Health training grant (T 32 GM007103) and from a generous gift from Paul Truex and family. The authors would like to also thank Brianna Stadsvold, who helped score colony color, and Zahilys Soto, who assisted with tetrad dissections.
Footnotes
Communicating editor: J. Nickoloff
Literature Cited
- Alberti A., Goffrini P., Ferrero I., Lodi T., 2000. Cloning and characterization of the lactate-specific inducible gene KlCYB2, encoding the cytochrome b2 of Kluyveromyces lactis. Yeast 16: 657–665. [DOI] [PubMed] [Google Scholar]
- Anand R. P., Tsaponina O., Greenwell P. W., Lee C. S., Du W., et al. , 2014. Chromosome rearrangements via template switching between diverged repeated sequences. Genes Dev. 28: 2394–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y., Kupiec M., 2004. DSB repair: the yeast paradigm. DNA Repair (Amst.) 3: 797–815. [DOI] [PubMed] [Google Scholar]
- Barry J. D., Ginger M. L., Burton P., McCulloch R., 2003. Why are parasite contingency genes often associated with telomeres. Int. J. Parasitol. 33: 29–45. [DOI] [PubMed] [Google Scholar]
- Barsoum E., Martinez P., Aström S. U., 2010. α3, a transposable element that promotes host sexual reproduction. Genes Dev. 24: 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechard L. H., Jamieson N., McEachern M. J., 2011. Recombination can cause telomere elongations as well as truncations deep within telomeres in wild-type Kluyveromyces lactis cells. Eukaryot. Cell 10: 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E. H., Epel E. S., Lin J., 2015. Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science 350: 1193–1198. [DOI] [PubMed] [Google Scholar]
- Blasco M., Lee H. W., Hande M. P., Samper E., Lansdorp P. M., et al. , 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91: 25–34. [DOI] [PubMed] [Google Scholar]
- Bobrowicz P., Wysocki R., Owsianik G., Goffeau A., Ułaszewski S., 1997. Isolation of three contiguous genes ACR1, ACR2, and ACR3, involved in resistance to arsenic compounds in the yeast Saccharomyces cerevisiae. Yeast 13: 819–828. [DOI] [PubMed] [Google Scholar]
- Brown C. A., Murray A. W., Verstrepen K. J., 2010. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr. Biol. 20: 895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholc M., Park Y., Lustig A. J., 2001. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. W. L., Blackburn E. H., 2003. Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Mol. Cell 11: 1379–1387. [DOI] [PubMed] [Google Scholar]
- Cech T. R., 2004. Beginning to understand the end of the chromosome. Cell 116: 273–279. [DOI] [PubMed] [Google Scholar]
- Chiruvella K. K., Liang Z., Wilson T. E., 2013. Repair of double-strand breaks by end joining. Cold Spring Harb. Perspect. Biol. 5: a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven R. J., Greenwell P. W., Dominska M., Petes T. D., 2002. Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR genes. Genetics 161: 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzentas N., 2010. Circoletto: visualizing sequence similarity with Circos. Bioinformatics 26: 2620–2621. [DOI] [PubMed] [Google Scholar]
- Deem A., Keszthelyi A., Blackgrove T., Vayl A., Coffey B., et al. , 2011. Break-induced replication is highly inaccurate. PLoS Biol. 9: e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lange T., Lundblad V., Blackburn E. H., 2006. Telomeres. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Dickson R. C., Markin J. S., 1980. Physiological studies of β-galactosidase induction in Kluyveromyces lactis. J. Bacteriol. 142: 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhead C., Dujon B., 2006. Structure of Kluyveromyces lactis subtelomeres: duplications and gene content. FEMS Yeast Res. 6: 428–441. [DOI] [PubMed] [Google Scholar]
- Garvik B., Carson M., Hartwell L., 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15: 6128–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover L., Jun J., Horn D., 2011. Microhomology-mediated deletion and gene conversion in African trypanosomes. Nucleic Acids Res. 39: 1372–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D. E., Aparicio O. M., Billington B. L., Zakian V. A., 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762. [DOI] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H., 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337. [DOI] [PubMed] [Google Scholar]
- Hackett J. A., Greider C. W., 2003. End resection initiates genomic instability in the absence of telomerase. Mol. Cell. Biol. 23: 8450–8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D., 2004. The molecular control of antigenic variation in Trypanosoma brucei. Curr. Mol. Med. 4: 563–576. [DOI] [PubMed] [Google Scholar]
- Horn D., Barry J. D., 2005. The central roles of telomeres and subtelomeres in antigenic variation in African trypanosomes. Chromosome Res. 13: 525–533. [DOI] [PubMed] [Google Scholar]
- Iyer S., Chadha A. D., McEachern M. J., 2005. A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol. Cell. Biol. 25: 8064–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul Z., Cesare A. J., Huschtscha L. I., Neumann A. A., Reddel R. R., 2012. Five dysfunctional telomeres predict onset of senescence in human cells. EMBO Rep. 13: 52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. M., Rivera M. A., Botchkina I. L., Shalaby R., Thor A. D., et al. , 2001. A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc. Natl. Acad. Sci. USA 98: 7982–7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraepelin G., 1984. Yeasts: characteristics and identification. Z. Allg. Mikrobiol. 24: 436. [Google Scholar]
- Krejci L., Altmannova V., Spirek M., Zhao X., 2012. Homologous recombination and its regulation. Nucleic Acids Res. 40: 5795–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Lustig A. J., 1996. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 10: 1310–1326. [DOI] [PubMed] [Google Scholar]
- Lieber M. R., 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79: 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou E. V., Williams E. M., Fan Y., Friedman C., Young J. M., et al. , 2005. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature 437: 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J., de Lange T., 2017. Telomeres in cancer: tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 18: 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M. J., 2008. Telomeres: Guardians of genomic integrity or double agents of evolution, pp. 100–113 in Origin and Evolution of Telomeres. Landes Bioscience, Austin, TX. [Google Scholar]
- McEachern M. J., Blackburn E. H., 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376: 403–409. [DOI] [PubMed] [Google Scholar]
- McEachern M. J., Blackburn E. H., 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 10: 1822–1834. [DOI] [PubMed] [Google Scholar]
- McEachern M. J., Haber J. E., 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 75: 111–135. [DOI] [PubMed] [Google Scholar]
- McEachern M. J., Iyer S., 2001. Short telomeres in yeast are highly recombinogenic. Mol. Cell 7: 695–704. [DOI] [PubMed] [Google Scholar]
- Mefford H. C., Trask B. J., 2002. The complex structure and dynamic evolution of human subtelomeres. Nat. Publ. Gr. 3: 91–102. [DOI] [PubMed] [Google Scholar]
- Mehta A., Haber J. E., 2014. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 6: a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D. H., Bailis A. M., 2007. Telomere dysfunction drives increased mutation by error-prone polymerases Rev1 and ζ in Saccharomyces cerevisiae. Genetics 175: 1533–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski P. A., Mieczkowska J. O., Dominska M., Petes T. D., 2003. Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisae. Proc. Natl. Acad. Sci. 100: 10854–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D., Reynolds G. E., Mejia R., Stark J. M., Murnane J. P., 2011. Subtelomeric regions in mammalian cells are deficient in DNA double-strand break repair. DNA Repair (Amst.) 10: 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon E. R., Rainey P. B., Nowak M. A., Lenski R. E., 1994. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 4: 24–33. [DOI] [PubMed] [Google Scholar]
- Naumov G. I., 2005. Domestication of dairy yeast Kluyveromyces lactis: transfer of the β-galactosidase (LAC4) and lactose permease (LAC12) gene cluster? Dokl. Biol. Sci. 401: 120–122. [DOI] [PubMed] [Google Scholar]
- Naumova E. S., Naumov G. I., Michailova Y. V., Martynenko N. N., Masneuf-Pomarède I., 2011. Genetic diversity study of the yeast Saccharomyces bayanus var. uvarum reveals introgressed subtelomeric Saccharomyces cerevisiae genes. Res. Microbiol. 162: 204–213. [DOI] [PubMed] [Google Scholar]
- Nickles K., McEachern M. J., 2004. Characterization of Kluyveromyces lactis subtelomeric sequences including a distal element with strong purine/pyrimidine strand bias. Yeast 21: 813–830. [DOI] [PubMed] [Google Scholar]
- Pâques F., Haber J. E., 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlevsky J. D., Chen J. J. L., 2012. It all comes together at the ends: telomerase structure, function, and biogenesis. Mutat. Res. Fundam. Mol. Mech. Mutagen. 730: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde F. E., Gorham H. Z., Louise E. J., 1997. Chromosome ends: all the same under their caps. Curr. Op. 7: 822–828. [DOI] [PubMed] [Google Scholar]
- Ricchetti M., Dujon B., Fairhead C., 2003. Distance from the chromosome end determines the efficiency of double strand break repair in subtelomeres of haploid yeast. J. Mol. Biol. 328: 847–862. [DOI] [PubMed] [Google Scholar]
- Rudd M. K., Friedman C., Parghi S. S., Linardopoulou E. V., Hsu L., et al. , 2007. Elevated rates of sister chromatid exchange at chromosome ends. PLoS Genet. 3: 0319–0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz J. F., Gómez-González B., Aguilera A., 2009. Chromosomal translocations caused by either pol32-dependent or pol32-independent triparental break-induced replication. Mol. Cell. Biol. 29: 5441–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini N., Ramakrishnan S., Elango R., Ayyar S., Zhang Y., et al. , 2013. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 502: 389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakofsky C. J., Ayyar S., Deem A. K., Chung W. H., Ira G., et al. , 2015. Translesion polymerases drive microhomology-mediated break-induced replication leading to complex chromosomal rearrangements. Mol. Cell 60: 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. E., Llorente B., Symington L. S., 2007. Template switching during break-induced replication. Nature 447: 102–105. [DOI] [PubMed] [Google Scholar]
- Steensma H. Y., de Jongh F. C. M., Linnekamp M., 1988. The use of electrophoretic karyotypes in the classification of yeasts: Kluyveromyces marxianus and K. lactis. Curr. Genet. 14: 311–317. [Google Scholar]
- Teng S. C., Zakian V. A., 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 8083–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood D. H., Zinzen R. P., McEachern M. J., 2004. Template requirements for telomerase translocation in Kluyveromyces lactis. Mol. Cell. Biol. 24: 912–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen K. J., Klis F. M., 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60: 5–15. [DOI] [PubMed] [Google Scholar]
- Wang Z.-R., Guo L., Chen L., McEachern M. J., 2009. Evidence for an additional base-pairing element between the telomeric repeat and the telomerase RNA template in Kluyveromyces lactis and other yeasts. Mol. Cell. Biol. 29: 5389–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters C. A., Strande N. T., Wyatt D. W., Pryor J. M., Ramsden D. A., 2014. Nonhomologous end joining: a good solution for bad ends. DNA Repair (Amst.) 17: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger R. J., Zakian V. A., 2012. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics 191: 1073–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray L. V., Witte M. M., Dickson R. C., Riley M. I., 1987. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., McEachern M. J., 2012. Maintenance of very long telomeres by recombination in the Kluyveromyces lactis stn1–M1 mutant involves extreme telomeric turnover, telomeric circles, and concerted telomeric amplification. Mol. Cell. Biol. 32: 2992–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Strains are available upon request.