Racolta et al. show two different impacts of CLE peptide treatment of Arabidopsis roots. In all genotypes tested, they find increased proliferative....
Keywords: receptor-like kinase, CLE, root apical meristem, root development
Abstract
Cell–cell communication is essential for plants to integrate developmental programs with external cues that affect their growth. Recent advances in plant signaling have uncovered similar molecular mechanisms in shoot, root, and vascular meristem signaling that involve receptor-like kinases and small, secreted peptides. Here, we report that the receptor-like kinases TOAD2/RPK2 and RPK1 regulate root growth by controlling cell proliferation and affecting meristem size. Two types of developmental alterations were observed upon exogenous CLE peptide application. The first type was detected in all plants treated, and comprise increased proliferative activity of cells in the stem cell niche and a delay of progression in differentiation of daughter cells. The second type was changes specific to the genotypes that are sensitive to CLE-driven root meristem inhibition and include a large decrease in the occurrence of cell divisions in longitudinal files, correlating with shorter meristems and cessation of root growth. The root meristems of toad2/rpk2 mutant plants are insensitive to the inhibitory effect of CLE17 peptide treatment, consistent with TOAD2/RPK2 function as a receptor for CLE peptides. In addition, a strong reduction in the expression of RPK1 protein upon CLE treatment, dependent on TOAD2/RPK2, suggests that these two RLKs mediate CLE signaling in a common pathway to control root growth.
UNDERSTANDING the molecular mechanisms of the cell fate decisions of cells arising from an undifferentiated meristematic state is key for understanding plant development. Increasing experimental evidence, compiled from research in Arabidopsis thaliana and other plant systems, has uncovered complex networks of interacting hormones, small peptides, RNAs, transcription factors, receptors, and other molecules regulating the patterning of meristems (Stahl and Simon 2010; Azpeitia and Alvarez-Buylla 2012; Petricka et al. 2012). However, less is known about how plants perceive external and internal signals, and how receptor–ligand interactions translate into controlled downstream molecular steps that will ultimately generate precise patterns of growth.
The paradigm of signaling through plasma membrane receptors implies that ligands bind to the extracellular domain of receptors and a signaling cascade triggers changes in post-translational and transcriptional programs, modulating plant growth. In Arabidopsis, a large monophyletic family of >400 genes encoding receptor-like protein kinases (RLKs), with a predicted extracellular domain [containing Leucine-Rich Repeat motifs (LRRs) in more than half of these RLKs], a single-pass transmembrane domain, and a cytoplasmic serine/threonine/tyrosine kinase domain (Shiu and Bleecker, 2001a,b; Diévart and Clark 2004; Oh et al. 2009). Despite the large number of identified RLKs, the specific functions are known for only a fraction of them (<50). The functions ascribed to these RLKs thus far indicate that they play key signaling roles in regulating cell fate specification or maintenance, cell growth, cell death, and pathogen response (Diévart and Clark 2004), and that they bind a variety of ligand molecules ranging from steroid hormones to peptides and small secreted proteins (Torii 2008). In addition to directly binding ligands, some RLKs also function as regulatory components of other RLK complexes (Li 2010; Halter et al. 2014; Imkampe et al. 2017).
One well-characterized signaling pathway includes, LRR-RLK CLAVATA1 (CLV1), which functions to control the size of the shoot apical meristem (SAM) by binding to a small secreted peptide, CLAVATA3 (CLV3) (Ogawa et al. 2008); this ultimately restricts the expression domain of the homeodomain transcription factor WUSCHEL (WUS), which defines a stem cell’s fate (Schoof et al. 2000). In addition, the receptor protein kinase CORYNE (CRN), lacking an extracellular domain, and the receptor-like protein CLAVATA2 (CLV2), lacking an intracellular kinase domain, form a heteromeric receptor complex that also binds CLV3 and regulates WUS in a separate pathway that is independent of the CLV1 pathway (Müller et al. 2008; Guo et al. 2010). The common phenotypic read-out of defects in the CLV pathway includes an enlarged SAM and supernumerary floral and fruit organs (Clark et al. 1997; Schoof et al. 2000; Durbak and Tax 2011).
In addition to its well-established role in modulating the maintenance of the SAM, CLV1 was recently identified to play a similar role in the root apical meristem (RAM) (Stahl et al. 2013). The RAM, located at the tip of the root, contains a group of frequently dividing stem cells (initials) surrounding three or four centrally located cells with low mitotic activity, called the quiescent center (QC). This stem cell niche of the RAM is the source of all cells that arise in layers and form concentrically arranged files of cell types. CLV1 expression in cells distal to the QC (toward the root tip) overlaps that of a non-LRR receptor kinase, ARABIDOPSIS CRINKLY4 (ACR4), which was previously shown to regulate formative cell divisions in lateral roots (LRs) and to control the integrity of the epidermal cell layer (Gifford et al. 2003; De Smet et al. 2008). Root phenotypes caused by ACR4 mutations, similar to mutations in a CLV3 homolog CLAVATA3/ENDOSPERM SURROUNDING REGION-LIKE (ESR)40 (CLE40), include the expanded expression of a WUS-RELATED HOMEOBOX 5 (WOX5) and supernumerary meristematic columella initials (De Smet et al. 2008; Stahl et al. 2009). Application of exogenous CLE 40 results in transcriptional upregulation of ACR4 but not CLV1, and even though a direct binding between ACR4 and CLE 40 was not demonstrated, CLV1 has the potential to directly bind CLE40 (Guo et al. 2010). These findings further demonstrate the importance of receptor complexes containing different receptors in modulating intricate signaling responses triggered by peptides in the CLV3 family.
Intercellular communication through small regulatory peptides, as described above, emerges as a key component of developmental programs (Fukuda and Higashiyama 2011; Delay et al. 2013). The regulatory peptides encoded by the CLE (CLAVATA3/ESR-related) family of 32 genes in Arabidopsis have been implicated not only in meristem maintenance, but also in a variety of developmental processes such as LR development, gravitopism, and protoxylem differentiation (Kiyohara and Sawa 2012; Qiang et al. 2013). The CLE proteins have a conserved 12–14-amino acid CLE motif at or near the C-terminus (Cock and McCormick 2001), and are proteolytically processed and further modified (Ni et al. 2011; Tamaki et al. 2013) to generate extracellular signaling molecules. Based on the response triggered by overexpression or exogenous treatment of Arabidopsis seedlings, CLE peptides are classified as type A and B CLE peptides. Treatment of roots with type A CLE peptides can induce early termination of meristem activity and cessation of growth (Fiers et al. 2005; Ito et al. 2006; Whitford et al. 2008), while type B CLE peptides can suppress tracheary element differentiation (Ito et al. 2006; Kinoshita et al. 2007; Hirakawa et al. 2008) but do not have an effect on root length. Overexpression and exogenous application experiments are used (Strabala et al. 2006; Kinoshita et al. 2007; Jun et al. 2010), largely due to the absence of visible phenotypes of CLE mutants (Jun et al. 2010). While functional assays clearly suggest that CLE peptides can signal through RLKs to regulate meristem size and activity in both SAM and RAM, few direct interactions have been demonstrated (Matsubayashi et al. 2002; Hirakawa et al. 2008; Ogawa et al. 2008).
TOADSTOOL 2 (TOAD2/RPK2, also called RECEPTOR-LIKE PROTEIN KINASE 2-RPK2, but for brevity hereafter we will use TOAD2) is another RLK that functions downstream of CLV3 in the regulation of SAM size (Kinoshita et al. 2010; Betsuyaku et al. 2011b). The mutant phenotypes of TOAD2 include the increased size of the SAM observed in CLV1 and CLV2 mutants, and those phenotypes are additive in higher-order mutants containing toad2, clv1, and clv2 (Kinoshita et al. 2010), suggesting that they may act in parallel pathways. In addition, a partial insensitivity to the effect of the CLV3-induced short root length (S) phenotype was reported for toad2 mutants, similar to crn and clv2. Unlike CRN (Müller et al. 2008), the overexpression of TOAD2 resembles the phenotypes of CLV3 overexpression and the wus loss-of-function mutant phenotype in which the size of the SAM is reduced (Kinoshita et al. 2010). Biochemical studies of CLV pathway component interactions using a transient gene expression system in Nicotiana benthamiana revealed that CLV1 is potentially forming multiprotein complexes with CLV2/CRN and with TOAD2 in a CRN-dependent manner (Betsuyaku et al. 2011b), but whether a single, large complex forms, or several independent complexes function in parallel, remains to be uncovered. In addition, TOAD2 physically interacts with BAM1 (BARELY ANY MERISTEM1, a member of the CLV1 family of LRR-RLKs), and both TOAD2 and BAM1 interact genetically with CLV2 in response to CLE peptide-mediated inhibition of root growth, but the specific CLE ligand is not known (Shimizu et al. 2015).
The LRR-RLK TOAD2 was also reported to be a key regulator of other developmental mechanisms that involve cell differentiation and specification of cell fates. Phenotypic analysis of toad2 mutants revealed enhanced shoot growth and male sterility due to pollen defects caused by abnormal differentiation of microspores and hypertrophy of the tapetum (Mizuno et al. 2007). TOAD2 also genetically interacts with another LRR-RLK, the RECEPTOR-LIKE PROTEIN KINASE 1 (RPK1), to coordinate central domain protoderm patterning during the late globular stage of embryogenesis (Nodine et al. 2007). Double homozygous rpk1 toad2 mutants are embryo lethal, arrest their development during early stages of embryogenesis, and lack the normal radial specification of cell types. Analysis of molecular markers indicates that outer layer specification is lost and that an outward expansion of inner markers is detected. However, whether the ground tissue cell fate of outer layers is an indirect consequence of misspecification of protoderm or directly due to a specific role of these RLKs in the ground tissue is still an unsolved issue. Interestingly, about half of the rpk1 toad2/+ embryos developing from rpk1 toad2/+ plants exhibit an arrest similar to that of the double-mutant embryos, while the other half are able to complete their development (Nodine et al. 2007). However, further analysis of rpk1 toad2/+ embryos that do not arrest at the globular stage revealed that additional developmental processes are affected at a lower penetrance in this mutant background, with ∼16% of rpk1 toad2/+ embryos developing only one cotyledon primordium and consequently emerging as seedlings with just one cotyledon (Nodine and Tax 2008). The single-cotyledon phenotype is also characteristic of rpk1 embryos but occurs at a much lower frequency (4.6%) (Nodine and Tax 2008). This phenotype indicates that failure to specify the outer layer at early stages perturbs subsequent patterning events. For instance, the accumulation of the phytohormone auxin, one of the key regulators of cotyledon patterning (Moller and Weijers 2009), is not detected at the site where cotyledon primordia should initiate in the rpk1 toad2/+ mutants (Nodine and Tax 2008). The auxin flux, and therefore the establishment of an auxin maxima, is regulated by the PIN-FORMED (PIN) efflux carrier proteins family through their polar localization in the plasma membrane (Friml 2010). The auxin efflux carrier PIN-FORMED1 (PIN1) is not expressed in the defective half of the embryos lacking one cotyledon primordium, correlating with an absence of auxin maxima. A link between misregulated polarity of PIN1 in the epidermis of rpk1 and the occurrence of plants with one cotyledon was also recently demonstrated in the rpk1 embryos with cotyledon defects (Luichtl et al. 2013).
Spatiotemporal regulation of RLK activity contributes to coordination of plant growth and development. The LRR-RLK BRASSINOSTEROID INSENSITIVE1 (BRI1) functions in brassinosteroid hormone perception, and mutations in BRI1 cause a dwarf phenotype due to reduced growth and development (Li and Chory 1997; Clouse and Sasse 1998). The growth defects of bri1 mutants, but not vascular tissue defects, are rescued by the expression of a functional BRI1 receptor from the A. thaliana MERISTEM LAYER 1 (AtmL1) promoter that has a restricted expression domain within the epidermis. This suggests that signaling in the epidermis is sufficient to restore cell–cell communication with inner layers to coordinate the growth of the entire plant. Additional experiments in which BRI1 expression is restricted to specific radial layers shows that signaling in the outer layer has the most significant effect on regulating root meristem size and QC identity (Hacham et al. 2011). These data suggest that signaling mechanisms taking place in the outer layers drive plant growth, although there is also evidence for “inside-out” signaling (Gallagher et al. 2004; Cui et al. 2007; Yadav et al. 2008).
Here, we report that signaling mediated by TOAD2 and RPK1 controls RAM activity. rpk1 mutants display an incompletely penetrant S phenotype that is enhanced when an additional toad2 allele is mutated. The short roots display misoriented cell divisions in the RAM that primarily affect cells of the LR cap (LRC), epidermis, cortex, and QC, and cause a decrease in the number of columella cell (CC) tiers. toad2 mutants are insensitive to the root growth arrest induced by exogenous application of CLE17 and CLE19 peptides. This implies that TOAD2 might function as a receptor for these or similar CLE peptides, alone or in a complex with CLV2 and CRN or other components. Transcript and protein levels of RPK1 are reduced upon CLE treatment and the protein reduction requires the presence of functional TOAD2 protein. Finally, the incompletely penetrant phenotype of rpk1 and the observed CLE responses suggest that additional unknown components of this regulation play an important role.
Materials and Methods
Plant material
The Arabidopsis seeds used in this study were wild-type Columbia-0 (Col-0) toad2-1/+, rpk1-1, rpk1-5 (Nodine et al. 2007), toad2-3/+ [a new loss-of-function insertion mutant in the fourth LRR, generated by the Wisconsin Arabidopsis Knockout Facility (Sussman et al. 2000), backcrossed five times into Col-0), rpk1-1 toad2-1/+, rpk1-5 toad2-3/+, crn-1, and clv2-8 (Durbak and Tax 2011). The seeds were surface-sterilized with a solution of 70% ethanol and 0.1% Triton X-100 for 20 min, washed three times in 95% ethanol, air dried, and plated at a distance of ∼4 mm on 1% (w/v) agar plates containing 0.5× Murashige and Skoog (MS) media and 0.05% 2-(N-Morpholino) ethane sulfonic acid (MES), pH 5.8. After stratification at 4° in the dark for 3 days, the seeds were grown vertically at 22° under a 16 hr light/8 hr dark cycle in a Conviron growth chamber. Seedlings used for CLE peptide assay were transferred to media containing different peptides 5 days after germination (DAG). Data were collected from at least three independent experiments in which different media and seed batches were used. Strains are available upon request.
Root growth inhibition assay
The root length was measured from the base of the hypocotyl to the tip of the primary root at the time of transfer, and then every 24 hr for the next 5 days. CLE peptides (Mimotopes, http://www.mimotopes.com/) with a purity of >70% were dissolved in 50 µl DMSO and then diluted to a final concentration of 2 mM using sterile sodium phosphate buffer (50 mM, pH 6.0). Peptides were added to the cooled sterilized media to a final concentration of 10 µM. Control plates were prepared by adding the same volume of DMSO/phosphate buffer that did not contain any CLE peptide. Plates were scanned and root lengths were measured on the captured images using ImageJ software.
Generation of transcriptional and translational fusions
To generate transcriptional fusions, the genomic regions upstream of the RPK1 (2913 bp) and TOAD2 (1314 bp) coding sequences were amplified using primers RPK1pF/RPK1pR and TOAD2pF/TOAD2pR, respectively (a list of primers used to generate transcriptional and translational fusions can be found in Table 1), and TOPO TA cloned into the GATEWAY entry vector pCR8 (Invitrogen, Carlsbad, CA). DNA sequences immediately upstream of CLE19 (2283 bp) and CLE17 (523 bp) start codons were amplified with primers CLE19pF/CLE19pR and CLE17pF/CLE17pR, respectively, and cloned into the pCR8 vector. The entry clones above were used in the Gateway LR reaction (Invitrogen) to insert the corresponding regulatory sequences into the Gateway-adapted T-DNA binary vector pFYTAG (GenBank Accession DQ370421, donated by C. Zhang and D.W. Galbraith). The resulting expression vectors carrying the promoter sequences driving the expression of enhanced yellow fluorescence protein (YFP) fused with the coding region of a histone 2A gene (HTA6; At5g59870) (Zhang et al. 2005) were used for Agrobacteium-mediated transformation of Col-0 Arabidopsis using the floral dip method (Clough and Bent 1998). Seeds (T1) were harvested, germinated on soil, and selected using 50 mg/liter BASTA (1:1000 dilution of Finale, Farnam Comp, Phoenix, AZ). At least five independently transformed lines were analyzed for the presence of transgenes using PCR genotyping and microscopy to visualize the YFP signal. The RPK1p::GFP-NLS and TOAD2::GFP-NLS translational fusions used in this study were previously described by Nodine et al. (2007).
Table 1. List of primers used to generate transcriptional and translational fusions and their 3′–5′sequence.
| Primer name | Primer sequence (5′–3′) |
|---|---|
| RPK1pF | ACCCGAGTTTTCTTTGTGTTGCTA |
| RPK1pR | CTTCTTTTTCTTCACAAGAG |
| TOAD2/RPK2pF | GATCCCTCTTCTTATGTGTAAATTG |
| TOAD2/RPK2pR | CTTCGTAACTTATCCCCAAAAATG |
| CLE19pF | CTCGAGGTAGTGTTTCAGGGATTGGA |
| CLE19pR | CTCGAGTTGTCTATTTTTGGTCAAAT |
| CLE17pF | GCCTCTATTTGTAGAAGAATGAGTGAGA |
| CLE17pR | CATCTCACAAAACCTTGTTCCGGA |
| RPK1cF | CTCGAGATGAAACTTCTGGGTTTGGT |
| RPK1cR | CTCGAGCAATCTAGAAGGCTGGATTC |
| TOAD2/RPK2cF | ACTAGTCAGATCTACCATACACGACGGAGGTTGTAGCTGCT |
| TOAD2/RPK2cR | ACTAGTATGACTTCTTTGCCTTCTTCAG |
| SCR F | AAGGGATAGAGGAAGAGGACT |
| SCR R | GGAGATTGAAGGGTTGTTGG |
| PNHGWF | TATTGTTGCGAACAGAATTG |
| PNHGWR | TTTTTGTTGTTTGGATTTTC |
Microscopy
For confocal images, fresh roots were counterstained with 10 μg/ml propidium iodide (PI) (Sigma [Sigma Chemical], St Louis, MO) for 1–2 min, rinsed, and mounted in water on microscope slides. GFP and YFP fluorescence was imaged by confocal microscopy using a Zeiss 510 Meta confocal microscope (Zeiss [Carl Zeiss], Thornwood, NY), equipped with 10× Plan Neofluar, 0.3 NA, 20× Plan Apo, 0.8 NA; 40× Plan Neofluar, 1.3 NA; and 63× Plan Apo, 1.4 NA objective lenses and a laser line with excitation at 488 nm. Images were captured using filter sets of BP505-530 for GFP and YFP, and LP560 for PI, and the AxioVision image processing software and Adobe Photoshop Elements 9.0 (Adobe Systems, San Jose, CA) for image processing. Staining of whole seedlings using a modified pseudo-Schiff PI (mPS-PI) procedure was carried out as described by Truernit et al. (2008), with the exception that seedlings were mounted in chloralhydrate solution. Wild-type and mutant seedlings containing QC184::GUS (Beta-Glucuronidase) and QC25::GUS markers were stained with X-Gluc (5-bromo-4-chloro-3-indolyl glucuronide) for 2–6 hr at 37°, mounted in 6:1 chloralhydrate: Lugol’s solution, and imaged using a Zeiss Axioplan microscope. Root lengths were measured using ImageJ software. Measurement of fluorescence intensity of GFP signals was done using the basic measuring tools feature of ImageJ software (Hartig 2013).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Root growth defects of rpk1 mutants are dominantly enhanced by toad2 mutations
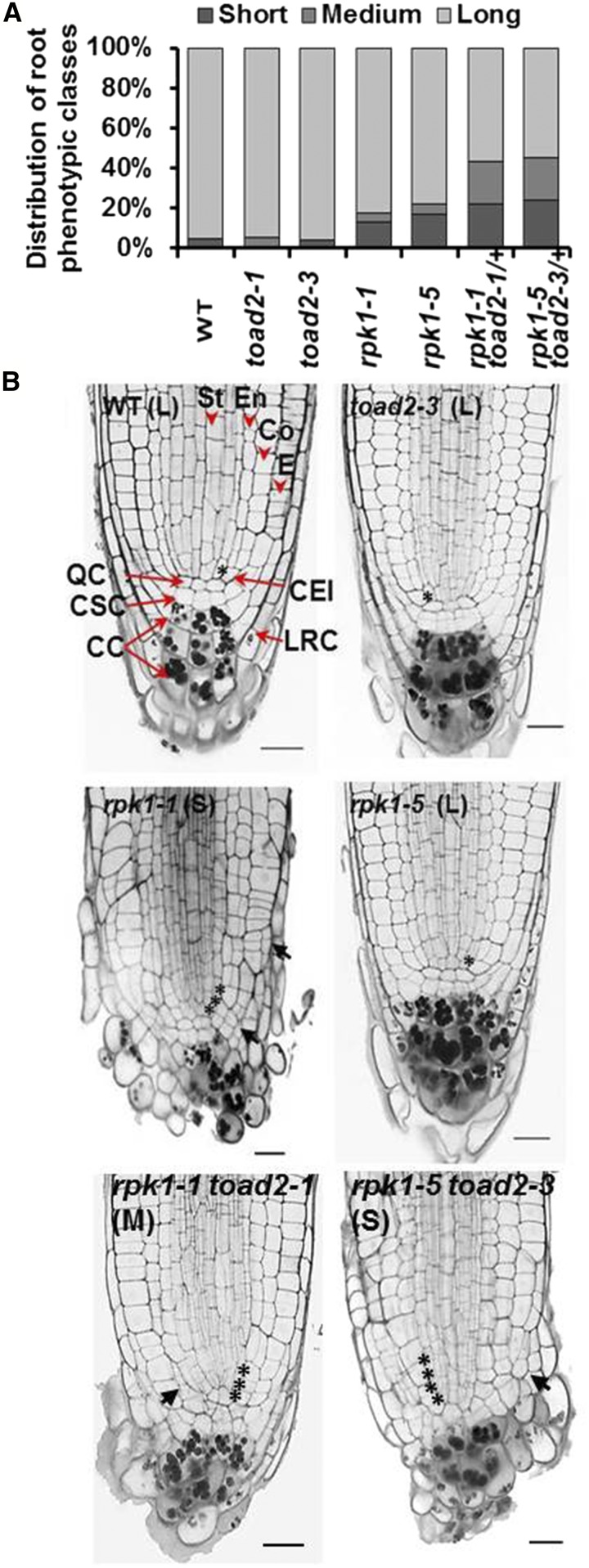
To investigate the role of RPK1 and TOAD2 in the regulation of primary root growth in Arabidopsis, wild-type (Col-0 ecotype) plants and rpk1-1, rpk1-5, toad2-1, toad2-3, rpk1-1 toad2-1/+, and rpk1-5 toad2-3/+ mutants were germinated and grown on MS plates, and their root growth was measured every 24 hr for 6 DAG. Homozygous rpk1 toad2 double mutants cannot be recovered due to their embryo-lethal phenotype (Nodine et al. 2007), and toad2-1 and toad2-3 are typically maintained as heterozygous lines due to their sterility. The plants hetero- and homozygous for the toad2 alleles were identified using PCR, and the root growth measurements were collected separately for individual plants of each genotype. While roots of toad2/+ and toad2 seedlings grow similarly to wild-type (Supplemental Material, Figure S1A), rpk1 mutants exhibit an incompletely penetrant root growth arrest leading to variable root length by 6 DAG on normal growth media (Figure S1A). In this study, 13% of rpk1-1 (and 16% of rpk1-5) roots do not measure ≥ 15% of the length of wild-type roots by 6 DAG (S), while 4% of rpk1-1 and 5% of rpk1-5 elongate to between 15 and 70% of the wild-type root length (M, medium root length). An increase (Fischer’s exact test, P < 0.05) in the frequency of S and M phenotypes is observed in the rpk1 toad2/+ seedlings independently of the allelic combinations analyzed (22% of rpk1-1 toad2-1/+ roots are S and 22% are M-type, while 24% of rpk1-5 toad2-3/+ roots are S and 22% are M-type) (Figure 1, A and B). A fraction of rpk1 and rpk1 toad2/+, and all toad2, seedlings elongate their roots similar to wild-type (Figure 1 and Figure S1) and constitute the L (long root length) class.
Figure 1.
Root growth defects of rpk1, toad2, and rpk1 toad2/+ mutants. (A) Distribution of root phenotypic classes of single and double mutants [rpk1, n = 130; toad2, rpk1 toad2/+, and wild-type (WT), n = 45]. S = short roots ≤ 15% of WT root length; M = medium roots, between 15 and 70% of WT root length; and L = long roots, average within WT range, ± 2 SD. Both rpk1 and rpk1 toad2/+ genotypes, independent of the alleles tested, show a significantly increased frequency of S-type roots compared to WT (Fisher’s exact test, P < 0.05). (B) Morphological defects of RAM (root apical meristem) of roots in the S and M phenotypic classes: representative pictures of longitudinal optical sections of 5 DAG (days after germination) roots stained using a modified pseudo-Schiff propidium iodide (mPS-PI) method). Asterisks indicate the position of cortical endodermal initial (CEI) cells and their daughters. Black arrows indicate regions of abnormal cell division. Red arrowheads mark specific cell files: stele (St), endodermis (En), cortex (Co), and epidermis (E), and red arrows indicate cell types: quiescent center (QC), columella stem cells (CSC), columella cells (CC), and lateral root cap cells (LRC). Bar, 20 µm; n > 10 for each genotype.
Root morphology is altered in the short roots of rpk1 and rpk1 toad2/+ mutants
In the RAM, the radially organized layers of the epidermis, cortex, endodermis, and the central stele (Figure 1B) are maintained by precisely oriented cell divisions of their corresponding initials located in the stem cell niche. To determine specific phenotypic defects in the RAM in the M and S phenotypic classes, we analyzed the patterning of different cell types in rpk1 and rpk1 toad2/+ mutants. In the S phenotypic class, and to a lesser extent in the M class, we observed that all plants display abnormally oriented cell division planes, a lack of organization of cells in distinguishable files, and irregular size of cells within files, indicating a loss of patterning in the mutants (arrows, Figure 1B). At 5 DAG, at least one cortical endodermal initial (CEI) was visible in a midsection (rarely present as an undivided daughter, asterisks in Figure 1B) in 12 out of 16 wild-type, 8 out of 18 L rpk1-1, and 6 out of 12 toad2-1 plants. In the mutants of the S phenotypic class, up to 3–4 cortical daughter cells are occasionally present (3 out of 18 rpk1-1 and 2 out of 10 rpk1-1 toad2-1/+ mutant roots) (asterisks in Figure 1B), forming a single-cell file distal to the cell that first undergoes the periclinal division that generates the two separate cortical layers (the cortex and endodermis).
The lack of typical organization and an easily discernable QC in the stem cell niche does not allow for clear cell type identification and quantification based on positioning. One exception is the markedly reduced number of the CC tiers containing starch-accumulating cells associated with the S and M phenotypes. At 5 DAG, the observed average number of CC tiers was 3.25 ± 0.4 in wild-type plants, and 3.25 ± 0.5 in toad2-1 and 2.3 ± 0.4 in rpk1-1 toad2/+ mutants (Figure 1B, compare S, M, and L). This phenotype correlates with an increased asymmetry of the root tip, as all short roots display an overproliferation of cells in the RAM, mostly in the radial dimension, affecting the epidermis, cortex, the LRC cells, and cells in the stem cell niche (Figure 1B, black arrows). In addition to aberrant cell division planes, the LRC and CC appear rounded in shape compared to wild-type and also lack the distinct organization of clearly defined cell tiers found in wild-type.
Marker analysis indicates that specification of cell types and organization of longitudinal cell files is disrupted in rpk1-1 toad2/+ mutant roots
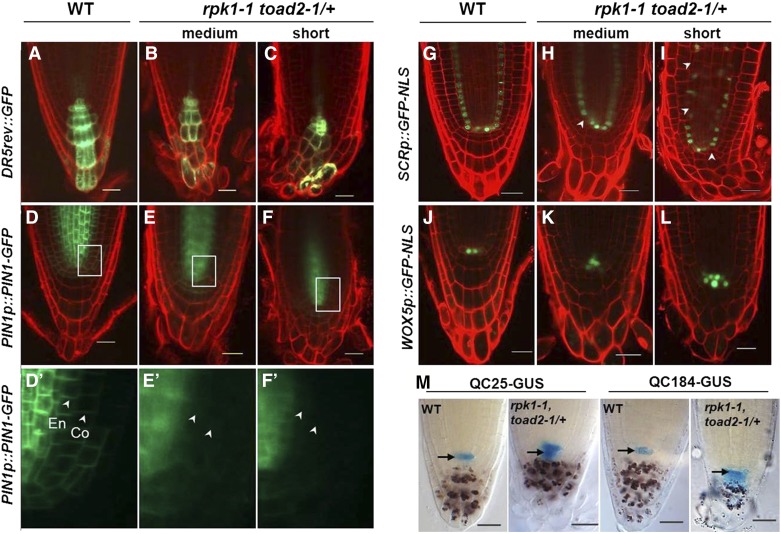
To determine the nature of root defects and the identity of cells affected in the abnormal patterning of the RAM, the activity of several molecular markers expressing GFP or GUS was analyzed in rpk1-1 toad2/+ double-mutant (L, M, and S) roots and compared to wild-type. The DR5rev::GFP construct comprises a synthetic auxin-responsive promoter (DR5) fused to the GFP reporter gene. The expression of DR5-driven reporters is therefore induced by the plant hormone auxin and has been previously used to indicate the auxin maxima in Arabidopsis (Friml et al. 2003). The basipetal transport of auxin from the CCs through the LRC cells toward the epidermis creates a gradient of the growth hormone that stimulates cell growth at lower concentrations (Swarup et al. 2005). Here, we analyzed whether the root apical auxin maxima was altered in rpk1-1 toad2/+ mutants by comparing DR5rev::GFP expression in the root apex of wild-type and mutant seedlings. The M and S phenotypic classes of rpk1-1 toad2/+ mutants (n = 12) express the DR5rev::GFP marker in the QC and columella stem cells (CSC) similar to wild-type (n = 15), but a clear expression gradient cannot be detected; often, CCs in the lowest tier contain the highest GFP signal (Figure 2, A–C). This aberrant distribution is more severely disrupted in the S phenotypic class than in the M class (n = 6 for each). This might indicate a defect in auxin transport through the misshapen CCs that lack clearly delineated apical and basal membranes, which affect the distribution of auxin and auxin carrier proteins (Swarup et al. 2005).
Figure 2.
Marker expression analysis indicates abnormal morphology and patterning of root cells in rpk1 toad2/+ mutants. Representative confocal images of DR5rev::GFP (A–C), PIN1p::PIN1-GFP (D–F and D′–F′), SCRp::GFP-NLS (G–I), and WOX5p::GFP-NLS (J–L) expression, and QC25::GUS and QC184::GUS localization (M) in wild-type (WT) and rpk1-1 toad2/+ roots. (D′–F′) represent a close-up view of the GFP channel in the region marked by boxes in (D–F), the white arrowheads indicate the localization of PIN1 at the plasma membrane in the cortex (Co) and endodermis (En). Mutant roots of the short and medium root length phenotypic classes are indicated. Seven-day-old seedling roots were imaged in (A–F) and (J–M), and 5-day-old roots were used in (G–I). White arrowheads in (G–I) indicate endodermal cells lacking a green fluorescent signal. The red counterstain is propidium iodide (PI) and the green is GFP fluorescence (A–L). Roots in M are stained with X-Gluc for GUS activity (blue) and with Lugol’s solution for starch granules (brown). Arrows in M mark the position of the QC. Bar, 20 µm.
To further analyze the root growth defects of mutant seedlings, we evaluated the localization of the auxin carrier protein, PIN1. In wild-type, PIN1 localizes mainly to the basal membrane of the vascular cells, but weak PIN1 signals can be also detected in the endodermis and the cortex (Blilou et al. 2005). PIN1p::PIN1-GFP expression in the wild-type was detected in the plasma membrane of vascular, endodermal, and cortex cells (Figure 2, D and D’). In contrast, rpk1-1 toad2/+ mutants show mainly cytoplasmic localization of the PIN1 protein in the vasculature, and very weak expression in the endodermis and cortex (Figure 2, E–F’). This distribution could affect the maintenance of instructive auxin gradients that are required for proper root growth. We did not detect an outward expansion of the expression pattern of these markers, as seen in the Toadstool phenotypic class of embryos (Nodine et al. 2007), indicating that patterning and cell fate specification occurred normally in the embryos that survived past the globular stage and that patterning is largely maintained postembryonically. The defects that we can detect in the short roots are subtler, affecting subcellular functions, rather than broad cell layer specification.
To analyze the identity of cell types in the RAM, the expression of the SCRp::GFP-NLS and WOX5p::GFP-NLS markers was compared in wild-type and rpk1-1 toad2/+ roots. The SCARECROW (SCR) transcription factor of the GRAS family is specifically expressed in the CEIs, the QC, and the endodermal cells. The nuclear-localized GFP driven by the SCR promoter was similar in wild-type and mutant roots (n = 47), indicating that the respective cell types are present in the mutant plants (Figure 2, G–I). In the S class of rpk1-1 toad2/+ mutants (n = 20), GFP expression is not detected in all expected cells and it is also not detected in a linear file of cells (arrowheads, Figure 2, H and I), indicating that the patterning and specification of some endodermal cells is aberrant in the double mutants.
The WUS-RELATED HOMEOBOX 5 (WOX5) homeodomain transcription factor acts downstream of SCR, functions in the QC to maintain columella stem cell signaling (Sarkar et al. 2007), and is often used as a marker for QC cell identity. In the wild-type roots, WOX5p::GFP-NLS is expressed in the QC cells and occasionally in one or a few vascular initials, with an average of 4.9 ± 0.9 (n = 18) cells expressing the GFP in their nuclei. In contrast, WOX5p::GFP-NLS expression in the rpk1-1 toad2/+ mutants is often detected in more cells, located mainly in the QC and in the cells above the putative QC cells (7.4 ± 1.4, n = 29, Student’s t-test P < 0.001) (Figure 2, J–L). This indicates that some aspects of QC cell fate are maintained in the cells generated through cell divisions of the QC and displaced proximally in the region of vascular initials. To further characterize the specification of cells in the stem cell niche, we analyzed the expression of the QC-specific markers QC184::GUS and QC25::GUS. These markers are expressed only in the QC cells of wild-type plants (arrows in Figure 2M, and (Sabatini et al. 2003). In the rpk1-1 toad2/+ roots, GUS activity is detected not only in the QC but also in the initials surrounding the QC, including in the vascular initials and in the CCs, indicating that select subsets of transcriptional programs are altered in the initials surrounding the QC in the double mutants (Figure 2M and Figure S2).
RPK1 and TOAD2 have partially overlapping expression domains in Arabidopsis roots
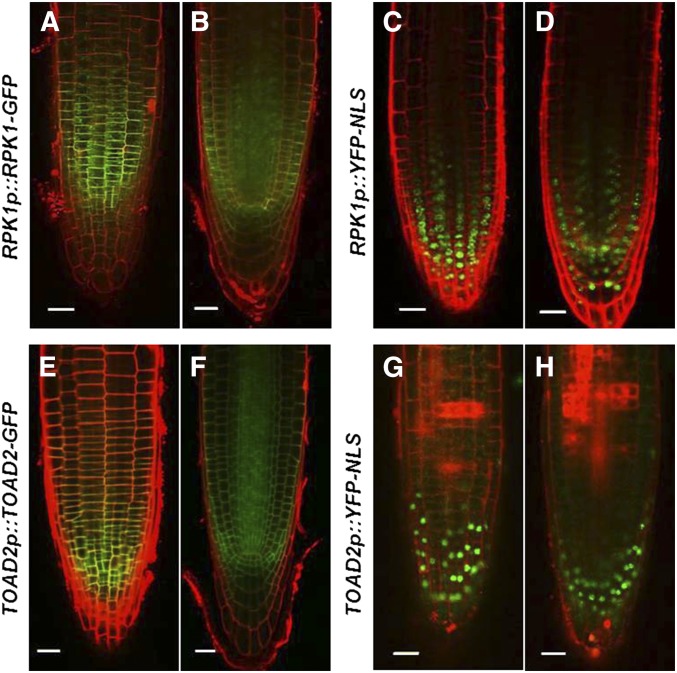
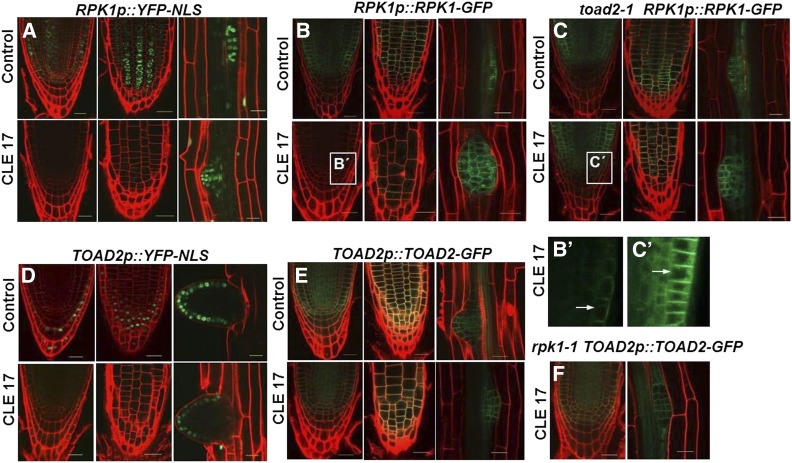
RPK1 and TOAD2 were previously reported to function redundantly in the early stages of Arabidopsis embryogenesis, where their partially overlapping expression was detected using GFP translational fusions (Nodine et al. 2007). Because we detected defects in root growth in rpk1 mutants and an increased frequency of defective roots in the rpk1, toad2/+ mutant seedlings, we tested the expression of these genes postembryonically by analyzing RPK1p::YFP-NLS and TOAD2p::YFP-NLS transcriptional fusions, and RPK1p::RPK1-GFP and TOAD2p::TOAD2-GFP translational fusions, in wild-type plants from 2 to 7 DAG. While fluorescent signals from transcriptional and translational fusions of both RLKs were detected throughout the roots, a more intense signal was detected in the RAM (Figure 3). RPK1 shows strong expression in the endodermis, cortex, and stele as well as in the QC and initial cells of the RAM (Figure 3, A–D). Reduced expression is also detected outside of the RAM and is not observed in the root vasculature or in the mature root cap cells. TOAD2 has a similar expression pattern as RPK1 in the root tip, with the exception of mature LRC cells where TOAD2 is expressed more strongly (Figure 3, E–H). Beyond the RAM, TOAD2 is also strongly expressed in stele cells throughout the entire root. Both fusion proteins appear to be plasma membrane-localized based on their fluorescent signal overlapping with the PI staining outlining the cells. While transcriptional and translational fusions have largely overlapping expression domains, we observed that the transcriptional fusion expression is detected at lower levels in the inner cell layers compared to the LRC and the RAM epidermis. We hypothesize that this could be due to the nature of the YFP marker protein and its stability in the cell.
Figure 3.
RPK1 and TOAD2 are expressed in partially overlapping domains in the root apical meristem. Representative confocal images of RPK1p::RPK1-GFP (A and B); RPK1p::YFP-NLS (C and D); TOAD2p::TOAD2-GFP (E and F); and TOAD2p::YFP-NLS (G and H). Images represent median optical sections (B, F, D, and H) and surface views (A, C, E, and G) of 7-day-old root tips counterstained with propidium iodide. Bar, 20 µm.
toad2 mutants are insensitive to in vitro treatment with CLE peptide
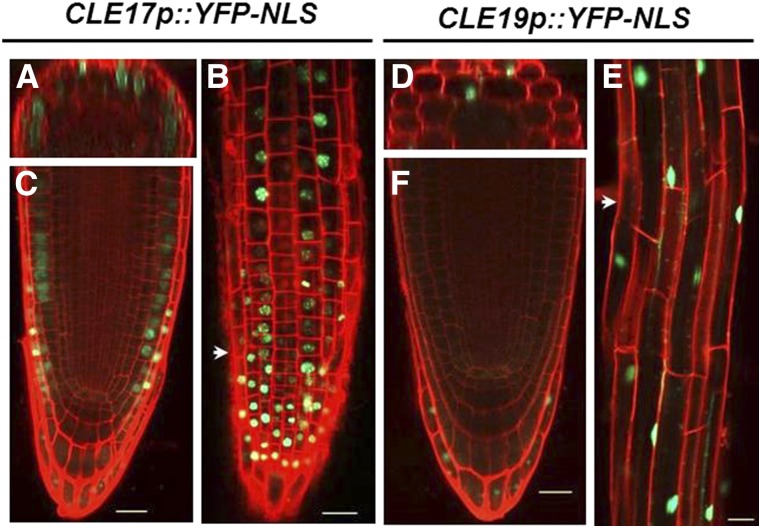
Recent functional studies of plant meristems indicate that small peptides from the CLE family play an important role in signaling through receptor kinases in the RAM, the SAM, and in vascular meristem development [reviewed in Betsuyaku et al. (2011a)]. In Arabidopsis, at least 18 of the 32 CLE genes are transcribed in specific or overlapping regions of the root (Jun et al. 2010). Wild-type roots overexpressing CLE genes or treated with exogenous CLE peptides (CLV3, CLE19, and CLE40) are shorter and have a reduced number of meristematic cells compared to untreated roots (Fiers et al. 2005). To verify the presence of root-expressed CLE genes, we generated transcriptional fusions with YFP and a nuclear localization signal for two genes, CLE17p::YFP-NLS and CLE19p::YFP-NLS, transformed them in wild-type plants, and analyzed their expression in four independent T3 generation lines (Figure 4). CLE17 is expressed in the RAM, the LRC cells, and in the epidermis (Figure 4, A–C), and CLE19 has a more restricted expression domain in the LRC cells (Figure 4, D–F). In the elongation and differentiation zone, both CLE genes are expressed mainly in the epidermal and cortical cells (Figure 4, B and E). The RAM expression of CLE17 and CLE19 therefore partially overlaps with the outer layer expression domains of RPK1 and TOAD2.
Figure 4.
CLE17 and CLE19 are expressed in the root apical meristem. Representative confocal pictures of roots counterstained with propidium iodide (red) expressing CLE17p::YFP-NLS (A–C) and CLE19p::YFP-NLS (D–F) (green) in 3-day-old root tips of wild-type Arabidopsis. Optical cross sections (A) and (D) show the region marked by arrowheads in (B and E), respectively. Images represent root tips in median sections (C and F), and surface views of the root tip (B) and of the differentiation zone (E and F). Bar, 20 µm.
Due to phenotypic similarities between rpk1 toad2/+ mutant roots and the CLE treated wild-type roots, we tested the effect of CLE17 and CLE19 peptides on plants lacking functional copies of RPK1 and TOAD2. Previously, the receptor-like protein CLV2 (Guo et al. 2010) and the receptor-like cytoplasmic kinase CRN (Miwa et al. 2008) were shown to be required for transmission of the CLE signals; therefore, in this study, we used plants carrying the mutant crn-1 and clv2-8 alleles as positive controls. In preliminary experiments, the root growth inhibitory effect of different amounts of peptide (0.5, 1.0, and 10.0 µM) was tested on wild-type plants. While the plants responded to all treatments, the 10 µM peptide concentration triggered the fastest response within 2–3 days after seedling transfer, which was similar to previous reports (Fiers et al. 2005) and was therefore the concentration selected for the root growth assays described below.
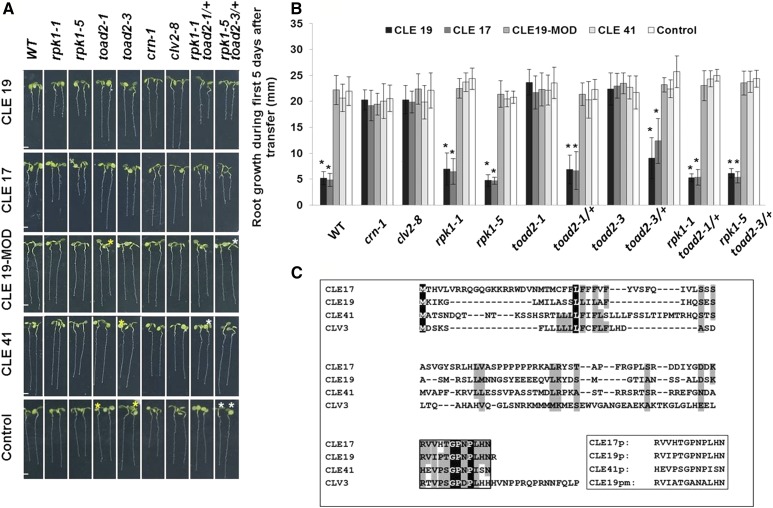
We generated the peptides corresponding to the 12-amino acid conserved CLE motif of two type A CLE peptides (CLE17 and CLE19), one type B CLE peptide (CLE41), and an additional peptide (CLE19-MOD) derived from the CLE19 sequence, in which all proline residues were replaced by alanine, to render the peptide nonfunctional (Song et al. 2012) (Figure 5C). Progeny of wild-type, crn-1, clv2-8, rpk1-1, rpk1-5, toad2-1/+, toad2-3/+, rpk1-1 toad2-1/+, and rpk1-5 toad2-3/+ plants were grown for 3 days on MS plates and then transferred and monitored for root growth on control or CLE peptide-containing MS media (Figure 5A). Wild-type seedlings, as well as toad2-1/+, toad2-3/+, rpk1-5, and rpk1-1 seedlings, showed sensitivity to CLE19 and CLE17 peptide treatment, their root growth ceased within 2–3 days after transfer to test plates (Figure 5, A and B), and significant differences in their root length were observed when compared to untreated plants. In contrast, the root lengths of homozygous toad2-1, toad2-3, crn-1, and clv2-8 seedlings grown on CLE17 and CLE19 peptide-containing MS media were not significantly different from those of plants grown on control plates after the 5-day treatment period, demonstrating insensitivity to CLE peptide treatment (Figure 5, A and B). In addition, all plant genotypes tested on CLE41, CLE19-MOD, and the no peptide control plates did not show altered root growth phenotypes when compared to wild-type seedlings, indicating that CLE41 and CLE19-MOD have no effect on root length.
Figure 5.
Root growth sensitivity after treatment with CLE peptides. (A) Root phenotypes of rpk1, toad2, and rpk1 toad2/+ mutants, and wild-type (WT), crn-1, and clv2-8 control plants shown after 5 days of treatment with exogenous CLE peptides. Progeny of toad2/+ plants and rpk1 toad2/+ heterozygote parents were used in the assays. Plants marked with white asterisks were genotyped as rpk1 mutants only, and with yellow asterisks were WT or heterozygous for toad2. Bar, 2 mm. (B) Distribution of root lengths 5 days after transfer to CLE treatment plates. Values represent average of root length measurements in three independent experiments. Error bars represent SD (* P < 0.001; n = 8 for toad2/+; n > 16 for all other genotypes, Student’s t-test, C.I. 95%). Significant changes with CLE treatment for each genotype relative to the untreated plants are marked with *. No significant differences are observed between WT and the other untreated genotypes (long phenotypic class). (C) Alignment of Arabidopsis CLV3 amino acid sequence with sequences of CLE peptides used in this study and the synthetic peptides (box) used for the root assay. Completely conserved amino acids are shaded in black; partially conserved or highly similar amino acids (groups of strongly similar properties scoring >0.5 in the Gonnet PAM 250 matrix ClustalW) are shaded in gray; and the conserved CLE motif is framed.
CLE17 treatment causes an S phenotype by negatively regulating the frequency of cell divisions in the proximal meristem
To analyze the mechanisms leading to the cessation of root growth in response to CLE treatment, progeny of heterozygous toad2-1/+ and toad2-3/+ and homozygous rpk1-5, rpk1-1, crn-1, and clv2-8 mutant seedlings, and control wild-type seedlings, were grown on plates containing added peptides, fixed and stained using a mPS-PI method, and visualized using confocal microscopy. The root growth and cell numbers of different RAM domains were quantified in these genotypic backgrounds and compared to wild-type when grown in the presence of 10.0 µM CLE17, CLE19, CLE41, and CLE19-MOD peptides, and on control plates.
We found that the roots of rpk1-5, rpk1-1, and wild-type seedlings respond to CLE17 and CLE19 treatment through a reduction of the overall length of their RAM, and no effect was observed on roots grown in the presence of CLE41 or CLE19-MOD, or on control plates (Figure 6, A and C). The size of the RAM is not decreased in the clv2-8, crn-1, or toad2 homozygous mutants after 5 days of treatment; therefore, toad2-1 and toad2-3 mutants, similar to crn-1 and clv2-8 mutants, are insensitive to the exogenously applied CLE peptide effect of reducing root meristem growth (Figure 6, A and C).
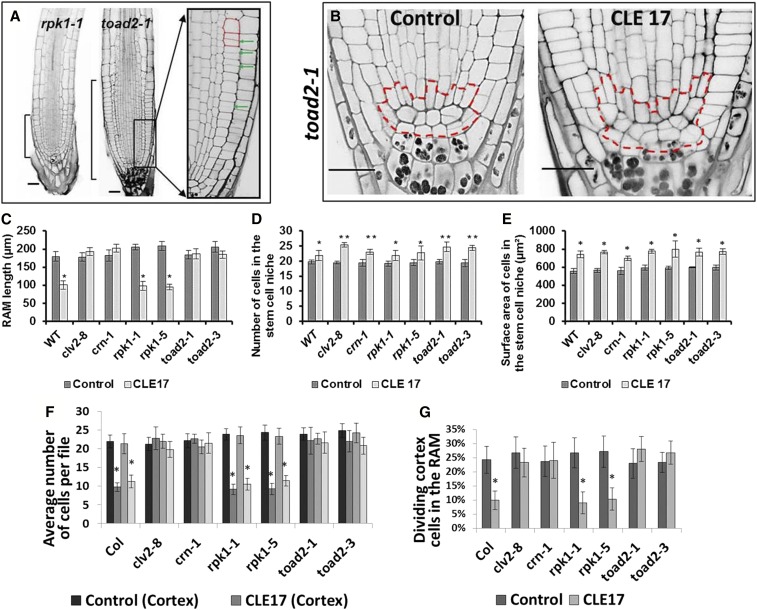
Figure 6.
Root apical meristem (RAM) changes in roots treated with CLE peptides. (A) Longitudinal median optical sections of CLE-treated roots stained using a modified pseudo-Schiff propidium iodide (mPS-PI) method. Brackets indicate the size of the RAM of roots grown in the presence of CLE17 for 5 days. Enlarged image of boxed area shows cortical cells (outlined in red) and dividing cells (marked by arrowheads). Bar, 20 µm. (B) Confocal image showing the organization of RAM cells with the stem cell niche area outlined by a dashed red line. (C–G) The effect of CLE17 treatment on root morphology (Student’s t- test, C.I. 95%). RAM length of seedlings grown on CLE17 plates for 5 days (* P < 0.001, n > 12) (C); the number of cells in the stem cell niche (D) (** P < 0.001 and * 0.005 < P < 0.05); and the surface area occupied by the stem cell niche (E) (* P < 0.001) in roots treated with CLE17 peptide. The average number of epidermal and cortical cells in RAM files (F) and the frequency of cell divisions occurring in the cortical file (G) measured in median optical sections of CLE17-treated and mPS-PI-fixed samples (* P < 0.001).
To further analyze the cause of the short root meristems, we quantified the number of cells in longitudinal epidermal and cortex cell files. The average numbers of epidermal and cortex cells observed in midlongitudinal optical sections of wild-type, rpk1-1, and rpk1-5 roots treated with CLE 17 peptide were found to be significantly less that the number of cells in crn-1, clv2-8, toad2-1, or toad2-3 roots (Figure 6F). Similar results were observed in CLE19-treated roots of toad2-1 and wild-type seedlings (Table S1). In the mPS-PI-stained roots, recently dividing cells are marked by a very thin cell wall having formed between the two daughter cells that have not yet elongated, therefore making them appear to be half the length of nondividing cells. This series of anticlinal (perpendicular to the long axis of the root) cell divisions in the distal meristem generates more cells and increases the root length; these are sometimes called transit-amplifying cell divisions. Upon analysis of dividing cells in longitudinal files, we found that the short roots of rpk1-5, rpk1-1, and wild-type seedlings present a decreased frequency of transit-amplifying cell divisions along the epidermal and cortical cell files after treatment with CLE17 peptide (Figure 6, A, F, and G), resulting in fewer cells comprising the RAM portion of these files. This effect is not observed in the long roots of toad2-1, toad2-3, crn-1, and clv2-8 seedlings upon CLE17 treatment, and the frequency of cell divisions is similar to that of untreated controls (Figure 6, A, F, and G).
To understand the effect of CLE peptide treatment on the stem cell niche, we measured the surface area occupied by the QC cells and the stem cells surrounding the QC, including the CEI daughters in midlongitudinal optical sections (Figure 6B). We found an increased number of cells comprising the stem cell niche and an increased surface area occupied by this region in all genotypes tested, regardless of the overall root length (Figure 6, D and E). When individual cell types within the stem cell niche were analyzed, the numbers of vascular initials and CSCs were not found to be significantly different between treated and nontreated roots (Figure 7, A and B), but increased numbers of CEIs and their daughters were detected in all roots. These results suggest that one effect of CLE treatment is a delay in the CEI divisions into the two separate types of cortex stem cells (endodermal and cortex stem cells), as well as a delay in the occurrence of periclinal divisions of their daughters, generating the separated initial cells in the respective files (Figure 7B). At 8–9 DAG, the majority of wild-type roots have already divided their CEI into endodermal and cortex stem cells. In all genotypes tested, we detected that <50% of the roots still have one or two CEIs in a medial optical section (n > 12). After CLE17 treatment, the number of CEIs detected, as well as their daughters that do not undergo a periclinal (parallel to the long axis) division, often forming a single cortical file of cells extending from the CEI, is increased. For instance, 60% of wild-type, 100% of crn-1, 80% clv2-8, 90% of rpk1, and 100% of toad2 (n > 12) roots have one or two visible CEIs, and 30% of wild-type, 70% of crn-1, 80% of clv2-8, 40% of rpk1, and 60% of toad2 roots have at least one undivided daughter, but the number of nonseparated CEI daughters varies from 1 to 8.
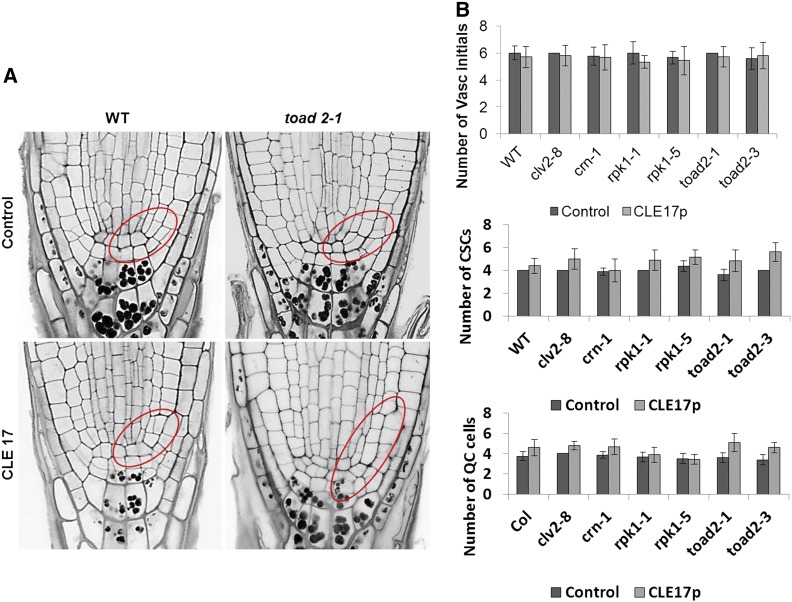
Figure 7.
The root apical meristem of roots treated with CLE peptides contains supernumerary CEI (cortical endodermal initial) and CEI daughter cells. (A) Representative pictures of longitudinal median optical sections of CLE17-treated roots stained using modified pseudo-Schiff propidium iodide (mPS-PI). Areas of CEI cells and their undivided daughters are outlined in red. Bar, 20 µm. (B) The number of vascular (vasc.) initials, the columella stem cells (CSCs), and quiescent center (QC) cells in plants treated with CLE17 (no significant differences, P > 0.05, Student’s t-test, C.I. 95%).
An increased number of presumptive QC cells is also detected, associated with increased cell divisions noted in the cells at the position of the QC (P < 0.05 between treated and nontreated controls). However, no statistically significant differences were found between different genotypes (Figure 7B). In conclusion, the increased frequency of QC cell division and the increase in the stem cell niche size, observed after CLE17 treatment, indicates a common mechanism by which CLE peptides act to control root development.
RPK1 is downregulated in CLE-treated roots and this process requires the presence of functional TOAD2
All toad2 roots respond to CLE treatment similarly, by continuous growth on the CLE plates (n = 142), while the rpk1 mutants, similarly to wild-type, cease their root growth; very rarely, we observed rpk1 mutant roots that elongated on CLE plates (4 out of 224). To unravel the potential genetic interactions between RPK1 and TOAD2 in root development and the regulation of the CLE-mediated signaling pathway in RAM growth, we analyzed the transcriptional and translational regulation of these receptor kinases upon CLE treatment. The expression of the RPK1p::YFP-NLS and TOAD2p::YFP-NLS transcriptional fusions was analyzed in wild-type plants treated with CLE17 and CLE19 peptides (Figure 8, A and D). The expression of RPK1p::YFP-NLS is significantly reduced in the CLE17-treated plants. We measured the YFP signal intensities in treated and untreated roots using ImageJ software, and found that in 80% of treated plants (n = 30) the YFP signal intensity is reduced to the background level, and in 20% of the plants the signal is reduced between 6- and 10-fold compared to untreated plants (Figure 8A). Interestingly, the reduction in fluorescence intensity is specific to the main RAM, and the fluorescence intensity is not changed in the LRs of treated plants compared to untreated controls (Figure 8A). In contrast, Image J analysis indicates that the fluorescence generated by TOAD2p::YFP-NLS activation is only partially reduced (two- to fourfold) in the roots treated with the CLE17 peptide (n = 24), while the signal from the fluorescence in the emerged LRs is not significantly different compared to control plants (Figure 8D, compare the range of change in the primary root to the LR).
Figure 8.
Regulation of RPK1 and TOAD2 by exogenous CLE17 treatment. Representative confocal pictures of propidium iodide (PI)-counterstained roots expressing RPK1p::YFP-NLS (A), TOAD2p::YFP-NLS (D), RPK1p::RPK1-GFP (B and C), or TOAD2p::TOAD2-GFP (E and F); wild-type (A, B, D, and E), toad2-1 mutant (C), and rpk1-1 mutant plants (F) express the transgenes. Each panel contains images of surface and median views of the root apical meristem, and an image of an emerging lateral root from the same plant (some of the lateral roots are not size matched, accounting for slight variation in the number of cells expressing the signal; selection was based on the age of the primary root). Close-up views of (B and C) are marked by white boxes and labeled (B′ and C′), respectively. Arrows indicate subcellular localization of GFP-labeled protein in intracellular vesicular compartments.
Analysis of RPK1p::RPK1-GFP and TOAD2p::TOAD2-GFP translational fusions in the wild-type plants treated with CLE17 and CLE19 peptides indicates a similar pattern. The signal from RPK1-GFP protein localized at the plasma membrane in the CLE17-treated plants is greatly reduced in the RAM, but not in the LR primordia (Figure 8, B and B´). The mean fluorescence intensity measured with ImageJ in the RPK1p::RPK1-GFP control was 32.52 ± 4.82, while in RPK1p::RPK1-GFP plants treated with CLE17 it was significantly weaker at 16.72 ± 3.71 (n = 16, P < 0.001). In contrast, the TOAD2-GFP signal is still detected in the plasma membrane of RAM cells of treated plants (Figure 8E). The expression of RPK1p::RPK1-GFP in toad2-1 plants is unchanged upon CLE17 treatment (Figure 8, C and C´), suggesting that transcription is still taking place. The mean fluorescence intensity of RPK1p::RPK1-GFP in toad2-1 control plants (32.64 ± 8.74) did not change significantly after CLE17 treatment (mean 33.9 ± 4.1, n = 8, P > 0.05). These results indicate that CLE17-induced downregulation of RPK1 is not activated in the toad2-1 mutant background. Also, RPK1-GFP is normally localized at the plasma membrane and in numerous intracellular vesicles (Figure 8, C and C´), and in CLE-treated roots we noted a residual fluorescent signal from internal vesicles, but not at the plasma membrane (Figure 8, B and B´). We also analyzed the expression of TOAD2p::TOAD2-GFP in rpk1-1 mutant plants and did not detect a change in the fluorescence intensities of membrane-localized TOAD2-GFP after CLE17 treatment (Figure 8F).
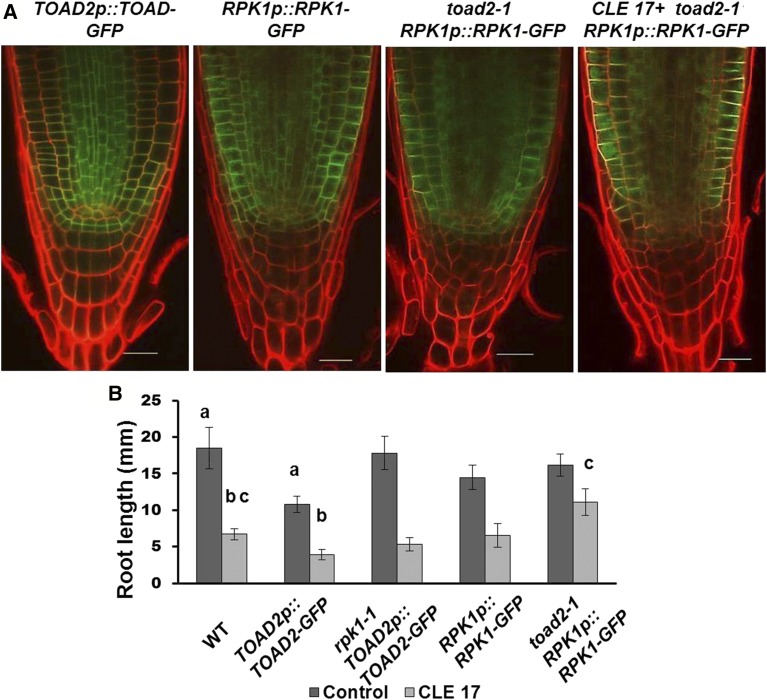
Transgenic homozygous plants expressing TOAD2p::TOAD2-GFP display a unique slower root growth phenotype over the period from 3 to 8 DAG compared to wild-type plants or plants expressing RPK1p::RPK1-GFP (Figure 9), indicating that the presence of more TOAD2 gene copies has the effect of slowing root growth, similar to exogenous CLE treatment. When the TOAD2p::TOAD2-GFP-expressing plants are treated with CLE17 or CLE19, their root growth ceases at least 24 hr sooner than that of the control plants, and their overall root length after 5 days of treatment is smaller compared to wild-type plants (Figure 9B). However, homozygous rpk1-1 plants containing the TOAD2p::TOAD2-GFP do not elongate differently than wild-type plants when grown on control, CLE17, or CLE19 plates (Figure 9B), indicating that RPK1 functions as an important regulatory component of the TOAD2-mediated response to both the endogenous and exogenously applied CLE peptides. Plants that express the RPK1p::RPK1-GFP transgene have similar root growth in the wild-type or toad2-1 mutant background, and their response to CLE treatment is similar to that observed in wild-type plants and toad2-1 mutants, respectively (Figure 9B). Interestingly, although the root length of RPK1p::RPK1-GFP-expressing plants is not significantly increased (at least until 8 DAG), the size and patterning of their distal meristem is different from in control plants. Wild-type plants at this stage have 4.5 ± 0.5 CC tiers, while RPK1p::RPK1-GFP have 6.0 ± 0.0 and toad2-1; RPK1p::RPK1-GFP have 6.7 ± 0.6 columella tiers. In addition, the cell alignment and the columella tiers appear disorganized in the distal meristem (Figure 9A).
Figure 9.
RPK1 overexpression induces TOAD2-dependent changes in root growth and distal meristem morphology. (A) Confocal images of 8-day-old root tips of wild-type (WT) and mutant rpk1-1 and toad2-1 expressing RPK1p::RPK1-GFP or TOAD2p::TOAD-GFP. (B) Root lengths of CLE17-treated and control plants overexpressing RPK1 or TOAD2. Letters indicate statistically significant differences (P < 0.001, Student’s t-test, n = 20 for each genotype) between the labeled pairs (compare the bars labeled “a” to each other; also the bars labeled “b” to each other and the bars labeled “c” to each other.).
Transcription of SCR and WOX5 is maintained upon CLE treatment
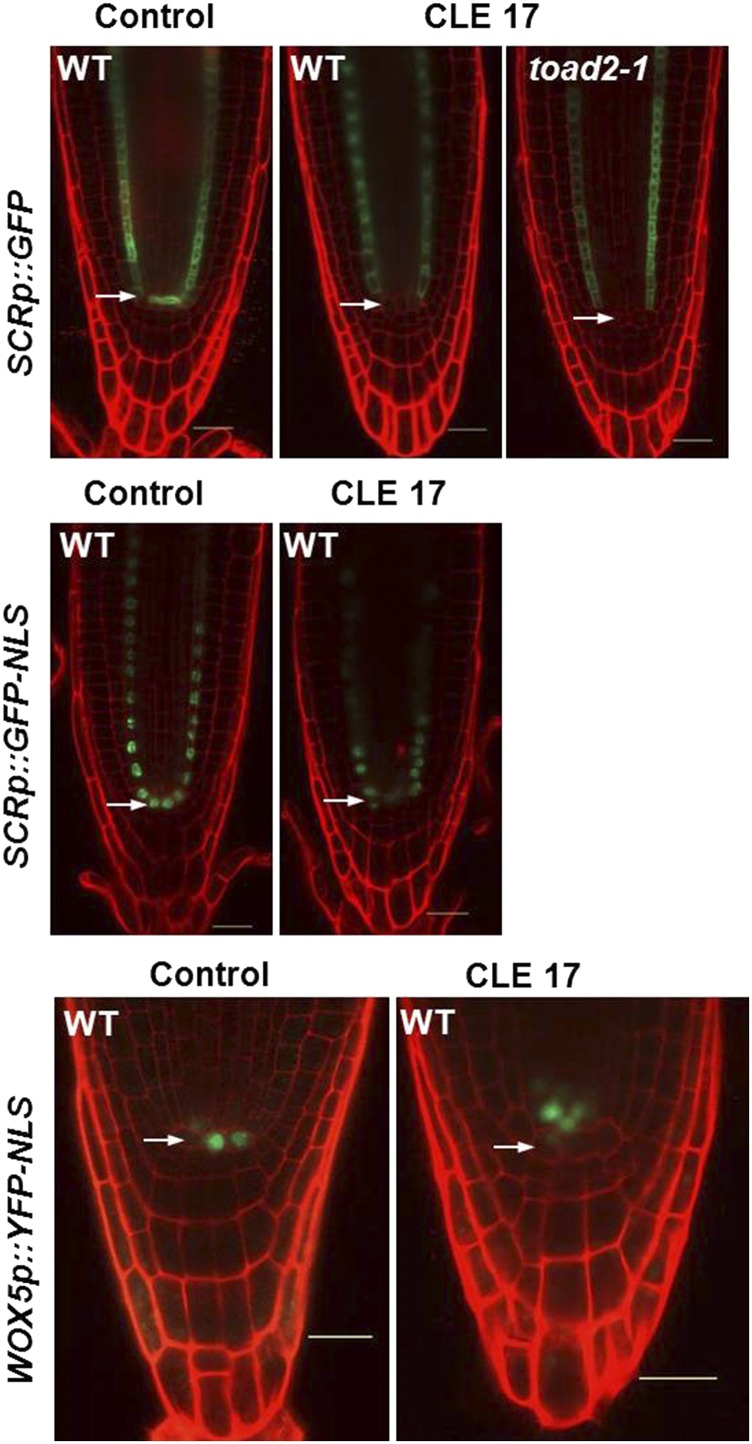
To test that downregulation of RPK1 gene expression after CLE treatment is specific to the RPK1 gene and not the result of more general transcriptional silencing occurring in the RAM, we analyzed the expression of the molecular markers SCRp::GFP, SCR::GFP-NLS, and WOX5::GFP-NLS after 5 days of growth in the presence of the CLE17 peptide.
The GFP signal from SCRp::GFP accumulates in the cytoplasm of QC, CEI, and endodermal cells of untreated plants (Figure 10). The expression is also detected in the endodermal cells of all wild-type and toad2-1 plants at similar levels as in the wild-type plants, but is often reduced in the QC and CEIs of CLE-treated plants (arrowheads in Figure 10, top panel), possibly indicating a downregulation of transcription. In contrast, when using a nuclear-localized version of GFP under the control of the same SCR promoter, we detected a GFP signal from QC cells (Figure 10, middle panel), indicating that transcription from the SCR promoter occurs in the QC, and leading to the conclusion that transcription from the SCR promoter is maintained. The difference in signal detected could be because the nuclear-localized version is concentrated in the nucleus, and the ER/cytoplasmic version of GFP is unstable in the QC upon CLE treatment. This might indicate that some intrinsic characteristics of QC cells are not maintained upon CLE treatment.
Figure 10.
Expression of SCR and WOX5 in CLE17-treated roots. Eight-day-old roots treated (CLE17) or untreated (control) imaged 5 days after transfer to treatment plates. SCRp::GFP (top panel, n = 12), SCRp::GFP-NLS (middle panel, n = 20 for each condition), and WOX5p::YFP-NLS (bottom panel, n = 18 treated and n = 12 untreated) expression (green) in roots counterstained with propidium iodide. Arrows indicate the position of the quiescent center. Bar, 20 µm.
WOX5 expression is also detected in the CLE17-treated wild-type plants, albeit the pattern is different from in the control plants. The number of cells expressing WOX5-GFP increases in the treated plants (6.2 ± 1.3) compared to the untreated controls (4.5 ± 1.2). Importantly, the cells expressing the WOX5 marker are localized above the putative QC cells (arrows in Figure 10, lower panel) in 67% of the analyzed plants, and 33% of the plants show WOX5 expression in both QC and vascular initial cells. These results indicate that the mechanisms restricting the WOX5-dependent QC cell fate are disrupted in the CLE17 treated roots, leading to misexpression of these markers in a larger group of cells, resulting from division of QC.
Discussion
Using analysis of mutants, transgene expression, and exogenous CLE treatment in Arabidopsis, we uncovered a role for the RLKs RPK1 and TOAD2 in the control of root growth and meristem patterning. Previously, TOAD2 was shown to function redundantly with RPK1 to maintain the protoderm cell fate in Arabidopsis embryos (Nodine et al. 2007); the loss of the regulatory role of the protoderm in the central domain of embryos leads to more defects, including a failure to specify cotyledon primordia (Nodine and Tax 2008). Genetic analyses also revealed that TOAD2 has a role in tapetum specification in the anther (Mizuno et al. 2007), and contributes to shoot apical homeostasis by transmitting the CLV3 signal (Kinoshita et al. 2010), possibly via its CRN-mediated interaction with CLV1 (Betsuyaku et al. 2011b). RPK1 was also shown to integrate environmental signals and to control abscisic acid (ABA)-dependent cell proliferation (Osakabe et al. 2005, 2010; Lee et al. 2011). Here, we show that RPK1 and TOAD2 have a role in the maintenance of root growth by controlling cell proliferation in the RAM; the main defects of mutant roots are arrested growth due to a short meristem, and aberrant cell divisions that result in supernumerary and disorganized radial cell layers. The penetrance of these rpk1 mutant phenotypes is enhanced in heterozygous toad2 mutants, indicating a dose-dependent requirement for both genes in the pathway controlling root meristem maintenance.
A precise balance between cell proliferation and cell differentiation determines root meristem size. The formation of auxin maxima in the RAM is an important signaling mechanism that controls cell division and differentiation, and therefore instructs morphogenesis and depends on the spatial distribution of PIN proteins (Grieneisen et al. 2007; Hacham et al. 2011). The interplay of different mechanisms that control root meristem size is supported by examples of BR signaling affecting the post-transcriptional regulation of PIN protein localization; this implies that brassinosteroid (BR)-mediated root meristem growth is controlled, to some extent, by auxin reallocation (Hacham et al. 2012). Here, we show that root proliferation defects of rpk1 toad2/+ mutants correlate with a disrupted auxin gradient in the root proximal meristem, and with a reduction in the cellular distribution of PIN1 protein. If cortical and endodermal cell specification and function is disrupted in rpk1 toad2/+ mutants that are defective in signaling, the abnormal distribution of PIN protein in these cells could, in turn, perturb the auxin flow and therefore the cell division patterns and differentiation.
Another level of control of root growth is represented by the patterning transcription factors. Mutations in SCR affect radial root patterning, cause an S phenotype (Scheres et al. 1995), and cause ectopic cell divisions of the QC, leading to a disorganized QC and LRC (Wysocka-Diller et al. 2000). The WOX5 transcription factor specifies some aspects of QC cell fate that are under the control of the SCR gene; in scr mutants, the expression of WOX5 is reduced or undetectable (Sarkar et al. 2007). Due to changes in cell morphology in rpk1 toad2/+ mutants, the QC cells are not easily distinguishable, and therefore we employed the analysis of specific markers for the QC and the endodermis. The RPK1 TOAD2 pathway does not appear to function upstream of SCR, as transcription of SCR was detected in the endodermal, CEI, and QC cells of rpk1-1 toad2/+ mutants, albeit that they were irregularly absent in some cells. It cannot be ruled out that RPK1 and TOAD2 are redundantly required postembryonically for SCR expression. In wild-type plants, the expression of the WOX5 marker is detected in the QC cells and occasionally at a lower intensity in the vascular initials. In the rpk1-1 toad2/+ plants, we did not detect ectopic WOX5 expression in cells other than the ones observed in wild-type plants; the expression of WOX5 indicates an increased division of QC cells, and the generation of small cells that reside at the center of the stem cell niche and continue to express the marker. A similar pattern was observed with the QC184 marker, which acts downstream of WOX5 (Sarkar et al. 2007), and QC25, indicating that cells around the presumptive QC still maintain some aspects of QC specification.
The role of the epidermis as a key regulator of inner cell fates was explored in the process of BR-mediated root growth (Hacham et al. 2011). Epidermis-restricted expression of BRI1 was shown to be sufficient to promote root meristem growth, providing evidence for signaling from the outer layers to the inner meristem. Both RPK1 and TOAD2 protein localize at the plasma membrane of epidermal cells, but are also expressed in inner cells throughout the RAM. In previous analysis of RPK1 and TOAD2 in embryos, we proposed that both receptors function in the epidermis to initiate radial patterning during embryogenesis (Nodine et al. 2007).
If these two receptor kinases function in signaling from the epidermis, what are the ligands for RPK1 and TOAD2? How is the signal transmitted from cell-to-cell? Does their activity follow a ligand sequestration model (Stahl and Simon 2009) or a ligand-induced trafficking and degradation of receptors model (Nimchuk et al. 2011)? Previous reports of CLV3 signal perception by TOAD2 prompted us to search for candidate ligands among root-specific CLE genes that have an expression pattern similar to, or overlapping, that of the receptors. CLE17 and CLE19, among others (Jun et al. 2010), are expressed in the RAM, so we tested the effect of the exogenously applied synthetic peptides corresponding to the CLE motif of these two genes on mutant roots. As previously reported (Fiers et al. 2005), treatment with type A CLE peptides caused an S phenotype of wild-type plants. We found that rpk1 as well as rpk1 toad2/+ mutants are also sensitive to CLE treatment, whereas toad2 mutants, similar to clv2-8 and crn-1, are insensitive to CLE19 and CLE17 treatment. We analyzed the effects of CLE treatment at a cell type-specific level, in both sensitive and insensitive plants, and found a common set of CLE-induced responses, as well as responses specific to the insensitive plants only. As a general response found in all genotypes, we observed an increase in the stem cell niche size due to an increased number of cell divisions in the QC, and an effect on the maintenance of CEI cells as well as their daughters as stem cells. A specific response to CLE treatment was the reduced proliferative activity of cells in the proximal meristem, resulting in fewer cells comprising the longitudinal files. In conclusion, despite the increased frequency of cell division of QC cells and the increased stem cell niche size of CLE-treated plants, this mechanism is not sufficient to overcome the reduction in the number of cells in the proximal meristem that accounts for shorter roots. Roots of toad2, clv2-8, and crn-1 mutants are insensitive to the CLE17-mediated reduction of transit-amplifying cell divisions and present longer roots, indicating a common mechanism by which CLE peptides act to control root growth. This implies that TOAD2 might function as a CLE receptor in a multi-protein receptor complex, containing CLV2 and CRN, which perceives the that CLE signals, or the individual contribution of TOAD2, CLV2, and CRN to root growth, converges on some unknown downstream general mechanism controlling root cell division and growth.
As RPK1 does not appear to function directly in CLE perception, we tested the regulation of both RPK1 and TOAD2 by exogenous CLE treatment. A strong downregulation of RPK1 transcriptional activity, as well as RPK1-GFP protein accumulation at the plasma membrane, was observed 5 days after CLE treatment. Normally, we observe RPK1-GFP localization in the plasma membrane and also in structures that we believe are part of the intracellular vesicular membrane system, but after CLE treatment, the residual protein detected was localized mostly to the inner membranes. A slight reduction in TOAD2 protein levels was also detected, but in all plants the protein was still present in the plasma membrane after treatment. However, in the absence of TOAD2, the level of RPK1 in the plasma membrane of treated plants remains unchanged. Our conclusion is that TOAD2 mediates this response by triggering RPK1 turnover at the transcript and protein level. The shorter root and increased sensitivity to CLE treatment of plants expressing additional TOAD2 phenocopy the CLE treatment, providing further support for a role for TOAD2 in perceiving the CLE signal. A similar phenotype of shorter roots and increased sensitivity to CLE peptides was reported for plants overexpressing TOAD2 under the control of the cauliflower mosaic virus 35S promoter (Kinoshita et al. 2010).
The response to CLE treatment by downregulating gene expression is not a general response, as other genes (SCR, WOX5) are still expressed at similar levels to untreated plants. The WOX5 misexpression in cells at the position of vascular initials and absence from the putative QC cells might indicate a proximal shift in QC activity. If QC cells lose their activity as a result of CLE action, it is possible that new QC cells are specified proximally from the presumptive QC. This observation also correlates with the increased cell division rates of cells at the center of the stem cell niche.
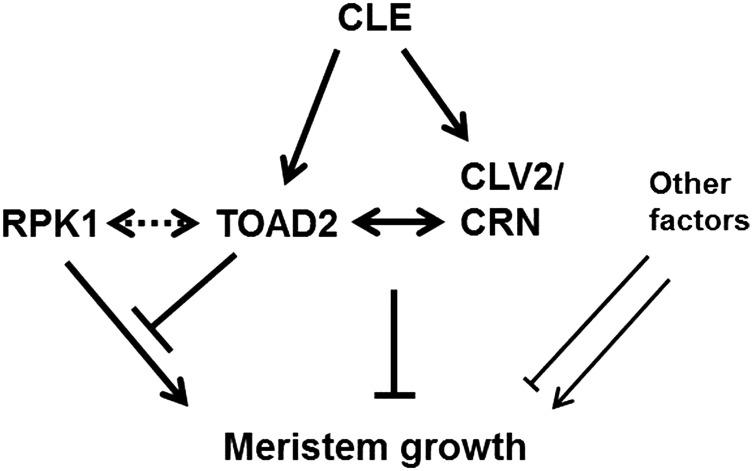
Our findings suggest that RPK1 and TOAD2 function to balance positive and negative regulation of root meristem growth, and the gene dosage or the level of activation by exogenous peptides disturbs root growth homeostasis. Our model indicates that CLE peptides are perceived by TOAD2 (or a complex containing TOAD2) (Figure 11). Upon ligand binding, a signaling cascade is initiated, resulting in the downregulation of RPK1 transcription and protein internalization. In the absence of RPK1, growth signals are no longer transmitted, resulting in short roots, but this is not an all-or-nothing process, as additional unidentified components also play a similar, redundant role. In rpk1 mutants, upon TOAD2 stimulation by exogenous CLE, the rpk1 pathway is already interrupted and therefore short roots are observed (similar to the phenotype of rpk1 mutants). Additional components of the RPK1 pathway that might act in parallel and could account for the low penetrance of the S phenotype in rpk1 mutants might also be downregulated upon TOAD2-ligand interaction (Figure 11).
Figure 11.
Model for RPK1 and TOAD2 interactions in controlling root growth. Diagram shows a proposed model of a regulatory network controlling root meristem growth by TOAD2, RPK1, CLV2, CRN, and other unknown components. Dotted double arrows indicate potential direct interaction, solid double arrows indicate known interactions. Arrows symbolize positive regulation and bars indicate negative regulation. Unknown components could have both positive and negative regulatory roles.
Further molecular and biochemical studies are needed to validate the potential binding of CLE peptides to the extracellular domain of TOAD2 and the mechanisms leading to either the sequestration of ligand molecules or the ligand–receptor internalization model of action. Processes reminiscent of ligand-induced receptor endocytosis in animals have been shown to occur in plants. For instance, LRR RLK FLAGELLIN-SENSING 2 (FLS2), which functions in plant innate immunity, has been shown to trigger an immune response after ligand binding and internalization in endocytic vesicles (Robatzek et al. 2006). Endocytosis of receptors and signaling from internal membranes was also described for the steroid receptor BRI1, where endocytic trafficking and signaling appear to be constitutive, and does not change with changes in ligand levels (Geldner et al. 2007). Contrary to sequestration models, signaling in the CLV1 pathway was also shown to be dependent on internalization of the receptor, regardless of the expanded diffusion of the CLV3 ligand (Nimchuk et al. 2011). In the case of ACR4, its internalization is dependent on its functionality, suggesting this as a mechanism of signaling regulation (Gifford et al. 2005). Is TOAD2 sequestering ligands that are expressed in the outer layers, such that their movement toward the stem cell niche is limited? Is TOAD2 binding CLE ligands and possibly being internalized, alongside RPK1? Future molecular dissection of this pathway is necessary to reveal the details of RPK1/TOAD2 interplay in CLE-mediated root growth.
Thus far, it appears that several RLKs and CLE ligands function in sometimes overlapping processes mediating root primary and secondary meristem growth. ACR4 and CLV1 regulate distal root meristem maintenance (Stahl et al. 2013) by responding to CLE40 signaling. LRR-RLK BARELY ANY MERISTEM (BAM3) and its putative ligand, CLE45, control the development of the protophloem, a secondary meristem (Depuydt et al. 2013). The CLE41/44 signal perceived by LRR-RLK PHLOEM INTERCALATED WITH XYLEM (PXY) in the procambial cells inhibits their differentiation and promotes their proliferation (Hirakawa et al. 2010). A more recent report indicates that TOAD2 and BAM1 physically interact and function with CLV2 in restricting the size of the root meristem (Shimizu et al. 2015).
In our study, RPK1 and TOAD2 are also implicated in controlling the balance between the proliferation and differentiation of root cell types. CLE treatment affects only the main root and not LR growth, even though TOAD2 and RPK1 are also expressed in the LRs; exploring the basis of the differential regulation of main roots and LRs might shed some light on the CLE regulation of root growth.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300148/-/DC1.
Acknowledgments
We thank the members of the Tax laboratory for technical support and comments on the manuscript and C. Zhang for providing the pWOX5::GFP-NLS construct and pFYTAG vector. This work was supported by National Science Foundation (NSF) grants MCB 0418946, NSF IOS-0922678, and NSF IOS 1257316 awarded to F.E.T., and A.R. received funding from the National Institutes of Health (grant T32 GM-08659) and the NSF (Integrative Graduate Education and Research Traineeship DGE-0114420).
Footnotes
Communicating editor: J. Birchler
Literature Cited
- Azpeitia E., Alvarez-Buylla E. R., 2012. A complex systems approach to Arabidopsis root stem-cell niche developmental mechanisms: from molecules, to networks, to morphogenesis. Plant Mol. Biol. 80: 351–363. [DOI] [PubMed] [Google Scholar]
- Betsuyaku S., Sawa S., Yamada M., 2011a The function of the CLE peptides in plant development and plant-microbe interactions. Arabidopsis Book 9: e0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsuyaku S., Takahashi F., Kinoshita A., Miwa H., Shinozaki K., et al. , 2011b Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol. 52: 14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., et al. , 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Clark S. E., Williams R. W., Meyerowitz E. M., 1997. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585. [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F., 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Clouse S. D., Sasse J. M., 1998. BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 427–451. [DOI] [PubMed] [Google Scholar]
- Cock J. M., McCormick S., 2001. A large family of genes that share homology with CLAVATA3. Plant Physiol. 126: 939–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Levesque M. P., Vernoux T., Jung J. W., Paquette A. J., et al. , 2007. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425. [DOI] [PubMed] [Google Scholar]
- Delay C., Imin N., Djordjevic M. A., 2013. Regulation of Arabidopsis root development by small signaling peptides. Front. Plant Sci. 4: 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S., Rodriguez-Villalon A., Santuari L., Wyser-Rmili C., Ragni L., et al. , 2013. Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3. Proc. Natl. Acad. Sci. USA 110: 7074–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I., Vassileva V., De Rybel B., Levesque M. P., Grunewald W., et al. , 2008. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322: 594–597. [DOI] [PubMed] [Google Scholar]
- Diévart A., Clark S. E., 2004. LRR-containing receptors regulating plant development and defense. Development 131: 251–261. [DOI] [PubMed] [Google Scholar]
- Durbak A. R., Tax F. E., 2011. CLAVATA signaling pathway receptors of Arabidopsis regulate cell proliferation in fruit organ formation as well as in meristems. Genetics 189: 177–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M., Golemiec E., Xu J., van der Geest L., Heidstra R., et al. , 2005. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17: 2542–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J., 2010. Subcellular trafficking of PIN auxin efflux carriers in auxin transport. Eur. J. Cell Biol. 89: 231–235. [DOI] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., et al. , 2003. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153. [DOI] [PubMed] [Google Scholar]
- Fukuda H., Higashiyama T., 2011. Diverse functions of plant peptides: entering a new phase. Plant Cell Physiol. 52: 1–4. [DOI] [PubMed] [Google Scholar]
- Gallagher K. L., Paquette A. J., Nakajima K., Benfey P. N., 2004. Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol. 14: 1847–1851. [DOI] [PubMed] [Google Scholar]
- Geldner N., Hyman D. L., Wang X., Schumacher K., Chory J., 2007. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21: 1598–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford M. L., Dean S., Ingram G. C., 2003. The Arabidopsis ACR4 gene plays a role in cell layer organisation during ovule integument and sepal margin development. Development 130: 4249–4258. [DOI] [PubMed] [Google Scholar]
- Gifford M. L., Robertson F. C., Soares D. C., Ingram G. C., 2005. ARABIDOPSIS CRINKLY4 function, internalization, and turnover are dependent on the extracellular crinkly repeat domain. Plant Cell 17: 1154–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen V. A., Xu J., Marée A. F., Hogeweg P., Scheres B., 2007. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013. [DOI] [PubMed] [Google Scholar]
- Guo Y., Han L., Hymes M., Denver R., Clark S. E., 2010. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 63: 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y., Holland N., Butterfield C., Ubeda-Tomas S., Bennett M. J., et al. , 2011. Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y., Sela A., Friedlander L., Savaldi-Goldstein S., 2012. BRI1 activity in the root meristem involves post-transcriptional regulation of PIN auxin efflux carriers. Plant Signal. Behav. 7: 68–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter T., Imkampe J., Mazzotta S., Wierzba M., Postel S., et al. , 2014. The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24: 134–143. [DOI] [PubMed] [Google Scholar]
- Hartig S. M., 2013. Basic image analysis and manipulation in ImageJ. Curr. Protoc. Mol. Biol. Chapter 14: Unit14.15. [DOI] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., et al. , 2008. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 105: 15208–15213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Kondo Y., Fukuda H., 2010. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22: 2618–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkampe J., Halter T., Huang S., Schulze S., Mazzotta S., et al. , 2017. The Arabidopsis leucine-rich repeat receptor kinase BIR3 negatively regulates BAK1 receptor complex formation and stabilizes BAK1 Plant Cell 29: 2285–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., et al. , 2006. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313: 842–845. [DOI] [PubMed] [Google Scholar]
- Jun J., Fiume E., Roeder A. H., Meng L., Sharma V. K., et al. , 2010. Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154: 1721–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A., Nakamura Y., Sasaki E., Kyozuka J., Fukuda H., et al. , 2007. Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48: 1821–1825. [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Betsuyaku S., Osakabe Y., Mizuno S., Nagawa S., et al. , 2010. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920. [DOI] [PubMed] [Google Scholar]
- Kiyohara S., Sawa S., 2012. CLE signaling systems during plant development and nematode infection. Plant Cell Physiol. 53: 1989–1999. [DOI] [PubMed] [Google Scholar]
- Lee I. C., Hong S. W., Whang S. S., Lim P. O., Nam H. G., et al. , 2011. Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol. 52: 651–662. [DOI] [PubMed] [Google Scholar]
- Li J., 2010. Multi-tasking of somatic embryogenesis receptor-like protein kinases. Curr. Opin. Plant Biol. 13: 509–514. [DOI] [PubMed] [Google Scholar]
- Li J., Chory J., 1997. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938. [DOI] [PubMed] [Google Scholar]
- Luichtl M., Fiesselmann B. S., Matthes M., Yang X., Peis O., et al. , 2013. Mutations in the Arabidopsis RPK1 gene uncouple cotyledon anlagen and primordia by modulating epidermal cell shape and polarity. Biol. Open 2: 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y., Ogawa M., Morita A., Sakagami Y., 2002. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296: 1470–1472. [DOI] [PubMed] [Google Scholar]
- Miwa H., Betsuyaku S., Iwamoto K., Kinoshita A., Fukuda H., et al. , 2008. The receptor-like kinase SOL2 mediates CLE signaling in Arabidopsis. Plant Cell Physiol. 49: 1752–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno S., Osakabe Y., Maruyama K., Ito T., Osakabe K., et al. , 2007. Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J. 50: 751–766. [DOI] [PubMed] [Google Scholar]
- Moller B., Weijers D., 2009. Auxin control of embryo patterning. Cold Spring Harb. Perspect. Biol. 1: a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R., Bleckmann A., Simon R., 2008. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Guo Y., Jin H., Hartsell J., Clark S. E., 2011. Characterization of a CLE processing activity. Plant Mol. Biol. 75: 67–75. [DOI] [PubMed] [Google Scholar]
- Nimchuk Z. L., Tarr P. T., Ohno C., Qu X., Meyerowitz E. M., 2011. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr. Biol. 21: 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine M. D., Tax F. E., 2008. Two receptor-like kinases required together for the establishment of Arabidopsis cotyledon primordia. Dev. Biol. 314: 161–170. [DOI] [PubMed] [Google Scholar]
- Nodine M. D., Yadegari R., Tax F. E., 2007. RPK1 and TOAD2 are two receptor-like kinases redundantly required for Arabidopsis embryonic pattern formation. Dev. Cell 12: 943–956. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Shinohara H., Sakagami Y., Matsubayashi Y., 2008. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294. [DOI] [PubMed] [Google Scholar]
- Oh M. H., Clouse S. D., Huber S. C., 2009. Tyrosine phosphorylation in brassinosteroid signaling. Plant Signal. Behav. 4: 1182–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y., Maruyama K., Seki M., Satou M., Shinozaki K., et al. , 2005. Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17: 1105–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y., Mizuno S., Tanaka H., Maruyama K., Osakabe K., et al. , 2010. Overproduction of the membrane-bound receptor-like protein kinase 1, RPK1, enhances abiotic stress tolerance in Arabidopsis. J. Biol. Chem. 285: 9190–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka J. J., Winter C. M., Benfey P. N., 2012. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 63: 563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang Y., Wu J., Han H., Wang G., 2013. CLE peptides in vascular development. J. Integr. Plant Biol. 55: 389–394. [DOI] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T., 2006. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Heidstra R., Wildwater M., Scheres B., 2003. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 1: 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A. K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., et al. , 2007. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814. [DOI] [PubMed] [Google Scholar]
- Scheres B., Di Laurenzio L., Willemsen V., Hauser M. T., Janmaat K., et al. , 1995. Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121: 53–62. [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K. F., Jürgens G., et al. , 2000. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644. [DOI] [PubMed] [Google Scholar]
- Shimizu N., Ishida T., Yamada M., Shigenobu S., Tabata R., et al. , 2015. BAM 1 and RECEPTOR-LIKE PROTEIN KINASE 2 constitute a signaling pathway and modulate CLE peptide-triggered growth inhibition in Arabidopsis root. New Phytol. 208: 1104–1113. [DOI] [PubMed] [Google Scholar]
- Shiu S. H., Bleecker A. B., 2001a Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98: 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S. H., Bleecker A. B., 2001b Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE 2001: re22. [DOI] [PubMed] [Google Scholar]
- Song X. F., Yu D. L., Xu T. T., Ren S. C., Guo P., et al. , 2012. Contributions of individual amino acid residues to the endogenous CLV3 function in shoot apical meristem maintenance in Arabidopsis. Mol. Plant 5: 515–523. [DOI] [PubMed] [Google Scholar]
- Stahl Y., Simon R., 2009. Is the Arabidopsis root niche protected by sequestration of the CLE40 signal by its putative receptor ACR4? Plant Signal. Behav. 4: 634–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y., Simon R., 2010. Plant primary meristems: shared functions and regulatory mechanisms. Curr. Opin. Plant Biol. 13: 53–58. [DOI] [PubMed] [Google Scholar]
- Stahl Y., Wink R. H., Ingram G. C., Simon R., 2009. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19: 909–914. [DOI] [PubMed] [Google Scholar]
- Stahl Y., Grabowski S., Bleckmann A., Kühnemuth R., Weidtkamp-Peters S., et al. , 2013. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr. Biol. 23: 362–371. [DOI] [PubMed] [Google Scholar]
- Strabala T. J., O’Donnell P. J., Smit A. M., Ampomah-Dwamena C., Martin E. J., et al. , 2006. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 140: 1331–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M. R., Amasino R. M., Young J. C., Krysan P. J., Austin-Phillips S., 2000. The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 124: 1465–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Kramer E. M., Perry P., Knox K., Leyser H. M., et al. , 2005. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7: 1057–1065. [DOI] [PubMed] [Google Scholar]
- Tamaki T., Betsuyaku S., Fujiwara M., Fukao Y., Fukuda H., et al. , 2013. SUPPRESSOR OF LLP1 1-mediated C-terminal processing is critical for CLE19 peptide activity. Plant J. 76: 970–981. [DOI] [PubMed] [Google Scholar]
- Torii K., 2008. Transmembrane receptors in plants: receptor kinases and their ligands, pp. 1–29 in Annual Plant Reviews, edited by Yang Z. Wiley-Interscience, New York. [Google Scholar]
- Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., et al. , 2008. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford R., Fernandez A., De Groodt R., Ortega E., Hilson P., 2008. Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc. Natl. Acad. Sci. USA 105: 18625–18630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka-Diller J. W., Helariutta Y., Fukaki H., Malamy J. E., Benfey P. N., 2000. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127: 595–603. [DOI] [PubMed] [Google Scholar]
- Yadav R. K., Fulton L., Batoux M., Schneitz K., 2008. The Arabidopsis receptor-like kinase STRUBBELIG mediates inter-cell-layer signaling during floral development. Dev. Biol. 323: 261–270. [DOI] [PubMed] [Google Scholar]
- Zhang C., Gong F. C., Lambert G. M., Galbraith D. W., 2005. Cell type-specific characterization of nuclear DNA contents within complex tissues and organs. Plant Methods 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.