Abstract
Meiotic crossovers must be properly patterned to ensure accurate disjunction of homologous chromosomes during meiosis I. Disruption of the spatial distribution of crossovers can lead to nondisjunction, aneuploidy, gamete dysfunction, miscarriage, or birth defects. One of the earliest identified genes involved in proper crossover patterning is Drosophila mei-41, which encodes the ortholog of the checkpoint kinase ATR. Analysis of hypomorphic mutants suggested the existence of crossover patterning defects, but it was not possible to assess this in null mutants because of maternal-effect embryonic lethality. To overcome this lethality, we constructed mei-41 null mutants in which we expressed wild-type Mei-41 in the germline after completion of meiotic recombination, allowing progeny to survive. We find that crossovers are decreased to about one-third of wild-type levels, but the reduction is not uniform, being less severe in the proximal regions of chromosome 2L than in medial or distal 2L or on the X chromosome. None of the crossovers formed in the absence of Mei-41 require Mei-9, the presumptive meiotic resolvase, suggesting that Mei-41 functions everywhere, despite the differential effects on crossover frequency. Interference appears to be significantly reduced or absent in mei-41 mutants, but the reduction in crossover density in centromere-proximal regions is largely intact. We propose that crossover patterning is achieved in a stepwise manner, with the crossover suppression related to proximity to the centromere occurring prior to and independently of crossover designation and enforcement of interference. In this model, Mei-41 has an essential function in meiotic recombination after the centromere effect is established but before crossover designation and interference occur.
Keywords: meiotic recombination, interference, centromere effect, Drosophila, ATR kinase
MEIOTIC crossovers are subject to numerous mechanisms of spatial control to ensure proper disjunction of homologous chromosomes and the generation of genetic diversity. Sturtevant (1913) described the phenomenon of crossover interference, where the presence of one crossover reduces the probability of crossovers nearby (reviewed in Berchowitz and Copenhaver 2010). Mather (1937) pointed out that for small chromosomes “the chiasma frequency equals one, no matter what the size”; Owen (1949) referred to this as the “obligate chiasma.” The phenomenon in which every pair of homologous chromosomes has at least one crossover that generates a chiasma to promote disjunction is commonly called crossover assurance (reviewed in Wang et al. 2015). Together with crossover homeostasis, which buffers crossover formation from increases or decreases in potential crossover precursors (Martini et al. 2006), assurance and interference demarcate the minimum and maximum number of crossovers per meiosis. Modeling suggests that crossover assurance, interference, and homeostasis are the result of a single patterning process (Wang et al. 2015). The mechanisms that achieve assurance, interference, and homeostasis remain obscure.
Less attention has been paid to the centromere effect; a spatial crossover patterning phenomenon first described by Beadle (1932). Crossovers are excluded from the vicinity of centromeres in many organisms, presumably because very proximal crossovers can interfere with homolog disjunction (Koehler et al. 1996; Lamb et al. 1996). There are two components to the reduction in crossovers near the centromere. First, Muller and Painter (1932) reported that crossing over is absent or extremely rare within the “inert regions,” now known to comprise heterochromatic, pericentromeric satellite sequence. The second component, which we refer to as the centromere effect, is the phenomenon Beadle described: the reduction in crossing over within crossover-competent regions of the genome as a function of proximity to the centromere. Beadle noticed that when regions with high crossover density were moved closer to the centromere by chromosome rearrangement, crossover frequency decreased. The converse—increased crossover density when centromere-proximal regions are moved away from the centromere—was shown by Mather (1939). The mechanisms underlying the centromere effect are also unknown.
Meiotic recombination is initiated by the formation of DNA double-strand breaks (DSBs). Each DSB can be repaired into a crossover or a noncrossover; the latter can be detected when they result in gene conversion, the unidirectional transfer of sequence from a donor (a homologous chromosome) to a recipient (the chromatid that received the DSB). In Drosophila, DSBs appear be to be excluded from the pericentric heterochromatin, explaining the absence of crossovers in those regions (Mehrotra and McKim 2006). The centromere effect could, in principle, be explained by decreased DSB density in proximal regions. However, recent whole-genome sequencing reveals that the density of noncrossover gene conversion is relatively constant across the assembled genome on each arm (Comeron et al. 2012; Miller et al. 2016). This suggests that DSB density is also fairly constant across the chromosome arm and that the centromere effect is exerted by regulating the outcome of DSB repair (crossover or noncrossover) in a manner that is dependent on distance to the centromere.
Mutations in the Drosophila mei-41 gene were first described by Baker and Carpenter (Baker and Carpenter 1972; Hari et al. 1995) who reported a polar reduction in crossovers, with a less severe effect on crossovers in proximal regions, and a possible decrease in interference. These observations suggest a potential role for Mei-41 in crossover patterning. Mei-41 is the Drosophila ortholog of ATR kinase, best known for regulating DNA damage-dependent cell cycle checkpoints (Hari et al. 1995). Consistent with this role, Mei-41 establishes a checkpoint that monitors progression of meiotic recombination (Ghabrial and Schüpbach 1999; Abdu et al. 2002). In addition, Mei-41 acts redundantly with ATM kinase to promote phosphorylation of histone H2AV at sites of meiotic DSBs (Joyce et al. 2011). However, it is unlikely that either of these functions explains the effects of crossover number or position noted by Baker and Carpenter. Understanding this role is further complicated by the finding that Mei-41 has an essential function in slowing the rapid nuclear cycles at the midblastula transition in embryonic development (Sibon et al. 1999). Females with null mutations in mei-41 are sterile because this function is lost, and thus the mutations used in previous studies of meiotic recombination are either hypomorphic or separation-of-function alleles (Laurençon et al. 2003).
We sought to investigate the possible function for Mei-41 in crossover patterning by analyzing crossover distribution in mei-41 null mutants. To overcome the requirement for maternal Mei-41, we used a transgene in which mei-41 expression is under control of a promoter that turns on only after recombination has been completed, thereby generating a fertile mei-41 “meiotic recombination null” mutant. We find that crossover and nondisjunction phenotypes are more severe in this mutant than in previously reported hypomorphic mutants. We observe a polar effect on chromosome 2L but not on the X; we suggest that this is due to retention of the centromere effect, which is weak on the X. However, interference and assurance are greatly decreased or lost. We propose that the centromere effect is established early in the meiotic recombination pathway and that Mei-41 has a recombination role after this establishment but before interference and assurance are achieved. Loss of Mei-41 leads to exit from the meiotic recombination pathway after establishment of the centromere effect and repair is then completed by alternative mechanisms that lack interference and assurance. These findings provide insight into the establishment of crossover patterning.
Materials and Methods
Drosophila stocks
Flies were maintained at 25° on standard medium. To overcome the maternal-effect embryonic lethality of mei-4129D null mutation (Sibon et al. 1999; Laurençon et al. 2003), wild-type genomic mei-41 was cloned into the P{attB, UASp::, w+m} vector (courtesy of Steve Rogers) via In-Fusion HD (Takara Bio, Mountain View, CA) and transformed into XL10-Gold Ultracompetent Cells (Agilent Technologies, Santa Clara, CA). This construct was injected via phiC31 integrase-mediated transgenesis into the X chromosome landing site M{3xP3-RFP.attP’}ZH-2A (BestGene, Chino Hills, CA). The resulting integrants, abbreviated herein as M{UASp::mei-41}, were crossed into a P{matα4::GAL4-VP16} background. All mei-41 null assays used the genotype:
The mei-41mei-P22 double mutant genotype was as above except the third chromosomes were mei-P22103 st/mei-P22103 BlmD2 Sb P{matα4::GAL4-VP16}. The mei-9mei-41 double mutant genotype was as above except the X chromosomes were y mei-9a mei-4129D/y M{UASp::mei-41} w mei-9a mei-4129D. The presence of the mei-9a mutation was confirmed by allele-specific PCR and by genetic tests (see Supplemental Material, Table S5 in File S1).
Hatch rates
To test M{UASp::mei-41} rescue efficiency, 60 virgin females of appropriate genotypes were crossed to 20 isogenized Oregon-Rm males (courtesy of Scott Hawley). Adults were mated in grape-juice agar cages containing yeast paste for 2 days prior to collection. Embryos were collected on grape-juice agar plates for 5 hr and scored for hatching 48 hr later.
Crossover assays and analyses
Meiotic crossovers on chromosome 2L were quantified by crossing netdppd-ho dp bprcn/+ virgin females of the appropriate mutant background to netdppd-ho dp bprcn males. All six markers were scored in progeny from each genotype, with the of exception mei-41; mei-P22. In that case, 731 XX females were scored for all six markers and an additional 1023 XXY females and XY males were scored for net–b; eye color markers pr and cn were excluded because of the presence of a w mutation in the mothers. These data were pooled for a final number of 1754 progeny scored.
Meiotic crossovers on X were quantified by crossing ysccvvgf · y+ virgin females of the appropriate background to ysccvvgf male. “· y+” is Dp(1;1)scV1, a duplication of the left end of the X, carrying y+, onto XR. All six markers were scored in all progeny.
To measure chromosome 4 crossovers, the mei-41 rescue genotype given above was made heterozygous for PBac{y+ w+m}(101F) and svspa-pol, which are near opposite ends of the assembled region of chromosome 4. These females were crossed with w1118; svspa-pol males and the progeny were scored for the poliert eye phenotype associated with svspa-pol homozygosity and the w+m of the PBac transgene. Although both the M{UASp::mei-41} and P{matα4::GAL4-VP16} transgenes also carry a w+m, both confer only mild eye coloration, so the strong red-eye phenotype of PBac{y+ w+m}(101F) is easily discerned.
Genetic distances, expressed here in centiMorgans (cM) rather than “map units,” as is traditionally used in Drosophila, were calculated using the equations of Stevens (1936). Crossover density was calculated by dividing the centiMorgans by the distance between markers (rounded to nearest 10 kb), using Drosophila melanogaster reference genome release 6.12 with transposable elements excluded, as described in Hatkevich et al. (2017). Including transposable elements in distances did not change any conclusions (see Table S4, a and b, in File S1).
The coefficient of coincidence (c) is calculated as where a and b are the number of single-crossover progeny in two intervals being compared, d is the number of double-crossover (DCO) progeny, and n is the total progeny scored. This is equivalent to observed DCOs divided by expected DCOs if the two intervals are independent (no interference). Interference (I) is 1 − c. Thus, I = 0 in the absence of interference and I = 1 if there is complete positive interference (no DCOs observed).
The centromere effect was quantified as in Hatkevich et al. (2017). The definition parallels that of I: CE = 1 − (O/E), where O is the number of crossovers observed and E is the number expected based on the average crossover density across the entire region assayed. CE therefore describes the deviation in crossover density in any interval from the mean density across all intervals.
For crossover assurance, we obtained the expected number of meioses in which a given region of the genome (X or net–cn) had no crossovers (E0) from the Poisson distribution, using mean number of crossovers in that region as the average rate of success. To convert observed crossover classes (parental, single, double, and triple crossover) to bivalent exchange classes (E0, E1, E2, and E3) we used the method of Weinstein (1936). This method accounts for the fact that an E1 “tetrad” gives two crossover chromatids and two parental chromatids, so the probability of recovering the crossover in the progeny is 0.5. Weinstein tested models with and without sister chromatid exchange and with and without chromatid interference (i.e., whether the chromatids involved in the two crossovers of a DCO are independent of one another). We used the model that he found to be the best fit to two large Drosophila data sets: no sister chromatid exchange and no chromatid interference.
X nondisjunction was scored by crossing virgin mutant females of the appropriate genotypes to ysccvvgf/Dp(1:Y)BS males. Exceptional progeny for X nondisjunction events originate from diplo-X and nullo-X ova, resulting in XXY (bar-eyed females) and XO (wild-eyed males) progeny, respectively. Numbers of exceptional progeny were doubled to account for those that do not survive to adulthood (XXX and YO).
Statistical analyses
For cM and c (and therefore I), 95% confidence intervals were calculated as where V(x) is the variance of parameter x. and (Stevens 1936). For nondisjunction, 95% confidence intervals and comparisons of rates across genotypes followed the statistical methods developed by Zeng et al. (2010).
For statistical analyses of interference, we conducted χ2 tests on two-by-two contingency tables of observed and expected DCOs for each genotype. A two-by-two table is appropriate for counts of events that are positive integer values and for which there is an expectation under the null hypothesis that mutant and wild type have the same levels of interference given their levels of recombination. This expected number of DCOs is derived by applying a model of the frequency of DCOs under no interference. Since the data do not have covariates or repeated measures, a χ2 test is the most straightforward. We applied Yates’ continuity correction because of low counts in some categories. χ2 tests were conducted using the GraphPad QuickCalcs online tool (https://www.graphpad.com/quickcalcs/contingency1.cfm).
For within-genotype analyses of interference, contingency tables were constructed in two ways. In the first method, columns had numbers with and without crossovers in the first interval, and rows had numbers with and without crossovers in the second interval. These P-values are reported in Figure 2. In the second method, columns had number of progeny with and without a DCO in the intervals being compared, and rows were observed and expected. Although P-values varied slightly between the two methods, none of the differences affected interpretations concerning statistical significance. For between-genotype comparisons, columns had observed and expected DCOs and rows had wild-type and mutant flies.
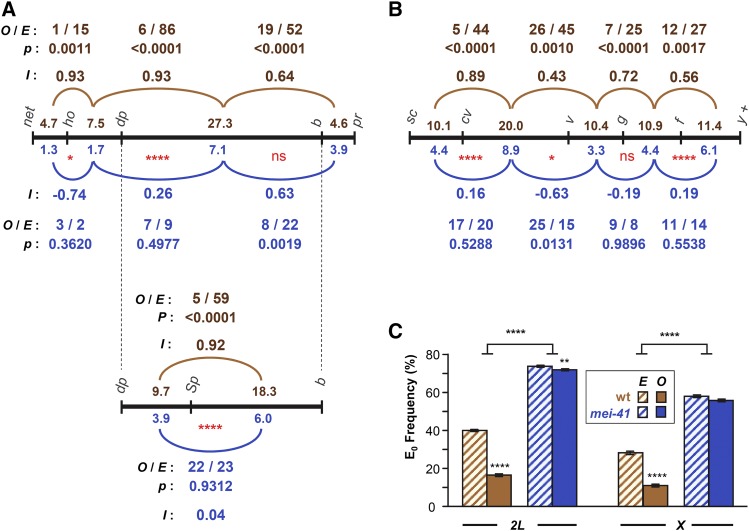
Figure 2.
Interference and assurance in mei-41 null mutants. (A) Interference on chromosome 2L. Black line represents genetic map of markers used, with size of each interval (in centiMorgan) listed above the line for wild-type females and below the line for mei-41 mutants. The pr–cn interval was omitted because it spans the centromere and because of low numbers of DCOs between this and the adjacent interval. Arcs represent pairs of adjacent intervals in which interference was assessed. Above (for wild type, brown) or below (mei-41, blue) each arc is listed the number of observed DCOs (O) and the number expected (E) if the two intervals are independent (no interference). P-values indicate the probability of observing these data under the null hypothesis, which is that the two intervals are independent (see Materials and Methods). Stevens’ interference (I), which equals 1 − (O/E), is also given. Red * below map lines are from Chi-squared analysis comparing O and E between wild type and mutant (see Materials and Methods). In a separate experiment, the large dp–b interval was further divided by the addition of Sp (wgSp-1). (B) Similar analysis of interference on the X chromosome. The f–y+ region spans the centromere, but since the marker on the right arm (y+) is hemizygous (i.e., a duplication of the tip of chromosome XL onto XR on one homolog), all crossovers must be to the left of the centromere. (C) Crossover assurance assessed by comparing frequencies of E0 bivalents. Expected E0 frequency is based on Poisson distribution from the average number of crossovers per meiosis; observed frequencies were calculated using the method of Weinstein (1936). Statistical significance between expected and observed E0 frequencies determined via Chi-square tests. Bars show 95% confidence intervals. Sample sizes for chromosomes 2L and X are given in Figure 1. For the dp–Sp–b experiment, n = 3325 flies, 928 crossovers for wild-type; n = 9740 flies, 972 crossovers for mutants. P-values reported were not corrected for multiple comparisons. Nonsignificance (ns) is for P > 0.05. * P < 0.05, ** P < 0.01, **** P < 0.0001.
A similar argument holds for assurance and CE. For assurance, columns were E0 and E>0, and rows had observed and expected counts. For CE, within-genotype comparisons had columns for with and without a crossover in the proximal interval and rows were observed and expected. For between-genotype comparisons, columns were observed and expected and rows were wild-type and mutant genotypes.
Data availability
All data necessary for confirming the conclusions presented in the article are presented within the article and the supplemental tables in File S1. Drosophila stocks are available upon request.
Results
Postgermarium expression of Mei-41 rescues embryonic lethality and creates a meiotic recombination null
Drosophila females homozygous for null mutations in mei-41 produce no viable progeny due to a requirement for maternally deposited Mei-41 at the midblastula transition (Sibon et al. 1999). Blm null mutants also exhibit maternal-effect embryonic lethality (McVey et al. 2007). To study meiotic recombination in Blm null mutants, Kohl et al. (2012) expressed wild-type Blm under indirect control of the α-tubulin 67C (matα) promoter via the Gal4>UASp system. This promoter is specific to the female germline, with expression initiating in the early vitellarium (Sanghavi et al. 2013), by which time recombination should be compete. In support of this expectation, crossover and nondisjunction assays on the occasional surviving progeny of Blm mutant females give similar results to those from embryos rescued by expressing UASp::Blm with the matα4::GAL4-VP16 driver in Blm null mothers (McVey et al. 2007; Kohl et al. 2012; Hatkevich et al. 2017).
We used the same system to overcome the maternal-effect inviability of embryos from mei-4129D homozygous null females (see Materials and Methods). To quantify the extent of maternal M{UASp::mei-41} rescue, we compared hatch rates of embryos from wild-type, mei-4129D, and P{UASp::mei-41} mei-4129D with and without P{matα4::GAL4-VP16} (Table 1). Embryos from females homozygous for mei-4129D with or without M{UASp::mei-41} but lacking P{matα4::GAL4-VP16} did not survive to hatching, whereas embryos from females with both components of the Gal4>UASp rescue system had a hatch rate of 52.8%. Most or all of the residual lethality is likely due to aneuploidy resulting from high nondisjunction in mei-41 mutants (13.6% X nondisjunction among progeny surviving to adulthood; Table S5 in File S1). Larvae that did hatch survived to adulthood, allowing for analysis of the crossover patterning landscape in a mei-41 null mutant. For simplicity, flies carrying this transgene system are denoted below as mei-4129D or mei-41 null mutants.
Table 1. Hatch rates for embryos from mei-41 mutants.
| Maternal genotype | Hatched (%) | Total (n) |
|---|---|---|
| Wild type | 73.1a | 2035 |
| mei-4129D | 0 | 527 |
| P{UASp::mei-41} mei-4129D | 0 | 837 |
| P{UASp::mei-41} mei-4129D; P{matα4::GAL4-VP16}/+ | 52.8b | 1187 |
This number is lower than expected for wild type. The cause of this is unknown.
The apparent lack of complete rescue may be the result of a high frequency of aneuploidy resulting from the absence of mei-41 during meiotic recombination.
Crossover reduction in mei-41 null mutants
Drosophila mei-41 was initially characterized as a meiotic mutant by Baker and Carpenter (1972). Hypomorphic mei-41 alleles resulted in an overall 46% decrease in crossovers relative to wild-type controls, and was measured in five adjacent intervals spanning the entirety of chromosome 2L and proximal 2R (∼20% of the euchromatic genome). We measured crossovers in this same region in mei-41 null mutant females and found a significantly more severe reduction of 67% (P < 0.0001; Figure 1, A and B). Given the many functions of Mei-41 in mitotically proliferating cells, we wanted to determine whether the remaining crossovers were meiotic or whether they possibly resulted from DNA damage within the premeiotic germline. As Mei-P22 is required to generate meiotic DSBs (Liu et al. 2002; Robert et al. 2016), any crossovers that are independent of Mei-P22 most likely result from damage occurring in premeiotic mitotic cell cycles or premeiotic S phase. Crossovers were completely abolished in mei-4129D; mei-P22103 double mutants (n = 1754). One vial had two female progeny that were mutant for all markers on the net–cn chromosome except pr. These may have arisen from a DCO in the adjacent b–pr and pr–cn regions, gene conversion of the pr mutation, or reversion of this mutation (an insertion of a 412 transposable element). Since these were in the same vial, they likely represent a single premeiotic event. We conclude that the vast majority of crossovers observed in the mei-41 null mutant females are meiotic in origin.
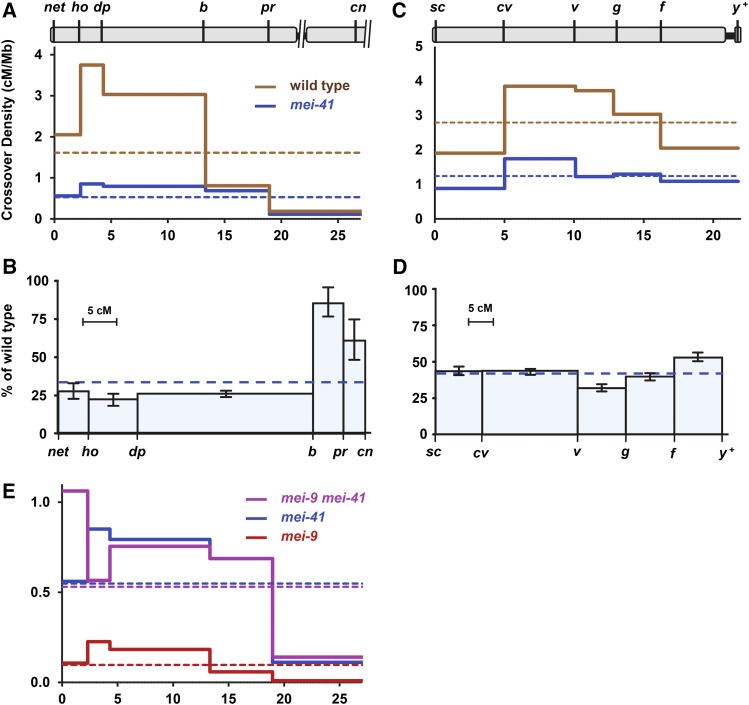
Figure 1.
Reduction of crossing over in mei-41 null mutants. (A and B) Crossover distribution on chromosomes 2L (A) and X (B) in mei-4129D mutants compared to wild type. Marker location indicated at top based on genome assembly position (megabase), excluding the centromere, unassembled pericentromeric satellite sequences, and transposable elements. Crossover density (solid lines) was determined for wild-type and mei-41 mutant females. Dotted lines show mean crossover density across the entire region. (C and D) Crossing over on chromosomes 2L (C) and X (D) in mei-4129D mutants as a percentage of wild type. The x-axis is scaled to genetic distance (centiMorgan) in wild-type females. Bars are 95% confidence intervals. (E) Crossover density in mei-9 and mei-41 single and double mutants. Note scale difference compared to (A). Wild-type chromosome 2L: n = 4222 progeny, 1943 crossovers. mei-41 chromosome 2L: n = 7801 progeny, 1175 crossovers. Wild-type X: n = 2179 progeny, 1367 crossovers. mei-41 X: n = 5174 progeny, 1396 crossovers. mei-9: n = 2433 progeny, 67 crossovers. mei-9mei-41: n = 1059 progeny, 165 crossovers. Wild-type and mei-9 single mutant data are from Hatkevich et al. (2017), used with permission. Full data sets are in Tables S1 and S2 in File S1. cM and cM/Mb, with 95% confidence intervals, are in Table S3 in File S1.
Baker and Carpenter (1972) described crossover reduction in mei-41 hypomorphic mutants as polar, with a more severe decrease in medial and distal regions of the chromosome than in proximal regions. This is also true in our null mutant: Although crossovers are significantly reduced in every interval, the average reduction in the three distal intervals is 75%, while in the two proximal intervals the decrease averages only 16% (Figure 1B). We also assayed crossing over across the entire X chromosome. Crossovers were reduced by an average of 57% on this chromosome; notably, the decrease was uniform across the entire chromosome, with no apparent polar effect (Figure 1, C and D).
One hypothesis to explain the polar effect on recombination on chromosome 2L is that there are region-specific requirements for Mei-41, with the protein being less important in proximal 2L. We tested this hypothesis by assessing the dependence of crossovers on Mei-9, the catalytic subunit of the putative meiotic resolvase (Sekelsky et al. 1995). Meiotic crossovers are reduced by ∼90% in mei-9 mutants, suggesting that most or all crossovers generated in wild-type flies require Mei-9 (Baker and Carpenter 1972; Sekelsky et al. 1995). However, in many mutants that affect meiotic recombination, including Blm, mei-218, and rec, crossovers are independent of Mei-9 (Sekelsky et al. 1995; Blanton et al. 2005; Hatkevich et al. 2017). Our interpretation is that when the meiotic crossover pathway is blocked because of loss of a critical component, repair is completed by alternative pathways that are independent of Mei-9 and other downstream meiotic recombination proteins. If Mei-41 is less important in proximal chromosome 2L, then crossovers in these regions may remain dependent on Mei-9. We scored crossovers along chromosome 2L in mei-9a mei-4129D double mutants (Figure 1E; we did not score the X chromosome because of the difficulty of recombining the mei-9 and mei-41 mutations and the UASp::mei-41 transgene onto the multiple-marked chromosome, and because the requirement for Mei-41 appeared to be similar across the X). The total genetic map length was similar between mei-9mei-41 double mutants and mei-41 single mutants (15.58 cM vs. 15.06 cM; P = 0.6679 by χ2 test comparing total crossovers and number of progeny scored), but significantly greater than that of mei-9 mutants (2.8 cM; P < 0.0001). There was no apparent difference in requirement for Mei-9 between the proximal and distal intervals. We conclude that all crossovers generated in mei-41 mutants are independent of mei-9, regardless of chromosomal location. This suggests that loss of Mei-41 disrupts progression through the meiotic crossover pathway at all sites along the chromosome.
The apparent polar effect on crossing over in mei-41 mutants can be explained by retention of the centromere effect
Compared to wild-type crossing over, the effects of loss of Mei-41 on meiotic crossing over is puzzling, as there seem to be substantially stronger effects in some regions of the genome than others, yet all crossovers in the mutant are independent of Mei-9. The conclusion that there is a polar effect on crossing over is based on comparing crossover frequencies in the mutant to those in wild-type females. Insight can also be gleaned by analyzing crossover distribution in the mutant in isolation. For example, Hatkevich et al. (2017) noted an apparently flat distribution of crossover in mutants lacking the Blm helicase. Their interpretation was that all crossover patterning is lost in Blm mutants, resulting in a distribution that reflects the DSB distribution.
Crossover distribution in mei-41 mutants does not mimic that of Blm mutants, at least in proximal chromosome 2L, suggesting that crossover patterning is not entirely lost in mei-41 mutants. In wild-type flies, crossover density is substantially lower in the pr–cn interval than in any of the other nine intervals that we assayed (0.11 ± 0.03 cM/Mb vs. 0.56 ± 0.09 cM/Mb in the next lowest interval, net–ho). The pr–cn interval is noteworthy because it spans the centromere, so recombination is strongly influenced by the centromere effect. To determine whether this phenomenon is affected by loss of Mei-41, we calculated CE as a measure of the centromere effect (Hatkevich et al. 2017). CE quantifies the difference in crossover density in a given interval relative to mean crossover density. It is thought that the significant decrease in the pr–cn interval is primarily due to the centromere effect. In wild-type females, if crossover density in the pr–cn interval were equal to the mean density across the entire region assayed, 643 crossovers would be expected; only 73 were observed (P < 0.0001), giving a CE value of 0.89. In mei-4129D mutants, 390 were expected but only 82 were observed (P < 0.0001), yielding a CE value of 0.79. This high value of CE is consistent with most or all of the centromere effect being intact in mei-4129D mutants, although the significant difference between mei-4129D and wild-type females (P = 0.0004) suggests that there may be mild amelioration in mei-41 mutants.
The decrease in crossing over on the X chromosome does not appear to be polar (Figure 1, C and D). The pericentric heterochromatin of the X chromosome spans ∼19 Mb, compared to ∼5 Mb on chromosome 2L and 7 Mb on 2R. This results in a much weaker centromere effect in the most proximal euchromatin of the X (Yamamoto and Miklos 1978). Thus, the lack of a polar decrease in crossing over on the X in mei-41 null mutants may be because the entire region being analyzed is, with respect to distance from the centromere, equivalent to the distal half of chromosome 2L.
The small chromosome 4 of D. melanogaster never has meiotic crossovers in wild-type females (reviewed in Hartmann and Sekelsky 2017), but does have crossovers in Blm mutants (Hatkevich et al. 2017). Hatkevich et al. argued that the absence of crossovers on chromosome 4 is due in large part to the centromere effect (∼4 Mb of heterochromatic satellite sequence between the centromere and the gene-containing region), and that it is loss of the centromere effect that permits crossing over on chromosome 4 in Blm mutants. We measured crossing over on chromosome 4 in mei-41 mutants. We recovered no crossovers between markers at opposite ends of the gene-containing region of chromosome 4 (n = 5555; P < 0.0001 compared to Blm), consistent with our interpretation that the centromere effect is not lost in mei-41 mutants.
Effects of loss of Mei-41 on crossover interference and assurance
Given the apparent retention of the centromere effect on crossing over, we asked whether the crossover patterning phenomena interference and assurance are affected by loss of Mei-41. We calculated interference (I) using the method of Stevens (1936). Stevens defined I as 1 − (O/E), where O is the number of DCOs observed and E is the number of DCO expected if the two intervals are independent of one another (see Materials and Methods). Thus, I = 1 indicates complete positive interference (no DCOs observed) and I = 0 indicates no interference (the two intervals are independent of one another).
Values for O, E, and I are given in Figure 2, A and B. On chromosome 2L, the only pairs of adjacent intervals that have enough DCOs to analyze interference are ho–dp/dp–b and dp–b/b–pr. In wild-type females, I was 0.93 ± 0.05 between the first pair and 0.64 ± 0.15 between the second pair (Figure 2A). In mei-41 mutants, we did not detect significant interference between the first pair (I = 0.26 ± 0.52, P = 0.4977), indicating a significant difference from the wild type (P < 0.0001). However, we did detect interference between the second pair of intervals in the mei-41 mutant (I = 0.63 ± 0.25, P = 0.0019), suggesting that interference is intact in this region (P = 0.9922 compared to the wild type). The dp–b interval is typically not used for measuring interference because of its large size (>27 cM in wild-type flies). Therefore, we reexamined interference within this region by subdividing it with another marker, wgSp-1. Again, interference was strong in wild-type females (I = 0.92 ± 0.07) but absent from mei-41 mutants (I = 0.04 ± 0.38; Figure 2A, bottom). Using the same analysis of interference across the X chromosome, we found significant positive interference between every pair of adjacent intervals in wild-type females, but no detectable interference in mei-41 mutants (Figure 2B; the P-value for cv–v and v–g is 0.0131, which would be considered significant, but this is because there were significantly more DCOs observed than expected, resulting in a negative value for I).
We used one additional method to assess the distribution of crossovers relative to one another. In many species, crossovers are distributed among bivalents such that the probability that any pair of homologous chromosomes does not receive a crossover is significantly lower than expected by chance, a phenomenon known as crossover assurance. It has been proposed that if there are sufficient well-spaced, crossover-eligible intermediates, then coupling interference with a mechanism to achieve a specific number of crossovers per meiosis (within a narrow range) will produce crossover assurance (Zhang et al. 2014; Wang et al. 2015). In this model, assurance is merely an outcome of interference and homeostasis.
Extrapolating from our measurements of crossovers on chromosomes X and 2L, we estimate approximately two crossovers per meiosis in mei-41 mutants. True assurance requires a minimum of three or five crossovers (one per major chromosome or arm, excluding chromosome 4); however, assurance among the residual crossovers could manifest as the two crossovers being on different chromosomes (or chromosome arms) more often than expected by chance. We compared the expected and observed frequency of meioses in which there were no crossovers (E0, for zero-exchange bivalent, frequency) on the X chromosome or on chromosome 2L. For expected E0 frequency we used the Poisson distribution expectation based on the average number of crossovers per meiosis. We used the method of Weinstein (1936) to transform counts of progeny that inherited parental, single crossover, DCO, etc., chromatids to observed bivalent exchange classes (see Materials and Methods). In wild-type flies, the expected E0 frequency for the X chromosome is 0.285, but the observed frequency was 0.112 (Figure 2C). This demonstrates crossover assurance that is significant (P < 0.0001) but incomplete (11% of meioses have no crossovers between the X chromosomes), as has been observed in previous studies (e.g., Weinstein 1936; Koehler et al. 1996). In mei-41 mutants, reduced crossing over results in a higher expected E0 frequency (0.582), but unlike the case in wild-type flies, the observed frequency (0.572) was not significantly different (P = 0.3008). Similar results were obtained with the chromosome 2L data (Figure 2C). For 2L, the difference between observed and expected in mei-41 mutants was significant (P = 0.0046), but given the small magnitude of the difference (0.740 expected, 0.720 observed) this may not be biologically meaningful.
Together, our data indicate that interference and assurance are significantly decreased or lost in mei-41 mutants, though it is possible that crossovers in proximal chromosome 2L retain interference.
Discussion
We have demonstrated that the Gal4>UASp rescue successfully overcomes maternal-effect embryonic lethality of mei-41 mutants, allowing us to perform meiotic crossover patterning analysis in mei-41 null mutants. The crossover reduction in null mutants is more severe than that previously reported for hypomophic mutants (Baker and Carpenter 1972), but the nonuniform reduction in crossing over on chromosome 2 is still present (Figure 1B).
We considered the hypothesis that the polar effect stems from differential requirement for Mei-41 in proximal and distal regions of the chromosome. However, in mei-41 mutants, crossovers in all regions are independent of the presumptive resolvase Mei-9 (Figure 1C). Our interpretation is that this reveals an essential role for Mei-41 in carrying out meiotic recombination throughout the genome. In the absence of Mei-41, the meiotic pathway is disrupted and repair is completed by alternative pathways that neither require functions specific to the meiotic pathway nor result in properties normally associated with meiotic recombination, such as crossover patterning.
Since the apparent polar effect is observed on chromosome 2 but not on the X, we hypothesized that the centromere effect is retained in mei-41 mutants. We calculated CE, a measure of how much crossover density in an interval deviates from the mean crossover density (Hatkevich et al. 2017), to compare the centromere effect between wild-type females and mei-41 mutants. Although every interval deviates significantly from the mean in wild-type flies, the very strong deviation in the pr–cn region (CE = 0.89) is probably due primarily to the suppression of crossovers associated with proximity to the centromere. Direct confirmation of the presence of a centromere effect requires moving the sequences to be analyzed away from the centromere through chromosome rearrangement, a difficult experiment because of the need to have structural homozygosity combined with heterozygosity for markers. Nonetheless, our data suggest that a strong centromere effect is retained in the absence of Mei-41.
In contrast to the absence of a strong impact on the centromere effect, our analysis suggests that interference and assurance are significantly disrupted when Mei-41 is absent. We did not detect any significant interference across the X chromosome (Figure 2B). On chromosome 2L, interference was lost between dp and b (when divided into two intervals), but was retained between this interval and b–pr (Figure 2A). It is notable that the only interval that appears to retain interference is b–pr, which is the closest interval to the centromere. This could indicate that Mei-41 does have different functions in proximal regions than in other parts of the genome. However, given that there appears to be no interference within the adjacent dp–b interval, the presence of interference between these intervals would require that crossover-eligible intermediates between dp and b should be subject to interference exerted by crossover designations in the proximal interval, while at the same time any crossovers designated between dp and b should not signal interference themselves. This seems unlikely, and perhaps indicates that there is some other effect or some idiosyncrasy associated with the b–pr interval.
Another argument regarding the reduced or absent interference in mei-41 mutants is that the strength of interference is inversely proportional to the genetic size of the interval in which it is measured. Therefore, since genetic intervals become shorter in mei-41 mutants, interference might be expected to become stronger. This expectation would not hold for the recombination proteins required to generate crossovers after interference has occurred, such as the proteins that resolve crossover-designated intermediates into crossovers [the “crossover maturation” step in the models of Zhang et al. (2014)]. Mei-9 and associated proteins are thought to be required for resolution (Baker and Carpenter 1972; Sekelsky et al. 1995; Yıldız et al. 2002). It is not meaningful to discuss interference in mei-9 mutants, since the number of crossovers per meiosis is well below one (0.06), but the uniform decrease in crossovers across chromosome 2L led Baker and Carpenter (1972) to conclude that Mei-41 acts earlier in crossover generation than Mei-9.
We believe the most parsimonious interpretation of our data is that loss of Mei-41 has little or no impact on the centromere effect but reduces or eliminates interference and assurance. We propose that crossover patterning in Drosophila occurs in a stepwise manner (Figure 3). Analysis of noncrossover gene conversion events mapped through whole-genome sequencing suggests that DSBs are, at a large scale, spread evenly throughout the assembled genome (Comeron et al. 2012; Miller et al. 2016; Hatkevich et al. 2017). The centromere effect is applied early by making some intermediates ineligible to enter the crossover pathway, with the probability of being affected in this way being related to distance to the centromere (Figure 3B). Subsequently, when any remaining crossover-eligible intermediate becomes designated for crossing over, interference precludes nearby intermediates from also adopting this fate (Figure 3C).
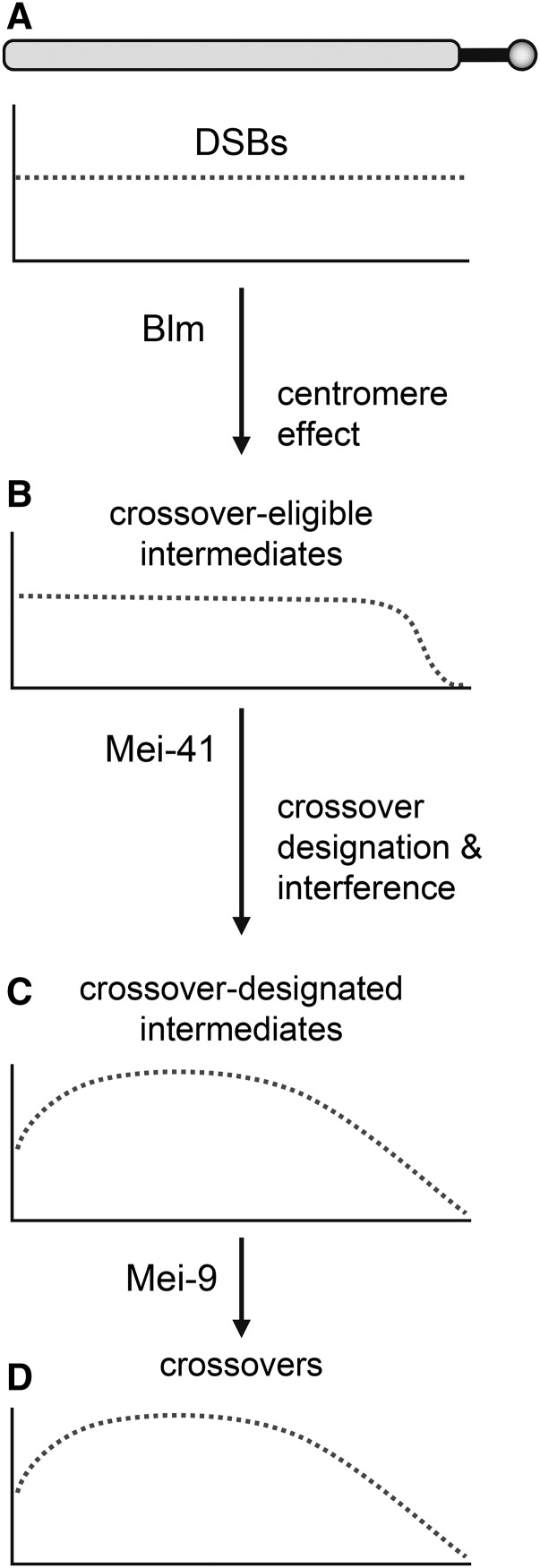
Figure 3.
Model for progressive enforcement of crossover patterning. The drawing at the top represents a chromosome arm. Thick solid line is pericentric satellite DNA, circle is centromere. (A) Based on whole-genome sequencing, the initial DSB distribution (dotted line) is flat at large scales (DSBs are excluded from the heterochromatic satellite DNA). The centromere effect revokes the eligibility of intermediates near the centromere to become crossovers. (B) This results in a distribution of crossover-eligible intermediates that is flat across much of the arm, then tailed as the centromere is approached. The shape of the tailing is unknown; a sigmoidal drop is shown here for illustrative purposes. Later, some intermediates are designated to become crossovers, and the resultant interference discourages other intermediates over large distances from achieving crossover designation. The resultant distribution of crossover-designated intermediates (C) and crossovers (D) is approximately skew normal, with the degree of skew being proportional to the length of satellite sequence that separates DSB-competent regions from the centromere. Blm, Mei-41, and Mei-9 are essential at different times in the crossover pathway, so crossovers generated in mutants that lack these proteins are made outside the normal meiotic pathway, and are therefore Mei-9 independent, and are either unpatterned (Blm mutants, distribution similar to A), are partially patterned (mei-41 mutant, resembles B), or are fully patterned (mei-9 mutant, resembles C and D).
Given a uniform distribution of DSBs and the fact that each of the chromosome arms in Drosophila has ∼1.0–1.3 crossovers per meiosis, interference alone will produce a crossover density that resembles a normal distribution (e.g., simulations in Zhang et al. 2014). The combination of a strong centromere effect and interference will yield a crossover density map that approximates a skew normal distribution (Figure 3D). Crossover distribution maps in Drosophila do resemble skew normal distributions, with much more skew on the major autosome arms than on the X chromosome, which also lacks a strong centromere effect (see figure S2 in Comeron et al. 2012).
Blm helicase has been proposed to have an essential function early in the meiotic recombination pathway (reviewed in Hatkevich and Sekelsky 2017). Loss of Blm results in an early exit from the meiotic pathway and completion of repair by alternative mechanisms. Since these alternative mechanisms do not involve patterning, the probability of becoming a crossover is the same for each intermediate, resulting in crossovers being evenly distributed across each chromosome arm. We propose that Mei-41 has some critical function after the centromere effect has been at least partially established. Loss of Mei-41 leads to exit from the meiotic pathway at this point. As with Blm mutants, every remaining intermediate has the same probability of becoming a crossover, so the crossover distribution in mei-41 mutants is similar to that in Figure 3B. Mei-9 is required only for maturation of crossover-designated intermediates into crossovers. Since this occurs after crossover designation, residual crossovers in a mei-9 mutant are patterned like crossovers in wild-type flies; but there are far fewer crossovers because most intermediates that had been designated to become crossovers are instead processed into noncrossover products.
In many model organisms, a subset of crossovers do not participate in interference and are generated by different resolvases than those that generate interfering crossovers (reviewed in Kohl and Sekelsky 2013). These “class II” crossovers are sometimes defined as lacking interference or being unpatterned. In the model discussed above, this distinction is not always appropriate, at least in mutant situations. Rather, crossovers generated outside of the primary pathway may be unpatterned (as in Blm mutants), partially patterned (as in mei-41 mutants), or patterned (as in mei-9 mutants). The only features that these crossovers have in common is that they are generated outside of the normal meiotic crossover pathway, presumably through general DSB repair pathways that act to ensure there are no unrepaired DNA structures persisting until the meiotic divisions begin.
Our data provide little insight into the molecular function of Mei-41 in the meiotic DSB repair pathway. In mitotic DSB repair, mei-41 mutants have no observable defects in the early steps of homologous repair of DSBs (e.g., resection, strand invasion, and repair synthesis), but Mei-41 appears to be required for the annealing or ligation steps of synthesis-dependent strand annealing (LaRocque et al. 2007). Korda Holsclaw and Sekelsky (2017) hypothesized that Mei-41 activates Marcal1, which then catalyzes annealing of complementary sequences. Synthesis-dependent strand annealing promotes formation of noncrossover products, in contrast to the apparent role for Mei-41 in promoting crossovers during meiotic recombination. Nonetheless, Mei-41 might have similar functions in mitotic and meiotic DSB repair if, in the latter, it activates a protein that catalyzes the annealing required for second-end capture, a process that might occur after an early requirement for Blm helicase but prior to crossover designation (e.g., models in Crown et al. 2014). Future studies to elucidate the role for Mei-41 might provide additional insights into meiotic crossover pathways and patterning.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300634/-/DC1.
Acknowledgments
We thank Nicole Crown, Michaelyn Hartmann, and Talia Hatkevich for comments on the manuscript and Corbin Jones and Nadia Singh for advice on statistical analyses. This work was supported by a grant from the National Institute of General Medical Sciences to J.S. under award 1R35 GM-118127.
Footnotes
Communicating editor: J. Birchler
Literature Cited
- Abdu U., Brodsky M., Schüpbach T., 2002. Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of Gurken through Chk2/Mnk. Curr. Biol. 12: 1645–1651. [DOI] [PubMed] [Google Scholar]
- Baker B. S., Carpenter A. T. C., 1972. Genetic analysis of sex chromosomal meiotic mutants in Drosophila melanogaster. Genetics 71: 255–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G. W., 1932. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc. Natl. Acad. Sci. USA 18: 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz L. E., Copenhaver G. P., 2010. Genetic interference: don’t stand so close to me. Curr. Genomics 11: 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton H. L., Radford S. J., McMahan S., Kearney H. M., Ibrahim J. G., et al. , 2005. REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genet. 1: e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown K. N., McMahan S., Sekelsky J., 2014. Eliminating both canonical and short-patch mismatch repair in Drosophila melanogaster suggests a new meiotic recombination model. PLoS Genet. 10: e1004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial A., Schüpbach T., 1999. Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nat. Cell Biol. 1: 354–357. [DOI] [PubMed] [Google Scholar]
- Hari K. L., Santerre A., Sekelsky J., McKim K. S., Boyd J. B., et al. , 1995. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell 82: 815–821. [DOI] [PubMed] [Google Scholar]
- Hartmann M. A., Sekelsky J., 2017. The absence of crossovers on chromosome 4 in Drosophila melanogaster: Imperfection or interesting exception? Fly (Austin) 11: 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatkevich T., Sekelsky J., 2017. Bloom syndrome helicase in meiosis: pro-crossover functions of an anti-crossover protein. Bioessays 39. DOI: 10.1002/bies.201700073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatkevich T., Kohl K. P., McMahan S., Hartmann M. A., Williams A. M., et al. , 2017. Bloom syndrome helicase promotes meiotic crossover patterning and homolog disjunction. Curr. Biol. 27: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce E. F., Pedersen M., Tiong S., White-Brown S. K., Paul A., et al. , 2011. Drosophila ATM and ATR have distinct activities in the regulation of meiotic DNA damage and repair. J. Cell Biol. 195: 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler K. E., Boulton C. L., Collins H. E., French R. L., Herman K. C., et al. , 1996. Spontaneous X chromosome MI and MII nondisjunction events in Drosophila melanogaster oocytes have different recombinational histories. Nat. Genet. 14: 406–414. [DOI] [PubMed] [Google Scholar]
- Kohl K. P., Sekelsky J., 2013. Meiotic and mitotic recombination in meiosis. Genetics 194: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl K. P., Jones C. D., Sekelsky J., 2012. Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science 338: 1363–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korda Holsclaw J., Sekelsky J., 2017. Annealing of complementary DNA sequences during double-strand break repair in Drosophila is mediated by the ortholog of SMARCAL1. Genetics 206: 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb N. E., Freeman S. B., Savage-Austin A., Pettay D., Taft L., et al. , 1996. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat. Genet. 14: 400–405. [DOI] [PubMed] [Google Scholar]
- LaRocque J. R., Jaklevic B., Su T. T., Sekelsky J., 2007. Drosophila ATR in double-strand break repair. Genetics 175: 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurençon A., Purdy A., Sekelsky J., Hawley R. S., Su T. T., 2003. Phenotypic analysis of separation-of-function alleles of MEI-41, Drosophila ATM/ATR. Genetics 164: 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Jang J. K., Kato N., McKim K. S., 2002. Mei-P22 encodes a chromosome-associated protein required for the initiation of meiotic recombination in Drosophila melanogaster. Genetics 162: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E., Diaz R. L., Hunter N., Keeney S., 2006. Crossover homeostasis in yeast meiosis. Cell 126: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather K., 1937. The determination of position in crossing-over. II. The chromosome length-chiasma frequency relation. Cytologia Fujii Jubilee Volume: 514–526. [Google Scholar]
- Mather K., 1939. Crossing over and heterochromatin in the X chromosome of Drosophila melanogaster. Genetics 24: 413–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Andersen S. L., Broze Y., Sekelsky J., 2007. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics 176: 1979–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S., McKim K. S., 2006. Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet. 2: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. E., Smith C. B., Yeganeh Kazemi N., Cockrell A. J., Arvanitakis A. V., et al. , 2016. Whole-genome analysis of individual meiotic events in Drosophila melanogaster reveals that noncrossover gene conversions are insensitive to interference and the centromere effect. Genetics 203: 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., Painter T. S., 1932. The differentiation of the sex chromosomes of Drosophila into genetically active and inert regions. Z. Induct. Abstammungs Vererbungsl. 62: 316–365. [Google Scholar]
- Owen A. R. G., 1949. A possible interpretation of the apparent interference across the centromere found by Callan and Montalenti in Culex pipiens. Heredity (Edinb) 3: 357–367. [DOI] [PubMed] [Google Scholar]
- Robert T., Nore A., Brun C., Maffre C., Crimi B., et al. , 2016. The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science 351: 943–949. [DOI] [PubMed] [Google Scholar]
- Sanghavi P., Laxani S., Li X., Bullock S. L., Gonsalvez G. B., 2013. Dynein associates with oskar mRNPs and is required for their efficient net plus-end localization in Drosophila oocytes. PLoS One 8: e80605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J. J., McKim K. S., Chin G. M., Hawley R. S., 1995. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics 141: 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibon O. C., Laurençon A., Hawley R., Theurkauf W. E., 1999. The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr. Biol. 9: 302–312. [DOI] [PubMed] [Google Scholar]
- Stevens W. L., 1936. The analysis of interference. J. Genet. 32: 51–64. [Google Scholar]
- Sturtevant A. H., 1913. The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J. Exp. Biol. 14: 43–59. [Google Scholar]
- Wang S., Zickler D., Kleckner N., Zhang L., 2015. Meiotic crossover patterns: obligatory crossover, interference and homeostasis in a single process. Cell Cycle 14: 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein A., 1936. The theory of multiple-strand crossing over. Genetics 21: 155–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Miklos G. L., 1978. Genetic studies on heterochromatin in Drosophila melanogaster and their implications for the functions of satellite DNA. Chromosoma 66: 71–98. [DOI] [PubMed] [Google Scholar]
- Yıldız Ö., Majumder S., Kramer B. C., Sekelsky J., 2002. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol. Cell 10: 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Li H., Schweppe N. M., Hawley R. S., Gilliland W. D., 2010. Statistical analysis of nondisjunction assays in Drosophila. Genetics 186: 505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Liang Z., Hutchinson J., Kleckner N., 2014. Crossover patterning by the beam-film model: analysis and implications. PLoS Genet. 10: e1004042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data necessary for confirming the conclusions presented in the article are presented within the article and the supplemental tables in File S1. Drosophila stocks are available upon request.